Structural insights to how mammalian capping enzyme reads the CTD code (original) (raw)
. Author manuscript; available in PMC: 2012 Jul 22.
Summary
Physical interaction between the phosphorylated RNA polymerase II carboxyl-terminal domain (CTD) and cellular capping enzymes is required for efficient formation of the 5' mRNA cap, the first modification of nascent mRNA. Here we report the crystal structure of the RNA guanylyltransferase component of mammalian capping enzyme (Mce) bound to a CTD phosphopeptide. The CTD adopts an extended β-like conformation that docks Tyr1 and Ser5-PO4 onto the Mce nucleotidyltransferase domain. Structure-guided mutational analysis verified that the Mce-CTD interface is a tunable determinant of CTD binding and stimulation of guanylyltransferase activity, and of Mce function in vivo. The location and composition of the CTD binding site on mammalian capping enzyme is distinct from that of a yeast capping enzyme that recognizes the same CTD primary structure. Thus, capping enzymes from different taxa have evolved different strategies to read the CTD code.
Introduction
The largest subunit of RNA polymerase II (Pol II) contains a unique C-terminal domain (CTD) composed of a tandemly repeated heptapeptide motif of a consensus sequence Y1S2P3T4S5P6S7 (Chapman et al., 2008; Egloff and Murphy, 2008). The CTD is essential, and it serves as a landing platform for factors that regulate and/or catalyze mRNA processing and gene expression (Buratowski, 2009; Phatnani and Greenleaf, 2006). The CTD undergoes successive rounds of phosphorylation and dephosphorylation at Ser2, Ser5 and Ser7 in coordination with the transcriptional cycle (Cho, 2007; Egloff et al., 2007; Zhang and Corden, 1991a). In the Pol II pre-initiation complex, the CTD is hypophosphorylated, but transitions to a hyperphosphorylated state in the elongation complex. The ensemble of phosphorylation sites shifts from Ser5 to Ser2 as Pol II moves through the transcription unit. Remodeling the CTD phosphorylation array, and accompanying changes in CTD conformation (Zhang and Corden, 1991b), are thought to regulate the ingress and egress of the proteins that modify the nascent RNA.
The m7GpppN 5' cap of eukaryal mRNA is essential for cell viability (Mao et al., 1996; Tsukamoto et al., 1997) and is the first modification of nascent pre-mRNA (Chiu et al., 2002; Halger and Shuman, 1992; Rasmussen and Lis, 1993). Capping entails three consecutive enzymatic reactions: i) the 5′ triphosphate terminus of pre-mRNA is cleaved to a diphosphate by RNA triphosphatase; ii) the diphosphate end is capped with GMP by RNA guanylyltransferase; iii) the GpppN cap is methylated by RNA guanine-N7 methyltransferase (Ghosh and Lima, 2010; Shuman, 2001). Capping enzymes interact directly with the Ser5-phosphorylated form of the Pol II CTD, which is generated immediately after transcription initiation (Cho et al., 1997; Ghosh and Lima, 2010; McCracken et al., 1997; Yue et al., 1997; Ho and Shuman, 1999). In particular, the phospho-specific binding of RNA guanylyltransferase (GTase) to the CTD is conserved among budding yeast, fission yeast, and mammals, notwithstanding major differences in the physical organization and domain architectures of their respective capping systems (reviewed by Ghosh and Lima, 2010). It is thought that the GTase-CTD contacts target cap addition specifically to nascent Pol II transcripts within an early temporal window, so that the capped pre-mRNA is protected from decay and shunted through downstream processing events (splicing and polyadenylylation) that are stimulated by the cap.
Metazoans encode a bifunctional capping enzyme composed of an N-terminal RNA triphosphatase domain fused to a C-terminal GTase domain (Ghosh and Lima, 2010; Shuman, 2002). The GTase enzymes comprise a branch of the covalent nucleotidyl transferase superfamily, which includes ATP-dependent and NAD+-dependent polynucleotide ligases (Gu and Lima, 2005; Shuman and Lima, 2004). GTases catalyze the transfer of GMP from GTP to the 5' diphosphate terminated RNA through a covalent GTase-(lysyl-Nζ)-GMP intermediate (Shuman and Hurwitz, 1981). Crystal structures of GTases and DNA ligases highlight a common fold consisting of an N-terminal nucleotidyl transferase (NT) domain and a C-terminal oligonucleotide-binding (OB) domain (Fabrega et al., 2003; Gu et al., 2010; Hakansson et al., 1997; Hakansson and Wigley, 1998; Lee et al., 2000; Odell et al., 2000; Subramanya et al., 1996). The GTase component of the mammalian capping enzyme interacts with the phosphorylated CTD; the triphosphate component does not (Ho et al., 1998; Ho and Shuman, 1999). Although mammalian GTase can bind CTDs that are phosphorylated at either Ser2 or Ser5 (Ho and Shuman, 1999), its guanylyltransferase activity is stimulated only when the CTD is phosphorylated at Ser5 (Ho and Shuman, 1999; Wen and Shatkin, 1999).
Atomic interactions between the phosphorylated CTD and a cellular GTase were first illuminated by the crystal structure of Candida albicans Cgt1 in complex with a CTD-Ser5 phosphopeptide (Fabrega et al., 2003). Mutational analyses validated that Cgt1 residues involved in CTD recognition were important for Cgt1 function in vivo. However, the lack of apparent conservation of the Cgt1 CTD-binding residues in the primary structures of mammalian GTases raised the prospect that capping enzymes from different taxa might recognize the same phosphorylated Pol II CTD primary structure in fundamentally different ways.
To uncover the basis for recognition of the phosphorylated CTD by mammalian capping enzyme Mce, we determined the crystal structure at a resolution of 2.8 Å of its GTase domain in complex with a CTD-Ser2/5 phosphopeptide. The structure reveals that the Mce NT domain recognizes a six amino acid segment of the CTD (S5PP6S7Y1S2PP3) in an extended β-like conformation. The Mce-CTD interface is distinct from that observed for Cgt1. Structure-guided mutational analyses confirm that the crystallographic contacts are important for Mce binding to CTD-PO4, for stimulation of Mce guanylyltransferase activity by a Ser5 phosphorylated CTD, and for Mce function in vivo.
Results and discussion
Overview of the mammalian RNA guanylyltransferase bound to RNA Poll II CTD
A segment of mouse Mce1 from amino acids (aa) 227 to 567 encompasses the GTase domain and retains guanylyltransferase activity in vitro (Figure S1). The Mce1-(227–567) protein (henceforth Mce) was purified (Figure S1A) and crystallized in the presence of an 18-aa synthetic CTD phosphopeptide, in which each Ser2 and Ser5 residue was phosphorylated (see below). The structure of the Mce-CTD complex was determined by molecular replacement using the NT domain of C. albicans Cgt1 (Fabrega et al., 2003) as a search model in Phaser (McCoy et al., 2007). Crystals contained two molecules of Mce in the asymmetric unit (protomers A and B). Additional electron densities were observed adjacent to the NT domain of both protomers (Figure S2A); these densities were identified as the phosphorylated Pol II CTD. The final model refined at 2.8 Å resolution had Rcryst and Rfree values of 0.23 and 0.28, respectively (Table 1).
Table 1.
