Clarifying lysosomal storage diseases (original) (raw)
. Author manuscript; available in PMC: 2012 Aug 1.
Published in final edited form as: Trends Neurosci. 2011 Jun 30;34(8):401–410. doi: 10.1016/j.tins.2011.05.006
Abstract
Lysosomal storage diseases (LSDs) are a class of metabolic disorders caused by mutations in proteins critical for lysosomal function. Such proteins include lysosomal enzymes, lysosomal integral membrane proteins, and proteins involved in the post-translational modification and trafficking of lysosomal proteins. There are many recognized forms of LSDs, and although individually rare, their combined prevalence is estimated to be 1 in 8000 births. Over two-thirds of LSDs involve central nervous system (CNS) dysfunction—progressive cognitive and motor decline—and these symptoms are often the most debilitating. Although the genetic basis for these disorders are clear and the biochemistry of the proteins well understood, the cellular mechanisms by which deficiencies in these proteins disrupts neuronal viability remain ambiguous. In this review, we provide an overview of the widespread cellular perturbations occurring in LSDs, how they may be linked, and interventions that may specifically or globally correct those defects.
Introduction
In all cells, including neurons, the primary degradative organelle is the lysosome. Lysosomes receive their ‘substrates’ through various pathways including endocytosis, phagocytosis, and autophagy. Substrates are degraded via a finely orchestrated network of membrane bound lysosomal proteins, soluble lysosomal hydrolases, lysosomal related organelles, and other cellular constituents. When lysosomal function is impaired, neuronal dysfunction and neurodegeneration can occur. Beginning with Gaucher disease in the mid-60s, the genetic and biochemical basis for many lysosomal storage diseases (LSDs) has been unraveled, and most have central nervous system (CNS) involvement (Table 1 & Table S1 in supplementary material online).
Table 1.
Therapies approved or in development for LSDs with CNS deficits*
| Disease | Protein deficiency | Gene1 or Cell2 Therapy | ERT | Small Molecules | |
|---|---|---|---|---|---|
| Clinical | Preclinical | Clinical | Preclinical | Clinical | Preclinical |
| Non-membrane bound lysosomal hydrolases | |||||
| Fabry | α-galactosidase A | [91]1 | Approved# | Phase 3NCT 00925301 | [92] |
| Gaucher Disease types II,III | β-glucosidase | Phase 2-32NCT 00176904 | [93]1 | Approved# | Approved# |
| GM1 Gangliosidosis | β-galactosidase | Phase 32EudraCT 2009-017026-39Phase 2-32NCT 00176904 | [94]1 | Phase 2NCT 00037830 | [95] |
| GM2 Gangliosidosis type I (Tay-Sachs) | β-Hexosaminidase A | Phase 32EudraCT 2009-017026 −39Phase 2-32NCT 00176904 | [96]1 | Phase 3NCT 00672022 | [97] |
| Mucolipidosis I (sialidosis type I, II) | α-acetyl neuraminidase | [98] | |||
| Mucopolysaccharidosis I (Hurler) | α-L-iduronidase | Phase 22NCT 00176917 | [99]1 | Approved# | |
| Mucopolysaccharidosis IIIA (Sanfilippo A) | Heparan N-sulphatase | [100]1 | Phase 1-2NCT 01155778 | [101] | |
| Mucopolysaccharidosis IIIB (Sanfilippo B) | α-N-acetyl-glucosaminidase | [102]1,2 | [103] | ||
| Mucopolysaccharidosis VII (Sly) | β-glucuronidase | Phase 22NCT 01043640 | [104] | ||
| Multiple sulfatase deficiency (Austin disease) | Multiple sulfatases | ||||
| Neuronal ceroid lipofuscinosis, late Infantile (LINCL) | tripeptidyl peptidase (TPP1) | Phase 11NCT 00151216, NCT00337636 | [105]1 | [106] | |
| Integral lysosomal membrane proteins | |||||
| Mucolipidosis IV | Mucolipin 1 | ||||
| Neuronal ceroid lipofuscinosis, juvenile (JNCL) | CLN3 | [107] | |||
| Niemann-Pick C (NPC) | NPC1 or NPC2 | Phase 2-32NCT 00176904 | Phase 1NCT 00410566 | Approved# |
All LSDs are recessively inherited monogenic disorders, of which two are X-linked. Loss-of-function mutations in one of the more than 50 known soluble enzymes (Table 1), including proteases, lipases, sulfatases, or proteins important in their synthesis or trafficking, cause substrate accumulation and lysosomal storage. Although individuals with LSD can display early symptoms, many are clinically normal at birth. This suggests that lysosomal dysfunction per se does not impact significantly the complex events of early brain development—neural induction, establishment of axis, neuronal differentiation and migration, and synapse formation. Furthermore, in the case of many different LSDs, children typically meet early developmental milestones, signifying that lysosomal storage does not affect neuronal function and maturation at early developmental stages.
Defects in proteins involved in lysosome regulation or function induce the accumulation of undigested molecules that can subsequently alter many cellular processes. These include lysosomal pH regulation, synaptic release, endocytosis, vesicle maturation, autophagy, exocytosis, and Ca2+ homeostasis [1–4]. Many of these phenotypes, some of which are discussed in detail below, can be markedly improved by substrate reduction therapy (SRT), or enzyme or gene replacement (Box 1). In animal models or in patient biopsies, efficacy can be monitored by concomitant resolution of storage material [5, 6]. However, such treatments are not yet available or feasible for most LSDs. Thus, understanding how the affected cellular pathways interconnect and impact the viability of neural cells is critical for future therapeutic development. A recently described transcriptional network, which controls the expression of many proteins involved in these various pathways, provides an important clue to the extent of their inter-relatedness [7]. This important discovery gives us a fresh opportunity to revisit how mutations in a single gene required for lysosomal function may impact secondary cellular functions beyond the lysosome.
BOX 1. Therapeutic Strategies for LSDs.
In addition to the therapies discussed in the main text, other small molecule, enzyme and gene replacement strategies are under intensive investigation for LSD treatment. For ERT, the list of recombinant enzymes approved by regulatory agencies is growing (see Table 1, Table S1 in supplementary material online). For gene therapy, the strategies to deliver enzymes to the brain include directed delivery using a variety of vector systems, or correction of hematopoietic stem cells for complementation in situ from cells which enter the brain (e.g., microglia) (see Table 1, Supplement 1 and reviews [63, 69–72] for more details on these approaches).
Other approaches to LSD therapy include:
Stop codon read through
Mammalian cells utilize three mRNA stop codons to terminate protein translation (UAA, UAG, and UGA). It has been shown that although these termination signals are highly efficient, “read through” of stop codons does occur, and this phenomenon is in part influenced by bases immediately downstream of the stop codon [73]. For genetic disorders resulting from a premature stop codon, inducing read through to increase expression of functional protein with gentamycin and its derivatives have shown early promise in cell culture and animal models [74, 75]. While these reagents have not yet shown clinical efficacy for the LSDs, newer derivatives may [76].
Substrate reduction and anti-inflammatories
Several candidates have emerged and are in early clinical testing in LSD patients, or have approved for clinical use. N-butyldeoxynojirimycin (miglustat or Zavesca®) is an inhibitor of glycosphingolipid synthesis that improves neurological phenotypes in patients with neuronopathic Gaucher disease and NPC. Other work in NPC mice indicated that allopregnanolone, a neurosteroid, could improve pathology, reduce inflammatory mediators and prolong lifespan [77]. However, more recent studies have suggested that it was the hydroxypropyl-β-cyclodextrin in the formulation buffer that provided benefit [78, 79]. Cyclodextrin is already in clinical testing for NPC therapy (Table 1). Finally, given the underlying neuroinflammation present in many neurodegenerative diseases [80], methods that reduce neuroinflammation have shown therapeutic promise [81–83], including the results with allopregnanolone discussed above [78, 85]. While such an approach may not address the fundamental defect as enzyme or gene replacement does, it may be sufficient to significantly prolong CNS function.
