HDAC4-regulated STAT1 activation mediates platinum resistance in ovarian cancer (original) (raw)
. Author manuscript; available in PMC: 2012 Jan 1.
Abstract
Ovarian cancer frequently acquires resistance to platinum chemotherapy, representing a major challenge for improving patient survival. Recent work suggests resistant clones exist within a larger drug sensitive cell-population prior to chemotherapy, implying that resistance is selected for rather than generated by treatment. We sought to compare clinically-derived, intra-patient paired models of initial platinum response and subsequent resistant relapse to define molecular determinants of evolved resistance. Transcriptional analysis of a matched cell-line series from three patients with high-grade serous ovarian cancer before and after development of clinical platinum resistance (PEO1/PEO4/PEO6, PEA1/PEA2, PEO14/PEO23) identified 91 up- and 126 down-regulated genes common to acquired resistance. Significantly enhanced apoptotic response to platinum treatment in resistant cells was observed following knockdown of HDAC4, FOLR2, PIK3R1 or STAT1 (p<0.05). Interestingly, HDAC4 and STAT1 were found to physically interact. Acetyl-STAT1 was detected in platinum sensitive but not HDAC4 over-expressing platinum resistant cells from the same patient. In resistant cells, STAT1 phosphorylation/nuclear translocation was seen following platinum exposure, whereas silencing of HDAC4 increased acetyl-STAT1 levels, prevented platinum induced STAT1 activation and restored cisplatin sensitivity. Conversely, matched sensitive cells were refractory to STAT1 phosphorylation on platinum treatment. Analysis of 16 paired tumor biopsies taken before and after development of clinical platinum resistance showed significantly increased HDAC4 expression in resistant tumors (n=7/16[44%]; p=0.04). Therefore, clinical selection of HDAC4 overexpressing tumor cells upon exposure to chemotherapy promotes STAT1 deacetylation and cancer cell survival. Together, our findings identify HDAC4 as a novel, therapeutically tractable target to counter platinum resistance in ovarian cancer.
Introduction
One of the greatest areas of unmet need compromising the successful treatment of ovarian cancer is the acquisition of clinical resistance to platinum chemotherapy. Platinum based compounds are standard first-line agents for ovarian cancer and initial response rates are high (1). However, subsequent relapse with acquired platinum resistance is frequent and closely linked to the poor survival associated with this cancer. Multiple mechanisms for platinum resistance have been described and are reviewed elsewhere (2-4).
A recent genomic analysis of a cell line series derived from three cases of serous ovarian cancer both before and after acquisition of clinical platinum resistance revealed that in addition to shared genomic features, sensitive and resistant tumor cells from the same patient also exhibit mutually exclusive genomic characteristics, indicating that rather than a direct linear evolution of resistance from sensitive disease in response to platinum challenge, platinum resistant clones are present from the outset at low abundance within the sensitive presenting tumor (5). In this model, the minor resistant clone persists despite effective killing of the dominant sensitive population and subsequently expands causing relapse. This is in contrast to alternative hypotheses of acquired resistance whereby mutations are proposed to arise in sensitive cells in response to treatment with chemotherapy. In vitro derivation of acquired resistance by treatment of a sensitive cancer cell line with platinum agents is likely to mimic this alternative hypothesis producing adaptive linear responses, which may not accurately reflect clinical resistance. As such we focused our analysis here on clinically derived models of resistance. Henceforth, for brevity we refer to this selection hypothesis as acquired platinum resistance, as it describes the known clinical entity of relapse within 6 months of last platinum therapy after previous remission/response.
Here we report the first linked gene expression profiling and functional analysis of intra-patient paired pre- and post- clinically acquired platinum resistance in ovarian cancer. Our analysis used ovarian cancer cell line series described previously (5, 6), identifying several novel modulators of platinum response and focuses on a previously un-reported functional mechanism that behaves in a fundamentally different manner between clinically platinum sensitive and resistant cells from the same patients. Additionally we noted that this mechanism operates to produce resistance independently of pre-existing established changes in platinum response caused by functional reversion of a germline BRCA2 truncating mutation (7). This work identifies therapeutic targets with implications for the management of ovarian cancer.
Materials and methods
Cell Lines and Reagents
The paired high grade serous ovarian carcinoma cell lines PEO1 vs PEO4/PEO6, PEA1 vs PEA2 and PEO14 vs PEO23 were obtained from Dr Simon Langdon (Edinburgh, UK) and have been described elsewhere (5-7). Cell lines verification was by Identifyler kit (Applied Biosystems). In the matched pairs the first set of cell lines (PEO1, PEA1, PEO14) were derived prior to, and the second set (PEO4/PEO6, PEA2, PEO23) following the onset of acquired clinical platinum resistance. SKOV3 cells were obtained from ECACC. Cisplatin response was measured by sulphorhodamine B (SRB) assay as described (8). All cell lines have confirmed TP53 mutations (5). BRCA1/2 sequencing was performed as described (9) (see also supplementary methods). All lines were maintained in RPMI1640 media with 10% foetal calf serum, penicillin, streptomycin, glutamine at 37°C/5%CO2. Antibodies: FOLR2 (Abcam), STAT1 (BD Biosciences), HDAC4, pSTAT1Y701, Acetyl-Lys (Cell Signalling), FAK, PIK3R1, Lamin A/C (Upstate), β-tubulin, HDAC4 (for IHC) (Santa Cruz).
Microarray Hybridisation and Data Analysis
RNA was prepared using TriReagent (Sigma) and hybridised to Sanger Hver1.2.1 10K cDNA microarrays as described elsewhere (10) (see also supplementary methods) and data analysed using the Genespring GX software package (Aglient) following Lowess normalisation and averaging of quadruplicate hybridisations. Genes were filtered based on expression change between sensitive and resistant cells after which t-tests were performed with Benjamini-Hochberg false discovery rate correction (see also supplementary figure S2).
siRNA transfection and apoptosis assay
Cells were grown to 60% confluence in 6-well plates prior to transfection with smartpool siRNAs directed to FAK, FOLR2, HDAC4, HSXIAPAF1, ITGB2, LAMA4, MYC, PIK3R1, PRKCBP1, STAT1, TAP1, VEGFA and control siRNAs, LAMIN A/C, non-targeting and siCONTROL-TOX (100nM final concentration) (Dharmacon, USA). Cells were re-transfected after 48 hours. SiRNAs in 1x siRNA buffer were mixed with 2μL transfection-reagent #1 (Dharmacon) per transfection in a total volume of 400μL with OptiMEM media. Following 30mins incubation siRNAs were added to 1600μL antibiotic free RPMI1640/10%FCS on cells. Twenty four hours after second transfection cells were reseeded. Cells reseeded in 6 well trays were incubated for 48hrs and protein and RNA samples prepared. RNA extraction was by RNeasy Mini Kit (Qiagen, UK). Cells reseeded into clear and opaque 96 well trays were treated identically: for each transfection condition, 24hrs after seeding, 3 replicate wells were treated with 25μM cisplatin and 3 wells left untreated. After 24 hours cells caspase activation was measured by caspaseGlo 3/7 (Promega, UK) assay and viable cell numbers inferred by 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay as described elsewhere (11). Caspase activity was normalised to cell density for each treatment.
