Crystal structure of the cell division protein FtsA from Thermotoga maritima (original) (raw)
Abstract
Bacterial cell division requires formation of a septal ring. A key step in septum formation is polymerization of FtsZ. FtsA directly interacts with FtsZ and probably targets other proteins to the septum. We have solved the crystal structure of FtsA from Thermotoga maritima in the apo and ATP-bound form. FtsA consists of two domains with the nucleotide-binding site in the interdomain cleft. Both domains have a common core that is also found in the actin family of proteins. Structurally, FtsA is most homologous to actin and heat-shock cognate protein (Hsc70). An important difference between FtsA and the actin family of proteins is the insertion of a subdomain in FtsA. Movement of this subdomain partially encloses a groove, which could bind the C-terminus of FtsZ. FtsZ is the bacterial homologue of tubulin, and the FtsZ ring is functionally similar to the contractile ring in dividing eukaryotic cells. Elucidation of the crystal structure of FtsA shows that another bacterial protein involved in cytokinesis is structurally related to a eukaryotic cytoskeletal protein involved in cytokinesis.
Keywords: cell division/FtsA/FtsZ/X-ray
Introduction
Prokaryotic cell division proceeds via formation of a septum at midcell, circumferential invagination of the cytoplasmic membrane and synthesis of a peptidoglycan layer, for bacteria that possess a cell wall. The majority of the proteins involved in cell division are encoded by the fts family of genes, named after the most common pheno type of their mutants, filamentous temperature-sensitive growth. At the non-permissive temperature, cells grow but septation is blocked, leading to long, filamentous cells. The proteins encoded by ftsA, ftsI (PBP3), ftsK, ftsL, ftsN, ftsQ, ftsW, ftsZ and zipA are thought to be directly involved in septation (for reviews see Donachie, 1993; Lutkenhaus, 1997; Rothfield et al., 1999). The main division and cell wall genes form a cluster (dcw) (Ayala et al., 1994) at 2.5 min of the genetic map of Escherichia coli (Bachmann, 1983). One key component of septation is the FtsZ protein, which is conserved in almost all bacteria (Erickson, 1997). It is the first constitutional protein of the divisome, a hypothetical polyprotein complex believed to carry out prokaryotic cell division (Nanninga, 1991). In rapidly growing cells, FtsZ forms a ring-like structure at midcell, shortly after the ‘birth’ of the cell, which contracts as the cell divides. Before contraction, other components of the divisome are sequentially recruited to the FtsZ ring. Shortly after the FtsZ ring has formed, FtsA and ZipA are located at midcell. The assembly of the other components of the divisome is dependent on the presence of FtsA and FtsZ. FtsA is, after FtsZ, the most highly conserved protein of the divisome, although it is lacking in archaea (Rothfield et al., 1999). At present, it is not known how bacteria without FtsA complete septal invagination since, for most bacteria in which the ftsA gene is inactivated, the FtsZ ring will form but is unable to contract (Addinall et al., 1996). There is strong evidence that FtsA and FtsZ interact in vivo and that this interaction is essential for bacterial cells to survive. The requirement of a stoichiometric balance of FtsA and FtsZ for proper cell division was the first indication that these proteins might interact. Septation is blocked by overexpression of either FtsA or FtsZ, and this inhibitory effect can be suppressed by concomitant overexpression of the other protein (Dai and Lutkenhaus, 1992; Dewar et al., 1992; Begg et al., 1998). Further evidence for this interaction came from fluorescence microscopy. The distribution of FtsA fused to green fluorescent protein (GFP) follows the same pattern as the spirals formed by some mutants of FtsZ (Ma et al., 1996). Cell division is impaired if the localization of FtsA to the FtsZ ring is prevented. Recently, 8–10 highly conserved residues at the C-terminus of FtsZ have been shown to be important for the interaction with FtsA (Din et al., 1998; Ma and Margolin, 1999; Yan et al., 2000). The association appears to have co-evolved, since the chimera GFP–FtsA from Rhizobium meliloti is unable to localize to the FtsZ ring formed by E.coli FtsZ, whereas a ring-like structure is observed in the presence of R.meliloti FtsZ (Ma et al., 1997). Once bound to the FtsZ ring, FtsA may form a bridge between FtsZ molecules and membrane-anchored proteins or integral membrane proteins of the septum. Genetic evidence suggests that FtsA interacts with FtsI (PBP3) (Tormo et al, 1986; Weiss et al., 1999) and that the localization of FtsK, FtsL, FtsN and FtsQ to the septum is dependent on FtsA (Addinall et al., 1997; Wang and Lutkenhaus, 1998; Chen et al., 1999; Ghigo et al., 1999).
Elucidation of the structure of FtsZ has shown that it shares its fold and nucleotide-binding motif with tubulin (Löwe and Amos, 1998). Like tubulin, FtsZ forms polymers in a GTP-dependent manner (Mukherjee and Lutkenhaus, 1998; Lu et al., 2000). The functional and structural resemblance to tubulin strongly suggest that FtsZ is the ancestral homologue of tubulin (Löwe and Amos, 1998; Nogales et al., 1998a,b). Based on 3D-structure prediction methods, it has been postulated that FtsA belongs to the actin family of proteins (Bork et al., 1992; Kabsch and Holmes, 1995), which includes actin (Kabsch et al., 1990), heat-shock cognate (Hsc) protein (Flaherty et al., 1990), hexokinase (Anderson et al., 1978) and glycerol kinase (Faber et al., 1989). Here, we present the crystal structure of FtsA from the thermophilic eubacterium Thermotoga maritima, at 1.9 Å resolution (for the apo form) and 2.6 Å resolution (for the ATP-bound form).
