Recognition of preproteins by the isolated TOM complex of mitochondria (original) (raw)
Abstract
A multisubunit complex in the mitochondrial outer membrane, the TOM complex, mediates targeting and membrane translocation of nuclear-encoded preproteins. We have isolated the TOM holo complex, containing the preprotein receptor components Tom70 and Tom20, and the TOM core complex, which lacks these receptors. The interaction of recombinant mitochondrial preproteins with both types of soluble TOM complex was analyzed. Preproteins bound efficiently in a specific manner to the isolated complexes in the absence of chaperones and lipids in a bilayer structure. Using fluorescence correlation spectroscopy, a dissociation constant in the nanomolar range was determined. The affinity was lower when the preprotein was stabilized in its folded conformation. Following the initial binding, the presequence was transferred into the translocation pore in a step that required unfolding of the mature part of the preprotein. This translocation step was also mediated by protease-treated TOM holo complex, which contains almost exclusively Tom40. Thus, the TOM core complex, consisting of Tom40, Tom22, Tom6 and Tom7, is a molecular machine that can recognize and partially translocate mitochondrial precursor proteins.
Keywords: mitochondria/protein folding/protein translocation/TOM complex
Introduction
The import of proteins into mitochondria is mediated by multisubunit translocases in the outer (TOM complex) and inner (TIM complexes) membranes of the organelles (Schatz and Dobberstein, 1996; Neupert, 1997). The TOM complex contains components that expose domains to the cytosol and function as preprotein receptors. The major receptor, Tom20, together with Tom22 is involved in the translocation of most precursors (Harkness et al., 1994; Lithgow et al., 1995). Another receptor, Tom70, forms a binding site for a more restricted set of preproteins, most notably the mitochondrial carrier family (Schlossmann et al., 1994; Brix et al., 1999). These receptors are loosely attached to the TOM core complex (Dekker et al., 1998; Rapaport et al., 1998a; Ahting et al., 1999). The subunits of the core complex (Tom40, Tom22, Tom7, Tom6 and Tom5) are embedded in the outer membrane and form the translocation pore (Künkele et al., 1998a; Ahting et al., 1999). Tom40 is the major component of the translocation pore (Hill et al., 1998; Künkele et al., 1998b), while Tom22 and Tom5 probably link the receptors to the pore (Dietmeier et al., 1997; van Wilpe et al., 1999).
Most mitochondrial precursor proteins are synthesized in the cytosol with an N-terminal extension, the presequence (or matrix-targeting sequence). Presequences have been shown to be necessary and sufficient to direct proteins to the mitochondria (Neupert, 1997). They are rich in positively charged amino acid residues, ∼15–50 residues long and have the potential to form amphiphilic α-helices in hydrophobic environments (Von Heijne, 1986; Roise and Schatz, 1988). Several studies, utilizing model systems, have suggested that the initial interaction of presequences occurs via direct association with membrane lipids (Tamm, 1991; Roise, 1992). The membrane environment would induce a regular structure in the presequence, which has a random-coil structure in aqueous solutions. On the other hand, cross-linking experiments have suggested that the presequence part of preproteins can interact tightly with various TOM components, in particular Tom5, Tom20, Tom22 and Tom40 (Gaikwad and Cumsky, 1994; Hönlinger et al., 1995; Dietmeier et al., 1997; Rapaport et al., 1997; Kanamori et al., 1999).
To investigate whether precursor proteins can bind directly to the TOM complex, we used an assay system containing soluble purified TOM complex and recombinant precursor proteins. The direct binding of precursor proteins to the TOM complex demonstrates that a lipid membrane is not essential for binding. Furthermore, the TOM core complex is sufficient for high-affinity binding and was found to contain several binding sites. Following the initial binding, the presequence part of the preproteins was transferred into the translocation pore in a step that required unfolding of the mature part of the preprotein when this part immediately followed the presequence.
Results
Binding of preproteins to isolated TOM complex
To study the binding of preproteins to isolated TOM holo complex, we analyzed the sedimentation behavior on sucrose gradients of the purified TOM complex, the precursor protein pSu9(1–69)-dihydrofolate reductase (DHFR) and the adduct between them. Free precursor was present at the top of the gradient while the TOM complex was found towards the bottom of the gradient (Figure 1, upper panel). Incubation of the precursor protein with the TOM complex prior to centrifugation resulted in co-migration of these components (Figure 1, lower panel). The specificity of binding was studied in the following experiments: (i) when excess precursor was incubated with the TOM complex, a major part of it remained unbound (Figure 1D), so it is possible to saturate the binding sites on the TOM complex; (ii) DHFR (without a presequence) did not bind to the TOM complex (not shown); (iii) unrelated proteins, whether folded like soybean trypsin inhibitor or unfolded like reduced carboxymethylated lactalbumin (RCMLA), did not bind to the TOM complex (not shown). Thus, the binding observed was dependent on the mitochondrial targeting signal and there was no binding to unrelated unfolded domains. We conclude that the soluble TOM complex can bind precursor proteins in a specific manner. Although some lipid molecules are attached to the purified TOM complex (see below), an intact bilayer structure appears not to be necessary for efficient binding of preproteins to the TOM complex.
