Id2 expression delineates differential checkpoints in the genetic program of CD8α+ and CD103+ dendritic cell lineages (original) (raw)
Abstract
Dendritic cells (DCs) have critical roles in the induction of the adaptive immune response. The transcription factors Id2, Batf3 and Irf-8 are required for many aspects of murine DC differentiation including development of CD8α+ and CD103+ DCs. How they regulate DC subset specification is not completely understood. Using an Id2-GFP reporter system, we show that Id2 is broadly expressed in all cDC subsets with the highest expression in CD103+ and CD8α+ lineages. Notably, CD103+ DCs were the only DC able to constitutively cross-present cell-associated antigens in vitro. Irf-8 deficiency affected loss of development of virtually all conventional DCs (cDCs) while Batf3 deficiency resulted in the development of Sirp-α− DCs that had impaired survival. Exposure to GM-CSF during differentiation induced expression of CD103 in Id2-GFP+ DCs. It did not restore cross-presenting capacity to _Batf3_−/− or CD103−Sirp-α−DCs in vitro. Thus, Irf-8 and Batf3 regulate distinct stages in DC differentiation during the development of cDCs. Genetic mapping DC subset differentiation using Id2-GFP may have broad implications in understanding the interplay of DC subsets during protective and pathological immune responses.
Keywords: antigen presentation, cross-presentation, differentiaton, Id2, priming, transcription factors
Introduction
CD11c+ dendritic cells (DCs) are essential in presenting antigen to initiate T-cell responses. They have critical roles in immunity because of their ability to recognize invading pathogens and mobilize immune cells to combat them. DCs can be categorized into a number of different subsets that largely reflect the pattern of expression of cell surface molecules and functional specializations (Geissmann et al, 2010; Steinman and Idoyaga, 2010).
One major division of DCs is conventional DCs (cDCs) and plasmacytoid DCs (pDCs). In murine spleen, cDCs can be further divided into CD8α+ DCs, CD4+ DCs and CD8α−CD4− (termed double-negative, DN) DCs. DCs enter the spleen and lymph nodes (LNs) through the blood as either mature pDCs or immature precursors of cDCs known as pro-DCs (reviewed in Liu and Nussenzweig (2010)). LNs contain multiple populations of DCs which include CD8α+ DCs and CD8α− DCs, together with DCs that migrate from the peripheral tissues (tissue-derived DCs). CD8α+ DCs contribute significantly to CD8+ T-cell activation via presentation of exogenous (den Haan and Bevan, 2002) or pathogen-derived antigens (Allan et al, 2003; Belz et al, 2004) while CD8α− DCs preferentially drive the activation of CD4+ T cells (Allenspach et al, 2008; Mount et al, 2008). Tissue-derived DCs differ depending on the type of peripheral tissues they drain. Langerhans cells (LCs) and the dermal DCs are found in skin while migratory tissue-derived CD103+ DCs originate from cutaneous and mucosal tissues such as the lamina propria of respiratory and gastrointestinal tracts (reviewed in Geissmann et al (2010)). The latter two DC subsets have a critical role in transporting antigens from body surfaces to LNs such that DCs resident in the LN can gain access to antigens (Liu and Nussenzweig, 2010).
Although a number of different DC subsets have been described, understanding how diverse DC subsets develop from a common progenitor is limited. Several factors have been identified as highly expressed in cDCs. These include inhibitor of DNA binding (Id)-2, interferon regulatory protein (Irf)-2 (Honda et al, 2004; Ichikawa et al, 2004), _Irf_-4 (Suzuki et al, 2004) and _Irf_-8 (Schiavoni et al, 2002, 2004; Tamura et al, 2005), PU.1 (Carotta et al, 2010), Ikaros, Gfi-1 (Rathinam et al, 2005), Batf3 (Hildner et al, 2008) and signal transducer and activator of transcription (Stat)-3 and _Stat_-5 (Wu and Liu, 2007; Merad and Manz, 2009).
