Elevated Rates of Protein Secretion, Evolution, and Disease Among Tissue-Specific Genes (original) (raw)
Abstract
Variation in gene expression has been held responsible for the functional and morphological specialization of tissues. The tissue specificity of genes is known to correlate positively with gene evolution rates. We show here, using large data sets, that when a gene is expressed highly in a small number of tissues, its protein is more likely to be secreted and more likely to be mutated in genetic diseases with Mendelian inheritance. We find that secreted proteins are evolving at faster rates than nonsecreted proteins, and that their evolutionary rates are highly correlated with tissue specificity. However, the impact of secretion on evolutionary rates is countered by tissue-specific constraints that have been held constant over the past 75 million years. We find that disease genes are underrepresented among intracellular and slowly evolving housekeeping genes. These findings illuminate major selective pressures that have shaped the gene repertoires expressed in different mammalian tissues.
The human body is assembled from >200 cell types present in a variety of tissue types. Variations in gene expression patterns are thought to underlie the morphological differences apparent between different tissue types (King and Wilson 1975; Alberts et al. 1994). Identifying the genes that specify morphological differentiation has been the subject of much research. However, a genome-scale analysis of these genes' characteristics has hitherto been lacking.
Comparative tissue gene-expression analysis can exploit high-throughput gene-expression data from expressed sequence tag (EST), serial analysis of gene expression (SAGE), and microarray gene-expression systems. In particular, high-quality data sets have been made available by Su and colleagues from 46 human and 45 mouse tissues obtained by use of high-density oligonucleotide microarrays (Su et al. 2002; http://expression.gnf.org). These have been normalized to enable informative comparisons among different tissues. These data show excellent reproducibility for repeat hybridizations of either the same sample, or different samples from the same tissue, when compared with SAGE or EST sets (Huminiecki et al. 2003).
Studying the evolution of genes has increased our understanding of the selective pressures that have shaped organism fitness (Hughes 1999; Wyckoff et al. 2000; Giraud et al. 2001; Jordan et al. 2001; Waterston et al. 2002; Zhang et al. 2002; Meiklejohn et al. 2003). Throughout gene evolution, different selective pressures have lead to fixation of coding-sequence changes in the population, which can be assessed by calculating the ratio of nonsynonymous to synonymous substitution rates (KA/KS) (Yang and Nielsen 2000). Genes with relatively low KA/KS ratios have been subject to negative (or purifying) selection; in contrast, genes with high ratios have been subject to positive (or adaptive) selection.
EST data have been used previously to show that substitution rates at nonsynonymous sites are strongly negatively correlated with tissue distribution breadth (Duret and Mouchiroud 2000). An additional observation reported was that, depending on the tissue source, different tissue-specific genes possess variable evolutionary rates. We decided to reinvestigate these findings and their implications in light of the recently sequenced human, mouse, and rat genomes, and by exploiting the reliable and independent source of microarray expression data made available by Su and colleagues (Su et al. 2002).
Several studies have considered the expression of single genes in multiple tissues from a single organism. In contrast, we wished to consider the expression of multiple genes in multiple tissues from two species (human and mouse) in order to investigate functional and evolutionary aspects of tissue biology. To link genetic, cellular, and tissue aspects with models of mammalian gene evolution, we have studied tissue-specific genes with respect to their involvement in disease, protein localization, and evolutionary rates.
RESULTS
Our initial studies investigated possible relationships between the following four quantities: (1) tissue specificity of gene expression, (2) protein secretion, (3) KA/KS ratio, and (4) association with human disease. Previous studies have suggested that the sequences of tissue-specific genes, and gene portions whose products are secreted, tend to be more divergent (Duret and Mouchiroud 2000; Waterston et al. 2002). It has been postulated (Duret and Mouchiroud 2000) that genes whose functions are critical for a larger number of tissues are more likely to be detrimental to the organism when mutated. To test this hypothesis, we sought to correlate tissue expression with disease genes that are annotated in the online Mendelian Inheritance in Man (OMIM) database (McKusick 2000).
Tissue Specificity of Gene Expression
We started by associating microarray expression data with gene sequences and calculating tissue-specificity values for these genes. Oligonucleotide tag sequences, and their associated expression profiles, were mapped to human genome coordinates and to 8159 human genes (see Methods). To limit the redundancy of tissue expression data sets, 27 tissues were chosen, such that no pair of tissue expression profiles was highly correlated (Supplemental Fig. 1 available online at www.genome.org; see Methods). Nevertheless, we note that brain and liver data from both fetal and adult organs were sufficiently different to be retained. Mouse one-to-one orthologs were obtained from the ENSEMBL database (http://www.ensembl.org), and consequently, KA/KS ratios could be calculated for 4960 of the 8159 genes. For each gene and each tissue, we calculated a tissue specificity value (T S), defined as the gene's fractional expression in that tissue relative to the sum of its expression in all tissues. For each gene, the maximum T S value (maxTS) among all tissues is thus an indicator of how much the expression of a gene is concentrated in few tissues.
To investigate possible relationships between T S and protein secretion, evolutionary rate, or disease, we divided the abovementioned 4960 genes into five partitions according to their maxTS (Table 1). Partition 1 (0 < maxTS < 0.1) contains housekeeping genes (Warrington et al. 2000) that are expressed relatively uniformly in most tissues, whereas partition 5 (maxTS > 0.4) contains genes that are highly expressed in one or two tissues.
Table 1.
