HIF1α–dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells (original) (raw)
HIF1α induction by mTOR represents a metabolic checkpoint for the differentiation of TH17 and Treg cells.
Abstract
Upon antigen stimulation, the bioenergetic demands of T cells increase dramatically over the resting state. Although a role for the metabolic switch to glycolysis has been suggested to support increased anabolic activities and facilitate T cell growth and proliferation, whether cellular metabolism controls T cell lineage choices remains poorly understood. We report that the glycolytic pathway is actively regulated during the differentiation of inflammatory TH17 and Foxp3-expressing regulatory T cells (Treg cells) and controls cell fate determination. TH17 but not Treg cell–inducing conditions resulted in strong up-regulation of the glycolytic activity and induction of glycolytic enzymes. Blocking glycolysis inhibited TH17 development while promoting Treg cell generation. Moreover, the transcription factor hypoxia-inducible factor 1α (HIF1α) was selectively expressed in TH17 cells and its induction required signaling through mTOR, a central regulator of cellular metabolism. HIF1α–dependent transcriptional program was important for mediating glycolytic activity, thereby contributing to the lineage choices between TH17 and Treg cells. Lack of HIF1α resulted in diminished TH17 development but enhanced Treg cell differentiation and protected mice from autoimmune neuroinflammation. Our studies demonstrate that HIF1α–dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells.
Upon antigen stimulation, naive T cells undergo extensive clonal expansion and differentiation for immune defense and regulation. A defining feature of T cell activation is the marked increase of the bioenergetic demands over the resting state. Activated T cells are highly anabolic and demonstrate a striking increase in glycolysis, as well as an increase in glucose and amino acid uptake (Fox et al., 2005; Jones and Thompson, 2007; Pearce, 2010). The reliance on glycolysis (even in the presence of high levels of oxygen) to generate ATP, which is far less efficient than oxidative phosphorylation, is an unusual metabolic aspect of proliferating T cells and cancer cells, the latter of which is known as the Warburg effect (Warburg, 1956). Fox et al. (2005) and Jones and Thompson (2007) have proposed that up-regulation of T cell metabolism is not merely a consequence of increased activation but rather a necessary step to facilitate activation. In support of this notion, proper regulation of glucose and sterol metabolism is required for the development of adaptive immune responses (Bensinger and Tontonoz, 2008; Bensinger et al., 2008; Cham et al., 2008). Conversely, anergic T cells fail to up-regulate the machinery necessary to support increased metabolism (Delgoffe et al., 2009; Zheng et al., 2009), whereas memory cell formation requires a lower metabolic activity (Araki et al., 2009; Pearce et al., 2009). Although a role for the metabolic pathways in T cell activation and responses is beginning to be appreciated, little information exists on their involvement in the differentiation of T cell functional subsets.
Discrete effector populations can develop from naive T cells to mediate specialized immune functions and are characterized by unique patterns of cytokine secretion. IFN-γ, IL-4, and IL-17 are the signature cytokines for TH1, TH2, and TH17 cells, respectively. In contrast, induced Foxp3+ regulatory T cells (Treg cells) act in synergy with natural Treg cells to promote immune tolerance and inhibit autoimmunity (Littman and Rudensky, 2010; Zhu et al., 2010). Induction of Treg cells in the peripheral immune compartment is closely related to the generation of TH17 cells, as the differentiation of both lineages is dependent on the pleiotropic cytokine TGF-β (Bettelli et al., 2006). Also, ROR-γt and Foxp3, the respective lineage-specific transcription factors for TH17 and Treg cells, are coexpressed in naive CD4 T cells exposed to TGF-β, but Foxp3 is dominant and antagonizes ROR-γt function unless IL-6 is present (Zhou et al., 2008). Thus, an inflammatory environment controls the balance between Treg and TH17 cell differentiation. The cytokines and environmental signals trigger a signaling cascade culminating in the transcriptional induction of lineage-specific cytokines and effector molecules. In particular, mTOR, a central regulator of cellular metabolism and protein translation, integrates various extracellular and intracellular signals to promote effector but not regulatory T cell differentiation (Delgoffe et al., 2009; Powell and Delgoffe, 2010). However, it remains unknown whether the basic metabolic machinery is actively regulated and contributes to T cell differentiation.
In this paper, we show that TH17 and Treg cells have marked differences in their glycolytic activity and expression of glycolytic enzymes. Combining pharmacological and genetic approaches, we found that glycolysis serves as a key metabolic checkpoint to direct the cell fate determination between TH17 and Treg cells. Specifically, the glucose analogue 2-deoxyglucose (2-DG), a prototypical inhibitor of the glycolytic pathway, dampened the development of T cells into TH17 cells while promoting Treg cell generation. In addition, deficiency in the transcription factor hypoxia-inducible factor 1α (HIF1α) in T cells diminished expression of the glycolytic molecules and altered the dichotomy between these two T cell lineages. Moreover, the HIF1α–dependent glycolytic pathway is downstream of mTOR signaling, thereby contributing to mTOR-mediated T cell differentiation. Our studies demonstrate that HIF1α–induced metabolic reprogramming orchestrates lineage differentiation of T cells.
RESULTS AND DISCUSSION
TH17-polarizing conditions induce HIF1α–dependent glycolytic activity
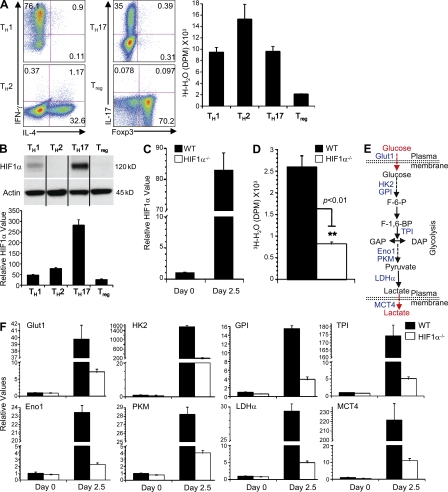
Although it has been well appreciated that T cell activation is accompanied by a metabolic switch to glycolysis (Fox et al., 2005; Jones and Thompson, 2007; Pearce, 2010), how glycolytic activity is regulated during T cell differentiation is poorly understood. We therefore measured glycolytic activity in various CD4 T cell lineages. To this end, we activated naive T cells under polarizing conditions in vitro and obtained TH1, TH2, TH17, and Treg cells with selective expression of IFN-γ, IL-4, IL-17, and Foxp3, respectively (Fig. 1 A). The glycolytic activity of differentiated cells was measured by the generation of 3H-labeled H2O from [3-3H]-glucose. In agreement with a very recent study (Michalek et al., 2011), TH1, TH2, and TH17 cells contained much higher glycolytic activity than Treg cells (Fig. 1 A), indicating strong up-regulation of glucose metabolism in effector T cells.
