D box and KEN box motifs in budding yeast Hsl1p are required for APC-mediated degradation and direct binding to Cdc20p and Cdh1p (original) (raw)
Abstract
The precise order of molecular events during cell cycle progression depends upon ubiquitin-mediated proteolysis of cell cycle regulators. We demonstrated previously that Hsl1p, a protein kinase that inhibits the Swe1p protein kinase in a bud morphogenesis checkpoint, is targeted for ubiquitin-mediated turnover by the anaphase-promoting complex (APC). Here, we investigate regions of Hsl1p that are critical both for binding to the APC machinery and for APC-mediated degradation. We demonstrate that Hsl1p contains both a destruction box (D box) and a KEN box motif that are necessary for Hsl1p turnover with either APCCdc20 or APCCdh1. In coimmunoprecipitation studies, the D box of full-length Hsl1p was critical for association with Cdc20p, whereas the KEN box was important for association with Cdh1p. Fusion of a 206-amino-acid fragment of Hsl1p containing these motifs to a heterologous protein resulted in APC-dependent degradation of the fusion protein that required intact D box and KEN box motifs. Finally, this bacterially expressed Hsl1p fusion protein interacted with Cdc20p and Cdh1p either translated in vitro or expressed in and purified from insect cells. Binding to Cdc20p and Cdh1p was disrupted completely by a D box/KEN box double mutant. These results indicate that D box and KEN box motifs are important for direct binding to the APC machinery, leading to ubiquitination and subsequent protein degradation.
Keywords: Cdc20p, Cdh1p, KEN box, destruction box, ubiquitination, anaphase-promoting complex
Ubiquitin-mediated protein degradation is necessary for the linear progression of molecular events during the cell cycle. Protein ubiquitination and subsequent degradation by the 26S proteasome is required for the initiation of DNA replication, the onset of anaphase, and for mitotic exit (for reviews, see King et al. 1996; Peters 1999; Zachariae and Nasmyth 1999). The ubiquitin pathway is a multi-step process in which the 76-amino-acid ubiquitin molecule is activated by an E1 enzyme, transferred to an E2 enzyme, and then attached covalently to the protein substrate either directly or in conjunction with an E3 enzyme (ubiquitin ligase) (for reviews, see Ciechanover 1994; Hochstrasser 1996). Proteins destined to be degraded are subject to multiple rounds of ubiquitin attachment (polyubiquitination) and are then proteolyzed by the 26S proteasome. The E3 or ubiquitin ligase is involved in protein substrate recognition and thereby confers specificity to the ubiquitination reaction (for reviews, see Ciechanover 1994; Hochstrasser 1996).
The SCF (Skp1p–cullin–F-box) ubiquitin ligases are a family of multi-subunit E3s that are responsible for ubiquitination of cell cycle regulators at the G1–S phase transition (for reviews, see King et al. 1996; Krek 1998; Peters 1998). SCF complexes have common Skp1 and cullin subunits, but distinct F-box protein subunits that contain either a WD40 domain or a leucine-rich repeat domain that binds directly to phosphorylated SCF substrates (for reviews, see Krek 1998; Patton et al. 1998; Peters 1998). In Saccharomyces cerevisiae, the G1-cyclins, Cln1p and Cln2p, bind SCFGrr1 (Deshaies et al. 1995; Skowyra et al. 1999), whereas the Cdc28p kinase inhibitor, Sic1p, has been shown to bind SCFCdc4 (Feldman et al. 1997; Skowyra et al. 1997; Verma et al. 1997), where Grr1p and Cdc4p are the F-box components of these SCFs.
The anaphase-promoting complex (APC), or cyclosome, another multi-subunit E3, is responsible for the ubiquitination of cell regulators at the metaphase–anaphase and mitosis–G1 transitions (for review, see Zachariae and Nasmyth 1999). The APC is comprised of a core complex of proteins and two WD40-containing proteins, Cdh1 (called Cdh1p or Hct1p in S. cerevisiae) and Cdc20, that bind to, and have been proposed to activate, the APC (Schwab et al. 1997; Visintin et al. 1997; Fang et al. 1998b; Kramer et al. 1998; Lim et al. 1998; Zachariae et al. 1998; Kotani et al. 1999). Cdc20 binds the APC (APCCdc20) during mitosis. APCCdc20 is required for the degradation of the anaphase inhibitors known as the securins (Pds1p in S. cerevisiae and Cut2p in Schizosaccharomyces pombe), thereby triggering sister chromatid separation (Cohen-Fix et al. 1996; Funabiki et al. 1996; Stratmann and Lehner 1996; Yamamoto et al. 1996; Ciosk et al. 1998; Zou et al. 1999; Zur and Brandeis 2001). Cdh1 binds to the APC (APCCdh1) in late mitosis and G1 and is responsible for ubiquitination of the mitotic cyclins (Clb1–4p in S. cerevisiae) during these cell cycle phases (Schwab et al. 1997; Visintin et al. 1997; Kramer et al. 1998; Zachariae et al. 1998; Jaspersen et al. 1999). APCCdh1 is also responsible for the degradation of Cdc20, thereby restricting APCCdc20 activity to a narrow window from G2 when Cdc20 is synthesized to late mitosis when Cdc20 is degraded by APCCdh1 (Fang et al. 1998b; Prinz et al. 1998; Shirayama et al. 1998). In contrast, Cdh1 levels remain relatively constant during the cell cycle, but phosphorylation of Cdh1 by the mitotic kinase Cdc2 (Cdc28p in S. cerevisiae) blocks Cdh1 binding to the APC, thereby preventing APCCdh1 activity during S, G2, and M phases when Cdc2/Cdc28p activity is high (Zachariae et al. 1998; Jaspersen et al. 1999; Shirayama et al. 1999; Blanco et al. 2000; Listovsky et al. 2000).
Two degradation motifs have been identified in APC substrates. The destruction box (D box), with the consensus sequence R-x-x-L-x-x-x-x-N/D/E, is important for the degradation of most APC substrates (Glotzer et al. 1991; for review, see Zachariae and Nasmyth 1999). Recently, human Cdc20 (hCDC20), which lacks a D box, was found to contain a new degradation signal called a ‘KEN box’ with the consensus sequence K-E-N-x-x-x-D/N that was critical for its degradation by APCCdh1 (Pfleger and Kirschner 2000). Subsequently, a few other APC substrates containing both KEN box and D box motifs have been identified. These APC substrates are human CDC6, a protein required for DNA replication, human securin, and the mitotic cyclin A in Drosophila melanogaster (Peterson et al. 2000; Jacobs et al. 2001; Zur and Brandeis 2001). Functional KEN boxes in yeast proteins have not yet been reported.
Because direct interactions between Cdc20 or Cdh1 with APC substrates have not been demonstrated, generally these proteins are believed to activate the APC, possibly by inducing allosteric changes in core APC subunits leading to specific substrate binding and ubiquitination. However, indirect evidence for substrate binding has led others to suggest, by analogy to the F-box proteins of the SCFs, that Cdc20 and Cdh1 may bind and target protein substrates to the APC (Schwab et al. 1997; Visintin et al. 1997; Shirayama et al. 1998; Burton and Solomon 2000; Ohtoshi et al. 2000; Sorensen et al. 2001).
We found previously that Hsl1p, a 170-kD budding yeast protein kinase that negatively regulates the Swe1p protein kinase in a bud morphogenesis checkpoint pathway (Lew and Reed 1995; Barral et al. 1999; McMillan et al. 1999; Shulewitz et al. 1999), is degraded via the APC (Burton and Solomon 2000). Like most APC-substrates, Hsl1p contains a D box that is important for its degradation. Hsl1p associates with both Cdc20p and Cdh1p based on both two-hybrid assays and coimmunoprecipitation from yeast extracts, indicating that it might interact directly with these proteins (Burton and Solomon 2000). Given these findings, we believed that Hsl1p might serve as a useful model substrate for understanding how proteins destined for degradation are recognized by the APC.
In this study we set out to understand which domains of Hsl1p are important for both recognition and turnover by the APC machinery. We noticed that Hsl1p contains a potential KEN box and were interested in addressing the following questions using Hsl1p as a model APC substrate: 1) Do KEN boxes serve as degradation signals in yeast? 2) Can Hsl1p bind directly to Cdc20p and/or Cdh1p? 3) What is the role of the D box and/or KEN box in APC recognition? We discuss a model for substrate recognition by the APC machinery.
Results
Hsl1p degradation requires intact D box and KEN box motifs
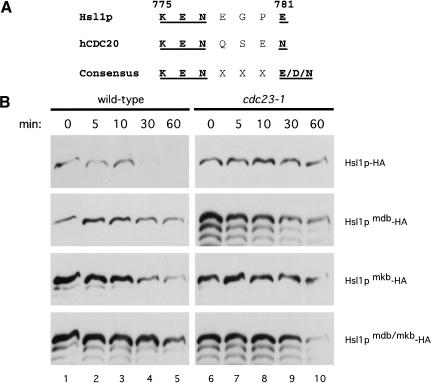
Previously, we demonstrated that the APC-dependent degradation of Hsl1p requires an intact destruction box (D box) motif (Burton and Solomon 2000). Following the recent identification of the KEN box (KENxxxN/D; bold indicates conserved and mutated residues) as a second APC-degradation signal important for APCCdh1-mediated degradation (Pfleger and Kirschner 2000), we scanned the Hsl1p sequence and found a potential KEN box motif that closely matched that of hCDC20 (Fig. 1A). To determine if this sequence influences APC-mediated degradation of Hsl1p, the conserved amino acid residues (underlined in Fig. 1A) were mutated to alanines. The stabilities of full length Hsl1p–HA or of Hsl1p–HA containing a mutated D box (RAALSDITN to AAAASDITA) (Hsl1pmdb–HA), a mutated KEN box (Hsl1pmkb–HA), or both mutations (Hsl1pmdb/mkb–HA) were investigated in G1, a stage of the cell cycle when APCCdh1 is active. Cells were arrested in G1 with α-factor and induced to express the different isoforms of Hsl1p–HA by galactose induction. Then, expression was terminated by the addition of glucose and cycloheximide to the medium. Levels of the different forms of Hsl1p–HA were monitored by immunoblot analysis with anti-HA antibodies (Fig. 1B). As shown previously (Burton and Solomon 2000), wild-type Hsl1p–HA was unstable in the presence of APCCdh1, but stable in a strain (cdc23-1) mutated for a core APC subunit (Fig. 1B, top row). In contrast, Hsl1pmdb–HA containing the mutated D box was relatively stable even in the presence of an active APC (Fig. 1B, second row, left panel). Similarly, mutation of the putative KEN box (Hsl1pmkb–HA) or of both motifs (Hsl1pmdb/mkb–HA) also resulted in Hsl1p stabilization (Fig. 1B, third and fourth rows, left panels). These results indicate that KEN boxes are recognized by the APC machinery in yeast and that both a D box and a KEN box are necessary for maximal rates of Hsl1p degradation by APCCdh1.
