Human Mediator Subunit Med26 Functions As A Docking Site For Transcription Elongation Factors (original) (raw)
. Author manuscript; available in PMC: 2012 Jul 8.
Summary
Promoter proximal pausing by initiated RNA polymerase II (Pol II) and regulated release of paused polymerase into productive elongation has emerged as a major mechanism of transcription activation. Reactivation of paused Pol II correlates with recruitment of SuperElongationComplexes (SECs) containing ELL/EAF family members, P-TEFb, and other proteins, but the mechanism of their recruitment is currently a major unanswered question. Here, we present evidence for a role of human Mediator subunit Med26 in this process. We identify in the conserved N-terminal domain of Med26 overlapping docking sites for SEC and a second ELL/EAF-containing complex, as well as general initiation factor TFIID. In addition, we present evidence consistent with the model that Med26 can function as a molecular switch that interacts first with TFIID in the Pol II initiation complex and then exchanges TFIID for complexes containing ELL/EAF and P-TEFb to facilitate transition of Pol II into the elongation stage of transcription.
Messenger RNA synthesis begins with recruitment of RNA polymerase II (Pol II) and other components of the transcription apparatus to the promoter to form a preinitiation complex, followed by ATP-dependent unwinding of the DNA and transcription initiation. Following initiation, Pol II moves away from the promoter, ultimately leading to establishment of a productive elongation complex. Although for many years most research focused on mechanisms that control the preinitiation and initiation stages of transcription, transcription of many genes is also regulated during transcript elongation.
Early studies showed that transcription of genes including the human c-MYC oncogene, the Drosophila heat shock gene Hsp70, and the integrated HIV-1 provirus is regulated by promoter-proximal pausing, in which Pol II initiates transcription but pauses or arrests downstream of the transcription start site (Saunders et al., 2006). Release from the pause is a rate-limiting step that can be regulated by DNA binding transcription factors or, in the case of HIV transcription, by the viral RNA binding transactivator Tat. Recently, genome-wide studies demonstrated the presence of promoter-proximally paused Pol II near the 5′-ends of a large fraction of genes, suggesting that regulated promoter-proximal pausing and release is a general feature of Pol II elongation (Guenther et al., 2007; Kininis et al., 2009; Muse et al., 2007; Nechaev et al., 2010; Rahl et al., 2010; Zeitlinger et al., 2007).
The duration of pausing or arrest during early elongation can be controlled by multiple transcription elongation factors that influence the elongation competence of Pol II. DRB-sensitivity inducing factor (DSIF) and negative elongation factor (NELF) function together to induce pausing (Wada et al., 1998; Yamaguchi et al., 1999). Release of paused Pol II in turn depends on phosphorylation of the Pol II CTD, SPT5, and perhaps other factors by positive transcription elongation factor b (P-TEFb, composed of the cyclin dependent kinase CDK9 and cyclin T1/T2) (Lis et al., 2000; Marshall et al., 1996; Peterlin and Price, 2006b; Wei et al., 1998). Also implicated in this process are additional transcription elongation factors, including members of the eleven nineteen lysine-rich in leukemia (ELL) family and their binding partners ELL-associated factor (EAF) 1 and EAF2, which together act directly to increase the rate of elongation by Pol II in vitro and regulate expression of Hsp70, HIV-1, and other genes (Byun et al., 2009; Kong et al., 2005; Lin et al., 2010; Shilatifard et al., 1996; Smith et al., 2008). Evidence suggests these factors contribute not only to release of paused Pol II but also to subsequent events occurring during transcription elongation, including cotranscriptional RNA processing (Martincic et al., 2009; Ni et al., 2004).
P-TEFb and ELL/EAF family members function as components of larger “super-elongation complexes” or SECs that contain the additional coregulators AFF4, AFF1, ENL, and AF9 (He et al., 2010; Lin et al., 2010; Sobhian et al., 2010; Yokoyama et al., 2010). In HIV-1 infected cells, Tat activates HIV-1 transcription by recruiting P-TEFb, ELL/EAF, and SEC family members through interactions with the TAR sequence in the 5′-end of the nascent HIV-1 transcript (He et al., 2010; Peterlin and Price, 2006a; Sobhian et al., 2010); however, mechanisms responsible for bringing these factors to host genes in uninfected cells remain poorly defined.
Mediator is an evolutionarily conserved coregulatory complex that was first identified in yeast (Kim et al., 1994; Koleske and Young, 1994). In metazoa, Mediator is composed of some 30 distinct subunits (Sato et al., 2004). Mediator exists in multiple, functionally distinct forms that share a common core of subunits and can be distinguished by the presence or absence of a kinase module composed of Cyclin C and isoforms of Med12, Med13, and CDK8. Kinase module has been implicated in both transcriptional repression and activation (Taatjes, 2010).
In metazoa, a subset of Mediator contains an additional subunit, Med26. Med26-containing Mediator copurifies with only a small amount of kinase module, but near-stoichiometric Pol II (Mittler et al., 2001; Sato et al., 2004; Taatjes et al., 2002). Med26-containing Mediator appears to play a key role in transcriptional activation (Mo et al., 2004; Naar et al., 2002; Ryu et al., 1999); however, mechanism(s) by which Med26 contributes to this process are not known. Here, we present evidence supporting the model that Med26 functions in part by recruiting ELL/EAF- and P-TEFb-containing complexes, including SEC, to a subset of human genes. We show that the human Mediator complex can recruit ELL/EAF- and P-TEFb-containing complexes to promoters via a direct interaction with the N-terminal domain (NTD) of Med26. The Med26 NTD also binds TFIID, and TFIID and elongation complexes interact with Med26 through overlapping binding sites. In addition, we show (i) that in cells wild-type Med26, but not a Med26 mutant that prevents binding of Mediator to elongation factors, regulates transcription of c-MYC and other genes and (ii) that Med26 knockdown interferes with elongation factor recruitment. We propose the Med26 NTD may function as a molecular switch that contributes to the transition of Pol II into productive elongation.
Results
Enrichment of Elongation Factors and TFIID in Mediator With Med26
As part of a MudPIT-based proteomic analysis of the human Mediator complex, we generated human cell lines expressing Mediator subunits with FLAG epitope tags, immunopurified (IPed) FLAG-Mediator subunits and their associated proteins, and identified proteins in anti-FLAG eluates by MudPIT mass spectrometry. In MudPIT datasets, the number of spectra corresponding to peptides from a particular protein correlates with the protein's abundance and length. The relative amount of a particular protein in a sample can be estimated from a normalized spectral abundance factor, or NSAF (Zhang et al., 2010).
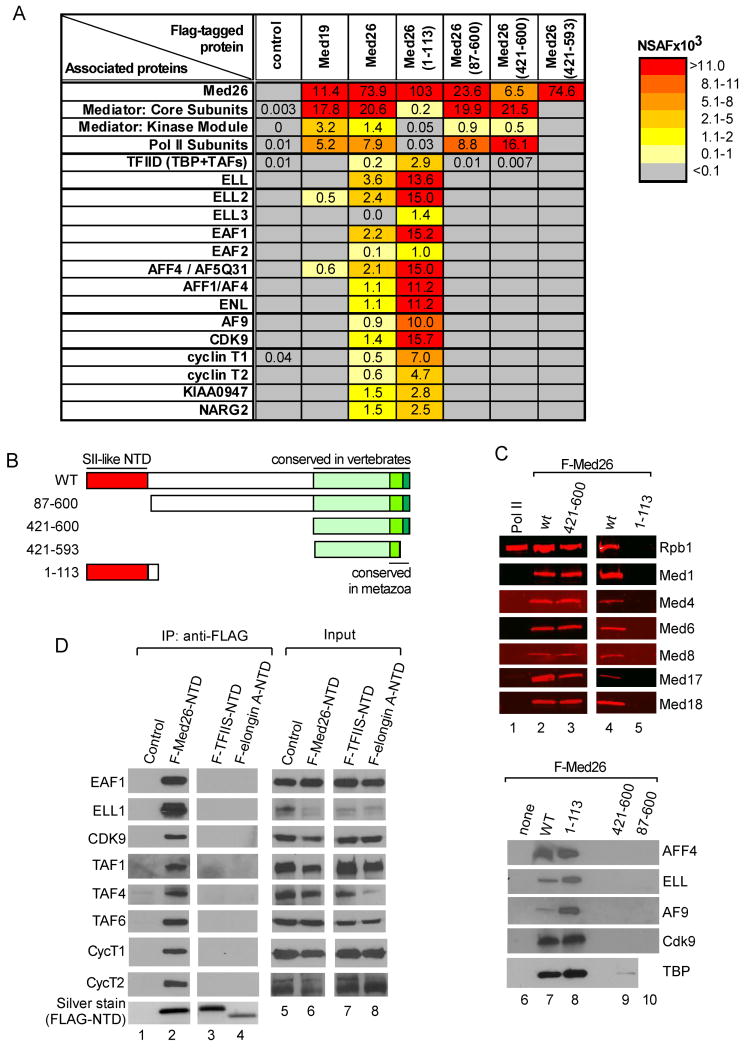
Proteomic analysis previously allowed us to define a complete set of human Mediator subunits (Sato et al., 2004). In-depth analysis of our MudPIT datasets revealed that additional proteins with roles in transcription were consistently enriched in FLAG-Med26 Mediator preparations, relative to Mediator purified through FLAG-Med19 (Figure 1A and Supplemental Table S1) or other core Mediator subunits (data not shown). Among these were initiation factor TFIID and transcription elongation factors including ELL and EAF family members, P-TEFb (CDK9, cyclin T1/T2), and MLL translocation partners AFF1, AFF4, AF9, and ENL, all of which are components of SEC (He et al., 2010; Lin et al., 2010; Sobhian et al., 2010). Also enriched were two previously uncharacterized proteins, KIAA0947 and NARG2. The NSAF values of these proteins were ∼5-20% of the average NSAF of core Mediator subunits, suggesting they bind only a fraction of Mediator in solution.