Tissue Specificity of Gene Expression Correlates Positively With Higher Evolutionary Rates (Median Values of KA, KS, and KA/KS), the Fraction of Genes Whose Products Are Secreted (f sec) and the Fraction of Genes That are Linked to Disease (f dis)
| Partition | maxTS range | Gene number | Median maxTS | Median KA/KS | Median KA | Median KS | fsec (%) | fdis (%) | fLe (%) | fAll (%) | fLe/fAll (%) | K-S test probability |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | ≤0.1 | 1571 | 0.082 | 0.059 | 0.034 | 0.530 | 14.3 | 9.2 | 7.6 | 34.8 | 22.0 | 1.4 × 10-4, 1.2 × 10-5, 7.2 × 10-7, 2.2 × 10-16, 2.0 × 10-3, 3.8 × 10-4, 2.2 × 10-16 |
| 2 | 0.1—0.2 | 2074 | 0.133 | 0.070 | 0.043 | 0.574 | 21.9 | 11.9 | 5.3 | 27.1 | 19.4 | |
| 3 | 0.2—0.3 | 645 | 0.243 | 0.078 | 0.054 | 0.594 | 32.7 | 12.5 | 2.3 | 18.5 | 12.6 | 9.1 × 10-2, 7.9 × 10-9 |
| 4 | 0.3—0.4 | 294 | 0.335 | 0.090 | 0.063 | 0.658 | 37.4 | 15.0 | 1.0 | 15.3 | 6.7 | 5.8 × 10-4 |
| 5 | ≥0.4 | 376 | 0.530 | 0.126 | 0.089 | 0.655 | 45.5 | 25.8 | 0.8 | 11.2 | 7.1 | --- |
Studies of All Genes
For each of the five maxTS partitions, we calculated three quantities as follows: the median of the genes' KA/KS values, the fraction of predicted (Nielsen et al. 1997) secreted gene products (f sec), and the fraction of disease genes (f dis) (see Methods). Each of these three quantities was found to exhibit positive correlations with maxTS (Table 1). Moreover, the five partitions display significantly different KA/KS distributions (Table 1; Fig. 1A). Thus, in general, a tissue-specific gene is more likely to be evolving faster, to have secreted products, and to be mutated in human disease than housekeeping genes.
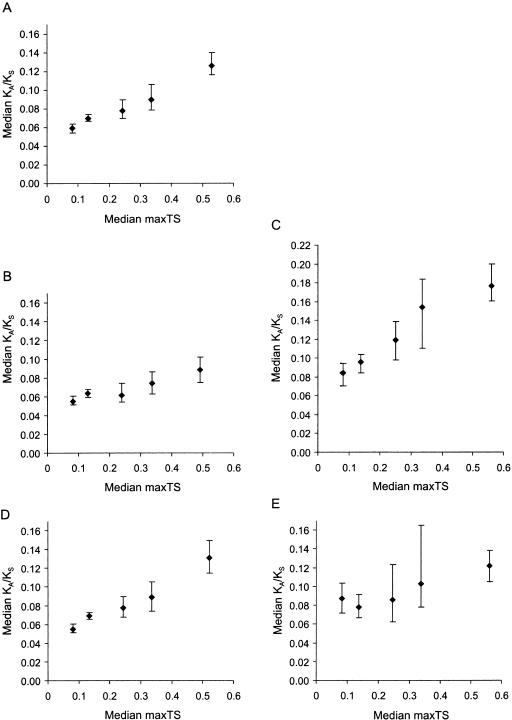
Figure 1.
Variation of the evolutionary rate (KA/KS) with maximum tissue specificity (maxTS) across five partitions. (A) All genes. (B) Genes whose products lack a detectable signal peptide. (C) Genes whose products contain a signal peptide, and are thus likely to be secreted. (D) Genes not known to be associated with human disease. (E) Disease genes. Error bars represent the 95% confidence interval for the median (as implemented by MINITAB, http://www.minitab.com). Regression formulae for partitions 1 to 5 are: y = 0.0467 + 0.143_x_; y = 0.0492 + 0.0773_x_; y = 0.0711 + 0.200_x_; y = 0.0415 + 0.163_x_; and, y = 0.0715 + 0.0869_x_, respectively, where y = median KA/KS and x = median maxTS. The probabilities (_P_-values) that equivalent partitions for secreted and nonsecreted gene products differ are as follows: partitions 1: P = 3 × 10-5; partitions 2: P = 4.25 × 10-11; partitions 3: P = 3.5 × 10-7; partitions 4: P = 4.8 × 10-6; and partitions 5: P = 8.8 × 10-13; and, _P_-values for disease and nondisease genes: partitions 1: P = 1.0 × 10-4; partitions 2: P = 0.14; partitions 3: P = 0.67; partitions 4: P = 0.10; and partitions 5: P = 0.21.
Increase of tissue specificity is also associated with elevated median values of both KA and KS (Table 1). Rate variation in synonymous site substitutions (KS) has been proposed previously to have arisen from nonsynonymous (KA) mutational influences of 5′- and 3′-flanking bases (Bains 1992; Duret and Mouchiroud 2000). Accounting for this effect abolishes the correlation between KS and tissue specificity (Duret and Mouchiroud 2000). Our use of the KA/KS ratio, instead of KA, takes account of the underlying variation in synonymous rates. Correlations between codon bias and gene-expression levels, which would cause KS variations, have also been suggested (Castillo-Davis and Hartl 2002; Duret 2002), but whether this effect is significant for mammalian genomes remains uncertain.