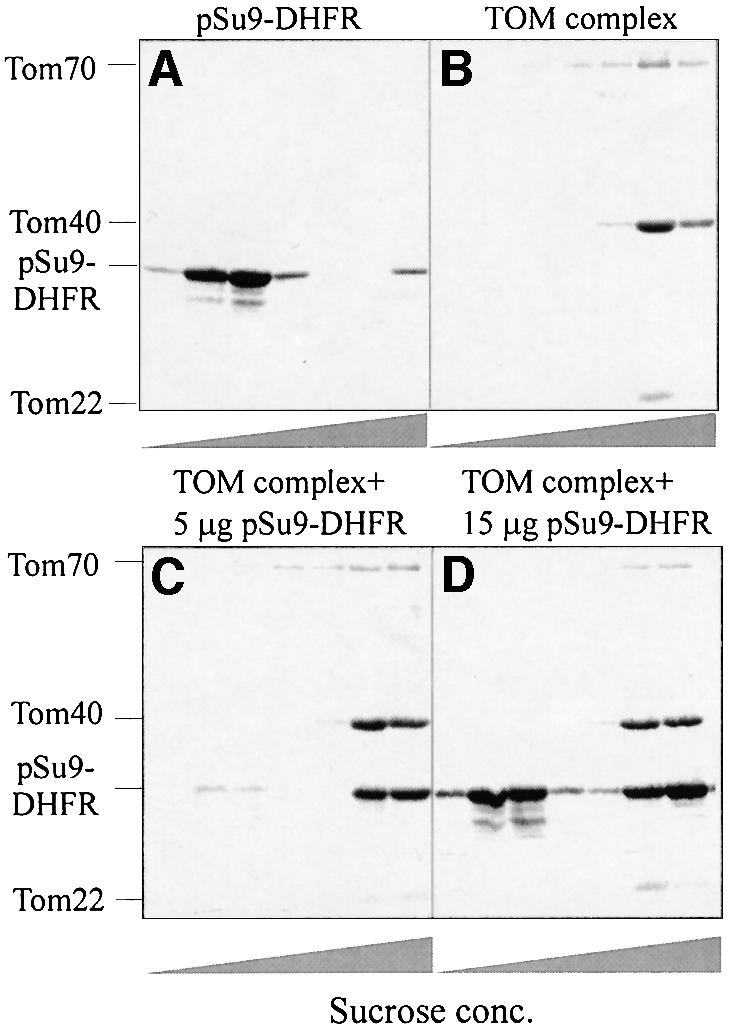
Fig. 1. Analysis of binding of mitochondrial precursor protein to isolated soluble TOM holo complex by sucrose gradient centrifugation. TOM holo complex and pSu9(1–69)-DHFR were incubated either separately (A and B) or together (C and D). In each case, samples were kept for 20 min at 4°C. There were 24 µg of TOM holo complex in each sample, and 10 µg of pSu9(1–69)-DHFR in (A), 5 µg in (C) and 15 µg in (D). After 16 h centrifugation at 140 000 g, 100 µl fractions were collected, trichloroacetic acid was added and precipitated proteins were analyzed by SDS–PAGE and Coomassie staining. The bands corresponding to the TOM components and the precursor protein are indicated.
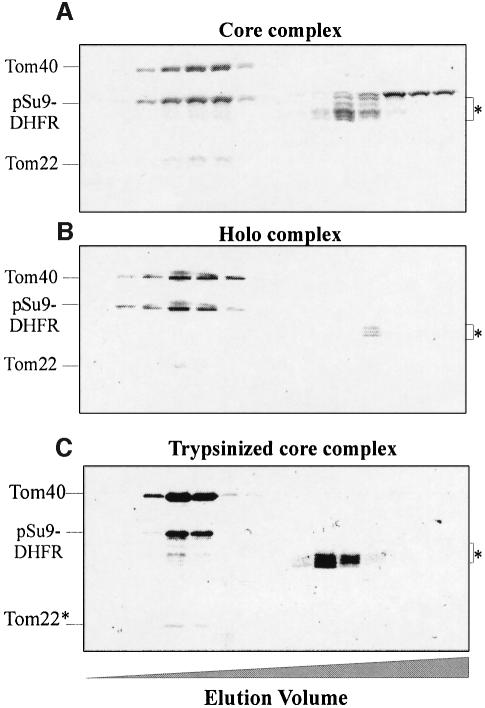
Are the receptor proteins, Tom20 and Tom70, required for precursor binding? We compared the binding capacity of the TOM holo complex, which contains these two proteins, to that of the core complex, which lacks them. Both complexes were incubated with pSu9(1–69)-DHFR and the reaction mixtures were analyzed by size-exclusion chromatography. Co-elution of the precursor with the TOM complex can be used as an assay for interaction of the two components since they have very different molecular masses. Under the conditions used, both the core and the holo TOM complexes bound the precursor (Figure 2A and B). Degradation products of the precursor that lack all or part of the presequence did not co-elute with the TOM complex but rather migrated as low molecular weight species. Therefore, in addition to the receptor proteins Tom20 and Tom70, other Tom components contain specific binding sites for precursor proteins.
Fig. 2. Binding of mitochondrial precursor protein to isolated soluble TOM complex analyzed by size-exclusion chromatography. TOM core complex (84 µg) (A) and holo complex (122 µg) (B) were incubated for 30 min at 4°C with pSu9(1–69)-DHFR (202 and 245 µg, respectively). The mixtures were then loaded on a size-exclusion column. Proteins were eluted, precipitated with trichloroacetic acid and analyzed by SDS–PAGE and Coomassie staining. Bands corresponding to the TOM components and the precursor protein are indicated. The preparations of precursor protein contained some degradation products (marked with an asterisk), which eluted in the lower molecular weight range. (C) Binding of pSu9(1–69)-DHFR to trypsinized TOM core complex. Purified TOM core complex (30 µg) was treated with trypsin, re-isolated by gel filtration and incubated for 30 min at 4°C with 60 µg of pSu9(1–69)-DHFR. Further treatment was as described above. Tom22*, trypsin-resistant fragment of Tom22.
As the hydrophilic domains of Tom22, Tom6 and Tom7 are possible candidates for such binding activity, we asked whether removal of these domains by mild trypsin treatment of the core complex would destroy its binding capacity. The trypsin-treated complex lacked these domains but still contained intact Tom40. The ability to bind preproteins was retained (Figure 2C). Protease treatment leaves only the single transmembrane anchors of the degraded Tom components, thus Tom40 appears to harbor presequence binding sites. What is the nature of the interactions between the TOM complex and precursors? Binding of pSu9(1–69)-DHFR to the core complex was performed at different salt concentrations. Binding was very efficient at 100 mM salt and moderate at 150 mM salt and there was practically no binding at a high salt concentration (250 mM) (not shown). These results suggest that electrostatic interactions play a role in the binding reaction.