Pharmacological chaperones
In some instances, a mutation in a particular lysosomal enzyme does not directly affect the functionality of the enzyme per se, but rather it leads to a misfolding of the enzyme in the ER following translation. The failure of the mutant enzyme to adopt a functional conformation, for example in cases of Fabry, Gaucher and Tay-Sachs disease, causes degradation via ER-associated protein degradation (ERAD). Disease in these cases is due to mutant enzymes not trafficking correctly to the lysosome. Chemical chaperones are synthetic molecules that promote correct protein folding in the ER. They are commonly active-site inhibitors and presumably provide a structure around which a nascent enzyme can fold. Once the enzyme folds correctly and is trafficked to the lysosome, it is fully functional. Based on earlier work [84, 85], several early phase clinical trials are currently underway testing the effects of chemical chaperones in Fabry, GM1 gangliosidoses [86] and Gaucher diseases. Notably, many chaperones, since they are small molecules, can cross the BBB. Another recent finding is that chaperones can stabilize wildtype enzymes used in ERT [70, 87]. This holds potential for lowering overall doses required for therapeutic benefit, and could substantially reduce costs associated with this therapy.
Traffic jams and lysosomal storage
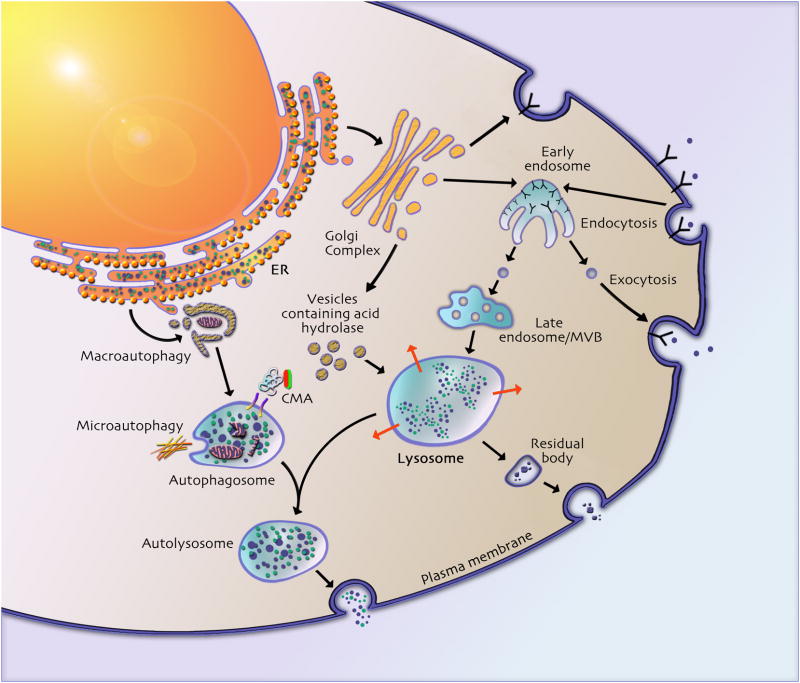
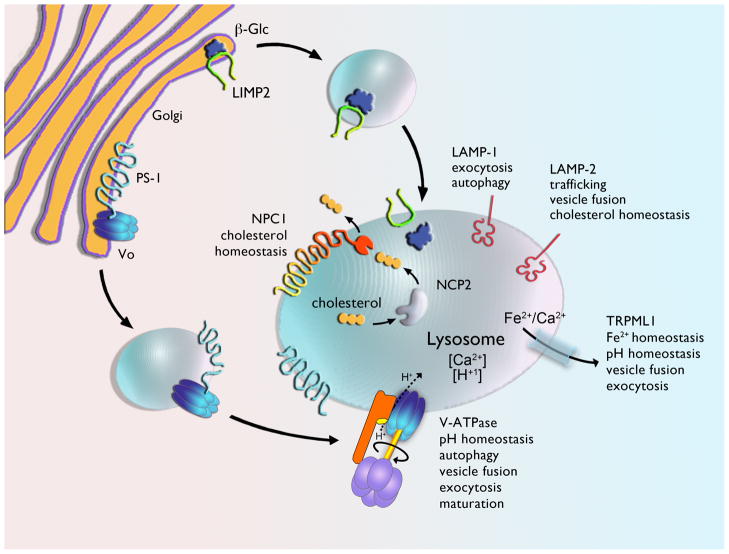
Lysosomal proteins traffic to the lysosome via distinct pathways, as described elegantly in recent reviews [8, 9] and highlighted in Figure 1. This schematic exemplifies the interplay between the endosome-lysosome system, and illustrates how various autophagic pathways are reliant on a functional lysosome for proper processing. Moreover, it places into context how roadblocks at specific steps may broadly impact cell viability. Recent studies illustrate how deletion of cargo carriers, rather than the cargo itself, has the potential for causing cellular pathologies. For example, the lysosomal integral membrane protein type 2 (LIMP-2) binds β-glucocerebrosidase (deficient in Gaucher disease) in the endoplasmic reticulum (ER) and facilitates the transport of β-glucocerebrosidase (β-Glc) to the lysosome (Figure 2) [10]. A consequence of LIMP-2 mutations is reduced β-glucocerebrosidase activity [11]. Also illustrative is the recent finding that the trafficking of the Vo subunit of vacuolar-type H+ ATPase (V-ATPase) to the lysosome requires presenilin-1 (PS1; Figure 2); furthermore, it was demonstrated that Alzheimer‘s disease (AD)-related mutations in PS1 disrupt this trafficking [12]. Without a functional V-ATPase, lysosomal pH is aberrant and autophagy is impaired. Irregular lysosomal pH and impaired autophagy may underlie amyloid-β (Aβ) accumulation in familial forms of early onset AD which involve mutations in PS1.
Figure 1.
Lysosomal proteins reach the lysosome directly, via the trans-Golgi network (TGN), or indirectly by way of the plasma membrane, as reviewed elsewhere [9]. In the direct pathway, upon trafficking to the TGN, some oligosaccharide side chains acquire the mannose-6-phosphate (M6P) recognition marker. Enzymes with this modification are recognized by cation-dependent and cation-independent mannose-6-phosphate receptors (M6PRs) in the TGN, mediating their sorting into the endosome-lysosome pathway [89, 90].
M6P-tagged molecules that escape M6PR recognition in the TGN are transported to the cell surface and secreted into the extracellular fluid. Secreted M6P labeled proteins are recognized by M6PR on the extracellular plasma membrane and delivered to the lysosome by receptor mediated endocytosis. This is the general mechanism underlying ERT for the soluble lysosomal hydrolase deficiencies found in several diseases (Table 1). In both direct and indirect pathways, acidification in early endosomes or lysosomes releases M6PR-hydrolase binding and the receptor is recycled back to the TGN for additional retrieval. Uptake of lysosomal hydrolases can also occur via the mannose receptor, as in the case of β-glucosidase replacement therapy for Gaucher disease.
Figure 2.
General organization and function of various integral membrane proteins important for trafficking of lysosomal enzymes (e.g., LIMP2 and PS1), lysosomal function (e.g., V-ATPase, TRPML1 and LAMP-1), cholesterol homeostasis (e.g., NPC1 NPC2, and LAMP2). Malfunction of these proteins have been implicated in multiple LSDs (Table 1). It is currently unknown if PS1 remains associated with Vo as it traffics to the lysosome, but this seems a likely possibility, given its presence in autophagic vesicles.
