Sorting of GPI-anchored proteins into ER exit sites by p24 proteins is dependent on remodeled GPI (original) (raw)
p24 complexes act as cargo receptors for sorting GPI-anchored proteins into COPII vesicles.
Abstract
Glycosylphosphatidylinositol (GPI) anchoring of proteins is a posttranslational modification occurring in the endoplasmic reticulum (ER). After GPI attachment, proteins are transported by coat protein complex II (COPII)-coated vesicles from the ER. Because GPI-anchored proteins (GPI-APs) are localized in the lumen, they cannot interact with cytosolic COPII components directly. Receptors that link GPI-APs to COPII are thought to be involved in efficient packaging of GPI-APs into vesicles; however, mechanisms of GPI-AP sorting are not well understood. Here we describe two remodeling reactions for GPI anchors, mediated by PGAP1 and PGAP5, which were required for sorting of GPI-APs to ER exit sites. The p24 family of proteins recognized the remodeled GPI-APs and sorted them into COPII vesicles. Association of p24 proteins with GPI-APs was pH dependent, which suggests that they bind in the ER and dissociate in post-ER acidic compartments. Our results indicate that p24 complexes act as cargo receptors for correctly remodeled GPI-APs to be sorted into COPII vesicles.
Introduction
Protein transport through the secretory pathway is mediated by membrane vesicles coated with cytoplasmic coat proteins. In the early secretory pathway, correctly folded proteins are transported from the ER into coat protein complex II (COPII)-derived vesicles (Bonifacino and Glick, 2004; Lee et al., 2004). Most secretory and membrane proteins are actively sorted and packaged into COPII vesicles, whereas a bulk flow mechanism also exists (Barlowe, 2003). One of the COPII components, Sec24, directly associates with cargo proteins and packages them into the vesicles depending on the sorting signals (Miller et al., 2003). Structural studies on COPII have revealed that Sec24 isoforms have multiple cargo recognition sites that bind to sorting signals and select cargo proteins (Mancias and Goldberg, 2008). Many membrane proteins contain sorting signals that are directly recognized by the intracellular machinery. However, in some cases, adaptor proteins, the so-called cargo receptors, which act as bridge between the cargo and COPII components, are needed for efficient transport (Dancourt and Barlowe, 2010). Additionally, because soluble secretory proteins cannot bind to Sec24, transmembrane cargo receptors are critical for linking to cytoplasmic COPII components (Appenzeller et al., 1999; Belden and Barlowe, 2001).
Glycosylphosphatidylinositol (GPI) anchoring of proteins is a highly conserved posttranslational modification that occurs in eukaryotes. To date, >150 proteins, including receptors, adhesion molecules, and enzymes, are known to be modified by GPI in mammalian cells. Biosynthesis of GPI and attachment to proteins are performed in the ER (Kinoshita et al., 2008). Once GPI-anchored proteins (GPI-APs) are formed, they are transported from the ER to the cell surface via the Golgi apparatus. For sorting GPI-APs that are localized in the lumen and cannot bind to COPII directly, cargo receptors are required for their efficient transport. The p24 family proteins are type-I membrane proteins that are recycled between the ER and the Golgi (Strating and Martens, 2009). These proteins form hetero-oligomeric complexes and bind to COPI and COPII. Members of the p24 family in yeast are involved in efficient packaging of GPI-APs into COPII vesicles (Schimmöller et al., 1995; Belden and Barlowe, 1996; Muñiz et al., 2000; Castillon et al., 2009). Genetic analyses also suggest that genes involved in structural remodeling of GPI anchors act together with p24 proteins in yeast (Elrod-Erickson and Kaiser, 1996; Haass et al., 2007). In mammalian cells, knockdown of p24 genes resulted in delayed transport of GPI-APs from the ER to the Golgi (Takida et al., 2008; Bonnon et al., 2010). The p23 and p24 proteins were coprecipitated with GPI-APs. The Sec24C and -D isoforms facilitated export of the p23–p24 complex from the ER, with the same preference for GPI-APs (Bonnon et al., 2010). Evidence suggests that the p24 family of proteins act as cargo receptors for GPI-APs from the ER, but it remains unclear how the modified proteins are sorted and how p24 proteins recognize GPI-APs.
During GPI-AP transport, the lipid and glycan parts of GPI are remodeled (Fujita and Kinoshita, 2010). Before exiting the ER, two structural remodeling reactions occur in mammalian cells; an acyl chain linked to inositol and a side-chain ethanolamine phosphate (EtNP) attached to the second mannose of the GPI anchor are removed by PGAP1 and PGAP5, respectively (Tanaka et al., 2004; Fujita et al., 2009). Both reactions are required for the efficient transport of GPI-APs from the ER to the Golgi. However, it is still not clear why the structural remodeling of GPI anchors is critical for efficient transport. Here, we elucidated that the two structural remodeling reactions of GPI anchors were critical for transporting GPI-APs to the ER exit sites (ERES). We also found that remodeled GPI anchors are efficiently recognized by the p24 family proteins. The association of p24 proteins with GPI-APs occurs at a pH found within the ER, whereas dissociation occurs at a pH found in ER–Golgi intermediate compartments (ERGIC) or the cis-Golgi. These results indicate that the remodeled structure of GPI after attachment to proteins acts as a sorting signal for ER exit that is recognized by p24 proteins.
Results
Structural remodeling of GPI-APs is required for sorting to the ERES
Before exiting of GPI-APs from the ER, two remodeling reactions—inositol deacylation by PGAP1 and removal of the second EtNP by PGAP5—occur in mammalian cells (Fig. 1). We previously established a method for screening mutant cells defective in transport of GPI-APs (Maeda et al., 2008). From the screening, we isolated C19 mutant cells defective in PGAP5 (Fujita et al., 2009). The remodeling of GPI glycan by PGAP5 is required for the efficient transport of GPI-APs from the ER. From the same screening, we obtained another mutant cell line, designated FPRC2, in which GPI-APs were resistant to phosphatidylinositol-specific phospholipase C (PI-PLC) because of a defect in PGAP1 (Fig. S1). Consistent with our previous finding that transport of GPI-APs, decay-accelerating factor (DAF), and CD59 from the ER to the Golgi was delayed in PGAP1-deficient cells (Tanaka et al., 2004), FPRC2 mutant clones also exhibited delayed transport of a reporter GPI-AP, VSVGex-Flag-GFP (VFG-GPI (Fig. 2 A). When the responsible genes were transfected into C19 and FPRC2 cells, the delay in transport was rescued (Fig. 2, B and C). In the steady state, the ER form of DAF was accumulated in pgap1 and pgap5 mutant cells (Fig. 2 D), which suggests that delayed transport was caused by due to a slow exit from the ER. The amount of the accumulated ER form of DAF in the steady state remained small relative to the mature forms of DAF because the mature forms are stably maintained at the plasma membrane. The accumulation of the ER form of DAF disappeared when the responsible gene was reintroduced (Fig. 2 D). These results suggest that both remodeling reactions mediated by PGAP1 and PGAP5 are required for the efficient transport of GPI-AP from the ER.
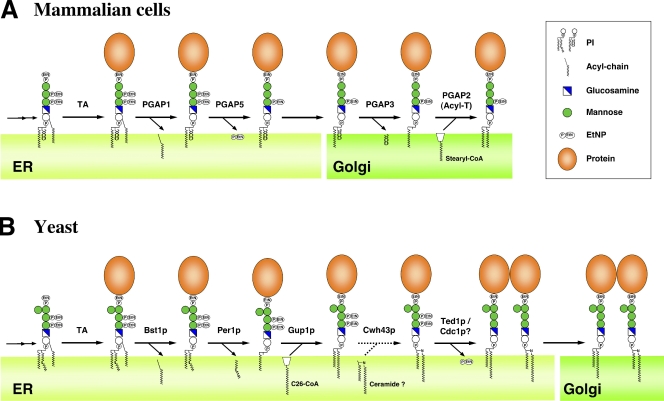
Figure 1.
Remodeling of GPI-APs in mammalian cells and yeast. (A) Remodeling of mammalian GPI-APs. Biosynthesis of GPI is performed in the ER through an enzymatic reaction pathway consisting of 10 steps (Kinoshita et al., 2008; Fujita and Kinoshita, 2010). Preformed GPI is attached to the C-terminus of a newly synthesized protein by GPI transamidase (TA). The acyl chain linked to inositol is eliminated by PGAP1. A side-chain EtNP attached to the second mannose is removed by PGAP5. After arrival of GPI-APs at the Golgi, fatty acid remodeling occurs; an unsaturated acid at the sn-2 position in the GPI lipid is removed by PGAP3 and then a saturated fatty acid (stearic acid) is transferred back. PGAP2 is a noncatalytic protein involved in the latter reaction. The putative catalytic component of the acyltransferase (Acyl-T) has yet to be discovered. (B) Remodeling of GPI-APs in the yeast Saccharomyces cerevisiae. After attachment of GPI to proteins, the acyl chain linked to inositol is eliminated by Bst1p (PGAP1 homologue). Then, the fatty acid remodeling of the GPI anchor is performed in the ER, mediated by Per1p (PGAP3 homologue) and Gup1p. In yeast, the diacylglycerol moiety in many GPI-APs is exchanged with ceramide by Cwh43p. Ted1p and Cdc1p, homologues of mammalian PGAP5, are localized in the ER. It was reported that Ted1p acts together with Emp24p and Erv25p, and that Cdc1p functions after Per1p and Gup1p in the same pathway, which suggests that they are involved in GPI remodeling. Side-chain EtNPs on the first and second mannoses are required for ceramide remodeling, whereas it remains unclear whether the side-chain EtNPs are eliminated.
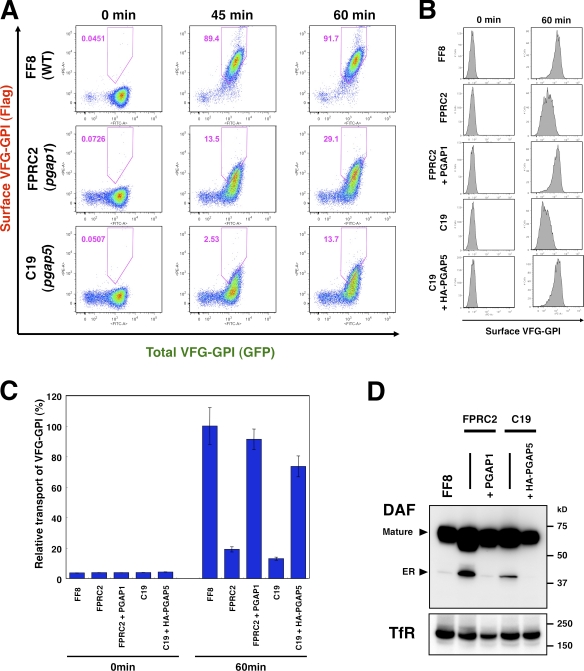
Figure 2.
