Cross-talk between Epidermal Growth Factor Receptor and Hypoxia-inducible Factor-1α Signal Pathways Increases Resistance to Apoptosis by Up-regulating Survivin Gene Expression (original) (raw)
. Author manuscript; available in PMC: 2011 Jul 10.
Published in final edited form as: J Biol Chem. 2006 Jul 17;281(36):25903–25914. doi: 10.1074/jbc.M603414200
Abstract
Although increasing evidence supports a link between epidermal growth factor receptor (EGFR) signaling and resistance to apoptosis, the mechanism by which the EGFR signaling pathway inhibits apoptosis is not well understood. In this study, we found that epidermal growth factor (EGF) stimulation increased the level of expression of the inhibitor of apoptosis protein survivin in breast cancer cells but not in normal mammary epithelial cells. We further demonstrated that activation of survivin gene expression is mediated by oxygen-independent hypoxia-inducible factor (HIF)-1_α_ up-regulation in EGF-treated cancer cells. EGFR signaling activated the phosphoinositide 3-kinase/AKT pathway, subsequently increasing the level of HIF-1_α_ under normoxic conditions. HIF-1_α_ then activated survivin gene transcription through direct binding to the survivin promoter. Furthermore, we found that overexpression of HIF-1_α_ small interfering RNA blocks EGF-induced survivin gene up-regulation and increases apoptosis induced by the chemotherapy drug docetaxel. However, transfection of a plasmid expressing HIF-1_α_ gene activates survivin gene expression and reduces the apoptotic response. Our results demonstrate a novel pathway for EGFR signaling-mediated apoptosis resistance in human cancer cells. Although the role of HIF-1_α_ in regulating cell survival under hypoxic conditions has been studied extensively, our results show that normoxic breast cancer cells utilize cross-talk between EGFR signals and HIF-1_α_ to up-regulate the anti-apoptotic survivin gene, providing a strong rationale for the targeting of HIF-1_α_ as a therapeutic approach for both hypoxic and normoxic tumor cells. Understanding key molecular events in EGFR signaling-induced apoptosis resistance should provide new information for the development of novel therapeutic agents targeting EGFR, HIF-1_α_, and/or survivin.
The EGFR2 signaling pathway plays a key role in the regulation of cell proliferation, survival, and differentiation (1, 2). It has been shown that the level of EGFR is up-regulated in many human tumor tissues. Activation of EGFR signaling has been associated with highly aggressive cancer types and poor responses to therapeutic agents (3–7). Prior preclinical and clinical studies have shown that blocking the EGFR signaling via monoclonal antibodies or inhibition of the EGFR tyrosine kinase with small molecules reduces the growth of breast cancers and sensitizes responses to chemotherapy (8–10).
Recently, we and others have shown that activation of the EGFR signaling pathway leads to the up-regulation of survivin, a member of the inhibitor of apoptosis (IAP) protein family (11–14). Survivin is broadly expressed in fetal tissues but is undetectable in the most normal adult tissues (15). However, a high level of survivin is found in most common tumor types, including over 70% of human breast cancer tissues at all stages of cancer development (16–18). It has been shown that in breast cancer cells, levels of survivin expression correlate with susceptibility to apoptosis (17). At present, the mechanism by which this up-regulation of survivin occurs in tumor cells having activated EGFR signaling is not fully understood. Recent studies have suggested, however, that activation of the phosphoinositide 3-kinase (PI3K)/AKT pathway by EGFR signaling causes up-regulation of survivin expression (12, 13). It is still unknown how PI3K/AKT signaling leads to survivin gene transcription.
Several studies have shown that under normoxic conditions, activation of EGFR signaling also increases the level of hypoxia-inducible factor 1_α_ (HIF-1_α_) through the PI3K/AKT pathway (19–21). HIF-1_α_, a member of the basic helix-loop-helix-PAS protein family (22), normally becomes highly up-regulated under hypoxic conditions, mostly as a result of inhibition of protein degradation. HIF-1_α_ can then activate transcription of many genes that are critical for continued cellular function under hypoxic conditions (22). Our previous study results have shown that survivin gene transcription is increased in hypoxic tumor cells (23). Due to the combined results of these studies, we speculated that the observed EGFR signaling- induced survivin gene expression might be mediated by transcriptional activity of HIF-1_α_ in an oxygen-independent manner.
In this study, we examined the effects of EGFR activation on the apoptotic response and survivin gene expression in human breast cancer cells. We found that EGF stimulation increases survivin gene expression specifically in breast cancer but does not in normal breast cells. Up-regulation of survivin gene expression reduces apoptosis induced by the chemotherapeutic drug docetaxel. We also discovered that in breast cancer cells, EGF up-regulates the level of HIF-1_α_ and that by down-regulation of HIF-1_α_ using HIF-1_α_ siRNA, we could significantly decrease those EGF-induced levels of survivin expression. Thus, cross-talk or a feedback loop between EGFR activation and HIF-1_α_ expression is implied. Furthermore, we demonstrated direct binding of HIF-1_α_ to the survivin promoter, which strongly suggests that EGF-activated survivin gene expression is indeed mediated by induction of transcriptional activity of HIF-1_α_ under normoxic conditions.
MATERIALS AND METHODS
Cell Lines
Normal immortalized human mammary epithelial cell line MCF-10A and breast cancer cell lines SK-BR-3, MDA-MB-231, and MCF-7 were obtained from the ATCC (Manassas, VA). SK-BR-3 and MDA-MB-231 cells were maintained in RPMI 1640, and MCF-7 cells were maintained in Dulbecco’s modified Eagle’s medium/F-12 medium (50:50; Mediatech) supplemented with 10 _μ_g/ml of insulin. All of the above media were supplemented with 10% fetal bovine serum (FBS) (Hyclone, Logan, UT) as well as 2 mML-glutamine, 100 IU/ml penicillin, and 100 _μ_g/ml streptomycin (Mediatech Herndon, VA). MCF-10A cells were cultured in Dulbecco’s modified Eagle’s medium/Ham’s F-12 medium supplemented with 20 ng/ml EGF, 500 ng/ml hydrocortisone, 100 ng/ml cholera toxin, 10 _μ_g/ml insulin, 2 mm l-glutamine, and 5% FBS.
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium Bromide (MTT) Cell Proliferation Assay
To measure the effects of EGF alone or in combination with docetaxel on breast cancer cells, 8 × 103 cells/well of MCF-7 and SK-BR-3 cells were plated in 96-well plates and cultured in the medium containing EGF and/or docetaxel (Aventis Pharma, Bridgewater, NJ) in various combinations of the drugs for 3 days. The percentage of viable cells in each well was examined by an MTT cell proliferation assay (Sigma), and the remaining viable cells were determined using Spectra Max Plus (Molecular Devices, Sunnyvale, CA).
Transfection
Plasmids expressing a full-length HIF-1_α_ cDNA gene were provided by Dr. Hua Zhong (Emory University, Atlanta, GA). The control empty pcDNA3 or pHIF-1_α_ plasmids were transfected into cultured tumor or normal cell lines in 6-well tissue culture plates using Lipofectamine 2000 (Invitrogen). Some groups of the plasmid-transfected cells were treated with docetaxel in the absence or presence of 100 ng/ml human EGF. Twenty-four hours after transfection, the cells were collected for Western blot analysis to determine the levels of HIF-1_α_ and survivin proteins or FACScan analysis for the percentage of the apoptotic cells.
Apoptosis Assay
Cellular apoptosis was determined using Annexin V-phycoerythrin (PE) and 7-amino-actinomycin D (7-AAD) (BD Biosciences). SK-BR-3 and MCF-7 cells were treated for 3 days with 25 or 50 nM of docetaxel in the absence or presence of 100 ng/ml EGF. Floating and adherent cells were labeled with Annexin V-PE and 7-AAD and then analyzed by FACScan (BD Biosciences) to determine the percentage of apoptotic cells.