Figure 1. Association of Med26 NTD with transcription elongation factors, MLL fusion partners, and TFIID.
(A) MudPIT of Med26-associated proteins. See also Table S1. (B) Schematic of wild type and mutant Med26 proteins. (C) Western blot of FLAG-IPed complexes from HeLa cells expressing wild type and mutant Med26 (lanes 2-5, 7-10) and of rat liver Pol II (lane 1) (Serizawa et al., 1992). Lanes 1-3 and 4-5, respectively, are from different blots. (D) Proteins copurifying with FLAG-Med26 NTD, TFIIS NTD, or Elongin A NTD, analyzed by western blotting. WT, wild type. Here and in subsequent figures, IP and input (3% of total) were analyzed on different blots; hence it is not possible to make quantitative comparisons between IP and input samples.
Docking Sites for TFIID and Elongation Factors in the Med26 NTD
TFIID, SEC components, and KIAA0947 and NARG2 could be enriched in FLAG-Med26 Mediator because they bind directly to Med26 or Med26-associated Mediator. Alternatively, because ELL/EAF can interact stably with Pol II in vitro (Banks et al., 2007; Shilatifard et al., 1997), and Med26-containing Mediator is enriched in Pol II, some or all of these proteins might be associated with Pol II rather than Mediator or Med26.
To begin to address these possibilities, we asked whether the same or distinct Med26 regions are necessary for interaction with Mediator and Pol II and with TFIID, SEC components, and KIAA0947 and NARG2. The most highly conserved region of Med26 is its NTD, which is similar to the NTDs of the elongation factors TFIIS and Elongin A and, based on an NMR structure of the TFIIS NTD, can be modeled as a globular domain composed of a four-helix bundle (Booth et al., 2000). In the Med26 C-terminus are approximately 10 amino acids that are conserved from insects to mammals, while a more extended C-terminal region of ∼100 amino acids is conserved in vertebrates (Bourbon, 2008).
We generated human cell lines stably expressing the series of FLAG-Med26 mutants diagrammed in Figure 1B and identified proteins associated with each by MudPIT and Western blotting (Figures 1A and 1C). The Med26 C-terminal domain (residues 421-600) is sufficient for its assembly into Mediator and interaction with Pol II. The most highly conserved C-terminal amino acids are critical for these interactions, since deletion of the last eight amino acids from the Med26 C-terminus disrupted Med26 binding to Mediator and Pol II. Whether Med26 makes direct contacts with Pol II and/or binds Pol II indirectly due to preferential association with a form of Mediator that binds Pol II is unclear; however, we believe the latter possibility is more likely, since we have not detected binding of purified Pol II to free Med26 in vitro (data not shown). Neither ELL nor EAF family members copurified with Pol II bound to Mediator containing Med26 lacking the NTD (Figures 1A and 1C, lanes 9 and 10), suggesting that the ELL/EAF binding site on Pol II is blocked when Pol II is bound to Mediator.
The Med26 NTD does not bind Mediator or Pol II. It does, however, copurify with TFIID, P-TEFb subunits, ELL and EAF family members, other SEC components, and NARG2 and KIAA0947 (Figures 1A and C, lanes 6-10). Binding of these proteins is specific for the Med26 NTD, as they do not copurify with the TFIIS- or Elongin A-NTDs (Figure 1D). Thus, the conserved Med26 N- and C- terminal regions are functional domains that support interactions with TFIID and transcription elongation factors and with Mediator and Pol II, respectively. Although we did not detect TFIID in association with Med26 mutants that lack the NTD in our MudPIT analyses, we detected small amounts of TFIID subunit TBP in association with these mutants by western blotting (Figure 1C), perhaps due to interactions between TBP and other Mediator subunits (Cai et al., 2010; Lariviere et al., 2006). We cannot, however, rule out the possibility that the Med26 C-terminus can also bind weakly to TFIID.
ELL and EAF are Components of at Least Two Med26 NTD-Associated Complexes
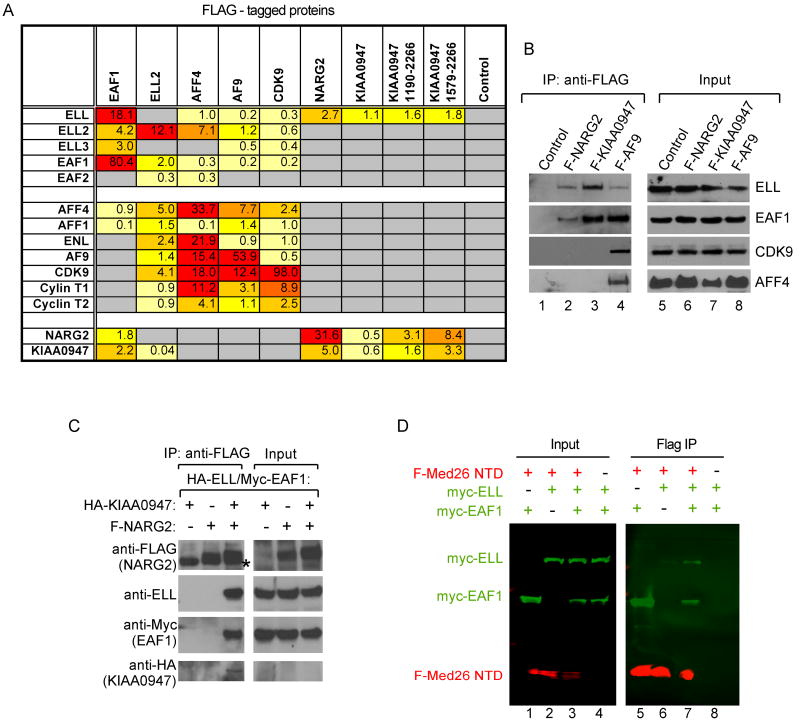
To define the complexes that interact with Med26, we generated cell lines stably expressing individual FLAG-tagged SEC components, NARG2, full-length KIAA0947, or KIAA0947 deletion mutants. Analysis of proteins copurifying with each protein argue that the Med26 NTD binds at least two types of complexes that share ELL and EAF family members. The first corresponds to SEC and includes P-TEFb, AFF4 and/or AFF1, and AF9 and/or ENL (Figures 2A and 2B, lane 4), and the second lacks SEC components, but includes KIAA0947 and NARG2 (Figures 2A and 2B, lanes 2-3). We detect only small amounts of Mediator subunits by MudPIT in FLAG IPs from cells expressing SEC subunits, most notably FLAG-AFF4 (data not shown), suggesting only a small fraction of SEC is stably associated with Mediator. Confirming the interaction of ELL/EAF1 with KIAA0947 and NARG2, recombinant ELL, EAF1, and KIAA0947 specifically co-IPed with FLAG-NARG2 when the 4 proteins were coexpressed in insect cells. The function of this novel complex will be the subject of a future study.
Figure 2. ELL/EAF-containing complexes bind the Med26 NTD.
(A) MudPIT of proteins bound to SEC components, NARG2, and KIAA0947. (B) Western blotting of proteins associated with FLAG-tagged NARG2, KIAA0947 and AF9. (C) Binding of NARG2 and KIAA0947 to ELL/EAF1. FLAG-IPed complexes from baculovirus-infected insect cells expressing the indicated proteins were analyzed by western blotting. Asterisk, IgH. (D) Binding of EAF1 to the Med26 NTD. Western blotting of FLAG-IPed complexes or lysates (Input) from baculovirus-infected insect cells, using anti-FLAG (M2) antibodies and Alexa Fluor 680-labeled anti-mouse IgG (light chain-specific) (red) or rabbit anti-cMyc antibodies and IR Dye™ 800-labeled goat anti rabbit IgG (green).
EAF Proteins Bind the Med26 NTD
Having observed that ELL and EAF family members are shared by at least two complexes that bind the Med26 NTD, we speculated that members of one or both families might directly interact with the Med26 NTD. As shown in Figure 2D, EAF1 bound the FLAG-Med26 NTD when the two proteins were coexpressed in insect cells in the absence of ELL; EAF2 also bound the Med26 NTD in the absence of ELL (data not shown). In contrast, ELL bound the Med26 NTD only in the presence of an EAF family member. These findings suggest EAF proteins function as adaptor molecules that link the Med26 NTD to ELL/EAF-containing complexes.