To further investigate possible correlations among maxTS, median KA/KS, f sec, and f dis, we analyzed the relationships between each pair of these quantities in turn. When considering protein secretion and evolutionary rate, the set of secreted gene products was found to exhibit a significantly higher median KA/KS value (0.115) than the set of nonsecreted gene products (0.065; Kolmogorov-Smirnov _P_-value < 2 × 10-16). When considering protein secretion and disease association, we observed that 39% of the complete set of disease genes encode predicted secreted proteins, compared with only 16.1% of genes that are not known to be associated with disease.
In addition, secreted proteins exhibit a greater correlation between median KA/KS and maxTS than do nonsecreted proteins (Fig. 1B,C). This suggests that genes encoding secreted products account for much of the dependency observed between tissue specificity and KA/KS. We also found that the median KA/KS differences between secreted and nonsecreted proteins are significant for each maxTS partition (Fig. 1, legend). This indicates that secretion and KA/KS are highly correlated, irrespective of tissue specificity.
We found no significant difference between the KA/KS distributions for disease genes and nondisease genes (Kolmogorov-Smirnov test probability for the difference = 0.36). However, when measuring the dependency between median KA/KS and tissue specificity, we found that for partition 1 only, disease genes exhibit significantly higher KA/KS values, on average, than nondisease genes (P = 1 × 10-4; Fig. 1D,E). It thus seems that slow-evolving housekeeping genes are underrepresented in disease. At first glance, this is surprising, because mutations in highly conserved and ubiquitously expressed genes might be thought to be more liable to cause disease. However, our previous finding that housekeeping genes are more likely to have been subject to strong purifying selection (i.e, have lower KA/KS values) suggests an alternative explanation. This is that housekeeping genes are underrepresented among disease genes, due to a higher chance of embryonic lethality when mutated. Thus, we predict that our results reflect prenatal pathology, rather than postnatal disease.
To test this hypothesis, we linked the human genes represented in the five partitions, with their probable orthologs in the nematode Caenorhabditis elegans (see Methods). No large-scale targeted-deletion data set is available for mammals. Results from an RNAi screen involving the majority of C. elegans genes (Kamath and Ahringer 2003) allowed us to associate 250 human genes in the five partitions with the RNAi phenotypes of their worm orthologs. For each partition, we calculated three parameters as follows: f Le, the fraction of human genes whose worm ortholog exhibited an embryonic or larval lethality phenotype; f ALL, the fraction of human genes in each partition in which a worm ortholog could be predicted; and their ratio f Le /f ALL, which represents the degree of over- or underrepresentation of embryonic or larval lethality phenotypes in the worm among their predicted human orthologs (Table 1). As predicted, we find a negative correlation between nematode lethality and mammalian tissue specificity (Table 1). In particular, partition 1 is overrepresented with human genes whose worm ortholog is associated with embryonic or larval lethality when deleted. If the tissue expression profiles and functions of these genes have remained under strong evolutionary constraints since the common ancestor of invertebrates and chordates 1 billion years ago, then their mammalian orthologs, when mutated, might also be more likely to result in embryonic lethality.
Studies of Tissue-Specific Genes
We then investigated whether single tissues exhibit variations in median KA/KS, f sec, and f dis. For this, we define human tissue-specific genes as those that possess TS values >0.1 (see Methods) for a particular tissue. For these genes, individual tissues exhibit up to threefold differences in median KA/KS values, 1.4-fold differences in median KS values, fourfold differences in f sec, and sevenfold differences in f dis (Table 2).
Table 2.
Variation of f sec , f dis and Median Evolution Rates for Tissue-Specific Genes From 27 Human Tissues, Sorted by Median KA/KS
| Tissue | Gene number | fsec (%) | fdis (%) | Median KA/KS | Median KA | Median KS | Median KA/KS nonsecreted | Median KA/KS secreted | ft (%) (gene number) |
|---|---|---|---|---|---|---|---|---|---|
| Fetal brain | 382 | 20.7 | 5.0 | 0.040 | 0.022 | 0.526 | 0.038 | 0.052 | ND |
| Amygdala | 350 | 25.1 | 7.4 | 0.041 | 0.024 | 0.553 | 0.036 | 0.058 | 60.5 (104) |
| Uterus | 128 | 32.0 | 14.1 | 0.061 | 0.036 | 0.595 | 0.044 | 0.081 | 29.3 (22) |
| Hep3b | 371 | 16.4 | 8.4 | 0.063 | 0.036 | 0.532 | 0.056 | 0.100 | ND |