Real Time Reverse Transcription-PCR
Total RNAs were isolated using the RNA Bee kit (Tel-test, Friendswood, TX). Each 2-_μ_g sample of RNA was amplified with the Omniscript RT kit using an oligo(dT) primer (Qiagen Inc., Valencia, CA) to generate 20 _μ_l of cDNAs. A 1–2-_μ_l sample of the cDNA was then quantified by real time PCR using primer pairs for survivin or _β_-actin with SYBR Green PCR Master mix. Real time PCR was performed using the ABI PRISM 7000 sequence detection system (Applied Biosystems, Foster City, CA). The primer pairs for detection of survivin gene expression were as follows: survivin forward, 5′-TCCACTGCCCCACTGAGAAC-3′; surviving reverse, 5′-TGGCTCCCAGCCTCCA-3′. These amplify a 77-nt PCR product located at nt 130–206 of the survivin mRNA. Amplification of the _β_-actin gene was used as an internal control for real time reverse transcription-PCR. The primer pair for the _β_-actin gene was as follows: forward, 5′-AAAGACCTGTACGCCAACACAGTGCTGTCTGG-3′; reverse, 5′-CGTCATACTCCTGCTTGCTGATCCACATCTGC-3′, which generates a 219-nt PCR product from nt 870 to 1089 of the _β_-actin mRNA sequence. The quantity of PCR product generated from amplification of the survivin gene was standardized using the quantity of _β_-actin product for each sample to obtain a relative level of gene expression.
Western Blot Analysis
For the various treatments described, cells were cultured in medium containing 10% serum and then changed to medium containing 2% serum or no serum when treated with 100 ng/ml human EGF (Invitrogen) and/or docetaxel. Inhibitors for PI3K (LY294002), MAPK (PD98059), and EGFR (AG1478) were obtained from Calbiochem. At the end of the assay, cells were lysed in lysis buffer, and Western blot analysis was performed as previously described (18). The detection antibodies for phosphoserine 473 Akt, p44/p42 MAPK, XIAP, and cleaved caspase-3 were from Cell Signaling Technology Inc. (Beverly, MA). Goat anti-human survivin (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) and mouse monoclonal anti-_β_-actin antibody (Sigma) were also used.
Caspase Activity Assay
Cells were treated with or without docetaxel, in the absence or presence of 100 ng/ml human EGF, for a period of 2 days. Cells were collected, and their lysates were examined for caspase-3-like activity using a specific substrate, Ac-DEVD-7-amino-4-trifluoromethylcoumarin, which detects the activities of caspase-3, caspase-7, caspase-10, or caspase-9 activity using Ac-LEHD-7-amino-4-trifluoromethylcoumarin, according to a standard protocol (Calbiochem). The results were measured using a Spectra Max fluorescence microplate reader (Molecular Devices). For each experiment, control groups with specific caspase inhibitors, including caspase-3 inhibitor (benzyloxycarbonyl-DEVD-aldehyde; BD Biosciences) and caspase-9 inhibitor (benzyloxycarbonyl-LEHD-aldehyde; Alexis Biochemicals, San Diego, CA), were done to ensure the specificity of the assay.
Chromatin Immunoprecipitation (ChIP) Assay
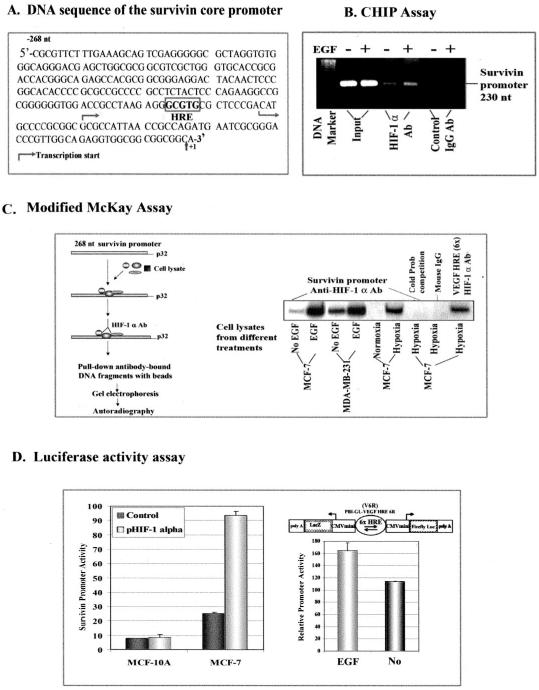
To demonstrate direct binding of HIF-1_α_ protein to the survivin promoter region in EGF-stimulated cancer cells, the SK-BR-3 cells were treated with 100 ng/ml EGF for 4 h. After cross-linking chromatin with proteins by 1% of formaldehyde, the assay was performed using a ChIP assay kit from Upstate (Charlottesville, VA), according to the company’s protocol. A monoclonal anti-HIF-1_α_ antibody (BD Biosciences) was added to precipitate the protein-chromatin complexes. A PCR primer pair for amplification of a 230-nt survivin promoter fragment is as follows: forward primer, 5′-GCGTTCTTTGAAAGCAGT-3′; reverse primer, 5′-ATCTGGCGGTTAATGGCG-3′.
Modified McKay Assay
A core survivin promoter containing 269 nt of the 5′-flanking region of the survivin gene was cut out from a survivin promoter-luciferase reporter plasmid (pluc-cyc1.2) (23). A DNA fragment containing six repeats of the hypoxia-responsive element (HRE) of vascular endothelial growth factor (VEGF) was also cut from pBI-GL V6R plasmid (23) as a positive control. The purified promoter fragments were 5′-end-labeled with [γ_-32P]dATP using T4 polynucleotide kinase (New England Biolabs, Ipswich, MA). The radiolabeled promoter fragments were then incubated with nuclear extracts obtained from breast cancer cells treated with or without 100 ng/ml EGF in a 1× binding buffer containing 10 mM Tris-HCl (pH 8.0), 150 mM KCl, 0.5 mM EDTA, 0.1% Triton X-100, 12.5% glycerol, and 0.2 mM dithiothreitol on ice for 1 h. Protein A-Sepharose beads conjugated with monoclonal anti-HIF-1_α antibody were added to precipitate the protein-DNA complexes. After washing for three times with a 1× TE buffer containing 10 mM Tris-HCl (pH 7.5) and 1 mM EDTA, the final pellet was resuspended in a 0.1 M NaHCO3, 0.1% SDS, 1× sample buffer and heated to 65 °C for 2 min. Then the protein A-Sepharose beads were pelleted, and the supernatant was analyzed by electrophoresis on a 1.4% agarose gel followed by autoradiography to determine whether the EGF-induced HIF-1_α_ protein had bound to the survivin promoter DNA fragments.
Luciferase Assay
The effect of EGF stimulation on survivin promoter activity was determined in breast cancer cells after transfecting pluc cyc-1.2 plasmid, a survivin promoter-luciferase reporter plasmid (23), for 24 h, followed by EGF treatment for 4 h. A pRL-SV-40 plasmid that expresses a Renilla luciferase gene (Promega, Madison, WI) was also cotransfected for all studies as an internal control. To determine whether direct overexpression of the HIF-1_α_ gene activates the survivin promoter, breast cancer and normal cells were cotransfected with pluc cyc-1.2 and pHIF-1_α_ plasmids for 24 h. The ability of direct activation of HRE by EGF-induced HIF-1_α_ was demonstrated using a MDA-MB-231 cell line, stably transfected with pBI-GL V6R plasmid containing six copies of HRE fragments of the VEGF gene and a firefly luciferase reporter gene (24) (provided by Dr. Hyunsuk Shim at Emory University). The transfected cells or MDA-MB-231 stable cells were cultured in the absence or presence of 100 ng/ml of EGF for 45 min. After various treatments as described above, the promoter activity of the cell lysates was determined using a dual luciferase activity assay kit from Promega.
Production of Adenoviral Vectors Expressing HIF-1_α_ siRNA
Plasmid vectors containing either HIF-1_α_ siRNA sequence 5′-CAGTGGATTACCACAGCTGA-3′ or survivin siRNA 5′-GGCTGGCTTCATCCACTGCCC-3′ were generated by cloning the synthesized oligonucleotide into pSilencer 2.1-U6 Neo plasmid (Ambion Inc., Austin, TX). Control pSilencer 2.1-U6 Neo plasmid vector containing a scrambled siRNA sequence, 5′-ACTACCGTTGTTATAGGTGT-3′, was obtained from the company. Adenoviral vectors expressing siRNA to HIF-1_α_ and control siRNA were produced by cloning a HindIII-EcoRI fragment from pSilencer 2.1-U6 Neo plasmid, which contains a U6-promoter-siRNA cassette (HIF-1_α_ or control siRNA), to a pcDNA3 HindIII-EcoRI site and then transferring the NotI-HindIII fragment from the pcDNA3 to the pAdtrack plasmid (25). After performing homologous recombination with an AdEasy adenoviral DNA backbone, the viral vectors were produced by transfecting into the human embryonic kidney cell line 293 (ATCC). The AdEasy system was provided by Dr. Bert Vogelstein at The Johns Hopkins University. Additional vector amplification was also performed in the 293 cell line. High titer viral vectors were purified by centrifugation and CsCI banding.