Affinity and stoichiometry of preprotein binding
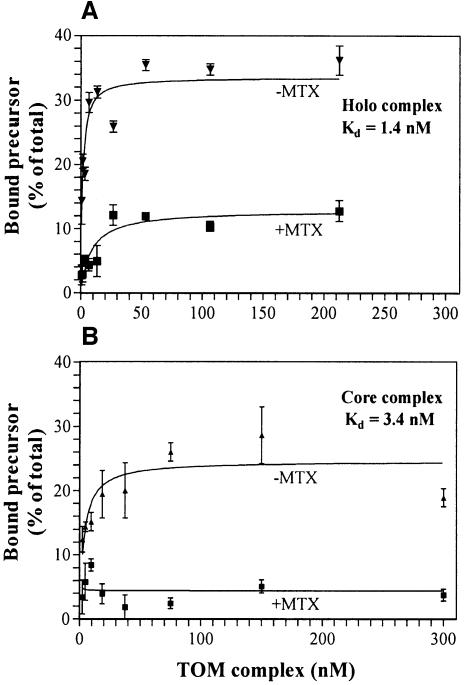
We used fluorescence correlation spectroscopy (FCS) to measure the equilibrium binding constants of fluorescently labeled pSu9(1–69)-DHFR with TOM holo and core complexes. FCS allows the determination of the diffusion constant of a fluorescent molecule in solution (Maiti et al., 1997). If the difference in the diffusion coefficients of the free and bound molecules is sufficiently large, binding of the fluorescent molecule to a target can be analyzed. In control experiments, fluorescently labeled DHFR (without a presequence) did not bind to the complex. The apparent dissociation constant for the equilibrium binding of the precursor to the TOM complex was determined to be in the lower nanomolar range: 1.4 nM for the holo complex and 3.4 nM for the core complex (Figure 3). This value suggests a relatively high affinity, significantly higher than the affinities of the same precursor to soluble domains of Tom20 or Tom70 that were measured with the surface plasmon resonance technique (Iwata and Nakai, 1998).
Fig. 3. Equilibrium binding of pSu9(1–69)-DHFR to the TOM holo and TOM core complexes measured by FCS. Fluorescently labeled pSu9(1–69)-DHFR (9 nM) was mixed with TOM holo (A) and TOM core (B) complexes at increasing concentrations in the absence (–MTX, triangles) or presence of 0.5 mM NADPH and 1 µM MTX (+MTX, squares). The data were fitted with a two-component model, free pSu9(1–69)-DHFR and pSu9(1–69)-DHFR bound to the TOM com plex. The ratio of fluorescence emitted by the bound precursor and total fluorescence was calculated for each sample and is presented as a function of TOM complex concentrations. Apparent binding constants of 1.4 ± 0.3 and 3.4 ± 1.0 nM were determined for the TOM holo complex and the TOM core complex, respectively.
Next, we measured the affinity of both complexes for a precursor in which the mature part was stabilized in its folded conformation by the addition of the specific ligand methotrexate (MTX). The affinity measured under these conditions, which correspond to the previously described cis site binding (Mayer et al., 1995a; Rapaport et al., 1998b), was strongly reduced (Figure 3). We could not calculate reliable dissociation constants for these conditions. This result confirms that stable interaction of mitochondrial precursor proteins with the TOM complex requires unfolding of the mature part of the preprotein when this part immediately followed the presequence.
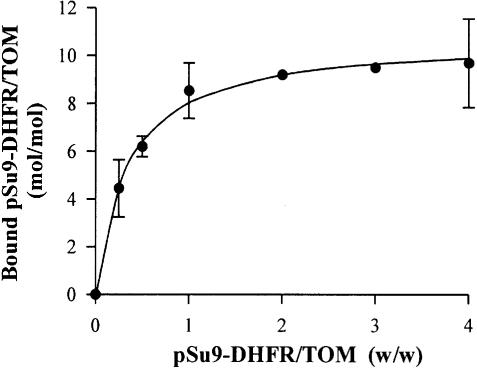
How many binding sites are present on the TOM core complex that lacks the receptors Tom20 and Tom70? The purified TOM core complex was incubated with increasing amounts of the precursor pSu9(1–45)-DHFR. The reaction mixtures were passed through a size-exclusion column and fractions containing co-eluted preprotein and TOM complex were analyzed by SDS–PAGE. The amounts of preprotein associated with the TOM complex were determined. To estimate the amount of bound proteins, increasing amounts of both precursor and TOM complex alone were loaded on the same gel for calibration. The number of precursor molecules bound per TOM complex is presented as a function of the amount of precursor added to the reaction mixture (Figure 4). Saturation of the precursor binding sites occurred when eight or nine molecules were bound to one TOM core complex.
Fig. 4. Titration of precursor-binding sites on the TOM core complex. Samples of TOM core complex (730 nM) were incubated in a 100 µl reaction volume for 30 min at 4°C with increasing amounts of pSu9(1–45)-DHFR. Further treatment was as described in the legend to Figure 2. The bands corresponding to Tom40 and pSu9(1–45)-DHFR that co-eluted from the size-exclusion column were quantified. By comparison with known amounts of these proteins loaded for calibration on the same gel, the amounts of the co-eluted proteins were calculated. The number of precursor molecules bound per TOM complex was calculated on the basis of molecular masses of 410 kDa for the TOM core complex and 25.6 kDa for pSu9(1–45)-DHFR.
Partial translocation and unfolding of preproteins
We asked whether, in addition to binding, the presequence is transferred into the import channel. As an assay for transfer, we determined protection of precursor against cleavage by added recombinant mitochondrial processing peptidase (MPP). Under our binding conditions, such protection was observed for preproteins containing presequences of different lengths, namely pSu9(1–45)-DHFR or pSu9(1–69)-DHFR. Incubation of precursor proteins with MPP in the absence of TOM complex or in the presence of bovine serum albumin (BSA) as controls resulted in complete or nearly complete removal of the presequence part of pSu9(1–69)-DHFR and pSu9(1–45)- DHFR, respectively (Figure 5A). In contrast, upon incubation of the preprotein with the TOM holo complex, a major part, if not all, of the precursor molecules were not cleaved by added MPP.