Results
The T.maritima ftsA gene was cloned into the pHis17 vector (B.Miroux, personal communication) and expressed as a C-terminal His-tagged protein in C41(DE3) cells (Miroux and Walker, 1996). The protein was purified over a Ni2+–NTA column, followed by size-exclusion chromatography. The protein eluted as a single peak and was concentrated to 80 mg/ml. This is in contrast to the FtsA protein from E.coli, which forms aggregates at higher protein concentrations (J.Mingorance and M.Vicente, personal communication). Selenomethionine (SeMet)-substituted protein was expressed in the same cells, which are non-methionine auxotrophs. Methionine synthesis was metabolically inhibited and SeMet incorporation was checked by electrospray mass spectrometry (Materials and methods). Approximately 15 mg of SeMet-containing protein were purified from a 1 l culture.
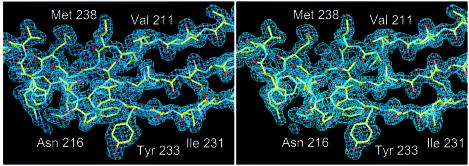
Native crystals were grown from 300 mM KCl, 50 mM MES pH 5.6, 5% polyethylene glycol (PEG) 6000. In order to grow crystals from SeMet-substituted protein, Mg-ADP was added and slightly different crystallization conditions were used, as described in Materials and methods. The crystals belonged to space group _P_21 and had variable cell constants (Table I). The phase problem was solved by multiple anomalous dispersion (MAD) methods, with crystals grown from SeMet-substituted protein. As the crystal form of the non-substituted protein (crystal form 1) was different from that of the SeMet-containing protein (crystal form 2), its structure was solved by molecular replacement using a preliminary model. Phases were improved with a mercury derivative of a crystal grown from the non-substituted protein. A high quality electron-density map based on crystal form 2 was obtained at 2.6 Å resolution. The model of crystal form 2 contains an Mg-ATP molecule, residues 6–319 and 327–392, and 71 water molecules, and was refined to an _R_-factor of 0.220 (Table II). The structure of crystal form 1 was solved at 1.9 Å resolution and refined to an _R_-factor of 0.221 (Table II). A representative portion of the final 2_F_o – _F_c map of crystal form 1 is shown in Figure 1.
Table I. Crystallographic data.
| Crystal form | λ (Å) | Resolution (Å) | I/σ_I_a | _R_mb (%) | Multiplicityc | Completeness (%)d | Occupanciese | _f_′/_f_″f |
|---|---|---|---|---|---|---|---|---|
| 1 | [space group _P_21 (4), a = 47.62, b = 53.52, c = 78.31 Å, β = 93.44°] | |||||||
| NATI | 0.9393 | 1.9 | 5.3 | 0.051 | 2.5 | 94.8 | ||
| PMA1 | 0.9393 | 2.4 | 7.2 | 0.056 | 3.0 | 99.0 | 0.457 (0.110) | |
| 2 | [space group _P_21 (4), a = 50.45, b = 73.74, c = 57.04 Å, β = 113.46°] | |||||||
| PEAK | 0.9787 | 2.6 | 4.3 | 0.057 | 6.0 (3.0) | 94.3 | 3.164 (4.697) | –5/6 |
| INFL | 0.9791 | 2.6 | 3.1 | 0.066 | 6.2 (3.1) | 97.4 | 0.000 (2.617) | –9/3 |
| HREM | 0.9393 | 2.6 | 3.3 | 0.063 | 6.3 (3.2) | 97.6 | 3.291 (2.159) | –5/2 |
Table II. Refinement statistics.
| | Crystal form | | | | ----------------------- | ----------------------- | ------------------------------- | | | 1 (empty) | 2 (ATP) | | | Model | 7–320, 327–390, 186 H2O | 6–319, 327–392, Mg, ATP, 71 H2O | | Diffraction data | NATI, 1.9 Å, all data | INFL, 2.6 Å, all data | | _R_-factor, _R_-freea | 0.221, 0.249 | 0.220, 0.282 | | B average/bonded (Å)b | 43.3, 2.0 | 26.3, 2.0 | | Geometry bonds/anglesc | 0.005 Å, 1.197° | 0.008 Å, 1.376° | | Ramachandrand (%) | 90.0/0.0 | 85.3/0.0 | | PDB IDe | 1E4F/r1E4FSF | 1E4G/R1E4GSF |
Fig. 1. Stereo drawing of the 2_F_o – _F_c electron-density map of crystal form 1 at 1.9 Å resolution. Residues 208–222 and 230–242 are shown. The figure was prepared with the program MAIN (Turk, 1992).