To determine the effect of down-regulation of HIF-1_α_ on tumor cells, 2 × 105 tumor cells/well were first cultured in 6-well plates and then transduced with Ad HIF-1_α_ siRNA or Ad Control siRNA vectors. 24 h after transduction, cells were cultured in serum-free medium overnight and treated with (or without) EGF for 45 min. Some groups received cotransfection of pSilencer HIF-1_α_ siRNA and pHIF-1_α_ plasmids to determine whether overexpression of HIF-1_α_ gene could rescue HIF-1_α_ siRNA-mediated down-regulation of survivin in EGF-treated cells. Last, collected cell lysates were examined for the levels of both HIF-1_α_ and survivin by Western blot analysis.
To examine the effect of EGF-induced HIF-1_α_ and survivin on apoptotic death in breast cancer cells after docetaxel treatment, 5 × 104/well of MCF-7, SK-BR-3, and MDA-MB-231 cells were plated in 12-well tissue culture plates. Cells were then transduced with AdControl siRNA or AdHIF-1_α_ siRNA vector. Some treatment groups were transfected with psilencer 2.1 U6-survivin siRNA plasmids. Following a 2-day treatment with docetaxel, the percentage of remaining viable cells was examined using an MTT cell proliferation assay.
RESULTS
EGF Stimulation Decreased Docetaxel-induced Apoptotic Response in Breast Cancer Cells
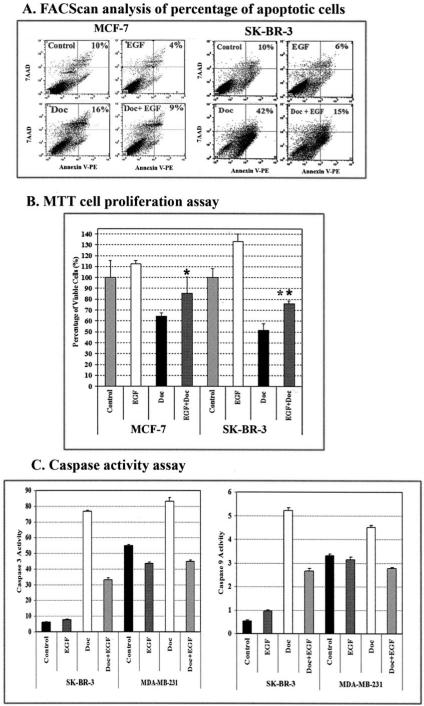
In order to determine the effect of activation of the EGFR signaling pathway on the apoptotic response in human cancer cells, we treated the human breast cancer cell lines SK-BR-3 and MCF-7 with a chemotherapeutic drug, docetaxel, in the absence or presence of human recombinant EGF. Previous reports have shown that SK-BR-3 cells have high levels of EGFR and Her-2/neu expression, whereas MCF-7 cells express low levels of EGFR and Her-2/neu (26). We chose those two cell lines to determine whether the levels of the EGF receptors present affect the EGF-induced apoptotic response. After treating the cancer cells with docetaxel in the absence or presence of EGF for 2 days, the percentage of apoptotic cells was determined using Annexin V-PE and 7-AAD staining followed by FACScan analysis. We found that the presence of EGF markedly decreased the percentage of apoptotic cell death induced by docetaxel. EGF treatment reduced the percentage of apoptotic cells present from 16 to 9% for MCF-7 cells and from 42 to 15% for SK-BR-3 cells (Fig. 1_A_). Additionally, we found that EGF decreased the rate of spontaneous apoptosis in both cell lines (Fig. 1_A_). Consistent with the results from the apoptosis assay, we found that EGF stimulation significantly decreased sensitivity of SK-BR-3 and MCF-7 cells to docetaxel as detected by the MTT cell proliferation assay (Fig. 1_B_). Furthermore, we found that EGF-induced resistance to apoptosis was detected in breast cancer cell lines expressing both a high level of EGFR (SK-BR-3) and a low level of EGFR (MCF-7).
FIGURE 1. Activation of the EGFR signaling pathway increases resistance to docetaxel-induced apoptosis in human breast cancer cells.
A, apoptosis assay using Annexin V staining and FACScan analysis. MCF-7 and SK-BR-3 cells were cultured for 2 days in medium containing 100 ng/ml EGF in the absence or presence of 25 nM (SK-BR-3) or 50 nM (MCF-7) docetaxel and 2% FBS. The cells were then stained with Annexin V-PE and 7-AAD and analyzed by FACScan. The percentages of apoptotic cells are shown in the upper panel, including both early (Annexin-V+, 7-AAD−) and late stage (Annexin+, 7-AAD+) apoptotic cells. B, EGF stimulation decreases the sensitivity of human breast cancer cells to docetaxel treatment. Cancer cells cultured in 96-well plates were treated for 3 days with the reagents as described above. The percentage of remaining viable cells was determined using a MTT cell proliferation assay. The results of this assay are expressed relative to the cell density of untreated cells. Each value in the graph represents the mean ± S.D. of five repeat samples. An asterisk indicates a significant difference when compared with the control value (*, p < 0.05; **, p < 0.001). C, activation of the EGFR signal inhibits docetaxel-induced caspase activity. The cells were treated with docetaxel in the absence or presence of EGF for 3 days. 25 _μ_g of proteins from their total cell lysates were examined for caspase-3-like or caspase-9 activity using substrates specific for these caspases. Fluorescence intensity was measured by a fluorescence microplate reader. The numbers in the figure represent mean values ± S.D. from three repeat groups.
To elucidate the mechanism of apoptosis resistance, we examined changes in regulation of apoptotic signals in EGF-stimulated cancer cells. Since caspase activation is a critical step in induction of apoptotic cell death, we first examined caspase-3-like and caspase-9 activities in MDA-MB-231 and SK-BR-3 cell lines 2 days after docetaxel treatment. As expected, docetaxel increased these caspase activities in both tumor cell lines. However, in the presence of EGF, the docetaxel-induced caspase-3-like and caspase-9 activities were markedly inhibited (Fig. 1_C_), suggesting that the EGF-induced resistance to apoptosis by docetaxel is mediated through inhibition of caspase activity.
EGF Up-regulates Survivin Expression in Human Breast Cancer Cells
It is well known that the IAPs block apoptotic signaling through inhibition of caspase activity (27). To determine whether IAPs play a role in inhibition of caspases in tumor cells having an activated EGFR signal, we examined changes in the level of an important IAP family protein, survivin, in both breast cancer and normal cell lines following EGF treatment. Our results from Western blot analysis demonstrated that EGF stimulation did increase the level of survivin protein in three breast cancer cell lines, including MDA-MB-231, SK-BR-3, and MCF-7 (Fig. 2_A_). Interestingly, survivin was not detected in the immortalized normal human mammary epithelial cell line MCF-10A, and EGF treatment failed to induce any survivin expression (Fig. 2_A_). To determine the mechanism of survivin up-regulation, we further examined survivin promoter activity and the level of survivin mRNA in EGF-treated cells. Using a luciferase reporter plasmid containing a 269-nt survivin core promoter fragment, we found a marked increase in survivin promoter activity in EGF-stimulated SK-BR-3 cells (Fig. 2_B_). Consistent with the level of survivin protein, survivin mRNA was elevated in the EGF-treated SK-BR-3 and MCF-7 cells (Fig. 2_C_) detected by real time reverse transcription-PCR. Additionally, we found that treatment of the cells by docetaxel increases the levels of survivin gene expression. A combined effect of docetaxel and EGF treatment further increased the level of survivin gene expression (Fig. 2_C_).
FIGURE 2. EGF stimulation increases the levels of survivin expression in breast cancer cells.
Cells were cultured in 2% FBS medium overnight, followed by treatment with human EGF at 100 ng/ml for 45 min to 4 h. A, examination of the level of survivin protein (16.5 kDa) in breast cancer and normal cell lines following EGF stimulation for 45 min by Western blot analysis. B, EGF stimulation activates survivin promoter activity. SK-BR-3 cells were transfected with a survivin promoter luciferase reporter plasmid (pluc cyc1.2) for 24 h and then treated with EGF for 4 h. The cell lysates were collected for the luciferase assay. C, total RNAs were isolated and analyzed by real time reverse transcription-PCR, as described under “Experimental Procedures,” to determine changes in the level of survivin mRNA after EGF treatment. EGF stimulation increases the levels of survivin mRNA in both MCF-7 and SK-BR-3 cell lines. Additionally, docetaxel treatment could also up-regulate survivin gene expression, and its expression level is further enhanced in the presence of EGF. The relative level of survivin mRNA is a ratio of the quantity of survivin to _β_-actin PCR products. A mean value of three repeat samples is shown.