Figure 1.
Hsl1p has both a D box and a KEN box and both are necessary for APC-mediated degradation in G1. (A) Hsl1p has a putative KEN box located at amino acids 775–781. The human CDC20 (hCDC20) KEN box and consensus sequence (Pfleger and Kirschner 2000) are shown for comparison. (B) APCCdh1-mediated degradation of Hsl1p–HA in G1-arrested cells requires both a D box and a KEN box. Cells were arrested in G1 with α-factor (100 ng/mL) and induced to express GAL–HSL1–HA (YJB123 and YJB125), GAL–HSL1mdb–HA (YJB229 and YJB270), GAL–HSL1mkb–HA (YJB257 and YJB271) and GAL–HSL1mdb/mkb–HA (YJB258 and YJB272) by the addition of galactose. Cells were shifted to 37°C to inactivate the APC in cdc23–1 strains. Proteins levels were monitored by immunoblotting with anti-HA antibodies at the indicated times following termination of Hsl1p expression.
Given the similarities between Cdh1p and Cdc20p, we were interested in whether APCCdc20 could also mediate Hsl1p degradation. Although normally Hsl1p is degraded late in mitosis (Burton and Solomon 2000), we found that Hsl1p–HA was stable in anaphase-arrested cells expressing a nondegradable form of Clb2p (data not shown). Therefore, we assessed Hsl1p stability in G1, a point in the cell cycle at which Hsl1p is known to be unstable (Fig. 1B; Burton and Solomon 2000). In order to eliminate APCCdh1 activity and look only at APCCdc20 activity in G1, we used _cdh1_Δ cdc28-13 cells with or without an integrated copy of _GAL–CDC20–myc. cdh1_Δ cells are viable, but have elevated levels of Clb2p, resulting in an inefficient G1 arrest with α-factor (Schwab et al. 1997; Visintin et al. 1997). The conditional cdc28-13 allele was used to arrest cells in G1 by shifting cells to the restrictive temperature (LoRincz and Reed 1986).
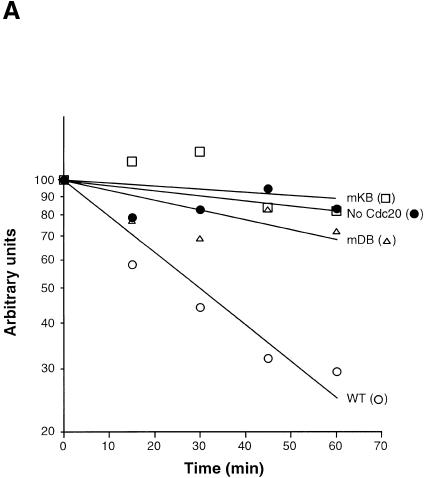
Using this strain background, we examined the stabilities of wild-type and mutant forms of Hsl1p–HA with or without Cdc20–pmyc expression. G1-arrested cells were induced to express GAL–HSL1–HA, GAL–HSL1mdb–HA, or GAL–HSL1mkb–HA either without or with GAL–CDC20–myc expression by the addition of galactose to the medium. Expression of the different Hsl1p–HA proteins (and of Cdc20p–myc in the indicated strains) was terminated by the addition of glucose/cycloheximide to the cells; protein levels were assessed by quantitative immunoblot analysis using anti-HA antibodies (Fig. 2A; see Materials and Methods). Wild-type Hsl1p–HA was relatively stable in the _cdh1_Δ cdc28-13 cells with most of the protein still present after 60 min (Fig. 2A, filled circles). However, Hsl1p–HA was unstable with a half-life of ∼30 min in cells overexpressing Cdc20p–myc, presumably due to APCCdc20-mediated degradation (Fig. 2A, cf. open circles with closed circles). In contrast, Hsl1pmdb–HA and Hsl1pmkb–HA were relatively stable in cells expressing Cdc20p–myc (Fig. 2A, open triangles and squares). FACS analysis confirmed that these cells remained arrested in G1 (Fig. 2B). We have also observed that most of the Hsl1p–HA is degraded as cells progress from mitosis to G1 in _cdh1_Δ cells (data not shown), indicating that endogenous Cdc20p is able to degrade Hsl1p in late mitosis. These results indicate that Hsl1p degradation can be mediated both by APCCdc20 and by APCCdh1 and that both D box and KEN box motifs are required in both cases.
Figure 2.
Cdc20p can promote the degradation of Hsl1p–HA in G1. (A) _cdh1_Δ cdc28-13 cells with (YJB367, YJB368 and YJB380) or without GAL–CDC20–myc (YJB366, YJB377 and YJB379) were arrested in G1 by inactivation of cdc28-13 at 37°C, then galactose was added to induce expression of GAL–HSL1–HA (YJB366 and YJB367), GAL–HSL1mdb–HA (YJB377 and YJB378) and GAL–HSL1mkb–HA (YJB379 and YJB380) and of GAL–CDC20–myc. Levels of wild-type and mutant forms of Hsl1p–HA were monitored at specified times after glucose/cycloheximide addition by quantitative immunoblotting with anti-HA antibodies (see Materials and Methods). Filled and open circles represent Hsl1p–HA without or with Cdc20–myc expression, respectively. The ratios of Hsl1pmdb–HA (open triangles) and Hsl1pmkb–HA (open squares) with versus without Cdc20–myc expression are shown. (B) FACS analysis of the indicated strains from the experiment in (A).
Hsl1p degradation motifs influence association with the APC machinery
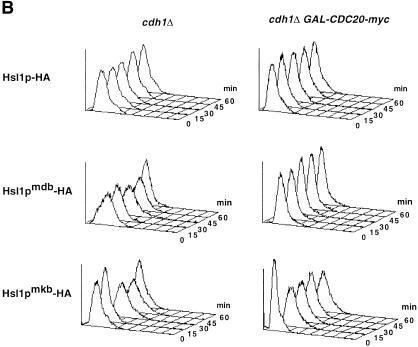
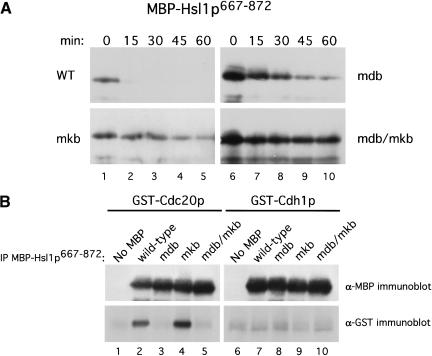
Next, we examined the importance of the D box and the KEN box for the association of Hsl1p with Cdc20p and Cdh1p in a coimmunoprecipitation assay. We showed previously that the D box in Hsl1p was important for association with Cdc20p, but not with Cdh1p (Burton and Solomon 2000). Full-length Hsl1p–HA, Hsl1pmdb–HA, Hsl1pmkb–HA, or Hsl1pmdb/mkb were co-overexpressed in yeast with either GST–Cdc20p or GST–Cdh1p. The different Hsl1p–HA proteins were then immunoprecipitated (IP) from cell extracts using anti-HA antibodies. Coimmunoprecipitation of GST–Cdc20p or GST–Cdh1p was examined by immunoblot analysis with anti-GST antibodies (Fig. 3A). As found previously, both GST–Cdc20p and GST–Cdh1p coimmunoprecipitated with wild-type Hsl1p–HA (Fig. 3A, lower panels, lanes 2 and 7), whereas little or no GST–Cdc20p or GST–Cdh1p was detected in control immunoprecipitations from strains lacking an HA-tag (Fig. 3A, lower panels, lanes 1 and 6). Mutation of the D box in Hsl1p compromised association with Cdc20p, but had little effect on Cdh1p association (Fig. 3A, lower panels, lanes 3 and 8; see also Figs. 5B, 6 and 8C, below). In contrast, mutation of the KEN box had little effect on Cdc20p association, but eliminated Cdh1p association (Fig. 3A, lower panels, lanes 4 and 9). Neither GST–Cdc20p nor GST–Cdh1p associated with Hsl1pmdb/mkb (Fig. 3A, lower panels, lanes 5 and 10). These results indicate that in the context of full-length Hsl1p–HA, the D box is important for association with Cdc20p, whereas the KEN box is important for association with Cdh1p. Since both motifs must be present for efficient degradation (Figs. 1B and 2A), these results also indicate that association, though necessary, is not sufficient to promote degradation. Though suggestive, it is important to note that these data do not show direct binding of Hsl1p to Cdc20p and Cdh1p. It is possible that another yeast protein, such as a core APC subunit, serves as a bridge between Hsl1p and Cdc20p or Cdh1p in the immunoprecipitates.
Figure 3.
Both Cdc20p and Cdh1p can coimmunoprecipitate with APC substrates. (A) Coimmunoprecipitation of GST–Cdc20p and GST–Cdh1p with full-length Hsl1p–HA depends on an intact D box and KEN box, respectively. The different forms of Hsl1p–HA were immunoprecipitated (IP) with anti-HA antibodies from extracts of cells coexpressing either GST–Cdc20p or GST–Cdh1p and then probed for the levels of precipitated Hsl1p–HA proteins (upper) or GST–Cdc20p and GST–Cdh1p (lower) by immunoblot analysis using anti-HA and anti-GST antibodies, respectively. (Lanes 1_–_10) Strains YJB221, YJB156, YJB230, YJB259, YJB260, YJB222, YJB218, YJB231, YJB261 and YJB262, respectively. (B) GST–Cdc20p and GST–Cdh1p can coimmunoprecipitate with Clb2p–HA. Clb2p–HA was immunoprecipitated with anti-HA antibodies from extracts of cells coexpressing GST–Cdc20p or GST–Cdh1p and proteins were monitored by immunoblot analysis as described in (A). (Lanes 1–6) Strains YJB221, YJB275, YJB276, YJB222, YJB277 and YJB278, respectively. Strains lacking an HA tag (YJB221 and YJB222) but expressing GST–Cdc20p or GST–Cdh1p, respectively, were used as negative controls (No HA) in A and B. mdb, mutant D box; mkb, mutant KEN box.
Figure 5.
Degradation of MBP–Hsl1p667–872 in G1 requires intact D box and KEN box motifs. (A) Cells were arrested in G1 with α-factor and then induced to express WT, mdb, mkb or mdb/mkb isoforms of MBP–Hsl1p667–872 by the addition of galactose (strains YJB306, YJB326, YJB327 and YJB328, respectively). MBP–Hsl1p667–872 levels were monitored by immunoblot analysis with anti-MBP antibodies at the indicated times after glucose/cycloheximide addition. (B) D box-dependent binding of GST–Cdc20p to MBP–Hsl1p667–872. MBP–Hsl1p667–872 was immunoprecipitated with anti-MBP antibodies from extracts of cells expressing GST–Cdc20p and GST–Cdh1p. Precipitated proteins were analyzed for the presence of GST–Cdc20p and GST–Cdh1p by immunoblot analysis with anti-GST antibodies (lower panels) and for MBP–Hsl1p667–872 fusion proteins by immunoblot analysis with anti-MBP antibodies (upper panels). (Lanes 1–10) Strains YJB221, YJB309, YJB338, YJB339, YJB340, YJB222, YJB310, YJB341, YJB342 and YJB343, respectively.