Structure
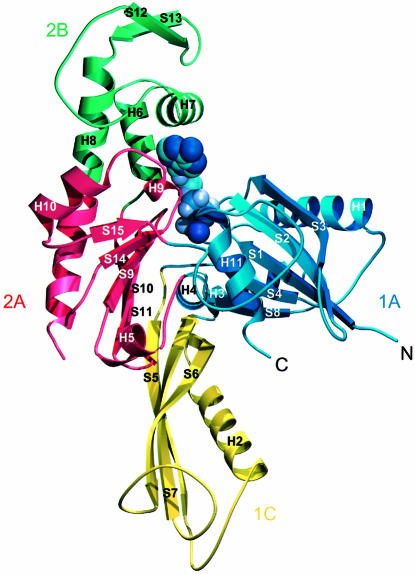
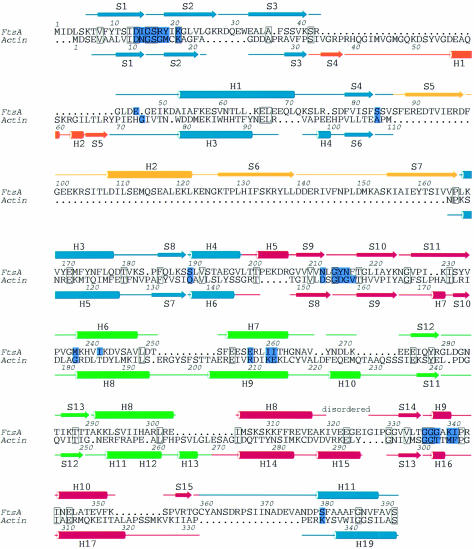
The 3D structure of FtsA resembles that of the actin family of proteins. Two domains with a common core form an interdomain cleft that contains a nucleotide-binding site. Each domain can be divided into two subdomains (Figure 2). The two larger subdomains, 1A and 2A, are similar to each other and consist of a five-stranded β-sheet, surrounded by three α-helices. The sheet is formed from a β-meander and a right-handed βαβ unit, and has strands in the order 8p4p1p2a3p for subdomain 1A and 15p14p9p10a11p for subdomain 2A. Both the topology and the size of the subdomains (109 and 90 residues for subdomain 1A and 2A, respectively) are similar, possibly because of evolution from a common ancestral domain (Bork et al., 1992; Kabsch and Holmes, 1995). The other two, smaller subdomains are more variable among the actin family and are composed of different combinations of strands and helices. The second subdomain of FtsA is located on the opposite side of the first domain from its position in actin. The position of the second subdomain was unexpected and was not recognized in the prediction of a 3D model of FtsA (Sánchez et al., 1994), which was based on a multiple sequence alignment of the actin family of proteins. It is composed of a three-stranded antiparallel β-sheet adjacent to an α-helix, in the order S5 H2 S6 S7. There is no clear homology between this subdomain (which we call subdomain 1C) and any known structure in the database. At the end of the insertion, strand 7 goes into helix 3 of subdomain 1A, which aligns perfectly with helix 5 of actin. Helix 3 is connected to strand 8 and, via helix 4, reaches subdomain 2A. This subdomain is again divided into two parts with regard to the primary sequence (Figure 4), but this time the insertion of subdomain 2B is in exactly the same position as in actin: it is inserted after helix 5 and three antiparallel β-strands of subdomain 2A. Helices 6 and 7 of subdomain 2B are both involved in co-ordinating the nucleotide. Subdomain 2B is completed with two additional, short antiparallel β-strands and the N-terminal part of helix 8. Helix 8 contributes to the structure of both subdomains 2A and 2B; its boundaries have been defined based on the homology to actin and Hsc70. A partially disordered loop connects helix 8 to strand 14, the latter being part of the five-stranded β-sheet of subdomain 2A. Strand 15 is connected by a long loop to the C-terminal helix, which belongs to subdomain 1A. The last 27 residues of the C-terminus of FtsA are disordered.
Fig. 2. Ribbon plot of the crystal structure of FtsA. The structure is divided into four domains, in analogy to the actin family of proteins, which are designated 1A (blue), the FtsA-specific domain 1C (yellow), subdomain 2A (red) and subdomain 2B. Secondary structure elements are labelled according to their order of appearance in the primary sequence of FtsA (Figure 4). Mg-ATP is depicted in purple. The figure was prepared using the program MOLSCRIPT (Kraulis, 1991).
Fig. 4. Structure-based sequence alignment between FtsA and actin (Protein Data Bank entry 1YAG-A) [using the DALI web server (Holm and Sander, 1993)]. Boundaries of the secondary structure elements were defined using the program DSSP (Kabsch and Sander, 1983). The colours of the secondary structure elements correspond to those of the domains shown in Figure 2. Active site residues are coloured in purple. Conserved residues are blocked. The figure was prepared using the program ALSCRIPT (Barton, 1993).
Active site
During model building, electron density for the nucleotide was recognized in the interdomain cleft between the two main domains of the SeMet-substituted protein. Since Mg-ADP was added prior to the crystallization trials, an ADP molecule was initially built into the electron density. After refinement, however, difference density strongly suggested the presence of a third phosphate. Since no ATP was added after the cells were lysed, the nucleotide must have bound to the protein during growth of the bacterial cells.
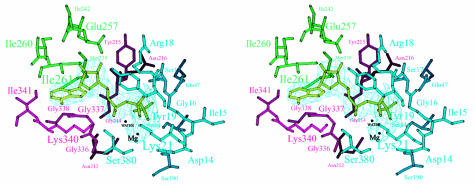
The nucleotide-binding pocket is located in a cleft formed by subdomains 1A, 2A and 2B. The adenosine is positioned in a hydrophobic pocket, composed of residues that belong to a short 310-helix (residues 337–341) of subdomain 2A and part of a helix in subdomain 2B (residues 257–261) (Figure 3). The pocket is closed at the back by SeMet238 and Ile242 of subdomain 2B, which form hydrogen bonds to O4* (by SeMet238) and O2* and O3* (by Ile261). The other side of the pocket is open to the outside.
Fig. 3. Stereo view of the active site of FtsA. Residues within hydrogen-bonding distance of Mg-ATP are labelled. Colours of the residues correspond to those of the subdomains as defined in Figure 2. The nucleotide is depicted in light green, within the 2_F_o – _F_c electron-density map. The figure was prepared using MOLSCRIPT (Kraulis, 1991).