Determination of Signaling Pathways Responsible for EGF-induced Survivin Gene Up-regulation
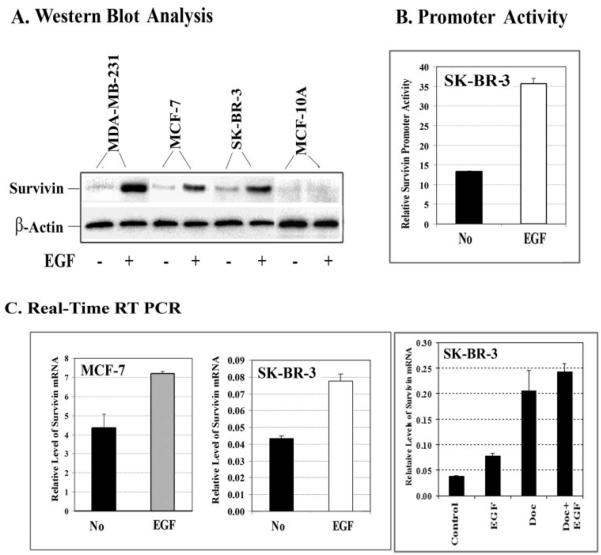
It is well known that EGFR signaling leads to the activation of both PI3K/AKT and MAPK pathways. We examined the roles of these pathways in the EGF-induced survivin expression in breast cancer cells. As expected, we observed increases in the levels of p-AKT and p44/42 MAPK in EGF-treated cells. Although the level of survivin expression was also up-regulated in those cells, the level of another IAP protein, XIAP, remained unaffected by EGF treatment (Fig. 3).
FIGURE 3. Analysis of signal transduction pathways regulating EGF-induced survivin gene expression.
Cells were pretreated with 10 _μ_M LY294002, PD98059, or AG1478 for 24 h in culture medium containing 2% FBS medium. Then 100 ng/ml EGF was added for 45 min. Total cell lysates (50 _μ_g of protein) were examined by Western blot analysis to determine the levels of phosphorylated serine 473 Akt (P-Ser473 Akt), phosphorylated p44/p42 MAPK, survivin, and XIAP. In both breast cancer cell lines, EGF stimulation activated the AKT and MAPK signal pathways, and blocking the PI3K/AKT signal prevented EGF-induced survivin expression. On the other hand, inhibition of the EGFR signal pathway with AG1478 reduced the level of survivin protein. The level of XIAP was not affected by EGF stimulation or the PI3K/AKT, MAPK, and EGFR inhibitors.
After pretreatment of the cancer cells with either the PI3K inhibitor LY294002 or the MAPK inhibitor PD98059, we were able to achieve marked reductions in the levels of p-AKT and p44/42 MAPK present in EGF-treated SK-BR-3 and MCF-7 cells. Furthermore, we found that inhibition of p-AKT completely prevented EGF-up-regulated survivin expression in both the SK-BR-3 and MCF-7 cell lines (Fig. 3). However, we believe that activation of AKT is probably not required for maintaining a basal level of survivin expression in these cancer cells, since similar levels of survivin expression were detected in both control cells and cells treated only with PI3K inhibitor (Fig. 3).
EGF stimulation also increased the level of p44/42 MAPK in SK-BR-3 and MCF-7 cell lines, with SK-BR-3 cells showing a very high level of up-regulation (Fig. 3). Blocking the MAPK pathway with PD98059 inhibited the level of p44/42 MAPK in both cell lines. The fact that a significant reduction of survivin was seen in the MAPK-blocked, EGF-treated SK-BR-3 cells suggests that both the AKT and MAPK pathways are involved in EGF-induced survivin up-regulation (Fig. 3).
After treating the cancer cells with EGF in the absence or presence of the EGFR inhibitor AG1478, we found that AG1478 counteracted the effects of EGF on survivin expression in both SK-BR-3 and MCF-7 cells. However, AG1478 treatment alone had no significant effects on the basal levels of survivin protein in these cells (Fig. 3).
EGF-induced HIF-1_α_ Up-regulates Survivin Expression in Breast Cancer Cells
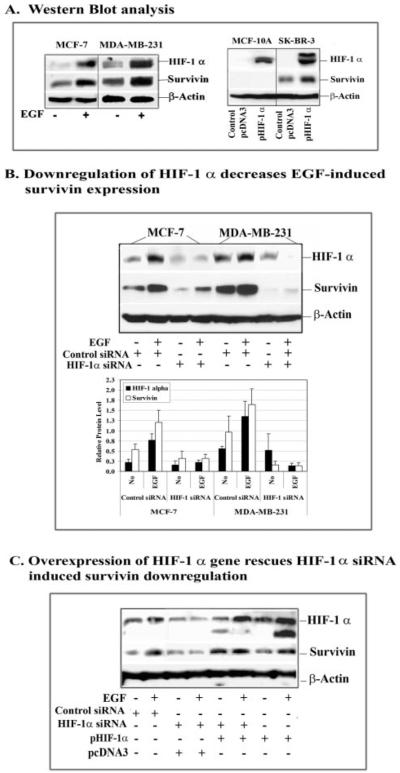
Previous reports have shown that activation of EGFR signaling can induce HIF-1_α_ in human tumor cells under normoxic conditions (20, 21). We previously found that survivin promoter activity is up-regulated in hypoxic tumor cells (23). To determine if the EGFR signaling-induced HIF-1_α_ is a mediator for survivin gene expression, we first examined the changes in both HIF-1_α_ and survivin levels in breast cancer cells after EGF treatment. Western blot analysis did show that EGF treatment induces high levels of both HIF-1_α_ and survivin expression in breast cancer cells (Fig. 4_A_). We further found that transfection of the plasmids expressing a HIF-1_α_ gene into cells increases the levels of HIF-1_α_ in both breast cancer SK-BR-3 and normal breast MCF-10A cell lines. However, up-regulation of the level of survivin protein is only detected in SK-BR-3 cells. This result provides direct evidence supporting HIF-1_α_-mediated survivin up-regulation. Additionally, the absence of survivin expression in HIF-1_α_-transfected MCF-10 A cells suggests that intrinsic transcriptional inhibitory factors prevent the basal level as well as HIF-1_α_-induced survivin transcription in normal cells.
FIGURE 4. Determination of the role of HIF-1_α_ in survivin up-regulation in normoxic breast cancer cell lines after EGF treatment.
A, Western blot analysis showed that EGF stimulation induces high levels of both HIF-1_α_ and survivin expression. Furthermore, transfection of HIF-1_α_ gene-expressing plasmids increases the level of survivin in breast cancer SK-BR-3 cells but not in normal breast epithelial MCF-10A cells. B, overexpression of HIF-1_α_ siRNA by transduction of the cells with a multiplicity of infection of 100 plaque-forming units/cell of adenoviral vectors inhibited EGF-induced as well as basal levels of HIF-1_α_ expression in MCF-7 and MDA-MB-231 cells. The levels of survivin protein were also markedly reduced. The relative levels of HIF-1_α_ and survivin proteins in the cells treated with control and HIF-1_α_ siRNAs were quantified by measuring the density of protein bands using Scion Image (Scion Corp., Frederick, MD). HIF-1_α_ or survivin protein level was calculated from a ratio of the density relative to β_-actin for each sample. The number in the figure represents the mean value of three repeat Western blots. C, over-expression of the HIF-1_α gene rescues HIF-1_α_ siRNA-induced survivin down-regulation. Breast cancer MCF-7 cells were co-transfected with HIF-1_α_ siRNA and pHIF-1_α_ plasmids for 24 h, and the levels of HIF-1_α_ and survivin proteins were examined by Western blot analysis. In the presence of an excessive amount of exogenous HIF-1_α_, expression of siRNA to HIF-1_α_ could no longer inhibit the level of EGF-induced survivin expression. Although transfection of pSilencer HIF-1_α_ siRNA plasmids showed less inhibitory effects on the levels of HIF-1_α_ and survivin proteins, compared with the effect produced by using AdHIF-1_α_ siRNA seen in this figure (B), it is appropriate for the design of this study, since the HIF-1_α_ gene is expressed from a plasmid vector.