Efficient transport of GPI-APs requires structural remodeling of GPI anchors by PGAP1 and PGAP5. (A) Flow cytometric analysis of transport of GPI-anchored reporter protein. Parental FF8 (WT), pgap1 mutant FPRC2, and pgap5 mutant C19 cells expressing VFG-GPI were stained with an anti-Flag antibody at the indicated times after a temperature shift from 40°C to 32°C. The percentages of cells in the pentagonal region are indicated. (B and C) The cell surface expression of VFG-GPI was analyzed by flow cytometry. FF8 (WT), FPRC2 (pgap1), C19 (pgap5), and FPRC2 cells stably expressing PGAP1 (+ PGAP1) and C19 cells stably expressing HA-tagged PGAP5 (+HA-PGAP5) were stained with anti-Flag antibody at the indicated times after the commencement of transport. The geometric mean fluorescence values of surface VFG-GPI in all cells shown in histograms (B) were quantified at each time point (C). The geometric mean of WT cells at 60 min was plotted as 100% relative transport. Values represent mean ± SD (error bars; n = 3). (D) Western blotting analysis of DAF in the steady state. Cell lysates of FF8 (WT), FPRC2 (pgap1), C19 (pgap5), and FPRC2 cells stably transfected with PGAP1, and C19 cells stably transfected with HA-PGAP5 were analyzed with an anti-DAF antibody under nonreducing conditions to evaluate the steady-state levels of the ER and mature forms. TfR, transferrin receptor for loading assessment.
We next investigated which step during transport from the ER to the Golgi was impaired in the mutant cells defective in GPI remodeling. To visualize the sorting of GPI-APs into the ERES, we used VFG-GPI, which could be monitored. For accumulation in the ER, VFG-GPI was expressed at 40°C. Sorting to the ERES was visualized by inhibiting budding using a temperature shift to 10°C (Kirk and Ward, 2007; Rivier et al., 2010). Immediately after the temperature shift, VFG-GPI could be observed throughout the ER, but did not merge with Sec13 in wild-type, pgap1, and pgap5 mutant cells (Fig. 3 A). After a 30-min incubation at 10°C, VFG-GPI was accumulated in dot-like structures that colocalized with Sec13 in wild-type cells (Fig. 3, B and C). In contrast, VFG-GPI exhibited reticular-like localization and did not colocalize with Sec13 in pgap1 and pgap5 mutant cells. The concentration of VFG-GPI at the ERES was significantly impaired in pgap1 and pgap5 cells (Fig. 3 D). These results indicate that structural remodeling of GPI anchors is critical for the sorting of GPI-APs to the ERES.
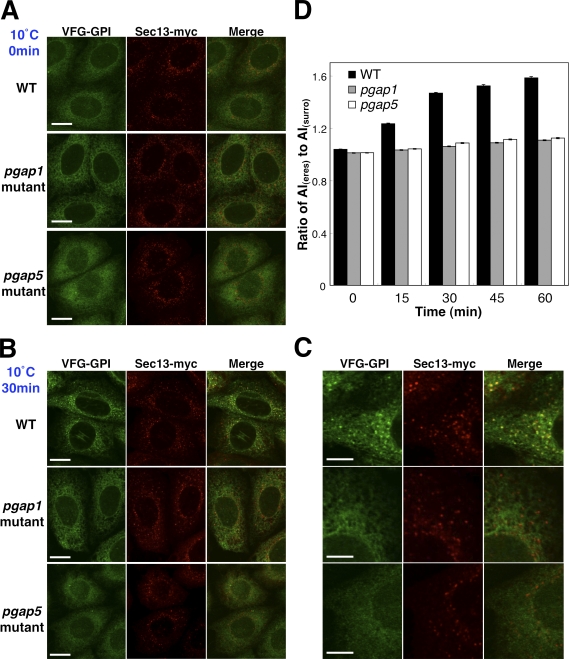
Figure 3.
Sorting of GPI-APs into the ERES is impaired in pgap1 and pgap5 mutant cells. (A–C) VFG-GPI were expressed and accumulated in the ER by incubating FF8 (WT), FPRC2 (pgap1), and C19 (pgap5) cells stably expressing myc-tagged Sec13 (Sec13-myc) with 1 µg/ml doxycycline at 40°C for 24 h. After incubation at 10°C for 0 min (A) or 30 min (B and C, high magnification of B), cells were fixed with 4% paraformaldehyde and stained with anti-myc antibody to label the ERES. Bars: (A and B) 10 µm; (C) 5 µm. (D) Relative intensity of VFG-GPI in the ERES. The ratios of average intensities of VFG-GPI in the ERES (AI(eres)) to those in the ERES surroundings (AI(surro)) were measured. The ratios between wild-type (WT) and pgap1 mutant cells and between WT and pgap5 mutant cells differed significantly at each time point. (n = 715–1,906; Student’s t test, P < 0.0001). Error bars indicate the standard error of the mean.
p23 and p24 associate with structurally remodeled GPI-APs
The impairment of GPI-AP sorting to the ERES seen in pgap1 and pgap5 mutant cells raised the question of why structural changes in GPI anchors are required for efficient sorting. We hypothesized that there are putative cargo receptor proteins that recognize the remodeled GPI anchors for packaging GPI-APs into vesicles. To verify this possibility, we determined proteins specifically associated with GPI-APs in the wild type, but not in pgap1 or pgap5 mutant cells. Cells with VFG-GPI accumulated in the ER were incubated at 32°C for 20 min to allow for transport, and then lysed in buffer containing 1% digitonin. VFG-GPI was collected with anti-Flag beads and proteins were analyzed by SDS-PAGE. Bands ∼20 kD were specifically coprecipitated from wild-type cells (Fig. 4 A). We analyzed the bands by mass spectrometry and identified them as Tmed10 (p23) and Tmed2 (p24; Fig. 4 B). The association of p23 and p24 with VFG-GPI in wild-type cells was also confirmed by Western blotting, whereas in the pgap1 and pgap5 mutant cells, association of p23 and p24 with VFG-GPI was greatly decreased (Fig. 4 C).
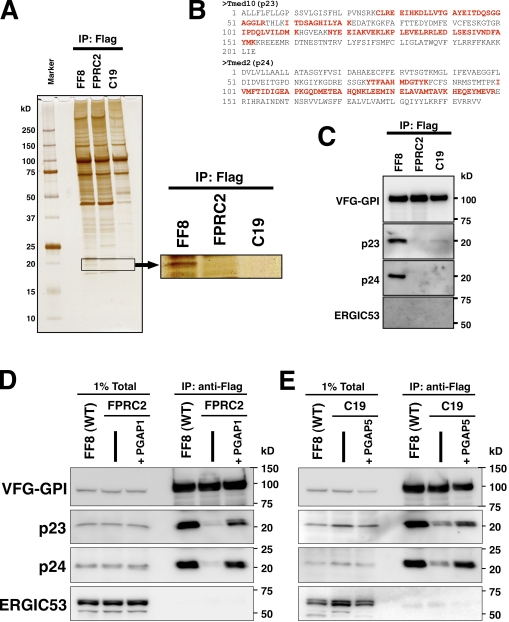
Figure 4.
Association of p23 and p24 with remodeled GPI-APs. (A) Detection of proteins specifically associated with VFG-GPI in FF8. VFG-GPI was expressed and accumulated in the ER in FF8, FPRC2, and C19 cells. The cells were then incubated at 32°C for 20 min to initiate transport of VFG-GPI. After cell lysis, VFG-GPI was precipitated with anti-Flag beads. Co-precipitated proteins were eluted using the Flag peptide and subjected to SDS-PAGE and silver staining. The boxed area at ∼20 kD is enlarged to the right. (B) Sequences of hamster Tmed10 (p23) and Tmed2 (p24). Protein bands at 20 kD in A were digested in-gel with trypsin and analyzed by mass spectrometry. The fragments detected by MS/MS analysis are shown in red. (C) Precipitated proteins in A were analyzed by Western blotting using a rabbit anti-p23, anti-p24, or anti-ERGIC53 polyclonal antibody. VFG-GPI was detected with an anti-GFP antibody. (D and E) Immunoprecipitation of VFG-GPI with p23 and p24. FF8, FPRC2, and FPRC2 stably expressing PGAP1 (D) or FF8, C19, and C19 stably expressing HA-PGAP5 (E) were cultured with 1 µg/ml doxycycline at 40°C for 24 h to induce VFG-GPI expression and its accumulation in the ER. The cells were then incubated at 32°C for 20 min to initiate VFG-GPI transport. After cell lysis, VFG-GPI was precipitated with anti-Flag beads and coprecipitated proteins were detected by immunoblotting using an anti-p23, anti-p24, or anti-ERGIC53 antibody. VFG-GPI was detected with an anti-GFP antibody. Total lysate corresponding to 1% and immunoprecipitates were used for analysis.
We next analyzed whether the decreased association of p23 and p24 with VFG-GPI in pgap1 and pgap5 mutant cells was caused by defects in remodeling of GPI anchors. Binding of VFG-GPI with p23 and p24 was recovered in PGAP1- or PGAP5-restored cells (Fig. 4, D and E). In contrast, binding of GPI-APs with p23 and p24 and sorting of GPI-APs to the ERES were normal in pgap2/pgap3 double mutant cells (Fig. S2, A and B). Thus, structural remodeling of GPI anchors by PGAP1 and PGAP5 in the ER, but not fatty acid remodeling of GPI anchors mediated by PGAP3 and PGAP2 in the Golgi, is specifically required for association with p23 and p24 and sorting GPI-APs to the ERES. This is consistent with sites where these remodeling reactions occur in mammalian cells (Fig. 1). The association of p23 and p24 with VFG-GPI was dependent on the GPI anchor because the association was significantly reduced when the GPI attachment signal was replaced with a transmembrane domain (Fig. S2 C). The association was time dependent and peaked 10–20 min after transport commenced (Fig. S2 D). In yeast and mammalian cells, it has been reported that the p24 family of proteins facilitate transport of GPI-APs from the ER to the Golgi, but the precise mechanisms are still unclear. Our results suggest that the p23–p24 complex recognizes the structurally remodeled GPI anchors.
