The rocks and shallows of deep RNA sequencing: Examples in the Vibrio cholerae RNome (original) (raw)
Abstract
New deep RNA sequencing methodologies in transcriptome analyses identified a wealth of novel nonprotein-coding RNAs (npcRNAs). Recently, deep sequencing was used to delineate the small npcRNA transcriptome of the human pathogen Vibrio cholerae and 627 novel npcRNA candidates were identified. Here, we report the detection of 223 npcRNA candidates in V. cholerae by different cDNA library construction and conventional sequencing methods. Remarkably, only 39 of the candidates were common to both surveys. We therefore examined possible biasing influences in the transcriptome analyses. Key steps, including tailing and adapter ligations for generating cDNA, contribute qualitatively and quantitatively to the discrepancies between data sets. In addition, the state of 5′-end phosphorylation influences the efficiency of adapter ligation and C-tailing at the 3′-end of the RNA. Finally, our data indicate that the inclusion of sample-specific molecular identifier sequences during ligation steps also leads to biases in cDNA representation. In summary, even deep sequencing is unlikely to identify all RNA species, and caution should be used for meta-analyses among alternatively generated data sets.
Keywords: RNA-seq, cDNA, ncRNA, npcRNA, transcriptome
INTRODUCTION
Transcriptomes are complete sets of cellular RNAs and consist of two RNA classes: messenger RNAs (mRNAs) and various types of nonprotein-coding RNAs (npcRNAs). The first provide templates for protein synthesis; the second are involved in various regulatory functions exerted by the RNA itself or in complex with proteins as ribonucleoprotein complexes. npcRNAs participate in regulating diverse biochemical pathways, including chromosome modification, transcription and translation, splicing, developmental timing, cell differentiation, proliferation, apoptosis, and organ development. Hence, a detailed knowledge of the global transcriptome (i.e., all functional RNAs) and its regulation are essential for understanding the majority of biochemical pathways at the molecular level. Two general approaches for investigating global cellular transcriptomes in depth are commonly used. The first is based on hybridization (microarrays, etc.); the second involves deep sequencing of cDNAs, also termed RNA-seq (Wang et al. 2009).
Depending on the RNA starting material, it is often advantageous to enrich the resulting cDNA libraries for certain RNA species: for instance, RNA isolation from subcellular fractions or RNPs (Rederstorff and Huttenhofer 2010), RNA size fractionation on denaturing polyacrylamide gels to separate small RNAs (Cavaille et al. 2000; Tang et al. 2002), counterselection of certain RNAs (e.g., rRNA, tRNA) (Liu et al. 2009) or affinity purification to enrich defined RNA subsets (Mrazek et al. 2007; Sultan et al. 2008), and cap trapper methods to generate RNA libraries specifically from capped RNAs (Carninci et al. 1996; Carninci 2007; Levin et al. 2010). The resulting cDNAs are cloned or directly sequenced with or without a primary amplification step. Depending on the deep-sequencing technology used (Illumina IG, ABI SOLiD, Roche 454, etc.), sequence read lengths vary from 30 to 400 bp. In recent years, many studies were performed using RNA-seq to identify and analyze small npcRNAs in all kingdoms of life (Wang et al. 2009; Sharma et al. 2010). Particularly in bacteria, the overwhelmingly high numbers of sequence reads in combination with comparably small genome sizes led to the assumption that we were dealing with virtually complete npcRNA transcriptome coverage (Liu et al. 2009; Perkins et al. 2009; Beaume et al. 2010). In a proof-of-principle attempt, Liu et al. (2009) applied RNA-seq to analyze the npcRNA transcriptome of the human bacterial pathogen Vibrio cholerae O1 El Tor clinical isolate N16961. They subtracted tRNAs and 5S rRNA prior to constructing the cDNA library, as these RNAs are major constituents of the small npcRNA transcriptome, and their subtraction is thought to significantly increase the coverage of novel npcRNAs. From 407,039 “perfect sequence reads” they identified 627 npcRNA candidates (Liu et al. 2009). It should be noted that close inspection of the data indicates that many npcRNA candidates represent overlapping fragments of the same RNAs. Hence, the number of 627 candidates is likely an overestimation. At the same time, we were conducting a survey of V. cholerae O1 El Tor clinical isolate VC3321 based on a cDNA library construction method that relied on 3′-C-tailing and 5′-adapter ligation of RNA (Abu-Qatouseh et al. 2010; Chinni et al. 2010; Raabe et al. 2010). Here, we present the results of our survey of V. cholerae small npcRNAs and compare them with those of Liu et al. (2009). Motivated by the low level of overlap between the two surveys, we conducted experiments to identify potential sources of bias involved in the tailing and ligation (including the chemistry and sequences of the adapters) steps.
RESULTS
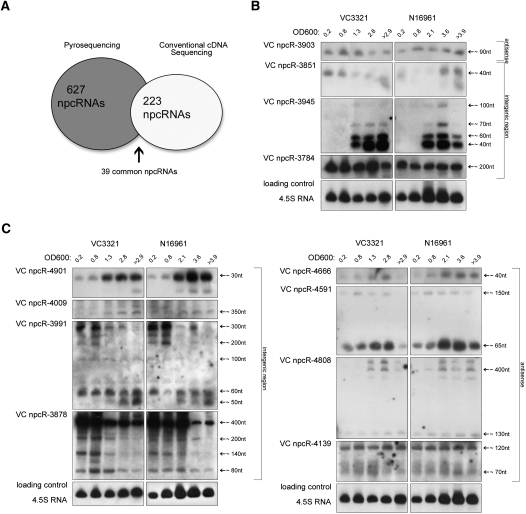
A total of 7500 randomly selected clones were sequenced using automated sequencing by chain termination (Supplemental Methods). From these we identified 223 npcRNA candidates (Supplemental Results; Supplemental Tables 1, 2; Supplemental Fig. 1A); 91 belong to the class of putative _cis_-antisense npcRNAs (Supplemental Table 1), and 132 are, according to the current genome annotation, intergenic (Supplemental Table 2). Surprisingly, out of 223 npcRNA candidates, only 39 were also present in the data set of Liu et al. (2009) (Fig. 1A; Supplemental Table 3).