Figure 1.
TH17-inducing conditions elicit HIF1α–dependent up-regulation of glycolytic activity. (A) Sorted naive T cells (CD4+CD62LhiCD44loCD25−) from WT mice were differentiated under TH1, TH2, TH17, or Treg cell–inducing conditions for 5 d, followed by intracellular staining of IFN-γ and IL-4 (TH1/TH2) or IL-17 and Foxp3 (TH17/Treg; left). The glycolytic activity of these cells was measured by the generation of 3H-labeled H2O from [3-3H]-glucose (right). (B) HIF1α expression was examined by Western blot (top) and real-time PCR analyses (bottom) in T cells activated under various conditions for 2.5 d. HIF1α mRNA levels in naive cells were set to 1. For Western blotting, samples were from different parts of the same gel with the same exposure. Black lines indicate that intervening lanes were spliced out. (C) HIF1α mRNA expression was examined in WT and HIF1α−/− naive T cells and in cells differentiated under TH17 conditions for 2.5 d. (D) Naive T cells from WT and HIF1α−/− mice were differentiated under TH17 conditions, followed by measurement of glycolytic activity. (E) Diagram of the glycolytic pathway, with the molecules measured highlighted in blue. (F) RNA was isolated from naive T cells from WT and HIF1α−/− mice, or those differentiated under TH17 conditions for 2.5 d, and used for real-time PCR analyses of glycolytic molecules. Expression levels in WT naive T cells were set to 1. Data in Fig. 1 are representative of two to three independent experiments. Data represent the mean ± SD.
We next tested whether the differential activity of glycolysis between effector and regulatory T cells is transcriptionally regulated. A key transcription factor orchestrating the expression of glycolytic enzymes is HIF1, a heterodimer comprised of α (HIF1α) and β (HIF1β) subunits (Nizet and Johnson, 2009). Although HIF1 has been implicated in T cell responses (Neumann et al., 2005; Lukashev et al., 2006), its function and regulation in T cell differentiation remain undefined. To determine the intrinsic function of HIF1 in T cells, we first examined expression of HIF1α, the regulated component of the complex. Although HIF1α is known to be induced by hypoxic conditions through a posttranslational-dependent mechanism, HIF1α can be up-regulated by NF-κB signaling in activated innate immune cells under normoxia (Rius et al., 2008; Nizet and Johnson, 2009). HIF1α was strongly induced at the protein level in T cells undergoing TH17 differentiation, whereas the lowest levels were found in Treg cells. Moreover, mRNA levels of HIF1α were also highly induced in T cells undergoing TH17 differentiation (Fig. 1 B). These results identify a selective up-regulation of HIF1α under TH17-polarizing conditions.
To investigate the role of HIF1α in T cell differentiation, we crossed mice carrying a conditional HIF1α allele (HIF1αflox/flox; Ryan et al., 2000) with CD4-Cre transgenic mice to delete the floxed HIF1α allele specifically in T cells (called HIF1α–/– mice thereafter). Real-time PCR analyses indicated efficient deletion of the HIF1α gene in T cells (Fig. 1 C). WT and HIF1α−/− mice contained similar numbers and distribution of thymocytes and peripheral CD4 and CD8 T cells (Fig. S1 A). Also, development and homeostasis of natural Treg cells appeared to be largely undisturbed (Fig. S1 B). Cell proliferation and apoptosis in response to TCR stimulation were also comparable between WT and HIF1α−/− T cells (Fig. S1, C and D). To examine whether HIF1α controls glycolytic activity in TH17 cells, we purified naive T cells from WT and HIF1α−/− mice and differentiated them under TH17-polarizing conditions. Deficiency in HIF1α resulted in greatly reduced glycolytic activity (Fig. 1 D), indicating an important role for HIF1α to promote glycolysis in TH17 differentiation.
Glucose utilization depends on a chain of reactions catalyzed by multiple enzymes, eventually leading to the generation of lactate and net production of two ATP molecules as the energy source. In addition, glucose transport 1 (Glut1) and monocarboxylic acid transporter member 4 (MCT4) serve as plasma membrane transporters for glucose uptake and lactate export, respectively (Fig. 1 E). Real-time PCR analyses of WT cells undergoing TH17 differentiation revealed marked up-regulation of genes encoding various molecules involved in glycolysis (Fig. 1 F). These include the transporters Glut1 and MCT4 and glycolytic enzymes HK2 (hexokinase 2), GPI (glucose-6-phosphate isomerase), TPI (triosephosphate isomerase), Eno1 (enolase 1), PKM (pyruvate kinase muscle), and LDH-α (lactate dehydrogenase), thus encompassing the entire spectrum of the glycolytic pathway. In contrast, expression levels of these molecules were much lower in HIF1α−/− cells cultured under similar conditions (Fig. 1 F). Therefore, HIF1α establishes the glycolytic gene expression program in TH17 cells.
HIF1α is required for TH17 differentiation
Having demonstrated a role for HIF1α in mediating glycolysis in TH17 cells, we next explored the functional requirement of HIF1α in effector T cell differentiation. Naive T cells from WT and HIF1α−/− mice were differentiated under TH17 conditions, and expression of IL-17 was examined by intracellular staining. As compared with WT cells, HIF1α−/− cells exhibited a reduction of IL-17+ cells (Fig. 2 A). CFSE dilution assays revealed a similar degree of proliferation between WT and HIF1α−/− cells, indicating that the defective IL-17 production was largely independent of cell expansion (Fig. S2 A). In addition, impaired IL-17 production was observed in HIF1α−/− cells, regardless of the inclusion of IL-23 or blocking antibodies to potentiate TH17 differentiation (Fig. S2 B). Furthermore, upon TCR restimulation, effector TH17 cells lacking HIF1α secreted a lower amount of IL-17, but a comparable level of IL-2, relative to WT cells (Fig. 2 B). Real-time PCR analyses revealed decreased expression of the TH17 family cytokines IL-17, IL-17F, IL-21, and IL-22 (Fig. 2 C). Thus, HIF1α deficiency results in impairment in the TH17 differentiation program. In contrast, in vitro differentiation of TH1 and TH2 effector cells was largely independent of HIF1α (Fig. S2 C).
Figure 2.