Crystallographic Data Statistics
| Data Statistics | |
|---|---|
| Source | APS 24ID-C |
| Wavelength (Å) | 0.979 |
| Resolution (Å) | 50 – 2.8 (2.9 – 2.8) |
| Space Group | I222 |
| Unit Cell (Å) a, b, c | 83.0, 114.7,150.2 |
| # of observations | 114760 |
| # of unique reflections | 17720 |
| Completeness (%) | 99.4 (97.7) |
| Redundancy | 6.5 (5.3) |
| Mean l/σl | 11.4 (2.7) |
| Rmergea | 8.4 (45.9) |
| Cut-off criteria l/σl | −1 |
| Refinement Statistics | |
|---|---|
| Resolution Limits (Å) | 50–2.8 (2.9–2.8) |
| # of reflections (working/test) | 17661 (16756/905) |
| Rworkb/Rfree | 0.224(0.33)/0.28(0.357) |
| # atoms protein/CTD/water/guanine | 4133/111/122/22 |
| Avg B-factors protein/CTD/water/guanine | 58.4/96.4/44.0/94.6 |
| Bond r.m.s deviations length (Å)/angles (°) | 0.005/1.0 |
| Ramchandran plotc | |
| Most favored | 374 (83.7%) |
| Additionally allowed | 72 (16.1%) |
| Generously allowed | 1 (0.2%) |
| Disallowed region | 0 (0%) |
| MolProbityd | |
| Favored | 483 (94.3%) |
| Allowed | 512 (100%) |
| Outliers | 0 (0%) |
| Clash Score | 80th percentile |
| MolProbity Score | 93rd percentile |
| PDB ID | 3RTX |
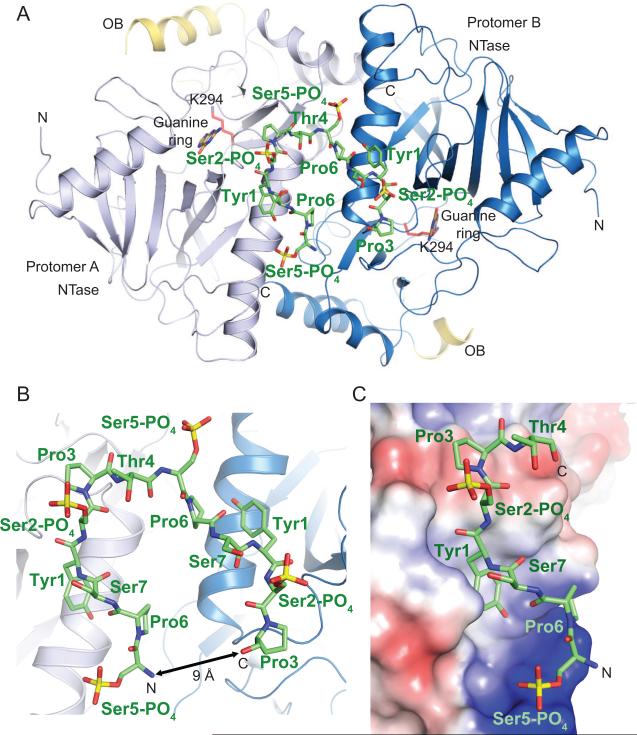
The Mce tertiary structure consists of NT and OB domains (Figure 1A) akin to the structures of fungal and viral GTases reported previously (Fabrega et al., 2003; Gu et al., 2010; Hakansson et al., 1997; Hakansson and Wigley, 1998). The NT domain includes aa 227–461 and a C-terminal helix (aa 553–567). The OB module spans aa 463–552 and projects away from the NT domain (Figure 1A). Electron density for the OB domains was very weak and precluded model building for most of the OB domains in the Mce structure. Recombinant capping enzymes purified from E. coli typically consist of a mixture of apoenzyme and enzyme-GMP and in the Mce crystal electron density was only observed for the guanine base in the nucleotide binding pocket in each of the two molecules in the asymmetric unit (Figure 1A).
Figure1. Structure of the mammalian capping enzyme Mce in complex with phosphorylated Pol II CTD.
A) A view of the Mce-CTD structure shown in ribbon representation with secondary structure elements indicated by arrows (β-strands) and ribbons (helices). A continuous thirteen amino acid CTD segment (in stick representation and colored green) interacts with two Mce protomers (protomer A and B). The NTase domains of the two protomers are labeled and colored in different shades of blue and the corresponding OB-fold domain elements that could be modeled are labeled and colored in yellow. N and C denote the location of the amino- and carboxy-terminus. The active site Lys294 and guanine base are labeled, shown in sticks and colored red and pale-yellow, respectively, to indicate the relative position of the Mce active site. All figures depicting structure were generated with PyMol (DeLano, 2002). B) A close-up view of the CTD associated with the two protomers of Mce. Mce is shown in ribbon representation with CTD residues labeled and represented by sticks. The distance between N and C termini of the CTD is denoted with an arrow. C) A close-up of Mce protomer A (surface representation colored by positive (blue) and negative (red) electrostatic potential). CTD residues are labeled and represented by sticks.
The Mce structure accounts for mutational data (Sawaya and Shuman, 2003) that identified sixteen essential and six nonessential side chains located within the seven signature peptide motifs of the GTase family (Figure S2B). Based on the crystal structure of Chlorella virus GTase, ten essential Mce side chains (Arg299, Arg315, Glu345, Tyr362, Lys458, Lys460, Asp468, Arg530, Lys533, and Asn537) were predicted to make direct contacts to the guanosine nucleotide. Six of the Mce side chains (Arg299, Arg315, Glu345, Tyr362, Lys458, and Lys460) could be included confidently in the Mce structure, which shows that they cluster around the nucleotide binding pocket (Figure S2B). The structure also confirms that the side chains of Asp343, Asp363, and Arg380 play essential structural roles in stabilizing the Mce active site via formation of salt bridges (Figure S2B). The structure explains why Asp296 and Arg452 were non-essential (Sawaya and Shuman, 2003), insofar as they are remote from the active site (Figure S2B).
Recognition of phosphorylated CTD by Mce
An 18 aa CTD phosphopeptide (T4S5PP6S7Y1S2PP3T4S5PP6S7Y1S2PP3T4S5PP6S7) was co-crystallized with Mce, of which 13 residues (S5PP6S7Y1S2PP3T4S5PP6S7Y1S2PP3) could be modeled into the electron density adjacent to the NT domains of the two Mce protomers in the asymmetric unit (Figure 1A and 1B and Figure S2A). Each protomer interacts with a 6 aa CTD segment (S5PP6S7Y1S2PP3) (Figure 1 and 2A). The two bound CTD segments are connected via Thr4 (Figure 1A, 1B and S2A), and each segment adopts an extended β-like conformation (Figure 1C and 2A). While it is interesting that two Mce protomers interact with a single CTD phosphopeptide, we do not think this `dimeric' architecture is physiologically relevant because residues in the Mce-Mce interface are not conserved in comparison to those in the Mce-CTD (see below). Our subsequent discussions of the atomic contacts at the Mce-CTD interface will refer to protomer A and its associated CTD segment because interactions between the CTD and Mce are similar in both complexes in the crystallographic asymmetric unit.
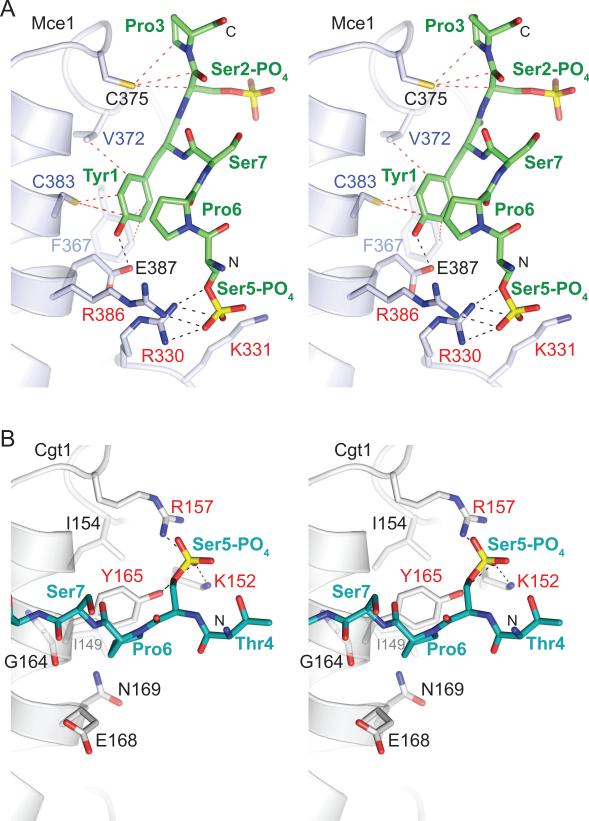
Figure 2. Capping enzyme interactions with phosphorylated CTD.
A) Stereo diagram of interactions between Mce and CTD. Selected side chains and phosphoserine residues are shown in stick representation and labeled. Potential hydrogen bonds and van der Waals contacts are denoted with black and red dashed lines, respectively. Mce residues that interact with CTD Tyr1 and Ser5-PO4 are labeled in blue and red, respectively. B) Stereo view of C. albicans guanylyltransferase Cgt1 (in light gray) bound to CTD (stick representation and colored cyan) depicting the same surface of Cgt1 as shown for Mce in panel A. Potential hydrogen bonds with Ser5-PO4 are shown with black dashes, and residues that interact with Ser5-PO4 are labeled in red.