A Role for The Med26 NTD in Mediator-Dependent Recruitment of Transcription Elongation Factors to Promoters In Vitro
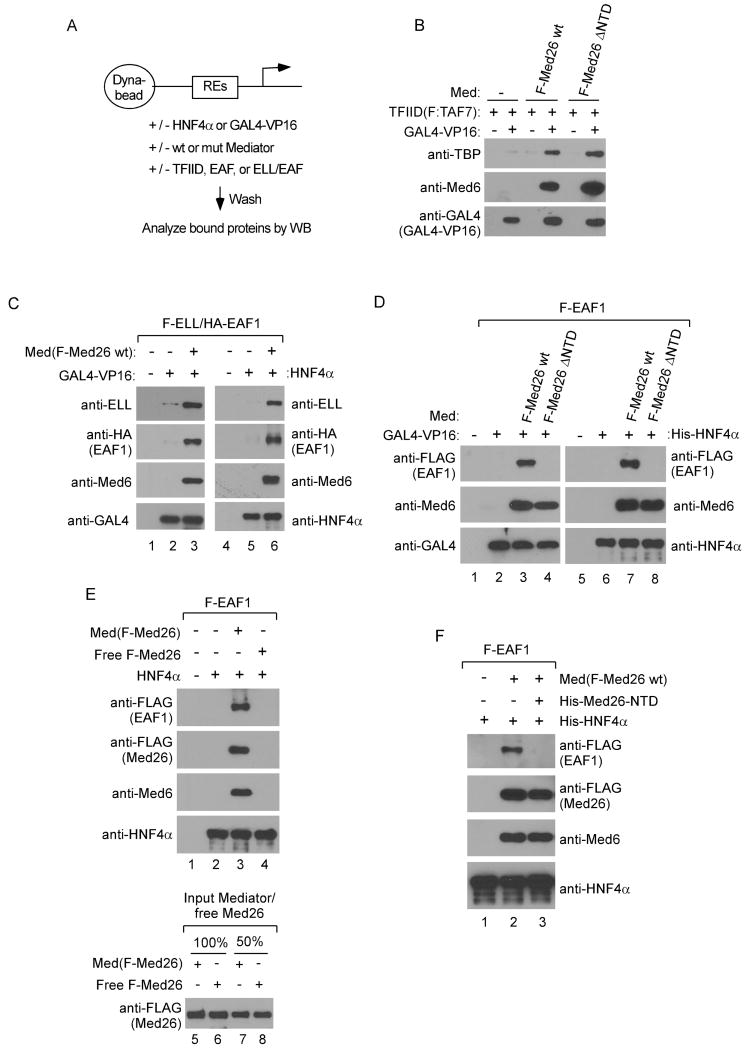
Our observation that the Med26 NTD directly binds both TFIID and ELL/EAF-containing complexes suggested that Med26 might contribute to transcriptional regulation at least in part by helping to recruit them to the genes they regulate. Carey and coworkers showed that Mediator enhances recruitment of TFIID to a promoter bound by a DNA binding transactivator, GAL4-VP16 (Johnson et al., 2002). To ask whether the Med26 NTD is responsible for this activity of Mediator, we employed similar immobilized template assays (Figure 3A), using Mediator complexes purified from cells expressing wild type FLAG-Med26 or Med26 that lacks the NTD but includes residues 421-600 (Med26ΔNTD). Both Med26- and Med26ΔNTD-containing Mediator enhanced GAL4-VP16-dependent binding of TFIID to an immobilized promoter (Figure 3B), arguing that the Med26 NTD is dispensable for TFIID recruitment in this assay and that additional contact(s) between Mediator and TFIID are responsible for this activity.
Figure 3. The Med26 NTD is needed for Mediator-dependent recruitment of ELL/EAF but not TFIID to promoters.
(A) Schematic of immobilized template assay. (B) Med26 NTD is dispensable for Mediator-dependent recruitment of TFIID to a promoter. Assays contained GAL4×5-MLT template and GAL4-VP16, TFIID, and Mediator (Med) with Med26 or Med26ΔNTD as indicated. (C) Mediator-dependent recruitment of ELL/EAF1 to GAL4- (lanes 1-3) or HNF4α-responsive promoters. Med(F-Med26 wt), Med26-Mediator. (D) Med26ΔNTD-Mediator [Med(F-Med26 ΔNTD)] does not recruit EAF1 to a promoter. (E) Free Med26 is not sufficient for EAF1 recruitment. Lanes 1-4, assays contained either free full length Med26 or Med26-Mediator. Lanes 5-8 show amounts of free Med26 and Med26-Mediator in binding assays. (F) Free Med26 NTD blocks Mediator-dependent EAF1 recruitment. Assays were performed with Med26-Mediator, with or without free Med26 NTD (His-Med26-NTD).
We next tested the ability of Mediator to recruit EAF-containing complexes to promoters in the presence of either GAL4-VP16 or the DNA binding transactivator HNF4α. As shown in Figures 3C and 3D, the ELL/EAF1 complex or EAF1 alone were recruited to promoters containing GAL4- or HNF4α-responsive elements in the presence of their cognate activator only when binding reactions included wild type Mediator. EAF1 was not recruited to either promoter, however, in binding reactions containing Med26ΔNTD-Mediator (Figure 3D).
To exclude the possibility that free Med26 protein in Mediator preparations is responsible for EAF1 recruitment, we compared the ability of free Med26 and FLAG-Med26-IPed Mediator to recruit EAF1 to an immobilized promoter. Unlike Mediator-associated Med26, free Med26 was not recruited by HNF4α to the immobilized template, and, accordingly, did not recruit EAF1 (Figure 3E). Furthermore, excess free Med26 NTD (residues 1-113) competes with Mediator for EAF1 recruitment (Figure 3F). Thus, Mediator can recruit EAF1 or EAF1-associated complexes to immobilized promoters via direct interaction of the Med26 NTD with EAF1.
Elongation Factors and TFIID Bind to Overlapping Surfaces of the Med26 NTD
As described above, the Med26 NTD is closely related in sequence to the TFIIS NTD and, based on an NMR structure of the TFIIS NTD, has been modeled as a four-helix bundle (Booth et al., 2000). To explore whether elongation factors and TFIID bind to distinct or overlapping surfaces of the Med26 NTD, we generated a series of Med26 NTD mutants in which various predicted surface residues were changed to alanine and asked whether we could identify mutations that differentially affect Med26 NTD interactions with ELL/EAF-containing complexes and with TFIID.
We observed that a GAL4-Med26 NTD fusion protein acts as a strong activator of a luciferase reporter driven by 5 GAL4 responsive elements upstream of the AdML promoter in transient transfection assays (Figure S1A). GAL4-Med26 NTD with alanines in place of predicted surface residues R61 and K62 or K74 and K75 were expressed well but failed to activate the luciferase reporter, whereas GAL4-Med26 NTD D13A was as active as the wild type GAL4-Med26 NTD. As shown in Figure S1B, GAL4-Med26 NTD and GAL4-Med26 NTD(D13A) could both recruit TFIID and EAF1 to immobilized DNA, whereas GAL4-Med26 NTD(R61A,K62A) and GAL4-Med26 NTD(K74A,K75A) could not. These residues are also critical for Med26 NTD interactions with both elongation factors and TFIID in cells, since neither EAF1, ELL, CDK9, Cyclin T1/T2 nor TFIID subunits were detected by western blotting in FLAG-IPs from cells stably expressing FLAG-Med26 NTD(R61A,K62A) or FLAG-Med26 NTD(K74A,K75A) (Figure 4A).
Figure 4. Med26 NTD mutants fail to bind ELL/EAF-containing complexes or TFIID.
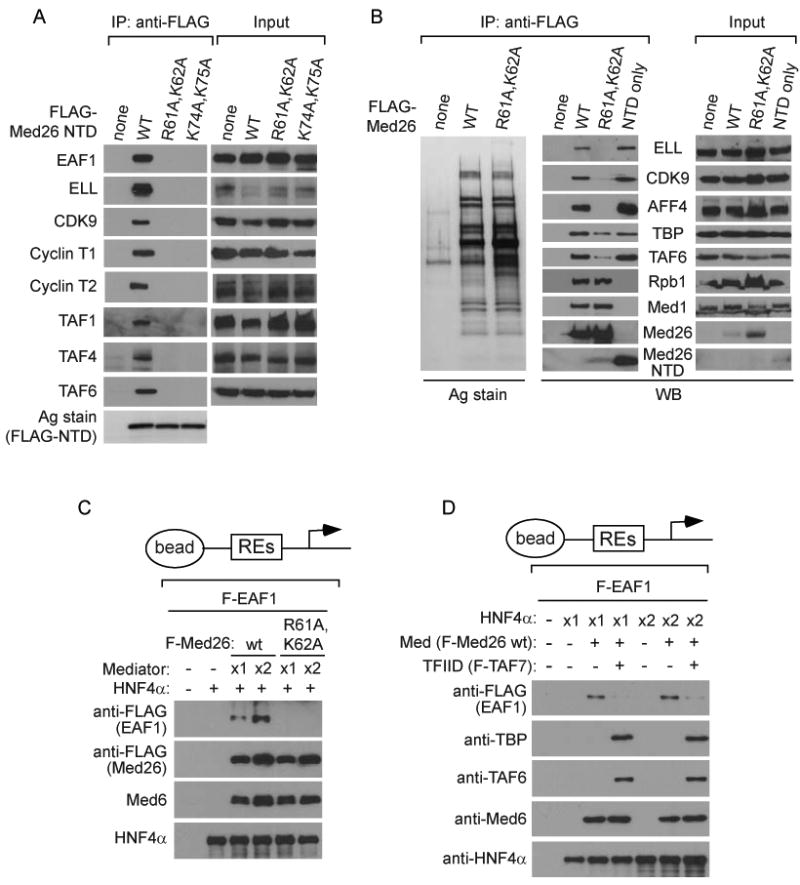
(A) Western blots of FLAG IPs or lysates (Input) from cells stably expressing the indicated FLAG-Med26 NTD proteins. Data is from the same experiment in Figure 1D. Negative and positive controls in the first two lanes of each panel are repeated from Figure 1D. (B) Defective binding of Med26(R61A,K62A)-Mediator to elongation factors. FLAG IPs from cells stably expressing the indicated FLAG-Med26 proteins analyzed by silver staining (Ag stain) or western blotting (WB). (C) Med26(R61A,K62A)- Mediator fails to recruit EAF1 to a promoter. Assays contained template, HNF4α, EAF1, and wild type or Med26(R61A,K62A)-Mediator. (D) TFIID blocks Mediator-dependent recruitment of EAF1 to a promoter. Assays contained template, HNF4α, EAF1, TFIID and Mediator.