p24 proteins are required for efficient transport of GPI-APs from the ER
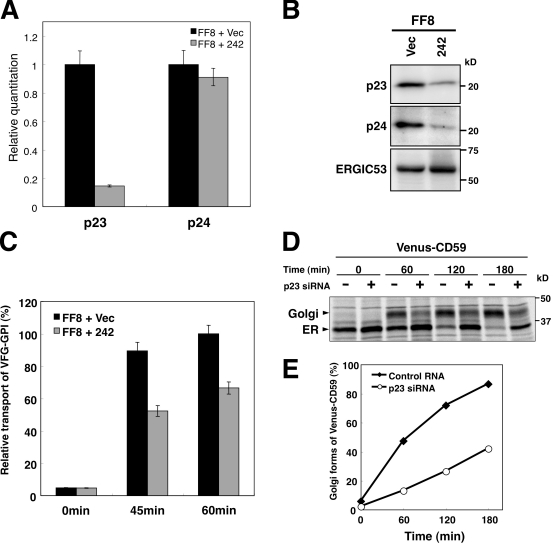
The involvement of p23 and p24 with respect to transport of GPI-APs from the ER was analyzed. The mRNA level of p23 in stable knockdown cells was decreased to 15% of that in control cells (Fig. 5 A). A substantial decrease in p23 protein was observed in the knockdown cells (Fig. 5 B). It has been reported that p24 proteins form a complex and stabilize each other (Belden and Barlowe, 1996; Marzioch et al., 1999; Denzel et al., 2000); therefore, we examined the status of p24 in p23 knockdown cells. The level of p24 was also decreased, whereas mRNA levels were not affected as expected (Fig. 5, A and B). Transport of VFG-GPI was significantly delayed in p23 knockdown cells (Fig. 5 C), as described previously (Takida et al., 2008; Bonnon et al., 2010). Transport from the ER to the Golgi of another GPI-AP was also examined. The p23 protein was transiently silenced in CHO-K1 cells stably expressing Venus-tagged human CD59 (Venus-CD59). Pulse-chase analysis with endoglycosidase-H (Endo-H) sensitivity revealed that p23 knockdown resulted in delayed transport of Venus-CD59 from the ER to the Golgi (Fig. 5, D and E). These results support the idea that p24 proteins are involved in transport of GPI-APs from the ER.
Figure 5.
p24 family is required for the GPI-AP transport from the ER. (A) Relative amount of p23 and p24 mRNA from FF8 cells stably transfected with empty vector (+Vec) or p23 siRNA vector (+242) as determined by quantitative RT-PCR analysis. (B) Western blotting of p23 and p24 in p23 knockdown cells. FF8 cells permanently transfected with empty vector (Vec) or p23 siRNA vector (242) were lysed and the proteins were resolved by SDS-PAGE, followed by immunoblotting using rabbit anti-p23, anti-p24, or anti-ERGIC53 polyclonal antibody. (C) The cell surface expression of VFG-GPI was traced by flow cytometry. FF8 cells stably transfected with empty vector (+Vec) or p23 siRNA vector (+242) were stained with anti-Flag antibody at the indicated times after the commencement of transport. The geometric mean fluorescent values of surface VFG-GPI were quantified at each time point. The geometric mean of FF8 + Vec at 60 min was plotted as 100% relative transport. Values represent the mean ± SD (error bars; n = 3). (D and E) Pulse-chase transport analysis of Venus-CD59. Cells stably expressing Venus-CD59 were transfected with siRNAs against p23 or control RNA. After 72 h, the cells were pulse-labeled with [35S]methionine/cysteine followed by chasing for the indicated times, and immunoprecipitated with an antibody against Venus. Immunoprecipitates were treated with Endo-H, separated by SDS-PAGE, and analyzed and quantified using a Cyclone Phosphor Imager (Packard). The proportion of Golgi form (Endo-H–resistant form) and Venus-CD59 were plotted in E.
Sorting of GPI-APs to COPII-coated vesicles was impaired in p23-knockdown cells
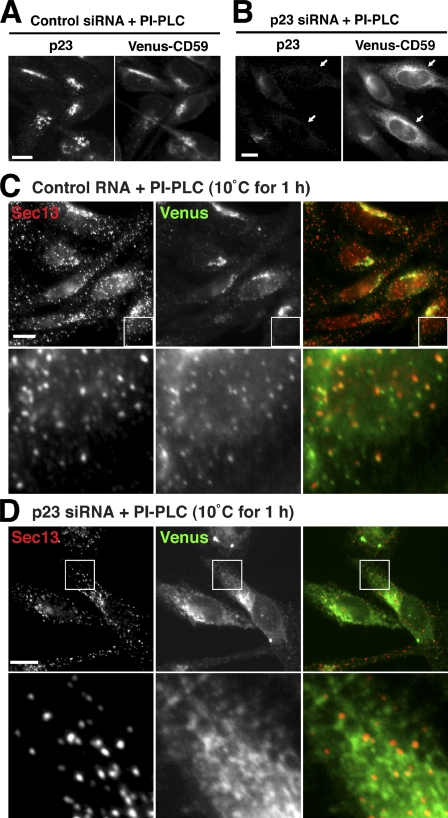
Because the transport of GPI-APs was delayed in p23 knockdown cells, we further analyzed cellular localization of GPI-APs. In the steady state, Venus-CD59 on the cell surface of knockdown cells was the same as in control cells (Fig. S3 A). Venus-CD59 on the cell surface was removed by PI-PLC to observe intracellular localization. In the control cells, the majority of the intracellular Venus-CD59 was localized at the Golgi apparatus (Fig. 6 A and S3 B), whereas in the p23 knockdown cells, ER reticular-like localization of Venus-CD59 was observed, colocalizing with calreticulin as well as at the Golgi (Fig. 6 B and S3 B). The ER localization of Venus-CD59 was more clearly observed in cells with an absence of p23 proteins (Fig. 6 B, arrows), which suggests that GPI-AP transport from the ER was impaired in p23 knockdown cells.
Figure 6.
Sorting of GPI-APs to the ERES is dependent on p24 family. (A and B) CHO-K1 cells stably expressing Venus-CD59 were transfected with control siRNA (A) or siRNA against p23 (B). After 72 h, the cells were treated with PI-PLC, followed by fixation, permeabilization, and immunostaining with anti-p23. Arrows indicate efficiently silenced cells. (C and D) CHO-K1 cells stably expressing Venus-CD59 were transfected with control RNA (C) or siRNA against p23 (D). After 72 h, the cells were treated with PI-PLC, incubated at 10°C for 1 h, fixed with 4% paraformaldehyde, and stained with anti-Sec13 to observe the ERES. In the merged images, Sec13 and Venus-CD59 were shown in red and green, respectively. Enlarged views of the boxed regions are shown below. Bars, 10 µm.
We next investigated whether p24 proteins are required for sorting of GPI-APs to the ERES. After PI-PLC treatment and incubation at 10°C for 1 h, Venus-CD59 had accumulated as punctuate structures that were largely colocalized with Sec13 in control cells (Fig. 6 C). Under these conditions, p23 also accumulated in punctuate structures, which were partially colocalized with Venus-CD59 and Sec13 (Fig. S3 C). In contrast, signals from Venus-CD59 remained in a typical reticular ER pattern and were mostly separated from Sec13 in the p23 knockdown cells (Fig. 6 D). These results indicate that the p24 family of proteins act as cargo receptors for GPI-APs by concentrating GPI-APs in the ERES, and facilitate efficient packaging into COPII vesicles.
Association of p23 and p24 with GPI-APs is pH dependent
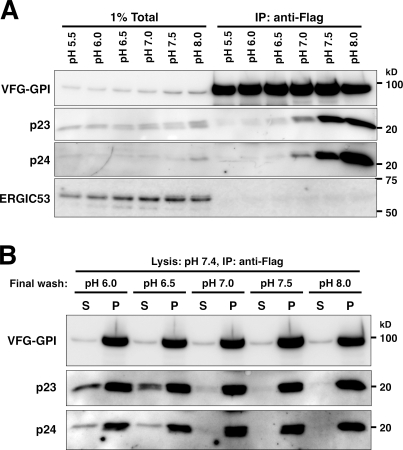
If the p24 proteins act as cargo receptors for GPI-APs, then mechanisms for their association with GPI-APs in the ER, and their dissociation in the ERGIC or cis-Golgi, are required. Calcium concentration is one of the factors for cargo binding and dissociation (Kawasaki et al., 2008; Dancourt and Barlowe, 2010). Leguminous (L)-type lectin-like cargo receptors such as ERGIC53 and VIP36 require calcium for their cargo binding. The binding of p23 and p24 with VFG-GPI, however, seemed to be metal independent because addition of EDTA in the lysis and washing buffers did not affect binding (Fig. S2 E). Cellular pH is another factor that regulates the interactions between sorting receptors and their cargo (Paroutis et al., 2004; Appenzeller-Herzog and Hauri, 2006); the neutral pH in the ER promotes binding of cargo molecules to their receptors, whereas the acidic pH in the ERGIC and the cis-Golgi induces dissociation of the cargo. We investigated whether the association of p23 and p24 with GPI-APs was affected by pH. VFG-GPI was extracted and precipitated in buffers at various pH values. The p23 and p24 proteins were coprecipitated with VFG-GPI in the buffer at a pH >7.0, whereas coprecipitation was not seen at a pH <6.5 (Fig. 7 A). VFG-GPI was still bound to the beads with the same efficiency under all pH conditions, which suggests that a lack of coprecipitation of p23 and p24 was not caused by loss of the binding ability of antibody beads. We further determined whether p23 and p24 that bound to VFG-GPI could be dissociated by lowering the pH. VFG-GPI was extracted and precipitated with anti-Flag beads in a pH 7.4 buffer. After washing the beads five times with pH 7.4 buffer, they were resuspended in buffers at various pH levels for 15 min at 4°C. Both p23 and p24 were released from the beads at a pH <6.5, but not at a pH >7.0 (Fig. 7 B). These results indicate that association and dissociation of p23 and p24 with GPI-APs was dependent on pH, and that p23 and p24 can be released from GPI-APs at a pH corresponding to the pH in the cellular organelle where dissociation is expected to occur.
Figure 7.
p24 proteins associated with GPI-APs in a pH-dependent manner. (A) Immunoprecipitation of VFG-GPI with p23 and p24 at various pH levels. FF8 cells were cultured with doxycycline at 40°C for 24 h to induce VFG-GPI expression and its accumulation in the ER. The cells were then incubated at 32°C for 20 min to initiate VFG-GPI transport. After cell lysis in lysis-IP buffer III of the indicated pH, VFG-GPI was precipitated with anti-Flag beads and washed five times in wash buffer III of the indicated pH, followed by immunoblotting using an anti-p23, anti-p24, or anti-ERGIC53 antibody. VFG-GPI was detected with an anti-GFP antibody. Total lysate corresponding to 1% and immunoprecipitates were used for analysis. Bands in 1% total fractions gradually increased with an increased pH partly because solubilization of proteins in 1% digitonin was slightly better at higher pH. (B) Release of p23 and p24 from VFG-GPI by lowering pH. FF8 cells prepared in a similar manner to A were lysed in lysis-IP buffer III, pH 7.4, and VFG-GPI was precipitated with anti-Flag beads. After washing five times with wash buffer III, pH 7.4, wash buffer III at the indicated pH was added and incubated at 4°C for 15 min. The supernatant (S) and bead pellets (P) were collected, followed by immunoblotting as in A.