Immunofluorescent microscopy
Coverslips (VWR International, UK) were treated with 1M HCl prior to cell seeding and incubation for 24hrs at 37°C /5% CO2. After indicated treatment, cells were washed with PBS then fixed/permeabilised at 37°C for 30min with 4% paraformaldehyde/1.8% TritonX-100/PBS. Coverslips were blocked in 10% goat serum/2% BSA/PBS for 30min then washed with PBS and incubated with primary antibodies overnight at 4°C. Coverslips were washed in PBS and incubated with 1:500 dilutions of fluorochrome-conjugated secondary antibodies (FITC anti-goat IgG, FITC anti-rabbit IgG, Alexa Fluor 555 anti-mouse IgG and Alexa Fluor 488 goat anti-rabbit IgG, Invitrogen, UK) and directly-labelled actin stain (Alexa Fluor 633 phalloidin, Invitrogen, UK) in blocking buffer for 1h. Cells were rinsed three times in PBS for 5 minutes and mounted onto slides using Vectashield media containing 4′,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories, Canada). Slides were visualised on an inverted confocal microscopy system (Axiovert 200M, Zeiss, US and TCS SP5, Leica, Germany).
Pharmacological inhibition of HDAC4
PEO4 and SKOV3 cells were seeded at 25,000 cells/well in 96 well plates in a volume of 50μl/well. After 24hrs, cells were either treated with 40μl of fresh culture media (untreated control) or 40μl fresh media containing HDAC inhibitor APHA4a at final concentration, calculated for 50μl final volume, of 5μM and 10μM. After one hour, 10μl of cisplatin was added to a final concentration of 25μM in 50μl; 10μl of fresh culture media was added to control cells. Caspase 3/7 and MTT assays were performed at 24hrs post cisplatin treatment as described above.
Additional methods
See Supplementary Data.
Results
Coupled gene expression and functional analysis of up-regulated transcripts in clinically acquired platinum resistance
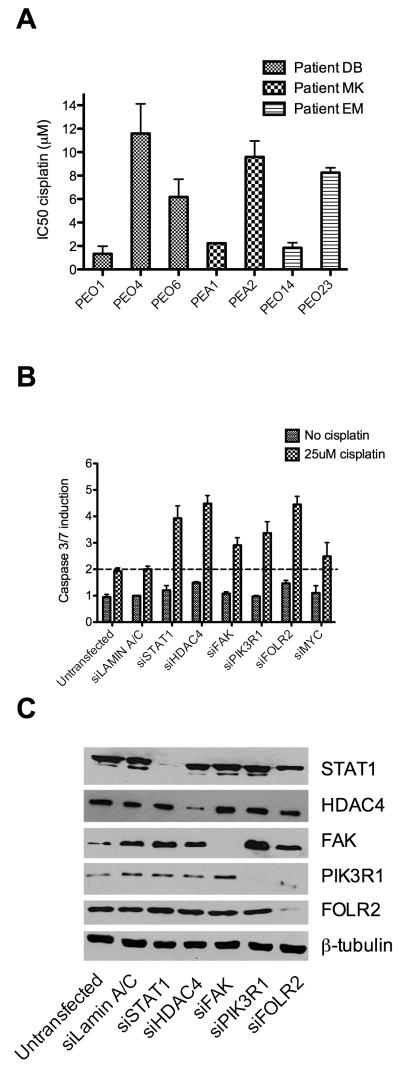
To confirm that acquired clinical resistance to platinum based chemotherapy was maintained in long-term platinum-free cell culture we carried out in vitro cisplatin sensitivity assays (Figure 1A). Average values from at least 3 replicate experiments revealed between 4 and 9 fold increase in cisplatin IC50 on acquisition of clinical resistance. Stable resistance in the absence of cisplatin suggests that clinical mechanisms of resistance are genetically or epigenetically determined: consistent with the description of distinct genomic differences between the sensitive and resistant paired lines studied here (5). Mutations in BRCA2 have been described previously in the cell lines PEO1 and PEO4 (7). We sequenced BRCA1 and 2 in all paired cell lines and identified previously described alterations in our stocks of PEO1 and PEO4 that are associated with functional BRCA2 (discussed in detail in supplementary figure S1). No further mutations were seen in BRCA1 or BRCA2 in other cell lines indicating that the clinical platinum resistance phenotype here is not accounted for by BRCA mutation/reversion. RNAs from cell line pairs were co-hybridised to cDNA microarrays. Normalised data were filtered as described identifying 91 unique upregulated and 126 downregulated genes in association with clinically acquired platinum resistance (supplementary figure S2 and supplementary tables S1A/B). We hypothesised that silencing of genes overexpressed in resistant cells might lead to resensitisation by re-engaging apoptotic response to cisplatin treatment and thus may directly reveal novel therapeutic targets for clinical reversal of resistance. Hence the overexpressed genes formed the focus of our onward strategy. Thirteen upregulated candidate genes were selected on the basis of either magnitude of overexpression or cellular function and were taken forward for functional assessment (supplementary table 2). We combined an siRNA based approach, using the platinum resistant PEO4 cell line, to a caspase 3/7 activation based apoptosis assay to evaluate the effect of each gene on the cellular response to platinum. In order to quantify the difference in platinum induced apoptosis between control and test genes the ratio of caspase3/7 induction in control transfected cells on cisplatin treatment was compared to that of target gene siRNA transfected cells. Six of the 13 genes demonstrated a 1.5 fold or greater increase in the ratio of caspase induction in the target gene siRNA compared to the control gene siRNA (platinum resensitisation ratio) (supplementary table 2), and these were taken forward for further analysis. Assays were repeated a further three times for these six genes and the average values plotted (figure 1B). The platinum re-sensitisation ratios were calculated and averages subjected to t-test for statistical significance (table 1). Knockdown of control and test genes was confirmed by western blot (figure 1C). Four genes, FOLR2, PIK3R1, HDAC4 and STAT1 emerged as significantly re-sensitising platinum resistant ovarian cancer cells. We focused here on two of these genes, HDAC4 and STAT1, as recent reports indicate that the transcription factor STAT1 can be regulated by acetyl modifications (12, 13) suggesting the possibility that STAT1 may transcriptionally control the switch from sensitivity to resistance in a manner that may be modulated clinically using HADC inhibitors, which are well developed and indeed approved for use in cutaneous T-cell lymphoma (14).
Figure 1. Identification of platinum resistance modulators.