The interface between Mce and CTD-PO4 buries approximately 675 A2 of the total surface area, mostly through packing of the Tyr1 side chain into a hydrophobic pocket in the NT domain formed by the side chains of Phe367 (Cδ and Cε), Val372 (Cγ), and Cys383 (Sγ) (Figure 1C and 2A). The hydroxyl group of Tyr1 side is also engaged in a H-bond interaction with the side chain carboxylate of Glu387 (Figure 2A). Direct contacts are also observed between Ser5-PO4 and side chain atoms of Mce residues Arg330 and Arg386 that form a positively charged pocket on the capping enzyme surface (Figure 1C and 2A). Additionally Lys331 side chain of Mce is in close proximity to make a direct contact to Ser5-PO4 (Figure 2A). The structural insights that Tyr1 and Ser5-PO4 dominate the Mce-CTD interface are fully consistent with earlier CTD structure-function studies showing that either mutation of CTD Tyr1 to alanine or subtraction of the phosphate from Ser5 effaced CTD binding to Mce and abolished CTD stimulation of its GTase activity (Pei et al., 2001). The same changes at Tyr1 and Ser5-PO4 ablate the interaction of the CTD with S. pombe guanylyltransferase Pct1 (Pei et al., 2001). Y1A or S5A substitutions in all heptad repeats of the CTD array are lethal in vivo in S. cerevisiae (Stiller et al., 2000; West and Corden, 1995).
The Mce-CTD complex shows that both Pro3 and Pro6 adopt a trans configuration analogous to that seen in crystal structures of other CTD-bound proteins, including: i) C. albicans guanylyltransferase Cgt1 (Fabrega et al., 2003; Figure 2B), ii) the peptidyl-proline isomerase Pin1-CTD (Verdecia et al., 2000), iii) the CTD-interacting domains (CIDs) of the 3' processing/termination factor Pcf11 and the splicing factor SCAF8 (Becker et al., 2008; Meinhart and Cramer, 2004), and iv) the small CTD phosphatase Scp1 (Zhang et al., 2006). Although each of these proteins recognizes the CTD in distinctly different conformations (see below), in silico modeling suggests that a cis proline would create steric clashes with each of the aforementioned complexes, including our structure of Mce. Consistent with our structural data in which Pro3 makes van der Waals (VDW) interactions with Mce Cys375 (Figure 1C and 2A), mutation of Pro3 to alanine lowers Mce-CTD interaction by 8-fold (Pei et al., 2001). Although the Pro6 side chain does not interact directly with Mce; the structure suggests that Pro6 could play an indirect role in positioning the Ser5-PO4 side chain via intramolecular VDW interactions with Try1 side chain (Cz) (Figure 2A). This could explain the 10-fold decreased affinity of Mce for a CTD substrate in which Pro6 was changed to alanine (Pei et al., 2001). To date, the only exception to the trans proline consensus of protein-bound CTDs is the crystal structure of CTD Ser5-specific phosphatase Ssu72, in which CTD Pro6 adopts cis configuration that positions Ser5-PO4 in the active site and avoids steric clashes with the enzyme that would be imposed by a Pro6 trans isomer (Xiang et al., 2010; Werner-Allen et al., 2010).
Mce1 interacts with a CTD that is phosphorylated either at Ser2 or Ser5 positions (Ho and Shuman, 1999). Competition experiments suggested the existence of a separate binding site for Ser2-PO4 CTD on Mce1 that does not overlap with the Ser5-PO4 interaction site because Mce1 can still be stimulated by Ser5-PO4 CTD in the presence of Ser2-PO4 CTD (Ho and Shuman, 1999). In the Mce-CTD structure, Ser2-PO4 is pointing away from the Mce-CTD interface (see above; Figure 1B, 1C and 2A). Although our structure does not provide insight to a second Ser2-PO4 CTD interaction site, it is consistent with available biochemical data which shows that a doubly Ser2/5-PO4 CTD does not bind to Mce1 any better than Ser2-PO4 or Ser5-PO4 CTD and was just as effective as Ser5-PO4 CTD in stimulating GTase activity (Ho and Shuman, 1999; Pei et al., 2001).
Thr4 does not contribute directly to the Mce-CTD interface (see above; Figure 1C and S2B); rather Thr4 bridges two CTD segments that are engaged in interactions with two Mce protomers (Figure 1 and S2A). This observation is consistent with the fact that substitution of Thr4 to alanine does not affect Mce interactions with the CTD (Pei et al., 2001) or CTD function in vivo (Stiller and Cook, 2004). It is worth noting that in the crystal structures of Cgt1-CTD (CDS1, Fabrega et al., 2003; Figure 2B) and Pin1-CTD (Verdecia et al., 2000) there are no Thr4 side chain contacts at the protein-CTD interface. Mutation of Thr4 to alanine does not affect CTD dephosphorylation by S. pombe Fcp1 or plant CTD Ser5 specific phosphatases Cpl1 and Cpl2 (Hausmann et al., 2004; 2005). However, the side chain of Thr4 is an important determinant for Ssu72's CTD Ser5 phosphatase activity (Hausmann et al., 2005). Consistent with the biochemical data, crystal structures of Ssu72-CTD complexes reveal that the side chain is involved in VDW interactions with the protein surface (Xiang et al., 2010; Werner-Allen et al., 2010).
CTD Ser7 is phosphorylated during early elongation and phosphorylation at this position persists until the Pol II reaches the 3' end of the transcription unit (Tietjen et al., 2010; Chapman et al., 2007). Serine occurs at the 7th position in the CTD heptad array most often but it is the most degenerate position and while phosphorylated in yeast, serine is not required at this position for cell viability (Kim et al., 2009; Stiller and Cook, 2004; West and Corden, 1995). However, Ser7-PO4 is required for optimal gene expression in mammals (Chapman et al., 2007) and for mammalian snRNA biognesis (Egloff et al., 2007). Our structure indicates that Ser7 does not contribute directly to the Mce-CTD interface.
Contacts to the CTD Tyr1 side chain contribute to CTD interaction
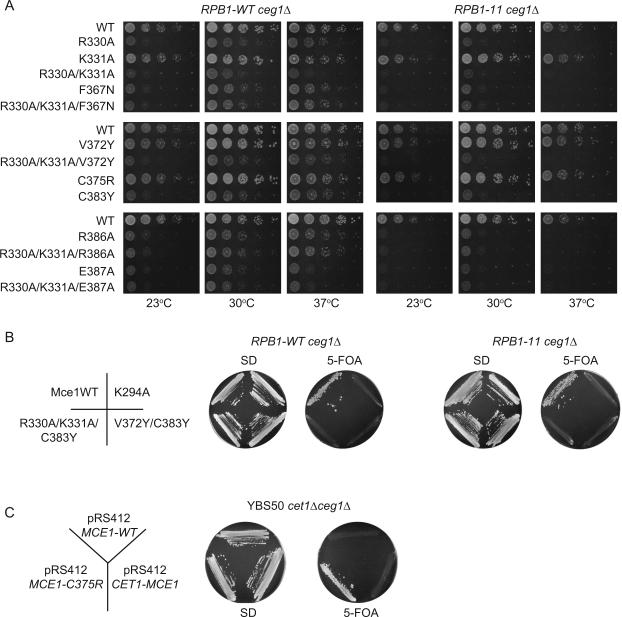
The CTD Tyr1 side chain makes contacts with four Mce side chains, which include VDW contacts with Phe367, Val372 and Cys383 and a hydrogen bond from the Tyr1 hydroxyl to the Glu387 carboxylate (Figure 1B and 2A). These interactions were probed by mutating Val372 and Cys383 to tyrosine, Phe367 to asparagine, and Glu387 to alanine. To gauge mutational effects on capping enzyme in vivo, we exploited the ability of Mce to substitute for the essential S. cerevisiae guanylyltransferase Ceg1, whereby MCE1 expression driven by the yeast TPI1 promoter complements a _ceg1_Δ strain in a plasmid shuffle assay (Figure 3; Sawaya and Shuman, 2003; Experimental Procedures). We found that whereas V372Y had little effect on yeast growth, the F367N, C383Y and E387A alleles resulted in slow growth phenotypes (Figure 3A).
Figure 3. Genetic analysis of Mce-CTD interactions.