We next tested the ability of FLAG-Med26(R61A,K62A) to assemble into Mediator and to interact with elongation factors and TFIID when expressed in human cells. As expected, both wild type and mutant Med26 assembled into Mediator, and Mediator containing the Med26 mutant failed to bind to elongation factors (Figure 4B). In addition, Mediator containing Med26(R61A,K62A) could not recruit EAF1 to a promoter (Figure 4C). Finally, Mediator containing Med26(R61A,K62A) could still interact with TFIID subunits (Figure 4B), consistent with our evidence that deletion of the Med26 NTD does not interfere with Mediator's ability to enhance recruitment of TFIID to a promoter in vitro (Figure 3B). We note that there was a modest decrease in the amount of TBP and TAF6 that co-IPed with Mediator purified from cells expressing FLAG-Med26(R61A,K62A); however, it is not clear whether this reflects a small decrease in the affinity of the mutant Mediator for TFIID or a decrease in expression of TFIID subunit(s) needed for the interaction, since TAF6 expression was decreased in these cells.
Taken together, these observations argue that ELL/EAF-containing complexes and TFIID bind to the same or overlapping surfaces of the Med26 NTD, even though interactions of TFIID with surface(s) outside of this domain are most important for its interaction with Mediator. In addition, they provide further support for the idea that EAF family proteins function as adapters that link ELL-containing complexes to Mediator via the Med26 NTD, since Med26 double point mutations that block interactions with EAF1 in vitro interfere with binding of the Med26 NTD or of intact Mediator to subunits of ELL/EAF-containing complexes in cells.
These observations suggested that Mediator might not be able to bind simultaneously to EAF1 and TFIID. Consistent with this possibility, EAF1 recruitment to an activator-bound promoter was substantially decreased when binding reactions contained TFIID (Figure 4D), raising the possibility that prior to initiation and/or the receipt of an appropriate signal, the Med26 NTD might be masked by interaction with TFIID and only become available to recruit ELL/EAF-containing complexes upon release of this interaction.
Med26 Regulates Cell Proliferation and Gene Expression
To study Med26 function in cells, we generated stable human 293T embryonic kidney cell lines expressing non-targeting or one of three different doxycycline (Dox) inducible short hairpin RNAs (shRNAs) targeting the 3′-UTR of endogenous MED26. Dox treatment silenced Med26 protein expression in cell lines expressing each of the Dox-inducible Med26 shRNAs (Figure 5A). Induction of any of the three Med26 shRNAs reduced cell proliferation, while non-targeting shRNA had no effect (Figures 5B and 5C). Proliferation of mouse ES cells was also inhibited by transfection of Med26 siRNAs, but not nontargeting siRNA or Med6, CDK8, or non-targeting siRNAs (Supplemental Figure S2A and B).
Figure 5. Effect of Med26 depletion on cell proliferation and gene expression.
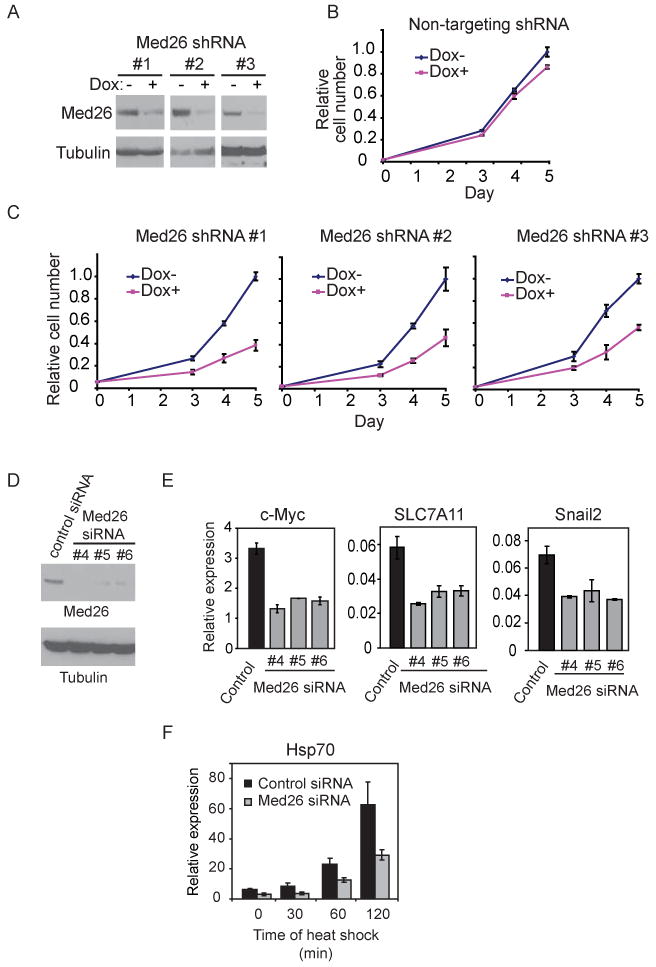
(A) Dox-inducible shRNA-mediated silencing of endogenous Med26 expression in 293T cells stably expressing different shRNAs. Cells were incubated with or without Dox for 48 hours, and cell lysates were analyzed by western blotting. (B)(C) Med26 depletion decreases cell proliferation in 293T cells stably expressing Dox-inducible shRNAs. Cell number is expressed relative to cell number at day 5 in cultures without Dox. (D) Western blots for endogenous Med26 or tubulin 48 hrs after transfection of non-targeting (control) or Med26 siRNAs. (E) Effect of siRNA-mediated Med26 depletion on gene expression. (F) Effect of Med26 depletion on Hsp70 induction by heat shock. Data points are the average of three independent experiments; error bars show standard deviation. See also Figure S2 and Table S2.
To identify Med26 target genes, we used Affymetrix expression arrays to analyze mRNA expression in 293T cells from which Med26 had been depleted by transient transfection of one of three different siRNAs (Figure 5D). Of the approximately 14,550 well-characterized genes on the array, more than 10% were significantly affected (adjusted p value ≤0.05) after Med26 knockdown by all three siRNAs. Interestingly, among the genes most negatively regulated by Med26 depletion were c_-MYC_ (MYC), HSP70 (HSPA1A), and SNAIL2 (SNAI2) (Table S2). As noted earlier, c_-MYC_ and HSP70 were among the first examples of genes shown to be regulated during the transition from early elongation to productive elongation (Saunders et al., 2006). In addition, results of a recent genome-wide analysis provide evidence that Pol II is enriched at the 5′-end of the SNAIL2 gene in mouse ES cells (Rahl et al., 2010), and the SNAIL2 ortholog Snail is one of many developmentally regulated genes controlled by promoter-proximal pausing in Drosophila (Zeitlinger et al., 2007).
Quantitative RT-PCR analysis confirmed that steady state expression of these and other genes was decreased following transfection of all three Med26 siRNAs (Figure 5E and Supplemental Figure S2C). Upon heat shock, synthesis of new Hsp70 mRNA was delayed in cells transfected with Med26 siRNA (Figure 5F), indicating that Med26 regulates not only steady state but also newly induced gene expression.
Effect of Med26 Depletion on SEC Recruitment and Pol II CTD Phosphorylation at the c-MYC and HSP70 Genes
To begin to investigate potential roles of Med26 in SEC recruitment in cells, we focused on two examples: steady state expression of the c-MYC gene and heat shock induced activation of the HSP70 gene. Endogenous Med26, P-TEFb kinase CDK9, and SEC component AFF4, as well as several exogenously expressed SEC components (FLAG-ELL, AFF1, and AF9) (Figures 6A, 6B, and S3A) are detected by ChIP at the c-MYC gene in human cells. These observations suggest that SEC contributes to c-MYC gene regulation and are consistent with previous evidence that P-TEFb regulates c-MYC expression (Glover-Cutter et al., 2009; Montanuy et al., 2008). Med26, like AFF4, ELL, AFF1, and AF9, is detected by ChIP throughout the body of the c-MYC gene in these cells, even though TBP occupancy is limited to the promoter region as expected (Figure S3A).