IC50s for platinum sensitive and resistant paired cell lines were calculated based on SRB assay data (n=3). This analysis confirmed an in vitro resistance to cisplatin in all clinically platinum resistant cell lines relative to their sensitive parent lines (Fold changes in IC50 between sensitive and matched resistant cells: PEO4- 8.7 [p=0.016], PEO6- 4.6 [p=0.004], PEA2- 4.1 [p=0.0002], PEO23- 4.5 [p=0.00002]) (A). Following microarray based expression analysis candidate genes were assessed for a role in acquired platinum resistance by a first round of siRNA knockdown followed by cisplatin treatment identifying six genes that enhanced apoptotic induction by cisplatin. Following replication, four of these genes (FOLR2 [p=0.004], HDAC4 [p=0.02], PIK3R1 [p=0.04] and STAT1 [p=0.0002]) passed statistical analysis (see also table 1) as enhancing apoptotic induction on cisplatin treatment when compared to control siRNA treated cells. Data are represented as means ±SEM of 4 replicate experiments (B). Specificity of siRNA was confirmed by western blot (C). All t-tests throughout are two-tailed and assume unequal variances.
Table 1.
Knockdown and resensitisation assays were carried out a total of four times using cisplatin resistant PEO4 cells for genes selected following first round analysis (see supplementary table S2 for results of first round analysis). Platinum resensitisation ratio is defined in table 1. Quadruplicate data was analysed by t-test for significant difference in cisplatin induced caspase 3/7 induction between test gene siRNA transfected cells and siRNA controls
| GeneSymbol | Platinum re-sensitisationratio | p-value |
|---|---|---|
| FAK | 1.38 | 0.07 |
| FOLR2 | 1.40 | 0.004 |
| HDAC4 | 1.55 | 0.02 |
| MYC | 1.40 | 0.25 |
| PIK3R1 | 1.97 | 0.04 |
| STAT1 | 1.73 | 0.0002 |
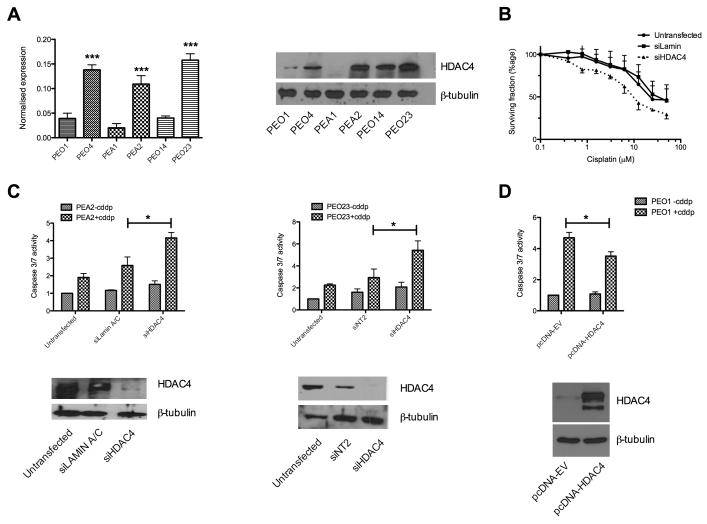
HDAC4 knockdown re-sensitises a panel of clinically-derived platinum resistant ovarian cancer cell lines
We used qRT-PCR and western blotting to validate the up-regulation of HDAC4 seen in the gene expression analysis (figure 2A). This indicated an increase in expression between sensitive and resistant cells in each pair tested. We demonstrated that the effect seen in the 24hr caspase3/7-activation assay is maintained at 72hr by SRB assay following HDAC4 siRNA knockdown (figure 2B). We then considered the effect of siRNA knockdown of HDAC4 in the clinically derived platinum resistant cell lines PEA2, PEO23 and SKOV3. Figure 2C and supplementary figure S4 show significantly increased apoptotic response to cisplatin following siRNA knockdown of HDAC4, compared to control siRNAs. Conversely, silencing of HDAC4 expression in sensitive cells (PEO1, PEA1, PEO14) did not significantly enhance platinum sensitivity, in keeping with their already relatively low HDAC4 expression levels (supplementary figure S3). However, over-expression of HDAC4 in platinum sensitive PEO1 cells resulted in decreased apoptotic response to cisplatin (p=0.032) indicating that the platinum resistant phenotype can be, at least partially, recapitulated by over-expression of HDAC4 alone (figure 2D). Deconvolution of the pool of four HDAC4 siRNAs confirmed target specific knockdown (supplementary figure S4B).
Figure 2. HDAC4 contributes to cisplatin resistance.
Over-expression of HDAC4 in platinum resistant cells was confirmed at the RNA (left) and protein level (right) (A) [***p<0.005 t-test]. The effect of HDAC4 knockdown on cisplatin response was validated by SRB assay, 72hr post cisplatin exposure of siRNA treated PEO4 cells. Data are mean ± SEM (n=3) (B). Clinically derived platinum resistant cell lines (PEA2/PEO23) were assessed for the contribution of HDAC4 to acquired resistance. Data in the upper panes represent the mean caspase 3/7 induction relative to control siRNA ±SEM (n=3) whereas lower pane shows confirmation of HDAC4 knockdown by western blot. *p<0.05 t-test comparing caspase 3/7 activation between siRNA treated and siRNA control cells following cisplatin exposure (C). Over-expression of HDAC4 in the platinum sensitive PEO1 cell line as described in the supplementary methods. Over-expression of protein (lower) was confirmed by western blot and accompanying caspase activation data (upper) demonstrates attenuated apoptotic response to cisplatin in cells over-expressing HDAC4 (n=4; p=0.032) comparing caspase 3/7 activation between pcDNA3.1-empty vector (EV) transfected and pcDNA3.1-HDAC4 treated cells (D).
Demonstration of a novel role for HDAC4 in acquired platinum resistance led us to consider possible targets for HDAC4 mediated deacetylation. Post-translational modification of the transcription factor STAT1 by acetylation at lysine residues 410 and 413 has been reported previously in melanoma cells (12) and shown to promote dephosphorylation at Tyr701 (13) thereby acting as a control mechanism for this protein in the interferon response. Since STAT1 was identified as upregulated in resistant cells in our analysis, and knockdown significantly re-sensitised cells to platinum (see above), we considered that STAT1 might be subject to acetyl regulation in acquired resistance and hence explored whether a functional interaction occurs between HDAC4 and STAT1.