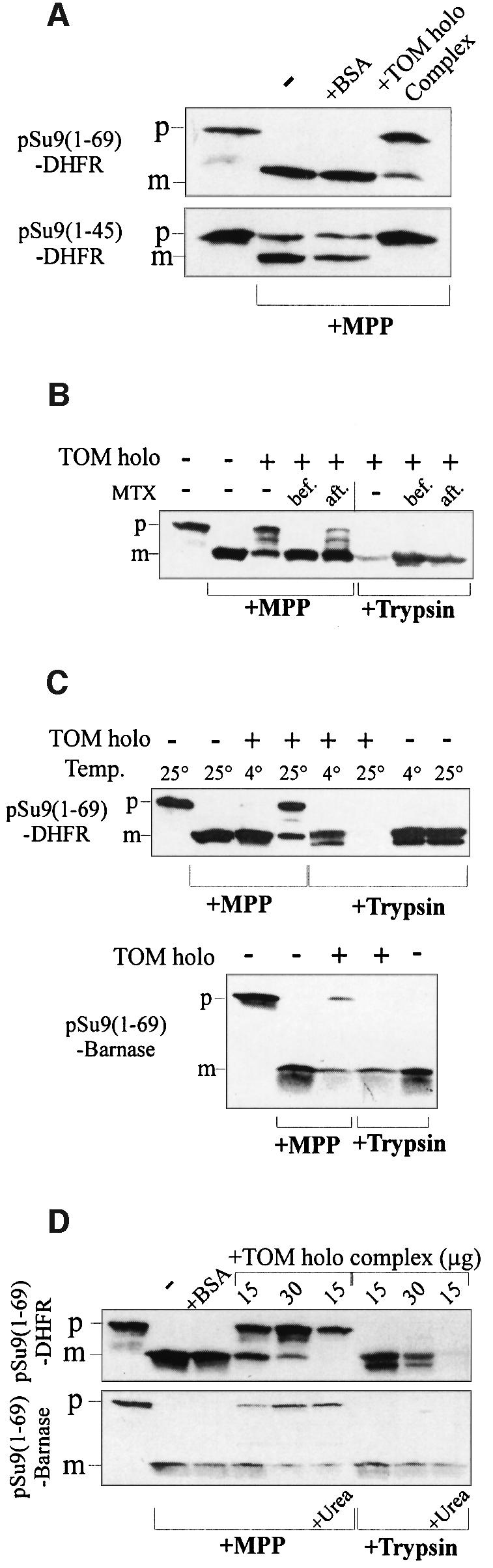
Fig. 5. TOM complex-mediated translocation of the presequence to a site inaccessible to MPP requires unfolding of the mature domain. (A) The isolated TOM holo complex can translocate the presequence of preproteins to a site inaccessible to MPP. Precursor proteins, pSu9(1–69)-DHFR and pSu9(1–45)-DHFR (3 µg each), were incubated with 15 µg of TOM holo complex (or 15 µg of BSA as a control) for 30 min at 25°C. MPP (2 µg) was added for 8 min at 25°C, followed by 4 mM EDTA to stop MPP activity. Proteins were analyzed by SDS–PAGE, blotted and immunodecorated with antibodies against mouse DHFR. (B) Stabilization of the mature part influences the protection against MPP. pSu9(1–69)-DHFR (3 µg) was incubated with or without TOM complex (60 µg) for 30 min at 25°C. MTX (1 µM) was present in this incubation mixture (bef.) or was added after incubation for 10 min at 0°C (aft.). Samples containing TOM complex were then split into two; MPP (15 µg/ml) was added to one half for 8 min at 25°C while trypsin (20 µg/ml) was added to the other for 15 min at 0°C. Trypsin was inhibited with 1 mM PMSF. Further treatment was as above. (C) Unfolding of the mature part is required to achieve protection against MPP. pSu9(1–69)-DHFR (3 µg) (upper panel) was incubated in the absence or presence of TOM complex (60 µg) for 30 min at 4°C or 10 min at 25°C. MPP was then added to the indicated samples for 8 min at 25°C, while trypsin (20 µg/ml) was added to the other samples for 15 min at 0°C. pSu9(1–69)-barnase (3 µg) (lower panel) was incubated in the absence or presence of TOM complex (30 µg) for 30 min at 30°C. MPP was then added to the indicated samples for 8 min at 25°C, while trypsin (20 µg/ml) was added to the other samples for 15 min at 0°C. Proteins were analyzed as above and immunodecorated with antibodies against barnase. (D) Unfolding of preproteins before their interaction with the TOM complex increases their extent of protection against MPP. Preproteins (3 µg) were incubated in the absence or presence of the indicated amounts of TOM holo complex for 30 min at 25°C. When indicated (+Urea) the preprotein was incubated first in 8 M urea for 30 min at 25°C and then diluted into the binding reaction (the concentration of urea in the binding reaction was 0.3 mM). Further treatment was as above. p and m, precursor and mature forms of preproteins, respectively.
This protection required the unfolding of the DHFR domain; precursor in which the DHFR domain was stabilized with MTX was not protected against MPP (Figure 5B). This unfolding was verified by trypsin degradation of the DHFR domain in the absence but not in the presence of MTX (Figure 5B). If MTX was added after binding of the precursor to the TOM complex, an intermediate situation was observed where a smaller fraction of the precursor was protected against MPP and unfolded.
The unfolding reaction was studied at various temperatures. Partial translocation and unfolding were observed when the initial incubation was at 25°C but not at 0°C (Figure 5C). Thus, the isolated TOM complex retains its ability to stabilize the unfolded conformation of the DHFR domain at 25°C in situ, i.e. in intact mitochondria and in outer membrane vesicles (Mayer et al., 1995b; Rapaport et al., 1998b). Can barnase, which is more stable than DHFR (Matouschek et al., 1997), also unfold by interacting with the isolated TOM complex? We found that, at 25°C, a fraction of pSu9(1–69)-barnase molecules was partially translocated and unfolded, as measured by protection against MPP treatment and trypsin treatment, respectively (Figure 5C, lower panel). In agreement with its higher stability, the precursor containing the barnase domain was protected less effectively than a DHFR-containing protein. To verify that the unfolding process is the limiting step in the translocation process, we presented the precursors to the TOM complex after denaturing them in 8 M urea. The unfolded precursors (as tested by trypsin degradation) were protected against MPP to a greater extent than the native precursors; no processed preprotein was observed after denaturation (Figure 5D). These results confirm the dependency of transfer to the high-affinity binding site on unfolding.
We next asked whether the receptors are essential for partial translocation and unfolding. Holo complex was treated with low concentrations of proteinase K, which resulted in the cleavage of Tom70, Tom22 and most of Tom6 (Figure 6). Under these conditions, full-length Tom20 was not detected, while residual amounts of characteristic proteolytic fragments were still observed (Figure 6). The treated complex was also able to provide protection against MPP (Figure 6). We conclude that the soluble complex is able to translocate the presequence part into the import pore and that Tom40 plays the central role in both recognition and translocation.