The phosphate moiety of the nucleotide is bound by two loops. Each loop connects the first and second strands of subdomain 1A and 2A, respectively (residues 16–19 of subdomain 1A and residues 212–216 of subdomain 2A). In the first loop, Ser17 binds to O1 of the β-phosphate and its main-chain nitrogen is at hydrogen-bonding distance from O1 and O3 of the β-phosphate and O3 of the γ-phosphate; Lys21 makes hydrogen bonds with the O2 of both the α-phosphate and the β-phosphate. Arg18 is in contact with O1 and O3 of the β-phosphate. The main-chain nitrogen of Tyr19 is hydrogen-bonded to O1 of the β-phosphate. In the second loop, Asn212 makes hydrogen bonds with the γ-phosphate and Gly214 with O1 of the α-phosphate; the main-chain nitrogen of Tyr215 is close to O2 of the γ-phosphate, O3 of the β-phosphate and O1 and O3 of the α-phosphate. Additional contributions to the binding of the phosphate moiety come from the main-chain nitrogens of Gly337 and Ser380 (both to the α-phosphate) and from Ser84 (to O3 of the γ-phosphate) and Gln47 (to O1 of the γ-phosphate). A magnesium ion is located in a hydrophilic pocket formed by the β- and γ-phosphates and by Asp14, Lys21, Ser190 and Asn212. A water molecule 3.5 Å from the γ-phosphate is a putative attacking nucleophile, positioned by hydrogen bonds with the main-chain nitrogen of Gly219 and the main-chain oxygen of Phe217. In actin, the structurally equivalent residues (Val159 and His161) have also been presumed to be involved in ATP hydrolysis (Flaherty et al., 1991).
Comparison of the FtsA structure with that of other proteins
A similarity search, using the DALI web server [EBI, Hinxton (Holm and Sander, 1993)], revealed that FtsA is more closely related to actin and Hsc70 than to any of the sugar kinases [actin: _Z_-score 21.3, with a root mean square deviation (r.m.s.d.) of superimposed Cα atoms of 3.2 Å over 262 equivalent residues (Protein Data Bank entry 1YAG-A); Hsc70: _Z_-score 17.3, r.m.s.d. of 3.0 Å over 247 equivalent residues (Protein Data Bank entry 1BA1); hexokinase: _Z_-score 15.3, r.m.s.d. of 3.5 Å over 259 equivalent residues (Protein Data Bank entry 1DGK-N); glycerol kinase: _Z_-score 13.4, with 238 equivalent residues (Protein Data Bank entry 1GLC-G)]. A closer look at the superimposition shows that the active site of FtsA (containing 25 amino acids) has more residues in common with actin and Hsc70 (seven and eight identical residues, respectively) than with hexokinase or glycerol kinase (six and four residues, respectively). As mentioned previously, FtsA, actin and Hsc70 are distinguished from sugar kinases by the sequence of the loop in domain 2A that binds the phosphate moiety (Bork et al., 1992). In sugar kinases, the residues of subdomain 2A that are in contact with the phosphate moiety are limited to a conserved Gly/Thr pair. In both FtsA and actin, this phosphate-binding loop is more extended and does not include a Gly/Thr pair (Figure 3). A structural sequence alignment based on the optimal superimposition of the structures of FtsA and actin is shown in Figure 4. It is immediately clear that, despite the low overall sequence similarity between actin and FtsA, the order of their secondary structure elements is very similar (Figure 4). The only exception is the second subdomain. As mentioned before, the length and the structure of this domain are variable among members of the actin family. In Hsc70 and actin, this subdomain is inserted after the third β-strand, whereas the third β-strand in FtsA is followed by a helix and the fourth β-strand of subdomain 1A, before the second subdomain is inserted. Interestingly, hexokinase also lacks the insertion after the third β-strand, and has two short, antiparallel β-strands inserted at the same place as subdomain 1C of FtsA. The fold and the topology of subdomain 1C in FtsA are unusual for the actin family and suggest that subdomain 1C has an important functional role, specific to FtsA. Some other, more minor differences exist between the structures of FtsA and actin or Hsc70. They probably also reflect specific functional differences between the proteins; examples are the fifth β-strand in subdomain 2A, which is present in actin and FtsA but absent in Hsc70, or the very long loop connecting subdomain 2A to the C-terminal helix of FtsA. Since this loop is in close proximity to subdomain 1C, it may be involved in a FtsA-specific function.
Discussion
Despite the absence of significant similarity in the amino acid sequences, FtsA shows the same architecture of its core domain and shares the nucleotide-binding site with actin and Hsc70. The common fold comprises two α/β domains enclosing a nucleotide-binding pocket. Since striking similarity has been found in proteins with entirely different functions, it has previously been postulated that these domains are the result of divergent evolution from a common ancestor (Bork et al., 1992; Kabsch and Holmes, 1995). A single domain, resembling the RNase H fold, could have improved its nucleotide-binding capacity by forming a dimer via gene duplication and fusion. This dimer could then have diverged into different proteins with specific functions, like the sugar kinases, Hsc70, FtsA and actin. These proteins would then have developed additional structural features to carry out their specific function, such as minor polymorphism in loops, strands and helices or insertions of entire domains. The divergence of the ancestor into members of the actin family must have occurred a long time ago, because of the low sequence similarity between the members.
At present, little is known about the function of FtsA. It is known that E.coli FtsA binds ATP and a few mutants have been reported (Sánchez et al., 1994). Some of them are impaired in ATP binding. For example, substitution of Gly336 (which is located in the glycine loop of domain 2A) with Asp reduces ATP binding, probably because the newly introduced side chain precludes binding of the nucleotide. Another mutant, Tyr215→Ala in E.coli FtsA, is impaired in phosphate binding. At the equivalent position in FtsA from T.maritima is Phe217. The main-chain oxygen of Phe217 forms a hydrogen bond to a water molecule, which may function as a nucleophile in ATP hydrolysis. Two mutants were reported to inactivate E.coli FtsA irreversibly at 42°C (Leu83→Phe and Gly205→Ser) (Robinson et al., 1988). The corresponding residues in T.maritima FtsA are Ile81 and Gly207. Ile81 is located in strand S4, which precedes one of the loops binding the γ-phosphate; Gly207 is located at the N-terminus of strand S9. Strand S9 contains Asn212, which is involved in chelating the Mg2+ ion and binding to the γ-phosphate. Both temperature-sensitive mutants are therefore probably structural mutants that indirectly affect the binding of the nucleotide.