STAT1 is activated and translocated to the nucleus in response to cisplatin treatment in acquired platinum resistant but not sensitive ovarian cancer cells
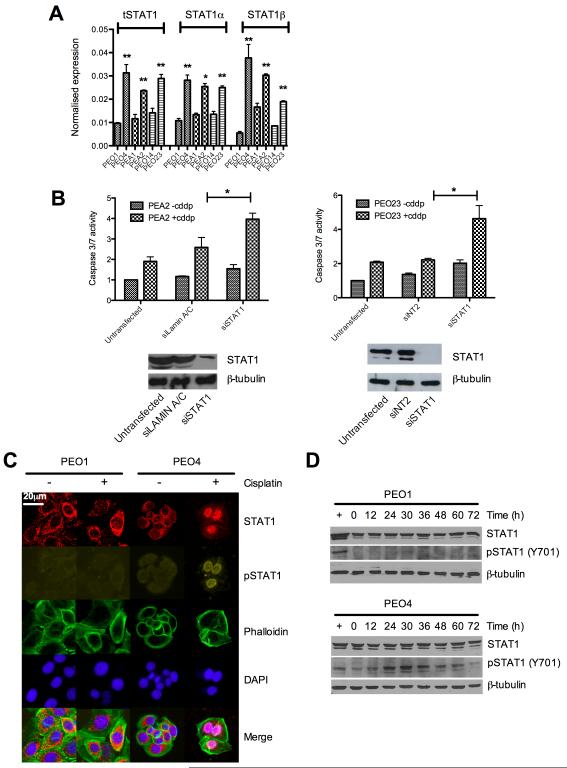
We examined the broader role of STAT1 in platinum resistance prior to more detailed mechanistic studies. STAT1 exists as two distinct isoforms, full length STAT1α and STAT1β, a truncated form considered to act as a dominant negative inhibitor of STAT1α. qRT-PCR in platinum sensitive and resistant lines demonstrated that both STAT1α and STAT1β were upregulated to the same extent in the resistant lines (figure 3A) suggesting that differences between isoforms did not explain the alteration in platinum sensitivity.
Figure 3. STAT1 contributes to cisplatin resistance and is phosphorylated at Y701 in resistant cells following platinum treatment.
cDNA from sensitive and resistant cell lines was quantified for expression of total STAT1 and the α and β isoforms of STAT1 individually (A) [*p<0.05, **p<0.01 t-tests comparing resistant cells to their sensitive matched line]. siRNA to STAT1 in further platinum resistant cell lines revealed a general effect of resensitisation to cisplatin following STAT1 knockdown as measured by caspase 3/7 induction. Effective knockdown was confirmed by western blot (See also supplementary figure S5) *p<0.05 t-test comparing caspase 3/7 activation between siRNA treated and siRNA control cells following cisplatin exposure (B). Subcellular location of total- and phospho-STAT1 was determined in sensitive and resistant cells by immunofluorescence microscopy as described. Nuclei were revealed using DAPI stain and actin cytoskeleton visualised by Alexa 633nm conjugated phalloidin stain (C) (see also supplementary figure S6). Identical matched cells were treated with cisplatin over 72hrs at 5μM for sensitive PEO1 cells and 25μM for resistant PEO4. Protein lysates were collected and total and phospho-STAT1 determined by western blot (D).
Next we performed RNAi knockdown of STAT1 in additional platinum resistant ovarian cancer cell lines: PEA2, PEO23 and SKOV3. Figure 3B and supplementary figure S5A depict the enhanced apoptotic response following STAT1 siRNA knockdown. Deconvolution of the pool of four siRNAs to STAT1 validated target specific phenotype (supplementary data figure S5B).
We explored whether STAT1 is activated in response to cisplatin and thus examined the subcellular localisation of STAT1. Immunofluorescence microscopy of platinum sensitive and matched, resistant cells showed that in the absence of platinum STAT1 is predominantly unphosphorylated and cytoplasmic. However nuclear STAT1, phosphorylated at Y701, is observed following cisplatin treatment in resistant cells (PEO4/PEA2), but not sensitive matched cells (PEO1/PEA1) where STAT1 remains largely unphosphorylated and cytoplasmic (figure 3C and supplementary figure S6A). This is in keeping with the canonical pathway of STAT1 activation by phosphorylation, dimerisation and nuclear translocation and suggests a platinum-induced, STAT1 mediated transcriptional response occurring specifically in acquired platinum-resistant cells. To corroborate these finding we also looked at pSTAT1 Y701 levels in the sensitive and resistant cells by western blot. Figure 3D shows striking discordance between the phospho-STAT Y701 levels in the paired sensitive/resistant cell lines PEO1 and PEO4, with no detectable Y701 phosphorylation in sensitive cells. In contrast phospho-STAT1 Y701 is detectable at baseline and is induced following platinum treatment in PEO4 cells. Expression of the STAT1 regulated gene IRF1 was measured in platinum resistant PEO4 cells and showed cisplatin mediated induction of IRF1 transcription which was abolished by STAT1 knockdown confirming functional activity of STAT1 in response to cisplatin exposure in resistant cells (figure S6B).
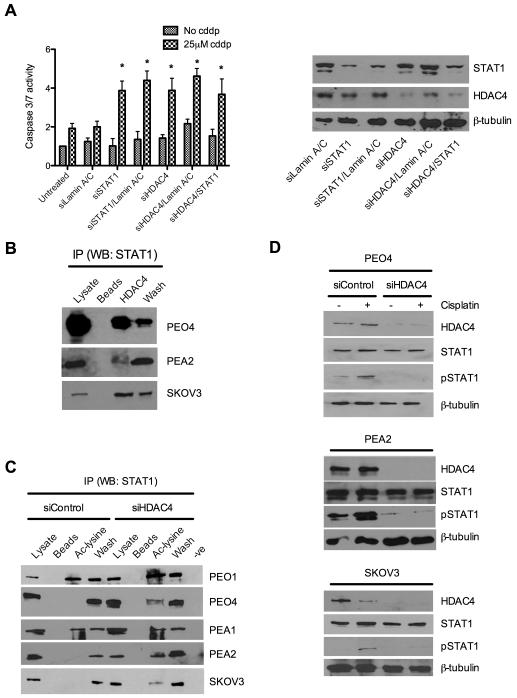
We hypothesised that if HDAC4 and STAT1 had independent roles in the platinum resistant phenotype we would expect additivity or synergy in platinum re-sensitisation if both were inhibited simultaneously. Therefore we performed dual knockdown of HDAC4 and STAT1 in resistant PEO4 cells. Data indicated no additional effect in apoptotic response to cisplatin over that seen following knockdown of either gene alone (figure 4A), implying redundancy between these two genes and supporting the notion that they may act in the same pathway in this phenotype.
Figure 4. HDAC4 and STAT1 act in a single, novel pathway in which HDAC4 is required for deacetylation and phosphorylation of STAT1 in response to cisplatin.
STAT1 and HDAC4 were knocked down alone and in combination using siRNA and induction of caspase 3/7 assessed ±cisplatin treatment in platinum resistant PEO4 cells *p<0.05 t-test comparing caspase 3/7 activation between siRNA treated and siRNA control cells following cisplatin exposure (left). Western blot confirmed knockdown at protein level (right) (A). Immunoprecipitates from untreated platinum resistant PEO4, PEA2 and SKOV3 cells were made using anti-HDCA4 antibody and probed for the presence of STAT1 (B). Immunoprecipitates from the platinum sensitive cell lines PEO1 and PEA1, the resistant paired lines PEO4 and PEA2 and the resistant line SKOV-3 were prepared using anti-acetyl lysine antibody and probed for the presence of STAT1 both before and after siRNA based knockdown of HDAC4 (C). Control and HDAC4 siRNA treated PEO4 (top), PEA2 (middle) and SKOV3 (bottom) cell lysates ± cisplatin were probed for phosphorylation of STAT1 at tyrosine 701. (D). Legend for (B) and (C): lysates- whole cell extract pre-immunoprecipitation; beads- protein G sepharose beads without primary antibody; HDAC4/Ac-lysine- specific immunoprecipitates; wash-whole-cell lysate column flow-through; −ve - primary antibody/lysate without protein G sepharose beads.