FIGURE 1.
Northern blot hybridization and statistical overview. (A) Graphical representation of candidate npcRNAs from Liu et al. (2009) (ultra-deep pyrosequencing) with those generated from our approach (conventional cDNA sequencing). (B) Examples of Northern blot hybridizations of npcRNA candidates that are shared in both data sets (Supplemental Table 3). (C) Northern blot hybridization analysis of npcRNA candidates found only in this study. (B,C) Northern blot analysis was performed on total RNA isolated from V. cholerae O1 El Tor clinical isolate VC3321 (VC3321) and V. cholerae O1 El Tor clinical isolate N16961 (N16961) at different OD600 values (Supplemental Fig. 1B), as shown at the top. npcRNA designations are indicated on the left and RNA sizes in nucleotides on the right.
To validate this unexpected result and exclude the remote possibility of V. cholerae clinical isolate-specific variations in npcRNA expression as a possible source of this discrepancy, we carried out Northern blot hybridization of npcRNA candidates on total RNA from both isolates at different stages of bacterial growth (Fig. 1B,C). From almost 200 RNA blots, we detected signals in 94. The remaining were devoid of signals for a number of possible reasons, including low abundance of the respective RNA species or hybridization probe performance. The 94 Northern blot-positive npcRNA candidates included 42 only identified by us, 38 only identified by Liu et al. (2009), and 14 that were present in both data sets (Fig. 1B,C; Supplemental Figs. 2–6). Of 12 npcRNAs that were selected for Northern blot analysis with total RNA from both isolates, all were positive (Fig. 1B,C). All 38 candidates that were specific to the Liu et al. (2009) data set gave positive signals on total RNA extracted from the V. cholerae O1 El Tor VC3321 isolate (Supplemental Figs. 4,5). Importantly, but not surprisingly, none of the Northern blots gave any indication of differential expression of any of the npcRNA candidates between the two V. cholerae isolates. At least for the 42 Northern-positive candidates that are specific to our survey, one could argue that their expression is restricted to early V. cholerae growth stages that Liu et al. (2009) did not use for library construction. However, except for four npcRNA candidates (VC npcR-3830, VC npcR-3852, VC npcR-3988, and VC npcR-4586), all others yielded signals at stages that should be represented in the Liu et al. (2009) libraries (Fig. 1C; Supplemental Figs. 2,3). Conversely, two of the 14 common candidates (VC npcR-4392 and VC npcR-4655) and seven of the 38 putative npcRNAs (IGR-201, IGR-510, IGR-1530, IGR-2243, IGR-6849, IGR-7595, and IGR-8196) that were specific to the Liu et al. (2009) set are only detected at early growth stages (Fig. 1B; Supplemental Figs. 4–6). This indicates that RNAs present below the Northern blot detection limit clearly have the potential to be covered by ultra-deep sequencing procedures. In summary, Northern blot analysis independently validated many npcRNA candidates of both data sets and excluded isolate-specific or growth-stage-dependent npcRNA expression as possible sources of the observed bias.
We then considered whether technical differences in cDNA library construction were the underlying cause of the relatively small intersection between the two data sets. The sources of possible bias for conventional and deep-sequencing protocols are summarized in Table 1. A number of steps during library construction can potentially lead to over- or under-representation of a given RNA. The procedures range from RNA preparation and selection (see the Introduction) to extensions of RNA 3′-ends (tailing or adapter ligation). Some of the variability that occurs during extensions are caused by differential accessibility of the RNA to reagents (e.g., adapters, enzymes), depending on RNA structure and modification of the 3′-terminal nucleotide(s) (Munafo and Robb 2010). On the other hand, variants of adapter sequences might ligate to the same RNA species with different efficiencies. The same may also be true of extensions at the 5′-ends of RNAs or at the 3′-ends of the first-strand cDNAs, depending on the cDNA synthesis protocol used. Often, these adapters include bar-code sequences for discriminating RNAs from different stages, or sources in parallel deep-sequencing runs. Even if they are as much as ∼20 nucleotides away from the end to be ligated, different bar-code tags might differentially influence ligation efficiency. Reverse transcription (RT) for first-strand DNA synthesis might be hampered by RNA structure and/or modification of nucleotides (Bakin and Ofengand 1993; Zhang et al. 2001; Hansen et al. 2002). Different enzyme preparations, buffer conditions, and reaction temperatures may all lead to different efficiencies of cDNA synthesis (Carninci et al. 1998). If cDNAs must be amplified prior to cloning or direct sequencing, bias can be generated by differential PCR efficiencies influenced by many parameters, including base composition, presence of repeats, and hairpin structures (Kieleczawa 2006). Finally, some cDNAs are less efficiently cloned into vectors than others for a variety of reasons. We would like to emphasize that for procedures 2–6 in Table 1, there is yet another possible source of variation that is more difficult to standardize from protocol to protocol, or from experiment to experiment, the denaturation step prior to RNA (or DNA) tailing or adapter ligation. Incomplete denaturation or fast renaturation of both RNA substrate and adapter might introduce great variation at these steps. In addition, alternative computational analyses of sequence reads, e.g., only allowing candidates from two independent sequencing runs (Liu et al. 2009), impact on overlap between data sets.