| Spinal cord | 150 | 36.7 | 14.0 | 0.069 | 0.042 | 0.575 | 0.053 | 0.103 | ND |
| Heart | 250 | 24.0 | 17.2 | 0.070 | 0.051 | 0.685 | 0.049 | 0.141 | 31.8 (49) |
| Salivary gland | 164 | 36.0 | 11.0 | 0.071 | 0.046 | 0.684 | 0.057 | 0.099 | 17.2 (15) |
| Ovary | 123 | 44.7 | 21.1 | 0.071 | 0.044 | 0.595 | 0.054 | 0.094 | 14.5 (11) |
| Prostate | 112 | 30.4 | 8.0 | 0.075 | 0.052 | 0.601 | 0.061 | 0.100 | 7.1 (4) |
| Placenta | 231 | 39.4 | 14.7 | 0.075 | 0.057 | 0.689 | 0.064 | 0.114 | 21.5 (26) |
| Thyroid | 97 | 35.1 | 16.5 | 0.076 | 0.044 | 0.566 | 0.070 | 0.133 | ND |
| Pancreas | 350 | 36.6 | 14.0 | 0.077 | 0.055 | 0.644 | 0.063 | 0.107 | ND |
| DOHH2 | 340 | 16.8 | 8.5 | 0.078 | 0.045 | 0.554 | 0.073 | 0.100 | ND |
| Pituitary gland | 133 | 35.3 | 12.0 | 0.083 | 0.047 | 0.553 | 0.076 | 0.098 | ND |
| THY- | 194 | 20.1 | 11.3 | 0.083 | 0.052 | 0.564 | 0.071 | 0.160 | ND |
| DRG | 131 | 37.4 | 14.5 | 0.085 | 0.048 | 0.565 | 0.060 | 0.098 | 40.9 (29) |
| HUVEC | 216 | 25.5 | 9.7 | 0.085 | 0.052 | 0.554 | 0.082 | 0.101 | ND |
| Spleen | 293 | 32.1 | 12.0 | 0.087 | 0.066 | 0.732 | 0.067 | 0.158 | 24.0 (43) |
| Adrenal gland | 132 | 25.8 | 17.4 | 0.091 | 0.060 | 0.607 | 0.083 | 0.131 | 24.7 (20) |
| Trachea | 78 | 50.0 | 14.1 | 0.100 | 0.068 | 0.704 | 0.070 | 0.225 | 22.7 (10) |
| Whole blood | 332 | 29.2 | 10.2 | 0.101 | 0.066 | 0.646 | 0.078 | 0.224 | ND |
| Lung | 119 | 46.2 | 21.9 | 0.101 | 0.086 | 0.715 | 0.082 | 0.156 | 29.4 (20) |
| Kidney | 171 | 34.5 | 26.3 | 0.102 | 0.064 | 0.629 | 0.091 | 0.120 | 47.3 (53) |
| Testis | 287 | 12.2 | 6.3 | 0.103 | 0.057 | 0.530 | 0.097 | 0.122 | 48.0 (59) |
| Liver | 314 | 42.0 | 33.1 | 0.114 | 0.083 | 0.652 | 0.086 | 0.164 | 54.0 (115) |
| Fetal liver | 214 | 45.8 | 38.8 | 0.126 | 0.086 | 0.636 | 0.088 | 0.177 | ND |
| Thymus | 121 | 40.5 | 9.1 | 0.127 | 0.082 | 0.622 | 0.067 | 0.241 | 46.8 (36) |
Duret and Mouchiroud (2000) have demonstrated previously, using EST data for tissue-specific genes, that gene evolutionary rates vary with tissue types. We have investigated this issue in depth using microarray gene-expression data. Pairwise comparisons of KA/KS frequency distributions of tissue-specific genes were used to identify human tissues whose expressed genes are evolving either significantly faster or slower than other tissues (using the median-log probability value, Table 3). We conclude that brain-, and fetal brain-specific genes are evolving relatively slowly, whereas genes specific for the liver, fetal liver, testis, thymus, and kidney are evolving rapidly. Similarly, using data from the mouse, we found that mouse genes specific for the brain, dorsal root ganglia, skeletal muscle, heart, and eye are evolving relatively slowly, whereas liver-, trachea- and gall bladder-specific genes are evolving rapidly. Duret and Mouchiroud (2000) found previously that KA values of brain-specific genes are significantly lower than those of most other tissues; and that, similarly, the KA values of liver-specific genes are significantly higher. Thus, our results here greatly extend the list of tissue pairs whose expressed genes' evolutionary rates differ significantly from those of other tissues.
Table 3.
Kolmogorov-Smirnov Test Probability Results for Significant Differences Between Pairs of Distributions of Tissue-Specific Genes KA/KS Values
| Ad | Am | DO | DR | Fb | Fl | HU | He | H3 | Ki | Li | Lu | Ov | Pa | Pg | Pl | Pr | Sg | Sc | Sp | TH | Te | Th | Ty | Tr | Ut | Wb | MP | Median Ka/Ks | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fb | + | — | + | + | — | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 8.25 | 0.040 |
| Am | + | — | + | + | — | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | — | + | 7.80 | 0.041 |
| Ut | + | — | + | — | + | + | + | — | — | + | + | + | — | — | — | — | — | — | — | + | — | + | + | — | + | — | + | 1.95 | 0.061 |
| H3 | + | + | — | — | + | + | — | — | — | + | + | + | — | — | — | — | — | — | — | + | — | + | + | — | + | — | + | 1.90 | 0.063 |
| Sc | — | + | — | — | + | + | — | — | — | + | + | + | — | — | — | — | — | — | — | — | — | + | + | — | + | — | + | 1.05 | 0.069 |
| He | + | + | — | — | + | + | — | — | — | + | + | + | — | — | — | — | — | — | — | — | — | + | + | — | + | — | + | 0.95 | 0.070 |
| Sg | — | + | — | — | + | + | — | — | — | + | + | + | — | — | — | — | — | — | — | — | — | + | + | — | — | — | — | 0.65 | 0.071 |
| Ov | — | + | — | — | + | + | — | — | — | + | + | + | — | — | — | — | — | — | — | — | — | + | + | — | + | — | + | 0.80 | 0.071 |
| Pl | — | + | — | — | + | + | — | — | — | + | + | + | — | — | — | — | — | — | — | — | — | + | + | — | — | — | — | 0.90 | 0.075 |
| Pr | — | + | — | — | + | + | — | — | — | + | + | — | — | — | — | — | — | — | — | — | — | + | + | — | + | — | + | 0.80 | 0.075 |