Having demonstrated a clear role for STAT1 in platinum resistance we considered the hypothesis outlined above, that HDAC4 may be acting as a STAT1 deacetylase, altering the impact of cisplatin on STAT1 signalling.
HDAC4 interacts with STAT1, modulating its acetylation, phosphorylation and nuclear translocation, thereby abrogating sensitivity to cisplatin
To assess STAT1 acetylation and the potential role of HDAC4 in STAT1 deacetlyation we performed co-immunoprecipitation and demonstrated an interaction between HDAC4 and STAT1 in resistant cells (PEO4, PEA2 and SKOV3: figure 4B). We assayed for the presence of acetyl-STAT1 by immunoprecipitation with anti-acetyl lysine antibody and western blot for STAT1. Ac-STAT1 was present in the platinum sensitive cell lines PEO1 and PEA1 (expressing low levels of HDAC4) but was undetectable in their HDAC4 over-expressing matched platinum resistant counterparts PEO4 and PEA2 and was similarly undetectable in platinum resistant SKOV-3 cells. However, following HDAC4 knockdown, acetyl-STAT1 was detectable in all resistant lines (figure 4C) indicating that HDAC4 is required for the deacetylation of STAT1 and providing a novel mechanistic link between these two modulators of platinum resistance. Next we considered the consequence of HDAC4 mediated deacetylation of STAT1 on its protein function and on the phenotype of clinically acquired platinum resistance.
We hypothesised that since acetylation of STAT1 has been shown to abrogate interferon induced phosphorylation at Y701 (13), and that since the knockdown of HDCA4 has been shown here to increase acetylation of STAT1, that HDAC4 may influence the phosphorylation, and hence nuclear accumulation, of STAT1. Figure 4D demonstrates that the cisplatin induced accumulation of phospho-STAT1 observed in the presence of HDAC4 is abrogated following HDAC4 knockdown in platinum resistant PEO4, PEA2 and SKOV3 cells. Immunofluorescence staining of PEO4 cells showed that knockdown of HDAC4 is linked to loss of cisplatin induced nuclear phospho-STAT1 accumulation (figure 5A).
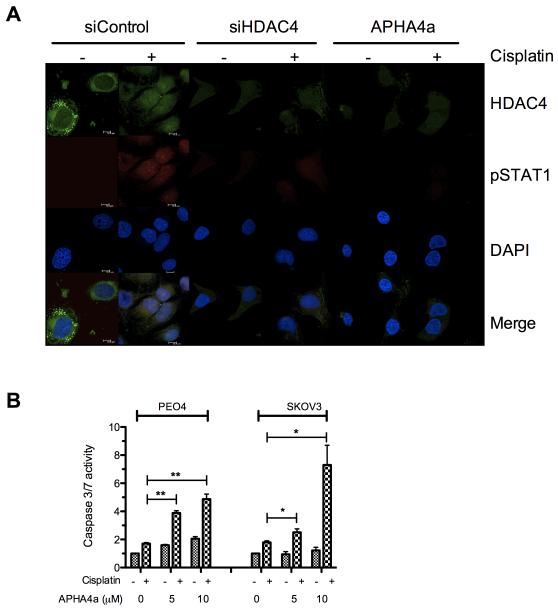
Figure 5. HDAC inhibition prevents nuclear localisation of phospho-STAT1 and resensitises resistant cells to cisplatin.
Immuno-fluorescent microscopy of platinum resistant SKOV3 cells reveals induction and nuclear localisation of pSTAT1-Y701 on 24hrs stimulation with 25μM cisplatin and shows that HDAC4 knockdown using siRNA or inhibition using 5μM APHA4a prevents accumulation of pSTAT1 (A). Treatment of the resistant cells PEO4 or SKOV3 with the HDAC inhibitor APHA4a at 5μM or 10μM enhances the apoptotic response to treatment with 25μM cisplatin measured after 24hr incubation (B) *p<0.05, **p<0.01 t-test comparing caspase 3/7 activation between vehicle and APHA4a treated cells following cisplatin exposure.
Acquired platinum resistance can be reversed by treatment with pharmacological inhibitors of HDACs
To consider the potential therapeutic utility of inhibiting HDAC4 or STAT1 we treated platinum resistant cells with the hydroxamic acid based HDAC inhibitor (HDACi) aroyl-pyrrolyl-hydroxy-amide 4a (APHA4a) (15). This treatment elicited a restoration of platinum sensitivity, as determined by induction of caspase 3/7 activity, in the resistant cell lines PEO4 and SKOV3 when combined with platinum at concentrations that had little or no single agent effect in the same assay (figure 5B). Treatment with HDACi resulted in a similar blockade on the phosphorylation of STAT1 shown at the morphological level by immunofluorescence microscopy. We also demonstrate loss of cisplatin induced STAT1 Tyr701 phosphorylation following co-treatment with cisplatin/HDACi or HDAC4 siRNA and a loss of nuclear phospho-STAT1 localisation indicating functional loss of transcription factor activity (Fig 5A).
HDAC4 is significantly over-expressed following acquired platinum resistant relapse in clinical samples
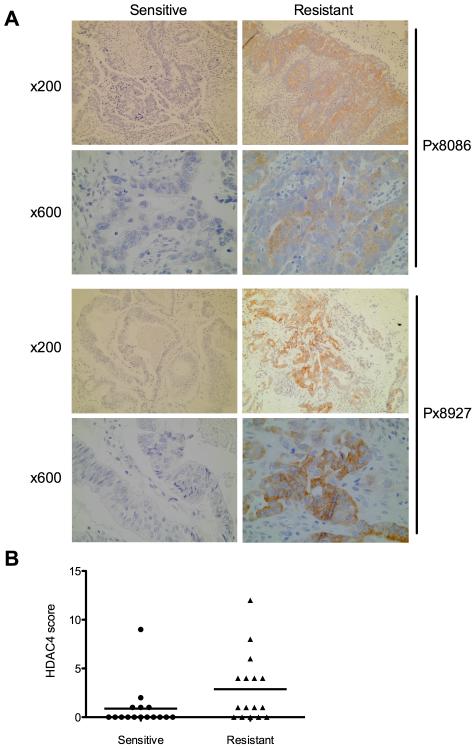
To validate the observed increase in HDAC4 expression in platinum resistant cells we measured HDAC4 expression by IHC in tumor biopsies from 16 clinically platinum sensitive ovarian cancer patients matched to second biopsies from the same patients subsequent to development of acquired platinum resistance. Additionally we utilised paired biopsies from 14 patients with platinum refractory disease; those biopsies obtained before and after platinum based chemotherapy. Sections were stained and scored, blind, by two pathologists. Intensity of staining was scored on a 0-3 scale and percentage cells stained scored on a 0-4 scale. The product score was derived and used for further statistical analysis. This revealed an increase in HDAC4 protein expression in 7/16 (44%) acquired platinum resistant biopsies compared with their matched platinum sensitive biopsies (figure 6A/B) (p=0.0413; Wilcoxon rank sum test). Interestingly, the analysis of 14 paired sections from patients with platinum refractory disease revealed no increase in HDAC4 expression between the paired biopsies taken before and after ineffective first-line platinum treatment indicating that up-regulation of HDAC4 is specifically an acquired resistance mechanism and is not involved in refractory disease (supplementary figure S7). Clinical parameters are shown in supplementary table S4. This analysis suggests that up to 44% of patients with acquired platinum resistance may have an HDAC4 mediated resistance pathway.