Historically, the structure and content of storage material was used for diagnosis and to group LSDs into subclasses. For many lysosomal hydrolases, microscopic evaluation of biopsied tissues and biochemical evaluation of urine provided evidence of accumulated cellular substrates and clues to the underlying enzymatic defects [13]. As a group, storage structure and content is diverse, as would be expected from the various functions of the proteins deficient in the LSDs. Once the role for these proteins is defined, the nature of the accumulated storage often makes sense, but not always. For example, deficiency in the proteins Niemann-Pick disease type C1 and C2 (NPC1, NPC2) (Figure 2), which are required for cholesterol egress from the lysosome, cause cholesterol accumulation in NPC disease (Table 1). In Mucopolysaccharidoses (MPS) diseases, proteoglycans accumulate as they are substrates for the defective enzymes (Table 1 and Table S1 in supplementary material online). However, in the Neuronal Ceroid Lipofuscinoses (NCLs), the cause of storage accumulation remains puzzling. In the case of the late infantile form of Batten disease (LINCL), its underlying cause is mutations in tripeptidyl peptidase I. This soluble lysosomal protease has broad substrate specificity, yet the mitochondrial membrane protein ATP synthase subunit C is the major storage constituent [14].
While their genesis may not always be clear, we can take advantage of storage burden as a biomarker for therapeutic efficacy in emerging treatment strategies. Resolution of storage detected in tissue sections, or diminution of excessive substrates in body fluids can be a reliable indicator of treatment efficacy for some LSDs. In Fabry disease, glycosphingolipid levels are elevated in plasma and urine, most notably in hemizygous affected males [5]. Enzyme replacement therapy (ERT) can dramatically lower these levels and reduce storage burden in tissues [5]. Likewise in MPS, elevated glycosaminoglycans (GAGs) in urine can be used to assess therapeutic efficacy [13].
The functions of some proteins involved in LSDs remain elusive, but perhaps storage constituents can predict how a protein contributes to normal endosomal-lysosomal function. Ceroid lipofuscinosis neuronal type 3 (CLN3), the protein defective in the common form of Juvenile Ceroid Lipofuscinosis (JNCL) is a case in point (Table 1). The exact molecular function of CLN3 has remained obscure since its discovery in 1995. JNCL storage material resembles finger prints under electron microscopy [15]. Placing the molecular function of CLN3 upstream of the lysosome is thus a reasonable assumption, since multilamellar structures could arise from defects occurring at the late endosome or multivesicular body (MVB) stage, before maturation into a lysosome (Figure 1). Intraluminal vesicles in MVBs are formed by internalization at the limiting membrane, and in the case of defects in CLN3, continued vesicle addition may cause the resultant multilamellar fingerprint.
Notably, not all storage is lysosomal. For example in cultured cortical neurons from α-N-acetyl-glucosaminidase deficient mice, which are a model of MPS IIIB, a disrupted Golgi structure is associated with the storage [16]. This material presumably arises from defective vesicle transfer from the Golgi to the lysosome. The finding that this storage co-localizes with the lysosomal associated membrane protein 1 (LAMP-1), a transmembrane protein that is commonly used as a marker to label the lysosome (Figure 2), suggests secondary effects on LAMP-1 trafficking. If intracellular movement of proteins and even organelles is impaired in LSDs, markers used commonly to identify cellular vesicles may be misleading. However for the wary eye, this could give important clues as to the pathways upstream and downstream affected by mutations in a single lysosomal protein.
The importance of vesicular pH
Considering the role of PS1 in V-ATPase trafficking (Figure 2) [12], it is interesting to speculate that the altered lysosomal pH in some LSDs [17, 18] may reflect global defects in trafficking. A recent genetic screen in yeast has demonstrated that mutations in proteins involved in MVB trafficking and vacuole fusion perturb vacuolar pH [19]. Surprisingly, this study showed that mutations in multiple proteins involved in trafficking to the lysosome resulted in decreased vacuolar pH. These changes could be due to enhanced activity of the V-ATPase. The V-ATPase is a holoenzyme consisting of a membrane bound Vo and cytosolic V1 components, and both V1 and Vo are composed of multiple subunits. The Vo subunit a1 is highly enriched, or even specific to neurons, in different animal models [20] and is required for acidification of degradative competent lysosomes [21]. Preservation of low pH is important for cargo release, lysosomal hydrolase maturation, vesicle maturation, autophagy, and neurotransmitter loading into synaptic vesicles [22–24]. Additionally, the pH gradient within the endomembrane system is required for intra-cellular trafficking [25] and its loss can impair mannose-6-phosphate receptor (M6PR) recycling back to the Golgi for reloading of newly synthesized lysosomal hydrolases.
In motor neurons, protons are pumped into synaptic vesicles by V-ATPase upon prolonged stimulation [26]. After fusion of the synaptic vesicle with the plasma membrane, V-ATPase remains resident in the plasma membrane and continues to extrude cytosolic protons to the extracellular space. The subsequent increase in cytosolic pH induces endocytosis and re-internalization of the V-ATPase, allowing neurotransmitter loading [26]. Based on the fundamental roles of V-ATPase in neurons it is not surprising that deletion of V100, the Drosophila homologue of the a1 subunit, causes synaptic vesicle buildup, prevents neurotransmitter release, and induces cell death [27] (See Box 2).
Box 2. Outstanding Questions.
- Is the storage pathogenic in LSDs? In some disorders, for example Huntington‘s disease, the aggregates within neurons are generally considered neuroprotective rather than pathogenic. It is the precursor material leading up to the aggregate that likely elicits neuronal death [88].
- Why are neurons particularly susceptible to dysfunction/neurodegeneration in many LSDs relative to cells from peripheral tissues?
- pH levels inside intracellular vesicles are known to be altered in some cell models of some LSDs [17, 18], but this has not been systematically evaluated for all LSDs. Does the pH of vesicles comprise the endolysosome system more globally and could this impair lysosomal function, autophagic flux, and exocytosis?
- Much of the data assessing the cellular effects of the noted deficiencies are from non-neural cells, but many of these diseases exhibit CNS defects predominantly. Thus, an important question to ask is how predictive are these findings? Will novel roles for brain-specific isoforms of proteins affected secondarily by these deficiencies yield new insights into disease pathogenesis?
- If storage is pathogenic, would treatments which enhance exocytosis be applicable to multiple LSDs?
- Would upregulation of CLEAR-responsive genes be sufficient for treating LSDs? How is this pathway activated and which signaling molecules are involved?
Mitochondrial ATP synthase is structurally and functionally similar to the V-ATPase. Recent data suggest that ATP synthase traffics to the plasma membrane from the Golgi under conditions of cell stress, and removes cytosolic protons [28, 29]. Given these findings, it is interesting to speculate that ATP synthase may aberrantly traffic to the plasma membrane in the setting of LSD. Mitochondrial ATP synthase subunit c is a component of the storage in several LSD’s, including LINCL and JNCL. In light of the possible role for ATP synthase in cytosolic pH regulation [28, 29] and data showing that defects in anterograde trafficking decreases lysosomal pH [19], pH disturbances and altered ATP synthase trafficking may be linked. Observations in yeast cells deficient in Btn1p, the ortholog to the JNCL-mutated protein CLN3, support this concept. Such mutants were observed to have decreased vacuolar pH, and the cells responded by enhancing proton removal from the cytosol to regulate this pH imbalance [30]. Without normalization of cytoplasmic pH, autophagy can be impaired [31].
Autophagy
Autophagy is a ubiquitous cellular process by which cytoplasmic proteins, and whole organelles such as mitochondria, are sequestered for lysosomal degradation [32, 33] (Figure 1). Autophagy can be divided into macroautophagy (organelle or cytoplasm enclosed in a double membrane), microautophagy, (cytoplasmic degradation following engulfment by the lysosome), and chaperone-mediated autophagy (CMA) (targeting of proteins to the lysosome via chaperones). Here, we focus on macroautophagy as CMA has been reviewed recently [34], and there is no evidence as yet for impaired microautophagy in LSDs. However, as microautophagy is responsive to comparable cellular signals and requires functioning lysosomes similar to macroautophagy, evidence linking microautophagy and LSDs is likely to emerge.