Figure 6. Effect of Med26 depletion on recruitment of SEC Components and Pol II CTD phosphorylation.
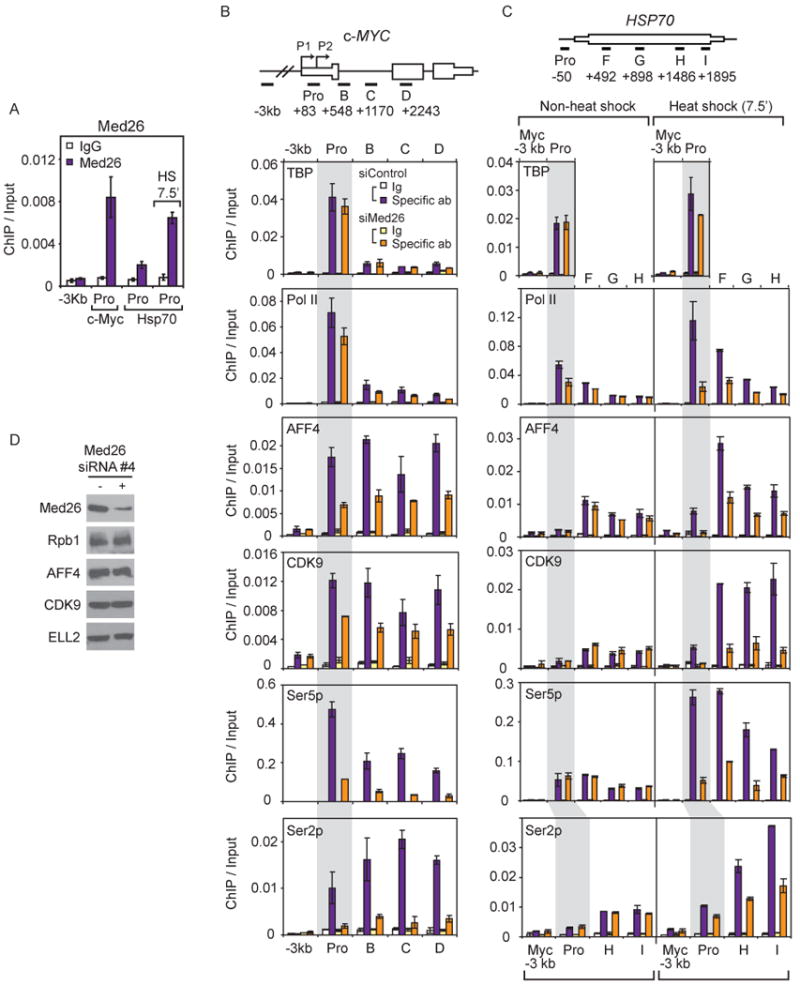
(A) Occupancy of Med26 on the promoter of c-MYC or HSP70 gene under non-heat or heat shocked condition (7.5 min). (B)(C) Effect of Med26 depletion on AFF4, CDK9 and Pol II occupancy on the c-MYC or HSP70 gene under steady state or heat shocked conditions in HEK293T cells. Ig, IgG (TBP, Pol II, AFF4, CDK9 and Ser5p ChIPs) or IgM (Ser2p ChIP) control; Spec ab, specific antibody. (A)(B)(C) ChIP/input is average from two biological replicates, error bars show data range. (D) Med26, Pol II and SEC components in HEK293T cells treated with non-targeting siRNA or Med26 siRNA #4.
Upon siRNA-mediated depletion of Med26, we observed little change in the amount of total Pol II (detected using an antibody that recognizes an epitope in the N-terminus of Rpb1) or TFIID subunit TBP at the c-MYC promoter region. About 2 kb downstream from the transcription start site, Pol II occupancy was decreased by about 50%, similar to the reduction in c-Myc mRNA after Med26 knockdown. Occupancy of SEC components AFF4 and CDK9 at the promoter and throughout the body of the gene (Figure 6B) was also decreased, consistent with evidence that the Med26 NTD contributes to recruitment of elongation factors, but not TFIID, to promoters in vitro. Promoter escape and transcript elongation by Pol II are associated with phosphorylation of the Pol II CTD by P-TEFb and other kinases (Bartkowiak et al., 2010; Buratowski, 2009; Kim et al., 2002; Marshall et al., 1996). Depletion of Med26 dramatically decreased the amount of Ser2 and Ser5 phosphorylated Pol II throughout the c-MYC gene (Figure 6B). In control experiments, we confirmed that Med26 knockdown had no significant effect on expression of Pol II subunit Rpb1 or any of the SEC components tested (Figure 6D).
Prior to heat shock, an initiated but paused Pol II is present in the promoter-proximal region of the Hsp70 gene (Core and Lis, 2008). Upon heat shock, Mediator (Park et al., 2001) and SEC components (Lin et al., 2010; Lis et al., 2000; Smith et al., 2008), are rapidly recruited to the Hsp70 gene, and the paused polymerase is released into productive elongation, allowing additional Pol II to be recruited to the gene (Boehm et al., 2003; Lis, 1998). Similarly, we detect Pol II and TBP at the promoter of the HSP70 gene prior to heat shock (Figures 6C and S3B). Consistent with our observation that there is detectable heat shock gene expression in non-heat shocked cells, a small amount of Pol II (Ser2- and Ser5-phosphorylated), as well as AFF4 and CDK9 could be detected in the body of the gene prior to heat shock (Figure 6C). Following 7.5 or 60 min of heat shock, Mediator subunits Med26 and Med1 and SEC components CDK9 and AFF4 were recruited (Figures 6A, 6C, and S3B). As observed at c-MYC, the ChIP signal for Med26 and, to a lesser extent, Med1 extended into the body of the HSP70 gene (Figure S3B). After 60 min of heat shock, we observed substantial release of Pol II into the body of the gene and recruitment of new Pol II to the promoter (Figure S3B).
To focus on the effect of Med26 depletion on early events in heat shock gene induction, we performed ChIP using chromatin from cells that had or had not been subjected to just 7.5 min of heat shock. Med26 depletion had little effect on the amount of total Pol II or of Ser 2- or Ser 5-phosphorylated Pol II, AFF4, or CDK9 at the HSP70 gene in non-heat shocked cells (Figure 6C), consistent with the presence of only a small amount of Med26 at the HSP70 promoter before heat shock (Figure 6A). After heat shock, however, Med26 depletion led to a significant reduction in new Pol II recruitment to the HSP70 promoter, accompanied by decreased occupancy of AFF4 and CDK9 and of Ser2 and Ser5 phosphorylated Pol II throughout the body of the HSP70 gene. Indeed, in heat shocked cells depleted of Med26 the amount of total and CTD-phosphorylated Pol II, AFF4, and CDK9 in the body of the gene remained similar to or only modestly higher than that observed in non-heat shocked cells (Figure 6C).
Together these findings suggest that Med26 contributes to Mediator activity at multiple stages of transcription. During transcription of c-MYC at steady state, Med26 seems particularly important for events after Pol II recruitment, including SEC recruitment and CTD phosphorylation. During activation of HSP70, however, Med26 knockdown not only decreases SEC recruitment, CTD phosphorylation, and release of Pol II into the body of the gene, but also leads to a significant decrease in new Pol II recruitment to the promoter. The effect of depleting Med26 on Pol II distribution at HSP70 is very similar to what was observed previously by Adelman et al. (2005) in experiments examining the effect of depleting the elongation factor TFIIS from Drosophila S2 cells. They attributed the decrease in new Pol II recruitment to failure of paused Pol II to move away from the promoter and make room for new Pol IIs to become associated with the gene; however, it seems likely that some of the effect we observe is due to defects in Mediator-dependent recruitment of new Pol II after Med26 knockdown.
A Role for the Med26 NTD in Regulating Cell Proliferation and Gene Expression
To assess the potential contribution of the Med26 NTD to regulation of cell proliferation and gene expression in cells, wild type Med26 or Med26(R61A,K62A) were stably expressed in HEK293T cell lines expressing Dox-inducible shRNAs targeting Med26. Since the Med26 shRNAs target sequences in the MED26 3′UTR, exogenously expressed Med26 mRNAs, which do not include the 3′-UTR, should not be affected by the Med26 shRNA.