TABLE 1.
Sources of potential bias in RNA sequencing
The small number of sequence reads generated from our library does not permit general and statistically valid conclusions on their own. However, considering the huge amount of reads obtained by Liu et al. (2009) and the relatively large number of npcRNA candidates from our survey that are absent from the RNA-seq data, we thought it pertinent to examine the underlying causes. Consequently, we systematically examined the hitherto largely neglected steps 2, 3, 4, and 6 outlined in Table 1, using three defined in vitro-transcribed RNA test substrates: Yyb RNA, identified by Liu et al. (2009) but not by us; npcRNA-4640, identified in our study but not by Liu et al. (2009); and rat BC1 RNA (Rozhdestvensky et al. 2001, 2007). Variabilities in steps 1, 5, and 7–9 are common knowledge in Molecular Biology protocols or have been described elsewhere (Sambrook et al. 1989; Saxena and Carninci 2010).
C-tailing reaction
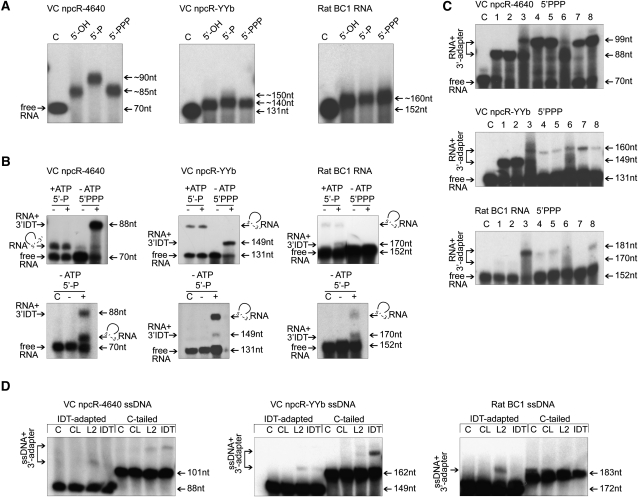
The three npcRNAs were radioactively labeled by incorporating [α32P]UTP during in vitro transcription of the PCR templates. In various cDNA construction protocols, the 5′-ends of the RNAs were differently modified or treated; hence, we monitored the influence of 5′-mono, 5′-triphosphate, and 5′-hydroxyl groups on 3′-end C-tailing efficiency and processivity. All templates examined were efficiently C-tailed (∼95% of RNA input). Interestingly, for each RNA substrate the resulting C-tail lengths displayed little variation, but among different RNA species these lengths ranged between ∼8 and ∼20 nucleotides (Fig. 2A). This contrasted remarkably with the poly(A)-tailing reaction catalyzed by the same enzyme, whereby the resulting tails were significantly longer and heterogeneous (data not shown). While we did not detect differences in C-tail lengths between the 5′-triphosphate and 5′-hydroxyl group-containing transcripts, the 5′-monophosphorylated VC npcR-4640 and VC YYb npcRNA substrates exhibited slightly longer C-tails (Fig. 2A). Addition of 10 μg of competing V. cholerae total RNA had no effects on the C-tailing processivity or its efficiency.
FIGURE 2.
Comparisons of C-tailing and adapter ligations to the 3′-ends of RNA or ssDNA templates. (A) 3′-end C-tailing reactions were performed with 5′-hydroxyl (5′-OH), 5′-monophosphate (5′-P), and 5′-triphosphate (5′-PPP) group-containing RNAs. The corresponding 5′-monophosphate RNAs served as controls (C). (B) IDT adapter (Liu et al. 2009) ligation to 3′-ends of 5′-monophosphorylated (5′-P) and 5′-triphosphorylated (5′-PPP) RNAs (Table 2). The presence or absence of the IDT adapter in the ligation reaction is indicated above the gel by “+” or “−”, respectively. +ATP or –ATP indicates the presence or absence, respectively, of ATP in the reactions. Potential RNA circularization products are depicted by half-circles. (C) Ligation of different adapters (1-IDT; 2-L2; 3-Ad1RNA; 4-Ad1; 5-UNC; 6-Ad7RNA; 7-Ad7; 8-Ad10) to 3′-ends of 5′-triphosphorylated (5′-PPP) RNAs (Table 2). The corresponding 5′-triphosphorylated RNAs served as controls (C). (D) DNA blot analysis for ligation of L2 and IDT adapters to tailed ssDNAs with ssDNA-specific probes. As controls, ssDNA was incubated in ligation buffer in the absence (C) or presence (CL) of T4 RNA ligase. (A_–_D) Values to the right represent molecular sizes.
Adapter ligation to the 3′-end of RNA
Next, we examined the efficiency of 3′-adapter ligation to 5′-mono- and 5′-triphosphorylated RNA templates. Three different RNA templates were ligated to a 5′-adenylated and 3′-blocked (dideoxycytidine, ddC) 18-mer linker (IDT-3′) harboring a 5′,5′-adenyl pyrophosphoryl moiety (App) (Fig. 2B; Table 2; Lau et al. 2001; Pak and Fire 2007; Liu et al. 2009). In the case of 5′-triphosphorylated npcRNAs, we observed the expected ligation for two of the three RNAs; however, 5′-triphosphorylated RNA templates that were efficiently ligated with the adapter did not perform well in the presence of ATP when they contained 5′-monophosphate groups. In contrast, for 5′-monophosphorylated BC1 RNA, we detected a ligation product, but the triphosphorylated transcript did not ligate with the IDT-3′ linker in our assay. When ATP was removed from the reactions, we observed RNA-adapter ligation with the 5′-monophosphate RNAs tested; however, the products of intramolecular ligation dominated (Fig. 2B). These data indicate that deadenylation of adapter by T4 RNA ligase is a reverse reaction of RNA adenylylation (Krug and Uhlenbeck 1982). Notably, in vivo rat BC1 and YYb RNAs harbor 5′-triphosphate groups, whereas the VC npcR-4640 RNA is likely the processing product of a bacterial operon (apo0230) (Argaman et al. 2001; Price et al. 2005; Rozhdestvensky et al. 2007). When individual ligation reactions were challenged by the addition of 10 μg of V. cholerae total RNA, no changes were observed. Currently, we are unable to explain why the phosphorylation state of the RNA 5′-end should influence ligation of the adapter to the 3′-end.