A) Serial 10-fold dilutions of _S. cerevisiaeRPB1-WTceg1_Δ and _RPB1-11ceg1_Δ strains bearing indicated MCE1 alleles spotted on YPAD agar grown at 23°C (left), 30°C (middle), and 37°C (right). B) _S. cerevisiaeRPB1-WTceg1_Δ and _RPB1-11ceg1_Δ strains bearing pYX-MCE1 plasmids containing wild-type or mutant alleles K294A, R330A/K331A/C383Y, and V372Y/C383Y were assessed for growth at 30°C on agar medium lacking (left) or containing (right) 5-FOA. C) C375R mutation in the Mce-CTD interface results in a gain of function. A C375R substitution rescues conditional lethality in yeast when S. cerevisiae YBS50 _cet1_Δ_ceg1_Δ strain is complemented with a centromeric pRS412 plasmid containing _MCE1_-C375R flanked by CET1 5' and 3' UTR elements but not when complemented with a similar plasmid bearing MCE1 wild-type (WT) or CET1(276–549)-linker-MCE1(227–567) (Experimental Procedures). Strains were streaked on agar medium lacking (left) or containing (right) 5-FOA.
To correlate growth defects with interactions between Mce and the RNA Pol II CTD, we utilized an additional _ceg1_Δ strain that differed with respect to the number of CTD repeats present in the Rpb1 subunit. The RPB1-WT strain contained a wild-type RPB1 gene with a full complement of CTD heptads whereas the other strain, which we designate RPB1-11, encodes only the 11 proximal CTD heptads (Experimental Procedures), a minimal number required to support growth (Figure 3). The growth defects for the detrimental mutations described above were exacerbated in the _RPB1-11ceg1_Δ strain (Figure 3A), consistent with the idea that mutations in Mce that are predicted to weaken interactions with the CTD synergize with a mutation in RNA polymerase II that eliminates a portion of the CTD.
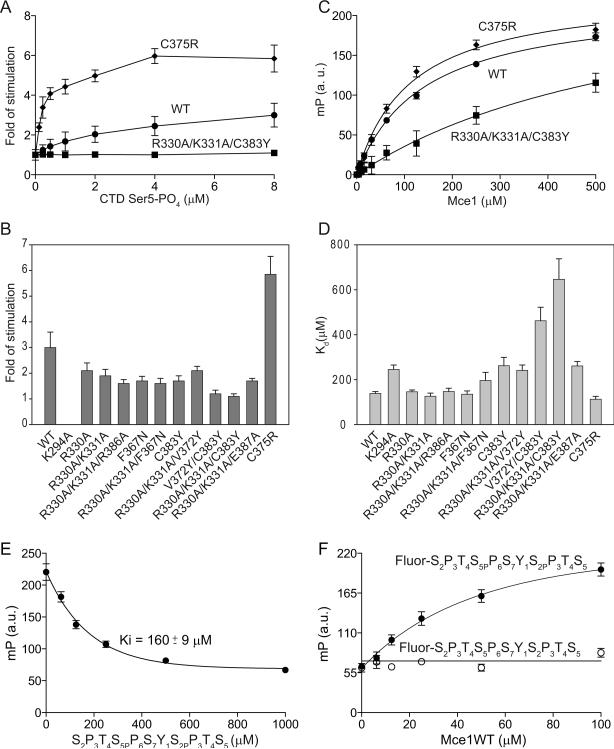
Interactions between Mce and the CTD were detected in vitro by monitoring Mce guanylyltransferase activity in the presence and absence of CTD Ser5-PO4 (Ho and Shuman, 1999). The formation of Mce-[α32P]GMP intermediate was stimulated 2.5-fold and 3-fold in the presence of tetrapeptide and hexaheptad CTD-Ser5-PO4, respectively. However, in the presence of two heptads the formation of covalent intermediate was enhanced by only 1.5-fold (Ho and Shuman, 1999). Consistent with these studies, we found that the yield of the covalent Mce-[α32P]GMP intermediate was stimulated 3-fold (with an apparent Kd of 1.5 μM) when wild-type Mce was incubated with a tetraheptad CTD-Ser5-PO4 peptide, but not with a tetraheptad CTD-Ser2-PO4 peptide (Figure 4A, 4B and S1C; Experimental Procedures). Under the condition tested, formation of the Mce-[α32P]GMP intermediate was stimulated ~1.3-fold in the presence of 18 aa synthetic CTD-PO4 (T4S5PP6S7Y1S2PP3T4S5PP6S7Y1S2PP3T4S5PP6S7, the same CTD substrate that was used for co-crystallization; data not shown). Consistent with previous results indicating that shorter CTD segments do not stimulate formation of the Mce-[α32P]GMP adduct, an 11 aa long phosphorylated CTD (S2P3T4S5PP6S7Y1S2PP3T4S5) that encompasses the 6 aa CTD residues (underlined) that contact Mce in the crystal structure was unable to stimulate Mce-[α32P]GMP formation (data not shown). These data suggest that avidity plays a substantive role in this assay, namely that stimulation of the Mce-[α32P]GMP adduct is achieved only when Mce is presented with multiple CTD heptad elements.
Figure 4. Stimulation of Mce guanylyltransferase activity by the CTD.
A) Stimulation of Mce wild-type, R330A/K331A/C383Y and C375R mutant proteins by increasing concentrations of CTD Ser5-PO4 (Experimental Procedures). Error bars (one standard deviation) were calculated from three independent experiments performed in triplicate. B) The fold increase in guanylylation activity was calculated by comparing the amount of enzyme-guanylate at 0 and 8 μM CTD Ser5-PO4 and depicted by bar graphs with error bars (one standard deviation). C) Data obtained from fluorescence polarization assays used to derive Kd values for Mce wild-type, R330A/K331A/C383Y and C375R mutant proteins at pH 7.0 (Experimental Procedures). Error bars (one standard deviation) obtained from three independent experiments performed in triplicate. D) Summary of dissociation constants in bar graph representation with error bars (one standard deviation) as determined by fluorescence polarization assays for Mce wild-type or mutant proteins and phosphorylated CTD. E) Displacement of fluorescein labeled CTD peptide (Fluor-S2P3T4S5PP6S7Y1S2PP3T4S5) bound to Mce wild-type protein with increasing quantities of unlabeled peptide (S2P3T4S5PP6S7Y1S2PP3T4S5) (Experimental Procedures). Fluorescence polarization was measured after incubating ternary complexes on ice for 20 minutes. Error bars (one standard deviation) were calculated from three independent experiments performed in triplicate. F) Mce1-CTD interaction depends on CTD phosphorylation at Ser2 and Ser5 positions. Fluorescein labeled Ser2 and Ser5 phosphorylated (Fluor-S2P3T4S5PP6S7Y1S2PP3T4S5; filled circles) and labeled dephosphorylated CTD (Fluor-S2P3T4S5P6S7Y1SPP3T4S5; open circles) peptides were incubated with increasing concentrations of Mce wild-type protein (Experimental Procedures).
In the presence of tetraheptad CTD-Ser5-PO4 peptide the F367N and C383Y mutants evinced reduced GTase stimulation (~1.7-fold), nearly half the level observed for the wild-type enzyme (Figure 4B and S1C). It is noteworthy that these mutated Mce proteins (and those described below) all displayed wild-type levels of enzyme guanylylation in the absence of the Ser5-PO4 CTD ligand, signifying that these amino acid substitutions did not have global effects on protein folding or catalysis (Figure S1B).
Although the 11 aa synthetic CTD peptide was unable to stimulate formation of the Mce-[α32P]GMP adduct, we could detect direct interactions by monitoring changes in fluorescence polarization in mixtures containing Mce and an analogous CTD peptide that was labeled at its N-terminus with fluorescein and phosphorylated on the central Ser2 and Ser5 positions (Fluor-S2P3T4S5PP6S7Y1S2PP3T4S5; Experimental Procedures). Fitting of polarization data to a hyperbolic curve showed that Mce interacts with this CTD peptide with a Kd of 139 μM (+/− 8.5) (Figure 4C, 4D and S3). Consistent with binding being dependent on the phosphorylated CTD peptide, the Fluor-CTD could be displaced with an unlabeled peptide equivalent in length and phase to the Fluor-labeled peptide (Experimental Procedures) with a calculated Ki of 160 μM (+/− 9) (Figure 4E). In addition, when the phosphorylated CTD peptide was dephosphorylated it no longer interacted with Mce (Figure 4F).
No effect in binding was observed for F367N in the fluorescence polarization assay, although C383Y exhibited a 1.9-fold decrease in affinity for the CTD (Figure 4D and S3). Because guanylylation assays were performed at pH 8.0 and fluorescence polarization measurements were conducted at pH 7.0, we queried if the higher pH might affect binding by accruing fluorescence polarization measurements for a subset of Mce mutants at pH 8.0. These data showed trends similar to those obtained at pH 7.0 (Figure S3).