Figure 6.
Cdc20p and Cdh1p translated in vitro can bind purified MBP–Hsl1p667–872 in a D box-dependent fashion. E. coli expressed MBP or MBP–Hsl1p667–872 proteins were purified on amylose resin and then mixed with [35S]methionine-labeled Cdc20p or Cdh1p (see Materials and Methods). Proteins bound to the resin were run on SDS-PAGE and visualized by fluorography (upper panels). Similar amounts of the MBP–Hsl1p667–872 proteins were present on the amylose beads as visualized by Coomassie staining following SDS-PAGE (lower panels).
Figure 8.
Direct binding between Cdh1p or Cdc20p and MBP–Hsl1p667–872 depends on D box and KEN box motifs. (A) Coomassie stained gel of binding assay with Cdh1p–6xHis beads (lanes 1–5) and control beads (lanes 6–10). Diluted E. coli extracts containing MBP or MBP–Hsl1p667–872 proteins were incubated with beads, washed and the bound proteins were eluted with 150 mM imidizole. Asterisks indicate positions of the MBP–Hsl1p667–872 proteins. (B) Immunoblot analysis of the samples shown in A. One tenth the amount of sample used for Coomassie staining was processed for immunoblot analysis using anti-MBP antibodies. (C) Immunoblot analysis of binding of MBP–Hsl1p667–872 to GST–Cdc20p beads (lanes 1–5) or uninfected cell beads (lanes 6–10). E. coli extracts were incubated with the glutathione beads as described in (A) except that proteins were not eluted from the beads. Immunoblot analysis was performed with anti-MBP (upper panel) and anti-GST antibodies (lower panel).
To test the generality of these findings, we tested whether Clb2p, a mitotic cyclin that is a well-documented APC substrate (Irniger et al. 1995; Schwab et al. 1997; Jaspersen et al. 1998; Zachariae et al. 1998) could associate also with Cdc20p and Cdh1p by coimmunoprecipitation studies. Clb2p–HA or Clb2pmdb–HA were co-overexpressed with GST–Cdc20p or GST–Cdh1p and analyzed as for Hsl1p–HA (Fig. 3B). Mutation of this D box stabilizes Clb2p (Irniger et al. 1995; Schwab et al. 1997; Jaspersen et al. 1998; Zachariae et al. 1998). Clb2p–HA was able to coimmunoprecipitate both GST–Cdc20p and GST–Cdh1p (Fig. 3B, lower panels, lanes 2 and 5), but these proteins were not present in immunoprecipitates from strains lacking an HA-tag (Fig. 3B, lower panels, lanes 1 and 4). However, coimmunoprecipitation of GST–Cdc20p and GST–Cdh1p was not affected by mutation of the Clb2p D box (Fig. 3B, lower panels, lanes 3 and 6). It is possible that a recently identified KEN box in Clb2p (S. Holloway, pers. comm.) may mediate association of Clb2pmdb with Cdc20p and Cdh1p. It is also possible that the interaction we observed between Cdc20p and Clb2p may reflect phosphorylation of Cdc20p by Cdc28p, as occurs with CDK2 and human CDC20 (Ohtoshi et al. 2000). These results indicate that the association of APC substrates with Cdc20p and Cdh1p is not unique to Hsl1p and that other APC substrates might associate with these proteins, although the requirements for the interactions may differ.
Hsl1p667–872 can act as a transposable degradation signal
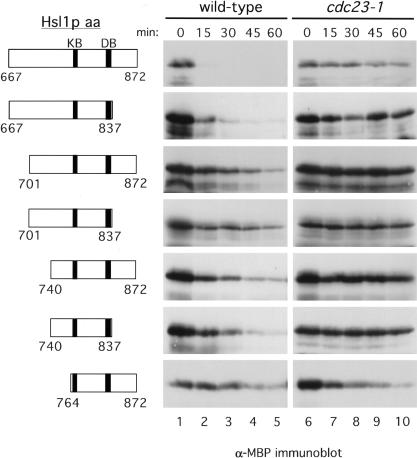
We investigated whether fragments of Hsl1p containing both the KEN box and the D box, located at amino acids 775–781 and 828–836, respectively, could serve as portable degradation signals. We started with a 206-amino-acid fragment containing both motifs plus 108 amino acids N-terminal to the KEN box (Hsl1p667–872; Fig. 4, top row, left panel). The upstream residues were included because of the observation that APC-mediated degradation of hCDC20 requires amino acid residues upstream of the KEN box (Pfleger and Kirschner 2000). Hsl1p667–872 was fused to the C terminus of the E. coli maltose binding protein (MBP) and expressed in yeast. The stabilities of this fusion protein and of progressively smaller fusion proteins comprising the KEN box and D box were tested in G1-arrested cells to examine APCCdh1p-mediated degradation (Fig. 4). MBP alone was found to be stable when expressed in yeast cells (data not shown). MBP–Hsl1p667–872 was very unstable and was undetectable 15 min after terminating its synthesis (Fig. 4, top row, middle panel). The instability of MBP–Hsl1p667–872 was due to APC-mediated degradation as the protein was stable in cdc23-1 cells (Fig. 4, top row). MBP–Hsl1p667–837, which removes 35 amino acid residues downstream of the D box, was also quite unstable; this instability was APC-dependent (Fig. 4, second row). However, truncation of the N-terminal domain upstream of the KEN box greatly stabilized the MBP–Hsl1p fusion proteins (MBP–Hsl1701–872, MBP–Hsl1p740–872, and MBP–Hsl1p764–872; Fig. 4, rows 3–7). These results suggest that sequences upstream of the KEN box motif are important for degradation of the fusion protein, although it is not yet clear if this region contains an additional degradation motif or if it provides a structural requirement for KEN box recognition. These results show that fragments of Hsl1p can act as transposable degradation signals when fused to MBP. Based on its rapid degradation, we chose MBP–Hsl1p667–872 for further analysis.
Figure 4.
Analysis of Hsl1p sequences that can act as transposable APC-dependent degradation signals when fused to the carboxyl terminus of the maltose binding protein (MBP). Left panels, schematic representations of amino acids of Hsl1p containing the KEN box and D box motifs (shaded boxes) that were fused to MBP (data not shown). Middle and right panels, stability of MBP–Hsl1p proteins in G1-arrested cells. Wild-type and cdc23-1 cells were arrested in G1 with α-factor (100 ng/mL) and then induced to express GAL–MBP–HSL1667–872 (YJB306 and YJB311), GAL–MBP–HSL1667–837 (YJB320 and YJB331), GAL–MBP–HSL1701–872 (YJB321 and YJB335), GAL–MBP–HSL1701–837 (YJB329 and YJB336), GAL–MBP–HSL1740–872 (YJB322 and YJB332), GAL–MBP–HSL1740–837 (YJB330 and YJB337) and GAL–MBP–HSL1764–872 (YJB307 and YJB311) fusions by the addition of galactose. Strains were subsequently shifted to 37°C to inactivate the APC in the cdc23–1 mutants and the levels of the MBP–Hsl1p fusion proteins were monitored by immunoblot analysis with anti-MBP antibodies at the indicated times after glucose/cycloheximide addition to terminate MBP–Hsl1p synthesis.
First, we wanted to confirm that the APC-mediated degradation of MBP–Hsl1p667–872 was D box- and KEN box-dependent. The stabilities of MBP–Hsl1p667–872, MBP–Hsl1p667–872mdb, MBP–Hsl1p667–872mkb, and MBP–Hsl1p667–872mdb/mkb were monitored in G1-arrested cells as described for full-length Hsl1p. Mutation of either the D box or KEN box resulted in stabilization of MBP–Hsl1p667–872 (Fig. 5A, mdb and mkb) and mutation of both motifs resulted in complete stabilization of the fusion protein (Fig. 5A, mdb/mkb). These findings indicate that APCCdh1-dependent degradation of MBP–Hsl1p667–872 and Hsl1p–HA both require a D box and a KEN box.
Next, we tested whether MBP–Hsl1p667–872, like Hsl1p, could associate with Cdc20p and Cdh1p by coimmunoprecipitation. The different forms of MBP–Hsl1p667–872 were co-overexpressed with either GST–Cdc20p or GST–Cdh1p. The MBP–Hsl1p667–872 fusions were immunoprecipitated and the levels of coimmunoprecipitating GST–Cdc20p or GST–Cdh1p were analyzed by immunoblot analysis (Fig. 5B, lower panels). We found that GST–Cdc20p had the same specificity of interaction with MBP–Hsl1p667–872 as was observed for full-length Hsl1p–HA (cf. Fig. 5B lower panel, lanes 1–5 with Fig. 3A). Mutation of the D box or of both the D box and the KEN box prevented the coimmunoprecipitation of GST–Cdc20p (Fig. 5B, lower panel, lanes 3 and 5), whereas GST–Cdc20p associated equally well with the wild-type and the KEN box mutant forms of MBP–Hsl1p667–872 (Fig. 5B, lower panel, lanes 2 and 4). In contrast, GST–Cdh1p did not associate with MBP–Hsl1p667–872 (Fig. 5B, lower panel, lanes 7–10), indicating either that Cdh1p cannot associate well with this domain of Hsl1p, or that the combination of the MBP and GST tags and anti-MBP–antibodies interferes with the association. We favor the latter possibility because MBP–Hsl1p667–872 is degraded in G1-arrested cells in which APCCdh1 activity predominates (Figs. 4A, 5A) and because of direct binding data between MBP–Hsl1p667–872 and Cdh1p presented below.
Cdc20p and Cdh1p bind directly to Hsl1p667–872 in a D box- and KEN box-dependent manner
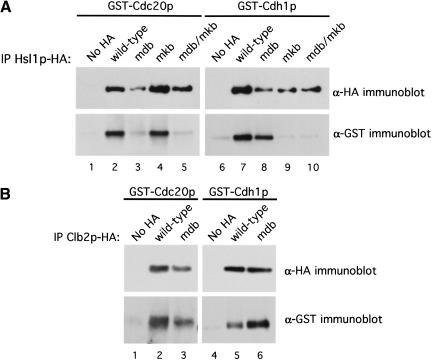
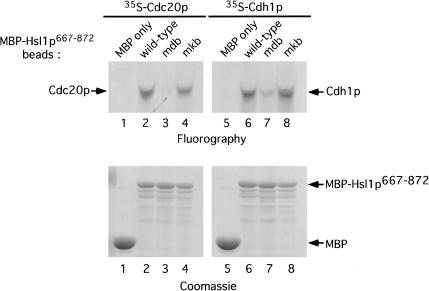
To test whether Hsl1p interacts directly with Cdc20p and Cdh1p, we used recombinant MBP–Hsl1p667–872 produced in E. coli and purified on amylose resin and [35S]methionine-labeled Cdc20p and Cdh1p translated in vitro in reticulocyte lysates (Fig. 6). MBP and the various MBP–Hsl1p proteins on amylose resin were incubated with 35S-labeled Cdc20p and Cdh1p and washed. The bound proteins were run on SDS-polyacrylamide gels and analyzed for Cdc20p or Cdh1p by fluorography (Fig. 6, top panels). 35S-labeled Cdc20p bound to MBP–Hsl1p667–872 and MBP–Hsl1p667–872mkb, but not to MBP alone or to MBP–Hsl1p667–872mdb (Fig. 6, top, left panel, lanes 1–4). 35S-labeled Cdh1p exhibited a similar binding profile for the different forms of MBP–Hsl1p667–872 (Fig. 6, top panels, cf. lanes 6–8 with lanes 2–4). Similar amounts of the recombinant MBP–Hsl1p667–872 proteins were present on the amylose beads, as visualized by Coomassie staining (Fig. 6, lower panels). Currently, we do not know why mutation of the KEN box has less of an effect on the association of MBP–Hsl1p667–872 with Cdh1p (see also below), than on that of full-length Hsl1p with Cdh1p.