TABLE 2.
Oligonucleotides used in ligation assays
To investigate the ligation reaction in broader detail, we monitored RNA-to-adapter ligation efficiencies with different RNA templates and various RNA, DNA, and RNA/DNA chimeric oligonucleotides (Fig. 2C; Table 2). Our results demonstrate that the efficiency of RNA 3′-end adapter ligation depends on the RNA species, possibly its terminal sequences and structures, and on the phosphorylation state of the RNA 5′-terminus. In addition, ligation efficiency is strongly influenced by the type and sequence of the adapter utilized (Fig. 2B,C; Table 2). It is important to emphasize that the RNA C-tailing efficiency and processivity in some cases depends on the npcRNA 5′-phosphorylation state and possibly also on the RNA sequence and structure; however, sufficient C-tailing was observed for all transcripts tested (Fig. 2A).
Adapter ligation to the 3′-end of first strand cDNA
In the protocol used by Liu et al. (2009), reverse transcription was followed by adapter ligation to the 3′-OH terminus of the resulting single-stranded cDNA (ssDNAs). To investigate potentially biasing influences on the ssDNA-adapter ligation, we examined this reaction with in vitro-synthesized ssDNA templates (Supplemental Table 4). For consistency, we utilized ssDNA templates that resembled the reverse complement strand with respect to either 3′-C-tailed or 3′IDT-ligated RNAs, and we chose two 5′-adenylated linkers (L2 and IDT) to monitor cDNA 3′-end ligation efficiency. Interestingly, both adapters were ligated to ssDNAs with less efficiency than to the 5′-triphosphorylated npcR-4640 and YYb RNA templates (Fig. 2B–D). We were surprised to find that the efficiency of ligation was strongly influenced by the remote 5′-end of the DNA adapter. For instance, the IDT-adapted VC npcR-4640 DNA template only ligated to the L2 oligonucleotide, while the otherwise identical cDNA generated from the C-tailed template ligated with either of the tested adapters (Fig. 2D). In contrast, no ligation products were observed for BC1 cDNA generated from the C-tailed template (Fig. 2D).
Adapter ligation to the 5′-end of RNA
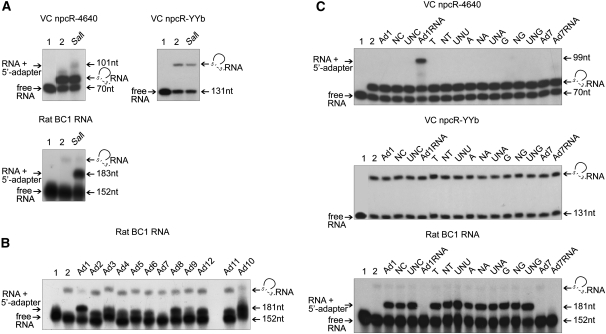
Our protocol relied on enriching the full-length cDNAs via DNA oligonucleotide ligation to the RNA 5′-ends. Therefore, we examined the efficiencies of ligating various 5′-adapters to the three RNA templates. Originally, BC1 RNA was chosen as an RNA control template in these reactions. The DNA SalI oligonucleotide adapter was empirically identified to perform best (data not shown); thus, our general cDNA library construction protocol was designed based on RNA 5′-adapter ligation with the SalI DNA oligonucleotide. When the 5′-monophosphorylated VC YYb and VC npcR-4640 RNAs were analyzed in ligation assays with the SalI adapter, the expected product was detected only for the VC npcR-4640 transcript (Fig. 3A; Table 2).
FIGURE 3.
Adapter ligation to 5′-ends of RNA templates with different oligonucleotides. (A) SalI adapter ligation to 5′-ends of 5′-monophosphorylated RNAs. (B) BC1 RNA ligation with fusion SalI adapters; the SalI restriction site was exchanged for the Roche MID tags (MID1–12), the resulting linkers being Ad1–12. (C) Different RNA templates ligated to Ad1 adapter derivates. Either 3′-nucleotides or sugars were altered (Table 2). (A–C) Lanes 1 and 2 represent control reactions. (Lane 1) Ligase and adapter were not added. (Lane 2) No adapter was added. Values to the right represent molecular sizes. Potential RNA circularization depicted as in Figure 2. (A–C) Oligonucleotides' sequence and abbreviations across the top of the gels are defined in Table 2.