We then transduced the cells for 48 h with adenoviral vectors expressing either HIF-1 α siRNA or control siRNA. Next, the transduced cells were treated with or without EGF for 45 min before they were collected. The resulting cell lysates were analyzed by Western blot for the levels of HIF-1_α_ and survivin. We found that overexpression of HIF-1_α_ siRNA markedly reduced EGF-induced HIF-1_α_. Meanwhile, EGF-induced up-regulation of survivin expression was completely blocked by the HIF-1_α_ siRNA (Fig. 4_B_). To further demonstrate the critical role for HIF-1_α_ in regulating survivin gene transcription, we co-expressed HIF-1_α_ siRNA and HIF-1_α_ gene plasmids in MCF-7 cells. 24 h after transfection, the cells were treated with EGF and collected for Western blot analysis. In the presence of excessive amounts of HIF-1_α_, expression of HIF-1_α_ siRNA could no longer block EGF-induced survivin down-regulation (Fig. 4_C_), supporting direct involvement of HIF-1_α_ in EGF-activated survivin expression.
HIF-1_α_ Activates Gene Transcription through Direct Interaction with the Survivin Promoter
At present, the mechanisms by which cellular factors may regulate survivin gene expression are not fully understood. Our previous study identifies a 269-nt DNA fragment located at the 5′-flanking region of the survivin gene that is able to activate tumor-specific gene transcription, exhibiting enhanced promoter activity under hypoxic conditions (23). To determine whether the EGF-activated survivin gene transcription is mediated by transcriptional activity of HIF-1_α_, we first analyzed the survivin core promoter sequences and found a putative HRE, 5′-GCGTG-3′, located at nt −81 to −85 of the 5′-flanking region of the survivin gene (Fig. 5_A_). To demonstrate the binding of HIF-1_α_ to the survivin promoter in living cells, we performed a ChIP assay in MCF-7 cells with or without EGF treatment. In the chromatin fraction pulled down by an anti-HIF-1_α_ antibody, we detected a higher level of the survivin promoter PCR fragments in EGF-treated cells than that in control cells (Fig. 5_B_). However, survivin promoter PCR fragments were not found in samples pulled down by a control IgG antibody.
FIGURE 5. Determination of the mechanism of HIF-1_α_-activated survivin gene expression.
A, DNA sequence of the survivin core promoter. A putative HRE site, located at −81 to −85 nt, is marked in the promoter sequence. B, detection of the binding of HIF-1_α_ to the survivin promoter in breast cancer cells using a ChIP assay. SK-BR-3 cells were treated with or without EGF for 4 h. A ChIP assay was then performed. 230-nt PCR products of the survivin promoter were only detectable in the samples pulled down by HIF-1_α_ antibody and not in control IgG samples. A marked higher level of the survivin promoter PCR products was seen in the EGF-treated sample compared with the no treatment sample. C, modified McKay assay for detection of the binding of HIF-1_α_ to the survivin promoter. Cancer cells were treated with or without EGF for 45 min, and nuclear extracts were incubated with 269-nt radiolabeled survivin promoter fragments, and the resulting HIF-1-DNA complexes were pulled down using anti-HIF-1_α_ antibody-conjugated Protein A beads. As shown, a high level of HIF-1_α_-survivin promoter complexes was found following incubation with nuclear extracts from EGF-treated breast cancer cells. The binding specificity was further demonstrated by the absence of survivin promoter bands in samples with a 10 times excess of unlabeled (cold) survivin promoter fragments or using nonspecific mouse IgG-conjugated Protein A beads. A DNA fragment containing six repeats of the HRE fragments of the human VEGF promoter was used as a positive control. D, luciferase activity assay. Overexpression of the HIF-1_α_ gene after co-transfection of pHIF-1_α_ and pluc cyc1.2 increases survivin promoter activity in MCF-7 tumor cells but fails to induce the promoter activity in normal MCF-10A cells. EGF-induced HIF-1_α_ was able to bind to the HRE and activate firefly luciferase gene expression under normoxic conditions, since luciferase activity was increased in the cell lysates from EGF-treated MDA-MB-231 cells stably transfected with pBI-GL-V6R plasmid, which has six repeats of VEGF HRE fragments.
To further confirm direct binding of HIF-1_α_ to the survivin core promoter, we used a modified McKay assay (28) to pull down survivin promoter DNA fragments (Fig. 5_C_). Our results demonstrated that the survivin promoter fragments did bind to the HIF-1_α_ protein and could be pulled down by HIF-1_α_ antibody. EGF-treated cell lysates showed much higher levels of these survivin promoter fragments as compared with control groups in both breast cancer cell lines (Fig. 5_C_). Since there is a moderate basal level of HIF-1_α_ found in those tumor cells, we also detected low to intermediate levels of survivin promoter fragments in our control groups (Fig. 5_C_).
To determine whether the binding of HIF-1_α_ to the survivin promoter actually could activate the promoter activity, we cotransfected pHIF-1_α_ plasmid with survivin promoter reporter pluc cyc1.2 plasmid into breast normal and cancer cell lines. We found that overexpression of the HIF-1_α_ gene markedly increased the survivin promoter activity in breast cancer MCF-7 cells but not in normal breast MCF-10A cells (Fig. 5_D_). Next, we wanted to determine if EGF-induced HIF-1_α_ interacts with the HRE site under normoxic conditions to activate the gene transcription. We used the human breast cancer cell line MDA-MB-231, stably transfected with a luciferase reporter plasmid containing six copies of the HRE fragment of the VEGF gene. These cells were treated with EGF in normoxia, and the resultant luciferase activity was measured in cell lysates. We found that EGF-induced HIF-1_α_ was able to activate the HRE-mediated transcription of the luciferase gene, suggesting that under normoxic conditions, HIF-1_α_ can bind to the HRE site and activate HRE-mediated gene transcription (Fig. 5_D_).
Overexpression of the HIF-1_α_ Gene Inhibits Docetaxel-induced Apoptosis under Normoxic Conditions
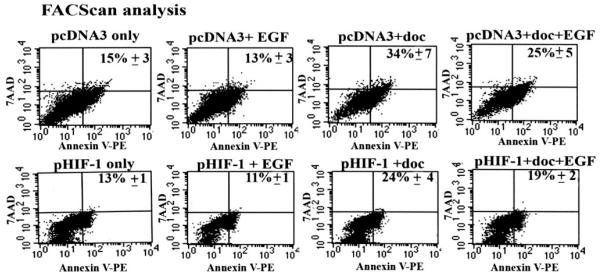
To establish a direct link between EGF-induced HIF-1_α_ up-regulation and resistance to apoptosis, we transfected SK-BR-3 cells with pHIF-1_α_ plasmids and then treated the cells with docetaxel in the absence or presence of EGF for 24 h. The cells were collected for Annexin V-PE staining and FACScan analysis of the percentage of apoptotic cells. We found that overexpression of the HIF-1_α_ gene reduced the percentage of docetaxel-induced apoptotic cells from 34 to 24%. The combination of overexpression of the HIF-1_α_ gene with EGF treatment further enhanced the inhibitory effect on apoptosis (Fig. 6), suggesting that HIF-1_α_ is a key mediator for EGF-induced resistance to apoptosis.
FIGURE 6. Examination of the effect of overexpression of HIF-1_α_ gene on docetaxel-induced apoptosis by FACScan analysis.
Breast cancer SK-BR-3 cells, transfected with pHIF-1_α_ or control pcDNA3 plasmids for 24 h, were treated with 25 nM docetaxel in the absence or presence of 100 ng/ml of EGF for an additional 24 h. Both floating and adhesive cells were collected for Annexin V-PE and 7-AAD staining followed by FACScan analysis. Expression of the HIF-1_α_ gene reduced the percentage of docetaxel-induced apoptosis. The combination of EGF activation with HIF-1_α_ gene expression further increased the inhibition effects. Since the cells were cultured in the medium containing 2% of FBS due to EGF treatment, we observed a relatively high level of the basal level of apoptosis in pcDNA3 control plasmid-transfected cells. The number in the figure is the mean value ± S.D. from three repeat samples.