| Ty | — | + | — | — | + | + | — | — | — | — | + | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | 0.40 | 0.076 |
| Pa | — | + | — | — | + | + | — | — | — | + | + | + | — | — | — | — | — | — | — | — | — | + | + | — | — | — | — | 0.70 | 0.077 |
| DO | — | + | — | — | + | + | — | — | — | + | + | + | — | — | — | — | — | — | — | — | — | + | + | — | — | + | — | 0.80 | 0.078 |
| TH | — | + | — | — | + | + | — | — | — | — | + | — | — | — | — | — | — | — | — | — | — | + | + | — | + | — | + | 1.00 | 0.083 |
| Pg | — | + | — | — | + | + | — | — | — | — | + | — | — | — | — | — | — | — | — | — | — | — | + | — | — | — | — | 0.45 | 0.083 |
| HU | — | + | — | — | + | + | — | — | — | — | + | — | — | — | — | — | — | — | — | — | — | — | + | — | — | + | — | 1.15 | 0.085 |
| DR | — | + | — | — | + | + | — | — | — | — | + | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | 0.45 | 0.085 |
| Sp | — | + | — | — | + | + | — | — | + | + | + | — | — | — | — | — | — | — | — | — | — | — | — | — | — | + | — | 0.95 | 0.087 |
| Ag | — | + | — | — | + | — | — | + | + | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | + | — | 0.90 | 0.091 |
| Tr | — | + | — | — | + | — | — | + | + | — | — | — | + | — | — | — | + | — | + | — | + | — | — | — | — | + | — | 1.50 | 0.100 |
| Wb | — | + | — | — | + | — | — | + | + | — | — | — | + | — | — | — | + | — | + | — | + | — | — | — | — | + | — | 1.75 | 0.101 |
| Lu | — | + | + | — | + | — | — | + | + | — | — | — | + | + | — | + | — | + | + | — | — | — | — | — | — | + | — | 1.70 | 0.101 |
| Ki | — | + | + | — | + | — | — | + | + | — | — | — | + | + | — | + | + | + | + | + | — | — | — | — | — | + | — | 2.00 | 0.102 |
| Te | — | + | + | — | + | + | — | + | + | — | + | — | + | + | — | + | + | + | + | — | + | — | — | — | — | + | — | 2.05 | 0.103 |
| Li | — | + | + | + | + | — | + | + | + | — | — | — | + | + | + | + | + | + | + | + | + | + | — | + | — | + | — | 4.00 | 0.114 |
| Fl | — | + | + | + | + | — | + | + | + | — | — | — | + | + | + | + | + | + | + | + | + | + | — | + | — | + | — | 4.35 | 0.126 |
| Th | — | + | + | — | + | — | + | + | + | — | — | — | + | + | + | + | + | + | + | — | + | — | — | — | — | + | — | 2.25 | 0.127 |
Tissue Specificity and Evolutionary Rates
For half of the human tissues considered, TS values vary uniformly with respect to KA/KS (Supplemental Fig. 2a). However, for other tissues, in particular from brain and liver, strong dependencies exist between TS and KA/KS (Supplemental Fig. 2b). For these two tissues, similar dependencies are apparent for either adult or fetal data (Table 3). In contrast, the distributions for the Hep3b-transformed liver cell line and normal liver cells differ significantly. This is likely to be due to the loss of liver-specific characteristics among Hep3b highly expressed genes, as seen by word stem analysis (Suppl. Table 1).
Tissue Biology
We have shown that secreted proteins possess a higher median KA/KS than nonsecreted proteins (see above). However, the median KA/KS values for secreted proteins vary greatly among different tissues (Table 2). We find that tissue-specific biology also influences gene evolution in a manner that is independent of protein secretion. At one extreme, brain-specific genes have lower KA/KS values than other tissues, regardless of whether secreted or nonsecreted proteins are considered (Table 2). At the other extreme, secreted genes expressed highly in spleen, whole blood, or thymus possess relatively high KA/KS values.
We sought to investigate whether tissues exhibit variations in their tissue-specific gene repertoires between human and mouse. We calculated the fraction (f T) of human genes that are specific to a tissue, whose mouse ortholog is also specific to that tissue. Values of f T (Table 2) demonstrate approximately eightfold variation among tissues, with brain and liver, the two tissues that exhibit most variation in protein-coding evolutionary rates, showing the greatest f T values. Thus, although on average, brain-specific proteins evolve most slowly and liver-specific proteins most rapidly, both tissues exhibit high conservation of their tissue-specific gene sets that have been maintained since the common ancestor of human and mouse.
Constancy of Selective Pressures
Selective pressures on protein-coding genes appear to have remained constant during mammalian evolution (Bulmer et al. 1991; Mouchiroud et al. 1995). We were interested in a related question, whether homologous tissues in different mammals express genes with different evolutionary rates. Using sets of mouse–rat orthologs, and mouse–human orthologs, and genes that are specific for mouse tissues, we found no such differences. The median KA/KS values for the longer evolutionary distance between human and mouse were highly correlated with median KA/KS values for the shorter evolutionary distance separating mouse from rat (Fig. 2; Table 4).
Figure 2.