Id2, a member of the helix-loop-helix (HLH) transcription factor family, is upregulated during DC development and is required for the development of CD8α+ DCs and LCs (Hacker et al, 2003). Id proteins act by antagonizing the DNA binding of activating E proteins and Id2 has been postulated to repress pDC development by suppressing HEB and E2A as overexpression of these factors in haematopoietic progenitors led to enhanced pDC development (Schiavoni et al, 2002; Tamura et al, 2005). More recently, it was shown that a third HLH protein, E2-2, specifically regulates generation and maintenance of pDCs (Cisse et al, 2008). Irf proteins have broad effects in DC development. Loss of either Irf-2 or Irf-4 results in defects in the development of CD8α− DCs while the absence of Irf-4 also disrupts pDC development in the spleen. In contrast, Irf-8 (also known as ICSBP) is required for CD8α+ DCs and pDC development (Schiavoni et al, 2002; Tamura et al, 2005). Similar to the impact of Irf-8 deficiency, inactivation of the Jun dimerization protein p21SNFT, Batf3, has also been reported to result in the loss of CD8α+ and tissue-derived CD103+ DCs, but does not impair pDC development (Hildner et al, 2008; Edelson et al, 2010).
CD103+ DCs found in peripheral LNs and lamina propria represent a heterogeneous group of cells in which those cells that express CD11b in the lamina propria do not appear to depend on Id2, Irf-8 or Batf3 for development (Ginhoux et al, 2009; Edelson et al, 2010). The analysis of compound knockout mice has led to a model in which conventional CD8α+ DCs and CD103+ DCs are thought to be developmentally related and possess similar functional and localization characteristics as both DC subsets are absent in mice lacking Id2, Irf-8 or Batf3 (Hildner et al, 2008; Ginhoux et al, 2009; Edelson et al, 2010). Such models, however, are unable to discriminate between the stepwise requirement for each of these transcription factors for differentiation nor their impact on modulating survival of DC subsets once DC precursors have developed.
While much effort has focussed on the murine DC subsets, it is now becoming clear that many aspects of the human and mouse DC systems are closely aligned; and thus, murine models are likely to be highly informative in understanding human disease influenced by DC behaviour. DC subsets in the human blood can be distinguished by their expression of the surface molecules BDCA-1 (CD1c), BDCA-2 (CD303) and BDCA-3 (CD141) (Dzionek et al, 2000). BDCA-1+ cells represent a population of myeloid DCs; BDCA-2 marks pDCs, while BDCA-3 identifies the human counterpart of murine CD8α+ DCs which both share the high capacity to capture exogenous antigens for cross-presentation and the expression of chemokine receptor XCR1 (Bachem et al, 2010; Crozat et al, 2010; Jongbloed et al, 2010; Poulin et al, 2010). The interplay between these different DC subsets is of considerable interest as detailed understanding of their generation and responses offers opportunities for exploiting them as targets for vaccines and therapeutic interventions in cancer, autoimmunity and infection (Palucka et al, 2010). However, despite this significant progress, recovering sufficient human DC of the various subsets for molecular and transcriptional profiling remains challenging.
To understand how the complex network of DC lineages is generated in a model system, we engineered an Id2-GFP mouse reporter strain that enabled us to track endogenous Id2 expression on a single cell level during DC differentiation. As Id proteins are concentration-dependent antagonists of E protein activity, we reasoned that Id2 expression would be tightly regulated in DCs. Indeed, this was the case as cDC subsets expressed a wide range of Id2-GFP with the highest expression in CD8α+ and CD103+ DC lineages in vivo. The equivalent DCs could also be identified in Flt3L-stimulated bone marrow cultures by correlating Id2 expression with surface expression of CD103 on DCs. By analysing Id2-GFP expression in the absence of Irf-8 and Batf3, we were able to delineate that Irf-8 is required for generation of both cDCs and pDCs, while in the absence of Batf3 Id2-GFP+Sirp-α− (CD8 equivalent) DCs develop but CD103+ DCs do not. Furthermore, GM-CSF is a potent stimulator of CD103 expression in vitro, even in the absence of Batf3; however, GM-CSF could not rescue cross-presenting potential from the same mice. Thus, the ability to map Id2-GFP expression in developing murine DC subsets has wide application in understanding the precise molecular regulation of DC differentiation.