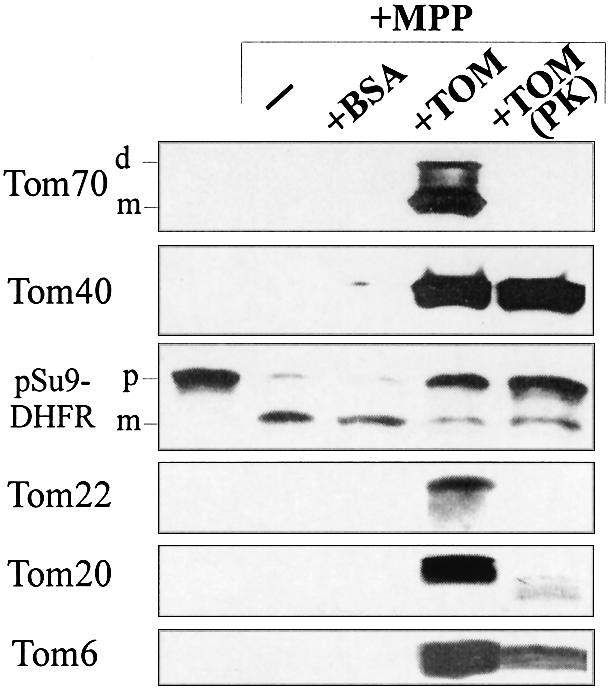
Fig. 6. Receptor proteins are not required for translocation to a site protected against MPP. pSu9(1–45)-DHFR (3 µg) was incubated for 30 min at 4°C with 15 µg of either TOM holo complex or TOM holo complex treated with proteinase K, or with BSA as a control. MPP was then added for 8 min at 25°C. Proteins were analyzed by SDS–PAGE, blotting and immunodecoration with antibodies against DHFR or TOM components. The bands corresponding to the monomeric (m) and dimeric (d) forms of Tom70, and of precursor and mature forms of the precursor protein (p and m, respectively) are indicated.
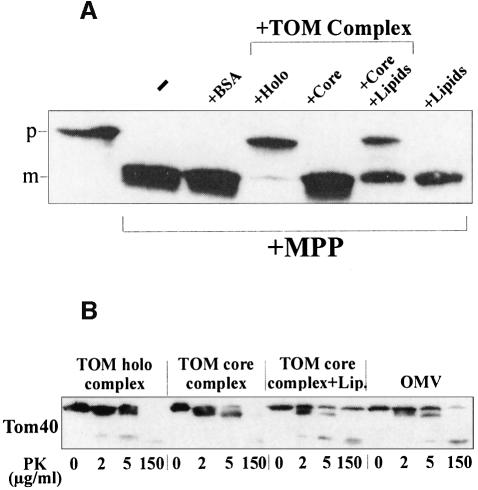
In the course of this study, we found that TOM core complex did not confer protection of preproteins against MPP (Figure 7A). The core complex is purified by solubilization of mitochondria using dodecylmaltoside (DDM) as a detergent rather than using digitonin, which is employed for the isolation of the holo complex. We found that the phospholipid content of the core complex is much lower that that of the holo complex. The amount of phospholipid present was 4 mol/mol TOM complex in the DDM-solubilized complex and 168 mol/mol TOM complex in the digitonin-solubilized TOM complex. As phospholipids have been shown to play an important role in protein translocation (van Voorst and de Kruijff, 2000), the low activity of the core complex may result from the low levels of structural lipid molecules. Such lipid molecules could be required for the proper function of the complex, similar to the finding that non-bilayer lipids stimulate the activity of the reconstituted bacterial protein translocase (van der Does et al., 2000). The addition of lipids extracted from the mitochondrial outer membrane restored the ability of the core complex to mediate partial translocation of the preprotein (Figure 7A). This restoration was paralleled by a change in the pattern of proteolytic fragments of Tom40 as a result of proteinase K treatment (Figure 7B). The restored pattern resembled more closely that of a native complex in outer membrane vesicles. Hence, lipids may induce a more ‘native-like’ conformation of the TOM complex and thus cause the complex to support translocation, which results in protection against MPP.
Fig. 7. (A) Addition of phospholipids restores the translocation activity of the TOM core complex. pSu9(1–69)-DHFR (3 µg) was incubated in the absence or presence of TOM complex (30 µg) for 30 min at 25°C. In the indicated sample, phospholipids (200 µg) were incubated with TOM core complex for 1 h at 4°C before addition of the preprotein. In the control sample, phospholipids without TOM complex were incubated with the preprotein. MPP was then added to the indicated samples for 8 min at 25°C. p and m, precursor and mature forms of preprotein, respectively. (B) Phospholipids affect the conformation of the TOM core complex. TOM holo complex (10 µg), TOM core complex (10 µg) with or without added phospholipids (67 µg), or outer membrane vesicles (OMV; 80 µg) were treated with the indicated amounts of proteinase K (PK) for 15 min at 0°C. Then proteinase K was inhibited by PMSF and proteins were analyzed by SDS–PAGE, blotting and immunodecoration with antibodies against Tom40.
Discussion
In order to understand better how precursor proteins are recognized by the mitochondria and translocated across the outer membrane, we have analyzed the interaction of mitochondrial precursor proteins with the TOM complex by studying isolated components. We found that purified precursors bound efficiently to the isolated complex in the absence of chaperones and lipids in a bilayer structure. Presequences were transferred into the translocation pore in such a manner that they became inaccessible to added MPP. Thus, the TOM complex, as isolated from mitochondria, represents the minimal machinery for the recognition and partial translocation of precursor proteins with an N-terminal extension. Similar binding characteristics have been described for a protein import complex obtained after solubilizing chloroplast outer envelopes with a mild detergent (Soll and Waegemann, 1992).
Previous reports suggested that purified domains of TOM components are able to bind precursor proteins (Brix et al., 1997; Schleiff et al., 1997; Komiya et al., 1998). In those experiments, recombinant soluble domains of TOM receptor proteins were incubated with sub-stoichiometric amounts of precursor proteins synthesized in cell-free systems that contained chaperones and potential presequence-binding factors. It has also been suggested that the presence of cytosolic chaperones is essential for the binding of mitochondrial precursors to the cytosolic domains of Tom70 and Tom20 (Komiya et al., 1997). Our results suggest that chaperones are not essential for targeting or translocation but rather have a stabilizing effect on precursor proteins.
In contrast to previous views, we conclude that interaction with a bilayer structure is not a prerequisite for translocation by the import machinery. TOM components may bind an amphipathic structure out of an equilibrium of ordered and disordered states. According to recent structural work, a presequence peptide binds in a hydrophobic groove formed by the cytosolic domain of Tom20 and, in this state, is present in an amphipathic helical structure (Abe et al., 2000).