Association of GPI-APs with p24 hetero-oligomers
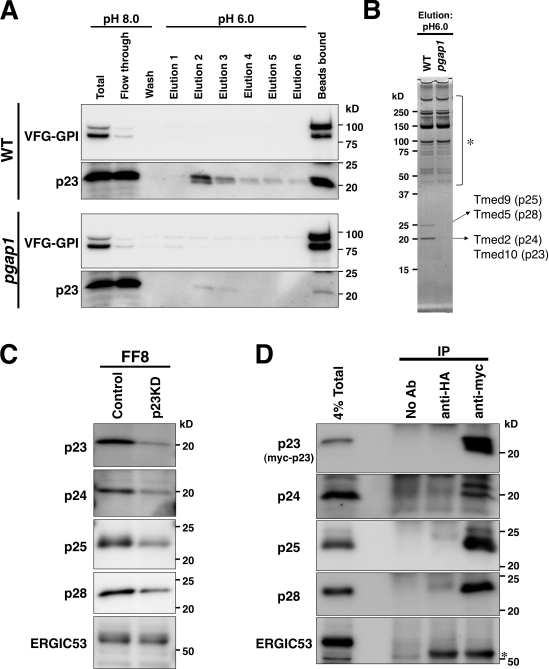
We further asked if additional factors are involved in complexes of GPI-APs with p23 and p24. The binding of p23 and p24 with VFG-GPI was dependent on the structure of the GPI anchor; therefore, we compared wild-type and pgap1 mutant cells. Cells were lysed in a pH 8.0 buffer, and VFG-GPI was purified with an anti-Flag column. After washing, proteins bound to the column were eluted with a buffer at pH 6.0. The p23 protein was eluted from the column loaded with the lysate from wild-type cells by decreasing the pH (Fig. 8 A). A small amount of p23 was eluted from the column loaded with the lysate of pgap1 mutant cells, which is consistent with the idea that p23 associates with remodeled GPI-APs effectively. Under these conditions, proteins eluted from these two columns were compared and two specific bands of ∼20 and ∼25 kD were found in the eluate of samples from the wild-type cells (Fig. 8 B). The protein bands were determined by mass spectrometry. The 20-kD band consisted of p23 and p24, which was the same as shown in Fig. 4. The 25-kD band was identified as a mixture of Tmed9 (p25) and Tmed5 (p28), members of the p24 family (Figs. 8 B and S4).
Figure 8.
p24 proteins form heteromeric complexes with subfamily members. (A and B) Identification of proteins associated with VFG-GPI depending on pH. FF8 (WT) and FPRC2 (pgap1) cells were cultured with doxycycline at 40°C for 24 h. The cells were then incubated at 32°C for 20 min. After cell lysis using lysis-IP buffer II, pH 8.0, VFG-GPI was purified using an anti-Flag column. After thorough washing in wash buffer II, pH 8.0, the binding proteins were eluted with elution buffer II, pH 6.0. After elution with six bed volumes of buffer, proteins bound to the column were extracted with SDS sample buffer. Each fraction was subjected to SDS-PAGE, followed by immunoblotting using an anti-GFP or anti-p23 antibody (A). Proteins in eluted fraction 2 from FF8 (WT) and FPRC2 (pgap1) were detected by silver staining (B). Protein bands at 20 and 25 kD were identified by mass spectrometry as Tmed10 (p23), Tmed2 (p24), Tmed9 (p25), and Tmed5 (p28). Detected fragments are shown in Fig. S4. Protein bands indicated by an asterisk (*) were observed through all fractions (elutions 1−6) at similar levels and were not specific in WT cells. (C) Knockdown of p23 destabilized other p24 proteins. FF8 cells permanently transfected with an empty vector (Control) or p23 siRNA vector (p23KD) were lysed, and proteins were resolved by SDS-PAGE, followed by immunoblotting using rabbit anti-p23, anti-p24, anti-p25, anti-p28, and anti-ERGIC53 polyclonal antibodies. (D) Coimmunoprecipitation of myc-p23 with p24 proteins. FF8 cells were stably transfected with a retrovirus vector expressing RNAi-resistant myc-tagged p23 (myc-p23) and shRNA against endogenous p23, as described in Fig. S5. After cell lysis, myc-p23 was precipitated with anti-HA (control) or anti-myc antibodies or without antibody (No Ab), and coprecipitated proteins were detected by immunoblotting against anti-p23, anti-p24, anti-p25, anti-p28, and anti-ERGIC53 antibodies. Total lysate corresponding to 4% and immunoprecipitates were used for analysis. *, IgG heavy chains.
The p24 family can be subdivided into four subfamilies: α, β, γ, and δ. The p25, p24, p28, and p23 proteins can be classified as p24α2, p24β1, p24γ2, and p24δ1, respectively (Strating and Martens, 2009). Our results suggest that p24 proteins form hetero-oligomers when they bind to GPI-APs. In the p23 knockdown cells, the amount of p25, p28, p23, and p24 was decreased, which suggests that p23 stabilizes other p24 subfamily members (Fig. 8 C). It has been reported that overexpression of members of the p24 family results in several artifacts, such as mislocalization and abnormal Golgi morphology (Blum et al., 1999; Rojo et al., 2000; Blum and Lepier, 2008). To analyze the hetero-oligomers between p24 proteins avoiding possible artifacts, we constructed a retrovirus vector to express myc-tagged p23 at low levels in combination with knockdown expression of endogenous p23 (Fig. S5, A–C). The myc-p23 was functional in stabilizing p24 and transporting GPI-APs (Fig. S5, C and D). Under these conditions, myc-tagged p23 was coimmunoprecipitated with p24, p25, and p28 (Fig. 8 D). Collectively, our results indicate that p23 forms a hetero-oligomeric complex with members of three other subfamilies.
Discussion
Most secretory proteins are sorted and enriched at the ERES for efficient transport from the ER. Cargo receptors play critical roles in linkage of their cargo to COPII components for enrichment in transport vesicles. In the case of GPI-APs, it has been postulated that the GPI anchor functions as an ER exit signal (Mayor and Riezman, 2004); however, molecular mechanisms for the sorting of GPI-APs into the ERES are not well understood. We previously determined that structural remodeling of the GPI anchor is required for the efficient transport of GPI-APs from the ER to the Golgi (Fujita et al., 2009). In this study, we elucidated the fact that GPI structural remodeling is critical for sorting GPI-APs to the ERES (Fig. 9). The remodeled GPI-APs are recognized by the p24 family of proteins that were required for efficient concentration of GPI-APs into COPII vesicles. These findings indicate that p24 proteins act as cargo receptors for correctly processed GPI-APs in the ER.
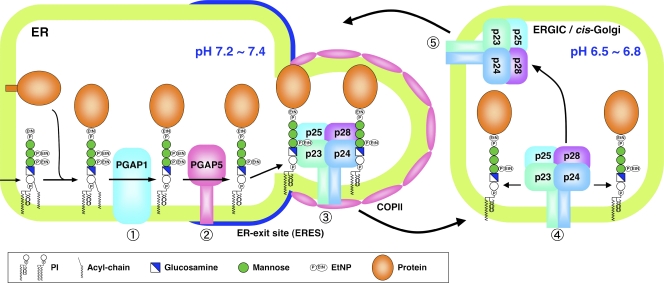
Figure 9.
A model of selective sorting and transport of GPI-APs from the ER. After GPI transfer to proteins by the GPI transamidase, the acyl chain linked to inositol in the GPI anchor is eliminated by PGAP1 (1), and a side-chain EtNP on the second mannose of the GPI anchor is removed by PGAP5 (2). These two GPI remodeling reactions in the ER are critical for the sorting of GPI-APs to the ERES. The remodeled GPI-APs are efficiently recognized by the p24 protein family complex that concentrates GPI-APs into the COPII-derived vesicles (3). After transport to the ERGIC or the cis-Golgi, GPI-APs dissociate from the p24 protein family complex because of decreased luminal pH in these compartments (4). The p24 complexes are retrieved from the Golgi to the ER by the COPI vesicles (5).
The association of p24 proteins with GPI-APs was dependent on environmental pH. Under neutral and mildly alkaline conditions, binding of p24 proteins to GPI-APs was clearly observed. Dissociation was observed under mildly acidic conditions. Ranges of pH for association (7.0–8.0) and dissociation (6.0–6.5) coincided with the pH in the ER and the ERGIC/cis-Golgi, respectively (Paroutis et al., 2004). Our results are consistent with a shuttling model for p24 proteins that bind to GPI-APs in the ER, transport them to and release them in the ERGIC/cis-Golgi, and are then returned to the ER (Fig. 9). It is possible that remodeling of the fatty acid moiety of the GPI anchor in the Golgi also favors dissociation of p24 proteins. It has yet to be determined which part of the GPI anchor is recognized by p24 proteins. We plan to study the interaction between p24 proteins and GPI-APs through biochemical and biophysical analyses in the near future.
There is a possibility that the association of p23 and p24 with GPI-APs is indirect and requires an adaptor protein linking p24 proteins with GPI-APs; however, there is some evidence to support the concept of direct binding. Chemical cross-linking experiments in yeast showed that Emp24p or Erv25p was directly cross-linked to a GPI-AP (Muñiz et al., 2000). Only the p24 family proteins were specifically eluted from VFG-GPI at pH 6.0 (Fig. 8), which suggests that association of p24 proteins with GPI-APs is direct. Homology modeling of the luminal domains of p23 and p24, termed GOLD (Golgi dynamics) domains, showed that they assume a jelly-roll fold, an antiparallel β sandwich structure consisting of two antiparallel β sheets each with three- or four-strands (Fig. S5, E–H). The jelly-roll β sandwich domains are often observed in sugar- and lipid-binding/processing proteins, such as bacterial sialidase for cell surface–attached carbohydrates (Gaskell et al., 1995), and the C2 domain of coagulation factor Va, which binds to phospholipids on the outside of the cell membrane (Macedo-Ribeiro et al., 1999). It seems possible that GOLD domains of p24 proteins recognize GPI anchors, which contain both lipid and carbohydrate portions.