HIF1α deficiency results in diminished TH17 differentiation. (A) Naive T cells from WT and HIF1α−/− mice were differentiated under TH17 conditions, followed by intracellular staining of IL-17. Data represent six independent experiments. (B) Effector TH17 cells, as described in A, were restimulated with 5 µg/ml of plate-bound anti-CD3) overnight, and supernatants were collected and subjected to Bioplex measurement of IL-17 and IL-2. (C) Effector TH17 cells, as described in A, were stimulated with anti-CD3/CD28 for 5 h for RNA and expression analyses. Data represent three independent experiments. (D and E) Naive T cells from WT and HIF1α−/− mice were differentiated under TH17 conditions for 2.5 d, and IL-23R (D) and Foxp3 (E) expression was examined by real-time PCR analyses. Data represent three independent experiments. (F and G) Naive T cells from WT and HIF1α−/− mice were differentiated under TH17 conditions for 2.5 d, and RNA was analyzed by microarrays to compare expression profiles between WT and HIF1α−/− cells (F) with selected genes in metabolism, transcription factors/signaling pathways, and surface/secreted molecules presented (G). Heat maps display gene expression relative to the WT mean on a log2 scale. (H) WT and HIF1α−/− mice were immunized with MOG/CFA and, 9 d later, DLN cells were stimulated with MOG for 5 d, followed by intracellular staining of IL-17. Data represent two independent experiments. (I–K) WT and HIF1α−/− mice were immunized with MOG/CFA and, 9 d later, DLN cells were expanded with MOG and IL-23 for 5 d, followed by adoptive transfer into C57BL/6 mice to induce TH17-polarized transfer EAE. Disease scores were recorded daily (I) and, 11–12 d after transfer, mice were euthanized for histological analysis of spinal cord inflammation (J) with pathology scores calculated (K). **, P < 0.01. Bars, 100 µm. Data represent two independent experiments. Data represent the mean ± SEM.
T cell differentiation is programmed by coordinated induction of transcription factors and cytokine receptors to facilitate the establishment of specific lineages (Littman and Rudensky, 2010; Zhu et al., 2010). To identify mechanisms of defective TH17 differentiation in HIF1α−/− cells, we examined the expression of transcription factors and cytokine receptors involved in TH17 differentiation. Expression of ROR-α, ROR-γt, AHR, IRF4, IL-6R, and IL-21R and the phosphorylation of Stat3 were comparable between differentiating WT and HIF1α−/− cells (unpublished data). In contrast, expression of IL-23R was significantly down-regulated (Fig. 2 D), whereas Foxp3 was up-regulated (Fig. 2 E), in HIF1α−/− cells under TH17-polarizing conditions. IL-23R is essential for terminal differentiation of TH17 cells (McGeachy et al., 2009), whereas Foxp3, the Treg cell master regulator, has been shown to antagonize ROR-γt function and TH17 differentiation (Zhou et al., 2008). Thus, dysregulated IL-23R and Foxp3 expression in HIF1α−/− cells likely contributes to their impaired TH17 differentiation.
To further understand how HIF1α regulates TH17 differentiation, we compared gene expression profiles of WT and HIF1α−/− cells differentiated toward TH17 cells using Affymetrix oligonucleotide arrays. A total of 581 probes showed equal or greater than twofold change (with false discovery rate <0.05) between WT and HIF1α−/− cells, including 194 up-regulated and 387 down-regulated entries upon HIF1α deficiency (Fig. 2 F and Table S1). We analyzed the gene list for enrichment of gene ontology and canonical pathways using the DAVID bioinformatics databases (Huang da et al., 2009). Remarkably, the 10 pathways with the most significant enrichment were all associated with glycolytic or related metabolic pathways (Fig. S3). Fig. 2 G showed the expression of a selective group of metabolic molecules, including the glycolytic enzymes described in Fig. 1 F, all of which were markedly down-regulated in HIF1α−/− cells. HIF1α−/− cells also showed altered expression of transcription factors, signaling molecules, and surface/secreted factors that have been implicated in T cell function and differentiation, including Gadd45β, Map3k1 (MEKK1), TLR2, Tnfrsf9 (4-1BB), and, as expected, IL-17. The canonical HIF1α target gene VEGF was also diminished in HIF1α−/− cells (Fig. 2 G). These findings reveal diverse HIF1α target genes in T cells that may collectively control T cell metabolism and differentiation.
To investigate the physiological significance of HIF1α in vivo, we immunized WT and HIF1α−/− mice with the myelin oligodendrocyte glycoprotein (MOG)35-55 peptide emulsified in CFA. After 9 d, we isolated draining LN (DLN) cells and stimulated them with the MOG peptide ex vivo. Tritiated thymidine incorporation assays revealed that antigen-specific T cells from WT and HIF1α−/− mice proliferated to a similar degree (unpublished data). However, IL-17 expression was reduced in T cells isolated from HIF1α−/− mice (Fig. 2 H). Moreover, we expanded the DLN cells with MOG and IL-23 and transferred the resulting cells into C57BL/6 mice. In this TH17-polarized transfer model of EAE (Kang et al., 2010), disease development was significantly delayed in mice receiving the HIF1α−/− cells comparing those receiving the WT cells (Fig. 2 I). Histological analyses showed less prominent leukocyte infiltration and inflammation in the spinal cords of mice transferred with HIF1α−/− cells (Fig. 2, J and K). Therefore, HIF1α is required for TH17 differentiation in vivo and contributes to TH17-dependent CNS inflammation.
The balance of TH17 and Treg cell differentiation depends on HIF1α
An abnormal up-regulation of Foxp3 mRNA expression in HIF1α−/− cells during TH17 differentiation (Fig. 2 E) prompted us to test whether HIF1α controls the balance between the closely regulated TH17 and Foxp3+ Treg cell lineages. Deficiency of HIF1α considerably increased the Foxp3+ population when naive T cells were activated in the presence of TGF-β, with more prominent changes observed under low nonsaturating TGF-β concentrations (Fig. 3 A). Consistent with these findings, mRNA expression of Foxp3, as well as Treg cell–associated surface molecules such as Ctla4 and Gpr83, was up-regulated in HIF1α−/− cells (Fig. 3 B). Furthermore, both in an in vitro Treg cell differentiation system mediated by splenic DCs (without exogenous cytokines) and in an ex vivo recall response after MOG immunization, HIF1α−/− T cells exhibited higher Foxp3 induction (Fig. 3, C and D). Finally, HIF1α−/− cells showed a reduction of IL-17 expression and a reciprocal increase in Foxp3 levels under TH17-polarizing conditions with titrated amounts of IL-6 (Fig. 3 E). This is in agreement with the notion that the dichotomy of TH17 and Treg cell is dependent on an inflammatory environment, especially the concentration of IL-6 (Bettelli et al., 2006). Thus, HIF1α is required to promote TH17 but to inhibit Treg cell differentiation.
Figure 3.