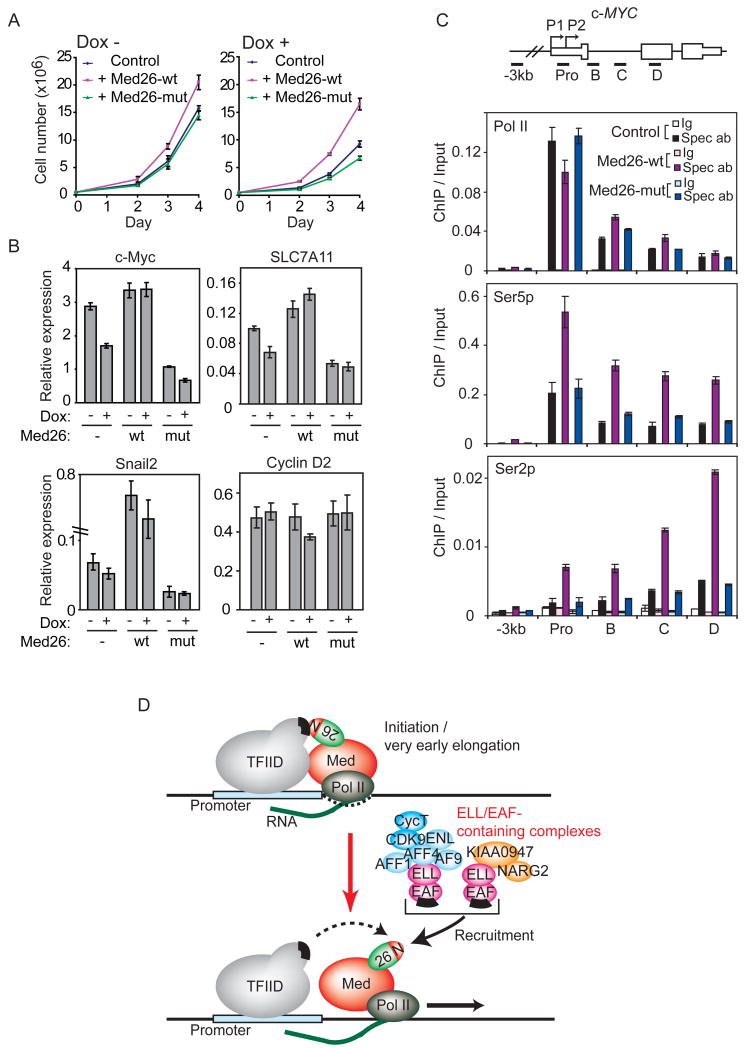
Even before depletion of endogenous Med26, the proliferation rate of cells expressing wild type Med26 was moderately higher than that of control cells or cells expressing Med26(R61A,K62A) (Figure 7A, left panel). After depletion of endogenous Med26, cells expressing wild type Med26 grew substantially faster than control cells or cells expressing Med26(R61A,K62A) (Figure 7A, right panel). Cells expressing mutant Med26 grew more slowly than control cells after shRNA induction, suggesting Med26(R61A,K62A) is a dominant negative inhibitor of Med26 function in cells when endogenous Med26 is limiting.
Figure 7. The Med26 NTD regulates cell proliferation and gene expression.
(A) Med26-wt (wild type), but not Med26-mut (R61A,K62A), rescues proliferation defect of Med26 depleted 293T cells expressing Dox-inducible Med26 shRNA #3 and exogenous wild type or mutant Med26, grown with or without Dox. (B) Wild type, but not Med26(R61A,K62A) rescues gene expression defect of Med26 depleted cells. (C) Effect of overexpressing Med26 or Med26(R61A,K62A) on Pol II occupancy at c-MYC in HeLa cells stably expressing wild type or mutant Med26. Ig, IgG (Pol II and Ser5p ChIPs) or IgM (Ser2p ChIP) control; Spec ab, specific antibody. (D) Model for Mediator and Med26 NTD function during the transition to productive elongation. In the initiation and/or early elongation complexes, the EAF binding site on the Med26 NTD is occluded by interaction with TFIID. As Pol II moves further from the initiation site and/or upon receipt of a suitable signal, the Med26 NTD is released from TFIID, allowing it to recruit ELL/EAF complexes. Green and red oval, Med26; N, Med26 NTD; black bars, binding sites for Med26 NTD on TFIID and EAF family members.
To investigate further the contribution of the Med26 NTD to gene expression, we used cell lines expressing Dox-inducible Med26 shRNAs to test the requirement for the NTD in expression of several genes that were down-regulated in cells transiently transfected with Med26 siRNAs. As a control, we also included CYCLIN D2, which was unaffected by Med26 knockdown in microarrays. The reduction in gene expression upon induction of Med26 shRNA in these cell lines was not as great as the reduction seen in cells transiently transfected with Med26 siRNAs, due either to less efficient depletion of Med26 by the shRNA in these cell lines or to leaky expression of shRNA even without Dox induction.
The results of these experiments are consistent with a role for the Med26 NTD in expression of each gene tested except for the control CYCLIN D2 gene; however, the responses of these genes to exogenously expressed Med26 differed (Figure 7B). Med26 shRNA reduced expression of c-MYC and SLC7A11 to a similar extent. In both cases, exogenously expressed wild type Med26 blocked the effect of depleting endogenous Med26, but led to only a modest increase in gene expression relative to control. In contrast, SNAIL2 expression was much higher in wild type Med26-expressing cells than in control cells even before induction of Med26 shRNA, suggesting that Med26 is limiting for SNAIL2 expression in human embryonic kidney cells. In each case, Med26(R61A,K62A) had a dominant negative effect, since its expression led to an approximately 2-fold decrease in mRNA levels whether or not the Med26 shRNA was induced.
To explore further the contribution of the Med26 NTD to gene regulation, we asked whether overexpression of wild type Med26 or Med26(R61A,K62A) differentially affects Pol II distribution or CTD phosphorylation at the c-MYC gene; these experiments were performed using the same HeLa cell lines in which we showed that both wild type and mutant Med26 are fully capable of assembling into Mediator. Overexpression of wild type but not mutant Med26 led to a small decrease in the amount of total Pol II at the c-MYC promoter and a concomitant small increase in total Pol II further downstream, consistent with the possibility that more Pol II is released from the promoter region in cells overexpressing wild type Med26 (Figure 7C). In addition, overexpression of wild type but not mutant Med26 increased the amount of Ser2- and Ser5-phosphorylated Pol II at the promoter and throughout the body of the gene. Notably, the effect of Med26 overexpression on Ser2-phosphorylation was more pronounced than the effect on Ser5-phosphorylation. Thus, a Med26 NTD mutation that prevents the interaction of Mediator with transcription elongation factors interferes with Med26 functions in cell proliferation and gene regulation.
Discussion
In this report, we show that the conserved N-terminal domain of Med26 functions as a docking site on Mediator for at least two types of complexes that contain members of the ELL/EAF family of transcription elongation factors: the multimeric SEC complexes, which also contain P-TEFb and the MLL translocation partners AFF4, AFF1, ENL, and AF9, and an additional ELL/EAF-containing complex of as yet unknown function.
In addition to ELL/EAF-containing complexes, the Med26 NTD contains a binding site for TFIID. Consistent with evidence that yeast Mediator, which lacks Med26, binds TBP through contacts with Med8 and other subunits in the head module (Cai et al., 2010; Lariviere et al., 2006), however, mutation of the Med26 NTD does not affect Mediator's ability to enhance binding of TFIID to a promoter, arguing that the Med26 NTD is not solely responsible for Mediator's interaction with TFIID. Together with our observation that prior binding of Mediator to TFIID at a promoter prevents Mediator from recruiting Pol II elongation factors via the Med26 NTD, this finding raises the possibility that the Med26 NTD is a molecular switch that interacts first with the Pol II initiation complex through direct interactions with TFIID and then exchanges TFIID for Pol II elongation factors to facilitate productive elongation and/or other elongation-associated processes modulated by these factors (Figure 7D). Consistent with this model, Med26 knockdown has little or no effect on TBP occupancy at the c-MYC and HSP70 promoters.
There is precedent for the idea that Mediator has a role in controlling events that occur during transcription elongation. Deletion of Med23 from mouse ES cells eliminates expression of the serum response gene EGR1 due to (i) loss of Mediator recruitment and (ii) failure to release Pol II from the promoter region (Balamotis et al., 2009; Wang et al., 2005). In yeast, Mediator subunits are detected by ChIP not only at promoters, but also in the coding regions of some genes (Andrau et al., 2006; Zhu et al., 2006). In human cells, the Mediator kinase module subunit CDK8 has been localized to coding regions of genes it activates (Donner et al., 2007; Donner et al., 2010). Similarly, we show that Med26 can be detected in the body of both the c-MYC and HSP70 genes. Thus, interactions between Med26 and the SEC might contribute not only to SEC recruitment near the transcription start site, but also to SEC recruitment or retention throughout elongation. Finally, several recent studies suggest a role for the Mediator kinase module in recruiting P-TEFb following activation of EGR1 and other serum response genes (Donner et al., 2010) and the thyroid hormone receptor-activated gene DioI (Belakavadi and Fondell, 2010). Although the relationship between Med26 and kinase module in gene regulation remains to be determined, there is little overlap between the genes most affected by manipulating Med26 expression in our studies and those found previously to be most affected by manipulating CDK8 expression (Donner et al., 2007; Donner et al., 2010). This, together with evidence for the existence of both free kinase module and of forms of Mediator containing either Med26 or kinase module, raises the possibility that Med26 and the kinase module act via distinct mechanisms to recruit elongation factors to different genes and/or under different conditions.