Proteins in the p24 family form a functional heteromeric complex, whereas it is still debatable whether they can exist as monomers, heterodimers, or heterotetramers, depending on the cellular compartments (Marzioch et al., 1999; Jenne et al., 2002). The p24 family of proteins is divided into four subfamilies: α, β, γ, and δ. In mammalian cells, there are three members in p24α and five members of p24γ, whereas the p24β and p24δ subfamilies each have a single member. We identified that p25, p24, p28, and p23 associated with VFG-GPI depending on pH. All subfamilies are contained within the complex associated with GPI-APs, because p25, p24, p28, and p23 belong to p24α, p24β, p24γ, and p24δ, respectively. Our results suggest that they could form a hetero-oligomer for their association with GPI-AP, although the stoichiometry among them remains to be determined. Additionally, our results also explain cargo selectivity. Our mass spectrometric analysis detected only one of each subfamily member; therefore, it is possible that heteromeric complexes consisting of different combinations of subfamily members recognize different cargoes from GPI-APs. These possibilities need to be addressed in future studies.
Mechanisms of sorting GPI-APs to the ERES are conserved in yeast and mammalian cells. Deletion of BST1, a yeast PGAP1 homologue, resulted in decreased concentration of the GPI-AP, Cwp2p, at the ERES (Castillon et al., 2009). Emp24p and Erv25p are yeast p24β (p24) and p24δ (p23) members, which bind to varieties of GPI-APs and facilitate exit of GPI-APs from the ER (Castillon et al., 2011). However, there are differences in the functions of p24. In mammalian cells, p24 proteins act to concentrate GPI-APs at the ERES for efficient packaging into COPII vesicles. In yeast, sorting of GPI-APs to ERES is independent of Emp24p. Instead, the yeast p24 complex appears to act as an adaptor proteins that facilitates vesicle formation by recruiting COPII components to the specific ERES containing already enriched GPI-APs (Castillon et al., 2011). This may be caused by differences in structure of the GPI anchor in the ER of yeast and mammalian cells (Fujita and Jigami, 2008). As illustrated in Fig. 1, inositol deacylation of the GPI anchor by PGAP1/Bst1p occurs in the ER in both yeast and mammalian cells. Removal of the side-chain EtNP by PGAP5 is also performed in the ER of mammalian cells, and most likely in yeast by PGAP5 homologues—Ted1p and/or Cdc1p—which are localized at the ER. However, in yeast, two additional types of lipid remodeling take place in the ER (Fig. 1 B): fatty acid remodeling by Per1p and Gup1p and ceramide remodeling by Cwh43p. This results in GPI-APs containing either ceramide- or diacylglycerol-based lipids with a very long chain fatty acid at the sn-2 position (Bosson et al., 2006; Fujita et al., 2006; Ghugtyal et al., 2007; Umemura et al., 2007). In contrast, fatty acid remodeling, which replaces an unsaturated fatty acid at the sn-2 position of phosphatidylinositol in the GPI anchor with a saturated stearic acid, occurs only after arrival at the Golgi in mammalian cells (Fig. 1 A; Tashima et al., 2006; Maeda et al., 2007). Fatty acid remodeling is essential for incorporation of GPI-APs into detergent-resistant membranes in yeast and mammalian cells. Different subcellular locations where fatty acid remodeling occurs coincide with different locations for association of GPI-APs with detergent-resistant membranes: the ER in yeast and the Golgi in mammalian cells (Brown and London, 1998; Bagnat et al., 2000). Additionally, ceramide is required for the efficient transport of GPI-APs in yeast, but not in mammalian cells (Watanabe et al., 2002; Rivier et al., 2010). In yeast, lipid-dependent concentration of GPI-APs seems more important for sorting to the ERES and Emp24p–Erv25p complexes then support packaging of GPI-APs into vesicles by bridging them with COPII components.
The p24 proteins play key roles in maintaining the fidelity of GPI-AP vesicular transport from the ER, but these are unlikely to be the only functions of p24 complexes (Strating and Martens, 2009). It has been reported that p24 proteins are involved in numerous cellular functions, including transport of other secretory proteins, the unfolded protein response, quality control, retrograde transport from the Golgi, and Golgi structure maintenance (Bremser et al., 1999; Aguilera-Romero et al., 2008; Strating and Martens, 2009; Dancourt and Barlowe, 2010). It should be clarified in future studies whether multiple phenotypes are caused by the direct or indirect function of p24 proteins. In this study, we elucidated that at least one of the direct functions of these proteins is to act as a cargo receptor in the ER and to package GPI-APs in COPII vesicles. Our current results provide evidence that GPI structures and their attachment to proteins are ensured and monitored by remodeling enzymes, with only the properly processed GPI-APs sorted and transported from the ER, and that cargo receptors recognize sorting signals within the modified GPI structure.
Materials and methods
Cells and culture
The 3B2A cells were established by stably transfecting CHO-K1 cells with pME-NEO plasmid bearing DAF and CD59 under an SRα promoter, and selecting by cell-sorting a clone expressing DAF and CD59 at high levels (Nakamura et al., 1997). The FF8 cells are 3B2A cells stably transfected with pTRE2puro-VSVGex-FF-mEGFP-GPI in conjunction with pUHrT62-1, an expression plasmid for reverse tetracycline-controlled transactivators (Maeda et al., 2008; Takida et al., 2008). C19 and FPRC2 mutant cells are derivatives of FF8 cells, which are defective in PGAP5 and PGAP1, respectively. C10 mutant cells derived from 3B2A cells are defective in PGAP1 (Tanaka et al., 2004). DM-C2 mutant CHO-K1 cells are defective in both PGAP3 and PGAP2 (Maeda et al., 2007). CHO-K1 cells expressing Venus-CD59 were obtained after limiting dilution of cells stably transfected with pME-puro-Venus–tagged FLAG-CD59. The cells were grown in Ham’s F12 medium supplemented with 10% (vol/vol) FCS, 600 µg/ml G418 and, if necessary, 6 µg/ml puromycin. Cells were maintained at 37°C/5% CO2 in a humidified atmosphere. FPRC2 + PGAP1 and C19 + HA-PGAP5 cells were selected with 800 µg/ml hygromycin after FPRC2 and C19 cells were transfected with pME-hyg-rat-PGAP1 and pME-hyg-HA-hPGAP5, respectively. For use in a retrovirus system, FF8, FPRC2, and C19 cells were transiently or stably transfected with plasmids that express mouse CAT1, a receptor for ecotropic retroviruses. FF8, FPRC2, and C19 cells stably expressing Sec13-myc were then established by infection with retroviruses produced in PLAT-E packaging cells (a gift from T. Kitamura, University of Tokyo, Tokyo, Japan) transfected with pLIB2-BSD-Sec13myc, followed by selection with 6 µg/ml blasticidin. FF8 cells stably expressing p23 siRNA or control siRNA were established by infection with a retrovirus produced in PLAT-E cells transfected with pSINsi-hU6-BSD-242 or pSINsi-hU6-BSD, followed by selection with 6 µg/ml blasticidin.
Antibodies and materials
The antibodies used were mouse monoclonal antibodies against Flag (clone M2; Sigma-Aldrich), myc (clone 9E10), HA (HA-7; Sigma-Aldrich), GFP (Roche), and transferrin receptor (Invitrogen); and rabbit polyclonal antibodies against ERGIC53 (Sigma-Aldrich), calreticulin (Thermo Fisher Scientific), GPP130 (Covance), and Venus (Rivier et al., 2010). Mouse monoclonal antibodies against CD59 (5H8) and DAF (IA10) were described in a previous study (Maeda et al., 2007). Rabbit anti-p23, anti-p24, and anti-p25 antibodies were provided by F. Wieland and I. Reckmann (Heidelberg University, Heidelberg, Germany), and J. Gruenberg (University of Geneva, Geneva, Switzerland). Rabbit anti-p28 antibody was provided by H.-P. Hauri and H. Farhan (University of Basel, Basel, Switzerland). Rabbit anti–human Sec13 antibody was provided by R. Schekman and B. Lesch (University of California, Berkeley, Berkeley, CA). The secondary antibodies used were horseradish peroxidase–conjugated anti–mouse immunoglobulin G (IgG), anti–rabbit IgG (GE Healthcare), anti–rabbit IgG-Fc specific (Jackson ImmunoResearch Laboratories, Inc.), anti–mouse IgG-Fcγ specific (Jackson ImmunoResearch Laboratories, Inc.), anti–mouse IgG Trueblot (eBioscience), phycoerythrin (PE)-conjugated goat anti–mouse IgG (BD), Alexa Fluor 594–conjugated goat anti–mouse IgG (Invitrogen), cyanine 5 (Cy5)-conjugated anti–rabbit IgG (Jackson ImmunoResearch Laboratories), and Cy3-conjugated anti–mouse IgG (Jackson ImmunoResearch Laboratories).
Plasmids
Plasmid pME-VSVGex-FF-mEGFP-GPI (VFG-GPI) encoded a reporter protein consisting of the extracellular domain of VSVGts (temperature-sensitive VSVG, a vesicular stomatitis virus G protein), a furin cleavage site, a Flag tag, mEGFP (modified EGFP) and a GPI attachment signal (Maeda et al., 2008; Takida et al., 2008). Plasmid pME-VSVGex-FF-mEGFP-TM (VFG-TM) contained a sequence for a non-GPI reporter protein consisting of the extracellular domain of VSVGts, a furin cleavage site, a Flag tag, mEGFP, and a carboxyl terminal region of human CD3ε containing a transmembrane domain. The construction of pME-puro-Venus-Flag-CD59 (Venus-CD59) has been described previously (Rivier et al., 2010). To construct pSINsi-hU6-BSD, which was an expression plasmid for siRNA and could be selected with blasticidin, the neomycin resistance gene in pSINsi-hU6 (Takara Bio Inc.) was replaced with the blasticidin resistance gene. Sequences of siRNA targeting p23 were selected using BLOCK-iT RNAi Designer (Invitrogen) and cloned into pSINsi-hU6-BSD, generating pSINsi-hU6-BSD-242 that contained the p23 siRNA sequence of 5′-GCCATATTCTGTATGCCAA-3′. This sequence was confirmed to interfere with p23 mRNA specifically as described previously (Takida et al., 2008). The DNA fragments bearing the hU6 promoter and the blasticidin resistance gene in pSINsi-hU6-BSD-242 were integrated into pLIB2, generating pLIB2-242BSD. To construct pLIB2-242BSD-myc-p23, myc-p23 was amplified by PCR from pCB6-mycp23 (Emery et al., 2000), which was provided by J. Gruenberg, and integrated into pLIB2-242BSD. To construct pLIB2-BSD-Sec13-myc, Sec13-myc was amplified by PCR with pCS2-Sec13-myc, which was provided by T. Lee (Carnegie Mellon University, Pittsburgh, PA), and integrated into pLIB2-BSD.