Figure 6. HDAC4 expression increases on acquisition of cisplatin resistance in matched clinical specimens.
Sections cut from FFPE ovarian tumor blocks from before and after acquisition of platinum resistance in individual patients were stained for HDAC4 expression and intensity and frequency of staining. Photomicrographs indicate increased expression of HDAC4 (brown staining) in resistant (right) compared to sensitive (left) biopsies from the same patient. Magnifications are indicated. (A). HDCA4 expression scores were produced for each section as described and the change in score on acquisition of platinum resistance analysed by Wilcoxon rank sum test. Scores for 16 paired sections, grouped as sensitive and resistant are indicated (p=0.0413) (B).
Discussion
Resistance to platinum chemotherapy continues to be a major obstacle in the treatment of ovarian and other cancers. A recent genomic analysis of the cell lines used in this study has shown that resistance to platinum appears not to arise by mutational adaptation of the sensitive tumor cells in the presence of chemotherapy: rather the resistant cells are present in the initial pre-chemotherapy tumor as a minor population (5). The implication is therefore that clinical mechanisms of acquired resistance may not be recapitulated in vitro by adaptive responses following exposure of homogeneous sensitive populations to chemotherapy. Mechanisms produced in vitro may reflect the artificial environment of the monolayer culture: a suggestion underscored by the lack of correlation between gene expression changes detected in a cisplatin resistant cell line, PEO1cddp (16), produced in vitro from sensitive PEO1 cells and those seen in the clinically-derived resistant PEO4 and PEO6 lines (r2 values: PEO1cddp vs PEO4= 0.25; PEO1cddp vs PEO6= 0.23; PEO4 vs PEO6= 0.81. Supplementary figure S8). Interestingly, most in vitro derived platinum resistant cell lines require periodic re-treatment with platinum in order to maintain resistance, indicating that the mechanisms affecting drug response are transient and adaptive rather than being genomically established. Conversely, the clinically derived, resistant cell lines used here exhibit stable platinum resistance in vitro, implying that the genomic differences reported between sensitive and resistant cells from the same patient (5) are likely to underpin the resistant phenotype. Such alterations between these cell-line pairs are extensive however and preclude efficient identification of resistance mechanisms by DNA level candidate selection. We therefore used transcriptional profiling to indicate changes in gene activity relating to acquired resistance.
Gene expression profiling has been used previously to attempt identification of better predictive biomarkers for clinically acquired platinum resistance and novel targets for therapy and has been applied to clinical material (17-19) and cell lines (20, 21). Jazaeri et al identified expression changes related to chemoresistance and, although underpowered for statistical significance, suggested that distinct mechanisms may be responsible for intrinsic and acquired chemoresistance, as we have also indicated here (17). Spentzos et al developed a 93-gene signature predictive of response to platinum based chemotherapy (18). Helleman et al identified a set of nine genes, predictive of intrinsic platinum resistance. These studies were focused on frozen clinical samples and as such were correlative rather than functional, however they suggest that gene expression measurements are informative in this context (19). Cheng et al compared global gene expression levels between isogenically matched, platinum sensitive and resistant ovarian cancer cell-line pairs (21). The analysis identified a number of dys-regulated genes however no functional validation was reported. Of note, the platinum-resistant cell-lines used were not clinically derived but were created by in vitro exposure of sensitive cells to cisplatin (21).
Our analysis identified four genes, HDAC4, STAT1, FOLR2 and PIK3R1 as over-expressed in clinically resistant cells, each of which also significantly potentiated cisplatin response when knocked down by siRNA. We were prospectively interested in whether our approach would identify unrelated mechanisms or common pathways of acquired resistance. It is of note that by using this discovery-based approach we identified a novel relationship between HDAC4 and STAT1 whereby they physically interact to create conditions under which STAT1 can be activated following cisplatin treatment in cells with acquired clinical platinum resistance but not in matched sensitive cells from the same patient. As a surrogate of this resistance-specific activation, we observed phosphorylation and nuclear translocation of STAT1 upon platinum treatment in resistant cells only. We showed that over-expression of HDAC4 promotes de-acetylation of STAT1 which facilitates STAT1 phosphorylation at Y701 and its subsequent nuclear translocation. In contrast, matched cisplatin sensitive cells have lower endogenous HDAC4 levels and readily detectable acetyl-STAT1: consequently cisplatin treatment is unable to induce phosphorylation of STAT1 (Y701) or STAT1 nuclear relocalisation (model summarised in supplementary figure S9).
The role of HDACs in de-acetylation of non-histone proteins is not a new concept. HDAC6, a class IIb HDAC found exclusively in the cytoplasm, deacetylates tubulin, contractin and Hsp90 affecting cell morphology, adhesion and migration (22). A sumoylation/acetylation crosstalk has been described for p53 that affects the transcriptional activity of this tumor suppressor (23) whereas activity of the FOXO proteins, downstream targets of AKT, can be modulated by p300/CBP mediated acetylation and deacetylation by the class III HDAC, SIRT1 (24). Kramer et al reported acetylation of STAT1 at lys410 and lys413 in melanoma cells and showed increased acetylation following HDAC inhibition or interferon α treatment (12) although HDAC4 was not analysed in their study. Following activation at the cell surface nuclear translocation of STAT1 occurs which has been reported to follow binding of STAT1 to importin-α5 via critical lysine residues 410 and 413 (25, 26): the same residues as identified by Kramer et al as the acetyl sites on STAT1 (12). This suggests a potential interference with the nuclear import machinery in platinum sensitive cell lines shown here to have constitutively acetylated STAT1 and to remain in the cytoplasm following platinum treatment (figure 3C/4C). Subsequent to their report of acetyl control of STAT1 (12), Kramer et al described a cyclical control system whereby phosphorylated, nuclear STAT1 is acetylated by the histone acetyl transferase CBP marking it for subsequent dephosphorylation by the phosphatase TCP45 resulting in decreased DNA binding and target gene transcription (13). Deacetylation by HDAC3 and nuclear-cytoplasmic translocation returns STAT1 to a latent cytoplasmic state in readiness for reactivation by subsequent interferon stimulation (13, 27). Our data suggests a similar but distinct set of effects. In contrast to previous studies, which identified the involvement of the class-I HDACs, HDAC1 and HDAC3 in STAT1 deacetylation, we identify the class-II deacetylase, HDAC4, as having a key role in the cisplatin-mediated activation of STAT1, specifically in platinum resistant cells that over-express this HDAC. Importantly we see that in both platinum sensitive and resistant lines, interferon treatment induces strong phosphorylation of STAT1 Y701 despite the observed differences in basal STAT1 acetylation levels between those platinum sensitive and resistant cells (supplementary figure S10). In contrast we only see STAT1 Y701 phsophorylation in response to cisplatin treatment in resistant lines. Our data indicates that both sensitive and resistant cells activate STAT1 in response to γ-IFN (supplementary figure S10) however only resistant cells activate STAT1 in response to cisplatin induced DNA damage (figure 3D), suggesting that this effect is not mediated via γ-IFN.