Macroautophagy is induced by cellular stresses and begins with the de novo formation of an autophagic membrane that expands and eventually sequesters a volume of cytoplasm or cellular organelle, forming an autophagosome. Once formed, fusion with the lysosome allows hydrolase degradation of autophagosomal contents (Figure 1). Reports demonstrating that genetic disruption of autophagy causes neurodegeneration in mice [35, 36] led to the hypothesis that the neurodegeneration in LSDs may be a consequence of impaired autophagy. Indeed, impaired autophagy has been reported in several models of LSDs including Pompe disease, NPC, the NCLs, multiple sulphatase deficiency (MSD), mucolipidosis type IV (MLIV), and GM1-gangliosidosis [37–42].
Work in HeLa cells has functionally linked cytosolic pH, lysosomal positioning, mammalian target of rapamycin complex 1(mTORC1) signaling, and autophagy [31]. Under nutrient rich conditions lysosomes are often near the plasma membrane, where associated mTORC1 is activated by proximal upstream signaling molecules at the plasma membrane. In this setting, autophagy is repressed. Under nutrient deprivation, elevated intracellular pH promotes perinuclear lysosomal repositioning, represses mTORC1 activity and stimulates autophagy. Perinuclear lysosomes are optimally positioned for fusion with incoming autophagosomes. Interestingly, promoting lysosomal trafficking to the perinuclear space conveyed a protective effect against mutant huntingtin and α-synuclein [31], two aggregation-prone proteins which are associated with Huntington‘s disease and Parkinson‘s Disease, respectively. Whether altered cytoplasmic pH underlies the noted impairments of autophagy in LSDs [37, 43] requires further investigation.
Defects in autophagy can affect the efficacy of ERT for LSD therapy. In a mouse model of Pompe disease, a non-neuronal LSD that causes fatal cardiomyopathy and skeletal muscle myopathy, administered recombinant enzymes accumulated in autophagic compartments, rather than in the lysosome [44]. Inactivation of ATG7, a vital component of the autophagosome, improved enzyme trafficking and normalized glycogen storage [44]. This strategy may similarly improve the bioavailability of enzymes for treatment of the neuronopathic LSDs [45].
Augmenting autophagy is also being tested as a therapy (Table 1). All-trans retinoic acid increases cation-independent M6PR trafficking from the Golgi to autophagosomes, thereby enhancing autophagosome maturation [46], however, such a therapy would have many non-specific targets. Lithium is another potential approach; addition of this drug to cerebellar cells cultured from CLN3 deficient mice was found to improve storage phenotypes and reduce neuronal cell death [46]. Lithium has also been tested as a potential therapy for other non-LSD neurodegenerative diseases. For example, lithium treatment of spinocerebellar ataxia type 1 (SCA1) mice, which harbor a polyglutamine repeat in ataxin 1 (ATXN1), was found to improved neurological function and hippocampal dendritic branching [47]. These effects were reported to be due to altered gene expression, however, the effects of lithium on autophagy were not tested in this study. Although results with lithium appear encouraging, it has many non-specific targets and is likely to have unwanted side-effects [48]. Finally, an important limitation with respect to LSDs that are caused by a primary lysosomal enzyme defect is that they may not be good candidates for strategies enhancing autophagy. While short-term improvements may occur, substrate accumulation is likely to ultimately continue in these diseases.
Exocytosis
Lysosomal function is intimately linked to exocytosis, and multiple LSDs such as MLIV, NPC, and Sialidosis have been shown to have impaired exocytosis [49–51]. Exocytosis is the removal of cellular cargo by fusion of vesicles with the plasma membrane (Figure 1). Synaptic vesicles and lysosomes both have exocytic activity. Activated exocytosis (eg. neurotransmitter release) and constitutive exocytosis (eg. of lysosomal contents) both result in secretion (eg. of signaling molecules such as neurotransmitters, or toxic metabolites) and plasma membrane expansion.
Recent data show that lysosomes and synaptic vesicles originate from a common early endosome, and preventing the maturation of either vesicle induces mistrafficking of their respective proteins to the opposite organelle [52]. Mechanisms for exocytic activation are also similar between the two types of vesicles. During synaptic vesicle release, neuronal action potentials reach the presynaptic terminal and activate voltage regulated Ca2+ channels causing an influx of Ca2+ into the presynaptic cytoplasm, vesicle fusion with the plasma membrane, and neurotransmitter release [53]. Synaptic vesicles are decorated with the Ca2+ sensor synaptotagmin and the Soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) synaptobrevin-2, which bind to the corresponding plasma membrane SNAREs syntaxin-1A and synaptosomal-associated protein 25 (SNAP-25) [54]. Lysosomal exocytosis utilizes Ca2+ influx into the cell, which drives interaction between the vesicular and target SNAREs synaptotagmin 7 (SYT7) and syntaxin 4 [55]. In cultures of embryonic fibroblast cells from mouse models of MSD and MPS IIIA, cholesterol accumulation was shown to result in SNARE mislocalization, preventing lysosome fusion and vesicle cycling. Reducing cholesterol levels by SRT with methyl-β-cyclodextran restored SNARE localization and lysosome function [56]. Cholesterol storage can also inhibit Rab GTPases, which promote membrane recycling; overexpressing Rab4 in cultured fibroblast cells derived from NPC1 deficient mice led to an activation of exocytosis and reduced lysosomal accumulation [57]. Thus SRT can not only reduce the storage burden, but also improve secondary phenotypes.
Advances in understanding exocytosis have emerged from defining the function of proteins deficient in lysosomal storage diseases. Recent data show that the ultimate fusogenic Ca2+ signal for lysosomes does not originate from voltage dependent Ca2+ channels, but rather the protein lysosomal transient receptor potential mucolipin-1 (TRPML1) [58]. Mutations in TRPML1 cause MLIV (Table 1). In the presence of the Ca2+ ionophore ionomycin, TRPML1 activates exocytosis [59]. When key residues of TRPML1 were mutated to create a constitutively active channel, enhanced endosomal and lysosomal Ca2+ mobilization induced LAMP-1 relocalization to the plasma membrane [60]. Whether LAMP-1 itself is important for exocytosis, or its presence on the plasma membrane is merely a result of lysosomal membrane fusion, is becoming clearer. In Sialidosis, which is caused by a deficiency in α-acetyl-neuraminidase, terminal sialic acid residues of LAMP-1 are not removed and its half-life at the plasma membrane is elevated [51]. The elevated exocytosis in these cells can be blocked by LAMP-1 knock-down [51].
Enhancing exocytosis could reduce storage in the short term, but a major concern with these approaches is that the products may not be adequately cleared in the brain, since even scavenger cells would harbor the enzymatic defect. Alternatively, autophagolysosome contents could be exocytosed and transported to the cerebrospinal fluid (CSF) and plasma. It would be interesting to test if the CSF from LSD patients contains elevated levels of accumulated storage material, or if enhancing exocytosis would cause accumulation in the neuropil and/or the CSF. LSD patients with CNS deficits do secrete storage products in urine [13, 14], but whether this occurs via exocytosis of autophagosome constituents from peripheral tissues or the brain is currently unknown.
The specialized vasculature system within the CNS may hinder extracellular clearance of exocytosed materials, and thus, contribute to cell death in this way. The familiar view of the blood brain barrier (BBB) is as a physical barrier that comprises the neurovascular system, and which blocks foreign particle entry. The BBB poses a major hurdle for drug delivery to brain, and it could also be a limiting force in substance removal after lysosomal exocytosis. An alternative, less neurocentric view to LSDs, is that defects within the neurovascular system may precede neuronal dysfunction [61]. Certainly, there is vascular remodeling within the CNS of LSD animal models [62] and obvious morphological changes in all cells of the neurovascular unit. Thus, drugs or other therapies capable of restoring BBB function may alleviate or slow disease progression, and as a bonus, would not be required to cross it.