In the structure of the SeMet-containing FtsA, an ATP molecule occupies the active site. The non-substituted protein does not contain this nucleotide. The most likely explanation for the presence of the ATP is that the substitution of Met238 by SeMet (which forms a hydrogen bond to the adenosine) stabilizes the nucleotide-binding state or prevents ATP hydrolysis. An indication of the absence of hydrolysis is that the position of the water molecule that could be involved in ATP hydrolysis is not exactly in line with the scissile bond of the nucleotide (Figure 3). The ATP could have been moved slightly out of the active site, because of substitution of Met238 by the larger SeMet.
The structural similarity between FtsA and members of the actin family may suggest that FtsA shares some of their functional properties as well. Although the overall similarity to the sugar kinases is not as high as that to actin or Hsc70, FtsA has the second subdomain inserted in the same place in the primary sequence as in hexokinase. Nevertheless, it is unlikely that FtsA has a kinase function, since there is no space adjacent to the active site to insert a substrate. In addition, the Gly/Thr pair that is characteristic of the active site of sugar kinases is missing in FtsA. The structural similarity between FtsA and Hsc70 lies in the ATPase domain of Hsc70. A chaperone function for FtsA is questionable, since FtsA does not have a substrate-binding domain like Hsc70. Thus, the closest structural homology exists between FtsA and actin. Analogous to this is the previously reported homology between FtsZ and tubulin. Although there is no biochemical evidence that FtsA forms actin-like filaments, it is remarkable to find two conserved proteins involved in the cytokinesis of bacteria that are structurally related to components of the eukaryotic cytoskeleton and have central roles in eukaryotic cytokinesis (Robinson and Spudich, 2000).
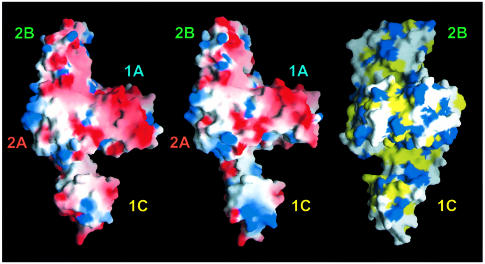
The most important difference between FtsA and other members of the actin family is the position and the topology of the second subdomain. This domain has the same topology as the antiparallel β-sheet comprising strands 2–5 and helix 1 of the first subdomain, but with the chain traced in the opposite direction. The connection of subdomain 1C to the rest of the protein is flexible, as reflected in the different crystal forms. The flexibility is not necessarily related to ATP binding, since different crystal forms were found of the empty form of the protein too. However, we cannot exclude the possibility that binding of the nucleotide may fix the second subdomain in a specific position and that the flexibility of subdomain 1C is functionally related to the nucleotide-binding state of the protein. Interestingly, movement of the second domain narrows the width of a cleft, located between subdomains 1A and 1C (demonstrated in the left and middle parts of Figure 5). The angle of rotation of subdomain 1C is 13.5°, with residue Glu89 acting as the mechanical hinge [as determined with the program DYNDOM (Hayward et al., 1998)]. The cleft is partially hydrophobic, as shown in the hydrophobicity plot (Figure 5, right). As mentioned in the Introduction, it has been reported that FtsA interacts with the C-terminus of FtsZ (Din et al., 1998; Ma and Margolin, 1999; Yan et al., 2000). It might be possible that the C-terminus of FtsZ binds to the groove that is partially enclosed by the movement of the second subdomain of FtsA (Figure 5). The binding to FtsZ may trigger a conformational change in FtsA that is associated with ATP conversion.
Fig. 5. Plot of the solvent-accessible surface of FtsA. Left and middle: electrostatic potential [–6 (red) to +6 kT (blue)] plotted on to the surface of FtsA in a slightly tilted orientation around the _y_-axis compared with Figure 2. The left picture shows the structure derived from crystal form 2 (with Mg-ATP bound); the middle picture shows the structure based on crystal form 1 (with an empty active site). The putative peptide-binding groove is located perpendicular to the plane between subdomains 1A and 1C (left and middle). The right-hand hydrophobicity plot is rotated 90° around the _y_-axis, with respect to the middle figure, to show the depth of the putative peptide-binding groove across the molecule. The colour code is as follows: yellow for hydrophobic residues (Ala, Val, Leu, Ile, Met, Phe, Pro), blue for hydrophilic residues (Lys, Arg, Glu, Asp) and white for Ser, Thr, Tyr, His, Cys, Asn, Gln, Trp and Gly. Domains are labelled as in Figure 2. This figure was prepared using GRASP (Nicholls, 1993).
Materials and methods
Bacterial expression and protein purification
The gene encoding the FtsA protein (SWALL accession No. Q9WZU0) from T.maritima (DSMZ No. 3109) was amplified by PCR using Pfu polymerase (Stratagene). Genomic DNA was isolated from cells provided by the DSMZ (Braunschweig, Germany). Two primers were used (forward primer: 5′-CGATTGGCATATGATTGACTTGTCAAAAAC TGTCTTTTA-3′ and reverse primer: 5′-GGCATAAGGATCCCT CCATCAATTCTTTGAAGAGTCTG-3′) to produce a DNA fragment of 1280 bp with unique restriction sites at the 5′-end (_Nde_I) and the 3′-end (_Bam_HI). After digestion, the fragment was cloned into _Nde_I–_Bam_HI-digested vector derived from pHis17 (B.Miroux, personal communication), resulting in plasmid pFE29. The protein was expressed under control of the T7 promoter, adding eight residues to the C-terminus (GSHHHHHH). The expressed protein contains 427 residues and has a molecular weight of 47.9 kDa. Plasmid pFE29 was transformed into C41(DE3) cells (Miroux and Walker, 1996) and expressed in early log phase after the addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). For large-scale expression, 12 l of 2× TY, containing 100 µg/ml ampicillin, were inoculated with a 100-fold diluted overnight culture and grown until an OD600 of 0.2–0.3 was reached at 37°C, before induction with IPTG.