RNA-seq technologies are used broadly to analyze differences in expression profiles during cell (organ) development, pathogen infections, different diseases, etc. In general, sample-specific molecular identifier (MID) sequences are introduced during cDNA construction so that different libraries can undergo high-throughput sequencing in parallel. We synthesized different fusion primers by exchanging the SalI restriction site with Roche MID tags (MID1–12) at the 5′-end of the SalI adapter (Table 2). The resulting DNA adapters (Ad1–Ad12) were ligated to the 5′-end of BC1 RNA. In response to sequence alterations in the 5′-ends of the adapter, we observed drastic variations in ligation efficiency (Fig. 3B). In the case of the MID7 tag (Ad7), we failed to detect any ligation. This variability was not only apparent with DNA-based adapters, but also with RNA-based oligonucleotides of the same sequence. Moreover, this variability was not restricted to BC1 RNA; other RNAs tested behaved similarly with Ad7 DNA- or RNA-based oligonucleotides (Fig. 3B,C). In contrast, the Ad1 adapter was ligated more efficiently than the original SalI linker (up to 60% of RNA input was ligated) (Fig. 3). These results demonstrate that changes, even at the 5′-end of the adapter, can drastically influence ligation efficiencies involving their 3′-ends. As a consequence, semiquantitative transcriptome analysis based on different MID identifiers that are fused to the 5′-end of the adapter might lead to false conclusions.
To examine the influences of the 3′-end compositions of adapters on ligation efficiencies, the 3′-terminal nucleotides of the AD1 adapter were permutated and compared in ligation reactions (Fig. 3C; Table 2). The AD7 adapter was included as a negative control. It is widely believed that RNA-based adapters greatly increase ligation efficiencies; however, Ad1 RNA- and Ad7 RNA-based adapters did not ligate to BC1 RNA at all, while various DNA or chimeric adapters performed well with BC1 RNA (Fig. 3C). Interestingly, reactions with the VC npcR-4640 RNA template were different again, as only the AD1 RNA-based oligonucleotide was ligated. No ligation products were detected for the VC YYb npcRNA template with any of the tested adapters.
DISCUSSION
We emphasize that the different results observed using the two V. cholerae small npcRNA sequencing approaches are not isolated examples. Recently, the bacterial small npcRNA transcriptomes of Salmonella typhi and Staphylococcus aureus were analyzed by Illumina-platform-based sequencing (Perkins et al. 2009; Beaume et al. 2010). Interestingly, the S. typhi meta-analysis identified only eight npcRNAs in common between the 40 novel Ilumina-based npcRNA candidates and a data set of 97 novel npcRNAs obtained by conventional sequencing methods (Perkins et al. 2009; Chinni et al. 2010). Similarly, the comparison of S. aureus intergenic npcRNAs revealed that only 17 candidates were recovered in common to both data sets (from a total of 65, versus 160 Ilumina-based intergenic candidates) (Abu-Qatouseh et al. 2010; Beaume et al. 2010).
Therefore, the RNA-seq methods commonly utilized (including the one reported here) might, on their own, be inefficient at completely covering even bacterial transcriptomes. Moreover, alternative methodologies of cDNA generation lead to different and, thus, incomparable outcomes. To achieve the highest possible RNA coverage, it is mandatory to utilize combinations of alternative and complementing methods of cDNA preparation. For instance, while various protocols aim to enrich for full-length cDNAs by RNA 5′-adapter ligation, our analysis of RNA 5′-adapter ligation efficiencies suggests a strong interdependence between adapter and RNA substrate. Hence, DNA, RNA, and chimeric 5′-adapters with identical sequences ligated with different efficiencies to given RNA templates. Therefore, a cocktail of several adapters should increase representation of different RNA species in deep-sequencing projects. Surprisingly, analysis of RNA 3′-adapter ligation revealed that even the phosphorylation state of the RNA 5′-ends drastically influences ligation efficiency (Fig. 2). It is suggested that the 5′-triphosphorylated form favors the RNA-adapter ligation by preventing self-ligation.
Beside these more technical considerations, it is obvious that the corresponding transcriptome analyses led to biased results due to enrichment for RNA primary transcripts, thereby leaving a number of processed 5′-monophosphorylated RNAs under-represented. However, it is impossible to draw general conclusions, because the phosphorylation state of the RNA influences 3′-terminal ligation efficiencies in a template-specific manner (Fig. 2). Additionally, the actual ratio of ligated product, nonligated template, and circular by-product is certainly a function of specific molecular concentrations and might vary accordingly. An alternative approach to modify RNA 3′-ends in preparation for subsequent cDNA synthesis is RNA C-tailing. Even though it is evident that the RNA C-tailing reaction catalyzed by poly(A) polymerase depends on 3′-end accessibility of the RNA, all three of our test RNAs were C-tailed equally well.
Alternative cDNA construction methods prefer A- over C-tailing, because both efficiency and processivity of the poly(A) polymerase are known to be higher in A-tailing reactions. However, 3′-poly(A) tails are common to eukaryotic mRNAs. In addition, a number of mammalian small npcRNAs harbor short oligo(A) stretches. Therefore, full-length sequences or natural polyadenylation would remain unidentified due to the addition of 3′-terminal poly(A) tails or internal priming during cDNA first-strand synthesis. RNA C-tailing offers a compromise between technically important RNA 3′-end modification and terminal nucleotide identification, with fewer possibilities of internal priming. Furthermore, gel purification is not required after RNA-tailing reactions, which is beneficial if the RNA starting material is available in limited amounts.
Because many methods of cDNA library construction favor cDNA 3′-adapter ligation, we examined this reaction for potential biases that might influence global transcriptome analyses. Our ssDNA ligation analysis revealed that the cDNA 5′-end sequence also impacts 3′-end ligation efficiencies. Beside obvious influences that stem from the cDNA sequence itself, the introduced RNA 3′-end modifications (tailing or adapters) might contribute to the efficiency of adapter ligation to cDNA 3′-ends (Fig. 2D). Here, too, a cocktail of different adapters might improve coverage of different RNA species.