Finally, our results have implications for mechanisms underlying misregulation of gene expression in mixed lineage leukemias caused by translocations of MLL gene to one of a large number of translocation partners. Among these translocation partners are the SEC components ELL, AFF4, AFF1, AF9, and ENL. The finding that in normal cells these proteins are found together in the SEC, together with recent evidence that MLL fusion proteins themselves can assemble into larger, SEC-like complexes (Lin et al., 2010; Yokoyama et al., 2010), is consistent with the model that inappropriate gene activation in mixed lineage leukemias may be due to mistargeting of SEC-like complexes by MLL fusion proteins (Lin et al., 2010; Mueller et al., 2009; Yokoyama et al., 2010). Our demonstration that SEC binds Mediator through the Med26 NTD suggests that mistargeting of the SEC by MLL fusion proteins could also lead to inappropriate activation of some genes by recruitment of Mediator and Pol II to aberrant chromosomal locations.
Experimental Procedures
Cell culture, cell lines
Parental HeLa S3 and Flp-In 293 cells (Invitrogen, Carlsbad, CA) and their derivatives were cultured as described (Cai et al., 2007; Sato et al., 2004). Construction of expression plasmids and generation of cell lines are described in Supplemental Information.
Western blotting, immunoprecipitation, and affinity purification
Antibodies used are described in Supplemental Information. Protein complexes were purified from cell lines stably expressing FLAG-tagged proteins using anti-FLAG (M2) agarose (Sigma) as described (Takahashi et al., 2009).
Mass spectrometry
Proteins were identified using MudPIT; for details see Supplemental Information. Normalized Spectral Abundance Factors (NSAFs) (Zhang et al., 2010) were used to estimate relative protein abundance.
Immobilized template assays
Biotinylated DNA fragments from the plasmids pG5MLT or pREx4MLT, which contain tandemly repeated GAL4- or HNF4α-responsive elements, respectively, were generated and bound to Dynabeads™ (Dynal). Recruitment assays were performed essentially as described (Takahashi et al., 2009).
Gene expression analysis and ChIP
Total RNA from HEK293T cells transfected with non-targeting control or Med26 siRNAs or from cell lines stably expressing Med26 shRNAs, with or without Med26 rescue constructs, was used to measure genome-wide gene expression with Affymetrix U133A plus 2.0 expression arrays or to measure expression of individual genes by qPCR. ChIP assays were performed with normal IgG or the indicated antibodies, and precipitated DNA was measured by qPCR. Primer sets, methods for gene expression and ChIP analyses, and siRNAs and shRNAs are detailed in Supplemental Information.
Supplementary Material
01
02
03
Highlights.
- Mediator subunit Med26 controls expression of c-MYC and other genes regulated during transcript elongation
- Med26 N-terminal domain (NTD) contributes to Med26-dependent gene regulation
- Med26 NTD recruits ELL- and P-TEFb-containing complexes to Mediator
- Med26-Mediator contributes to recruitment of ELL/P-TEFb complexes to targets in vitro and in cells
Acknowledgments
We thank T. Suganuma for helpful discussions, M. Thirman for a generous gift of the anti-EAF1 antibody, K. Zueckert-Gaudenz and A. Peak for help with microarray experiments, and M. Katt and V. Neubauer for tissue culture. C.L. is a Ph.D. thesis student registered with the Open University. This work was supported by funds from the Stowers Institute and by grant GM41628 from NIGMS, NIH to J.W.C. and R.C.C.
Footnotes
Accession Numbers: Microarray data are deposited in GEO under accession number GSE28715.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adelman K, Marr MT, Werner J, Saunders A, Ni Z, Andrulis ED, Lis JT. Efficient release from promoter-proximal stall sites requires transcript cleavage factor TFIIS. Mol Cell. 2005;17:103–112. doi: 10.1016/j.molcel.2004.11.028. [DOI] [PubMed] [Google Scholar]
- Andrau JC, van de PL, Lijnzaad P, Bijma T, Koerkamp MG, van de PJ, Werner M, Holstege FC. Genome-wide location of the coactivator mediator: Binding without activation and transient Cdk8 interaction on DNA. Mol Cell. 2006;22:179–192. doi: 10.1016/j.molcel.2006.03.023. [DOI] [PubMed] [Google Scholar]
- Balamotis MA, Pennella MA, Stevens JL, Wasylyk B, Belmont AS, Berk AJ. Complexity in transcription control at the activation domain-mediator interface. Sci Signal. 2009;2:ra20. doi: 10.1126/scisignal.1164302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks CA, Kong SE, Spahr H, Florens L, Martin-Brown S, Washburn MP, Conaway JW, Mushegian A, Conaway RC. Identification and Characterization of a Schizosaccharomyces pombe RNA Polymerase II Elongation Factor with Similarity to the Metazoan Transcription Factor ELL. J Biol Chem. 2007;282:5761–5769. doi: 10.1074/jbc.M610393200. [DOI] [PubMed] [Google Scholar]
- Bartkowiak B, Liu P, Phatnani HP, Fuda NJ, Cooper JJ, Price DH, Adelman K, Lis JT, Greenleaf AL. CDK12 is a transcription elongation-associated CTD kinase, the metazoan ortholog of yeast Ctk1. Genes Dev. 2010;24:2303–2316. doi: 10.1101/gad.1968210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belakavadi M, Fondell JD. Cyclin-dependent kinase 8 positively cooperates with Mediator to promote thyroid hormone receptor-dependent transcriptional activation. Mol Cell Biol. 2010;30:2437–2448. doi: 10.1128/MCB.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm AK, Saunders A, Werner J, Lis JT. Transcription factor and polymerase recruitment, modification, and movement on dhsp70 In Vivo in the minutes following heat shock. Mol Cell Biol. 2003;23:7628–7637. doi: 10.1128/MCB.23.21.7628-7637.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth V, Koth CM, Edwards AM, Arrowsmith CH. Structure of a conserved domain common to the transcription factors TFIIS, elongin A, and CRSP70. J Biol Chem. 2000;275:31266–31268. doi: 10.1074/jbc.M002595200. [DOI] [PubMed] [Google Scholar]
- Bourbon HM. Comparative genomics supports a deep evolutionary origin for the large, four-module transcriptional mediator complex. Nucleic Acids Res. 2008;36:3993–4008. doi: 10.1093/nar/gkn349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski S. Progression through the RNA polymerase II CTD cycle. Mol Cell. 2009;36:541–546. doi: 10.1016/j.molcel.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun JS, Wong MM, Cui W, Idelman G, Li Q, De SA, Bilke S, Haggerty CM, Player A, Wang YH, Thirman MJ, Kaberlein JJ, Petrovas C, Koup RA, Longo D, Ozato K, Gardner K. Dynamic bookmarking of primary response genes by p300 and RNA polymerase II complexes. Proc Natl Acad Sci U S A. 2009;106:19286–19291. doi: 10.1073/pnas.0905469106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai G, Imasaki T, Yamada K, Cardelli F, Takagi Y, Asturias FJ. Mediator head module structure and functional interactions. Nat Struct Mol Biol. 2010;17:273–279. doi: 10.1038/nsmb.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Jin J, Yao T, Gottschalk AJ, Swanson SK, Wu S, Shi Y, Washburn MP, Florens L, Conaway RC, Conaway JW. YY1 functions with INO80 to activate transcription. Nat Struct Mol Biol. 2007;14:872–874. doi: 10.1038/nsmb1276. [DOI] [PubMed] [Google Scholar]
- Core LJ, Lis JT. Transcription regulation through promoter-proximal pausing of RNA polymerase II. Science. 2008;319:1791–1792. doi: 10.1126/science.1150843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donner AJ, Ebmeier CC, Taatjes DJ, Espinosa JM. CDK8 is a positive regulator of transcriptional elongation within the serum response network. Nat Struct Mol Biol. 2010;17:194–201. doi: 10.1038/nsmb.1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donner AJ, Szostek S, Hoover JM, Espinosa JM. CDK8 is a stimulus-specific positive coregulator of p53 target genes. Mol Cell. 2007;27:121–133. doi: 10.1016/j.molcel.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover-Cutter K, Larochelle S, Erickson B, Zhang C, Shokat K, Fisher RP, Bentley DL. TFIIH-associated Cdk7 kinase functions in phosphorylation of C-terminal domain Ser7 residues, promoter-proximal pausing, and termination by RNA polymerase II. Mol Cell Biol. 2009;29:5455–5464. doi: 10.1128/MCB.00637-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He N, Liu M, Hsu J, Xue Y, Chou S, Burlingame A, Krogan NJ, Alber T, Zhou Q. HIV-1 Tat and host AFF4 recruit two transcription elongation factors into a bifunctional complex for coordinated activation of HIV-1 transcription. Mol Cell. 2010;38:428–438. doi: 10.1016/j.molcel.2010.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KM, Wang J, Smallwood A, Arayata C, Carey M. TFIID and human mediator coactivator complexes assemble cooperatively on promoter DNA. Genes Dev. 2002;16:1852–1863. doi: 10.1101/gad.995702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Bjorklund S, Li Y, Sayre MH, Kornberg RD. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- Kim YK, Bourgeois CF, Isel C, Churcher MJ, Karn J. Phosphorylation of the RNA polymerase II carboxyl-terminal domain by CDK9 is directly responsible for human immunodeficiency virus type 1 tat-activated transcriptional elongation. Mol Cell Biol. 2002;22:4622–4637. doi: 10.1128/MCB.22.13.4622-4637.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kininis M, Isaacs GD, Core LJ, Hah N, Kraus WL. Postrecruitment regulation of RNA polymerase II directs rapid signaling responses at the promoters of estrogen target genes. Mol Cell Biol. 2009;29:1123–1133. doi: 10.1128/MCB.00841-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koleske AJ, Young RA. An RNA polymerase II holoenzyme responsive to activators. Nature. 1994;368:466–469. doi: 10.1038/368466a0. [DOI] [PubMed] [Google Scholar]