In an attempt to fully occlude the pocket that coordinates the CTD Tyr1 side chain, we combined V372Y and C383Y substitutions in the V372Y/C383Y double mutant. This mutant allele failed to support growth in either _RPB1-WTceg1_Δ or _RPB1-11ceg1_Δ strains, as determined by the absence of colonies after 10 days during 5-FOA selection at 18°C, 30°C or 37°C (Figure 3B and data not shown). Ni-NTA pull-down and western blot analyses revealed that Mce-V372Y/C383Y was expressed in yeast at levels comparable to wild-type Mce (Figure S4). Consistent with this result, Mce-V372Y/C383Y guanylyltransferase activity could not be stimulated by up to 8 μM Ser5-PO4 CTD (~6-fold above the estimated Kd for the wild-type protein; Figure 4B and S1C), despite the fact that its basal guanylyltransferase activity was normal. In addition, Mce-V372Y/C383Y exhibited a 4-fold higher Kd (462 μM +/− 60) for the CTD in the fluorescence polarization assay (Figure 4D and S3) compared to wild-type Mce.
Contacts to the Ser5-PO4 contribute to Mce function
Contacts to the CTD Ser5-PO4 phosphate are established by a positively charged pocket on the Mce surface composed by side chains of Arg330, Lys331 and Arg386 (Figure 1B and 2A). Disruption of this charged surface was achieved by introducing individual alanine substitutions at positions 330, 331 and 386 or by combining these mutations pairwise (R330A/K331A) or all together R330A/K331A/R386A. Yeast R330A and R386A strains grew slower than wild-type MCE1 cells, whereas K331A had no effect on growth (Figure 3A). The K331A/R330A and R330A/K331A/R386A compound mutants had growth defects similar to R330A or R386A. In each case the growth phenotypes of these MCE1 mutants were more severe in _RPB1-11ceg1_Δ when compared to _RPB1-WTceg1_Δ. While little difference in CTD binding versus wild-type Mce were detected by fluorescence polarization for the R330A, R330A/K331A and R330A/K331A/R386A proteins (Figure 4D and S3A), each showed decreased stimulation of guanylylation in comparison to wild-type (2.1-, 1.9-, 1.6-fold, respectively; Figure 4B and S1C).
Contacts to Tyr1 and Ser5-PO4 synergize to facilitate CTD interaction
The R330A/K331A mutation was combined with F367N, V372Y, E387A or C383Y mutations to simultaneously perturb contacts with Ser5-PO4 and Tyr1 (Figure 3A, 4B, 4D, S1 and S3). The most instructive findings emerged from the combination of C383Y and R330A/K331A. The R330A/K331A/C383Y protein exhibited a Kd of 646.5 μM (± 91) for the CTD, a value 4.7-fold higher than observed for wild-type Mce (Figure 4C, 4D and S3). Consistent with this reduction in affinity, Mce-R330A/K331A/C383Y guanylyltransferase activity could not be stimulated by Ser5-PO4 CTD at up to 8 μM CTD concentration (Figure 4A, 4B and S1C) although basal guanylyltransferase activity was normal. Indeed, the mce1-R330A/K331A/C383Y allele did not support growth in either _RPB1-WTceg1_Δ or _RPB1-11ceg1_Δ strains (Figure 3B). Ni-NTA pull-down and western blot analysis revealed this mutant protein was expressed in yeast strains at levels comparable to wild-type (Figure S4). Thus, lesions at the Mce–CTD interface that cause the greatest decrement in CTD affinity and GTase activation (without impacting basal GTase activity; Figure S1B) also have the greatest impact on Mce function in vivo.
A C375R mutation in the Mce-CTD interface results in a gain-of-function
Mce Cys375 side chain Sγ interacts via VDW contacts with the CTD Ser2 Cα and Pro3 (Cδ) (Figure 1C and 2A). We attempted to perturb these interactions by changing Cys375 to a bulkier arginine side chain. Counter to our expectations, the C375R substitution increased the affinity of Mce for the CTD (Kd of 112.7 μM +/− 13; Figure 4C, 4D and S3) and resulted in a higher stimulation of guanylylation in the presence of CTD Ser5-PO4 (5.9-fold) compared to wild-type Mce (Figure 4A, 4B and S3C). Mce-C375R displayed wild-type levels of guanylylation in the absence of CTD and was unaffected by inclusion of a CTD Ser2-PO4 ligand (Figure S1C). These results attest to a specific gain-of-function in response to CTD Ser5-PO4. As one might expect, the mce1-C375R allele grew as well as wild-type in _RPB1-WTceg1_Δ and _RPB1-11ceg1_Δ (Figure 3A).
We next queried whether C375R could positively influence Mce function in vivo. The MCE1 wild-type allele does not support growth in S. cerevisiae YBS50 _cet1_Δ_ceg1_Δ strain when expressed under control of the CET1 promoter on a centromeric plasmid (Figure 3C; see Experimental Procedures) although this defect can be overcome either by placing the analogous MCE1 construct on a 2μ plasmid (Figure S5A; Experimental Procedures) or by driving MCE1 transcription from a CEN plasmid using a constitutive TPI1 promoter (Figure S5A; Experimental Procedures; Sawaya and Shuman, 2003). Consistent with the gain of function effects observed for the Mce-C375R protein in vitro, the mce1-C375R allele rescued conditional lethality in yeast when its expression was driven by the CET1 promoter on a CEN plasmid (Figure 3C); albeit with a slower growth phenotype compared to _TPI1_-MCE1 (Figure S5B). It is difficult to fully account for this gain of function in the absence of a structure of the C375R protein, but in silico modeling suggests that the arginine side chain could be accommodated in a CTD complex and that Arg375 might make additional VDW or electrostatic contacts to the CTD.
During the course of these studies an inactive Mce mutant was generated, Mce-K294A, in which the catalytic lysine was changed to alanine. As expected (Ho et al., 1998), Mce-K294A was incapable of guanylylation (Figure S1B). Fluorescence polarization data revealed that Mce-K294A affinity for CTD was reduced by ~1.8 fold in comparison to wild-type Mce (Figure 4D and S3). These results suggest a reciprocal relationship between CTD interaction and guanylyltransferase activity via an allosteric effect in which CTD stimulation of Mce-(lysyl-Nζ)-GMP formation also works in reverse, namely that in the absence of the enzyme guanylate, interactions with the CTD are weaker. The added value of such a feedback loop would be that guanylylated Mce, already primed for mRNA capping, is loaded on the phosphorylated CTD.
Distinct CTD interfaces of metazoan and fungal capping enzymes
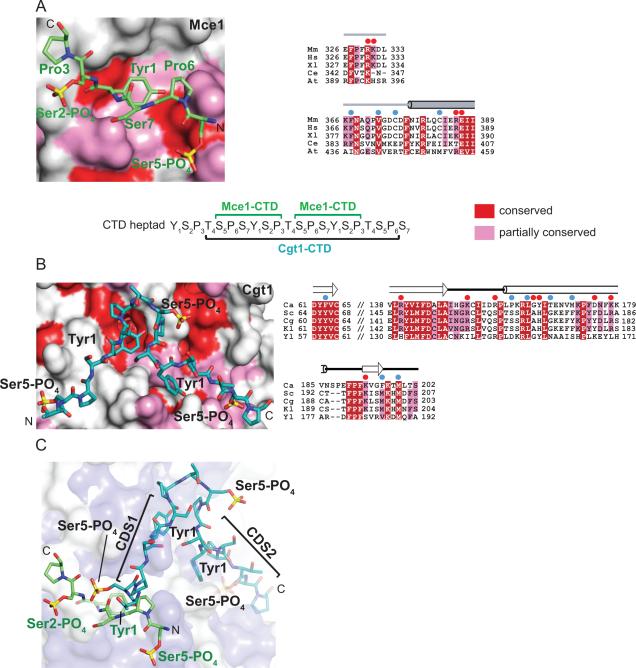
The NTase domains of Mce and Cgt1 recognize CTD peptides in an extended β-like conformation (see above; Figure 1C and 2A); however, the CTD moieties that interact with the respective capping enzyme surfaces are different (Figure 5). Comparison of CTD complexes with Mce and Cgt1 along with structure-based sequence alignments reveals that these two GTases utilize non-overlapping surfaces to recognize the phosphorylated CTD (Figure 5). Mce side chains involved in contacts to the phosphorylated CTD are not conserved in Cgt1 and are substituted by side chains with altered physiochemical properties, and vice versa (Figure 5A and 5B). The distinct properties of these non-overlapping surfaces in mammalian and fungal GTases that interact with the phosphorylated CTD suggest that these enzymes may have converged during evolution, evolving different surfaces to interact with and differentiate between phosphorylated states of the CTD. It is noteworthy that the chemical natures of Mce side chains involved in CTD interaction are either fully conserved (Arg330, Val372, Glu387) or partially conserved (Lys331, Phe367, Cys383, and Arg386) among metazoans and plants (Figure 5A) although the side chain at position 375 in Mce in Arabidopsis thaliana is replaced by arginine, an interesting observation given the gain of function observed for the Mce C375R substitution (Figure 3C, 4 and 5A).