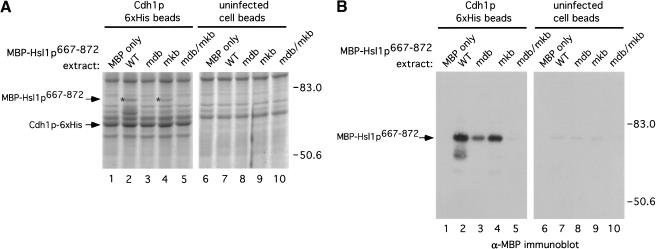
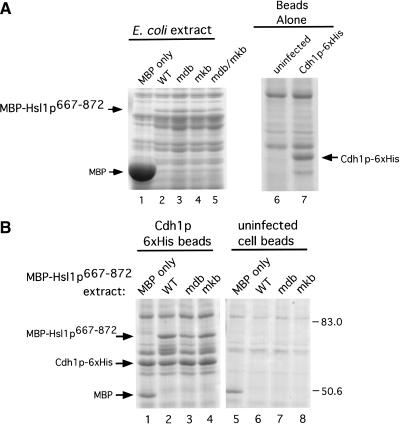
We used purified proteins to rule out the possibility that the interaction of Hsl1p with these proteins was mediated by a core APC subunit or other protein from the reticulocyte lysate. We used purified Cdh1p–6xHis recombinant protein from insect cells (Jaspersen et al. 1999) in binding assays with MBP–Hsl1p667–872. Extracts from uninfected control cells or from cells infected with a baculovirus encoding Cdh1p–6xHis were incubated with Talon metal affinity resin (Clonetech) to purify Cdh1p–6xHis. A band corresponding to Cdh1p–6xHis was observed (Fig. 7A, lane 7 arrow) in the sample derived from baculovirus-infected cells that migrated at the appropriate molecular weight and was absent in the uninfected cell control (Fig. 7A, cf. lanes 6 and 7). Extracts from E. coli cells expressing MBP, MBP–Hsl1p667–872, MBP–Hsl1p667–872mdb, MBP–Hsl1p667–872mkb, or MBP–Hsl1p667–872mdb/mkb were prepared (Fig. 7A, lanes 1–5). The different E. coli extracts were incubated either with resin bound to Cdh1p–6xHis or with beads incubated with uninfected cell extracts. Beads were washed and bound proteins were examined by Coomassie staining of SDS-polyacrylamide gels (Fig. 7B). Protein bands corresponding to the MBP–Hsl1p667–872 fusion proteins were observed clearly in eluates from the Cdh1p–6xHis beads, but not with the uninfected cell bead eluates (Fig. 7B, cf. lanes 2–4 with lanes 6–8). MBP–Hsl1p667–872mdb appeared to bind Cdh1p somewhat less well than MBP–Hsl1p667–872 (Fig. 7B, cf. lane 3 with lane 2). The nonspecific binding of MBP to both of the resins (Fig. 7B, lanes 1 and 5) is likely due to the massive amounts of MBP in the E. coli lysates (Fig. 7A, cf. lane 1 with lanes 2–5). Given the high stoichiometry of MBP–Hsl1p–Cdh1p binding, it is unlikely that their association is mediated by a contaminant in the Cdh1p–6xHis preparation because these are all present at very low stoichiometries relative to Cdh1p–6xHis and are also present in the preparation from uninfected cells (Fig. 7A lanes 6 and 7).
Figure 7.
Direct binding of Cdh1p to MBP–Hsl1p667–872. (A) Coomassie stained gels of starting materials used for the direct binding assays in B and in Figure 8. (Left panel) 10 μL of total E. coli extracts expressing MBP or the different forms of MBP–Hsl1p667–872. (Right panel) 25 μL of cobalt resin from uninfected cell extracts and Cdh1p–6xHis baculovirus-infected Sf9 cell extracts. (B) Cobalt resin was incubated with extracts from insect cells infected with Cdh1p–6xHis baculovirus (lanes_1–4_) or from uninfected cells (lanes 5–8). Resins were washed and then incubated with 1 mL of the indicated E. coli extracts containing MBP or one of the MBP–Hsl1p667–872 fusion proteins and washed. Bound proteins were eluted with 150 mM imidizole and visualized by Coomassie staining following SDS-PAGE.
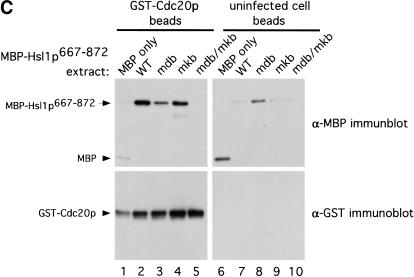
We improved the sensitivity of this assay to D box and KEN box mutations by diluting the E. coli lysates sufficiently (20-fold) that binding of MBP–Hsl1p667–872 proteins began to decline. We also reduced the amount of MBP alone (by 320-fold) so that it was similar to the amount of the MBP–Hsl1p667–872 fusion proteins used. By Coomassie staining we observed binding of MBP–Hsl1p667–872 and MBP–Hsl1p667–872mkb to Cdh1p–6xHis (Fig. 8A, lanes 2 and 4). A faint band corresponding to MBP–Hsl1p667–872mdb was detected by Coomassie staining and confirmed by immunoblot analysis (Fig. 8A,B, lane 3). Mutation of both the D box and the KEN box abolished detectable binding to Cdh1p–6xHis completely (Fig. 8A,B, lane 5). MBP and the MBP–Hsl1p667–872 proteins did not bind to the control uninfected cell resin (Fig. 8A,B, lanes 6–10).
Direct binding of Hsl1p667–872 to Cdc20p was tested using the diluted bacterial extracts expressing MBP or the MBP–Hsl1p667–872 proteins and GST–Cdc20p expressed and purified from insect cells (Fig. 8C). Like Cdh1p–6xHis, GST–Cdc20p bound to MBP–Hsl1p667–872 and MBP–Hsl1p667–872mkb, but showed no detectable binding to MBP–Hsl1p667–872mdb/mkb (Fig. 8C, lanes 2, 4, and 5). Reduced but detectable binding to MBP–Hsl1p667–872mdb was observed (Fig. 8C, cf. lanes 3 and 8). These results indicate that MBP–Hsl1p667–872 binds Cdh1p–6xHis and GST–Cdc20p directly and that the interactions require intact D box and KEN box motifs within Hsl1p.
Discussion
We found that Hsl1p contains both a D box and a KEN box degradation motif, both of which are important for APC-mediated turnover with either APCCdc20 or APCCdh1. A 206-amino-acid segment (Hsl1p667–872) containing these degradation signals was sufficient to promote APC-mediated degradation of an otherwise stable protein in yeast. Finally, this domain of Hsl1p was able to interact directly with Cdh1p and Cdc20p, dependent on an intact D box and KEN box. The identification of a yeast protein with a functional KEN box indicates that this motif is conserved evolutionarily.
The D box and the KEN box form a bipartite degradation signal in Hsl1p
Hsl1p is a member of a growing subfamily of APC substrates that contain KEN boxes. The KEN box was first identified in human CDC20 and Nek2 and in mouse B99 as a motif essential for the Cdh1-dependent degradation of these proteins (Pfleger and Kirschner 2000). Subsequently, essential KEN boxes have been found in human CDC6 (Peterson et al. 2000), human securin (Zur and Brandeis 2001), Drosophila cyclin A (Jacobs et al. 2001), and now budding yeast Hsl1p. In these four cases, a functional D box is located within 53 amino acids of the KEN box (counting from the lysine of the KEN box to the arginine of the D box). A simple database search of budding yeast sequences reveals 92 proteins containing matches to both a basic D box (RxxLxxxx(NDEQ)) and a basic KEN box (KENxxx(NDEQ)), in 32 of which the two motifs are separated by fewer than 100 amino acids. Although only some of these proteins will be authentic APC substrates, the exercise indicates that dual recognition may be a common theme (discussed in more detail below). It is interesting to note that Cut2p and Pds1p, the securin proteins in fission and budding yeast, respectively, each contain a potential KEN box within the first 15 amino acids of the protein, situated upstream of their characterized D boxes. This positioning is virtually identical to that of the characterized KEN box in human securin (Zur and Brandeis 2001), indicating that the positioning of these motifs is critical, despite the low overall similarity between these proteins.
Both the D box and the KEN box are essential for efficient Hsl1p degradation by both APCCdc20 and APCCdh1. The efficient degradation of Hsl1p in G1 cells containing only one form of the APC (either APCCdh1, Figs. 1 and 5, or APCCdc20, Fig. 2) indicates that these signals are recognized in concert, not by parallel pathways. The ability of both APCCdc20 and APCCdh1 to recognize both motifs suggests that many proteins will be substrates for both forms of the APC, although at different times. For Hsl1p it is likely that APCCdc20 initiates Hsl1p degradation during mitosis, whereas any remaining Hsl1p may be degraded by APCCdh1 during mitotic exit and entry into G1. In addition to temporal control, it is possible that differences in substrate specificity between these APC forms could arise from differences in affinity for particular degradation motifs within a substrate, or from weak interactions with other regions of the substrate.