Evolutionary constraints on mouse tissues have been constant since the divergence of human and rodent lineages (∼75 million years ago). Median KA/KS values have been calculated for mouse tissue-specific genes using the yn00 method of Yang and Nielsen (2000; see Table 4). The 1:1 rat–mouse and human–mouse orthology relationships were obtained from Ensembl's Compara database.
Table 4.
Human–Mouse and Mouse–Rat Median KA/KS Values for Mouse-Tissue-Specific Genes (see Fig. 2 legend)
| Mouse tissue | Human gene number | Human—mouse median KA/KS | Rat gene number | Rat—mouse median KA/KS |
|---|---|---|---|---|
| Amygdala | 273 | 0.040 | 268 | 0.053 |
| DRG | 240 | 0.048 | 234 | 0.054 |
| Eye | 124 | 0.064 | 126 | 0.065 |
| Skeletal muscle | 253 | 0.065 | 237 | 0.078 |
| Heart | 199 | 0.066 | 175 | 0.083 |
| Umbilical cord | 190 | 0.066 | 172 | 0.093 |
| Tongue | 71 | 0.066 | 77 | 0.084 |
| Thymus | 216 | 0.067 | 206 | 0.093 |
| Epidermis | 72 | 0.070 | 79 | 0.060 |
| Salivary gland | 243 | 0.076 | 241 | 0.082 |
| Uterus | 98 | 0.076 | 95 | 0.104 |
| Thyroid | 189 | 0.076 | 176 | 0.083 |
| Digits | 99 | 0.080 | 98 | 0.089 |
| Adrenal gland | 117 | 0.081 | 106 | 0.112 |
| Brown fat | 259 | 0.081 | 240 | 0.101 |
| Prostate | 136 | 0.083 | 131 | 0.090 |
| Lung | 128 | 0.084 | 114 | 0.098 |
| Placenta | 198 | 0.089 | 192 | 0.122 |
| Stomach | 116 | 0.091 | 120 | 0.126 |
| Testis | 338 | 0.092 | 343 | 0.133 |
| Bone marrow | 223 | 0.094 | 210 | 0.126 |
| Large intestine | 167 | 0.095 | 167 | 0.122 |
| Ovary | 121 | 0.096 | 115 | 0.104 |
| Bone | 179 | 0.096 | 166 | 0.122 |
| Small intestine | 175 | 0.096 | 168 | 0.116 |
| Mammary gland | 171 | 0.098 | 177 | 0.118 |
| Spleen | 167 | 0.099 | 152 | 0.124 |
| Adipose tissue | 91 | 0.105 | 79 | 0.134 |
| Kidney | 181 | 0.108 | 186 | 0.160 |
| Liver | 268 | 0.122 | 257 | 0.165 |
| Trachea | 73 | 0.132 | 74 | 0.192 |
| Gall bladder | 197 | 0.133 | 185 | 0.171 |
DISCUSSION
Three factors that are not mutually exclusive appear to predominate in shaping the repertoires of genes expressed in restricted tissue sets. First, the most rapidly evolving genes have a greater likelihood of being expressed in fewer tissues. Our results, which used large-scale microarray gene-expression data confirm and extend those from a previous EST-based study (Duret and Mouchiroud 2000).
A second factor affecting gene expression in tissues is protein cellular localization. We have shown that there is a strong correlation among tissue specificity, protein secretion, and evolutionary rates. Secreted gene products have significantly higher KA/KS values than nonsecreted gene products and are enriched among tissue specific genes. To some degree, these observations are consistent with the `genetic arms race' hypothesis (Dawkins and Krebs 1979) operating between mammalian hosts and their pathogens. The high KA/KS values of secreted gene products in spleen, whole blood, or thymus might be a direct consequence of these tissues' participation in immune functions. Plasma proteins produced by the liver may also be more accessible to pathogens, and hence, more susceptible to positive selection forces.
Lastly, gene-expression profiles are largely influenced by tissue-specific biology. For example, brain-specific genes are poorly represented among disease genes, and purifying selection appears to predominate in preserving brain-specific functions, such as cognition and information processing. This is consistent with a model in which mutation of brain-specific genes is more likely to result in embryonic lethality, compared with mutation of genes highly expressed in other tissues (Table 2). Tissue-specific biology and protein-secretion effects are largely independent. For instance, brain-specific genes, which encode secreted proteins, evolve more slowly than genes encoding secreted proteins from other tissues. Testis-specific genes are associated with elevated KA/KS values, even for intracellular gene products (Table 2). This may be a consequence of sexual selection (Darwin 1871; Wyckoff et al. 2000). Such factors in tissue biology appear to have been held relatively constant over the past 75 million years, since the divergence of human and rodent lineages (Fig. 2; Table 4).
Our analysis of disease genes has resulted in three findings. Firstly, the frequency of Mendelian-type diseases positively correlates with gene tissue specificity (Table 1). Secondly, the set of disease genes is enriched in genes whose products are secreted. Thirdly, although no significant differences between the evolutionary rates of disease and nondisease genes were detected, we found significant differences between these rates among genes with low tissue-specificity (i.e., housekeeping genes, partition 1, Fig. 1D,E). These three findings highlight a complex association between disease genes, tissue specificity, and evolution rates.
We find that slowly evolving housekeeping and intracellular proteins' genes are underrepresented in human disease. This is likely to be due to higher degrees of purifying selection forces acting upon them and the greater chance of embryonic lethality when mutated. These genes can thus be regarded as essential to the organism's course of development. To test this assertion, we considered the association between tissue specificity of human genes and the embryonic and larval lethality exhibited by targeted deletion of their probable orthologs in the nematode C. elegans. Consistent with the positive correlation between tissue specificity and disease, we find that nematode lethal phenotypes are negatively correlated with human-tissue specificity (Table 1).