Cells were harvested and frozen in liquid nitrogen. The cells were lysed by sonication after the addition of lysozyme in 50 mM Tris pH 8.0. After centrifugation, the lysate was loaded on to a 12 ml Ni2+–NTA column. After an extensive wash with 300 mM NaCl, 20 mM imidazole, 50 mM Tris pH 6.0, the protein was eluted with 300 mM imidazole, 50 mM Tris pH 6.0. Peak fractions were pooled and concentrated before loading on to a Sepharose 6B gel filtration column (Amersham–Pharmacia), equilibrated in 50 mM Tris, 1 mM EDTA, 1 mM sodium azide pH 7.5. The protein eluted as a single peak. It can be stored for several months at 4°C.
SeMet-substituted FtsA
SeMet-substituted protein was expressed in the same strain, C41(DE3), which is not auxotrophic for methionine. Methionine biosynthesis was inhibited by growth conditions as described previously (Van Duyne et al., 1993). A preculture, grown in 2× TY, was used at a 1000-fold dilution to inoculate 200 ml of minimal medium containing 1× M9 supplemented with 0.4% glucose, 2 mM MgSO4, 100 µg/ml ampicillin, 25 µg/ml FeSO4, 10 ng/ml CaCl2 and 1 µg/ml each of the vitamins riboflavin, niacinamide, pyridoxine and thiamine. The overnight culture was diluted 50-fold into 10 l of prewarmed minimal medium and grown for 4 h to an OD600 of 0.35. A mixture of l-amino acids was added as solids [per litre of culture: 50 mg of SeMet (Sigma), 50 mg of leucine, isoleucine and valine and 100 mg of lysine, threonine and phenylalanine (all from Fluka)]. After 15 min, protein expression was induced by the addition of 1 mM IPTG and the culture was grown for an additional 6 h. Protein was purified as described for the non-substituted protein, except that 5 mM β-mercaptoethanol was added to the buffers for the Ni2+–NTA column and 1 mM dithiothreitol was used in all other buffers. A typical yield was 15 mg of protein per litre of cells. Electrospray mass spectrometry measurements of the non-substituted protein and the SeMet-containing protein were used to check SeMet incorporation (FtsA: observed, 47 990.3 Da; calculated, 47 984.0 Da; SeMet FtsA: observed, 48 310.2 Da; calculated, 48 312.3 Da).
Crystallization and data collection
Crystals were grown by the sitting-drop vapour diffusion technique. The crystallization solution for the non-substituted protein was 300 mM KCl, 50 mM MES pH 5.6, 5% PEG 6000. SeMet-substituted protein required a slightly different crystallization solution: 200 mM ammonium acetate, 100 mM sodium acetate pH 5.5, 15% PEG 4000. Droplets were composed of 2 µl of protein (13 mg/ml), 2 µl of crystallization solution and 0.1–0.3 µl of PEG 400 (Hampton Research). In order to crystallize the SeMet-containing protein, 1 mM ADP and 2 mM MgCl2 were added to the protein before use in crystallization trials. The protein precipitated slightly, probably because of a change in pH, after which the solution cleared again. Crystals were grown at 18°C and were used when they were between 1 week and 4 months old. Crystals were frozen in mother liquor with 30% PEG 400. A single mercury derivative was obtained after soaking a native crystal overnight in cryobuffer, containing 5 mM phenylmercury acetate (pma). Crystals belong to space group _P_21, with variable cell constants. The structures of two crystal forms were determined; the cell dimensions are given in Table I.
Structure determination and refinement
All data were indexed and integrated with the MOSFLM package (Leslie, 1991) and further processed using the CCP4 suite of programs (CCP4, 1994). An initial MAD density map was obtained by locating five selenium atoms in datasets PEAK, INFL and HREM of crystal form 2 using the program SOLVE (Terwilliger and Berendzen, 1999). Initial phases were calculated using MLPHARE and SOLOMON (Abrahams and Leslie, 1996). The first model was built manually using the program MAIN (Turk, 1992). With this model, crystal form 1 was solved by molecular replacement using AMoRe (Navaza, 1994). Crystal form 1 also yielded a single site mercury derivative, PMA1. A difference Fourier map against the molecular replacement solution aligned both sources of phase information. Two-crystal averaging using DMMULTI (CCP4, 1994) including experimental phases for the MAD data and experimental phases from the SIR mercury data improved map quality dramatically. A final run of SOLOMON with blurred phase probability distributions produced a map of exceptional quality for crystal form 2 at 2.6 Å resolution and the model for crystal form 2 was completed. One molecule of Mg-ATP was located. Crystallographic refinement was carried out using CNS (Brunger et al., 1998) with Engh and Huber parameters (Engh and Huber, 1991). All data to maximum resolution were used during refinement and water molecules were picked automatically as implemented in CNS and checked manually afterwards. Crystal form 1 was again solved by molecular replacement using AMoRe; it immediately became clear that domain 1C needed to be moved slightly in order to fit into the 2_F_o – _F_c map. The active site in crystal form 1 contains only water molecules. After moving subdomain 1C, the model for crystal form 1 was refined using CNS against all available data to 1.9 Å. Waters were picked as implemented in CNS and checked. All manual model building was carried out using MAIN. The parameters of the final models are summarized in Table II.