Down-regulation of the Levels of HIF-1_α_ or Survivin Gene Expression Reverses EGF-induced Resistance to Apoptosis
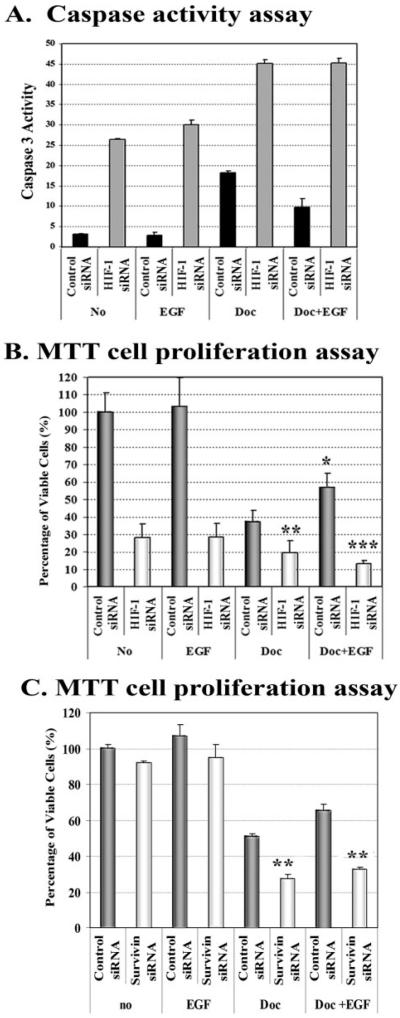
To further demonstrate that EGF-induced resistance to apoptosis is indeed the result of up-regulation of HIF-1_α_, which consequently activates survivin gene expression, we examined the effects of HIF-1_α_ down-regulation, using HIF-1_α_ or survivin siRNA, on overall sensitivity to apoptosis induction. We found that expression of HIF-1_α_ siRNA using adenoviral vectors in the no treatment control and the EGF-treated SK-BR-3 cells induced 8–10-fold increases in caspase-3-like activity as compared with control siRNA vector-containing cells (Fig. 7_A_). Furthermore, the presence of HIF-1_α_ siRNA enhanced 2.5 times the level of active caspase-3 induced by docetaxel treatment. Although co-treatment with EGF reduced the docetaxel-induced caspase-3 activity, down-regulation of HIF-1_α_ with siRNA completely reversed the ability of exogenous EGF to induce resistance to docetaxel (Fig. 7_A_). The changes in caspase-3 activity among the different treatment groups were also correlated with the percentage of apoptotic cell death in the cells. As shown in Fig. 7_B_, overexpression of HIF-1_α_ siRNA significantly increased apoptotic cell death in docetaxel-treated cells by 18% (p = 0.006, Student’s t test). EGF treatment protected the cells from docetaxel-induced apoptosis and increased the percentage of viable cells from 37% in the group without EGF to 57% in EGF-treated cells (Fig. 7_B_, p = 0.01). Importantly, overexpression of HIF-1_α_ siRNA significantly reduced the EGF-induced resistance to docetaxel, since there was a 44% decrease in the percentage of viable cells in the AdHIF-1_α_ siRNA-transduced cells when compared with control siRNA-transduced cells (p < 0.0001, Student’s t test). We also examined the effect of direct inhibition of survivin gene expression on EGF-induced resistance to apoptosis. We found that expression of survivin siRNA significantly sensitized SK-BR-3 cells to docetaxel treatment (Fig. 7_C_, p = 0.0004, Student’s t test). Down-regulation of survivin expression also increased docetaxel-induced cell death even in the presence of EGF stimulation (Fig. 7_C_, p = 0.0003, Student’s t test). Therefore, our results strongly supported a role for HIF-1_α_-mediated survivin up-regulation in EGF-induced resistance to apoptosis in breast cancer cells.
FIGURE 7. Down-regulation of HIF-1_α_ or survivin gene expression using siRNAs enhances docetaxel-induced apoptosis and reverses EGF signal-mediated resistance to apoptosis.
SK-BR-3 cells were transduced with AdControl siRNA or HIF-1_α_ siRNA vectors for 24 h. The cells were then treated with 25 nM docetaxel, in the absence or presence of 100 ng/ml EGF for an additional 24 h. A, caspase activity was examined in the cell lysates using a caspase-3-like specific substrate. The numbers in the figure represent mean values of three repeat groups. Similar results were obtained from repeat experiments. As shown, inhibition of HIF-1_α_ expression increases caspase-3 activity in tumor cells for all treatment groups. Additionally, EGF-induced inhibition of caspase-3 activity in docetaxel-treated cells was reversed by the down-regulation of HIF-1_α_ with siRNA. B, results from the MTT cell proliferation assay show that down-regulation of HIF-1_α_ significantly increased docetaxel-induced cell death and completely blocked EGF-induced resistance to docetaxel treatment in SK-BR-3 cells. On the other hand, a significant reduction of docetaxel-induced cell death is found in EGF-treated, control siRNA vector-transduced cells. C, transfection of plasmids expressing survivin siRNA significantly increases docetaxel-induced cell death and counteracts the protection effect of EGF stimulation. The absorbance value of cells transduced with AdControl siRNA vector or plasmids expressing control siRNA serves as a relative cell number of 100%. Each value in the bar graph represents a mean value ± S.D. of four repeat samples. *, p < 0.01; **, p < 0.001; ***, p < 0.0001.
DISCUSSION
Overexpression of EGFR and activation of the EGFR signaling pathway are found in a high percentage of human breast cancer tissues and have been associated with poor prognosis for the patients (3, 5, 7, 29). Although EGFR signaling, including activation of EGFR and Her-2/neu, is known to confer resistance to apoptosis in cancer cells (30, 31), the exact mechanism by which EGFR signaling regulates the apoptotic pathway has yet to be elucidated.
In this study, we have examined the effects of the activation of EGFR signaling on the apoptotic response in human breast cancer cell lines. We have shown that EGF stimulation reduces the sensitivity of breast cancer cells to docetaxel, a chemotherapeutic drug that is commonly used for treatment of breast cancer. We have further demonstrated that decreased drug sensitivity is the result of inhibition of the activities of caspase-3 and caspase-9, which consequently causes the observed resistance to apoptosis in EGF-treated breast cancer cells.
IAP proteins, including survivin and XIAP, are known to act as caspase inhibitors, blocking caspase activation and further inhibiting activities of the activated caspases (27, 32, 33). Recent studies, including ours, have shown that activation of EGFR leads to up-regulation of survivin expression (11–14). Additionally, it has been demonstrated that overexpression of Her-2/neu and EGFR genes increases survivin expression and apoptosis resistance in breast cancer cell lines (11). Results from examination of 195 cases of human invasive breast cancer tissues indicated that up-regulation of survivin by EGFR signaling is not just a phenomenon seen in cancer cell lines in vitro. In fact, about 80% of the tumors were also found to be positive for survivin, and its expression level was correlated with co-expression of Her-2/neu and EGFR (11).
Our current study has further demonstrated that EGF stimulation induces survivin expression at both the mRNA and protein levels in breast cancer cells but not in normal mammary epithelial cells. It is possible that the much higher levels of EGFR seen in many breast cancer cell lines as compared with normal cells contribute in part to the difference in the response to EGF treatment. However, lower levels of EGFR may not be the only reason for these differences, since the MCF-7 cell line expresses a low level of EGFR but exhibits a similar effect as two other tumor cell lines expressing high levels of EGFR and/or Her-2/neu (34). The intrinsic properties of breast cancer cells may determine the responsiveness to activation of EGFR and up-regulation of survivin gene expression, since survivin is not detected in normal breast tissues, and cultured normal mammary epithelial cells either express a very low level or lack survivin gene expression (23).
It has been shown that the XIAP, a strong caspase inhibitor, prevents activation of caspase-3 by blocking caspase-9 activity as well as binding to activated caspase-3 (35). Interestingly, we found that EGFR activation did not affect the level of XIAP. Thus, up-regulated survivin expression in EGF-treated cells may play a key role in inhibiting caspase activity and creating resistance to apoptosis. Although the role of survivin in blocking caspase-9 activity has been elucidated, there is no structural evidence showing a direct interaction between survivin and caspase-3 (33). It is possible that survivin may interact with XIAP or other proteins, such as p21, to enhance the inhibitory effect on caspase-3 (36, 37).
A recent study also showed that treatment of human ovarian cells with paclitaxel transiently induced EGFR phosphorylation and PI3K activation, resulting in an increase in the level of survivin expression. Inhibition of either the EGFR or PI3K pathway was found to enhance the apoptotic cell death induced by paclitaxel (13). Up-regulation of survivin gene expression in human cancer cells after paclitaxel or docetaxel treatment has been reported by several laboratories, including ours (13, 14, 38). It was also demonstrated that treatment of human cancer cells by way of survivin siRNA down-regulated survivin expression and was able to sensitize cells to paclitaxel-induced cell death (38). Establishment of a clear link between EGFR signaling and survivin up-regulation in the apoptotic response to chemotherapy drugs will provide us with new information and a justifiable rationale for targeting this signaling pathway in the development of novel therapeutic approaches.