Figure 5. CTD interactions with yeast and mammalian capping enzymes.
A) A close-up view and surface representation of the NTase domain of Mce (colored light gray) centered at the CTD binding site. Fully or partially conserved amino acids are colored in red or pink, respectively. Sequence conservation for Mce was generated by structure based sequence alignment of the corresponding metazoan NTase domains from M. musculus (Mm), H. sapiens (Hs), X. laevis (Xl), C. elegans (Ce), and A. thaliana (At). CTD residues are labeled, shown in stick representation and colored green. B) A closeup view of C. albicans Cgt1 surface (colored light gray) centered at the CTD binding site. Conserved residues are color coded as described in panel A. Sequence conservation was mapped on Cgt1 surface by aligning sequences corresponding to the NTase domains of C. albicans (Ca), S. cerevisiae (Sc), C. glabrata (Cg), K. lactis (Kl), and Y. lipolytica (Yl). CTD residues are shown in sticks, colored cyan and select positions are labeled. Aligned primary amino acid sequences are shown in the right panel and shaded according to sequence conservation. Secondary structure elements of Mce and Cgt1 are shown above the alignments. Residues in Mce or Cgt1 that are involved in H-bond and hydrophobic interactions are specified by red and blue circles above the sequence alignments, respectively. C) Surfaces of the NTase domains of Mce (blue) and Cgt1 (light gray) were superimposed to illustrate that the respective CTD elements (in stick representation colored as in panels A and B) are mostly non-overlapping and involve distinct surfaces of the Mce or Cgt1 NTase domains.
Reading the CTD code: Diversity in CTD conformation and recognition
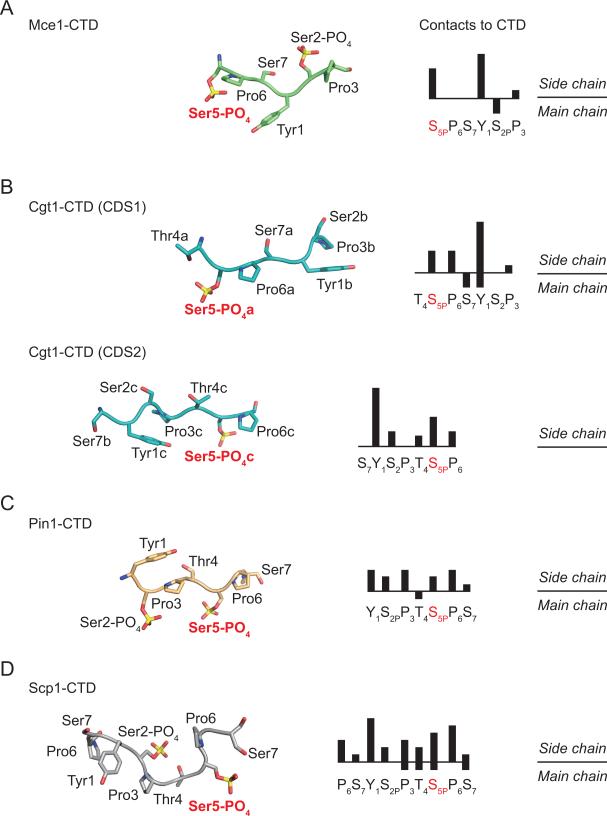
Despite being composed of a repeating heptapeptide sequence, the CTD is able to interact with a multitude of RNA processing factors in a coordinated fashion during various stages of transcription (Buratowski, 2009). Structural studies on protein-CTD complexes indicate that in solution the CTD is predominately disordered, and adopts defined structures only after binding to CTD interacting proteins (Noble et al., 2005; Lima, 2005). The Mce-CTD structure reveals that the S5PP6S7Y1S2PP3 ligand (see above; Figure 2A and 7A) adopts an extended β-like conformation that allows Tyr1 and Ser5-PO4 side chains to constitute the majority of interactions with Mce surface (see above; Figure 2A and 7A), although VDW contacts between Mce and CTD Ser2-PO4 main chain (C and Cα atoms) and Pro3 side chain are also observed (Figure 2A and 7A).
The structures of Candida albicans guanylyltransferase Cgt1 in complex with CTD reveals that the fungal enzyme also recognizes phosphorylated CTD in an extended β-turn conformation in both CTD docking sites, CDS1 and CDS2 (Fabrega et al., 2003; Figure 6B). This conformation of CTD allows every other side chain of the CTD to orient toward or away from the Cgt1-CTD interface. Whereas the CTD is recognized by Mce, Cgt1-CDS1 and Cgt1-CDS2 in different heptad phases, the Cgt1-CTD interfaces are also dominated by Cgt1 contacts to CTD Tyr1 and Ser5-PO4 side chains (Figure 6A and 6B). Additional VDW contacts are also observed between Cgt1-CDS1 and CTD Pro3, Pro6 and Ser7 (see above) and Cgt1-CDS2 with CTD Ser2, Thr4 and Pro6 (Figure 6B).
Figure 6. Conformational plasticity in CTD structures and mode of CTD recognition.
CTD heptad arrays extracted from structures of A) Mce (in green), B) Cgt1- CDS1 and CDS2 (in cyan; PDB: 1P16), C) Pin1 (in orange; PDB: 1F8A), and D) Scp1 (in gray; PDB: 1GHQ). CTD segments shown in ribbon (main chain) and stick representation (side chain) were aligned vertically based on the position of the Ser5-PO4 Cα atom. Although approximate, the orientation of the CTD structures also places the respective binding partners below the CTD. Right panels indicate phased CTD heptad sequences in bar graph format to illustrate the number of contacts (magnitude of the bar height) between the respective binding partner and CTD side chains (above) and main chain (below).
The peptidyl-proline isomerase Pin1 binds to CTD via its WW domain as illustrated by the crystal structure of a Pin-CTD complex in which both Ser2 and Ser5 were phosphorylated (Verdecia et al., 2000). In contrast to capping enzymes, the Pin1-bound CTD encompasses a single CTD heptad, which adopts an extended type-II polyproline helix (Figure 6C). This conformation orients every third residue of the CTD to a unique side of the coil such that both Ser2 and Ser5 phosphorylated side chains project from one face, Pro3 and Pro6 on another, and Tyr1, Thr4 and Ser7 project from the third. In contrast to interactions with the capping enzymes, all seven residues of the CTD are contacted by residues on the Pin1 surface; however, Pin1 establishes more contacts to CTD Tyr1, Pro3 and Pro6 side chains (Figure 6C).
Studies using a catalytically inactive form of the CTD phosphatase Scp1 revealed a structure of a nine amino acid CTD peptide which adopts a β-turn conformation (Figure 6D) in complex with Scp1 (Zhang et al., 2006). This conformation allows the CTD to fit into a groove within Scp1 that leads to its active site. Similar to Pin1 and in contrast to capping enzymes, Scp1 contacts each of the CTD residues in the complex; however more contacts were observed to Tyr1, Ser5-PO4, and Pro6 residues in comparison to other residues within the CTD substrate.
Conclusion
The complex between Mce and the phosphorylated CTD provides yet another example of how proteins differentially interact with a conformationally plastic CTD structure, a property that likely underlies reading, writing and erasing of the CTD code (Buratowski, 2003; Buratowski, 2009). While Mce interacts with a an extended β-like conformation of the CTD that projects the side chains of Tyr1 and Ser5-PO4 into the Mce-CTD interface (Figure 6A), it is interesting to note that Mce and C. albicans capping enzymes differ in their interactions with the phosphorylated CTD despite reading the same CTD code. Furthermore, capping enzyme-CTD complexes are distinct from other available structures of protein-CTD complexes (Verdecia et al., 2000; Meinhart and Cramer, 2004; Becker et al., 2008; Zhang et al., 2006) although it is interesting that contacts to CTD Tyr1 and phosphorylated Ser5 side chains predominate in most of the CTD-protein complexes resolved thus far, consistent with the observation that these two positions in the CTD heptad array are the least degenerate in Nature (Chapman et al., 2008).