HIF1α deficiency promotes Treg cell differentiation and alters TH17/Treg cell balance. (A) Naive T cells from WT and HIF1α−/− mice were activated in the presence of 1 or 10 ng/ml TGF-β for 5 d, followed by Foxp3 staining. Data represent four independent experiments. (B) Naive T cells from WT and HIF1α−/− mice were activated in the presence of IL-2 and varying amount of TGF-β for 5 d, followed by real-time PCR analyses of Foxp3, Ctla4, and Gpr83 expression. (C) Naive T cells from WT and HIF1α−/− mice were stimulated with splenic DCs and anti-CD3 for 5 d, and Foxp3 mRNA was analyzed. (D) WT and HIF1α−/− mice were immunized with MOG and, 9 d later, DLN cells were stimulated with MOG for 5 d, followed by Foxp3 staining. (E) Naive T cells from WT and HIF1α−/− mice were differentiated under TH17 conditions with varying doses of IL-6 for 5 d, followed by intracellular staining of IL-17 and Foxp3. Data represent four independent experiments. (F) WT naive T cells were differentiated under TH17 or Treg cell–inducing conditions, followed by real-time PCR analyses of glycolytic molecules. Expression levels in naive T cells were set to 1. Data represent two independent experiments. Data represent the mean ± SEM (B and C) or the mean ± SD (F).
Blocking glycolysis reciprocally alters TH17 and Treg cell differentiation
Having established an essential role for HIF1α in controlling the balance between TH17 and Treg cell differentiation, we next asked whether HIF1α–dependent metabolic reprogramming is instrumental to such cell fate decision. Consistent with the selectively high glycolytic activity and HIF1α expression in TH17 relative to Treg cells (Fig. 1, A and B), real-time PCR analyses revealed that the expression of metabolic genes involved in glycolysis was much higher in differentiating TH17 cells than in Treg cells (Fig. 3 F).
To directly test the importance of the metabolic reprogramming in T cell differentiation, we cultured naive T cells under TH17-polarizing conditions in the presence or absence of 2-DG, a prototypical inhibitor of the glycolytic pathway via blocking hexokinase, the first rate-limiting enzyme of glycolysis. As expected, 2-DG treatment of T cells reduced the glycolytic activity (Fig. 4 A). Importantly, treatment of T cells with 2-DG resulted in diminished production of IL-17 (Fig. 4 B). Although high concentrations of 2-DG (10–50 mM) have been shown to inhibit T cell proliferation (Cham et al., 2008), at the lower dose of 2-DG used in this study (1 mM), we observed minimal inhibitory effects of 2-DG on cell proliferation, as indicated by the indistinguishable CSFE dilution between control and 2-DG–treated samples (Fig. 4 B). As a comparison, we treated T cells with the mTOR inhibitor rapamycin, which blocks mTOR-dependent metabolic pathways including glycolysis (Düvel et al., 2010; Powell and Delgoffe, 2010). Similar to the effects of 2-DG, treatment of T cells with rapamycin inhibited the glycolytic activity and the production of IL-17 (Fig. 4, A and B). Moreover, mRNA expression levels of TH17-associated cytokines, including IL-17, IL-17F, IL-21, and IL-22, were all inhibited by 2-DG and rapamycin treatments (Fig. 4 C). Notably, because the cells were treated with the pharmacological inhibitors during their differentiation but not at the time of acute restimulation of effector TH17 cells, the diminished production of these TH17 signature cytokines reflected a specific effect of glycolytic inhibition on the differentiation.
Figure 4.
Blocking glycolysis reciprocally reduces TH17 but promotes Treg cell differentiation. (A) Naive T cells were differentiated under TH17 conditions for 2.5 d in the presence of vehicle (Ctrl), 1 mM 2-DG, or 50 nM rapamycin (Rapa). Glycolytic activity was measured as described in Fig. 1 A. Data represent two independent experiments. (B) Naive T cells were labeled with CFSE and differentiated under TH17 conditions in the presence of vehicle, 1 mM 2-DG, or 50 nM rapamycin, followed by intracellular staining of IL-17. Data represent six independent experiments. (C) Differentiated TH17 cells, as described in B, were stimulated with anti-CD3 for 5 h and used for real-time PCR analyses of cytokine gene expression. Data represent three independent experiments. (D) Naive OT-II T cells (Thy1.1+) cells were transferred into C57BL/6 mice and immunized with OVA, with daily treatment of 2-DG or vehicle controls. DLN cells were analyzed 6 d later for the proportion of the IL-17+ population among donor T cells. Data represent two independent experiments. (E) Naive T cells were treated with vehicle, 2-DG, or rapamycin and activated in the presence of 1 ng/ml TGF-β for 5 d, followed by Foxp3 staining. Data represent four independent experiments. (F) Naive T cells were treated with vehicle, 2-DG, or rapamycin and differentiated under TH17 conditions, except for the lower dose of IL-6 used (0.2 ng/ml), followed by intracellular staining of IL-17 and Foxp3. Data represent four independent experiments. (G–I) WT mice were immunized with MOG and, 9 d later, DLN cells were expanded with MOG and IL-23 in the presence or absence of 2-DG for 5 d. An aliquot of cells were analyzed by intracellular staining for Foxp3 and IL-17 expression (G), with the remaining cells transferred into C57BL/6 mice for the induction of TH17-polarized transfer EAE (H). Spinal cord was analyzed for pathology scores at day 12 after transfer (I). *, P < 0.05; **, P < 0.01. Data represent two independent experiments. Data represent the mean ± SD (A) or the mean ± SEM (C, D, H, and I).
To test the effects of 2-DG on the development of TH17 cells in vivo, we transferred naive T cells (CD62LhiCD44lo) from OT-II TCR-transgenic mice (specific for OVA323-339; Thy1.1+) into C57BL/6 mice and immunized them with antigen, followed by daily 2-DG treatment in vivo. At day 6 after immunization, we isolated DLN cells and measured IL-17 production in donor-derived cells by intracellular cytokine staining. Treatment of 2-DG resulted in significantly reduced IL-17–expressing cells (Fig. 4 D). Moreover, mRNA expression and secretion of IL-17 were considerably reduced after 2-DG treatment (Fig. S4, A and B). We therefore conclude that TH17 differentiation depends on the glycolytic activity.
We next examined the effects of 2-DG and rapamycin on Treg cell generation. Rapamycin has been shown to potentiate Treg cell differentiation (Haxhinasto et al., 2008; Sauer et al., 2008; Cobbold et al., 2009). Similar to the effect of rapamycin, 2-DG also promoted Treg cell generation (Fig. 4 E). Furthermore, under conditions that allowed simultaneous generation of TH17 and Treg cells, 2-DG and rapamycin diminished IL-17 production but promoted Foxp3 induction (Fig. 4 F). Collectively, these results indicate that inhibition of glycolysis blocks TH17 development while promoting Treg cell generation.