Mammalian global transcriptome analyses based on tiling arrays suggested that major parts of the genomes were transcribed, indicating that they might encode many more as yet unidentified RNAs (Kapranov et al. 2002; Bertone et al. 2004; Cheng et al. 2005). Recent meta-analyses of deep sequencing and tiling array data challenged this assumption (van Bakel and Hughes 2009; van Bakel et al. 2010). The RNAs detected by deep sequencing only accounted for a minor fraction of the genome-wide transcription observed by microarrays. Of course, microarrays suffer from their own technical limitations: The low signal-to-noise ratio in hybridization experiments accompanied by high false-positive rates in transcript identification might explain the differences in meta-analyses to a certain extent, and biases introduced during cDNA construction should be seriously considered as an alternative explanation for the reported discrepancies.
CONCLUSION
Our data present an urgent warning that our understanding of transcriptomes and their corresponding regulation are still incomplete. It is an illusion to think that we can cover an entire transcriptome with a single library for ultra-deep sequencing, as they are all restricted by hitherto poorly addressed variations in methods and reagents (adapter sequences). Furthermore, comparisons of transcriptomes from different data sets generated by different methods or adapters should be made with caution—if at all.
MATERIALS AND METHODS
V. cholerae culture conditions
V. cholerae El Tor clinical isolates VC3321 (Ogawa) and N16961 (Inaba) were obtained from the Department of Medical Microbiology, School of Medical Sciences, Universiti Sains Malaysia. One hundred microliters of V. cholerae culture were aseptically inoculated into 10 mL of Lysogeny Broth (LB) medium and incubated at 37°C at 220 rpm. We then inoculated 250 mL of LB medium with 2.5 mL of overnight culture. Based on the observed growth curves (Supplemental Fig. 1B), cells were harvested during the lag, mid-log, and stationary phases. Total RNA from bacterial cells was extracted using the Trizol reagent (Invitrogen) according to the manufacturer's instructions.
cDNA library construction
Total RNA (200 μg) of the three aforementioned growth stages of V. cholerae El Tor clinical strain VC3321 were equally combined and size fractionated (10–500 nt) on 8% denaturing polyacrylamide gels (7 M urea, 1 X TBE buffer). Passive elution was conducted in 0.3 M NaOAc (pH 5.3) overnight at 4°C and size-selected RNA was ethanol precipitated. Subsequently, 5 μg of size-fractionated, heat-denatured RNA was treated with 5 U of tobacco acid pyrophosphatase (Epicenter) for 1 h at 37°C and C-tailed with poly(A) polymerase (15 U; Invitrogen) for 2 h at 37°C. C-tailing reactions were performed in volumes of 100 μL containing 50 mM Tris-HCl (pH 8.0), 300 mM NaCl, 10 mM MgCl2, 1 mM DTT, 0.4 mM EDTA, 2.5 mM MnCl2, 50 μg/mL BSA, 2 mM CTP, and 80 U of ribonuclease inhibitor (RiboLock, Fermentas) (DeChiara and Brosius 1987). The C-tailed RNAs were purified by phenol-chloroform extraction, followed by ethanol precipitation. Subsequently, the SalI adapter was ligated as described by Raabe et al. (2010). First-strand cDNA synthesis was carried out with the Transcriptor Reverse Transcriptase (Roche) and a NotI DNA primer (5′-GACTAGTTCTAGATCGCGAGCGGCCGCCCGGGGGGGGGGGGGGG-3′), followed by single-stranded PCR reactions using the SalI oligonucleotide. The PCR product was then double digested with SalI (Roche) and NotI (Roche) restriction endonucleases, ligated (T4 DNA ligase, Roche) into the pSPORT1 vector (Invitrogen), and then transformed into E. coli Top10 (Invitrogen) electrocompetent cells.
Northern blot analysis
Total RNA from V. cholerae El Tor clinical strains VC3321 and N16961 was separated on 8% polyacrylamide/7 M urea gels and electro-transferred to positively charged nylon membranes (Ambion BrightStar-Plus membranes). After UV cross-linking, the membranes were hybridized with npcRNA-specific, 32P-end-labeled oligonucleotide probes (Supplemental Table 4) overnight at temperatures ranging from 46°C to 58°C in 1 M sodium phosphate (pH 6.2), 7% sodium dodecyl sulfate (SDS). After hybridization, the membranes were washed twice with 0.2 M sodium phosphate, 1% (w/v) SDS buffer for 10 min at 40°C, and exposed to MS film (Kodak) at –80°C overnight.
Ligation of different adapters to 5′-ends of RNAs
npcRNA candidates were in vitro transcribed from PCR templates containing a T7 promoter using 250 U of T7 RNA polymerase (Fermentas), and gel purified. npcRNAs were radioactively labeled by incorporating [α-32P]UTP during the in vitro transcription. Subsequently, eluted npcRNAs were treated with Antarctic phosphatase (New England Biolabs) for 1 h at 37°C and 5′-phosphorylated using 10 U of T4 polynucleotide kinase (New England Biolabs) for 1 h at 37°C. Alternatively, to synthesize 5′-monophosporylated RNAs, in vitro transcripts were treated with 10 U of tobacco acid pyrophosphatase (Epicenter) for 1 h at 37°C. Each enzymatic reaction, apart from specific buffers, was supplemented with 40 U of ribonuclease inhibitor.
Different adaptors (Table 2) were ligated to the 5′-ends of heat-denatured RNAs using T4 RNA ligase (Fermentas). Reactions were performed in volumes of 30 μL containing 50 mM HEPES-NaOH (pH 8.0), 10 mM MgCl2, 10 mM DTT, 1 mM ATP, 15 U of T4 RNA ligase, ∼0.5 pmol of npcRNA templates, 100 pmol of adapter, and 40 U of ribonuclease inhibitor. Ligation reactions were initially incubated for 1 h at room temperature, then transferred to 30°C for 2 h. All samples were analyzed on denaturing 8% (w/v) polyacrylamide, 7 M urea gels in 1 X TBE buffer. Gels were dried and exposed to MS film (Kodak) at –80°C.