- Kong SE, Banks CAS, Shilatifard A, Conaway JW, Conaway R. ELL-associated factors 1 and 2 are positive regulators of RNA polymerase II elongation factor ELL. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:10094–10098. doi: 10.1073/pnas.0503017102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lariviere L, Geiger S, Hoeppner S, Rother S, Strasser K, Cramer P. Structure and TBP binding of the Mediator head subcomplex Med8-Med18-Med20. Nat Struct Mol Biol. 2006;13:895–901. doi: 10.1038/nsmb1143. [DOI] [PubMed] [Google Scholar]
- Lin C, Smith ER, Takahashi H, Lai KC, Martin-Brown S, Florens L, Washburn MP, Conaway JW, Conaway RC, Shilatifard A. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol Cell. 2010;37:429–437. doi: 10.1016/j.molcel.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lis J. Promoter-associated pausing in promoter architecture and postinitiation transcriptional regulation. Cold Spring Harb Symp Quant Biol. 1998;63:347–356. doi: 10.1101/sqb.1998.63.347. [DOI] [PubMed] [Google Scholar]
- Lis JT, Mason P, Peng J, Price DH, Werner J. P-TEFb kinase recruitment and function at heat shock loci. Genes Dev. 2000;14:792–803. [PMC free article] [PubMed] [Google Scholar]
- Marshall NF, Peng J, Xie Z, Price DH. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J Biol Chem. 1996;271:27176–27183. doi: 10.1074/jbc.271.43.27176. [DOI] [PubMed] [Google Scholar]
- Martincic K, Alkan SA, Cheatle A, Borghesi L, Milcarek C. Transcription elongation factor ELL2 directs immunoglobulin secretion in plasma cells by stimulating altered RNA processing. Nat Immunol. 2009;10:1102–1109. doi: 10.1038/ni.1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler G, Kremmer E, Timmers HTh, Meisterernst M. Novel critical role of a human Mediator complex for basal RNA polymerase II transcription. EMBO Rep. 2001;2:808–813. doi: 10.1093/embo-reports/kve186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo X, Kowenz-Leutz E, Xu H, Leutz A. Ras induces mediator complex exchange on C/EBPβ. Mol Cell. 2004;13:241–250. doi: 10.1016/s1097-2765(03)00521-5. [DOI] [PubMed] [Google Scholar]
- Montanuy I, Torremocha R, Hernandez-Munain C, Sune C. Promoter influences transcription elongation: TATA-box element mediates the assembly of processive transcription complexes responsive to cyclin-dependent kinase 9. J Biol Chem. 2008;283:7368–7378. doi: 10.1074/jbc.M706243200. [DOI] [PubMed] [Google Scholar]
- Mueller D, Garcia-Cuellar MP, Bach C, Buhl S, Maethner E, Slany RK. Misguided transcriptional elongation causes mixed lineage leukemia. PLoS Biol. 2009;7:e1000249. doi: 10.1371/journal.pbio.1000249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naar AM, Taatjes DJ, Zhai W, Nogales E, Tjian R. Human CRSP interacts with RNA polymerase II and adopts a specific conformation. Genes Dev. 2002;16:1339–1344. doi: 10.1101/gad.987602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechaev S, Fargo DC, dos Santos G, Liu L, Gao Y, Adelman K. Global analysis of short RNAs reveals widespread promoter-proximal stalling and arrest of Pol II in Drosophila. Science. 2010;327:335–338. doi: 10.1126/science.1181421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Z, Schwarts BE, Werner J, Suarez JR, Lis T. Coordination of transcription, RNA processing, and surveillance by P-TEFb kinase on heat shock genes. Mol Cell. 2004;13:55–65. doi: 10.1016/s1097-2765(03)00526-4. [DOI] [PubMed] [Google Scholar]
- Park JM, Werner J, Kim JM, Lis JT, Kim YJ. Mediator, Not holoenzyme, is directly recruited to the heat shock promoter by HSF upon heat shock. Mol Cell. 2001;8:9–19. doi: 10.1016/s1097-2765(01)00296-9. [DOI] [PubMed] [Google Scholar]
- Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol Cell. 2006a;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol Cell. 2006b;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu S, Zhou S, Ladurner AG, Tjian R. The transcriptional cofactor complex CRSP is required for activity of the enhancer binding protein Sp1. Nature. 1999;397:446–450. doi: 10.1038/17141. [DOI] [PubMed] [Google Scholar]
- Sato S, Tomomori-Sato C, Parmely TJ, Florens L, Zybailov B, Swanson SK, Banks CA, Jin J, Cai Y, Washburn MP, Conaway JW, Conaway RC. A set of consensus mammalian mediator subunits identified by multidimensional protein identification technology. Mol Cell. 2004;14:685–691. doi: 10.1016/j.molcel.2004.05.006. [DOI] [PubMed] [Google Scholar]
- Saunders A, Core LJ, Lis JT. Breaking barriers to transcription elongation. Nat Rev Mol Cell Biol. 2006;7:557–567. doi: 10.1038/nrm1981. [DOI] [PubMed] [Google Scholar]
- Serizawa H, Conaway RC, Conaway JW. A carboxyl-terminal-domain kinase associated with RNA polymerase II transcription factor delta from rat liver. Proc Natl Acad Sci U S A. 1992;89:7476–7480. doi: 10.1073/pnas.89.16.7476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A, Haque D, Conaway RC, Conaway JW. Structure and function of RNA polymerase II elongation factor ELL: Identification of two overlapping ELL functional domains that govern its interaction with polymerase and the ternary elongation complex. J Biol Chem. 1997;272:22355–22363. doi: 10.1074/jbc.272.35.22355. [DOI] [PubMed] [Google Scholar]
- Shilatifard A, Lane WS, Jackson KW, Conaway RC, Conaway JW. The human ELL gene encodes a novel RNA polymerase II elongation factor. Science. 1996;271:1873–1876. doi: 10.1126/science.271.5257.1873. [DOI] [PubMed] [Google Scholar]
- Smith ER, Winter B, Eissenberg JC, Shilatifard A. Regulation of the transcriptional activity of poised RNA polymerase II by the elongation factor ELL. Proc Natl Acad Sci U S A. 2008;105:8575–8579. doi: 10.1073/pnas.0804379105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobhian B, Laguette N, Yatim A, Nakamura M, Levy Y, Kiernan R, Benkirane M. HIV-1 Tat assembles a multifunctional transcription elongation complex and stably associates with the 7SK snRNP. Mol Cell. 2010;38:439–451. doi: 10.1016/j.molcel.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taatjes DJ. The human Mediator complex: a versatile, genome-wide regulator of transcription. Trends Biochem Sci. 2010;35:315–322. doi: 10.1016/j.tibs.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taatjes DJ, Naar AM, Andel F, Nogales E, Tjian R. Structure, function, and activator-induced conformations of the CRSP coactivator. Science. 2002;295:1058–1062. doi: 10.1126/science.1065249. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Martin-Brown S, Washburn MP, Florens L, Conaway JW, Conaway RC. Proteomics reveals a physical and functional link between hepatocyte nuclear factor 4alpha and transcription factor IID. J Biol Chem. 2009;284:32405–32412. doi: 10.1074/jbc.M109.017954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose S, Sugimoto S, Yano K, Hartzog GA, Winston F, Buratowski S, Handa H. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 1998;12:343–356. doi: 10.1101/gad.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Balamotis MA, Stevens JL, Yamaguchi Y, Handa H, Berk AJ. Mediator requirement for both recruitment and postrecruitment steps in transcription initiation. Mol Cell. 2005;17:683–694. doi: 10.1016/j.molcel.2005.02.010. [DOI] [PubMed] [Google Scholar]
- Wei P, Garber ME, Fang S, Fischer WH, Jones KA. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell. 1998;92:451–462. doi: 10.1016/s0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, Handa H. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell. 1999;97:41–51. doi: 10.1016/s0092-8674(00)80713-8. [DOI] [PubMed] [Google Scholar]
- Yokoyama A, Lin M, Naresh A, Kitabayashi I, Cleary ML. A higher-order complex containing AF4 and ENL family proteins with P-TEFb facilitates oncogenic and physiologic MLL-dependent transcription. Cancer Cell. 2010;17:198–212. doi: 10.1016/j.ccr.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, Young RA. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet. 2007;39:1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wen Z, Washburn MP, Florens L. Refinements to label free proteome quantitation: how to deal with peptides shared by multiple proteins. Anal Chem. 2010;82:2272–2281. doi: 10.1021/ac9023999. [DOI] [PubMed] [Google Scholar]
- Zhu X, Wiren M, Sinha I, Rasmussen NN, Linder T, Holmberg S, Ekwall K, Gustafsson CM. Genome-wide occupancy profile of mediator and the Srb8-11 module reveals interactions with coding regions. Mol Cell. 2006;22:169–178. doi: 10.1016/j.molcel.2006.03.032. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01
02
03