The interplay between the D box and the KEN box is not as clear for the other characterized KEN box-containing proteins. Human CDC20 does not contain an obvious D box and is degraded solely by APCCdh1 (Pfleger and Kirschner 2000). No D box was reported for either Nek2 or B99, both of which can be degraded via APCCdh1, although it was not reported whether they can also be degraded via APCCdc20 (Pfleger and Kirschner 2000). These studies led to the suggestion that the KEN box was required for recognition by APCCdh1 but not APCCdc20. Intriguingly, potential D boxes have been identified in both Nek2 and B99 (C. Pfleger and M. Kirschner, pers. comm.). In human CDC6, mutation of either the D box or the KEN box could stabilize CDC6 partially, whereas the double mutant was stabilized fully (Peterson et al. 2000). For Drosophila cyclin A, both motifs must be intact for full instability (Jacobs et al. 2001). In contrast, either motif was capable of destabilizing human securin (Zur and Brandeis 2001). Human CDC6, Drosophila cyclin A, and human securin were reported to be degraded by APCCdh1, APCCdc20, and either APCCdh1 or APCCd20p, respectively (Peterson et al. 2000; Jacobs et al. 2001; Zur and Brandeis 2001). In these studies, however, it was not tested whether human CDC6 and Drosophila cyclin A could also be degraded via APCCdc20 and APCCdh1, respectively (Peterson et al. 2000; Jacobs et al. 2001). Taken together, these findings suggest that some proteins require a bipartite KEN box/D box signal for APC-mediated proteolysis and that both APCCdh1 and APCCdc20 can recognize the KEN box degradation signal. Other substrates seem to have both motifs, though the roles of each have not yet been tested. Finally, it is possible that still other APC substrates that appear to lack one of these motifs actually have highly degenerate ones that are not recognizable by crude primary sequence analysis (see below).
Direct binding of APC substrates to Cdc20 and Cdh1
Hsl1p binds directly to Cdc20p and Cdh1p and these interactions are mediated by the D box and KEN box within Hsl1p. This finding simultaneously identifies a biochemical role for these degradation motifs and establishes Cdh1p and Cdc20p as substrate targeting proteins for the APC. The simplest model would be that these proteins bind directly to the degradation motifs within the APC substrate. However, it remains a formal possibility that intact D box and KEN box signals are not the actual sites of Cdc20 and Cdh1 binding but rather influence protein conformation at a distant site(s) that interacts with these proteins.
The D box was most important for the interaction of full-length Hsl1p with Cdc20p, whereas the KEN box was more important for the interaction with Cdh1p. Currently, it is unclear why the KEN box was critical for Cdh1p-binding to full-length Hsl1p whereas the D box was important for Cdh1p-binding to MBP–Hsl1p667–872. It is possible that other sequences in the full-length protein interfere with the ability of Cdh1p to recognize the D box. Nevertheless, elimination of both motifs was required to disrupt the interaction between MBP–Hsl1p667–872 and Cdh1p in vitro. Due to the previous absence of demonstrable direct binding to APC substrates, Cdh1 and Cdc20 have been widely viewed as APC activators with one or more of the APC core subunits serving as a substrate receptor. Although we cannot exclude an additional role for Cdc20p and Cdh1p in APC activation, our results establish that one essential function of these proteins is to bind Hsl1p. In agreement with our findings, a direct interaction between the N termini of hCDC20 and hCDH1 with several APC substrates has been shown (Pfleger et al. 2001).
Given the conservation of the APC ubiquitin ligase and of D boxes and KEN boxes, we suggest that Cdc20 and Cdh1 proteins serve to target all substrates to the APC. For many substrates, this interaction may be too weak to survive coimmunoprecipitation regimens. Nonetheless, even a transient interaction, as in other enzyme-substrate interactions, may suffice for presentation to the APC. Hsl1p was identified via its two-hybrid interaction with Cdc20p (Burton and Solomon 2000) and therefore may represent the high-affinity end of a spectrum of interactions of substrates with Cdc20p and Cdh1p. Although direct binding of APC substrates has not been reported previously, the different substrate specificities of APCCdc20 and APCCdh1 (Schwab et al. 1997; Visintin et al. 1997; Fang et al. 1998a; Shirayama et al. 1998; Pfleger and Kirschner 2000), plus the different effects of a D box mutation in full-length Hsl1p on association with Cdc20p and Cdh1p (Burton and Solomon 2000), had provided indirect evidence for a targeting function for Cdc20 and Cdh1.
A model for substrate presentation by Cdc20 and Cdh1
An important aspect of our findings is that substrate binding to Cdc20p and Cdh1p is not necessarily sufficient for efficient degradation of that substrate. For example, mutation of the D box or the KEN box of Hsl1p compromised its degradation severely, but still allowed strong binding to Cdh1p or Cdc20p, respectively. Similarly, mutation of the D box stabilizes Clb2p (Amon et al. 1994; Irniger et al. 1995), but had little effect on its binding to Cdc20p or Cdh1p (Fig. 3). Although a KEN box was sufficient for Hsl1p to bind to Cdh1p in vivo, a D box was still needed for what can be termed the “presentation” of Hsl1p to the APC in an orientation that leads to efficient degradation. Our results suggest that both Cdc20p and Cdh1p need to bind both degradation motifs in order to present Hsl1p to the APC properly. A prediction of this model is that Cdc20p and Cdh1p each have two binding sites, one for a D box and one for a KEN box.
Do other APC substrates interact with Cdc20 and/or Cdh1 via both a D box and a KEN box? The simple answer would seem to be no as only a minority of APC substrates contain both motifs. Interestingly, the two motifs are usually very close together, being separated by 25 to 53 amino acids (from the start of one motif to the start of the second) in the four characterized proteins with both motifs (human CDC6, human securin, Drosophila cyclin A, and yeast Hsl1p). A large number of potential substrates seem to have both motifs close together (see above). This striking juxtaposition is consistent with simultaneous interaction of the two degradation motifs with two binding pockets on Cdc20p or Cdh1p. In a number of APC substrates, only one motif (usually a D box) has been identified and found necessary for degradation. Perhaps in these cases binding via just one motif can present the substrate properly to the APC. Dual motif binding may be more efficient, but one motif can suffice. Indeed, single mutations in Hsl1p reduced degradation severely, but did not eliminate it. An alternative possibility is that both the D box- and the KEN box-binding regions of Cdc20 and Cdh1 must be engaged for efficient substrate presentation. In such a model, substrates that contain only one discernable degradation motif still must contain sequences that are compatible with the second binding pocket of Cdc20 and/or Cdh1 in order to be presented for ubiquitination by the APC. The main binding energy for the interaction would come from the obvious degradation motif, whereas important positional information could come from a weak interaction provided by a highly degenerate motif. Structural insights will be required to address these issues in further detail.
Materials and methods
Yeast strains and plasmid constructions
All yeast strains are derivatives of W303 (ade2-1 trp1-1 leu2-3,112 his3-11,15 ura3-1 can1-100); their relevant genotypes are listed in Table 1. Plasmids are denoted by brackets. Yeast transformations were performed using published methods (Gietz et al. 1995). All PCR products were sequenced to confirm that no extraneous mutations were introduced. Underlined residues in oligonucleotides indicate introduced restriction sites or nucleotide changes, as appropriate. All mutagenesis was performed by Quikchange (Stratagene). Yeast media (YPD and complete minimal [CM]) were prepared as described (Ausubel et al. 1995).
Table 1.
Yeast strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| YJB14 | MATa bar1Δ | Burton and Solomon 2000 |
| YJB115 | MATa bar1Δ::URA3 cdc23-1 | Burton and Solomon 2000 |
| YJB123 | MATa bar1Δ GAL-HSL1-HA::TRP1 | Burton and Solomon 2000 |
| YJB125 | MATa bar1Δ::URA3 cdc23-1 GAL-HSL1-HA::TRP1 | Burton and Solomon 2000 |
| YJB156 | YJB123 [pEG-KT-_CDC20_] | Burton and Solomon 2000 |
| YJB218 | YJB123 [pEG-KT-_CDH1_] | Burton and Solomon 2000 |
| YJB221 | YJB14 [pEG-KT-_CDC20_] | Burton and Solomon 2000 |
| YJB222 | YJB14 [pEG-KT-_CDH1_] | Burton and Solomon 2000 |
| YJB229 | MATa bar1Δ GAL-HSL1mdb-HA::LEU2 | Burton and Solomon 2000 |
| YJB230 | YJB229 [pEG-KT-_CDC20_] | Burton and Solomon 2000 |
| YJB231 | YJB229 [pEG-KT-_CDH1_] | Burton and Solomon 2000 |
| YJB257 | MATa bar1Δ GAL-HSL1mkb-HA::LEU2 | This study |
| YJB258 | MATa bar1Δ GAL-HSL1mdb/mkb-HA::LEU2 | This study |
| YJB259 | YJB257 [pEG-KT-_CDC20_] | This study |
| YJB260 | YJB258 [pEG-KT-_CDC20_] | This study |
| YJB261 | YJB257 [pEG-KT-_CDH1_] | This study |
| YJB262 | YJB258 [pEG-KT-_CDH1_] | This study |
| YJB265 | MATa bar1Δ::URA3 cdc23-1 GAL-HSL1mdb/mkb-HA::LEU2 | This study |
| YJB266 | MATa bar1Δ GAL-HSL1-HA::LEU2 | This study |
| YJB270 | MATa bar1Δ::URA3 cdc23-1 GAL-HSL1mdb-HA::LEU2 | This study |
| YJB271 | MATa bar1Δ::URA3 cdc23-1 GAL-HSL1mkb-HA::LEU2 | This study |
| YJB272 | MATa bar1Δ::URA3 cdc23-1 GAL-HSL1mdb/mkb-HA::LEU2 | This study |
| YJB273 | MATa bar1Δ GAL-CLB2-HA::LEU2 | This study |
| YJB274 | MATa bar1Δ GAL-CLB2mdb-HA::LEU2 | This study |
| YJB275 | YJB273 [pEG-KT-_CDC20_] | This study |
| YJB276 | YJB274 [pEG-KT-_CDC20_] | This study |
| YJB277 | YJB273 [pEG-KT-_CDH1_] | This study |
| YJB278 | YJB274 [pEG-KT-_CDH1_] | This study |
| YJB306 | YJB14 [_GAL-MBP-HSL1667-872-_YEplac181] | This study |
| YJB307 | YJB14 [_GAL-MBP-HSL1764-872-_YEplac181] | This study |
| YJB308 | YJB115 [_GAL-MBP-HSL1667-872-_YEplac181] | This study |
| YJB309 | YJB306 [pEG-KT-_CDC20_] | This study |
| YJB310 | YJB306 [pEG-KT-_CDH1_] | This study |
| YJB311 | YJB115 [_GAL-MBP-HSL1764-872-_YEplac181] | This study |
| YJB320 | YJB14 [_GAL-MBP-HSL1667-837-_YEplac181] | This study |
| YJB321 | YJB14 [_GAL-MBP-HSL1701-872-_YEplac181] | This study |
| YJB322 | YJB14 [_GAL-MBP-HSL1740-872-_YEplac181] | This study |
| YJB326 | YJB14 [_GAL-MBP-HSL1mdb667-872-_YEplac181] | This study |
| YJB327 | YJB14 [_GAL-MBP-HSL1mkb667-872-_YEplac181] | This study |
| YJB328 | YJB14 [_GAL-MBP-HSL1mdb/mkb667-872-_YEplac181] | This study |
| YJB329 | YJB14 [_GAL-MBP-HSL1701-837-_YEplac181] | This study |
| YJB330 | YJB14 [_GAL-MBP-HSL1740-837-_YEplac181] | This study |
| YJB331 | YJB115 [_GAL-MBP-HSL1667-837-_YEplac181] | This study |
| YJB332 | YJB115 [_GAL-MBP-HSL1740-872-_YEplac181] | This study |
| YJB335 | YJB115 [_GAL-MBP-HSL1701-872-_YEplac181] | This study |
| YJB336 | YJB115 [_GAL-MBP-HSL1701-837_-YEplac181] | This study |
| YJB337 | YJB115 [_GAL-MBP-HSL1740-837-_YEplac181] | This study |
| YJB338 | YJB326 [pEG-KT-_CDC20_] | This study |
| YJB339 | YJB327 [pEG-KT-_CDC20_] | This study |
| YJB340 | YJB328 [pEG-KT-_CDC20_] | This study |
| YJB341 | YJB326 [pEG-KT-_CDH1_] | This study |
| YJB342 | YJB327 [pEG-KT-_CDH1_] | This study |
| YJB343 | YJB328 [pEG-KT-_CDH1_] | This study |
| YJB366 | W303a cdc28-13:TRP1 cdh1Δ::LEU2 GAL-HSL1-HA::HIS3 | This study |
| YJB367 | YJB366 GAL-CDC20-myc::URA3 | This study |
| YJB368 | W303a cdc28-13::TRP1 cdh1Δ::LEU2 | This study |
| YJB377 | YJB368 GAL-HSLlmdb-HA::HIS3 | This study |
| YJB387 | YJB377 GAL-CDC20-myc::URA3 | This study |
| YJB379 | YJB368 GAL-HSL1mkb-HA::HIS3 | This study |
| YJB380 | YJB379 GAL-CDC20-myc::URA3 | This study |
The _GAL–HSL1–HA_–YIplac128 and _GAL–HSL1mdb_–_HA_–YIplac128 constructs were described previously (Burton and Solomon 2000). _GAL–HSL1mkb_–_HA_–YIplac128 and _GAL–HSL1mdb/mkb–HA_–YIplac128 were made from _GAL–HSL1–HA_–YIplac128 or _GAL–HSL1mdb–HA_–YIplac128, respectively, using oligonucleotides MSO880 (5′-CG/ATC/TCT/GGG/GTG/TCT/ACA/AAT/GCG/GCA/GCT/GAG/GGC/CCG/GCG/TAT/CC A/ACC/AAA/ATT/GAG-3′) and MSO881 (5′-CTC/AAT/TTT/GGT/TGG/ATA/CGC/CGG/GCC/CTC/AGC/TGC/CGC/AT T/TGT/AGA/CAC/CCC/AGA/GAT/CG-3′). The nucleotide changes result in the mutagenesis of the KEN box motif (bold indicates mutated amino acids) K775E776N777XXXE781 within Hsl1p to A775A776A777XXXA781.