In addition to recognition, the purified complex is able to transfer the presequence part from an exposed location at the complex to a location inaccessible to added MPP, probably into the translocation pore. Insertion of the presequence into the import pore can occur in the absence of an external energy source and receptors. Increasing the affinity of presequence-binding sites may lead to a vectorial movement of the precursor from the surface to the pore (Mayer et al., 1995b; Komiya et al., 1998; Rapaport et al., 1998b). Precursor molecules approaching the mitochondrial outer membrane are normally first bound to low-affinity binding sites on the cis side of the membrane and then transferred to sites with higher affinity on the trans side of the membrane (Mayer et al., 1995a,b; Rapaport et al., 1998b). The observed lower affinity of precursors for soluble receptor domains, as compared with the high affinity for the TOM complex measured in the present study, would support this view (Iwata and Nakai, 1998; Abe et al., 2000). Stable interaction of the presequence with the trans side and inaccessibility of the MPP cleavage site on the cis side of the outer membrane are accompanied by unfolding of the mature part of the preprotein if this is situated close to the targeting sequence (Mayer et al., 1995b; Rapaport et al., 1998b). Likewise, when in the soluble complex the cleavage site was inaccessible to the added MPP, the mature part was unfolded. The TOM complex has to be in a particular conformation to perform this partial translocation, as the core complex (which contains the required components) was inactive in the absence of lipids. When the soluble domains of the components of the TOM complex were removed by treatment with protease, binding of the preprotein occurred so that the MPP cleavage site was still protected. The environment of the presequence in the protected location is provided mainly by the membrane-embedded parts of the TOM complex. The bulk of these parts are composed by Tom40. These findings are in line with previous studies, which identified Tom40 as the major component involved in both the formation of the translocation pore and binding of precursor proteins (Vestweber et al., 1989; Rapaport et al., 1997, 1998a; Hill et al., 1998; Künkele et al., 1998b).
Interestingly, we found the number of precursor-binding sites per TOM complex to be similar to the estimated number of eight or nine Tom40 molecules present in each TOM complex (Künkele et al., 1998a). Thus, each Tom40 molecule may bind one precursor molecule. Binding to the soluble TOM core complex could occur from either the cytosolic or the intermembrane face. It is unclear how many preprotein molecules can bind simultaneously to the TOM complex embedded in the mitochondrial outer membrane. The high number of binding sites in the isolated complex could be the result of enabling the presequence to bind at sites that are not accessible when the membrane-integrated complex is analyzed. Another possibility is that all these binding sites are available in vivo too, but that, due to a different topological arrangement of the native complex, as compared with the soluble complex, the interaction of preprotein in transit with one of these sites prevents the binding of other preproteins by steric hindrance. A clear answer can come only from information on the structure of the TOM complex in the presence of preproteins.
Materials and methods
General biochemical procedures
Mitochondria and outer membrane vesicles from Neurospora crassa were isolated as described (Mayer et al., 1993). TOM holo or core complexes were purified according to published procedures (Künkele et al., 1998a; Ahting et al., 1999). The purity of both complexes was >95%. Trypsinized or proteinase K-treated complexes were obtained by addition of trypsin (20 µg/ml) or proteinase K (2 µg/ml) for 15 min at 4°C, respectively. Trypsin was inhibited with soybean trypsin inhibitor (400 µg/ml). Proteinase K was inactivated with 1 mM phenylmethylsulfonyl fluoride (PMSF). Proteins were analyzed by gel electrophoresis, immunodecoration and visualization by the enhanced chemiluminescence (ECL) method (Amersham). All protein concentrations were determined using the Bradford assay and immunoglobulin Gs as standard (Bio-Rad). Lipids were prepared from mitochondrial outer membrane vesicles of N.crassa as previously described (de Kroon et al., 1997).
Preparation and labeling of precursor proteins
C-terminally His-tagged versions of pSu9-DHFR consisting of the presequence of subunit 9 of the F0-ATPase (amino acid residues 1–69 or 1–45) and DHFR were constructed. A cysteine codon was inserted by PCR between the region encoding the DHFR domain and the six His residues. This construct was cloned in a pQE40 vector (Qiagen), expressed in Escherichia coli strain BL21 and purified by Ni-NTA chromatography according to the manufacturer’s protocol. A C-terminally His-tagged version of pSu9(1–69)-barnase was also constructed. It was overexpressed and purified as described above.
Su9(1–69)-DHFR was fluorescently labeled by reacting it with 0.01 mM 5-(and -6)-carboxytetramethylrhodamine, succinimidyl ester (Molecular Probes), for 12 h at 4°C at a molar protein:dye ratio of 10:1. Another labeling procedure involved reducing the preprotein with tris(2-carboxyethyl)phosphine–HCl, then incubating it with 0.16 mM 5-carboxytetramethylrhodamine maleimide for 12 h at 4°C in the presence of 1 µM MTX and 0.5 mM NADPH. In both procedures, unbound dye was removed by passing the fluorescently labeled protein over a size-exclusion column (PD-10; Amersham–Pharmacia Biotech).
Gel filtration analysis and gradient centrifugation
Purified TOM complex in buffer containing 50 mM potassium acetate, 0.1% DDM, 10% glycerol pH 7.0 was incubated in a total reaction volume of 100 µl for 1 h on ice with precursors or control proteins. The mixtures were applied to a TSK G4000 PWXL gel filtration column (30 cm × 7.8 mm; TosoHaas) and eluted with a buffer containing 10 mM MOPS, 50 mM potassium acetate, 10% glycerol, 0.5% digitonin pH 7.0, at a flow rate of 0.45 ml/min. Fractions (0.3 ml) were collected and analyzed by SDS–PAGE and Coomassie staining. As standards for quantification, increasing amounts of either preprotein or the TOM complex alone were loaded on the same gel. The gels were scanned and bands were quantified by densitometry. For analysis of binding by gradient centrifugation, the total reaction volume was 190 µl and the incubation buffer contained 50 mM potassium acetate, 0.1% DDM pH 7.0. The samples were loaded on the top of sucrose step gradients. The steps (100 µl) contained 11, 17, 22, 27 and 35% (w/v) sucrose in 0.5 mM EDTA, 10 mM MOPS–KOH and 0.5% digitonin. The gradients were centrifuged for 16 h at 140 000 g in an SW60 Ti rotor (Beckman). Fractions of 100 µl were collected and analyzed by SDS–PAGE and Coomassie staining.