Mutational analyses of Mce revealed the importance of contacts to Tyr1 and Ser5-PO4 positions within the CTD heptad array as mutations designed to disrupt these interactions impair growth in yeast, display lower stimulation of guanylylation, and lower affinities for phosphorylated CTD in comparison to the wild-type enzyme. The importance of Mce interactions with Ser5-PO4 CTD are further supported by our finding that Mce(C375R) binds to phosphorylated CTD better than the wild-type, displays increased stimulation of guanylylation in the presence of a CTD Ser5-PO4 substrate, and that complements loss of yeast capping enzyme when this mutant allele is provided on a single copy plasmid under control of the endogenous CET1 promoter. Taken together, these data underscore the importance of contacts between the capping enzyme and phosphorylated CTD, interactions that spatially and temporally regulate recruitment of capping enzyme to RNA polymerase II during early stages of transcription elongation.
Experimental Procedures
Cloning, Expression, and Purification of Recombinant Proteins
Procedures for cloning, expression, and purification of recombinant proteins utilized for crystallization trials and in vitro biochemistry are described in the Supplemental Data.
Crystallographic Analysis
Procedures for crystallographic analysis are provided in the Supplemental Data. Refinement and data statistics are documented in Table 1.
Complementation by plasmid shuffle in yeast
Plasmids and strains for complementation analyses are described in the main text or in the Supplemental Data.
Guanylyltransferase Assay
Procedures for basal and CTD dependent stimulated guanylylation activities are described in the Supplemental Data. In summary, Mce wild-type and mutant proteins were incubated with radiolabeled GTP [10% α32P] with or without CTD phosphopeptides at 37°C and quenched after 10 min with 1% SDS. The extent of gunylylation was quantified as described in the Supplemental Data.
Fluorescence polarization
Procedures for fluorescence polarization are described in the Supplementary Data. Briefly, fluorescein labeled phosphorylated CTD (Fluor-S2P3T4S5PP6S7Y1S2PP3T4S5) and dephosphorylated CTD (Fluor-S2P3T4S5P6S7Y1SPP3T4S5) peptides were incubated with increasing concentrations of Mce wild-type and polarization was measured at 23°C. For displacement assay fluorescein labeled CTD peptide (Fluor-S2P3T4S5PP6S7Y1S2PP3T4S5) bound to 100 μM Mce wild-type protein was incubated with increasing quantities (62.5 μM to 1000 μM) of unlabeled peptide (S2P3T4S5PP6S7Y1S2PP3T4S5).
Structure factors and coordinates are deposited in the RCSB Protein Data Bank with the accession code 3RTX.
Supplementary Material
01
Highlights
- Structure of a mammalian capping enzyme (CE) RNA Pol II CTD phosphopeptide complex
- Mammalian and yeast CEs use distinct surfaces to bind the Pol II CTD
- Mammalian and yeast CEs exploit different strategies to read the CTD code
- Interactions between mammalian CE and Pol II CTD are required for function in vivo
Acknowledgements
We thank Poulami Samai, Pravin Nair, and Shuman and Lima Lab members for helpful discussions. This work is based in part upon research conducted at the Advanced Photon Source on the Northeastern Collaborative Access Team beamlines, which are supported by award RR-15301 from the National Center for Research Resources at the National Institutes of Health. Use of the Advanced Photon Source is supported by the U.S. Department of Energy, Office of Basic Energy Sciences, under Contract No. DEAC02-06CH11357. A.G. was supported in part by a long-term fellowship by the HFSP. This work was supported in part by NIH grants GM052470 (S.S.) and GM061906 (A.G. and C.D.L).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Becker R, Loll B, Meinhart A. Snapshots of the RNA processing factor SCAF8 bound to different phosphorylated forms of the carboxyl-terminal domain of RNA polymerase II. J Biol Chem. 2008;283:22659–22669. doi: 10.1074/jbc.M803540200. [DOI] [PubMed] [Google Scholar]
- Buratowski S. The CTD code. Nat Struct Biol. 2003;10:679–680. doi: 10.1038/nsb0903-679. [DOI] [PubMed] [Google Scholar]
- Buratowski S. Progression through the RNA polymerase II CTD cycle. Mol Cell. 2009;36:541–546. doi: 10.1016/j.molcel.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman RD, Heidemann M, Albert TK, Mailhammer R, Flatley A, Meisterernst M, Kremmer E, Eick D. Transcribing RNA polymerase II is phosphorylated at CTD residue serine-7. Science. 2007;318:1780–1782. doi: 10.1126/science.1145977. [DOI] [PubMed] [Google Scholar]
- Chapman RD, Heidemann M, Hintermair C, Eick D. Molecular evolution of the RNA polymerase II CTD. Trends Genet. 2008;24:289–296. doi: 10.1016/j.tig.2008.03.010. [DOI] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu YL, Ho CK, Saha N, Schwer B, Shuman S, Rana TM. Tat stimulates cotranscriptional capping of HIV mRNA. Mol Cell. 2002;10:585–597. doi: 10.1016/s1097-2765(02)00630-5. [DOI] [PubMed] [Google Scholar]
- Cho EJ. RNA polymerase II carboxy-terminal domain with multiple connections. Exp Mol Med. 2007;39:247–254. doi: 10.1038/emm.2007.28. [DOI] [PubMed] [Google Scholar]
- Cho EJ, Takagi T, Moore CR, Buratowski S. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 1997;11:3319–3326. doi: 10.1101/gad.11.24.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific, San Carlos; CA, USA: 2002. [Google Scholar]
- Egloff S, Murphy S. Cracking the RNA polymerase II CTD code. Trends Genet. 2008;24:280–288. doi: 10.1016/j.tig.2008.03.008. [DOI] [PubMed] [Google Scholar]
- Egloff S, O'Reilly D, Chapman RD, Taylor A, Tanzhaus K, Pitts L, Eick D, Murphy S. Serine-7 of the RNA polymerase II CTD is specifically required for snRNA gene expression. Science. 2007;318:1777–1779. doi: 10.1126/science.1145989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrega C, Shen V, Shuman S, Lima CD. Structure of an mRNA capping enzyme bound to the phosphorylated carboxy-terminal domain of RNA polymerase II. Mol Cell. 2003;11:1549–1561. doi: 10.1016/s1097-2765(03)00187-4. [DOI] [PubMed] [Google Scholar]
- Ghosh A, Lima CD. Enzymology of RNA cap synthesis. Wiley Interdisciplinary Reviews: RNA. 2010;1:152–172. doi: 10.1002/wrna.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu M, Lima CD. Processing the message: structural insights into capping and decapping mRNA. Curr Opin Struct Biol. 2005;15:99–106. doi: 10.1016/j.sbi.2005.01.009. [DOI] [PubMed] [Google Scholar]
- Gu M, Rajashankar KR, Lima CD. Structure of the Saccharomyces cerevisiae Cet1-Ceg1 mRNA capping apparatus. Structure. 2010;18:216–227. doi: 10.1016/j.str.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakansson K, Doherty AJ, Shuman S, Wigley DB. X-ray crystallography reveals a large conformational change during guanyl transfer by mRNA capping enzymes. Cell. 1997;89:545–553. doi: 10.1016/s0092-8674(00)80236-6. [DOI] [PubMed] [Google Scholar]
- Hakansson K, Wigley DB. Structure of a complex between a cap analogue and mRNA guanylyl transferase demonstrates the structural chemistry of RNA capping. Proc Natl Acad Sci U S A. 1998;95:1505–1510. doi: 10.1073/pnas.95.4.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halger J, Shuman S. A freeze-frame view of eukaryotic transcription during elongation and capping of nascent mRNA. Science. 1992;255:983–986. doi: 10.1126/science.1546295. [DOI] [PubMed] [Google Scholar]
- Hausmann S, Koiwa H, Krishnamurthy S, Hampsey M, Shuman S. Different strategies for carboxyl-terminal domain (CTD) recognition by serine 5-specific CTD phosphatases. J Biol Chem. 2005;280:37681–3. doi: 10.1074/jbc.M505292200. [DOI] [PubMed] [Google Scholar]