Isolation of FPRC2 mutant cells defective in PGAP1
FF8 cells were treated with 1 µg/ml _N_-methyl-_N_’-nitro-_N_-nitrosoguanidine for 20 h and cultured for 1 wk. The cell sorting was performed based on a transport assay, as described in “Transport assay of reporter proteins by flow cytometry,” in the first and second rounds of cell sorting using a cell sorter (FACSAria; BD). The cells in which GPI-APs were resistant to treatment with PI-PLC were sorted in the third round. The population obtained by this sorting procedure was subjected to limiting dilution, and a clone in which GPI-APs, CD59, and DAF were resistant to PI-PLC was selected and designated the FPRC2 cell line. Transfection of PGAP1 fully restored PI-PLC resistance of GPI-APs in FPRC2 cells.
Transport assay of reporter proteins by flow cytometry
Cells derived from FF8 cells were cultured in complete medium containing 1 µg/ml doxycycline at 40°C for 24 h, harvested with trypsin-EDTA solution (Sigma-Aldrich), and incubated in complete medium at 32°C. The cells were stained with an M2 anti-Flag antibody and PE-conjugated goat anti–mouse IgG, and analyzed using a FACSCanto (BD) or FACSAria. In some cases, cells were transiently transfected with expression plasmids for reporter proteins using electroporation. After 1.5 d, the temperature was increased to 40°C and the cells were cultured for a further 24 h, followed by incubation at 32°C for the required times. In addition to the first gating (forward scatter and side scatter), a second gating to select cells expressing the same amount of reporter protein (brightness of EGFP in FL1) was performed for analysis by FlowJo software (Tree Star).
Sorting of cargo proteins into the ERES
FF8, FPRC2, and C19 cells stably expressing Sec13-myc were cultured on gelatin-coated and acid-washed 12-mm-diameter glass coverslips at 37°C for 1 d, followed by incubation with 1 µg/ml doxycycline at 40°C for 24 h to induce VFG-GPI expression and accumulation in the ER. The media were then changed to pre-chilled Ham’s F12 containing 10% FCS, 20 mM Hepes, pH 7.4, and 100 µg/ml cycloheximide. After incubating at 10°C in a water bath for the required time, the cells were washed once with PBS, fixed with 4% paraformaldehyde in PBS for 30 min, then washed twice with PBS and incubated with 10 mM ammonium chloride in PBS. The cells were permeabilized and blocked with blocking buffer A (0.1% saponin, 1% BSA, and 0.1% sodium azide in PBS) for 1 h. The cells were then incubated with anti-myc antibody diluted 1:300 in blocking buffer B (0.1% saponin, 2.5% goat serum, and 0.05% sodium azide in PBS) for 1 h. After washing three times with blocking buffer A, the cells were incubated with Alexa Fluor 594–conjugated goat anti–mouse IgG (1:800) in blocking buffer A for 1 h. After washing three times with blocking buffer A, the coverslips were mounted in Prolong Gold antifade reagent (Invitrogen). Images of the stained cells were acquired by a FluoView FV1000 confocal microscope (Olympus). VFG-GPI and Alexa Fluor 594 bound to Sec13-myc were excited by wavelengths of 488 nm and 559 nm, and the emitted fluorescence was captured at 505–540 nm and 575–675 nm, respectively. Regions of interest (ROI) indicating ERES (ROI(eres)) were automatically set using MetaMorph software Ver. 3.0 (Molecular Devices). The ROI(eres)s were transferred to VFG-GPI images, and the average emission intensities of ROI(eres) (AI(eres)) and those of the surrounding regions (AI(surro)) were measured. A journal was programmed for the automatic measurements on MetaMorph. The ratios of AI(eres) to AI(surro) were calculated at each ERES and time point.
To observe Venus-CD59 in p23 knockdown cells, clonal CHO-K1 cells expressing Venus-CD59 were transfected with siRNA or negative siRNA (QIAGEN) using Lipofectamine 2000 (Invitrogen). After 24 h, cells were detached and cultured for 2d on acid-washed 12-mm-diameter glass coverslips at 37°C. To visualize internal Venus-CD59 signals, the cells were washed with Opti-MEM twice and incubated with 0.3 U/ml PI-PLC (Invitrogen) in Opti-MEM at 37°C for 40 min. After incubating at 10°C for 1 h in complete Ham’s F12 medium supplemented with 20 mM Hepes-HCl, pH 7.4, the cells were washed twice with PBS containing Ca2+ and Mg2+, PBS(+), fixed in 4% paraformaldehyde in PBS for 8 min, then washed three times with 10 mM glycine in PBS(+) to block residual paraformaldehyde. The cells were permeabilized and blocked with 3% BSA in PBS (+) containing 0.05% (wt/vol) saponin for 30 min at room temperature. After washing with 0.1% BSA in PBS (+), the cells were incubated with anti-Sec13 antibody (1: 10,000), anti-p23 antibody (1:1,000), and/or anti-myc antibody (1:50) in 1% BSA in PBS (+) for 1 h. For staining with anti-calreticulin (1:100) and anti-GPP130 (1:800), cells were permeabilized with 0.1% Triton X-100. After washing three times with 0.1% BSA in PBS (+), the cells were incubated with Cy5-conjugated anti–rabbit IgG (1:200) and/or Cy3-conjugated anti–mouse IgG (1:200) in 1% BSA in PBS (+) for 1 h. After washing three times with 0.1% BSA in PBS (+), the coverslips were mounted in Prolong Gold antifade reagent. Micrographs were acquired with a 100× 1.4 numerical aperture oil objective lens with a microscope (AXIOZ1) and a charge-coupled device camera (AxioCam MRm) controlled by the software AxioVisionRel.4.6 (all from Carl Zeiss). The data were processed with ImageJ software.
Identification of proteins that associated with VFG-GPI
FF8, FPRC2, and C19 cells (4 × 107) were cultured in complete medium containing 1 µg/ml doxycycline at 40°C for 24 h, harvested with trypsin-EDTA solution, and incubated in complete medium at 32°C for 20 min. The cells were then centrifuged, washed once with PBS, and lysed in 1.2 ml of lysis-IP buffer I (1% digitonin, 20 mM MES/Hepes, pH 7.4, 100 mM NaCl, and protease inhibitor mixture) for 1 h. Insoluble material was removed by centrifugation at 20,000 g for 15 min, and the VFG-GPI was precipitated from the supernatants with anti-Flag beads. The VFG-GPI and coprecipitated proteins were washed five times with wash buffer I (0.5% digitonin, 20 mM MES/Hepes, pH 7.4, and 100 mM NaCl), and eluted with wash buffer I containing 500 µg/ml Flag peptide for 2 h. Proteins were analyzed by silver staining or Western blotting. Protein samples were digested in-gel with trypsin, and analyzed by nanocapillary reversed-phase Liquid chromatography– tandem mass spectrometry (LC-MS/MS) using a C18 column (inner diameter of 75 mm) on a nanoLC system (Ultimate, LC Packing) coupled to a quadruple time-of-flight mass spectrometer (QTOF Ultima; Waters). Proteins were identified by database searching using Mascot Daemon (Matrix Science).
For the pH elution, FF8 and FPRC cells (8 × 107) were cultured in complete medium containing 1 µg/ml doxycycline at 40°C for 24 h, harvested with trypsin-EDTA solution, and incubated in complete medium at 32°C for 20 min. The cells were then centrifuged, washed once with PBS, and lysed in 4.8 ml of lysis-IP buffer II (1% digitonin, 20 mM MES/Hepes, pH 8.0, 100 mM NaCl, and protease inhibitor mixture) for 1 h. Insoluble material was removed by centrifugation at 20,000 g for 15 min, and VFG-GPI was purified with an anti-Flag column, followed by washing with 50 column volumes of wash buffer II (0.5% digitonin, 20 mM MES/Hepes, pH 8.0, and 100 mM NaCl), and eluted with elution buffer II (0.5% digitonin, 20 mM MES/Hepes, pH 6.0, and 100 mM NaCl). The eluted proteins were analyzed by silver staining and Western blotting. Proteins were determined by LC-MS/MS as described earlier in this section.
Immunoprecipitation of VFG-GPI
Cells (107) were cultured in complete medium containing 1 µg/ml doxycycline at 40°C for 24 h, harvested with trypsin-EDTA solution, and incubated in complete medium at 32°C for 20 min. The cells were then centrifuged, washed once with PBS, and lysed in 600 µl of lysis-IP buffer III (1% digitonin, 20 mM MES/Hepes, pH 7.4, or indicated pH, 100 mM NaCl, and protease inhibitor mixture) for 1 h. Insoluble material was removed by centrifugation at 20,000 g for 15 min, and the VFG-GPI was precipitated from the supernatants with anti-Flag beads. The VFG-GPI and coprecipitated proteins were washed five times with wash buffer III (0.5% digitonin, 20 mM MES/Hepes, pH 7.4 or indicated pH, and 100 mM NaCl), boiled in SDS sample buffer, and analyzed by Western blotting. For the pH release experiments, cells (6 × 107) were lysed in Lysis-IP buffer III, pH 7.4. After washing with wash buffer III, pH 7.4, five times, the beads were divided into six aliquots and incubated in 80 µl of wash buffer III (at the indicated pH) for 15 min at 4°C, and the supernatant was then collected.
Immunoprecipitation of p24 complex
FF8 cells expressing myc-tagged p23 (2 × 107 cells/sample) were quickly harvested with trypsin-EDTA solution, centrifuged, washed once with PBS, and lysed in 600 µl of lysis-IP buffer I for 1 h. Insoluble material was removed by centrifugation at 20,000 g for 15 min, and the myc-tagged p23 was precipitated from the supernatants with or without 1.5 µg of anti-HA or anti-myc antibody, followed by addition of 40 µl of protein G–Sepharose (GE). After 2 h of incubation at 4°C, precipitated proteins were washed five times with wash buffer I, boiled in SDS sample buffer, and analyzed by Western blotting.