A CLEARly integrated network
The pathways outlined in Figure 1 are interdependent, supporting a coordinated network. A commonly reported observation in LSDs is that decreases in the activity of a disease-associated enzyme is concomitant with increases in other lysosomal enzymes, suggesting that gene expression required for their interdependence is similarly orchestrated. In a recent study, this theory was bioinformatically and experimentally validated [7]. Motif sequence analysis identified a 10 base pair sequence near the transcription start site of many known lysosomal genes and some genes of unknown function. The motif, named the coordinated lysosomal expression and regulation (CLEAR) element was found in, among other genes, the lysosomal marker genes LAMP-1 and CLN3, CLN5, NPC1, NPC2, and β-galactosidase. The transcription factor EB (TFEB) was found to enter the nucleus and bind the CLEAR element. Furthermore, TFEB localized primarily in the nucleus of cultured cell models of MPS II, MPS IIIA, and MSD, but was cytoplasmic in control cells, suggesting that this pathway is activated under lysosomal storage conditions [7].
Over-expression of TFEB in HeLa cells was shown to increase the expression of 291 genes [7], including genes in the autophagosome pathway, lysosomal hydrolases, lysosomal biogenesis genes, and genes encoding V-ATPase and TRPML1. Enhanced storage clearance was observed as a consequence of TFEB overexpression [7]. Although not yet tested in vivo, the CLEAR network appears to provide cells with a compensatory response mechanism to pathogenic protein accumulation and lysosomal dysfunction.
Discovery of the CLEAR element brings the field of LSDs together and provides a unified regulatory pathway of lysosomal stress. As discussed above, many LSDs have common defects in autophagy, calcium regulation, pH regulation, and exocytosis. Overexpression of TFEB results in up-regulation of proteins involved in all of these processes. The possibility exists that utilizing the CLEAR network, or some of its key proteins, may restore cellular defects in many of the LSDs. Moreover, the ability to amplify lysosome biogenesis via the CLEAR network provides a method for augmenting the effects of enzyme replacement or gene therapy [63, 64]. Additionally, the therapeutic utility of the CLEAR network may extend beyond LSDs. Chinese hamster ovary cells rendered NPC1 null display increased β-amyloid accumulation, and in other cells, addition of spingolipids impairs amyloid precursor protein processing [65, 66]. Thus, restoration of cholesterol levels by CLEAR network activation could be therapeutic not only for LSDs, but also for AD and other neurodegenerative diseases where these fundamental cellular processes are impaired.
Finally, it is important to mention that not all lysosomal and non-lysosomal genes involved in LSDs are regulated by CLEAR elements [7]. It is possible that other lysosomal gene regulatory pathways exist that have yet to be identified. In fact, the exact lysosomal stress pathways responsible for TFEB activation, which induces its nuclear translocation, are unknown. Elucidating the events upstream of TFEB activation may lead to a more coherent understanding of how cells first sense and respond to lysosomal stress, providing opportunities for therapeutic intervention beyond those in current use (Box 1).
Summary
Strides in cell biology over the last 50 years have been critical in expanding our view of the lysosome from a degradative organelle to one intimately linked to multiple cellular processes. Genetic mutations in specific proteins, particularly those involved in cellular trafficking pathways, have been associated with many LSDs. Yet how these deficiencies induce cellular dysfunction, and in many cases, CNS deficits, remains unsettled (Box 2). What the field requires are methods to assess vesicle, protein and lipid trafficking in situ to dissect the interplay between neurons, glia and brain vasculature – all harboring deficiencies of the same protein but possibly reacting in different manners to those disruptions. Even among neurons there are differences in their responses to storage or defects in endosomal and autophagic pathways. Myelinated, large sensory neurons accumulate large vacuoles and ubiquitinated aggregates, promoting degeneration, while smaller unmyelinated sensory neurons induce lysosome biogenesis [67]. This enhanced lysosomal biogenesis may underlie the relatively slower degeneration of small sensory neurons in the setting of LSD, and may reflect an in vivo example of CLEAR network activation [7, 67]. Moreover, neuronal classes extend well beyond differences in size [68], and it will be important to understand the response of each cell type to the specific LSD under study.
We envision that advances in imaging methods merged with new animal models will elucidate the temporal progression of disease in specific brain regions and cell types. In addition, identifying sites of initial vulnerability will aide in the development of directed therapies where focal small molecule or gene replacement holds the most promise.
Supplementary Material
01
Acknowledgments
Preparation of this review was supported by grants from the National Institutes of Health and the Roy J Carver Trust. Figures were illustrated by Mary Moye-Rowley.
Glossary
Blood brain barrier (BBB)
The blood brain barrier limits access of proteins, small molecules, cells and pathogens do the brain. The primary barrier is formed by the endothelial cells lining brain vessels, where tight junctions prohibit passage. Only selected molecules are transported across the BBB
Enzyme Replacement Therapy (ERT)
ERT takes advantage of plasma membrane-resident M6PRs or mannose receptors for the uptake of enzyme into cells after exogenous delivery intravenously. This enzyme replacement therapy has improved the lives of many individuals with genetic deficiency of one of many soluble hydrolases, albeit restricted to the treatment of peripheral symptoms so far
ERAD
A cellular process where misfolded proteins in the ER are tagged with ubiquitin molecules, exported from the ER, and degraded in the proteasome
Gene Therapy
Addition of exogenous or removal of endogenous genes utilizing viral or non-viral methods. For the LSDs, gene addition directly to affected tissues, or into hematopoietic stem cells are in early clinical testing (Table 1)
Lysosome
First described by Christina de Duve in 1955 as a particle containing enzymes with lytic properties [108]. It is membrane enclosed compartment containing hydrolytic enzymes that digest macromolecules
Lysosomal Storage
The accumulation of material into large cytoplasmic vesicles which are the result of insufficient lysosomal degradation
Sphingolipids
In contrast to glycerolphospholipids which contain a glycerol backbone, sphingolipids contain a sphingosine backbone which is found in sphingomyelin, gangliosides, and sulfatides
Substrate Reduction Therapy (SRT)
SRT utilizes inhibitors of substrate synthesis to reduce substrate levels and hence storage burden. These inhibitors are commonly small molecules, many of which can cross the BBB
Trans-Golgi Network (TGN)
Part of the Golgi complex required for sorting and transport of proteins to the appropriate site
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Vitner EB, et al. Common and uncommon pathogenic cascades in lysosomal storage diseases. J Biol Chem. 2010;285:20423–20427. doi: 10.1074/jbc.R110.134452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bellettato CM, Scarpa M. Pathophysiology of neuropathic lysosomal storage disorders. J Inherit Metab Dis. 2010;33:347–362. doi: 10.1007/s10545-010-9075-9. [DOI] [PubMed] [Google Scholar]