We observed that disease genes are more likely to be highly expressed in tissues such as liver, kidney, or lung, and to have products that are secreted. Consequently, these correlations should assist in the prioritization of candidate disease genes for genetic-association studies. Moreover, the identification of tissue specificity, protein secretion, and tissue-specific biology as main factors influencing gene evolutionary rates should assist investigations into the evolution of individual mammalian tissues and organs.
METHODS
Mapping Expression Data to Sequence
We used NOVARTIS microarray data (http://expression.gnf.org) from hybridizations of RNA from human and mouse tissues to Affymetrix 25-mer oligonucleotide tags (U95A and U74A, respectively). GenBank sequences linked to these Affymetrix tags were mapped to the genome assemblies using BLAT (http://genome.ucsc.edu; Kent 2002) and a 95% identity lower threshold. Subsequently, the tag was associated with an ENSEMBL gene (human ENSEMBL mart version 14.1, using NCBI build 31 and mouse ENSEMBL mart version 12.1, using the February 2002 mouse build 2) if the best-scoring BLAT alignment overlapped at least one exon, or else lay within 1 kb of an exon, of that gene. A total of 8159 human and 6911 mouse ENSEMBL genes were then assigned the expression levels of its associated Affymetrix tag in all of the tissues investigated. These expression levels represent the Average Difference (AD) between sense tags and missense tags (Su et al. 2002). A total of 23% of human genes and 12% of mouse genes were represented by multiple tags on the Affymetrix oligonucleotide array. For these genes, we summed the expression levels of their tags. Lastly, to avoid artefactual negative AD values, we have set the minimal value of expression for a gene in a tissue to 20 AD, as previously (Su et al. 2002).
Tissue Specificity and KA/KS Values
We define the tissue specificity (TS_i_) value for a gene expressed in tissue i as its AD expression value in i divided by the sum of its AD values in all tissues. This definition would not be appropriate if data from multiple tissues with significantly similar expression profiles were used. Consequently, we calculated the correlation coefficients between every mouse or human tissue pair. The correspondent distance matrix was then subjected to single-linkage hierarchical clustering. We discarded one of each pair of tissues associated with a Euclidian distance of <0.21 (human, see Suppl. Fig. 1) or 0.33 (mouse). A total of 12,481 mouse and human 1:1 ortholog pairs were obtained from ENSEMBL (Clamp et al. 2003; http://www.ensembl.org). The ratio of KA (the number of nonsynonymous substitutions per nonsynonymous site) to KS (the number of synonymous substitutions per synonymous site) was calculated using the yn00 method of Yang and Nielsen (2000). We used a threshold of TS >0.1 to define tissue-specific genes in human. This threshold is approximately three times the median TS values in tissues. For mouse data, due to a greater number of tissues, this threshold was set to 0.08.
fsec,fdis
Predictions of protein secretion and association with human disease were obtained from ENSEMBL mart 14.1. Disease annotation is based on OMIM (www.ncbi.nlm.nih.gov/Omim; McKusick 2000). Secretion annotation is based on the SignalP v1.2 method (Nielsen et al. 1997). SignalP v1.1 is associated with a false negative rate of 0.9% and a false positive rate of 17.6% (Menne et al. 2000). Type I and multipass transmembrane proteins with signal peptides are assigned to the secreted category, whereas type II and multipass transmembrane proteins without signal peptides are assigned to the nonsecreted category (Menne et al. 2000).
C. elegans RNAi Phenotype Study
We gathered from Wormbase (wormpep110) (www.wormbase.org) the protein sequences of C. elegans genes that have been associated with an embryonic or larval lethality phenotype by the RNAi study of Kamath et al. (Kamath and Ahringer 2003). Reciprocal best-match sequences between these C. elegans proteins and human gene products were identified using BLASTP (Altschul et al. 1997) and a minimum alignment coverage of 60%. These 3044 reciprocal best matches are considered as candidate human and nematode orthologs.
Statistics
The two-sided Kolmogorov-Smirnov test was used to investigate whether two data sets may reasonably be assumed to sample the same distribution. The use of this test was indicated, as it does not require the assumption that data are distributed normally. The Pearson's product moment correlation coefficient was used as a measure of the linear association between two variables.
Acknowledgments
We thank the Medical Research Council (UK) for financial support.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1924004.
Footnotes
[Supplemental material is available online at www.genome.org.\]
References
- Alberts, B., Bray, D., Lewis, J., Raff, M., Roberts, K., and Watson, J.D. 1994. Chapter 1. From single cells to multicellular organisms. In Molecular biology of the cell, pp. 26-39. Garland Publishing, Inc., New York.
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25**:** 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bains, W. 1992. Local sequence dependence of rate of base replacement in mammals. Mutat. Res. 267**:** 43-54. [DOI] [PubMed] [Google Scholar]
- Bulmer, M., Wolfe, K.H., and Sharp, P.M. 1991. Synonymous nucleotide substitution rates in mammalian genes: Implications for the molecular clock and the relationship of mammalian orders. Proc. Natl. Acad. Sci. 88**:** 5974-5978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo-Davis, C.I. and Hartl, D.L. 2002. Genome evolution and developmental constraint in Caenorhabditis elegans. Mol. Biol. Evol. 19**:** 728-735. [DOI] [PubMed] [Google Scholar]
- Clamp, M., Andrews, D., Barker, D., Bevan, P., Cameron, G., Chen, Y., Clark, L., Cox, T., Cuff, J., Curwen, V., et al. 2003. Ensembl 2002: Accommodating comparative genomics. Nucleic Acids Res. 31**:** 38-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin, C. 1871. The descent of man and selection in relation to sex. D. Appleton, New York.