At present, how EGFR signaling leads to the expression of the survivin gene is still unclear. It has been shown that activation of the PI3K pathway by EGFR signaling leads to survivin up-regulation (12, 13). Results of our study also showed that inhibition of the PI3K pathway blocked EGF-induced survivin up-regulation in human breast cancer cell lines. However, the basal level of survivin expression was not affected even when the level of p-AKT was completely inhibited by a PI3K inhibitor, suggesting that the mechanism for constitutive expression of survivin may be different from EGF-induced survivin gene transcription. Although the role of PI3K/AKT in up-regulation of survivin expression has been established, we found that the effect of MAPK on survivin expression differs among breast cancer cell lines. Inhibition of the MAPK pathway blocked survivin gene up-regulation in SK-BR-3 but not in MCF-7 cells.
The PI3K/AKT pathway has been associated with important cellular pathways controlling cell proliferation and survival (39). To develop therapeutic approaches targeting EGFR signaling-induced apoptosis-resistant cancer cells, it is crucial to determine how activation of PI3K/AKT activity leads to survivin gene transcription. Previous studies have revealed another link between EGFR signaling and up-regulation of HIF-1_α_ protein synthesis mediated by activation of PI3K/AKT (19, 40). Our laboratory recently demonstrated that survivin promoter activity was up-regulated in hypoxic tumor cells (23). By further analysis of the survivin core promoter sequences, we have now identified a putative HRE consensus 5′-GCGTG-3′ region located in −81 to −85 nt of the 5′-flanking region of the survivin gene. Therefore, we believed it possible that transcriptional activation of survivin gene expression by EGF is mediated by HIF-1_α_. Our present study results demonstrated that the levels of both HIF-1_α_ and survivin are significantly increased in EGF-treated, normoxic tumor cells. We further determined that HIF-1_α_ is indeed a key transcription factor for EGFR signaling-activated survivin gene expression, since down-regulation of HIF-1_α_ using HIF-1_α_ siRNA significantly reduced the level of survivin expression in human cancer cells. Maintaining the level of HIF-1_α_-activated survivin seems to be very important for survival of normoxic tumor cells, since overexpression of HIF-1_α_ siRNA alone resulted in activation of caspase activity and apoptosis in about 70% of the breast cancer cells. In addition, HIF-1_α_ siRNA also markedly enhanced apoptotic cell death in docetaxel-treated tumor cells, even after EGF stimulation.
The results of our study have demonstrated, for the first time, that there is cross-talk between EGFR and HIF-1_α_ signaling pathways that can up-regulate survivin gene expression and increase resistance to apoptosis. First, we showed that overexpression of the HIF-1_α_ gene activates survivin promoter activity and the level of the protein. Using ChIP and modified McKay assays, we determined that there is a direct interaction between HIF-1_α_ and the survivin promoter, thus finding that the transcriptional activity of HIF-1_α_ is highly likely to activate survivin gene transcription. HIF-1_α_ has been previously defined as a hypoxia-inducible transcriptional factor, and its role in normoxic tumor cells remained largely unclear. Our results provide direct evidence that HIF-1_α_ is able to bind to HRE sites and activate HRE-mediated gene transcription under normoxic conditions in human tumor cells. In another study, we found that hypoxia-induced HIF-1_α_ up-regulation could also activate survivin gene expression using a similar mechanism.3 Currently, the precise mechanism for HIF-1_α_-mediated transcriptional activation of the survivin gene is under investigation in our laboratory.
In this study, we found that EGF stimulation does not induce survivin expression in normal mammary epithelial cells. Moreover, overexpression of the HIF-1_α_ gene activated survivin promoter activity and increased the level of survivin protein in breast cancer cell lines but not in normal cells, suggesting the presence of transcriptional inhibitory factor(s) in normal cells, preventing activation of the survivin promoter by HIF-1_α_. At present, we are conducting studies to identify the transcriptional factors involved in the tumor cell-specific activation of survivin gene expression.
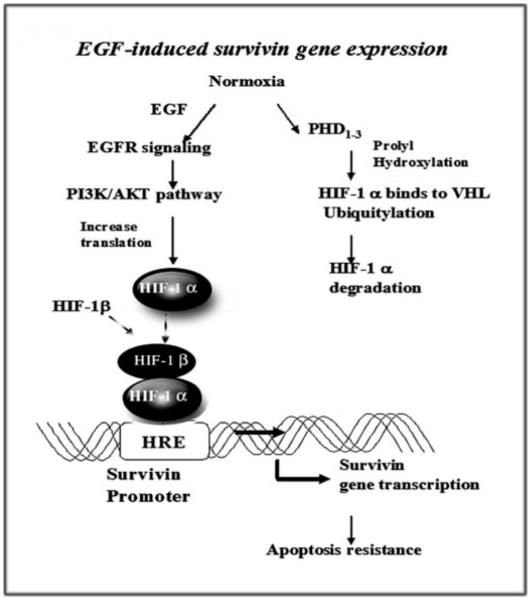
In conclusion, we have identified a novel pathway that mediates resistance to apoptosis in EGFR signal-activated human tumor cells. Our results have demonstrated that activation of the EGFR signaling pathway leads to the up-regulation of HIF-1_α_ through the PI3K/AKT pathway. We revealed that HIF-1_α_ directly binds to the survivin promoter to activate gene transcription, resulting in resistance to apoptosis in normoxic, EGFR signal-activated tumor cells (Fig. 8). Since activation of the EGFR signaling pathway and the presence of hypoxic tumor regions are commonly found in solid tumors, it is possible that coexistence of activation of those two conditions makes tumor cells highly resistant to apoptosis through HIF-1_α_-mediated up-regulation of survivin as well as other factors. Results from our study further high-light the importance of HIF-1_α_-mediated expression of survivin in tumor progression and resistance to therapy. Additionally, our results provide a strong rationale for the development of novel approaches targeting HIF-1_α_ for prevention of tumor progression as well as for cancer therapy, alone or in combination with other chemotherapeutic agents. Thus, the benefits from elucidation of this most likely common pathway that can function in both normoxia and hypoxia should eventually lead to much improved control and treatment of breast and other cancers.
FIGURE 8. Schematic illustration of a cross-talk between the EGFR and HIF-1_α_ signal pathways showing up-regulation of survivin gene expression and induction of resistance to apoptosis.
Activation of EGFR signaling increases HIF-1_α_ protein synthesis under normoxic conditions through the PI3K/AKT signal pathway. HIF-1_α_ then directly interacts with the survivin promoter, up-regulating the level of survivin gene expression, which results in resistance to apoptosis in tumor cells.
Acknowledgments
We thank Dr. Hyunsuk Shim for kindly providing the pBI-V6R stable MDA-MB-231 cell line, Dr. Fengzhi Li for the pluc cyc1.2 plasmid and survivin siRNA sequence, Dr. Bert Vogelstein for the AdEasy system, Dr. Erwin Van Meir for the pBI-V6R plasmid, and Drs. Hua Zhong and Ruoxiang Wang for the pHIF-1α plasmid. We thank Nicholyn Hutchinson and Kathleen Kite-Powell for manuscript editing.
Footnotes
*
This work was supported by NCI, National Institutes of Health Grants R29 CA 80017 and R01 CA95643, an Idea Award of the Department of Defense Breast Cancer Research Program, and the Avon Breast Cancer Research Foundation.
2
The abbreviations used are: EGFR, epidermal growth factor receptor; IAP, inhibitor of apoptosis; EGF, epidermal growth factor; HIF, hypoxia-inducible factor; Doc, docetaxel; HRE, hypoxia-responsive element; 7-AAD, 7-amino-actinomycin D; PI3K, phosphoinositide 3-kinase; siRNA, small interfering RNA; FBS, fetal bovine serum; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; PE, phycoerythrin; nt, nucleotide(s); MAPK, mitogen-activated protein kinase; VEGF, vascular endothelial growth factor.