- 3.Ballabio A, Gieselmann V. Lysosomal disorders: from storage to cellular damage. Biochimica et biophysica acta. 2009;1793:684–696. doi: 10.1016/j.bbamcr.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 4.Bezprozvanny I. Calcium signaling and neurodegenerative diseases. Trends Mol Med. 2009;15:89–100. doi: 10.1016/j.molmed.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eng CM, et al. A phase 1/2 clinical trial of enzyme replacement in fabry disease: pharmacokinetic, substrate clearance, and safety studies. American journal of human genetics. 2001;68:711–722. doi: 10.1086/318809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu G, et al. Functional correction of CNS phenotypes in a lysosomal storage disease model using adeno-associated virus type 4 vectors. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2005;25:9321–9327. doi: 10.1523/JNEUROSCI.2936-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sardiello M, et al. A gene network regulating lysosomal biogenesis and function. Science. 2009;325:473–477. doi: 10.1126/science.1174447. [DOI] [PubMed] [Google Scholar]
- 8.Saftig P, et al. Lysosomal membrane proteins: life between acid and neutral conditions. Biochem Soc Trans. 2010;38:1420–1423. doi: 10.1042/BST0381420. [DOI] [PubMed] [Google Scholar]
- 9.Saftig P, Klumperman J. Lysosome biogenesis and lysosomal membrane proteins: trafficking meets function. Nat Rev Mol Cell Biol. 2009;10:623–635. doi: 10.1038/nrm2745. [DOI] [PubMed] [Google Scholar]
- 10.Reczek D, et al. LIMP-2 is a receptor for lysosomal mannose-6-phosphate-independent targeting of beta-glucocerebrosidase. Cell. 2007;131:770–783. doi: 10.1016/j.cell.2007.10.018. [DOI] [PubMed] [Google Scholar]
- 11.Blanz J, et al. Disease-causing mutations within the lysosomal integral membrane protein type 2 (LIMP-2) reveal the nature of binding to its ligand beta-glucocerebrosidase. Hum Mol Genet. 2010;19:563–572. doi: 10.1093/hmg/ddp523. [DOI] [PubMed] [Google Scholar]
- 12.Lee JH, et al. Lysosomal proteolysis and autophagy require presenilin 1 and are disrupted by Alzheimer-related PS1 mutations. Cell. 2010;141:1146–1158. doi: 10.1016/j.cell.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cox TM. Biomarkers in lysosomal storage diseases. In: Mehta A, et al., editors. Fabry Disease: Perspectives from 5 Years of FOS. Oxford: Oxford PharmaGenesis; 2006. [PubMed] [Google Scholar]
- 14.Wisniewski KE, et al. Increased urine concentration of subunit c of mitochondrial ATP synthase in neuronal ceroid lipofuscinoses patients. J Inherit Metab Dis. 1994;17:205–210. doi: 10.1007/BF00711619. [DOI] [PubMed] [Google Scholar]
- 15.Goebel HH. The neuronal ceroid-lipofuscinoses. Seminars in pediatric neurology. 1996;3:270–278. doi: 10.1016/s1071-9091(96)80031-3. [DOI] [PubMed] [Google Scholar]
- 16.Vitry S, et al. Storage vesicles in neurons are related to Golgi complex alterations in mucopolysaccharidosis IIIB. Am J Pathol. 2010;177:2984–2999. doi: 10.2353/ajpath.2010.100447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raychowdhury MK, et al. Molecular pathophysiology of mucolipidosis type IV: pH dysregulation of the mucolipin-1 cation channel. Human molecular genetics. 2004;13:617–627. doi: 10.1093/hmg/ddh067. [DOI] [PubMed] [Google Scholar]
- 18.Pearce DA, et al. Action of BTN1, the yeast orthologue of the gene mutated in Batten disease. Nature genetics. 1999;22:55–58. doi: 10.1038/8861. [DOI] [PubMed] [Google Scholar]
- 19.Brett CL, et al. Genome-wide analysis reveals the vacuolar pH-stat of Saccharomyces cerevisiae. PLoS One. 2011;6:e17619. doi: 10.1371/journal.pone.0017619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang W, et al. V-ATPase V0 sector subunit a1 in neurons is a target of calmodulin. J Biol Chem. 2008;283:294–300. doi: 10.1074/jbc.M708058200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williamson WR, et al. A dual function of V0-ATPase a1 provides an endolysosomal degradation mechanism in Drosophila melanogaster photoreceptors. J Cell Biol. 2010;189:885–899. doi: 10.1083/jcb.201003062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marshansky V, Futai M. The V-type H+-ATPase in vesicular trafficking: targeting, regulation and function. Curr Opin Cell Biol. 2008;20:415–426. doi: 10.1016/j.ceb.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weimer RM, Jorgensen EM. Controversies in synaptic vesicle exocytosis. J Cell Sci. 2003;116:3661–3666. doi: 10.1242/jcs.00687. [DOI] [PubMed] [Google Scholar]
- 24.Mijaljica D, et al. V-ATPase engagement in autophagic processes. Autophagy. 2011:7. doi: 10.4161/auto.7.6.15812. [DOI] [PubMed] [Google Scholar]
- 25.Sobota JA, et al. Inhibitors of the V0 subunit of the vacuolar H+-ATPase prevent segregation of lysosomal- and secretory-pathway proteins. J Cell Sci. 2009;122:3542–3553. doi: 10.1242/jcs.034298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Z, et al. Vesicular ATPase inserted into the plasma membrane of motor terminals by exocytosis alkalinizes cytosolic pH and facilitates endocytosis. Neuron. 2010;68:1097–1108. doi: 10.1016/j.neuron.2010.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hiesinger PR, et al. The v-ATPase V0 subunit a1 is required for a late step in synaptic vesicle exocytosis in Drosophila. Cell. 2005;121:607–620. doi: 10.1016/j.cell.2005.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmidt C, et al. Amyloid precursor protein and amyloid beta-peptide bind to ATP synthase and regulate its activity at the surface of neural cells. Mol Psychiatry. 2008;13:953–969. doi: 10.1038/sj.mp.4002077. [DOI] [PubMed] [Google Scholar]
- 29.Xing SL, et al. Neuronal cell surface ATP synthase mediates synthesis of extracellular ATP and regulation of intracellular pH. Cell Biol Int. 2011;35:81–86. doi: 10.1042/CBI20090441. [DOI] [PubMed] [Google Scholar]
- 30.Padilla-Lopez S, Pearce DA. Saccharomyces cerevisiae lacking Btn1p modulate vacuolar ATPase activity to regulate pH imbalance in the vacuole. J Biol Chem. 2006;281:10273–10280. doi: 10.1074/jbc.M510625200. [DOI] [PubMed] [Google Scholar]
- 31.Korolchuk VI, et al. Lysosomal positioning coordinates cellular nutrient responses. Nature cell biology. 2011;13:453–460. doi: 10.1038/ncb2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yue Z, et al. The cellular pathways of neuronal autophagy and their implication in neurodegenerative diseases. Biochim Biophys Acta. 2009;1793:1496–1507. doi: 10.1016/j.bbamcr.2009.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bejarano E, Cuervo AM. Chaperone-mediated autophagy. Proc Am Thorac Soc. 2010;7:29–39. doi: 10.1513/pats.200909-102JS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hara T, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 36.Komatsu M, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 37.Cao Y, et al. Autophagy is disrupted in a knock-in mouse model of juvenile neuronal ceroid lipofuscinosis. J Biol Chem. 2006;281:20483–20493. doi: 10.1074/jbc.M602180200. [DOI] [PubMed] [Google Scholar]
- 38.Fukuda T, et al. Autophagy and lysosomes in Pompe disease. Autophagy. 2006;2:318–320. doi: 10.4161/auto.2984. [DOI] [PubMed] [Google Scholar]
- 39.Jennings JJ, Jr, et al. Mitochondrial aberrations in mucolipidosis Type IV. J Biol Chem. 2006;281:39041–39050. doi: 10.1074/jbc.M607982200. [DOI] [PubMed] [Google Scholar]
- 40.Pacheco CD, et al. Autophagy in Niemann-Pick C disease is dependent upon Beclin-1 and responsive to lipid trafficking defects. Hum Mol Genet. 2007;16:1495–1503. doi: 10.1093/hmg/ddm100. [DOI] [PubMed] [Google Scholar]