- Dawkins, R. and Krebs, J.R. 1979. Arms races between and within species. Proc. R. Soc. Lond. B. Biol. Sci. 205**:** 489-511. [DOI] [PubMed] [Google Scholar]
- Duret, L. 2002. Evolution of synonymous codon usage in metazoans. Curr. Opin. Genet. Dev. 12**:** 640-649. [DOI] [PubMed] [Google Scholar]
- Duret, L. and Mouchiroud, D. 2000. Determinants of substitution rates in mammalian genes: Expression pattern affects selection intensity but not mutation rate. Mol. Biol. Evol. 17**:** 68-74. [DOI] [PubMed] [Google Scholar]
- Giraud, A., Matic, I., Tenaillon, O., Clara, A., Radman, M., Fons, M., and Taddei, F. 2001. Costs and benefits of high mutation rates: Adaptive evolution of bacteria in the mouse gut. Science 291**:** 2606-2608. [DOI] [PubMed] [Google Scholar]
- Hughes, A.L. 1999. Adaptive evolution of genes and genomes. Oxford University Press, New York.
- Huminiecki, L., Lloyd, A.T., and Wolfe, K.H. 2003. Congruence of tissue expression profiles from Gene Expression Atlas, SAGEmap and TissueInfo databases. BMC Genomics 4**:** 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan, I.K., Kondrashov, F.A., Rogozin, I.B., Tatusov, R.L., Wolf, Y.I., and Koonin, E.V. 2001. Constant relative rate of protein evolution and detection of functional diversification among bacterial, archaeal and eukaryotic proteins. Genome Biol. 2**:** RESEARCH0053. [DOI] [PMC free article] [PubMed]
- Kamath, R.S. and Ahringer, J. 2003. Genome-wide RNAi screening in Caenorhabditis elegans. Methods 30**:** 313-321. [DOI] [PubMed] [Google Scholar]
- Kent, W.J. 2002. BLAT–the BLAST-like alignment tool. Genome Res. 12**:** 656-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, M.C. and Wilson, A.C. 1975. Evolution at two levels in humans and chimpanzees. Science 188**:** 107-116. [DOI] [PubMed] [Google Scholar]
- McKusick, V.A. 2000. Online mendelian inheritance in man, OMIM (TM). [DOI] [PubMed]
- Meiklejohn, C.D., Parsch, J., Ranz, J.M., and Hartl, D.L. 2003. Rapid evolution of male-biased gene expression in Drosophila. Proc. Natl. Acad. Sci. 100**:** 9894-9899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menne, K.M., Hermjakob, H., and Apweiler, R. 2000. A comparison of signal sequence prediction methods using a test set of signal peptides. Bioinformatics 16**:** 741-742. [DOI] [PubMed] [Google Scholar]
- Mouchiroud, D., Gautier, C., and Bernardi, G. 1995. Frequencies of synonymous substitutions in mammals are gene-specific and correlated with frequencies of nonsynonymous substitutions. J. Mol. Evol. 40**:** 107-113. [DOI] [PubMed] [Google Scholar]
- Nielsen, H., Engelbrecht, J., Brunak, S., and von Heijne, G. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10**:** 1-6. [DOI] [PubMed] [Google Scholar]
- Su, A.I., Cooke, M.P., Ching, K.A., Hakak, Y., Walker, J.R., Wiltshire, T., Orth, A.P., Vega, R.G., Sapinoso, L.M., Moqrich, A., et al. 2002. Large-scale analysis of the human and mouse transcriptomes. Proc. Natl. Acad. Sci. 99**:** 4465-4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrington, J.A., Nair, A., Mahadevappa, M., and Tsyganskaya, M. 2000. Comparison of human adult and fetal expression and identification of 535 housekeeping/maintenance genes. Physiol. Genomics 2**:** 143-147. [DOI] [PubMed] [Google Scholar]
- Waterston, R.H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J.F., Agarwal, P., Agarwala, R., Ainscough, R., Alexandersson, M., An, P., et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420**:** 520-562. [DOI] [PubMed] [Google Scholar]
- Wyckoff, G.J., Wang, W., and Wu, C.I. 2000. Rapid evolution of male reproductive genes in the descent of man. Nature 403**:** 304-309. [DOI] [PubMed] [Google Scholar]
- Yang, Z. and Nielsen, R. 2000. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol. Biol. Evol. 17**:** 32-43. [DOI] [PubMed] [Google Scholar]
- Zhang, J., Zhang, Y.P., and Rosenberg, H.F. 2002. Adaptive evolution of a duplicated pancreatic ribonuclease gene in a leaf-eating monkey. Nat. Genet. 30**:** 411-415. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://expression.gnf.org; Gene Expression Atlas.
- http://www.ensembl.org; Project ENSEMBL.
- http://genome.ucsc.edu; UCSC Genome Server.
- http://www.ncbi.nlm.nih.gov/Omim; Online Mendelian Inheritance in Man (OMIM).
- http://www.minitab.com; MINITAB statistical software.
- http://www.wormbase.org; WORMBASE database.