Ligation of different adapters to 3′-ends of RNAs
3′-end adapter ligation was performed as above. To prepare 5′-monophosporylated RNAs, in vitro-transcribed npcRNAs were treated with Antarctic phosphatase (New England Biolabs) for 1 h at 37°C and 5′-phosphorylated using [γ-32P]GTP and 10 U of T4 polynucleotide kinase (New England Biolabs) for 1 h at 37°C. Prior to ligation, adapters (Table 2) with 5′-OH groups were phosphorylated using 10 U of T4 polynucleotide kinase (New England Biolabs) for 1 h at 37°C.
Ligation of adapters to 3′-ends of ssDNAs
3′IDT and L2 adapters were ligated to the single-stranded DNAs (ssDNAs) that were in vitro synthesized by MWG and Biolegio (Supplemental Table 4). All obtained ssDNAs contained 5′-hydroxyl groups. Ligation was performed as described above; no ATP was added to the reaction. The resulting reaction products were resolved on denaturing 8% (w/v) polyacrylamide, 7 M urea gels in 1 X TBE and electro-transferred to positively charged nylon membranes (Ambion BrightStar-Plus membranes). After UV cross-linking, the membranes were hybridized with ssDNA-specific, 32P-end-labeled DNA oligonucleotide probes (Table 2; Supplemental Table 4) overnight at 56°C in 1 M sodium phosphate (pH 6.2), 7% SDS buffer. After hybridization, the membranes were washed twice with 0.2 M sodium phosphate, 1% (w/v) SDS buffer for 10 min at 50°C, and exposed to MS film (Kodak) at –80°C.
C-tailing of RNAs
C-tailing reactions with in vitro- transcribed, heat-denatured RNAs were performed as described above. RNAs containing 5′-monophosphate were prepared by tobacco acid pyrophosphatase treatment (see above). Notably, similar conditions were used when A-tailing control experiments were performed, except CTP was replaced with 2 mM ATP.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Ravichandran Manickam for providing V. cholerae O1 El Tor VC3321 and N16961 isolates and Marsha Bundman for editorial assistance. This work was supported by the National Genome Research Network (grant #NGFNIII 01GS0808 to J.B. and T.S.R.) as well as by the Deutsche Forschungsgemeinschaft (grant #BR 754/4-1 to J.B.). T.H.T. is supported by the Malaysia Ministry of Science, Technology and Innovation National E-Science Fund Grant No. 02-01-05-SF0156.
Footnotes
REFERENCES
- Abu-Qatouseh LF, Chinni SV, Seggewiss J, Proctor RA, Brosius J, Rozhdestvensky TS, Peters G, von Eiff C, Becker K 2010. Identification of differentially expressed small non-protein-coding RNAs in Staphylococcus aureus displaying both the normal and the small-colony variant phenotype. J Mol Med 88: 565–575 [DOI] [PubMed] [Google Scholar]
- Argaman L, Hershberg R, Vogel J, Bejerano G, Wagner EG, Margalit H, Altuvia S 2001. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr Biol 11: 941–950 [DOI] [PubMed] [Google Scholar]
- Bakin A, Ofengand J 1993. Four newly located pseudouridylate residues in Escherichia coli 23S ribosomal RNA are all at the peptidyltransferase center: analysis by the application of a new sequencing technique. Biochemistry 32: 9754–9762 [DOI] [PubMed] [Google Scholar]
- Beaume M, Hernandez D, Farinelli L, Deluen C, Linder P, Gaspin C, Romby P, Schrenzel J, Francois P 2010. Cartography of methicillin-resistant S. aureus transcripts: detection, orientation and temporal expression during growth phase and stress conditions. PLoS ONE 5: e10725 doi: 10.1371/journal.pone.0010725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertone P, Stolc V, Royce TE, Rozowsky JS, Urban AE, Zhu X, Rinn JL, Tongprasit W, Samanta M, Weissman S, et al. 2004. Global identification of human transcribed sequences with genome tiling arrays. Science 306: 2242–2246 [DOI] [PubMed] [Google Scholar]
- Carninci P 2007. Constructing the landscape of the mammalian transcriptome. J Exp Biol 210: 1497–1506 [DOI] [PubMed] [Google Scholar]
- Carninci P, Kvam C, Kitamura A, Ohsumi T, Okazaki Y, Itoh M, Kamiya M, Shibata K, Sasaki N, Izawa M, et al. 1996. High-efficiency full-length cDNA cloning by biotinylated CAP trapper. Genomics 37: 327–336 [DOI] [PubMed] [Google Scholar]
- Carninci P, Nishiyama Y, Westover A, Itoh M, Nagaoka S, Sasaki N, Okazaki Y, Muramatsu M, Hayashizaki Y 1998. Thermostabilization and thermoactivation of thermolabile enzymes by trehalose and its application for the synthesis of full length cDNA. Proc Natl Acad Sci 95: 520–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavaille J, Buiting K, Kiefmann M, Lalande M, Brannan CI, Horsthemke B, Bachellerie JP, Brosius J, Huttenhofer A 2000. Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc Natl Acad Sci 97: 14311–14316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S, Long J, Stern D, Tammana H, Helt G, et al. 2005. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science 308: 1149–1154 [DOI] [PubMed] [Google Scholar]