FCS
FCS measurements were performed with a ConfoCor fluorescence correlation spectrometer (Carl Zeiss Jena, Jena and EVOTEC Biosystems, Hamburg, Germany). In this configuration, laser light from a He/Ne laser was passed through a 543 nm filter and focused into the sample solution via a Zeiss C-Apochromat objective (40×, 1.2 W korr). Emitted fluorescence was collected by the same objective; laser scattering was blocked by a dichroic mirror. Fluorescence was detected with an avalanche photodiode and processed by a 288-channel hardware correlator PC-card. Experimental autocorrelation curves were then fitted by theoretical autocorrelation functions using the FCS-plus 1.0 software package (EVOTEC Biosystems) yielding the diffusion times of free and bound fluorescent preprotein. To determine the dimensions of the excitation volume, a measurement was taken on a molecule with a known diffusion coefficient (rhodamine 6G). Measurements were taken in typical sample volumes of 40 µl in a 384-well chamber and diffusion times of individual fluorescent molecules were calculated from the time-dependent fluctuations of the fluorescent signal recorded in a confocal volume element. TOM complex concentrations were in the range 1–300 nM and the concentration of fluorescently labeled pSu9(1–69)-DHFR ranged between 5 and 20 nM. All measurements were taken at room temperature. The buffer for measurements contained 10 mM MOPS, 20 mM KCl, 5 mM MgCl2, 0.25% BSA, 0.5% digitonin, 5% glycerol pH 7.0. The sampling time was 15 s.
Protection of preprotein bound to the TOM complex against cleavage by MPP
Chemical amounts of mitochondrial precursors were incubated in buffer containing 50 mM potassium acetate, 20 mM Tris–HCl, 0.1% digitonin, 1 mM dithiothreitol, 2 mM MnCl2 pH 7.4, alone or in the presence of either purified TOM complex or BSA for 30 min at various temperatures. Then recombinant MPP purified from an E.coli strain overexpressing this protein [a kind gift of Dr V.Geli (Luciano et al., 1997)] was added and incubation was continued for 8 min at 25°C. MPP was then inhibited with 4 mM EDTA. Proteins in the reaction mixtures were precipitated with trichloroacetic acid and analyzed by SDS–PAGE, and immunodecoration with antibodies against Tom components and DHFR or barnase was performed.
Acknowledgments
Acknowledgements
The excellent technical assistance of M.Braun, P.Heckmeyer and U.Staudinger is gratefully acknowledged. We thank Dr V.Geli for the E.coli strain expressing MPP, Dr C.Koehler for the antibody against barnase, Dr A.Matouschek for the barnase construct, and Dr K.Kaluza (Roche Diagnostics GmbH) for making possible the use of the FCS equipment. This research was supported by the Sonderforschungsbereich 184 of the Deutsche Forschungsgemeinschaft (S.N. and W.N.), and the Münchener Medizinische Wochenschrift (S.N.).
References
- Abe Y., Shodai,T., Muto,T., Mihara,K., Torii,H., Nishikawa,S., Endo,T. and Kohda,D. (2000) Structural basis of presequence recognition by the mitochondrial protein import receptor Tom20. Cell, 100, 551–560. [DOI] [PubMed] [Google Scholar]
- Ahting U., Thun,C., Hegerl,R., Typke,D., Nargang,F.E., Neupert,W. and Nussberger,S. (1999) The TOM core complex: the general protein import pore of the outer membrane of mitochondria. J. Cell Biol., 147, 959–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brix J., Dietmeier,K. and Pfanner,N. (1997) Differential recognition of preproteins by the purified cytosolic domains of the mitochondrial import receptors Tom20, Tom22 and Tom70. J. Biol. Chem., 272, 20730–20735. [DOI] [PubMed] [Google Scholar]
- Brix J., Rudiger,S., Bukau,B., Schneider-Mergener,J. and Pfanner,N. (1999) Distribution of binding sequences for the mitochondrial import receptors Tom20, Tom22 and Tom70 in a presequence-carrying preprotein and a non-cleavable preprotein. J. Biol. Chem., 274, 16522–16530. [DOI] [PubMed] [Google Scholar]
- Dekker P.J.T., Ryan,M.T., Brix,J., Müller,H., Hönlinger,A. and Pfanner,N. (1998) Preprotein translocase of the outer mitochondrial membrane: molecular dissection and assembly of the general import pore complex. Mol. Cell. Biol., 18, 6515–6524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kroon A.I.P.M., Dolis,D., Mayer,A., Lill,R. and de Kruijff,B. (1997) Phospholipid composition of highly purified mitochondrial outer membranes of rat liver and Neurospora crassa. Is cardiolipin present in the mitochondrial outer membrane? Biochim. Biophys. Acta, 1325, 108–116. [DOI] [PubMed] [Google Scholar]
- Dietmeier K., Hönlinger,A., Bömer,U., Dekker,P.J.T., Eckerskorn,C., Lottspeich,F., Kübrich,M. and Pfanner,N. (1997) Tom5 functionally links mitochondrial preprotein receptors to the general import pore. Nature, 388, 195–200. [DOI] [PubMed] [Google Scholar]
- Gaikwad A.S. and Cumsky,M.G. (1994) The use of chemical cross-linking to identify proteins that interact with a mitochondrial presequence. J. Biol. Chem., 269, 6437–6443. [PubMed] [Google Scholar]
- Harkness T.A., Nargang,F.E., van der Klei,I., Neupert,W. and Lill,R. (1994) A crucial role of the mitochondrial protein import receptor MOM19 for the biogenesis of mitochondria. J. Cell Biol., 124, 637–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill K., Model,K., Ryan,M.T., Dietmeier,K., Martin,F., Wagner,R. and Pfanner,N. (1998) Tom40 forms the hydrophilic channel of the mitochondrial import pore for preproteins. Nature, 395, 516–521. [DOI] [PubMed] [Google Scholar]
- Hönlinger A. et al. (1995) The mitochondrial receptor complex: Mom22 is essential for cell viability and directly interacts with preproteins. Mol. Cell. Biol., 15, 3382–3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata K. and Nakai,M. (1998) Interaction between mitochondrial precursor proteins and cytosolic soluble domains of mitochondrial import receptors, Tom20 and Tom70, measured by surface plasmon resonance. Biochem. Biophys. Res. Commun., 253, 648–652. [DOI] [PubMed] [Google Scholar]