_GAL–CLB2–HA_–YIplac128 was made by PCR amplification of CLB2 using MSO899 (5′-CCC/GGA/TCC/ATG/TCC/AAC/CCA/ATA/GAA/AAC/ACA/G-3′) and MSO898 ( 5′-CCC/GTC/GAC/TTC/ATG/CAA/GGT/CAT/TAT/ATC/ATA/GC C-3′), digesting with _Bam_HI and _Sal_I and ligating to GAL_–YIplac128–_HA cut with the same enzymes. _GAL–CLB2mdb–HA_–YIplac128 was created by mutagenesis of _GAL–CLB2–HA_–YIplac128 using oligonucleotides MSO891 (5′-GG/TTT/TTG/AGG/AAT/GTA/CAA/GCT/TTG/GCC/GCA/AAC/AAT/GT A/ACA/GCT/ACG/ACA/TTT/CAA/AAG/AGT/AAT/GCG-3′) and MSO892 (5′-CGC/ATT/ACT/CTT/TTG/AAA/TGT/CGT/AGC/TGT/TAC/ATT/GTT/TGC/GGC/CAA/AGC/TTG/TA C/ATT/CCT/CAA/AAA/CC-3′). Nucleotide changes result in the mutagenesis of the R25XXL28XXXXN33 destruction box within Clb2p to A25XXA28XXXXA33.
To make maltose-binding protein (MBP)–HSL1 fusions in yeast, the _GAL–MBP_–YEplac181 vector was constructed. The malE gene and MCS from pMAL-c2 (New England Biolabs) was amplified using MSO910 (5′-CCC/GAT/ATC/ATG/AAA/ATC/GAA/GAA/GGT/AAA/CTG/G-3′) and MSO911 (5′-CCC/AAG/CTT/GCC/TGC/AGG/TCG/ACT/CTA/GAG/G-3′), cut with _Eco_RV and _Hin_dIII, and ligated to pEG-[KT] (Mitchell et al. 1993) that was cut with _Sac_I, end-filled with T4 DNA polymerase, and then cut with _Hin_dIII. This construct was then digested with _Sca_I and _Hin_dIII to isolate a _GAL–malE_-containing fragment with the MCS of pMAL-c2. This fragment was ligated to YEplac181 that had been cut with _Eco_RI, blunted, and then cut with _Hin_dIII to yield _GAL–MBP_–YEplac181. Various fragments of HSL1 (see below) were amplified by PCR, cut with _Bam_HI and _Sal_I, and ligated to _GAL–MBP_–YEplac181 cut with the same enzymes to yield constructs encoding MBP–Hsl1p fusions for expression in yeast cells. _GAL–MBP–HSL1_667–872–YEplac181 and mutant derivatives were constructed by PCR from _GAL–HSL1–HA_–YIplac128 and its mutant versions using MSO919 (5′-CCC/GGA/TCC/ATG/CAA/AAC/TCG/GCT/TCA/AAG/TCC-3′) and MSO920 (5′-CCC/GTC/GAC/CTG/AAT/AGG/TTT/GAG/TGG/TG-3′). The following GAL–MBP–HSL1 derivatives were made by PCR with the indicated oligonucleotides: _GAL–MBP–HSL1667–837–YEplac181, MSO919 and MSO940 (5′-CCC/GTC/GAC/TGA/GTT/CGT/GAT/ATC/TGA/AAG-3′); GAL–MBP–HSL1701–872_–YEplac181, MSO942 (5′-CCC/GGA/TCC/ATG/AAG/AAA/CCA/GCA/TCC/GAA/AAT/GTG-3′) and MSO920; _GAL–MBP–HSL1_701–837–YEPlac181, MSO942 and MSO940; _GAL–MBP–HSL1_740–872–YEplac181, MSO943 (5′-CCC/GGA/TCC/ATG/GAA/GAG/GAA/GAG/GAC/AAT/G-3′) and MSO920; _GAL–MBP–HSL1_740–837–YEPlac181, MSO943 and MSO940; _GAL–MBP–HSL1_764–872–YEplac181, MS0918 (5′-CCC/GGA/TCC/ATG/GAC/ACT/TTT/ACG/ATC/TCT/GGG-3′) and MSO920.
_HSL1_667–872–pMAL-c2 and mutant derivatives were constructed for expression of MBP–Hsl1p667–872 in E. coli by cutting the appropriate _GAL–MBP–HSL1_667–872–YEplac181 plasmids with _Bam_HI and _Sal_I to isolate the DNA fragment containing _HSL1_667–872, which was then ligated to pMAL-c2 cut with the same enzymes.
_GAL–HSL1–HA_–pRS303 was described previously (Burton and Solomon 2000). _GAL–HSL1mdb–HA_–pRS303 and _GAL–HSL1mkb–HA_–pRS303 were made by cutting the corresponding constructs in YIplac128 with _Hin_dIII, blunting the ends with Klenow (New England Biolabs), digesting with _Bam_HI, isolating the HSL1–HA isoforms, and ligating into pRS303 cut with _Xba_I, end-filled with Klenow, and then cut with _Bam_HI.
pEG–[KT]–CDC20 and pEG–[KT]–CDH1 were described previously (Burton and Solomon 2000). GAL–CDC20–myc_–YIplac211 was constructed by isolating CDC20 from pEG–[KT]–_CDC20 by cutting with _Sma_I and Sal_I. The CDC20 fragment was then ligated to YIplac211–_myc, cut with _Bam_HI, end-filled with Klenow, and then cut with _Sal_I. CDC20_– and CDH1_–Bluescript constructs used for in vitro translations (see below) were obtained by PCR using pEG–[KT]–_CDC20 and pEG–[KT]–_CDH1 as PCR templates. For CDC20, oligonucleotides MSO954 (5′-CCC/ATG/GCA/ATG/CCA/GAA/AGC/TCT/AGA/G-3′) and MSO955 (5′-CCC/CTC/GAG/CCT/GAT/CAA/ATA/TTG/GCT/GG-3′) were used, and for CDH1, oligonucleotides MSO956 (5′-CCC/ATG/GCA/ATG/TCC/ACA/AAC/CTG/AAC/CC-3′) and MSO957 (5′-CCC/CTC/GAG/ACG/TAT/TTG/ATT/AAA/TGC/GTC-3′) were used. The resulting PCR products were cut with _Nco_I and Xho_I and ligated to Bluescript II KS− (Stratagene) cut with the same enzymes. The GST–_CDC20 baculovirus construct was made by subcloning CDC20 into the pVT4 vector (a kind gift from Vasiliki Tsakraklides, Yale University, New Haven, CT). pVT4 was constructed by PCR amplification of GST using MSO810 (5′-GGG/GGA/TCC/ATG/TCC/CCT/ATA/CTA/GGT/TAT-3‘) and MSO811 (5′-G/GGC/GGA/CCG/CTT/AAG/ATC/GAT/TCC/CGG/GCC/CAT/GGA/GCC/ACG/CGG/AAC/CAG/GGA/GGA/GGA/TTT/TGG/AGG/ATG/GTC/GCC/ACC-3′), digestion with _Bam_HI and _Rsr_II, and ligation to pFASTBAC1 (GIBCO BRL) cut with the same enzymes. The complete CDC20 coding region was cloned into pVT4 by a blunt-end ligation using the _Afl_II site in pVT4. The _CDH1_–6xHis baculoviral construct (Jaspersen et al. 1999) was a kind gift from Dr. David Morgan (University of California, San Francisco).