The work presented here demonstrates a novel mechanism involving HDAC4 and places this interaction at the centre of the response to platinum chemotherapy revealing clinically relevant phenotypic differences resulting from within-patient changes in HDAC4 gene expression. Our data indicate that an HDAC4 threshold level exists that creates a dichotomous STAT1 response that differentiates clinical platinum sensitivity from acquired clinical platinum resistance. Pharmacological and clinical data (figures 5 and 6) suggest this mechanism may be a frequent event and can be targeted to reverse resistance using HDAC inhibitors, many of which are either in or near the clinic. It further suggests that HDAC4/classII HDAC inhibitors may be more appropriate than pan-HDAC inhibitors in this indication, with potentially lower toxicity/non-specificity.
This has implications for management of platinum resistant disease as a relatively modest reduction in HDAC4 can cause resistant cells to behave like their sensitive parental cells with respect to both the control of STAT1 and the cytotoxic effects of cisplatin. HDAC4 has not previously been shown to functionally modulate clinically acquired resistance to platinum based treatment.
Although STAT1 is most commonly associated with pro-apoptotic signalling it is also associated with cell survival in certain contexts. STAT1 is re-localised to the nucleus of breast cancer cells following doxorubicin treatment and is associated with increased apoptotic response (28) and enhances apoptotic response to DNA damage in mouse fibroblasts via downregulation of mdm2, a negative regulator of p53 (29). Conversely, STAT1 can induce docetaxel resistance in prostate cancer cells (30) and cisplatin resistance in A2780 ovarian cancer cells (20). Roberts et al carried out expression profiling of ovarian cancer cell lines with predefined response to a number of chemotherapeutics (20). Gene expression was correlated with drug IC50 across the cell lines used and this identified an association between STAT1 and response to cisplatin and AMD473. Overexpression of STAT1 by transfection was shown to increase resistance to both agents however inhibition of upstream Janus kinase by treatment with AG490 (31) increased sensitivity to AMD473 but not cisplatin, perhaps suggesting that cisplatin mediated activation of STAT1 may be independent of JAK2/3. The study included the cell line pair PEO1 and PEO4, however interestingly the analysis did not take advantage of the matched nature of these lines. Our study demonstrates that the behaviour of STAT1 can differ fundamentally, even within a single patient’s tumor before and after the onset of clinical resistance to platinum, thus highlighting the importance of the cellular context in understanding STAT1 responses following stimulus.
In summary, we have demonstrated that molecular profiling of appropriate, clinically derived model systems coupled to functional assays can reveal novel biological mechanisms underlying acquired platinum resistance. The identification of an HDAC4 mediated STAT1 response ‘switch’ represents a novel mechanism of in-vivo acquired platinum resistance that is demonstrably amenable to therapeutic modulation and can result in re-sensitisation to platinum based chemotherapy. The recent report that platinum resistant clones exist within the platinum sensitive presenting tumor also raises the possibility of therapeutically targeting these cells in front-line therapy to increase survival and/or delay the onset of resistance. Further work will determine the most appropriate predictive biomarker(s) for identifying patients who might benefit from this approach. Drug development and clinical trials around HDAC4 or STAT1 inhibition will address the therapeutic potential of re-engaging response to platinum based treatment in resistant cells by disrupting this specific mechanism in ovarian and potentially other platinum treated cancers. Further work is required to identify the downstream mechanistic functions of STAT1 that confer this resistant phenotype and to further integrate this novel mechanism in the context of other parallel signalling changes in acquired resistance identified here and elsewhere. Additionally it remains unclear how platinum induced DNA damage leads to STAT1 activation. A comprehensive screen of ligands, receptors and associated kinases (JAKs and others) in the context of platinum treatment would help to address this question. Establishment of a systems-level understanding of acquired platinum resistance will allow us to better predict the behaviour of each tumor and identify rational means of targeting it for re-engagement of apoptotic response to platinum chemotherapy.
Supplementary Material
1
2
3
4
Acknowledgements
We thank Ovarian Cancer Action, Cancer Research UK, the Josephine Webber trust and the Saudi Arabian Ministry of Higher Education for funding; the Wellcome Trust Sanger Institute for microarrays; the ECMC/NIHR Biomedical Research Centre and Imperial College London Bioinformatics core and Edinburgh ECMC for infrastructural support; Kirsten Timms and Mark Carey for BRCA1 and BRCA2 sequencing; Katherine Stemke Hale for cell line authentication; Caroline Michie for identifying and supplying ovarian tumor FFPE blocks; and Giorgia Trevisan for 2nd scoring of IHC slides.