Quantitative reverse-transcription PCR analysis
For RT-PCR, total RNA was prepared using RNeasy Mini kit (QIAGEN) and reverse transcribed using SuperScript VILO reverse transcription (Invitrogen). TaqMan probes-conjugated with FAM and primers were designed by Primer Express software (Applied Biosystems). For hamster β-actin (ACTB), the TaqMan probe sequence was 5′-CCTTCCTTCCTGGGTATG-3′, and the primer sequences were 5′-TGCCCTGAGGCTCTTTTCC-3′ (forward) and 5′-TCGTGGATGCCACAGGATT-3′ (reverse). For hamster p23, the TaqMan probe sequence was 5′-CTGTATGCCAAGGAAG-3′, whereas the primer sequences were 5′-ACAGATTCTGCGGGCCATAT-3′ (forward) and 5′-AGCAAATTTCCCCTTAGTTGCA-3′ (reverse). For hamster p24, the TaqMan probe sequence was 5′-CACACGCAGAGGAGT-3′, and the primer sequences were 5′-TCGGGCTACTTCGTCAGCAT-3′ (forward) and 5′-GGTGACCCGCTCGAAGAA-3′. The qPCR was performed and analyzed with the StepOne Real-Time PCR System (Applied Biosystems). RNA expression was normalized to the expression of ACTB and relative expression was calculated by the −ΔΔCT method.
RNAi by oligonucleotides
Oligonucleotides for RNAi were purchased from QIAGEN (p23 siRNA sequence: 5′-CAGGCCATATTCTGTATGCCA-3′). AllStars negative control siRNA (QIAGEN) was used as a nonsilencing negative control. Oligonucleotides were transfected using Lipofectamine 2000 according to the manufacturer’s instructions.
Pulse-chase metabolic labeling
The CHO-K1 cells expressing Venus-CD59 transfected with siRNA or negative control siRNA were cultured at 80–100% confluence in 6 cm dishes. Cells were preincubated with DME lacking l-methionine and l-cysteine for 30 min and then pulsed with 100 µCi/ml 35S-methionine and 35S-cysteine (Easy Tag Expre35S35S; PerkinElmer) for 30 min. The chase reaction was started by exchange to complete Ham’s F12 medium supplemented with excess amount of cold l-methionine and l-cysteine (1.5 mM each). At each time point, cell metabolism was immediately stopped by addition of 10 mM NaF/NaN3. Cells were collected by scraping and solubilized with RIPA buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1% Triton X-100, 0.5% deoxycholate, and 0.1% SDS) plus protease inhibitors (10 µg/ml leupeptin, 5 µg/ml pepstatin A, and 2 µg/ml aprotinin) and immunoprecipitated with anti–FLAG-agarose beads. The beads were incubated with 0.5 mU of Endo-H (Roche) in 100 mM NaOAc, pH 5.2, 0.8% 2-mercaptoethanol, and 0.2% SDS for overnight.
PI-PLC treatment
Samples containing 1−2 × 106 cells were incubated in 50 µl of Ham’s F12 medium containing 0.1% BSA, 0.5 mM EDTA, and 50 mU PI-PLC (final concentration, 1 U/ml; Invitrogen) at 37°C for 1 h. The cells were stained with anti-CD59 and PE-conjugated anti–mouse IgG, then analyzed by flow cytometry.
Homology modeling of GOLD domain
Structural models of GOLD domains of p23 and p24 were constructed using the homology modeling software application MODELLER ver.9.8 (Martí-Renom et al., 2000; Eswar et al., 2006). Threading was performed using Phyre server (Kelley and Sternberg, 2009), and the coordinate of Sec14-like protein 2, supernatant protein factor (PDB accession no. 1OLM) was used as a template (Stocker et al., 2002; Stocker and Baumann, 2003). The quality of the resultant models was evaluated by PROCHECK (Laskowski et al., 1993), which gave Ramachandran plots and the geometric parameters within the allowed conformational space. Figures of the structures were prepared using PyMOL (http://www.pymol.org/).
Online supplemental material
Fig. S1 shows isolation of FPRC2 cells defective in PGAP1. Fig. S2 shows immunoprecipitation of VFG-GPI with p23 and p24. Fig. S3 shows colocalization of Venus-CD59 with p23. Fig. S4 shows identification of proteins associated with VFG-GPI in pH-dependent manner. Fig. S5 shows construction of myc-tagged p23 and modeling of p23 and p24. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201012074/DC1.
Acknowledgments
We would like to thank Manuel Muñiz (University of Seville) and Guillaume A. Castillon (University of Geneva) for helpful discussions and sharing data; Yasu S. Morita (Osaka University), Yoshiko Murakami (Osaka University), and Shinya Hanashima (RIKEN Advanced Study Institute) for helpful discussion; Tina Lee, Felix Wieland, Inge Reckmann, Jean Gruenberg, Randy Schekman, Bob Lesch, Hans-Peter Hauri, Hesso Farhan, and Toshio Kitamura for reagents; Keiko Kinoshita and Noriyuki Kanzawa (Osaka University) for technical help; Masafumi Koyama (Olympus) for microscopic analysis; Kazunobu Saito (Osaka University) for mass spectrometry; and Kohjiro Nakamura and Yuko Kabumoto (Osaka University) for assistance in cell sorting.
This work was supported by grant-in-aids from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to T. Kinoshita and M. Fujita), the Naito foundation (to M. Fujita), the Senri Life Science Foundation (to M. Fujita), a Swiss National Science Foundation professorship (to R. Watanabe), and support from the Swiss National Science Foundation (to H. Riezman). M. Fujita was supported by the Osaka University Global Center of Excellence Program.
Footnotes
Abbreviations used in this paper:
COPII
coat protein complex II
DAF
decay-accelerating factor
Endo-H
endoglycosidase-H
ERES
ER exit sites
ERGIC
ER–Golgi intermediate compartment
EtNP
ethanolamine phosphate
GPI
glycosylphosphatidylinositol
GPI-AP
GPI-anchored protein
PE
phycoerythrin
PI-PLC
phosphatidylinositol-specific phospholipase C
VFG
VSVGex-Flag-GFP
References
- Aguilera-Romero A., Kaminska J., Spang A., Riezman H., Muñiz M. 2008. The yeast p24 complex is required for the formation of COPI retrograde transport vesicles from the Golgi apparatus. J. Cell Biol. 180:713–720 10.1083/jcb.200710025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appenzeller C., Andersson H., Kappeler F., Hauri H.P. 1999. The lectin ERGIC-53 is a cargo transport receptor for glycoproteins. Nat. Cell Biol. 1:330–334 10.1038/14020 [DOI] [PubMed] [Google Scholar]
- Appenzeller-Herzog C., Hauri H.P. 2006. The ER-Golgi intermediate compartment (ERGIC): in search of its identity and function. J. Cell Sci. 119:2173–2183 10.1242/jcs.03019 [DOI] [PubMed] [Google Scholar]
- Bagnat M., Keränen S., Shevchenko A., Shevchenko A., Simons K. 2000. Lipid rafts function in biosynthetic delivery of proteins to the cell surface in yeast. Proc. Natl. Acad. Sci. USA. 97:3254–3259 10.1073/pnas.060034697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlowe C. 2003. Signals for COPII-dependent export from the ER: what’s the ticket out? Trends Cell Biol. 13:295–300 10.1016/S0962-8924(03)00082-5 [DOI] [PubMed] [Google Scholar]
- Belden W.J., Barlowe C. 1996. Erv25p, a component of COPII-coated vesicles, forms a complex with Emp24p that is required for efficient endoplasmic reticulum to Golgi transport. J. Biol. Chem. 271:26939–26946 10.1074/jbc.271.43.26939 [DOI] [PubMed] [Google Scholar]
- Belden W.J., Barlowe C. 2001. Role of Erv29p in collecting soluble secretory proteins into ER-derived transport vesicles. Science. 294:1528–1531 10.1126/science.1065224 [DOI] [PubMed] [Google Scholar]
- Blum R., Lepier A. 2008. The luminal domain of p23 (Tmp21) plays a critical role in p23 cell surface trafficking. Traffic. 9:1530–1550 10.1111/j.1600-0854.2008.00784.x [DOI] [PubMed] [Google Scholar]
- Blum R., Pfeiffer F., Feick P., Nastainczyk W., Kohler B., Schäfer K.H., Schulz I. 1999. Intracellular localization and in vivo trafficking of p24A and p23. J. Cell Sci. 112:537–548 [DOI] [PubMed] [Google Scholar]
- Bonifacino J.S., Glick B.S. 2004. The mechanisms of vesicle budding and fusion. Cell. 116:153–166 10.1016/S0092-8674(03)01079-1 [DOI] [PubMed] [Google Scholar]
- Bonnon C., Wendeler M.W., Paccaud J.P., Hauri H.P. 2010. Selective export of human GPI-anchored proteins from the endoplasmic reticulum. J. Cell Sci. 123:1705–1715 10.1242/jcs.062950 [DOI] [PubMed] [Google Scholar]
- Bosson R., Jaquenoud M., Conzelmann A. 2006. GUP1 of Saccharomyces cerevisiae encodes an O-acyltransferase involved in remodeling of the GPI anchor. Mol. Biol. Cell. 17:2636–2645 10.1091/mbc.E06-02-0104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremser M., Nickel W., Schweikert M., Ravazzola M., Amherdt M., Hughes C.A., Söllner T.H., Rothman J.E., Wieland F.T. 1999. Coupling of coat assembly and vesicle budding to packaging of putative cargo receptors. Cell. 96:495–506 10.1016/S0092-8674(00)80654-6 [DOI] [PubMed] [Google Scholar]
- Brown D.A., London E. 1998. Structure and origin of ordered lipid domains in biological membranes. J. Membr. Biol. 164:103–114 10.1007/s002329900397 [DOI] [PubMed] [Google Scholar]
- Castillon G.A., Watanabe R., Taylor M., Schwabe T.M., Riezman H. 2009. Concentration of GPI-anchored proteins upon ER exit in yeast. Traffic. 10:186–200 10.1111/j.1600-0854.2008.00857.x [DOI] [PubMed] [Google Scholar]
- Castillon G.A., Aguilera-Romero A., Manzano J., Epstein S., Kajiwara K., Funato K., Watanabe R., Riezman H., Muñiz M. Mol. Biol. Cell. 2011 doi: 10.1091/mbc.E11-04-0294. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dancourt J., Barlowe C. 2010. Protein sorting receptors in the early secretory pathway. Annu. Rev. Biochem. 79:777–802 10.1146/annurev-biochem-061608-091319 [DOI] [PubMed] [Google Scholar]