Acknowledgments
Acknowledgements
We thank the staff at ID14-4 of ESRF (Grenoble, France) for assistance with the MAD data collection and Ian Fearnley (MRC-LMB, Cambridge, UK) for performing mass spectrometry. We appreciate the stimulating discussions with Miguel Vicente and Jesus Mingorance (CSIC, Madrid, Spain).
References
- Abrahams J.P. and Leslie,A.G.W. (1996) Methods used in the structure determination of bovine mitochondrial F1-ATPase. Acta Crystallogr. D, 52, 30–42. [DOI] [PubMed] [Google Scholar]
- Addinall S.G., Bi,E. and Lutkenhaus,J. (1996) FtsZ ring formation in fts mutants. J. Bacteriol., 178, 3877–3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addinall S.G., Cao,C. and Lutkenhaus,J. (1997) FtsN, a late recruit to the septum in Escherichia coli. Mol. Microbiol., 25, 303–309. [DOI] [PubMed] [Google Scholar]
- Anderson C.M., McDonald,R.C. and Steitz,T.A. (1978) Sequencing a protein by X-ray crystallography. Interpretation of yeast hexokinase B at 2.5 Å resolution by model building. J. Mol. Biol., 123, 1–13. [DOI] [PubMed] [Google Scholar]
- Ayala J.A., Garrido,T., de Pedro,M.A. and Vicente,M. (1994) Molecular biology of bacterial septation. In Hakenbeck,R. and Ghuysen,J.M. (eds), New Comprehensive Biochemistry, Vol. 27: Bacterial Cell Wall. Elsevier, Amsterdam, The Netherlands, pp. 73–101. [Google Scholar]
- Bachmann B.J. (1983) Linkage map of Escherichia coli K-12, edition 7. Microbiol. Rev., 47, 180–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton G.J. (1993) ALSCRIPT: a tool to format multiple sequence alignments. Protein Eng., 6, 37–40. [DOI] [PubMed] [Google Scholar]
- Begg K., Nikolaichik,Y., Crossland,N. and Donachie,W.D. (1998) Roles of FtsA and FtsZ in activation of division sites. J. Bacteriol., 180, 881–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork P., Sander,C. and Valencia,A. (1992) An ATPase domain common to prokaryotic cell-cycle proteins, sugar kinases, actin, and Hsp70 heat-shock proteins. Proc. Natl Acad. Sci. USA, 89, 7290–7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger A.T. et al. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Chen J.C., Weiss,D.S., Ghigo,J.M. and Beckwith,J. (1999) Septal localization of FtsQ, an essential cell division protein in Escherichia coli. J. Bacteriol., 181, 521–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Dai K. and Lutkenhaus,J. (1992) The proper ratio of FtsZ to FtsA is required for cell division to occur in Escherichia coli. J. Bacteriol., 174, 6145–6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewar S.J., Begg,K.J. and Donachie,W.D. (1992) Inhibition of cell division initiation by an imbalance in the ratio of FtsA to FtsZ. J. Bacteriol., 174, 6314–6316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Din N., Quardokus,E.M., Sackett,M.J. and Brun,Y.V. (1998) Dominant C-terminal deletions of FtsZ that affect its ability to localize in Caulobacter and its interaction with FtsA. Mol. Microbiol., 27, 1051–1063. [DOI] [PubMed] [Google Scholar]
- Donachie W.D. (1993) The cell cycle of Escherichia coli. Annu. Rev. Microbiol., 47, 199–230. [DOI] [PubMed] [Google Scholar]
- Engh R.A. and Huber,R. (1991) Accurate bond and angle parameters for X-ray protein-structure refinement. Acta Crystallogr. A, 47, 392–400. [Google Scholar]
- Erickson H.P. (1997) FtsZ, a tubulin homologue in prokaryote cell division. Trends Cell Biol., 7, 362–367. [DOI] [PubMed] [Google Scholar]
- Faber H.R., Pettigrew,D.W. and Remington,S.J. (1989) Crystallization and preliminary X-ray studies of Escherichia coli glycerol kinase. J. Mol. Biol., 207, 637–639. [DOI] [PubMed] [Google Scholar]
- Flaherty K.M., Delucaflaherty,C. and McKay,D.B. (1990) 3-Dimensional structure of the ATPase fragment of a 70k heat-shock cognate protein. Nature, 346, 623–628. [DOI] [PubMed] [Google Scholar]
- Flaherty K.M., McKay,D.B., Kabsch,W. and Holmes,K.C. (1991) Similarity of the 3-dimensional structures of actin and the ATPase fragment of a 70-kDa heat-shock cognate protein. Proc. Natl Acad. Sci. USA, 88, 5041–5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghigo J.M., Weiss,D.S., Chen,J.C., Yarrow,J.C. and Beckwith,J. (1999) Localization of FtsL to the Escherichia coli septal ring. Mol. Microbiol., 31, 725–737. [DOI] [PubMed] [Google Scholar]
- Hayward S. and Berendsen,H.J.C. (1998) Systematic analysis of domain motions in proteins from conformational change; new results on citrate synthase and T4 lysozyme. Proteins, 30, 144–154. [PubMed] [Google Scholar]
- Holm L. and Sander,C. (1993) Protein-structure comparison by alignment of distance matrices. J. Mol. Biol., 233, 123–138. [DOI] [PubMed] [Google Scholar]
- Kabsch W. and Holmes,K.C. (1995) Protein motifs 2. The actin fold. FASEB J., 9, 167–174. [DOI] [PubMed] [Google Scholar]
- Kabsch W. and Sander,C. (1983) Dictionary of protein secondary structure: pattern recognition of hydrogen bonded and geometrical features. Biopolymers, 22, 2577–2637. [DOI] [PubMed] [Google Scholar]