To evaluate the contribution of glycolysis to autoimmune inflammation in vivo, we used the TH17-polarized transfer model of EAE, with 2-DG added during the in vitro expansion of DLN cells. Such treatment resulted in reduced IL-17+ cells and simultaneous increase of Foxp3+ Treg cells (Fig. 4 G). When these cells were transferred into C57BL/6 recipients, 2-DG–treated cells exhibited markedly reduced ability to cause EAE (Fig. 4 H). This was associated with diminished leukocyte infiltration and spinal cord inflammation, as revealed by histological analyses (Fig. 4, H and I; and Fig. S4 C). Therefore, blocking glycolysis alters TH17 and Treg cell differentiation and protects mice from EAE.
mTOR is required for HIF1α induction in TH17 cell differentiation
A key role for HIF1α to direct reciprocal TH17 and Treg cell differentiation prompted us to identify upstream molecular pathways responsible for HIF1α induction in differentiating TH17 cells (Fig. 1 B). Because cell fate decision between TH17 and Treg cells is critically dependent on IL-6, we analyzed HIF1α expression in T cells stimulated with varying amount of IL-6. HIF1α was induced by IL-6 in a dose-dependent manner (Fig. 5 A), indicating an important role for IL-6 signaling in HIF1α induction.
Figure 5.
mTOR activity is required for HIF1α induction in TH17-differentiating cells. (A) WT naive T cells were activated by anti-CD3/CD28 in the presence of various concentrations of TGF-β and IL-6 for 2.5 d, and HIF1α expression was analyzed by real-time PCR (levels in activated cells without TGF-β/IL-6 were set to 1). (B) Naive T cells were treated with vehicle (Ctrl) or 50 nM rapamycin (Rapa) and differentiated under TH17 conditions. HIF1α expression was examined by real-time PCR analyses (levels in naive cells were set to 1). (C) Naive T cells were differentiated as described in B, and expression of glycolytic molecules was examined by real-time PCR analyses. Data in Fig. 5 are representative of at least two independent experiments. Data represent the mean ± SEM (A) or the mean ± SD (B and C). (D) HIF1α–induced metabolic reprogramming in T cell differentiation.
Recent experiments have shown that HIF1α is responsible for the glycolytic response downstream of mTOR (Düvel et al., 2010). As rapamycin treatment was similar to HIF1α deficiency at altering TH17 and Treg cell differentiation, we hypothesized that rapamycin-sensitive mTOR signaling is important for HIF1α induction in T cells. To test this, we treated T cells with rapamycin and examined HIF1α expression. Blocking mTOR activity with rapamycin in differentiating TH17 cells markedly inhibited the induction of HIF1α (Fig. 5 B). Consistent with these findings, the induction of transporters Glut1 and MCT4, as well as various glycolytic enzymes, was substantially diminished by rapamycin treatment (Fig. 5 C). In contrast, 2-DG treatment, at the dose that was sufficient to alter TH17 and Treg cell differentiation, had a minimal effect on the RNA levels of HIF1α and the metabolic molecules (Fig. S5, A and B). This result confirmed a role for 2-DG to directly inhibit glycolytic enzyme activity and excluded a potential indirect effect of 2-DG on regulating mTOR activity. Consistent with a high glycolytic activity in TH1 and TH2 lineages (Fig. 1 A), 2-DG treatment was also capable of diminishing their differentiation (Fig. S5 C). Although mTOR is required for the generation of TH1 and TH2 cells (Delgoffe et al., 2009), HIF1α was not strongly induced under TH1- and TH2-polarizing conditions (Fig. 1 B) or involved in their functional differentiation (Fig. S2 C). Collectively, HIF1α is selectively expressed in TH17 cells and its induction depends on IL-6 signaling and mTOR activity, whereas the glycolytic pathway contributes to TH1 and TH2 differentiation in a HIF1α–independent manner.
Collectively, our studies demonstrate that an elevated glycolytic activity is not only associated with TH17 cell differentiation but that the glycolytic machinery is actively regulated to direct the differentiation of TH17 and Treg cells. This process is programmed by the transcription factor HIF1α through stimulating a glycolytic gene expression program. Therefore, the HIF1α–dependent glycolytic pathway serves as a metabolic checkpoint to distinguish cell fate decision between these two closely related T cell lineages. We provide further evidence that mTOR signaling is essential to induce HIF1α expression in TH17 cells, suggesting that the HIF1α–dependent glycolytic pathway is an integral component downstream of mTOR to mediate TH17 and Treg cell differentiation (Fig. 5 D; Delgoffe et al., 2009; Powell and Delgoffe, 2010). In contrast, deficiency of HIF1α did not substantially alter TH1 and TH2 differentiation, indicating a HIF1α–independent glycolytic pathway for the generation of these effector lineages. Interestingly, depletion of amino acids with the small molecule halofuginone has recently been shown to selectively inhibit TH17 cell differentiation (Sundrud et al., 2009). Therefore, TH17 cell differentiation has selective bioenergetic requirements for glucose and amino acid metabolism. Conversely, generation of antigen-specific Treg cells is enhanced by amino acid depletion (Cobbold et al., 2009) or inhibition of glucose metabolism (this study). Recently added evidence further suggests that Treg cells preferentially use fatty acids and inhibition of fatty acid β-oxidation impairs Treg cell differentiation (Michalek et al., 2011). Notably, although overexpression of HIF1α has been shown to enhance Foxp3 expression (Ben-Shoshan et al., 2008), our results with HIF1α–deficient and 2-DG–treated T cells have clearly established a negative role for HIF1α–induced metabolic reprogramming in Treg cell development. To our knowledge, these studies identify the first evidence that the glycolytic pathway and helper T cell differentiation are linked by the HIF1α–mediated gene expression program. Given the evolutionarily conserved nature of the hypoxic response and glycolytic pathway, we propose that the adaptive immune system has evolved to adopt these basic metabolic machineries to regulate cell fate decision of helper T cells. Furthermore, the selective dependence of TH17 differentiation on HIF1α–mediated metabolic reprogramming has provided novel targets for therapeutic intervention of autoimmune and inflammatory diseases elicited by TH17 cells.
MATERIALS AND METHODS
Mice.
HIF1αflox/flox and OT-II TCR-transgenic mice on the C57BL/6 background were purchased from The Jackson Laboratory (Ryan et al., 2000). Mice at 7–12 wk of age were used. All mice were kept in specific pathogen-free conditions within the Animal Resource Center at St. Jude Children’s Research Hospital. Animal protocols were approved by the Institutional Animal Care and Use Committee of St. Jude Children’s Research Hospital.
Flow cytometry.