- Denzel A., Otto F., Girod A., Pepperkok R., Watson R., Rosewell I., Bergeron J.J., Solari R.C., Owen M.J. 2000. The p24 family member p23 is required for early embryonic development. Curr. Biol. 10:55–58 10.1016/S0960-9822(99)00266-3 [DOI] [PubMed] [Google Scholar]
- Elrod-Erickson M.J., Kaiser C.A. 1996. Genes that control the fidelity of endoplasmic reticulum to Golgi transport identified as suppressors of vesicle budding mutations. Mol. Biol. Cell. 7:1043–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery G., Rojo M., Gruenberg J. 2000. Coupled transport of p24 family members. J. Cell Sci. 113:2507–2516 [DOI] [PubMed] [Google Scholar]
- Eswar N., Webb B., Marti-Renom M.A., Madhusudhan M.S., Eramian D., Shen M.Y., Pieper U., Sali A. 2006. Comparative protein structure modeling using Modeller. Curr. Protoc. Bioinformatics. Chapter 5:Unit 5.6 10.1002/0471250953.bi0506s15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M., Jigami Y. 2008. Lipid remodeling of GPI-anchored proteins and its function. Biochim. Biophys. Acta. 1780:410–420 [DOI] [PubMed] [Google Scholar]
- Fujita M., Kinoshita T. 2010. Structural remodeling of GPI anchors during biosynthesis and after attachment to proteins. FEBS Lett. 584:1670–1677 10.1016/j.febslet.2009.10.079 [DOI] [PubMed] [Google Scholar]
- Fujita M., Umemura M., Yoko-o T., Jigami Y. 2006. PER1 is required for GPI-phospholipase A2 activity and involved in lipid remodeling of GPI-anchored proteins. Mol. Biol. Cell. 17:5253–5264 10.1091/mbc.E06-08-0715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M., Maeda Y., Ra M., Yamaguchi Y., Taguchi R., Kinoshita T. 2009. GPI glycan remodeling by PGAP5 regulates transport of GPI-anchored proteins from the ER to the Golgi. Cell. 139:352–365 10.1016/j.cell.2009.08.040 [DOI] [PubMed] [Google Scholar]
- Gaskell A., Crennell S., Taylor G. 1995. The three domains of a bacterial sialidase: a beta-propeller, an immunoglobulin module and a galactose-binding jelly-roll. Structure. 3:1197–1205 10.1016/S0969-2126(01)00255-6 [DOI] [PubMed] [Google Scholar]
- Ghugtyal V., Vionnet C., Roubaty C., Conzelmann A. 2007. CWH43 is required for the introduction of ceramides into GPI anchors in Saccharomyces cerevisiae. Mol. Microbiol. 65:1493–1502 10.1111/j.1365-2958.2007.05883.x [DOI] [PubMed] [Google Scholar]
- Haass F.A., Jonikas M., Walter P., Weissman J.S., Jan Y.N., Jan L.Y., Schuldiner M. 2007. Identification of yeast proteins necessary for cell-surface function of a potassium channel. Proc. Natl. Acad. Sci. USA. 104:18079–18084 10.1073/pnas.0708765104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenne N., Frey K., Brugger B., Wieland F.T. 2002. Oligomeric state and stoichiometry of p24 proteins in the early secretory pathway. J. Biol. Chem. 277:46504–46511 10.1074/jbc.M206989200 [DOI] [PubMed] [Google Scholar]
- Kawasaki N., Ichikawa Y., Matsuo I., Totani K., Matsumoto N., Ito Y., Yamamoto K. 2008. The sugar-binding ability of ERGIC-53 is enhanced by its interaction with MCFD2. Blood. 111:1972–1979 10.1182/blood-2007-06-097022 [DOI] [PubMed] [Google Scholar]
- Kelley L.A., Sternberg M.J. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4:363–371 10.1038/nprot.2009.2 [DOI] [PubMed] [Google Scholar]
- Kinoshita T., Fujita M., Maeda Y. 2008. Biosynthesis, remodelling and functions of mammalian GPI-anchored proteins: recent progress. J. Biochem. 144:287–294 10.1093/jb/mvn090 [DOI] [PubMed] [Google Scholar]
- Kirk S.J., Ward T.H. 2007. COPII under the microscope. Semin. Cell Dev. Biol. 18:435–447 10.1016/j.semcdb.2007.07.007 [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., Moss D.S., Thornton J.M. 1993. Main-chain bond lengths and bond angles in protein structures. J. Mol. Biol. 231:1049–1067 10.1006/jmbi.1993.1351 [DOI] [PubMed] [Google Scholar]
- Lee M.C., Miller E.A., Goldberg J., Orci L., Schekman R. 2004. Bi-directional protein transport between the ER and Golgi. Annu. Rev. Cell Dev. Biol. 20:87–123 10.1146/annurev.cellbio.20.010403.105307 [DOI] [PubMed] [Google Scholar]
- Macedo-Ribeiro S., Bode W., Huber R., Quinn-Allen M.A., Kim S.W., Ortel T.L., Bourenkov G.P., Bartunik H.D., Stubbs M.T., Kane W.H., Fuentes-Prior P. 1999. Crystal structures of the membrane-binding C2 domain of human coagulation factor V. Nature. 402:434–439 10.1038/46594 [DOI] [PubMed] [Google Scholar]
- Maeda Y., Tashima Y., Houjou T., Fujita M., Yoko-o T., Jigami Y., Taguchi R., Kinoshita T. 2007. Fatty acid remodeling of GPI-anchored proteins is required for their raft association. Mol. Biol. Cell. 18:1497–1506 10.1091/mbc.E06-10-0885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda Y., Ide T., Koike M., Uchiyama Y., Kinoshita T. 2008. GPHR is a novel anion channel critical for acidification and functions of the Golgi apparatus. Nat. Cell Biol. 10:1135–1145 10.1038/ncb1773 [DOI] [PubMed] [Google Scholar]
- Mancias J.D., Goldberg J. 2008. Structural basis of cargo membrane protein discrimination by the human COPII coat machinery. EMBO J. 27:2918–2928 10.1038/emboj.2008.208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martí-Renom M.A., Stuart A.C., Fiser A., Sánchez R., Melo F., Sali A. 2000. Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 29:291–325 10.1146/annurev.biophys.29.1.291 [DOI] [PubMed] [Google Scholar]
- Marzioch M., Henthorn D.C., Herrmann J.M., Wilson R., Thomas D.Y., Bergeron J.J., Solari R.C., Rowley A. 1999. Erp1p and Erp2p, partners for Emp24p and Erv25p in a yeast p24 complex. Mol. Biol. Cell. 10:1923–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor S., Riezman H. 2004. Sorting GPI-anchored proteins. Nat. Rev. Mol. Cell Biol. 5:110–120 10.1038/nrm1309 [DOI] [PubMed] [Google Scholar]
- Miller E.A., Beilharz T.H., Malkus P.N., Lee M.C., Hamamoto S., Orci L., Schekman R. 2003. Multiple cargo binding sites on the COPII subunit Sec24p ensure capture of diverse membrane proteins into transport vesicles. Cell. 114:497–509 10.1016/S0092-8674(03)00609-3 [DOI] [PubMed] [Google Scholar]
- Muñiz M., Nuoffer C., Hauri H.P., Riezman H. 2000. The Emp24 complex recruits a specific cargo molecule into endoplasmic reticulum-derived vesicles. J. Cell Biol. 148:925–930 10.1083/jcb.148.5.925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura N., Inoue N., Watanabe R., Takahashi M., Takeda J., Stevens V.L., Kinoshita T. 1997. Expression cloning of PIG-L, a candidate N-acetylglucosaminyl-phosphatidylinositol deacetylase. J. Biol. Chem. 272:15834–15840 10.1074/jbc.272.25.15834 [DOI] [PubMed] [Google Scholar]
- Paroutis P., Touret N., Grinstein S. 2004. The pH of the secretory pathway: measurement, determinants, and regulation. Physiology (Bethesda). 19:207–215 [DOI] [PubMed] [Google Scholar]
- Rivier A.S., Castillon G.A., Michon L., Fukasawa M., Romanova-Michaelides M., Jaensch N., Hanada K., Watanabe R. 2010. Exit of GPI-anchored proteins from the ER differs in yeast and mammalian cells. Traffic. 11:1017–1033 10.1111/j.1600-0854.2010.01081.x [DOI] [PubMed] [Google Scholar]
- Rojo M., Emery G., Marjomäki V., McDowall A.W., Parton R.G., Gruenberg J. 2000. The transmembrane protein p23 contributes to the organization of the Golgi apparatus. J. Cell Sci. 113:1043–1057 [DOI] [PubMed] [Google Scholar]
- Schimmöller F., Singer-Krüger B., Schröder S., Krüger U., Barlowe C., Riezman H. 1995. The absence of Emp24p, a component of ER-derived COPII-coated vesicles, causes a defect in transport of selected proteins to the Golgi. EMBO J. 14:1329–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocker A., Baumann U. 2003. Supernatant protein factor in complex with RRR-alpha-tocopherylquinone: a link between oxidized Vitamin E and cholesterol biosynthesis. J. Mol. Biol. 332:759–765 10.1016/S0022-2836(03)00924-0 [DOI] [PubMed] [Google Scholar]
- Stocker A., Tomizaki T., Schulze-Briese C., Baumann U. 2002. Crystal structure of the human supernatant protein factor. Structure. 10:1533–1540 10.1016/S0969-2126(02)00884-5 [DOI] [PubMed] [Google Scholar]
- Strating J.R., Martens G.J. 2009. The p24 family and selective transport processes at the ER-Golgi interface. Biol. Cell. 101:495–509 10.1042/BC20080233 [DOI] [PubMed] [Google Scholar]
- Takida S., Maeda Y., Kinoshita T. 2008. Mammalian GPI-anchored proteins require p24 proteins for their efficient transport from the ER to the plasma membrane. Biochem. J. 409:555–562 10.1042/BJ20070234 [DOI] [PubMed] [Google Scholar]
- Tanaka S., Maeda Y., Tashima Y., Kinoshita T. 2004. Inositol deacylation of glycosylphosphatidylinositol-anchored proteins is mediated by mammalian PGAP1 and yeast Bst1p. J. Biol. Chem. 279:14256–14263 10.1074/jbc.M313755200 [DOI] [PubMed] [Google Scholar]
- Tashima Y., Taguchi R., Murata C., Ashida H., Kinoshita T., Maeda Y. 2006. PGAP2 is essential for correct processing and stable expression of GPI-anchored proteins. Mol. Biol. Cell. 17:1410–1420 10.1091/mbc.E05-11-1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umemura M., Fujita M., Yoko-O T., Fukamizu A., Jigami Y. 2007. Saccharomyces cerevisiae CWH43 is involved in the remodeling of the lipid moiety of GPI anchors to ceramides. Mol. Biol. Cell. 18:4304–4316 10.1091/mbc.E07-05-0482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe R., Funato K., Venkataraman K., Futerman A.H., Riezman H. 2002. Sphingolipids are required for the stable membrane association of glycosylphosphatidylinositol-anchored proteins in yeast. J. Biol. Chem. 277:49538–49544 10.1074/jbc.M206209200 [DOI] [PubMed] [Google Scholar]