- Kabsch W., Mannherz,H.G., Suck,D., Pai,E.F. and Holmes,K.C. (1990) Atomic structure of the actin–DNase I complex. Nature, 347, 37–44. [DOI] [PubMed] [Google Scholar]
- Kraulis P.J. (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- Laskowski R.A., Macarthur,M.W., Moss,D.S. and Thornton,J.M. (1993) Procheck—a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- Leslie A.G.W. (1991) Recent Changes to the MOSFLM Package for Processing Film and Image Plate Data. Newsletter, SERC Laboratory, Daresbury, Warrington WA4 4AD, UK. [Google Scholar]
- Löwe J. and Amos,L.A. (1998) Crystal structure of the bacterial cell-division protein FtsZ. Nature, 391, 203–206. [DOI] [PubMed] [Google Scholar]
- Lu C.L., Reedy,M. and Erickson,H.P. (2000) Straight and curved conformations of FtsZ are regulated by GTP hydrolysis. J. Bacteriol., 182, 164–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutkenhaus J. (1997) Bacterial cell division and the Z ring. Annu. Rev. Biochem., 66, 93–116. [DOI] [PubMed] [Google Scholar]
- Ma X.L. and Margolin,W. (1999) Genetic and functional analyses of the conserved C-terminal core domain of Escherichia coli FtsZ. J. Bacteriol., 181, 7531–7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X.L., Ehrhardt,D.W. and Margolin,W. (1996) Co-localization of cell division proteins FtsZ and FtsA to cytoskeletal structures in living Escherichia coli cells by using green fluorescent protein. Proc. Natl Acad. Sci. USA, 93, 12998–13003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X.L., Sun,Q., Wang,R., Singh,G., Jonietz,E.L. and Margolin,W. (1997) Interactions between heterologous FtsA and FtsZ proteins at the FtsZ ring. J. Bacteriol., 179, 6788–6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miroux B. and Walker,J.E. (1996) Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol., 260, 289–298. [DOI] [PubMed] [Google Scholar]
- Mukherjee A. and Lutkenhaus,J. (1998) Dynamic assembly of FtsZ regulated by GTP hydrolysis. EMBO J., 17, 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanninga N. (1991) Cell division and peptidoglycan assembly in _Escherichia coli._Mol. Microbiol., 5, 791–795. [DOI] [PubMed] [Google Scholar]
- Navaza J. (1994) AMoRe—an automated package for molecular replacement. Acta Crystallogr. A, 50, 157–163. [Google Scholar]
- Nicholls A., Bharadwaj,R. and Honig,B. (1993) Grasp: graphical representation and analysis of surface properties. Biophys. J., 64, A166. [Google Scholar]
- Nogales E., Downing,K.H., Amos,L.A. and Löwe,J. (1998a) Tubulin and FtsZ form a distinct family of GTPases. Nature Struct. Biol., 5, 451–458. [DOI] [PubMed] [Google Scholar]
- Nogales E., Wolf,S.G. and Downing,K.H. (1998b) Structure of the αβ-tubulin dimer by electron crystallography. Nature, 391, 199–203. [DOI] [PubMed] [Google Scholar]
- Robinson A.C., Begg,K.J. and Donachie,W.D. (1988) Mapping and characterization of mutants of the Escherichia coli cell-division gene, FtsA. Mol. Microbiol., 2, 581–588. [DOI] [PubMed] [Google Scholar]
- Robinson D.N. and Spudich,J.A. (2000) Towards a molecular understanding of cytokinesis. Trends Cell Biol., 10, 228–237. [DOI] [PubMed] [Google Scholar]
- Rothfield L., Justice,S. and Garcia Lara,J. (1999) Bacterial cell division. Annu. Rev. Genet., 33, 423–448. [DOI] [PubMed] [Google Scholar]
- Sánchez M., Valencia,A., Ferrándiz,M.J., Sander,C. and Vicente,M. (1994) Correlation between the structure and biochemical activities of FtsA, an essential cell-division protein of the actin family. EMBO J., 13, 4919–4925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger T.C. and Berendzen,J. (1999) Automated MAD and MIR structure solution. Acta Crystallogr. D, 55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tormo A., Ayala,M.A., de Pedro,M.A., Aldea,M. and Vicente,M. (1986) Interaction between FtsA and PBP3 proteins in the Escherichia coli septum. J. Bacteriol., 166, 985–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk D. (1992) Weiterentwicklung eines Programms für Molekülgrafik und Elektrondichte-Manipulation und seine Anwendung auf verschiedene Protein-Strukturaufklärungen. PhD Thesis, Technische Universität München, Germany.
- Van Duyne G.D., Standaert,R.F., Karplus,P.A., Schreiber,S.L. and Clardy,J. (1993) Atomic structures of the human immunophilin Fkbp-12 complexes with Fk506 and rapamycin. J. Mol. Biol., 229, 105–124. [DOI] [PubMed] [Google Scholar]
- Wang L.L. and Lutkenhaus,J. (1998) FtsK is an essential cell division protein that is localized to the septum and induced as part of the SOS response. Mol. Microbiol., 29, 731–740. [DOI] [PubMed] [Google Scholar]
- Weiss D.S., Chen,J.C., Ghigo,J.M., Boyd,D. and Beckwith,J. (1999) Localization of FtsI (PBP3) to the septal ring requires its membrane anchor, the Z ring, FtsA, FtsQ, and FtsL. J. Bacteriol., 181, 508–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan K., Pearce,K.H. and Payne,D.J. (2000) A conserved residue at the extreme C-terminus of FtsZ is critical for the FtsA–FtsZ interaction in Staphylococcus aureus. Biochem. Biophys. Res. Commun., 270, 387–392. [DOI] [PubMed] [Google Scholar]