3
X. Peng, P. Karna, Z. Cao, B. Jiang, M. Zhou, and L. Yang, unpublished results.
REFERENCES
- 1.Danielsen AJ, Maihle NJ. Growth Factors. 2002;20:1–15. doi: 10.1080/08977190290022185. [DOI] [PubMed] [Google Scholar]
- 2.Chrysogelos SA, Dickson RB. Breast Cancer Res. Treat. 1994;29:29–40. doi: 10.1007/BF00666179. [DOI] [PubMed] [Google Scholar]
- 3.Harris AL. Breast Cancer Res. Treat. 1994;29:1–2. doi: 10.1007/BF00666176. [DOI] [PubMed] [Google Scholar]
- 4.Bucci B, D’Agnano I, Botti C, Mottolese M, Carico E, Zupi G, Vecchione A. Anticancer Res. 1997;17:769–774. [PubMed] [Google Scholar]
- 5.Buchholz TA, Tu X, Ang KK, Esteva FJ, Kuerer HM, Pusztai L, Cristofanilli M, Singletary SE, Hortobagyi GN, Sahin AA. Cancer. 2005;104:676–681. doi: 10.1002/cncr.21217. [DOI] [PubMed] [Google Scholar]
- 6.Navolanic PM, Steelman LS, McCubrey JA. Int. J. Oncol. 2003;22:237–252. [PubMed] [Google Scholar]
- 7.Reis-Filho JS, Milanezi F, Carvalho S, Simpson PT, Steele D, Savage K, Lambros MB, Pereira EM, Nesland JM, Lakhani SR, Schmitt FC. Breast Cancer Res. 2005;7:1028–1035. doi: 10.1186/bcr1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baselga J. Ann. Oncol. 2002;13:8–9. doi: 10.1093/annonc/mdf092. [DOI] [PubMed] [Google Scholar]
- 9.Mendelsohn J, Baselga J. Oncogene. 2000;19:6550–6565. doi: 10.1038/sj.onc.1204082. [DOI] [PubMed] [Google Scholar]
- 10.Normanno N, De Luca A, Maiello MR, Mancino M, D’Antonio A, Macaluso M, Caponigro F, Giordano A. Front. Biosci. 2005;10:2611–2617. doi: 10.2741/1725. [DOI] [PubMed] [Google Scholar]
- 11.Asanuma H, Torigoe T, Kamiguchi K, Hirohashi Y, Ohmura T, Hirata K, Sato M, Sato N. Cancer Res. 2005;65:11018–11025. doi: 10.1158/0008-5472.CAN-05-0491. [DOI] [PubMed] [Google Scholar]
- 12.Wang Q, Greene MI. Exp. Mol. Pathol. 2005;79:100–107. doi: 10.1016/j.yexmp.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 13.Qiu L, Wang Q, Di W, Jiang Q, Schefeller E, Derby S, Wanebo H, Yan B, Wan Y. Int. J. Oncol. 2005;27:823–830. [PubMed] [Google Scholar]
- 14.Peng XH, Cao ZH, Xia JT, Carlson GW, Lewis MM, Wood WC, Yang L. Cancer Res. 2005;65:1909–1917. doi: 10.1158/0008-5472.CAN-04-3196. [DOI] [PubMed] [Google Scholar]
- 15.Altieri DC. Nat. Rev. Cancer. 2003;3:46–54. doi: 10.1038/nrc968. [DOI] [PubMed] [Google Scholar]
- 16.Ambrosini G, Adida C, Altieri DC. Nat. Med. 1997;3:917–921. doi: 10.1038/nm0897-917. [DOI] [PubMed] [Google Scholar]
- 17.Tanaka K, Iwamoto S, Gon G, Nohara T, Iwamoto M, Tanigawa N. Clin. Cancer Res. 2000;6:127–134. [PubMed] [Google Scholar]
- 18.Yang L, Cao Z, Yan H, Wood WC. Cancer Res. 2003;63:6815–6824. [PubMed] [Google Scholar]
- 19.Zhong H, Chiles K, Feldser D, Laughner E, Hanrahan C, Georgescu MM, Simons JW, Semenza GL. Cancer Res. 2000;60:1541–1545. [PubMed] [Google Scholar]
- 20.Phillips RJ, Mestas J, Gharaee-Kermani M, Burdick MD, Sica A, Belperio JA, Keane MP, Strieter RM. J. Biol. Chem. 2005;280:22473–22481. doi: 10.1074/jbc.M500963200. [DOI] [PubMed] [Google Scholar]
- 21.Laughner E, Taghavi P, Chiles K, Mahon PC, Semenza GL. Mol. Cell Biol. 2001;21:3995–4004. doi: 10.1128/MCB.21.12.3995-4004.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Semenza GL, Agani F, Feldser D, Iyer N, Kotch L, Laughner E, Yu A. Adv. Exp. Med. Biol. 2000;475:123–130. doi: 10.1007/0-306-46825-5_12. [DOI] [PubMed] [Google Scholar]
- 23.Yang L, Cao Z, Li F, Post DE, Van Meir EG, Zhong H, Wood WC. Gene Ther. 2004;11:1215–1223. doi: 10.1038/sj.gt.3302280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Post DE, Van Meir EG. Gene Ther. 2001;8:1801–1807. doi: 10.1038/sj.gt.3301605. [DOI] [PubMed] [Google Scholar]
- 25.He TC, Zhou S, da Costa LT, Yu J, Kinzler KW, Vogelstein B. Proc. Natl. Acad. Sci. U. S. A. 1998;95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Camirand A, Zakikhani M, Young F, Pollak M. Breast Cancer Res. 2005;7:570–579. doi: 10.1186/bcr1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deveraux QL, Roy N, Stennicke HR, Van Arsdale T, Zhou Q, Srinivasula SM, Alnemri ES, Salvesen GS, Reed JC. EMBO J. 1998;17:2215–2223. doi: 10.1093/emboj/17.8.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoffman WH, Biade S, Zilfou JT, Chen J, Murphy M. J. Biol. Chem. 2002;277:3247–3257. doi: 10.1074/jbc.M106643200. [DOI] [PubMed] [Google Scholar]
- 29.Abd El-Rehim DM, Pinder SE, Paish CE, Bell JA, Rampaul RS, Blamey RW, Robertson JF, Nicholson RI, Ellis IO. Br. J. Cancer. 2004;91:1532–1542. doi: 10.1038/sj.bjc.6602184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kothari S, Cizeau J, McMillan-Ward E, Israels SJ, Bailes M, Ens K, Kirshenbaum LA, Gibson SB. Oncogene. 2003;22:4734–4744. doi: 10.1038/sj.onc.1206666. [DOI] [PubMed] [Google Scholar]
- 31.Gibson S, Tu S, Oyer R, Anderson SM, Johnson GL. J. Biol. Chem. 1999;274:17612–17618. doi: 10.1074/jbc.274.25.17612. [DOI] [PubMed] [Google Scholar]
- 32.Tamm I, Wang Y, Sausville E, Scudiero DA, Vigna N, Oltersdorf T, Reed JC. Cancer Res. 1998;58:5315–5320. [PubMed] [Google Scholar]
- 33.Riedl SJ, Renatus M, Schwarzenbacher R, Zhou Q, Sun C, Fesik SW, Liddington RC, Salvesen GS. Cell. 2001;104:791–800. doi: 10.1016/s0092-8674(01)00274-4. [DOI] [PubMed] [Google Scholar]
- 34.Rae JM, Scheys JO, Clark KM, Chadwick RB, Kiefer MC, Lippman ME. Breast Cancer Res. Treat. 2004;87:87–95. doi: 10.1023/B:BREA.0000041585.26734.f9. [DOI] [PubMed] [Google Scholar]
- 35.Huang Y, Park YC, Rich RL, Segal D, Myszka DG, Wu H. Cell. 2001;104:781–790. [PubMed] [Google Scholar]
- 36.Dohi T, Okada K, Xia F, Wilford CE, Samuel T, Welsh K, Marusawa H, Zou H, Armstrong R, Matsuzawa S, Salvesen GS, Reed JC, Altieri DC. J. Biol. Chem. 2004;279:34087–34090. doi: 10.1074/jbc.C400236200. [DOI] [PubMed] [Google Scholar]
- 37.Suzuki A, Ito T, Kawano H, Hayashida M, Hayasaki Y, Tsutomi Y, Akahane K, Nakano T, Miura M, Shiraki K. Oncogene. 2000;19:1346–1353. doi: 10.1038/sj.onc.1203429. [DOI] [PubMed] [Google Scholar]
- 38.Ling X, Bernacki RJ, Brattain MG, Li F. J. Biol. Chem. 2004;279:15196–15203. doi: 10.1074/jbc.M310947200. [DOI] [PubMed] [Google Scholar]
- 39.Fry MJ. Breast Cancer Res. 2001;3:304–312. doi: 10.1186/bcr312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang BH, Jiang G, Zheng JZ, Lu Z, Hunter T, Vogt PK. Cell Growth Differ. 2001;12:363–369. [PubMed] [Google Scholar]