Hsl1p half-life studies
Half-life experiments for Hsl1p–HA and the various MBP–Hsl1p fusions were essentially as described previously (Burton and Solomon 2000). Briefly, cells were arrested in G1 with α-factor (100 ng/mL), induced to express Hsl1p–HA or MBP–Hsl1p by the addition of galactose (2%), and shifted to 37°C to inactivate the APC where appropriate. Expression was terminated by the addition of glucose (2%) and cycloheximide (0.5 mg/mL). For half-life studies in cdh1Δcdc28-13 cells expressing Hsl1p–HA isoforms, a G1 arrest was achieved by shifting to 37° C for 3 h and then treating cells as above except that cells were kept at 37°C for the remainder of the experiment. Levels of protein were monitored by immunoblot analysis as described previously (Burton and Solomon 2000), using either rabbit anti-HA antibodies (50 ng/mL; Santa Cruz) or anti-MBP antibodies (0.36 μg/mL). Quantitative immunoblot analysis was performed on scanned autoradiographs using the program NIH Image 1.62 (developed at the U.S. National Institutes of Health and available on the Internet at http://rsb.info.nih.gov/nih-image/). Experimental samples and serial dilutions of Hsl1p–HA from the same autoradiograph were scanned and assigned arbitrary units based on the dilution standard. Plots of wild-type Hsl1p–HA are in arbitrary units, whereas the plots for Hsl1pmdb–HA and Hsl1pmkb–HA represent the ratios of Hsl1p levels with and without Cdc20–myc expression to adjust for Cdc20p independent degradation of these proteins. Samples for FACS were prepared as described previously (Burton and Solomon 2000).
Protein extract preparation
Protein extracts for Hsl1p half-life studies using full-length Hsl1p–HA were prepared as described previously (Burton and Solomon 2000). For MBP–Hsl1p fusions, extracts were prepared by bead-beating for 4 min in 1× sample buffer (SB; 16.6% SDS, 26% glycerol, 262 mM Tris base, 150 mM DTT) and boiling for 10 min. Samples were spun for 5 min in a microcentrifuge and the supernatants were centrifuged for 10 min at 70,000 rpm at 15°C in a Beckman Optima ultracentrifuge in a TLA-100.2 rotor.
Coimmunoprecipitation analysis
For coexpression of wild-type, mdb, mkb, or mdb/mkb forms of full-length Hsl1p–HA with either GST–Cdc20p or GST–Cdh1p, cells were grown overnight in 100 mL of CM-Ura raffinose (4%) to an OD600 of 0.35–0.40. Galactose (4%) was added for 6 h at 30°C to induce the expression of Hsl1p–HA. Analysis of MBP–Hsl1p fusions, coexpressed with GST–Cdc20p or GST–Cdh1p was as described above, except that CM-Ura-Leu medium was used. Extract preparation and immunoprecipitations were performed as described previously (Burton and Solomon 2000), except that anti-MBP antibodies (18 μg) were used for immunoprecipitating MBP–Hsl1p fusions.
In vitro binding assays
Recombinant MBP or MBP–Hsl1p667–872 isoforms were produced by induction in E. coli with 0.3 mM IPTG (Sigma) for 2 h at 37°C at an OD600 = 0.5. Cells were pelleted and then disrupted in Lysis buffer (20 mM Tris-HCl at pH 7.5, 200 mM NaCl) containing protease inhibitors (1 mM PMSF, 10 μg each leupeptin, chymostatin, and pepstatin [Chemicon]) by sonication for three times 1 min on ice with a 5 min rest between pulses (setting 5, Branson sonifier 450). Lysates were clarified by centrifugation at 10,000 rpm for 30 min in an SA600 rotor (Sorvall) at 4°C. Lysates were used either directly or diluted 20-fold for MBP–Hsl1p667–872 isoforms or 320-fold for MBP for use in binding assays using Cdh1p–6xHis bound to cobalt resin (Talon resin, Clonetech) or GST–Cdc20p bound to glutathione resin (Sigma; see below). For binding assays using in vitro translated 35S-Cdc20p or 35S-Cdh1p (see below), lysates were prepared as above except that the Lysis buffer also contained 1 mM EDTA. MBP and MBP–Hsl1p667–872 isoforms were purified from the E. coli lysates by incubation with amylose resin (New England Biolabs) for 2 h and washed three times with Wash buffer (20 mM Tris-HCl at pH 7.5, 500 mM NaCl, 1 mM EDTA). A final wash was performed in Lysis buffer with 1mM EDTA.
_CDC20_– and _CDH1_–Bluescript plasmids were translated in vitro using the TNT® T7-coupled reticulocyte lysate system (Promega) for 3 h at 30°C. Proteins were labeled using 0.4 μCi/μL [35S]methionine (NEN) in the reaction. 10 μL of the above reaction was incubated with 10 μL of amylose resin containing bound MBP or MBP–Hsl1p667–872 isoforms (see above) in 1 mL of Lysis buffer plus 1 mM EDTA for 2 h at 4°C with rotation. Beads were then washed three times in 1 mL Wash buffer and samples were run on SDS-PAGE and processed for fluorography.
Cdh1p–6xHis and GST–Cdc20p baculoviruses were used to infect 108 Sf9 insect cells at a multiplicity of infection of 5 for 48 h. Infected and uninfected cells were pelleted at 500_g_ for 5 min at 4°C, frozen in liquid nitrogen, and stored at −80°C for future use. Cells were lysed in 15 mL Lysis buffer as described (Fisher et al. 1995). Lysates were clarified by ultracentrifugation at 40,000 rpm for 30 min at 4°C in a 60Ti rotor (Beckman). Lysates were then incubated with either 500 μL of Talon cobalt resin (50% slurry, Clonetech) or 400 μL glutathione agarose (50% slurry, Sigma) for 1.5 h at 4°C with rotation. Talon beads were washed three times with 5 mL Buffer D (50 mM sodium phosphate, 300 mM NaCl, 10% glycerol at pH 7.8) plus 1 mM DTT and 10 mM imidizole and then transferred to a 1.7-mL tube in Buffer D plus 1 mM DTT. Beads were pelleted and resuspended in 250 μL of Buffer D plus 1 mM DTT. Glutathione beads were washed three times with 5 mL of wash buffer (10 mM sodium phosphate at pH 7.5, 500 mM NaCl, 1% Triton X-100) and once with 1 mL 1× PBS buffer (10 mM sodium phosphate at pH 7.5, 150 mM NaCl). Beads were transferred to a 1.7-mL tube, pelleted, and resuspended in 200 μL of 1× PBS. Fifty microliters of either bead slurry were then incubated with 1 mL of E. coli extract expressing MBP or MBP–Hsl1p667–872 isoforms for 2 h at 4°C with rotation. Beads were pelleted, washed three times in 1 mL immunoprecipitation buffer (50 mM potassium HEPES at pH 7.6, 1 mM MgCl2, 0.1% Tween-20, 10% glycerol), and then either eluted with 30 μL of Buffer D plus 1 mM DTT and 150 mM imidizole for 30 min at 4°C with rotation for Cdh1p–6xHis beads or resuspended directly in 25 μL of 2.5× SB (7.5% SDS, 287.5 mM sucrose, 162.5 mM Tris-HCl at pH 6.8, 357.5 mM β-mercaptoethanol) for GST–Cdc20p beads. 6 μL of 5× SB was added to the eluted samples. For Cdh1p–6xHis samples, 25 μL were run on SDS-PAGE for Coomassie staining; 2.5-μL samples were run on SDS-PAGE for immunoblot analysis with α-MBP antibodies (72 ng/mL). For GST–Cdc20p samples, first 5 μL were processed for immunoblot analysis with anti-MBP antibodies to detect MBP–Hsl1p667–872 binding. Then the immunoblot was stripped as described previously (Burton and Solomon 2000) and re-probed with anti-GST antibodies to detect GST–Cdc20p present on the glutathione beads.
Acknowledgments
We thank David Morgan for the Cdh1p–6xHis baculovirus; Zachary Pitluck for affinity-purified anti-MBP and anti-GST antibodies; Rocco Carbone from The Yale Cancer Center Flow Cytometry Shared Resource (U.S. Public Health Service grant CA-16359) for performing FACS analysis; Vasiliki Tsakraklides for help with the baculovirus work; Adrienne Natrillo for technical assistance; Aiyang Cheng, Mark Hochstrasser, Denis Ostapenko and Vasiliki Tsakraklides for critical reading of the manuscript; and Philipp Kaldis and Karen Ross for insightful discussions. We are grateful to Cathie Pfleger and Marc Kirschner for communication of unpublished results.
This work was supported by a Jane Coffin Childs Fellowship (J.L.B.), grant T32-CA-09159-24 from the NIH awarded to the Molecular & Oncologic Virology Training Program (J.L.B.), grant GM47830 from the NIH (M.J.S.), and grant RPG-98-270 from the American Cancer Society (M.J.S.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL Mark.Solomon@Yale.edu; FAX (203) 432-3104.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.917901.