- Hausmann S, Schwer B, Shuman S. An encephalitozoon cuniculi ortholog of the RNA polymerase II carboxyl-terminal domain (CTD) serine phosphatase Fcp1. Biochemistry. 2004;43:7111–7120. doi: 10.1021/bi0499617. [DOI] [PubMed] [Google Scholar]
- Ho CK, Shuman S. Distinct roles for CTD Ser-2 and Ser-5 phosphorylation in the recruitment and allosteric activation of mammalian mRNA capping enzyme. Mol Cell. 1999;3:405–411. doi: 10.1016/s1097-2765(00)80468-2. [DOI] [PubMed] [Google Scholar]
- Ho CK, Sriskanda V, McCracken S, Bentley D, Schwer B, Shuman S. The guanylyltransferase domain of mammalian mRNA capping enzyme binds to the phosphorylated carboxyl-terminal domain of RNA polymerase II. J Biol Chem. 1998;273:9577–9585. doi: 10.1074/jbc.273.16.9577. [DOI] [PubMed] [Google Scholar]
- Kim M, Suh H, Cho EJ, Buratowski S. Phosphorylation of the yeast Rpb1 C-terminal domain at serines 2, 5 and 7. J Biol Chem. 2009;284:26241–26426. doi: 10.1074/jbc.M109.028993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Chang C, Song HK, Moon J, Yang JK, Kim HK, Kwon ST, Suh SW. Crystal structure of NAD(+)-dependent DNA ligase: modular architecture and functional implications. EMBO J. 2000;19:1119–1129. doi: 10.1093/emboj/19.5.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima CD. Inducing interactions with the CTD. Nature structural & molecular biology. 2005;12:102–103. doi: 10.1038/nsmb0205-102. [DOI] [PubMed] [Google Scholar]
- Mao X, Schwer B, Shuman S. Mutational analysis of the Saccharomyces cerevisiae ABD1 gene: cap methyltransferase activity is essential for cell growth. Mol Cell Biol. 1996;16:475–480. doi: 10.1128/mcb.16.2.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken S, Fong N, Rosonina E, Yankulov K, Brothers G, Siderovski D, Hessel A, Foster S, Shuman S, Bentley DL. 5'-Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes Dev. 1997;11:3306–3318. doi: 10.1101/gad.11.24.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinhart A, Cramer P. Recognition of RNA polymerase II carboxy-terminal domain by 3'-RNA-processing factors. Nature. 2004;430:223–226. doi: 10.1038/nature02679. [DOI] [PubMed] [Google Scholar]
- Noble CG, Hollingworth D, Martin SR, Ennis-Adeniran V, Smerdon SJ, Kelly G, Taylor IA, Ramos A. Key features of the interaction between Pcf11 CID and RNA polymerase II CTD. Nature structural & molecular biology. 2005;12:144–151. doi: 10.1038/nsmb887. [DOI] [PubMed] [Google Scholar]
- Odell M, Sriskanda V, Shuman S, Nikolov DB. Crystal structure of eukaryotic DNA ligase-adenylate illuminates the mechanism of nick sensing and strand joining. Mol Cell. 2000;6:1183–1193. doi: 10.1016/s1097-2765(00)00115-5. [DOI] [PubMed] [Google Scholar]
- Pei Y, Hausmann S, Ho CK, Schwer B, Shuman S. The length, phosphorylation state, and primary structure of the RNA polymerase II carboxyl-terminal domain dictate interactions with mRNA capping enzymes. J Biol Chem. 2001;276:28075–28082. doi: 10.1074/jbc.M102170200. [DOI] [PubMed] [Google Scholar]
- Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20:2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- Rasmussen EB, Lis JT. In vivo transcriptional pausing and cap formation on three Drosophila heat shock genes. Proc Natl Acad Sci U S A. 1993;90:7923–7927. doi: 10.1073/pnas.90.17.7923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawaya R, Shuman S. Mutational analysis of the guanylyltransferase component of Mammalian mRNA capping enzyme. Biochemistry. 2003;42:8240–8249. doi: 10.1021/bi034396d. [DOI] [PubMed] [Google Scholar]
- Shuman S. Structure, mechanism, and evolution of the mRNA capping apparatus. Prog Nucleic Acid Res Mol Biol. 2001;66:1–40. doi: 10.1016/s0079-6603(00)66025-7. [DOI] [PubMed] [Google Scholar]
- Shuman S. What messenger RNA capping tells us about eukaryotic evolution. Nat Rev Mol Cell Biol. 2002;3:619–625. doi: 10.1038/nrm880. [DOI] [PubMed] [Google Scholar]
- Shuman S, Hurwitz J. Mechanism of mRNA capping by vaccinia virus guanylyltransferase: characterization of an enzyme--guanylate intermediate. Proc Natl Acad Sci U S A. 1981;78:187–191. doi: 10.1073/pnas.78.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuman S, Lima CD. The polynucleotide ligase and RNA capping enzyme superfamily of covalent nucleotidyltransferases. Curr Opin Struct Biol. 2004;14:757–764. doi: 10.1016/j.sbi.2004.10.006. [DOI] [PubMed] [Google Scholar]
- Stiller JW, Cook MS. Functional unit of the RNA polymerase II C-terminal domain lies within heptapeptide pairs. Eukaryot. Cell. 2004;3:735–740. doi: 10.1128/EC.3.3.735-740.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiller JW, McConaughy BL, Hall BD. Evolutionary complementation for polymerase II CTD function. Yeast. 2000;16:57–64. doi: 10.1002/(SICI)1097-0061(20000115)16:1<57::AID-YEA509>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Subramanya HS, Doherty AJ, Ashford SR, Wigley DB. Crystal structure of an ATP-dependent DNA ligase from bacteriophage T7. Cell. 1996;85:607–615. doi: 10.1016/s0092-8674(00)81260-x. [DOI] [PubMed] [Google Scholar]
- Tietjen JR, Zhang DW, Rodriguez-Molina JB, White BE, Akhtar MS, Heidemann M, Li X, Chapman RD, Shokat K, Keles S, et al. Chemical-genomic dissection of the CTD code. Nature structural & molecular biology. 2010;17:1154–1161. doi: 10.1038/nsmb.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto T, Shibagaki Y, Imajoh-Ohmi S, Murakoshi T, Suzuki M, Nakamura A, Gotoh H, Mizumoto K. Isolation and characterization of the yeast mRNA capping enzyme beta subunit gene encoding RNA 5'-triphosphatase, which is essential for cell viability. Biochem Biophys Res Commun. 1997;239:116–122. doi: 10.1006/bbrc.1997.7439. [DOI] [PubMed] [Google Scholar]
- Verdecia MA, Bowman ME, Lu KP, Hunter T, Noel JP. Structural basis for phosphoserine-proline recognition by group IV WW domains. Nat Struct Biol. 2000;7:639–643. doi: 10.1038/77929. [DOI] [PubMed] [Google Scholar]
- Wen Y, Shatkin AJ. Transcription elongation factor hSPT5 stimulates mRNA capping. Genes Dev. 1999;13:1774–1779. doi: 10.1101/gad.13.14.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner-Allen JW, Lee CJ, Liu P, Nicely NI, Wang S, Greenleaf AL, Zhou P. Cis proline-mediated pSer5-dephosphorylation by the RNA polymerase II CTD phosphatase Ssu72. J Biol Chem. 2010 doi: 10.1074/jbc.M110.197129. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West ML, Corden JL. Construction and analysis of yeast RNA polymerase II CTD deletion and substitution mutations. Genetics. 1995;140:1223–1233. doi: 10.1093/genetics/140.4.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang K, Nagaike T, Xiang S, Kilic T, Beh MM, Manley JL, Tong L. Crystal structure of the human symplekin-Ssu72-CTD phosphopeptide complex. Nature. 2010 doi: 10.1038/nature09391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Z, Maldonado E, Pillutla R, Cho H, Reinberg D, Shatkin AJ. Mammalian capping enzyme complements mutant Saccharomyces cerevisiae lacking mRNA guanylyltransferase and selectively binds the elongating form of RNA polymerase II. Proc Natl Acad Sci U S A. 1997;94:12898–12903. doi: 10.1073/pnas.94.24.12898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Corden JL. Identification of phosphorylation sites in the repetitive carboxyl-terminal domain of the mouse RNA polymerase II largest subunit. J Biol Chem. 1991a;266:2290–2296. [PubMed] [Google Scholar]
- Zhang J, Corden JL. Phosphorylation causes a conformational change in the carboxyl-terminal domain of the mouse RNA polymerase II largest subunit. J Biol Chem. 1991b;266:297–2302. [PubMed] [Google Scholar]
- Zhang Y, Kim Y, Genoud N, Gao J, Kelly JW, Pfaff SL, Gill GN, Dixon JE, Noel JP. Determinants for dephosphorylation of the RNA polymerase II C-terminal domain by Scp1. Mol Cell. 2006;24:759–770. doi: 10.1016/j.molcel.2006.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01