For analysis of surface markers, cells were stained in PBS containing 2% (wt/vol) BSA and the appropriate antibodies from eBioscience. Foxp3 staining was performed according to the manufacturer’s instructions (eBioscience). For intracellular cytokine staining, T cells were stimulated for 4–5 h with phorbol 12-myristate 13-acetate (PMA) and ionomycin in the presence of monensin before being stained according to the manufacturer’s instructions (BD). Simultaneous Foxp3 and IL-17 staining was performed using Foxp3 staining procedure after 4 h of stimulation with PMA and ionomycin. Flow cytometry data were acquired on an upgraded five-color FACScan or multi-color LSRII (BD) and were analyzed with FlowJo software (Tree Star).
Cell purification and culture.
Lymphocytes were isolated from spleen and peripheral LNs and naive T cells were sorted on a MoFlow (Beckman Coulter) or Reflection (i-Cyt), as described previously (Liu et al., 2010). Sorted naive T cells (CD4+CD62LhiCD44loCD25−) were used for in vitro culture in Click’s medium (plus β-mercaptoethanol) supplemented with 10% (vol/vol) FBS and antibiotics. T cells were activated with 2 µg/ml anti-CD3 (2C11; Bio X Cell), 2 µg/ml anti-CD28 (37.51; Bio X Cell), and 100 U/ml human IL-2. For Treg cell differentiation, cultures were supplemented with 1 or 10 ng/ml human TGF-β1. For TH1 conditions, 3.5 ng/ml IL-12 and 10 µg/ml anti–IL-4 (11B11; Bio X Cell) were added. For TH2 conditions, 10 ng/ml IL-4 and 10 µg/ml anti–IFN-γ were added. For TH17 conditions, 10 µg/ml anti–IL-2, 10 µg/ml anti–IL-4, 10 µg/ml anti–IFN-γ, 2 ng/ml TGF-β1, 20 ng/ml IL-23, and 20, 2, or 0.2 ng/ml IL-6 were included in cultures, and for certain experiments, only TGF-β1 and IL-6 were used, as indicated. For DC–T cell co-culture, 5 × 104 CD11c+ DCs purified from spleens and 2.5 × 105 naive T cells were cultured in the presence of anti-CD3 without exogenous cytokines. For pharmacological inhibitor treatments, cells were incubated with vehicle, 0.5–1 mM 2-DG (Sigma-Aldrich), or 10–50 nM rapamycin (EMD) at the onset of cultures.
RNA and immunoblot analysis.
RNA was extracted with an RNeasy kit (QIAGEN), and cDNA was synthesized with Superscript III/II reverse transcription (Invitrogen). Real-time PCR was performed as described previously (Liu et al., 2009), and the expression of each target gene is presented as the fold change relative to the expression of WT control samples. Primer sequences used to determine expression of glycolytic molecules were: HIF1α (forward, 5′-AGCTTCTGTTATGAGGCTCACC-3′; reverse, 5′-TGACTTGATGTTCATCGTCCTC-3′), Glut1/Slc2a1 (forward, 5′-CAGTTCGGCTATAACACTGGTG-3′; reverse, 5′-GCCCCCGACAGAGAAGATG-3′), HK2 (forward, 5′-TGATCGCCTGCTTATTCACGG-3′; reverse, 5′-AACCGCCTAGAAATCTCCAGA-3′), LDH-α (forward, 5′-CATTGTCAAGTACAGTCCACACT-3′; reverse, 5′-TTCCAATTACTCGGTTTTTGGGA-3′), PKM (forward, 5′-GCCGCCTGGACATTGACTC-3′; reverse, 5′-CCATGAGAGAAATTCAGCCGAG-3′), MCT4 (forward, 5′-TCACGGGTTTCTCCTACGC-3′; reverse, 5′-GCCAAAGCGGTTCACACAC-3′), GPI (forward, 5′-TCAAGCTGCGCGAACTTTTTG-3′; reverse, 5′-GGTTCTTGGAGTAGTCCACCAG-3′), TPI (forward, 5′-CCAGGAAGTTCTTCGTTGGGG-3′; reverse, 5′-CAAAGTCGATGTAAGCGGTGG-3′), and Eno1 (forward, 5′-TGCGTCCACTGGCATCTAC-3′; reverse, 5′-CAGAGCAGGCGCAATAGTTTTA-3′). Other molecules were examined using primer and probe sets purchased from Applied Biosystems. Immunoblot analysis was performed as described previously (Liu et al., 2010) using polyclonal anti–HIF1α antibody (Cayman Chemicals) and anti–β-actin (Sigma-Aldrich).
In vivo antigen-specific TH17 differentiation.
C57BL/6 mice were injected i.p. with 2-DG (2 g/kg body weight) or solvent alone (PBS). After 6 h, approximately one million naive T cells (CD4+CD62LhiCD44loCD25–) from OT-II TCR-transgenic mice (Thy1.1+) were injected into these mice via tail vein. After 24 h, they were immunized s.c. with 100 µg OVA323-339 in the presence of incomplete Freund’s adjuvant (Difco). Mice were sacrificed at day 6 after immunization, and DLNs were isolated and either stimulated with the cognate peptide for 2–3 d for RNA assay or secreted cytokine measurement or pulsed with PMA and ionomycin for 5 h for intracellular staining for donor-derived T cells (Thy1.1+). 2-DG/PBS was given daily until the day before the mice were euthanized.
MOG immunization and TH17-polarized transfer EAE.
Mice were immunized with 100 µg MOG peptide in CFA (Difco) with 500 µg Mycobacterium tuberculosis. After 9 d, DLNs were prepared and cultured for 5 d with MOG35-55 and IL-23 and, where applicable, 1 mM 2-DG was included. Live cells were harvested at the end of culture, and 2 × 107 cells were transferred into mice that were irradiated sublethally (500 Rad) 4 h before transfer, supplemented by i.p. injections of 200 ng pertussis toxin on the day of transfer and 2 d later, as previously described (Kang et al., 2010). The mice were observed daily for clinical signs and scored as described previously (Chi and Flavell, 2005).
Histopathology and immunohistochemistry.
Mice were euthanized on day 11–12 after transfer, and spinal cords were fixed by immersion with 10% neutral buffered formalin solution and decalcified. Fixed tissues were embedded in paraffin, sectioned, and stained with H&E, with serial histological sections stained immunohistochemically to determine the distribution and types of inflammatory cells in the brain and spinal cord. For immunohistochemical detection of T cells, goat polyclonal primary antibodies against CD3 (Santa Cruz Biotechnology, Inc.) was used at a 1:350 on tissue sections subjected to antigen retrieval for 30 min at 98°C (Target Retrieval, pH 6; Dako). Similarly, to detect macrophages and other myeloid cells in the CNS, a rat monoclonal primary antibody against Mac2 (Accurate Chemical) was used at a 1:20,000 on tissue sections subjected to antigen retrieval for 30 min at 98°C (Citrate buffer, pH 6; Invitrogen). Spinal cord pathology was assigned scores by an experienced pathologist (P. Vogel) as follows: spinal column was divided into cervical (two to three sections), thoracic (three to five sections), and lumbar (three to five sections) and was scored for inflammatory and degenerative lesions (mononuclear perivascular cuffing, gliosis, vacuolation caused by swollen axon sheaths, and polymorphonuclear infiltration) according to the predetermined qualitative and semiquantitative criteria of 0 = absence of lesions, 1 = minimal to mild inconspicuous lesions, 2 = conspicuous lesions, 3 = prominent multifocal lesions, and 4 = marked coalescing lesions. Sections from animals in different groups were scored in staggered order to prevent score bias.