References
- Amon A, Irniger S, Nasmyth K. Closing the cell cycle circle in yeast: G2 cyclin proteolysis initiated at mitosis persists until the activation of G1 cyclins in the next cycle. Cell. 1994;77:1037–1050. doi: 10.1016/0092-8674(94)90443-x. [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. New York: John Wiley & Sons; 1995. [Google Scholar]
- Barral Y, Parra M, Bidlingmaier S, Snyder M. Nim1-related kinases coordinate cell cycle progression with the organization of the peripheral cytoskeleton in yeast. Genes & Dev. 1999;13:176–187. doi: 10.1101/gad.13.2.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco MA, Sanchez-Diaz A, de Prada JM, Moreno S. APCste9/srw1 promotes degradation of mitotic cyclins in G1 and is inhibited by cdc2 phosphorylation. EMBO J. 2000;19:3945–3955. doi: 10.1093/emboj/19.15.3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton JL, Solomon MJ. Hsl1p, a Swe1p inhibitor, is degraded via the anaphase-promoting complex. Mol Cell Biol. 2000;20:4614–4625. doi: 10.1128/mcb.20.13.4614-4625.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A. The ubiquitin-proteasome proteolytic pathway. Cell. 1994;79:13–21. doi: 10.1016/0092-8674(94)90396-4. [DOI] [PubMed] [Google Scholar]
- Ciosk R, Zachariae W, Michaelis C, Shevchenko A, Mann M, Nasmyth K. An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell. 1998;93:1067–1076. doi: 10.1016/s0092-8674(00)81211-8. [DOI] [PubMed] [Google Scholar]
- Cohen-Fix O, Peters JM, Kirschner MW, Koshland D. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes & Dev. 1996;10:3081–3093. doi: 10.1101/gad.10.24.3081. [DOI] [PubMed] [Google Scholar]
- Deshaies RJ, Chau V, Kirschner MW. Ubiquitination of the G1 cyclin Cln2p by a Cdc34p-dependent pathway. EMBO J. 1995;14:303–312. doi: 10.1002/j.1460-2075.1995.tb07004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang G, Yu H, Kirschner MW. The checkpoint protein MAD2 and the mitotic regulator CDC20 form a ternary complex with the anaphase-promoting complex to control anaphase initiation. Genes & Dev. 1998a;12:1871–1883. doi: 10.1101/gad.12.12.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang G, Yu H, Kirschner MW. Direct binding of CDC20 protein family members activates the anaphase-promoting complex in mitosis and G1. Mol Cell. 1998b;2:163–171. doi: 10.1016/s1097-2765(00)80126-4. [DOI] [PubMed] [Google Scholar]
- Feldman RM, Correll CC, Kaplan KB, Deshaies RJ. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- Fisher RP, Jin P, Chamberlin HM, Morgan DO. Alternative mechanisms of CAK assembly require an assembly factor or an activating kinase. Cell. 1995;83:47–57. doi: 10.1016/0092-8674(95)90233-3. [DOI] [PubMed] [Google Scholar]
- Funabiki H, Yamano H, Kumada K, Nagano K, Hunt T, Yanagida M. Cut2 proteolysis required for sister-chromatid separation in fission yeast. Nature. 1996;381:438–441. doi: 10.1038/381438a0. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH, Willems AR, Woods RA. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- Glotzer M, Murray AW, Kirschner MW. Cyclin is degraded by the ubiquitin pathway. Nature. 1991;349:132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. Ubiquitin-dependent protein degradation. Annu Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- Irniger S, Piatti S, Michaelis C, Nasmyth K. Genes involved in sister chromatid separation are needed for B-type cyclin proteolysis in budding yeast. Cell. 1995;81:269–278. doi: 10.1016/0092-8674(95)90337-2. [DOI] [PubMed] [Google Scholar]
- Jacobs HW, Keidel E, Lehner CF. A complex degradation signal in Cyclin A required for G1 arrest, and a C-terminal region for mitosis. EMBO J. 2001;20:2376–2386. doi: 10.1093/emboj/20.10.2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen SL, Charles JF, Tinker-Kulberg RL, Morgan DO. A late mitotic regulatory network controlling cyclin destruction in Saccharomyces cerevisiae. Mol Biol Cell. 1998;9:2803–2817. doi: 10.1091/mbc.9.10.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen SL, Charles JF, Morgan DO. Inhibitory phosphorylation of the APC regulator Hct1 is controlled by the kinase Cdc28 and the phosphatase Cdc14. Curr Biol. 1999;9:227–236. doi: 10.1016/s0960-9822(99)80111-0. [DOI] [PubMed] [Google Scholar]
- King RW, Deshaies RJ, Peters JM, Kirschner MW. How proteolysis drives the cell cycle. Science. 1996;274:1652–1659. doi: 10.1126/science.274.5293.1652. [DOI] [PubMed] [Google Scholar]
- Kotani S, Tanaka H, Yasuda H, Todokoro K. Regulation of APC activity by phosphorylation and regulatory factors. J Cell Biol. 1999;146:791–800. doi: 10.1083/jcb.146.4.791. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kramer ER, Gieffers C, Holzl G, Hengstschlager M, Peters JM. Activation of the human anaphase-promoting complex by proteins of the CDC20/Fizzy family. Curr Biol. 1998;8:1207–1210. doi: 10.1016/s0960-9822(07)00510-6. [DOI] [PubMed] [Google Scholar]
- Krek W. Proteolysis and the G1–S transition: The SCF connection. Curr Opin Genet Dev. 1998;8:36–42. doi: 10.1016/s0959-437x(98)80059-2. [DOI] [PubMed] [Google Scholar]
- Lew DJ, Reed SI. A cell cycle checkpoint monitors cell morphogenesis in budding yeast. J Cell Biol. 1995;129:739–749. doi: 10.1083/jcb.129.3.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim HH, Goh P-Y, Surana U. Cdc20 is essential for the cyclosome-mediated proteolysis of both Pds1 and Clb2 during M phase in budding yeast. Curr Biol. 1998;8:231–234. doi: 10.1016/s0960-9822(98)70088-0. [DOI] [PubMed] [Google Scholar]
- Listovsky T, Zor A, Laronne A, Brandeis M. Cdk1 is essential for mammalian cyclosome/APC regulation. Exp Cell Res. 2000;255:184–191. doi: 10.1006/excr.1999.4788. [DOI] [PubMed] [Google Scholar]
- LoRincz A, Reed SI. Sequence analysis of temperature-sensitive mutations in the Saccharomyces cerevisiae gene CDC28. Mol Cell Biol. 1986;6:4099–4103. doi: 10.1128/mcb.6.11.4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan JN, Longtine MS, Sia RAL, Theesfeld CL, Bardes ESG, Pringle JR, Lew DJ. The morphogenesis checkpoint in Saccharomyces cerevisiae: Cell cycle control of Swe1p degradation by Hsl1p and Hsl7p. Mol Cell Biol. 1999;19:6929–6939. doi: 10.1128/mcb.19.10.6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell DA, Marshall TK, Deschenes RJ. Vectors for the inducible overexpression of glutathione S-transferase fusion proteins in yeast. Yeast. 1993;9:715–722. doi: 10.1002/yea.320090705. [DOI] [PubMed] [Google Scholar]
- Ohtoshi A, Maeda T, Higashi H, Ashizawa S, Hatakeyama M. Human p55/Cdc20 associates with cyclin A and is phosphorylated by the cyclin A–Cdk2 complex. Biochem Biophys Res Comm. 2000;268:530–534. doi: 10.1006/bbrc.2000.2167. [DOI] [PubMed] [Google Scholar]
- Patton EE, Willems AR, Tyers M. Combinatorial control in ubiquitin-dependent proteolysis: Don’t Skp the F-box hypothesis. Trends Genet. 1998;14:236–243. doi: 10.1016/s0168-9525(98)01473-5. [DOI] [PubMed] [Google Scholar]
- Peters JM. SCF and APC: The Yin and Yang of cell cycle regulated proteolysis. Curr Opin Cell Biol. 1998;10:759–768. doi: 10.1016/s0955-0674(98)80119-1. [DOI] [PubMed] [Google Scholar]
- Peters JM. Subunits and substrates of the anaphase-promoting complex. Exp Cell Res. 1999;248:339–349. doi: 10.1006/excr.1999.4443. [DOI] [PubMed] [Google Scholar]
- Peterson BO, Wagener C, Marinoni F, Kramer ER, Melixetian M, Denchi EL, Gieffers C, Matteucci C, Peters JM, Helin K. Cell cycle- and cell growth-regulated proteolysis of mammalian CDC6 is dependent on APC-CDH1. Genes & Dev. 2000;14:2330–2343. doi: 10.1101/gad.832500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfleger CM, Kirschner MW. The KEN box: An APC recognition signal distinct from the D box targeted by Cdh1. Genes & Dev. 2000;14:655–665. [PMC free article] [PubMed] [Google Scholar]
- Pfleger CM, Lee E, Kirschner MW. Substrate recognition by the Cdc20 and Cdh1 components of the anaphase-promoting complex. Genes & Dev. 2001;15:2396–2407. doi: 10.1101/gad.918201. (this issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz S, Hwang ES, Visintin R, Amon A. The regulation of Cdc20 proteolysis reveals a role for APC components Cdc23 and Cdc27 during S phase and early mitosis. Curr Biol. 1998;8:750–760. doi: 10.1016/s0960-9822(98)70298-2. [DOI] [PubMed] [Google Scholar]
- Schwab M, Lutum AS, Seufert W. Yeast Hct1 is a regulator of Clb2 cyclin proteolysis. Cell. 1997;90:683–693. doi: 10.1016/s0092-8674(00)80529-2. [DOI] [PubMed] [Google Scholar]
- Shirayama M, Zachariae W, Ciosk R, Nasmyth K. The Polo-like kinase Cdc5p and the WD-repeat protein Cdc20p/fizzy are regulators and substrates of the anaphase promoting complex in Saccharomyces cerevisiae. EMBO J. 1998;17:1336–1349. doi: 10.1093/emboj/17.5.1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama M, Toth A, Galova M, Nasmyth K. APCCdc20 promotes exit from mitosis by destroying the anaphase inhibitor Pds1 and cyclin Clb5. Nature. 1999;402:203–207. doi: 10.1038/46080. [DOI] [PubMed] [Google Scholar]
- Shulewitz MJ, Inouye CJ, Thorner J. Hsl7p localizes to a septin ring and serves as an adaptor in a regulatory pathway that relieves tyrosine phosphorylation of Cdc28 protein kinase in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:7123–7137. doi: 10.1128/mcb.19.10.7123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowyra D, Craig KL, Tyers M, Elledge SJ, Harper JW. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- Skowyra D, Koepp DM, Kamura T, Conrad MN, Conaway RC, Conaway JW, Elledge SJ, Harper JW. Reconstitution of G1 cyclin ubiquitination with complexes containing SCFGrr1 and Rbx1. Science. 1999;284:662–665. doi: 10.1126/science.284.5414.662. [DOI] [PubMed] [Google Scholar]
- Sorensen CS, Lukas C, Kramer ER, Peters JM, Bartek J, Lukas J. A conserved cyclin-binding domain determines functional interplay between anaphase-promoting complex–Cdh1 and Cyclin A–Cdk2 during cell cycle progression. Mol Cell Biol. 2001;21:3692–3703. doi: 10.1128/MCB.21.11.3692-3703.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratmann R, Lehner CF. Separation of sister chromatids in mitosis requires the Drosophila pimples product, a protein degraded after the metaphase/anaphase transition. Cell. 1996;84:25–35. doi: 10.1016/s0092-8674(00)80990-3. [DOI] [PubMed] [Google Scholar]
- Verma R, Annan RS, Huddleston MJ, Carr SA, Reynard G, Deshaies RJ. Phosphorylation of Sic1p by G1 Cdk required for its degradation and entry into S-phase. Science. 1997;278:455–460. doi: 10.1126/science.278.5337.455. [DOI] [PubMed] [Google Scholar]
- Visintin R, Prinz S, Amon A. CDC20 and CDH1: A family of substrate-specific activators of APC-dependent proteolysis. Science. 1997;278:460–463. doi: 10.1126/science.278.5337.460. [DOI] [PubMed] [Google Scholar]
- Yamamoto A, Guacci V, Koshland D. Pds1p is required for faithful execution of anaphase in the yeast, Saccharomyces cerevisiae. J Cell Biol. 1996;133:85–97. doi: 10.1083/jcb.133.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachariae W, Nasmyth K. Whose end is destruction: Cell division and the anaphase-promoting complex. Genes & Dev. 1999;13:2039–2058. doi: 10.1101/gad.13.16.2039. [DOI] [PubMed] [Google Scholar]
- Zachariae W, Schwab M, Nasmyth K, Seufert W. Control of cyclin ubiquitination by CDK-regulated binding of Hct1 to the anaphase promoting complex. Science. 1998;282:1721–1724. doi: 10.1126/science.282.5394.1721. [DOI] [PubMed] [Google Scholar]
- Zou H, McGarry TJ, Bernal T, Kirschner MJ. Identification of a vertebrate sister-chromatid separation inhibitor involved in transformation and tumorigenesis. Science. 1999;285:418–422. doi: 10.1126/science.285.5426.418. [DOI] [PubMed] [Google Scholar]
- Zur A, Brandeis M. Securin degradation is mediated by fzy and fzr, and is required for complete chromatid separation but not for cytokinesis. EMBO J. 2001;20:792–801. doi: 10.1093/emboj/20.4.792. [DOI] [PMC free article] [PubMed] [Google Scholar]