Results
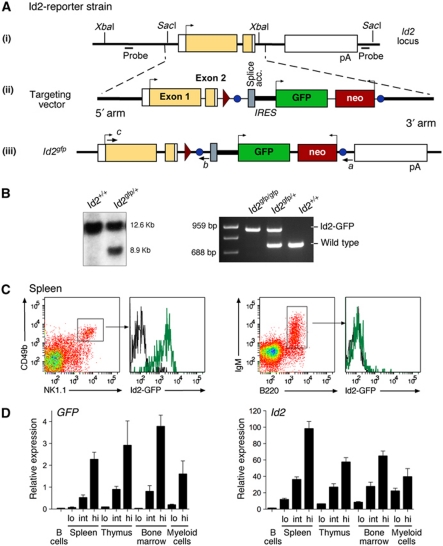
To define the expression of Id2 and its role in different haematopoietic lineages, we generated a reporter allele by inserting an internal ribosome entry site (IRES)-GFP cassette into the 3′ untranslated region of the Id2 gene (Figure 1A). The targeted reporter allele, _Id2_gfp, resulted in the transcription of a bicistronic mRNA that produced wild-type Id2 protein and GFP. This targeting strategy predicted that the IRES-GFP cassette would not affect the upstream Id2 mRNA transcript. To confirm this, homozygous _Id2_gfp/gfp mice were generated (Figure 1B). _Id2_gfp/gfp mice were indistinguishable in survival, haematopoietic cellularity and lineage composition from C57BL/6 controls (NK cells: C57BL/6, 2.3 × 105±6.7 × 104/spleen; Id2 gfp/+, 1.94 × 105±4.2 × 104/spleen; Id2 gfp/gfp, 1.94 × 105±1.9 × 104/spleen; total DCs: C57BL/6, 2.1 × 106±6.4 × 104/spleen; Id2 gfp/+, 1.9 × 106±8.3 × 104/spleen; Id2 gfp/gfp, 2.1 × 106±5.8 × 104/spleen; and data not shown). As predicted, Id2-GFP was abundantly expressed in NK cells and silenced in B cells (Figure 1C). Moreover, the expression of GFP correlated exactly with Id2 transcription in a variety of different haematopoietic lineages (Figure 1D).
Figure 1.
Generation and validation of _Id2_gfp reporter mouse strains. (A) The genomic locus of Id2. Exons are represented by boxes; introns are represented as black lines; coding regions are shaded yellow; non-translated regions are in white; arrows indicate the direction of translation. The alleles derived from the integration of the targeting vector and subsequent manipulations are shown. pA, polyadenylation signal sequence; circles, frt sites; triangles, loxP sites. The Id2-GFP reporter line was derived from an embryonic stem cell (ES) clone that lacked the 5′ LoxP site and was identified by PCR. The position and direction of the genotyping primers (a–c) and the _Sac_I and _Xba_I sites used for Southern blotting are indicated. (B) Southern blot analysis of ES cell _Sac_I-digested DNA showing the wild-type (12.6 kb) and targeted (8.9 kb) alleles (left panel). PCR genotyping of tail DNA using the primer set a/b/c showing the correct amplification of the wild-type (688 bp) and _Id2_gfp (959 bp) alleles (right panel). (C) Id2-GFP expression in B220+IgM+ B cells derived from peripheral LNs and splenic NK1.1+CD49b+ NK cells from naive wild-type (black line) and _Id2_gfp/gfp (green line) mice. (D) Quantitative PCR analysis for the indicated transcripts of live (PI−) mixed populations of cells from spleen, thymus and bone marrow purified on the basis of their expression of Id2-GFP. Data are the mean±s.e.m. of two experiments.
Id2 expression in DCs in vivo
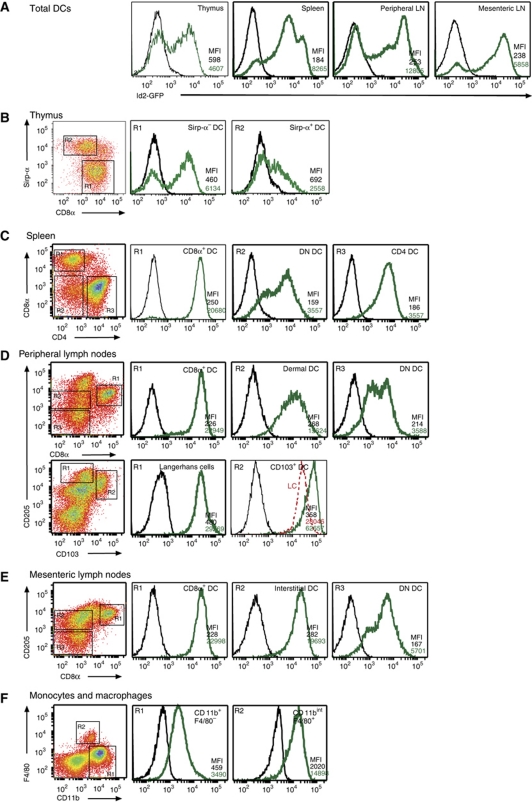
Next, we investigated Id2-GFP in DCs in vivo to determine if the level of expression identified individual DC subsets. To track the expression of Id2 in cDCs and pDCs in vivo, we isolated DCs from thymus, spleen and peripheral and mesenteric LNs of _Id2_gfp/gfp mice (Figure 2A–E). This approach allowed the delineation of several populations of DCs not previously thought to express Id2 and showed that DCs isolated from different tissues expressed distinct amounts of Id2 (Figure 2A). Thymic DCs were divided into four populations with discrete fractions of both CD8α+Sirp-α− and Sirp-α+ DCs expressing Id2-GFP (Figure 2B). pDCs, which were identified by their intermediate expression of CD11c and high expression of CD45RA, were uniformly very low for Id2-GFP (Supplementary Figure S1). In spleen and LN, all cDCs (defined as CD11chigh) expressed Id2-GFP but varied in the level of expression among the different DC subsets (Figure 2C–E). Unexpectedly, CD4+ DCs in spleen and DN DCs in spleen and LN also expressed Id2-GFP (Figure 2C). Id2-GFP fluorescence in cells of the monocyte/macrophage lineages was at a level that was similar to DN DCs and dermal DCs, respectively (Figure 2C, D and F).