Gene expression profiling by microarray analysis.
RNA samples in biological triplicates were analyzed using the GeneTitan peg array (HT_MG-430_PM; Affymetrix) and expression signals were summarized using the RMA algorithm (Expression Console version 1.1; Affymetrix). Differentially expressed transcripts were identified by ANOVA (Genomics Suite version 6.5; Partek), and the Benjamini-Hochberg method was used to estimate the false discovery rate (Benjamini and Hochberg, 1995). Gene lists were analyzed for enrichment of Gene Ontology and canonical pathways using the DAVID bioinformatics databases (Huang da et al., 2009). The entire microarray data was deposited into the GEO series database under accession number GSE29765.
Bioplex assay.
Differentiated effector cells were restimulated with anti-CD3 overnight and supernatants were harvested. Cytokine levels in the supernatants were measured using MILLIPLEX kits for mouse cytokine/chemokine according to the manufacturer’s instruction (Millipore).
Glycolysis flux assay.
Glycolysis of naive or effector cells was determined by measuring the detritiation of [3-3H]-glucose (Hue et al., 1984). In brief, the assay was initiated by adding 1 µCi [3-3H]-glucose (PerkinElmer) and, 2 h later, medium was transferred to microcentrifuge tubes containing 50 µl 5 N HCL. The microcentrifuge tubes were then placed in 20-ml scintillation vials containing 0.5 ml water and the vials capped and sealed. 3H2O was separated from un-metabolized [3-3H]-glucose by evaporation diffusion for 24 h at room temperature. Cell-free sample containing 1 µCi 3H-glucose was included as a background control.
Statistical analysis.
P-values were calculated with Student’s t test. P-values <0.05 were considered significant.
Online supplemental material.
Fig. S1 shows the normal development, proliferation, and apoptosis of T cells in HIF1α−/− mice. Fig. S2 shows the effects of HIF1α deficiency on effector T cell differentiation. Fig. S3 shows the enrichment of metabolic pathways in HIF1α gene targets. Fig. S4 shows the in vivo effects of 2-DG on IL-17 expression and EAE pathogenesis. Fig. S5 shows the effects of 2-DG on metabolic gene expression and TH1/TH2 differentiation. Table S1 lists the 581 probes that were differentially expressed between WT and HIF1α−/− TH17 cells. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20110278/DC1.
Acknowledgments
We thank Caryn Cloer, Christopher Dillion, Laura McCormick, and Sharad Shrestha for help with animal colony management and other support. We thank members of the St. Jude Immunology FACS core facility and the St. Jude Hartwell Center for their technical support.
This work is supported by US National Institutes of Health grants NS064599 and AR053573 (H. Chi) and AI47891, AI44828, AI40646, and GM52735 (D.R. Green), the George J. Mitchell fellowship from St. Jude (R. Wang), and the American Lebanese Syrian Associated Charities.
We declare no competing financial interests.
Footnotes
Abbreviations used:
2-DG
2-deoxyglucose
DLN
draining LN
Glut1
glucose transport 1
HIF1α
hypoxia-inducible factor 1α
MCT4
monocarboxylic acid transporter member 4
MOG
myelin oligodendrocyte glycoprotein
PMA
phorbol 12-myristate 13-acetate
Treg cell
regulatory T cell
References
- Araki K., Turner A.P., Shaffer V.O., Gangappa S., Keller S.A., Bachmann M.F., Larsen C.P., Ahmed R. 2009. mTOR regulates memory CD8 T-cell differentiation. Nature. 460:108–112 10.1038/nature08155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shoshan J., Maysel-Auslender S., Mor A., Keren G., George J. 2008. Hypoxia controls CD4+CD25+ regulatory T-cell homeostasis via hypoxia-inducible factor-1alpha. Eur. J. Immunol. 38:2412–2418 10.1002/eji.200838318 [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J.R. Stat. Soc. Series B Stat Methodol. 57:289–300 [Google Scholar]
- Bensinger S.J., Tontonoz P. 2008. Integration of metabolism and inflammation by lipid-activated nuclear receptors. Nature. 454:470–477 10.1038/nature07202 [DOI] [PubMed] [Google Scholar]
- Bensinger S.J., Bradley M.N., Joseph S.B., Zelcer N., Janssen E.M., Hausner M.A., Shih R., Parks J.S., Edwards P.A., Jamieson B.D., Tontonoz P. 2008. LXR signaling couples sterol metabolism to proliferation in the acquired immune response. Cell. 134:97–111 10.1016/j.cell.2008.04.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettelli E., Carrier Y., Gao W., Korn T., Strom T.B., Oukka M., Weiner H.L., Kuchroo V.K. 2006. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 441:235–238 10.1038/nature04753 [DOI] [PubMed] [Google Scholar]
- Cham C.M., Driessens G., O’Keefe J.P., Gajewski T.F. 2008. Glucose deprivation inhibits multiple key gene expression events and effector functions in CD8+ T cells. Eur. J. Immunol. 38:2438–2450 10.1002/eji.200838289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi H., Flavell R.A. 2005. Cutting edge: regulation of T cell trafficking and primary immune responses by sphingosine 1-phosphate receptor 1. J. Immunol. 174:2485–2488 [DOI] [PubMed] [Google Scholar]
- Cobbold S.P., Adams E., Farquhar C.A., Nolan K.F., Howie D., Lui K.O., Fairchild P.J., Mellor A.L., Ron D., Waldmann H. 2009. Infectious tolerance via the consumption of essential amino acids and mTOR signaling. Proc. Natl. Acad. Sci. USA. 106:12055–12060 10.1073/pnas.0903919106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgoffe G.M., Kole T.P., Zheng Y., Zarek P.E., Matthews K.L., Xiao B., Worley P.F., Kozma S.C., Powell J.D. 2009. The mTOR kinase differentially regulates effector and regulatory T cell lineage commitment. Immunity. 30:832–844 10.1016/j.immuni.2009.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Düvel K., Yecies J.L., Menon S., Raman P., Lipovsky A.I., Souza A.L., Triantafellow E., Ma Q., Gorski R., Cleaver S., et al. 2010. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol. Cell. 39:171–183 10.1016/j.molcel.2010.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox C.J., Hammerman P.S., Thompson C.B. 2005. Fuel feeds function: energy metabolism and the T-cell response. Nat. Rev. Immunol. 5:844–852 10.1038/nri1710 [DOI] [PubMed] [Google Scholar]