Footnotes
Conflict of Interest The authors declare no conflict of interest
References
- 1.Herzog TJ. Recurrent ovarian cancer: how important is it to treat to disease progression? Clin Cancer Res. 2004;10:7439–49. doi: 10.1158/1078-0432.CCR-04-0683. [DOI] [PubMed] [Google Scholar]
- 2.Agarwal R, Kaye SB. Ovarian cancer: strategies for overcoming resistance to chemotherapy. Nat Rev Cancer. 2003;3:502–16. doi: 10.1038/nrc1123. [DOI] [PubMed] [Google Scholar]
- 3.Kartalou M, Essigmann JM. Mechanisms of resistance to cisplatin. Mutat Res. 2001;478:23–43. doi: 10.1016/s0027-5107(01)00141-5. [DOI] [PubMed] [Google Scholar]
- 4.Rabik CA, Dolan ME. Molecular mechanisms of resistance and toxicity associated with platinating agents. Cancer Treat Rev. 2007;33:9–23. doi: 10.1016/j.ctrv.2006.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cooke SL, Ng CKY, Melnyk N, Garcia MJ, Hardcastle T, Temple J, et al. Genomic analysis of genetic heterogeneity and evolution in high-grade serous ovarian carcinoma. Oncogene. 2010;29:4905–13. doi: 10.1038/onc.2010.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Langdon SP, Lawrie SS, Hay FG, Hawkes MM, McDonald A, Hayward IP, et al. Characterization and properties of nine human ovarian adenocarcinoma cell lines. Cancer Res. 1988;48:6166–72. [PubMed] [Google Scholar]
- 7.Sakai W, Swisher EM, Jacquemont C, Chandramohan KV, Couch FJ, Langdon SP, et al. Functional restoration of BRCA2 protein by secondary BRCA2 mutations in BRCA2-mutated ovarian carcinoma. Cancer Res. 2009;69:6381–6. doi: 10.1158/0008-5472.CAN-09-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ross DT, Scherf U, Eisen MB, Perou CM, Rees C, Spellman P, et al. Systematic variation in gene expression patterns in human cancer cell lines. Nat Genet. 2000;24:227–35. doi: 10.1038/73432. [DOI] [PubMed] [Google Scholar]
- 9.Hennessy BTJ, Timms KM, Carey MS, Gutin A, Meyer LA, Flake DD, et al. Somatic mutations in BRCA1 and BRCA2 could expand the number of patients that benefit from poly (ADP ribose) polymerase inhibitors in ovarian cancer. J Clin Oncol. 2010;28:3570–6. doi: 10.1200/JCO.2009.27.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pellagatti A, Vetrie D, Langford CF, Gama S, Eagleton H, Wainscoat JS, et al. Gene expression profiling in polycythemia vera using cDNA microarray technology. Cancer Res. 2003;63:3940–4. [PubMed] [Google Scholar]
- 11.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 12.Krämer OH, Baus D, Knauer SK, Stein S, Jäger E, Stauber RH, et al. Acetylation of Stat1 modulates NF-kappaB activity. Genes Dev. 2006;20:473–85. doi: 10.1101/gad.364306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krämer OH, Knauer SK, Greiner G, Jandt E, Reichardt S, Guhrs K-H, et al. A phosphorylation-acetylation switch regulates STAT1 signaling. Genes & Development. 2009;23:223–35. doi: 10.1101/gad.479209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Federico M, Bagella L. Histone deacetylase inhibitors in the treatment of hematological malignancies and solid tumors. J Biomed Biotechnol. 2011;2011:475641. doi: 10.1155/2011/475641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mai A, Valente S, Rotili D, Massa S, Botta G, Brosch G, et al. Novel pyrrole-containing histone deacetylase inhibitors endowed with cytodifferentiation activity. Int J Biochem Cell Biol. 2007;39:1510–22. doi: 10.1016/j.biocel.2007.03.020. [DOI] [PubMed] [Google Scholar]
- 16.Sellar GC, Li L, Watt KP, Nelkin BD, Rabiasz GJ, Stronach EA, et al. BARX2 induces cadherin 6 expression and is a functional suppressor of ovarian cancer progression. Cancer Res. 2001;61:6977–81. [PubMed] [Google Scholar]
- 17.Jazaeri AA, Awtrey CS, Chandramouli GVR, Chuang YE, Khan J, Sotiriou C, et al. Gene expression profiles associated with response to chemotherapy in epithelial ovarian cancers. Clin Cancer Res. 2005;11:6300–10. doi: 10.1158/1078-0432.CCR-04-2682. [DOI] [PubMed] [Google Scholar]
- 18.Spentzos D, Levine DA, Kolia S, Otu H, Boyd J, Libermann TA, et al. Unique gene expression profile based on pathologic response in epithelial ovarian cancer. J Clin Oncol. 2005;23:7911–8. doi: 10.1200/JCO.2005.02.9363. [DOI] [PubMed] [Google Scholar]
- 19.Helleman J, Jansen MPHM, Span PN, van Staveren IL, Massuger LFAG, Meijer-van Gelder ME, et al. Molecular profiling of platinum resistant ovarian cancer. Int J Cancer. 2006;118:1963–71. doi: 10.1002/ijc.21599. [DOI] [PubMed] [Google Scholar]
- 20.Roberts D, Schick J, Conway S, Biade S, Laub PB, Stevenson JP, et al. Identification of genes associated with platinum drug sensitivity and resistance in human ovarian cancer cells. Br J Cancer. 2005;92:1149–58. doi: 10.1038/sj.bjc.6602447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng TC, Manorek G, Samimi G, Lin X, Berry CC, Howell SB. Identification of genes whose expression is associated with cisplatin resistance in human ovarian carcinoma cells. Cancer Chemother Pharmacol. 2006;58:384–95. doi: 10.1007/s00280-005-0171-8. [DOI] [PubMed] [Google Scholar]
- 22.Valenzuela-Fernández A, Cabrero JR, Serrador JM, Sánchez-Madrid F. HDAC6: a key regulator of cytoskeleton, cell migration and cell-cell interactions. Trends Cell Biol. 2008;18:291–7. doi: 10.1016/j.tcb.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 23.Wu S-Y, Chiang C-M. Crosstalk between sumoylation and acetylation regulates p53-dependent chromatin transcription and DNA binding. Embo J. 2009;28:1246–59. doi: 10.1038/emboj.2009.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–5. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 25.Fagerlund R, Melen K, Kinnunen L, Julkunen I. Arginine/lysine-rich nuclear localization signals mediate interactions between dimeric STATs and importin alpha 5. J Biol Chem. 2002;277:30072–8. doi: 10.1074/jbc.M202943200. [DOI] [PubMed] [Google Scholar]
- 26.McBride KM, Banninger G, McDonald C, Reich NC. Regulated nuclear import of the STAT1 transcription factor by direct binding of importin-alpha. Embo J. 2002;21:1754–63. doi: 10.1093/emboj/21.7.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kramer OH, Heinzel T. Phosphorylation-acetylation switch in the regulation of STAT1 signaling. Mol Cell Endocrinol. 2010;315:40–8. doi: 10.1016/j.mce.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 28.Thomas M, Finnegan CE, Rogers KM-A, Purcell JW, Trimble A, Johnston PG, et al. STAT1: a modulator of chemotherapy-induced apoptosis. Cancer Res. 2004;64:8357–64. doi: 10.1158/0008-5472.CAN-04-1864. [DOI] [PubMed] [Google Scholar]
- 29.Townsend PA, Scarabelli TM, Davidson SM, Knight RA, Latchman DS, Stephanou A. STAT-1 interacts with p53 to enhance DNA damage-induced apoptosis. J Biol Chem. 2004;279:5811–20. doi: 10.1074/jbc.M302637200. [DOI] [PubMed] [Google Scholar]
- 30.Patterson SG, Wei S, Chen X, Sallman DA, Gilvary DL, Zhong B, et al. Novel role of Stat1 in the development of docetaxel resistance in prostate tumor cells. Oncogene. 2006;25:6113–22. doi: 10.1038/sj.onc.1209632. [DOI] [PubMed] [Google Scholar]
- 31.Wang LH, Kirken RA, Erwin RA, Yu CR, Farrar WL. JAK3, STAT, and MAPK signaling pathways as novel molecular targets for the tyrphostin AG-490 regulation of IL-2-mediated T cell response. J Immunol. 1999;162:3897–904. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2
3
4