Figure 2.
Multiple DC subsets express Id2 in vivo. _Id2_gfp/gfp (green line) and wild-type (black line) cells were analysed by flow cytometry for GFP expression in different DC and myeloid populations. The different cell types were defined as described in Materials and methods. (A) Total DC (CD11c+) populations from thymus, spleen, peripheral LN and mesenteric LNs; (B) thymic DC populations; (C) splenic DC populations; (D) peripheral (pooled DCs from inguinal, brachial, axillary, superficial cervical LNs) and (E) mesenteric LN DCs; and (F) monocyte and macrophage (from peritoneal lavage) lineages. All plots are gated on CD11c+ cells and markers as indicated in the dot plots (left panels). In (D), the fluorescence intensity of Langerhans cells has been shown in red for comparison. Mean fluorescence intensity (MFI) is shown for GFP expression for each gated population. Data are representative of at least two to three independent experiments.
Id2 expression in haematopoietic progenitors
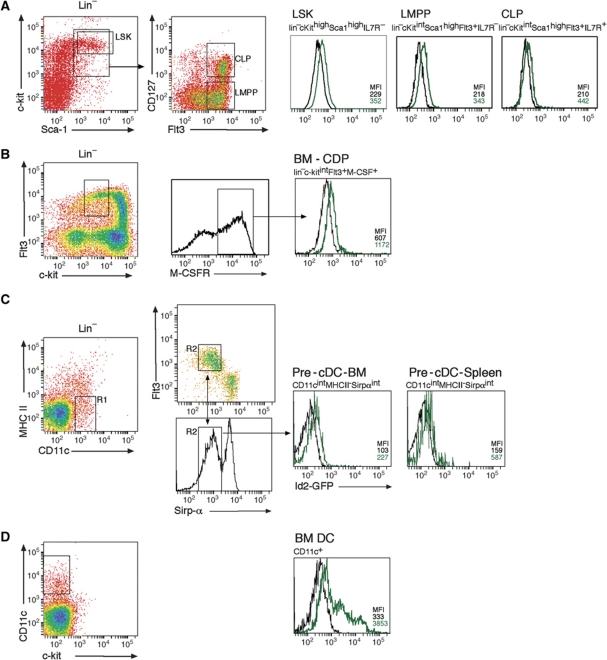
Given that all cDC subsets expressed some level of Id2-GFP, we wished to examine whether Id2 was induced during DC differentiation or alternately might be constitutively expressed in precursor DCs and down modulated as populations matured. Id2-GFP expression was negligible in lineage-negative scahighc-kithigh (LSK) cells (including the Flt3+ lymphoid primed multipotent progenitor, LMPP), common lymphoid progenitors and common DC progenitors (CDPs, also known as pro-DCs) (Figure 3A and B). Similarly, BM and splenic pre-cDCs had showed little upregulation of Id2 (Figure 3C) and analysis of CD11c+ bone marrow DCs revealed that a large fraction of cells were low or negative for Id2-GFP (Figure 3D) demonstrating that activation of the Id2 gene occurred relatively late in the differentiation of DCs. Thus, Id2 expression is induced in precursors that have been committed to the cDC pathway.
Figure 3.