- Kanamori T., Nishikawa,S.-I., Nakai,M., Shin,I., Schultz,P.G. and Endo,T. (1999) Uncoupling of transfer of the presequence and unfolding of the mature domain in precursor translocation across the mitochondrial outer membrane. Proc. Natl Acad. Sci. USA, 96, 3634–3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiya T., Rospert,S., Schatz,G. and Mihara,K. (1997) Binding of mitochondrial precursor proteins to the cytoplasmic domains of the import receptors Tom70 and Tom20 is determined by cytoplasmic chaperones. EMBO J., 16, 4267–4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiya T., Rospert,S., Koehler,C., Looser,R., Schatz,G. and Mihara,K. (1998) Interaction of mitochondrial targeting signals with acidic receptor domains along the protein import pathway: evidence for the ‘acidic chain’ hypothesis. EMBO J., 17, 3886–3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Künkele K.-P. et al. (1998a) The preprotein translocation channel of the outer membrane of mitochondria. Cell, 93, 1009–1019. [DOI] [PubMed] [Google Scholar]
- Künkele K.-P., Juin,P., Pompa,C., Nargang,F.E., Henry,J.-P., Neupert,W., Lill,R. and Thieffry,M. (1998b) The isolated complex of the translocase of the outer membrane of mitochondria. J. Biol. Chem., 273, 31032–31039. [DOI] [PubMed] [Google Scholar]
- Lithgow T., Glick,B.S. and Schatz,G. (1995) The protein import receptor of mitochondria. Trends Biochem. Sci., 20, 98–101. [DOI] [PubMed] [Google Scholar]
- Luciano P., Geoffroy,S., Brandt,A., Hernandez,J.F. and Geli,V. (1997) Functional cooperation of the mitochondrial processing peptidase subunits. J. Mol. Biol., 272, 213–225. [DOI] [PubMed] [Google Scholar]
- Maiti S., Haupts,U. and Webb,W.W. (1997) Fluorescence correlation spectroscopy: diagnostics for sparse molecules. Proc. Natl Acad. Sci. USA, 94, 11753–11757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matouschek A., Azem,A., Ratliff,K., Glick,B.S., Schmid,K. and Schatz,G. (1997) Active unfolding of precursor proteins during mitochondrial protein import. EMBO J., 16, 6727–6736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer A., Lill,R. and Neupert,W. (1993) Translocation and insertion of precursor proteins into isolated outer membranes of mitochondria. J. Cell Biol., 121, 1233–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer A., Nargang,F.E., Neupert,W. and Lill,R. (1995a) MOM22 is a receptor for mitochondrial targeting sequences and cooperates with MOM19. EMBO J., 14, 4204–4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer A., Neupert,W. and Lill,R. (1995b) Mitochondrial protein import: reversible binding of the presequence at the trans side of the outer membrane drives partial translocation and unfolding. Cell, 80, 127–137. [DOI] [PubMed] [Google Scholar]
- Neupert W. (1997) Protein import into mitochondria. Annu. Rev. Biochem., 66, 863–917. [DOI] [PubMed] [Google Scholar]
- Rapaport D., Neupert,W. and Lill,R. (1997) Mitochondrial protein import. Tom40 plays a major role in targeting and translocation of preproteins by forming a specific binding site for the presequence. J. Biol. Chem., 272, 18725–18731. [DOI] [PubMed] [Google Scholar]
- Rapaport D., Künkele,K.-P., Dembowski,M., Ahting,U., Nargang,F.E., Neupert,W. and Lill,R. (1998a) Dynamics of the TOM complex of mitochondria during binding and translocation of preproteins. Mol. Cell. Biol., 18, 5256–5262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapaport D., Mayer,A., Neupert,W. and Lill,R. (1998b) cis and trans sites of the TOM complex in unfolding and initial translocation of preproteins. J. Biol. Chem., 273, 8806–8813. [DOI] [PubMed] [Google Scholar]
- Roise D. (1992) Interaction of a synthetic mitochondrial presequence with isolated yeast mitochondria: mechanism of binding and kinetics of import. Proc. Natl Acad. Sci. USA, 89, 608–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roise D. and Schatz,G. (1988) Mitochondrial presequences. J. Biol. Chem., 263, 4509–4511. [PubMed] [Google Scholar]
- Schatz G. and Dobberstein,B. (1996) Common principles of protein translocation across membranes. Science, 271, 1519–1526. [DOI] [PubMed] [Google Scholar]
- Schleiff E., Shore,G.C. and Goping,I.S. (1997) Interactions of the human mitochondrial protein import receptor, hTom20, with precursor proteins in vitro reveal pleiotropic specificities and different receptor domain requirements. J. Biol. Chem., 272, 17784–17789. [DOI] [PubMed] [Google Scholar]
- Schlossmann J., Dietmeier,K., Pfanner,N. and Neupert,W. (1994) Specific recognition of mitochondrial preproteins by the cytosolic domain of the import receptor MOM72. J. Biol. Chem., 269, 11893–11901. [PubMed] [Google Scholar]
- Soll J. and Waegemann,K. (1992) A functionally active protein import complex from chloroplasts. Plant J., 2, 253–256. [Google Scholar]
- Tamm L.K. (1991) Membrane insertion and lateral mobility of synthetic amphiphilic signal peptides in lipid model membranes. Biochim. Biophys. Acta, 1071, 123–148. [DOI] [PubMed] [Google Scholar]
- van der Does C., Swaving,J., Van Klompenburg,W. and Driessen,A.J.M. (2000) Non-bilayer lipids stimulate the activity of the reconstituted bacterial protein translocase. J. Biol. Chem., 275, 2472–2478. [DOI] [PubMed] [Google Scholar]
- van Wilpe S. et al. (1999) Tom22 is a multifunctional organizer of the mitochondrial preprotein translocase. Nature, 401, 485–489. [DOI] [PubMed] [Google Scholar]
- van Voorst F. and de Kruijff,B. (2000) Role of lipids in the translocation of proteins across membranes. Biochem. J., 347, 601–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vestweber D., Brunner,J., Baker,A. and Schatz,G. (1989) A 42K outer-membrane protein is a component of the yeast mitochondrial protein import site. Nature, 341, 205–209. [DOI] [PubMed] [Google Scholar]
- von Heijne G. (1986) Mitochondrial targeting sequences may form amphiphilic helices. EMBO J., 5, 1335–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]