- 41.Settembre C, et al. A block of autophagy in lysosomal storage disorders. Hum Mol Genet. 2008;17:119–129. doi: 10.1093/hmg/ddm289. [DOI] [PubMed] [Google Scholar]
- 42.Venkatachalam K, et al. Motor deficit in a Drosophila model of mucolipidosis type IV due to defective clearance of apoptotic cells. Cell. 2008;135:838–851. doi: 10.1016/j.cell.2008.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Curcio-Morelli C, et al. Macroautophagy is defective in mucolipin-1-deficient mouse neurons. Neurobiol Dis. 2010;40:370–377. doi: 10.1016/j.nbd.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fukuda T, et al. Autophagy and mistargeting of therapeutic enzyme in skeletal muscle in Pompe disease. Molecular therapy : the journal of the American Society of Gene Therapy. 2006;14:831–839. doi: 10.1016/j.ymthe.2006.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Raben N, et al. Suppression of autophagy permits successful enzyme replacement therapy in a lysosomal storage disorder--murine Pompe disease. Autophagy. 2010;6:1078–1089. doi: 10.4161/auto.6.8.13378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang JW, et al. Lithium rescues the impaired autophagy process in CbCln3(Deltaex7/8/Deltaex7/8) cerebellar cells and reduces neuronal vulnerability to cell death via IMPase inhibition. Journal of neurochemistry. 2011;116:659–668. doi: 10.1111/j.1471-4159.2010.07158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Watase K, et al. Lithium therapy improves neurological function and hippocampal dendritic arborization in a spinocerebellar ataxia type 1 mouse model. PLoS medicine. 2007;4:e182. doi: 10.1371/journal.pmed.0040182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pasquali L, et al. Intracellular pathways underlying the effects of lithium. Behav Pharmacol. 2010;21:473–492. doi: 10.1097/FBP.0b013e32833da5da. [DOI] [PubMed] [Google Scholar]
- 49.LaPlante JM, et al. Lysosomal exocytosis is impaired in mucolipidosis type IV. Mol Genet Metab. 2006;89:339–348. doi: 10.1016/j.ymgme.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 50.Xu S, et al. Defects of synaptic vesicle turnover at excitatory and inhibitory synapses in Niemann-Pick C1-deficient neurons. Neuroscience. 2010;167:608–620. doi: 10.1016/j.neuroscience.2010.02.033. [DOI] [PubMed] [Google Scholar]
- 51.Yogalingam G, et al. Neuraminidase 1 is a negative regulator of lysosomal exocytosis. Dev Cell. 2008;15:74–86. doi: 10.1016/j.devcel.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Newell-Litwa K, et al. Roles of BLOC-1 and adaptor protein-3 complexes in cargo sorting to synaptic vesicles. Mol Biol Cell. 2009;20:1441–1453. doi: 10.1091/mbc.E08-05-0456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nizami S, et al. Presynaptic roles of intracellular Ca(2+) stores in signalling and exocytosis. Biochem Soc Trans. 2010;38:529–535. doi: 10.1042/BST0380529. [DOI] [PubMed] [Google Scholar]
- 54.Lang T, Jahn R. Core proteins of the secretory machinery. Handb Exp Pharmacol. 2008:107–127. doi: 10.1007/978-3-540-74805-2_5. [DOI] [PubMed] [Google Scholar]
- 55.Rao SK, et al. Identification of SNAREs involved in synaptotagmin VII-regulated lysosomal exocytosis. J Biol Chem. 2004;279:20471–20479. doi: 10.1074/jbc.M400798200. [DOI] [PubMed] [Google Scholar]
- 56.Fraldi A, et al. Lysosomal fusion and SNARE function are impaired by cholesterol accumulation in lysosomal storage disorders. EMBO J. 2010;29:3607–3620. doi: 10.1038/emboj.2010.237. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 57.Choudhury A, et al. Elevated endosomal cholesterol levels in Niemann-Pick cells inhibit rab4 and perturb membrane recycling. Mol Biol Cell. 2004;15:4500–4511. doi: 10.1091/mbc.E04-05-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dong XP, et al. TRP channels of intracellular membranes. J Neurochem. 2010;113:313–328. doi: 10.1111/j.1471-4159.2010.06626.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.LaPlante JM, et al. Identification and characterization of the single channel function of human mucolipin-1 implicated in mucolipidosis type IV, a disorder affecting the lysosomal pathway. FEBS Lett. 2002;532:183–187. doi: 10.1016/s0014-5793(02)03670-0. [DOI] [PubMed] [Google Scholar]
- 60.Dong XP, et al. Activating mutations of the TRPML1 channel revealed by proline-scanning mutagenesis. J Biol Chem. 2009;284:32040–32052. doi: 10.1074/jbc.M109.037184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zlokovic BV. Neurodegeneration and the neurovascular unit. Nat Med. 2010;16:1370–1371. doi: 10.1038/nm1210-1370. [DOI] [PubMed] [Google Scholar]
- 62.Chen YH, et al. Molecular signatures of disease brain endothelia provide new sites for CNS-directed enzyme therapy. Nat Med. 2009;15:1215–1218. doi: 10.1038/nm.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sands MS, Davidson BL. Gene therapy for lysosomal storage diseases. Mol Ther. 2006;13:839–849. doi: 10.1016/j.ymthe.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 64.Sands MS, Haskins ME. CNS-directed gene therapy for lysosomal storage diseases. Acta Paediatr Suppl. 2008;97:22–27. doi: 10.1111/j.1651-2227.2008.00660.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Malnar M, et al. Niemann-Pick type C cells show cholesterol dependent decrease of APP expression at the cell surface and its increased processing through the beta-secretase pathway. Biochim Biophys Acta. 2010;1802:682–691. doi: 10.1016/j.bbadis.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tamboli IY, et al. Sphingolipid storage affects autophagic metabolism of the amyloid precursor protein and promotes Abeta generation. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2011;31:1837–1849. doi: 10.1523/JNEUROSCI.2954-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhou X, et al. Deletion of PIK3C3/Vps34 in sensory neurons causes rapid neurodegeneration by disrupting the endosomal but not the autophagic pathway. Proc Natl Acad Sci U S A. 2010;107:9424–9429. doi: 10.1073/pnas.0914725107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Doyle JP, et al. Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell. 2008;135:749–762. doi: 10.1016/j.cell.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rohrbach M, Clarke JT. Treatment of lysosomal storage disorders : progress with enzyme replacement therapy. Drugs. 2007;67:2697–2716. doi: 10.2165/00003495-200767180-00005. [DOI] [PubMed] [Google Scholar]
- 70.Parenti G. Treating lysosomal storage diseases with pharmacological chaperones: from concept to clinics. EMBO Mol Med. 2009;1:268–279. doi: 10.1002/emmm.200900036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mingozzi F, High KA. Therapeutic in vivo gene transfer for genetic disease using AAV: progress and challenges. Nature reviews Genetics. 2011;12:341–355. doi: 10.1038/nrg2988. [DOI] [PubMed] [Google Scholar]
- 72.Naldini L. Ex vivo gene transfer and correction for cell-based therapies. Nature reviews Genetics. 2011;12:301–315. doi: 10.1038/nrg2985. [DOI] [PubMed] [Google Scholar]
- 73.McCaughan KK, et al. Translational termination efficiency in mammals is influenced by the base following the stop codon. Proc Natl Acad Sci U S A. 1995;92:5431–5435. doi: 10.1073/pnas.92.12.5431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Keeling KM, et al. Gentamicin-mediated suppression of Hurler syndrome stop mutations restores a low level of alpha-L-iduronidase activity and reduces lysosomal glycosaminoglycan accumulation. Hum Mol Genet. 2001;10:291–299. doi: 10.1093/hmg/10.3.291. [DOI] [PubMed] [Google Scholar]
- 75.Sleat DE, et al. Aminoglycoside-mediated suppression of nonsense mutations in late infantile neuronal ceroid lipofuscinosis. Eur J Paediatr Neurol. 2001;5(Suppl A):57–62. doi: 10.1053/ejpn.2000.0436. [DOI] [PubMed] [Google Scholar]