Expression of Id2 in lymphoid progenitors. (A) Bone marrow progenitor cells were analysed by depletion of lineage expressing cells then stained for sca-1, c-kit, M-CSFR and Flt3 and analysed by flow cytometry for lin−sca-1+c-kit+ (LSK) cells, lymphoid primed multipotent progenitors (LMPPs, defined as LSKFlt3high), common lymphoid progenitors (CLPs) and (B) common DC progenitor (CDPs) as indicated. (C) Bone marrow and splenic DC progenitors were analysed by depletion of lineage expressing cells (CD19, NK1.1, CD3 and Ter119) then stained for CD11c, Flt3, MHC II and Sirp-α and analysed for the pre-cDC population by flow cytometry (Liu and Nussenzweig, 2010). Profiles show the gating strategy in which CD11c+MHC II− cells (region 1, R1) were then selected for expression of Flt3 and Sirp-α (region 2, R2). (D) CD11c+ cells were isolated from bone marrow by density gradient centrifugation. Gated populations from _Id2_gfp/gfp (green line) and wild-type (black line) mice were then assessed for GFP fluorescence. Data are representative of at least two experiments.
Id2 expression in DCs in vitro
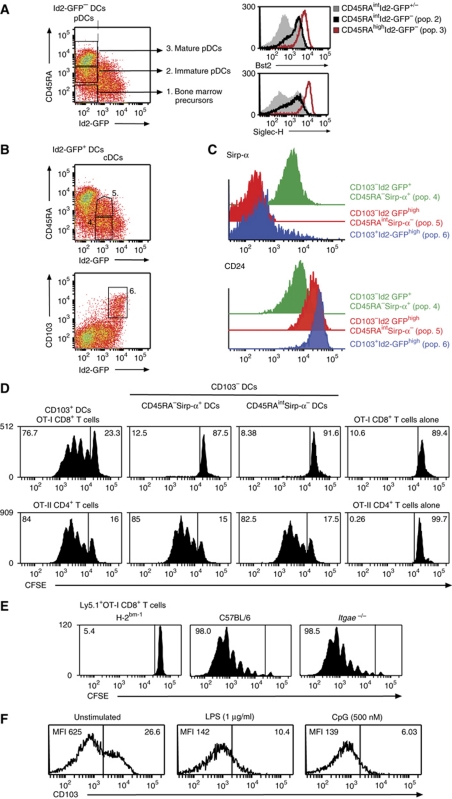
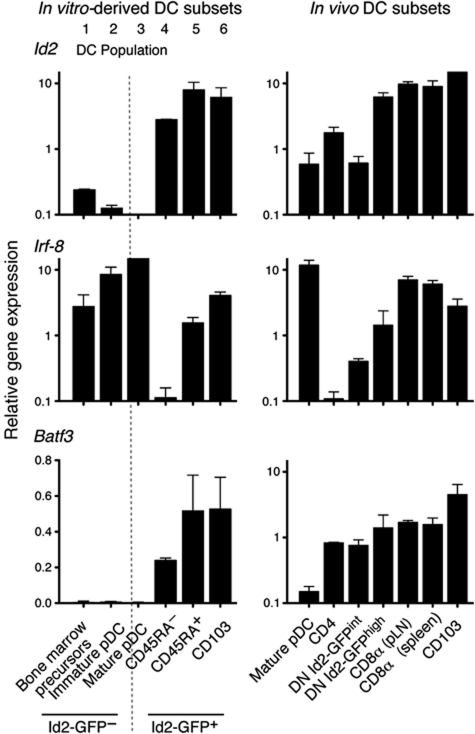
The different amounts of Id2-GFP observed in different DC subsets in vivo led us to propose that distinct levels of Id2 expression could be used to define DC subsets and this hierarchy may also be maintained in vitro. If so, Id2-GFP mapping of DC subsets in vitro would enable the recovery of DC subsets in sufficient numbers for the detailed dissection of the functional and molecular aspects of DC development. It should be noted that although the surface molecule CD8α is expressed on DCs isolated directly ex vivo, this marker is not expressed on _in vitro_-derived CD8α-equivalent DCs. These cells have been previously identified by their lack of expression of Sirp-α, and as shown below, intermediate expression of CD45RA. As observed in vivo, DCs generated in vitro expressed graded levels of Id2-GFP allowing us to discriminate six distinct DC populations (Figure 4A–C). These could be divided into the Id2-GFP negative populations that also lack CD103 expression (1) CD45RA− DCs; (2) CD45RAint DCs that expressed the pDC markers Bst-2 and Siglec-H (Figure 4A, right panels); (3) CD45RAhigh (analogous to the previously described mature pDC); and the Id2-GFP expressing populations; (4) CD45RA− DCs; and (5) CD45RAint (Figure 4B). Further dissection of the Id2-GFP+ cDC populations revealed the presence of CD45RA−CD103+ DCs that expressed the highest amounts of Id2-GFP (population 6, Supplementary Table SI). Tissue-derived CD103+ cDCs have not been previously identified in vitro but this staining was specific as it was not detected in similar cultures derived from _Itgae_−/− (CD103-deficient) mice (Supplementary Figure S2). This analysis also showed that the Id2-GFP+CD45RA− population (pop. 4) of DCs was Sirp-α+ while the Id2-GFP+CD45RAint population (pop. 5) was predominantly Sirp-α− and these populations expressed distinct levels of CD24 consistent with the previously described in vitro phenotype of CD8α− DCs and CD8α+ DCs (Figure 4C; Naik et al, 2005).