- Haxhinasto S., Mathis D., Benoist C. 2008. The AKT–mTOR axis regulates de novo differentiation of CD4+Foxp3+ cells. J. Exp. Med. 205:565–574 10.1084/jem.20071477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W., Sherman B.T., Lempicki R.A. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4:44–57 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Hue L., Sobrino F., Bosca L. 1984. Difference in glucose sensitivity of liver glycolysis and glycogen synthesis. Relationship between lactate production and fructose 2,6-bisphosphate concentration. Biochem. J. 224:779–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones R.G., Thompson C.B. 2007. Revving the engine: signal transduction fuels T cell activation. Immunity. 27:173–178 10.1016/j.immuni.2007.07.008 [DOI] [PubMed] [Google Scholar]
- Kang Z., Altuntas C.Z., Gulen M.F., Liu C., Giltiay N., Qin H., Liu L., Qian W., Ransohoff R.M., Bergmann C., et al. 2010. Astrocyte-restricted ablation of interleukin-17-induced Act1-mediated signaling ameliorates autoimmune encephalomyelitis. Immunity. 32:414–425 10.1016/j.immuni.2010.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littman D.R., Rudensky A.Y. 2010. Th17 and regulatory T cells in mediating and restraining inflammation. Cell. 140:845–858 10.1016/j.cell.2010.02.021 [DOI] [PubMed] [Google Scholar]
- Liu G., Burns S., Huang G., Boyd K., Proia R.L., Flavell R.A., Chi H. 2009. The receptor S1P1 overrides regulatory T cell-mediated immune suppression through Akt-mTOR. Nat. Immunol. 10:769–777 10.1038/ni.1743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G., Yang K., Burns S., Shrestha S., Chi H. 2010. The S1P(1)-mTOR axis directs the reciprocal differentiation of T(H)1 and T(reg) cells. Nat. Immunol. 11:1047–1056 10.1038/ni.1939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukashev D., Klebanov B., Kojima H., Grinberg A., Ohta A., Berenfeld L., Wenger R.H., Ohta A., Sitkovsky M. 2006. Cutting edge: hypoxia-inducible factor 1alpha and its activation-inducible short isoform I.1 negatively regulate functions of CD4+ and CD8+ T lymphocytes. J. Immunol. 177:4962–4965 [DOI] [PubMed] [Google Scholar]
- McGeachy M.J., Chen Y., Tato C.M., Laurence A., Joyce-Shaikh B., Blumenschein W.M., McClanahan T.K., O’Shea J.J., Cua D.J. 2009. The interleukin 23 receptor is essential for the terminal differentiation of interleukin 17-producing effector T helper cells in vivo. Nat. Immunol. 10:314–324 10.1038/ni.1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalek R.D., Gerriets V.A., Jacobs S.R., Macintyre A.N., MacIver N.J., Mason E.F., Sullivan S.A., Nichols A.G., Rathmell J.C. 2011. Cutting edge: distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J. Immunol. 186:3299–3303 10.4049/jimmunol.1003613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann A.K., Yang J., Biju M.P., Joseph S.K., Johnson R.S., Haase V.H., Freedman B.D., Turka L.A. 2005. Hypoxia inducible factor 1 alpha regulates T cell receptor signal transduction. Proc. Natl. Acad. Sci. USA. 102:17071–17076 10.1073/pnas.0506070102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nizet V., Johnson R.S. 2009. Interdependence of hypoxic and innate immune responses. Nat. Rev. Immunol. 9:609–617 10.1038/nri2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce E.L. 2010. Metabolism in T cell activation and differentiation. Curr. Opin. Immunol. 22:314–320 10.1016/j.coi.2010.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce E.L., Walsh M.C., Cejas P.J., Harms G.M., Shen H., Wang L.S., Jones R.G., Choi Y. 2009. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature. 460:103–107 10.1038/nature08097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell J.D., Delgoffe G.M. 2010. The mammalian target of rapamycin: linking T cell differentiation, function, and metabolism. Immunity. 33:301–311 10.1016/j.immuni.2010.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rius J., Guma M., Schachtrup C., Akassoglou K., Zinkernagel A.S., Nizet V., Johnson R.S., Haddad G.G., Karin M. 2008. NF-kappaB links innate immunity to the hypoxic response through transcriptional regulation of HIF-1alpha. Nature. 453:807–811 10.1038/nature06905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan H.E., Poloni M., McNulty W., Elson D., Gassmann M., Arbeit J.M., Johnson R.S. 2000. Hypoxia-inducible factor-1alpha is a positive factor in solid tumor growth. Cancer Res. 60:4010–4015 [PubMed] [Google Scholar]
- Sauer S., Bruno L., Hertweck A., Finlay D., Leleu M., Spivakov M., Knight Z.A., Cobb B.S., Cantrell D., O’Connor E., et al. 2008. T cell receptor signaling controls Foxp3 expression via PI3K, Akt, and mTOR. Proc. Natl. Acad. Sci. USA. 105:7797–7802 10.1073/pnas.0800928105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundrud M.S., Koralov S.B., Feuerer M., Calado D.P., Kozhaya A.E., Rhule-Smith A., Lefebvre R.E., Unutmaz D., Mazitschek R., Waldner H., et al. 2009. Halofuginone inhibits TH17 cell differentiation by activating the amino acid starvation response. Science. 324:1334–1338 10.1126/science.1172638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O. 1956. On the origin of cancer cells. Science. 123:309–314 10.1126/science.123.3191.309 [DOI] [PubMed] [Google Scholar]
- Zheng Y., Delgoffe G.M., Meyer C.F., Chan W., Powell J.D. 2009. Anergic T cells are metabolically anergic. J. Immunol. 183:6095–6101 10.4049/jimmunol.0803510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Lopes J.E., Chong M.M., Ivanov I.I., Min R., Victora G.D., Shen Y., Du J., Rubtsov Y.P., Rudensky A.Y., et al. 2008. TGF-beta-induced Foxp3 inhibits T(H)17 cell differentiation by antagonizing RORgammat function. Nature. 453:236–240 10.1038/nature06878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Yamane H., Paul W.E. 2010. Differentiation of effector CD4 T cell populations (*). Annu. Rev. Immunol. 28:445–489 10.1146/annurev-immunol-030409-101212 [DOI] [PMC free article] [PubMed] [Google Scholar]