- 76.Nudelman I, et al. Repairing faulty genes by aminoglycosides: development of new derivatives of geneticin (G418) with enhanced suppression of diseases-causing nonsense mutations. Bioorg Med Chem. 2010;18:3735–3746. doi: 10.1016/j.bmc.2010.03.060. [DOI] [PubMed] [Google Scholar]
- 77.Langmade SJ, et al. Pregnane X receptor (PXR) activation: a mechanism for neuroprotection in a mouse model of Niemann-Pick C disease. Proc Natl Acad Sci U S A. 2006;103:13807–13812. doi: 10.1073/pnas.0606218103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Davidson CD, et al. Chronic cyclodextrin treatment of murine Niemann-Pick C disease ameliorates neuronal cholesterol and glycosphingolipid storage and disease progression. PLoS One. 2009;4:e6951. doi: 10.1371/journal.pone.0006951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu B, et al. Reversal of defective lysosomal transport in NPC disease ameliorates liver dysfunction and neurodegeneration in the npc1−/− mouse. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:2377–2382. doi: 10.1073/pnas.0810895106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Glass CK, et al. Mechanisms underlying inflammation in neurodegeneration. Cell. 2010;140:918–934. doi: 10.1016/j.cell.2010.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Settembre C, et al. Systemic inflammation and neurodegeneration in a mouse model of multiple sulfatase deficiency. Proc Natl Acad Sci U S A. 2007;104:4506–4511. doi: 10.1073/pnas.0700382104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Oswald MJ, et al. Glial activation spreads from specific cerebral foci and precedes neurodegeneration in presymptomatic ovine neuronal ceroid lipofuscinosis (CLN6) Neurobiol Dis. 2005;20:49–63. doi: 10.1016/j.nbd.2005.01.025. [DOI] [PubMed] [Google Scholar]
- 83.Wu YP, Proia RL. Deletion of macrophage-inflammatory protein 1 alpha retards neurodegeneration in Sandhoff disease mice. Proc Natl Acad Sci U S A. 2004;101:8425–8430. doi: 10.1073/pnas.0400625101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fan JQ, et al. Accelerated transport and maturation of lysosomal alpha-galactosidase A in Fabry lymphoblasts by an enzyme inhibitor. Nat Med. 1999;5:112–115. doi: 10.1038/4801. [DOI] [PubMed] [Google Scholar]
- 85.Sawkar AR, et al. Gaucher disease-associated glucocerebrosidases show mutation-dependent chemical chaperoning profiles. Chem Biol. 2005;12:1235–1244. doi: 10.1016/j.chembiol.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 86.Suzuki Y. Chemical chaperone therapy for GM1-gangliosidosis. Cellular and molecular life sciences : CMLS. 2008;65:351–353. doi: 10.1007/s00018-008-7470-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lieberman RL, et al. Effects of pH and iminosugar pharmacological chaperones on lysosomal glycosidase structure and stability. Biochemistry. 2009;48:4816–4827. doi: 10.1021/bi9002265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Arrasate M, et al. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431:805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- 89.Braulke T, Bonifacino JS. Sorting of lysosomal proteins. Biochim Biophys Acta. 2009;1793:605–614. doi: 10.1016/j.bbamcr.2008.10.016. [DOI] [PubMed] [Google Scholar]
- 90.Luzio JP, et al. Lysosomes: fusion and function. Nat Rev Mol Cell Biol. 2007;8:622–632. doi: 10.1038/nrm2217. [DOI] [PubMed] [Google Scholar]
- 91.Choi JO, et al. Characterization of Fabry mice treated with recombinant adeno-associated virus 2/8-mediated gene transfer. J Biomed Sci. 2010;17:26. doi: 10.1186/1423-0127-17-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Marshall J, et al. Substrate reduction augments the efficacy of enzyme therapy in a mouse model of Fabry disease. PLoS One. 2010;5:e15033. doi: 10.1371/journal.pone.0015033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Enquist IB, et al. Successful low-risk hematopoietic cell therapy in a mouse model of type 1 Gaucher disease. Stem Cells. 2009;27:744–752. doi: 10.1634/stemcells.2008-0844. [DOI] [PubMed] [Google Scholar]
- 94.Baek RC, et al. AAV-mediated gene delivery in adult GM1-gangliosidosis mice corrects lysosomal storage in CNS and improves survival. PLoS One. 2010;5:e13468. doi: 10.1371/journal.pone.0013468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Elliot-Smith E, et al. Beneficial effects of substrate reduction therapy in a mouse model of GM1 gangliosidosis. Mol Genet Metab. 2008;94:204–211. doi: 10.1016/j.ymgme.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 96.Cachon-Gonzalez MB, et al. Effective gene therapy in an authentic model of Tay-Sachs-related diseases. Proc Natl Acad Sci U S A. 2006;103:10373–10378. doi: 10.1073/pnas.0603765103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Maegawa GH, et al. Pyrimethamine as a potential pharmacological chaperone for late-onset forms of GM2 gangliosidosis. J Biol Chem. 2007;282:9150–9161. doi: 10.1074/jbc.M609304200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang D, et al. Short-term, high dose enzyme replacement therapy in sialidosis mice. Mol Genet Metab. 2005;85:181–189. doi: 10.1016/j.ymgme.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 99.Ellinwood NM, et al. Safe, efficient, and reproducible gene therapy of the brain in the dog models of Sanfilippo and Hurler syndromes. Mol Ther. 2011;19:251–259. doi: 10.1038/mt.2010.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fraldi A, et al. Functional correction of CNS lesions in an MPS-IIIA mouse model by intracerebral AAV-mediated delivery of sulfamidase and SUMF1 genes. Hum Mol Genet. 2007;16:2693–2702. doi: 10.1093/hmg/ddm223. [DOI] [PubMed] [Google Scholar]
- 101.Hemsley KM, et al. Effect of cisternal sulfamidase delivery in MPS IIIA Huntaway dogs--a proof of principle study. Mol Genet Metab. 2009;98:383–392. doi: 10.1016/j.ymgme.2009.07.013. [DOI] [PubMed] [Google Scholar]
- 102.Heldermon CD, et al. Therapeutic efficacy of bone marrow transplant, intracranial AAV-mediated gene therapy, or both in the mouse model of MPS IIIB. Mol Ther. 2010;18:873–880. doi: 10.1038/mt.2010.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ficko-Blean E, et al. Structural and mechanistic insight into the basis of mucopolysaccharidosis IIIB. Proc Natl Acad Sci U S A. 2008;105:6560–6565. doi: 10.1073/pnas.0711491105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bielicki J, et al. Comparison of ventricular and intravenous lentiviral-mediated gene therapy for murine MPS VII. Mol Genet Metab. 2010;101:370–382. doi: 10.1016/j.ymgme.2010.08.013. [DOI] [PubMed] [Google Scholar]
- 105.Hackett NR, et al. Safety of direct administration of AAV2(CU)hCLN2, a candidate treatment for the central nervous system manifestations of late infantile neuronal ceroid lipofuscinosis, to the brain of rats and nonhuman primates. Hum Gene Ther. 2005;16:1484–1503. doi: 10.1089/hum.2005.16.1484. [DOI] [PubMed] [Google Scholar]
- 106.Chang M, et al. Intraventricular enzyme replacement improves disease phenotypes in a mouse model of late infantile neuronal ceroid lipofuscinosis. Mol Ther. 2008;16:649–656. doi: 10.1038/mt.2008.9. [DOI] [PubMed] [Google Scholar]
- 107.Seehafer SS, et al. Immunosuppression alters disease severity in juvenile Batten disease mice. J Neuroimmunol. 2011;230:169–172. doi: 10.1016/j.jneuroim.2010.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.De Duve C, et al. Tissue fractionation studies. 6. Intracellular distribution patterns of enzymes in rat-liver tissue. Biochem J. 1955;60:604–617. doi: 10.1042/bj0600604. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01