- Chinni SV, Raabe CA, Zakaria R, Randau G, Hoe CH, Zemann A, Brosius J, Tang TH, Rozhdestvensky TS 2010. Experimental identification and characterization of 97 novel npcRNA candidates in Salmonella enterica serovar Typhi. Nucleic Acids Res 38: 5893–5908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeChiara TM, Brosius J 1987. Neural BC1 RNA: cDNA clones reveal nonrepetitive sequence content. Proc Natl Acad Sci 84: 2624–2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen MA, Kirpekar F, Ritterbusch W, Vester B 2002. Post-transcriptional modifications in the A-loop of 23S rRNAs from selected archaea and eubacteria. RNA 8: 202–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Cawley SE, Drenkow J, Bekiranov S, Strausberg RL, Fodor SP, Gingeras TR 2002. Large-scale transcriptional activity in chromosomes 21 and 22. Science 296: 916–919 [DOI] [PubMed] [Google Scholar]
- Kieleczawa J 2006. Fundamentals of sequencing of difficult templates–an overview. J Biomol Tech 17: 207–217 [PMC free article] [PubMed] [Google Scholar]
- Krug M, Uhlenbeck OC 1982. Reversal of T4 RNA ligase. Biochemistry 21: 1858–1864 [DOI] [PubMed] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG, Bartel DP 2001. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294: 858–862 [DOI] [PubMed] [Google Scholar]
- Levin JZ, Yassour M, Adiconis X, Nusbaum C, Thompson DA, Friedman N, Gnirke A, Regev A 2010. Comprehensive comparative analysis of strand-specific RNA sequencing methods. Nat Methods 7: 709–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JM, Livny J, Lawrence MS, Kimball MD, Waldor MK, Camilli A 2009. Experimental discovery of sRNAs in Vibrio cholerae by direct cloning, 5S/tRNA depletion and parallel sequencing. Nucleic Acids Res 37: e46 doi: 10.1093/nar/gkp080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrazek J, Kreutmayer SB, Grasser FA, Polacek N, Huttenhofer A 2007. Subtractive hybridization identifies novel differentially expressed ncRNA species in EBV-infected human B cells. Nucleic Acids Res 35: e73 doi: 10.1093/nar/gkm244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munafo DB, Robb GB 2010. Optimization of enzymatic reaction conditions for generating representative pools of cDNA from small RNA. RNA 16: 2537–2552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak J, Fire A 2007. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science 315: 241–244 [DOI] [PubMed] [Google Scholar]
- Perkins TT, Kingsley RA, Fookes MC, Gardner PP, James KD, Yu L, Assefa SA, He M, Croucher NJ, Pickard DJ, et al. 2009. A strand-specific RNA-Seq analysis of the transcriptome of the typhoid bacillus Salmonella typhi. PLoS Genet 5: e1000569 doi: 10.1371/journal.pgen.1000569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Huang KH, Alm EJ, Arkin AP 2005. A novel method for accurate operon predictions in all sequenced prokaryotes. Nucleic Acids Res 33: 880–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raabe CA, Sanchez CP, Randau G, Robeck T, Skryabin BV, Chinni SV, Kube M, Reinhardt R, Ng GH, Manickam R, et al. 2010. A global view of the nonprotein-coding transcriptome in Plasmodium falciparum. Nucleic Acids Res 38: 608–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rederstorff M, Huttenhofer A 2010. cDNA library generation from ribonucleoprotein particles. Nat Protoc 6: 166–174 [DOI] [PubMed] [Google Scholar]
- Rozhdestvensky TS, Kopylov AM, Brosius J, Huttenhofer A 2001. Neuronal BC1 RNA structure: Evolutionary conversion of a tRNA(Ala) domain into an extended stem-loop structure. RNA 7: 722–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozhdestvensky TS, Crain PF, Brosius J 2007. Isolation and posttranscriptional modification analysis of native BC1 RNA from mouse brain. RNA Biol 4: 11–15 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T 1989. Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor,NY [Google Scholar]
- Saxena A, Carninci P 2010. Whole transcriptome analysis: what are we still missing? WIREs Syst Biol Med doi: 10.1002/wsbm.135 [DOI] [PubMed] [Google Scholar]
- Sharma CM, Hoffmann S, Darfeuille F, Reignier J, Findeiss S, Sittka A, Chabas S, Reiche K, Hackermuller J, Reinhardt R, et al. 2010. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature 464: 250–255 [DOI] [PubMed] [Google Scholar]
- Sultan M, Schulz MH, Richard H, Magen A, Klingenhoff A, Scherf M, Seifert M, Borodina T, Soldatov A, Parkhomchuk D, et al. 2008. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science 321: 956–960 [DOI] [PubMed] [Google Scholar]
- Tang TH, Bachellerie JP, Rozhdestvensky T, Bortolin ML, Huber H, Drungowski M, Elge T, Brosius J, Huttenhofer A 2002. Identification of 86 candidates for small non-messenger RNAs from the archaeon Archaeoglobus fulgidus. Proc Natl Acad Sci 99: 7536–7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bakel H, Hughes TR 2009. Establishing legitimacy and function in the new transcriptome. Brief Funct Genomics Proteomics 8: 424–436 [DOI] [PubMed] [Google Scholar]
- van Bakel H, Nislow C, Blencowe BJ, Hughes TR 2010. Most “dark matter” transcripts are associated with known genes. PLoS Biol 8: e1000371 doi: 10.1371/journal.pbio.1000371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M 2009. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10: 57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YJ, Pan HY, Gao SJ 2001. Reverse transcription slippage over the mRNA secondary structure of the LIP1 gene. Biotechniques 31: 1286–1294 [DOI] [PubMed] [Google Scholar]