Figure 4.
In vitro cross-presentation in Flt3L-stimulated cultures is limited to CD103-expressing Id2-GFPhigh DCs. (A–C) Flt3L-derived DCs from _Id2_gfp/gfp BM were analysed on day 5 of culture. Six different DC populations were discriminated based on their expression of CD11c, CD45RA and CD103. Right panels: CD11c+CD45RA+Id2-GFP− cells expressed markers of pDCs. Histograms show expression of Bst2 (upper right panel) and Siglec-H (lower right panel) of total CD45RAint cells (grey shading), CD45RAintId2-GFP− immature pDCs (black line). The expression of markers for CD103−CD45RAhigh (mature) pDC is indicated in red. (B) Populations 4, 5 and 6 could be discriminated based on their expression of Sirp-α or CD103. Profiles are representative of at least 10 independent experiments with similar results. (C) Id2-GFP DC subsets express distinct levels of Sirp-α and CD24. (D) In vitro generated _Id2_gfp/gfp DCs were flow cytometrically sorted 5 days after initiation of cell culture according to their expression of CD103, CD45RA, Sirp-α and Id2-GFP and analysed for their ability to cross-present cell-associated OVA to CFSE-labelled OVA-specific CD8+ T cells (upper panels). The ability of these subsets of present exogenous antigen to CFSE-labelled OVA-specific CD4+ T cells was evaluated as a control (lower panels). Data are representative of four independent experiments. T-cell proliferation was analysed in 1–3 replicates for each DC subset/responder population for each experiment. (E) Ly5.1+CFSE-labelled CD8+ OVA-specific T cells were adoptively transferred into H-2Kbm-1, B6 or _Itgae_−/− mice 1 day before transfer of 2 × 107 OVA-coated H-2bm-1 splenocytes. Proliferation of Ly5.1+Vα2+CD8+ T cells in spleen was analysed by flow cytometry after 60 h. Data are representative of two independent experiments with seven individuals analysed in each group. (F) The expression level of surface CD103 was monitored by flow cytometry 18 h after exposure to TLR ligands LPS or CpG. Data are representative of at least five independent experiments and show MFI expression levels.
Differentiation of in vitro generated DC subsets
To determine the relationship between different populations of DCs observed in vitro, DC subsets from day 5 cultures were purified into the six fractions described in Figure 4 and re-cultured in Flt3L-conditioned medium for further 3 days (Supplementary Figure S3). We hypothesized that Id2-GFP−CD45RA− DCs (pop. 1) contained multipotent DC precursors. Concordant with this notion, this subset gave rise to all identified in vitro subsets of DCs. Id2-GFP−CD45RAhi (pop. 3) generated only pDCs while Id2-GFP−CD45RAint DCs (pop. 2) predominantly gave rise to Id2-GFP−CD45RAhi cells, suggesting that they contain the major immature population of pDCs. CD103−Id2-GFP+CD45RA−Sirp-α+ DCs (pop. 4) maintained their phenotype while CD103−Id2-GFP+CD45RAintSirp-α− DCs (pop. 5) generated Sirp-α− DCs that expressed varying levels of CD103 and CD103−Sirp-α+ DCs. In contrast, CD103+ DCs (pop. 6) almost exclusively gave rise to CD103+ DCs, suggesting that these cells are terminally differentiated. Thus, it appears that some in vitro subsets represent end-state DC populations (e.g., mature pDCs and CD103+ DCs) while other subsets retain differentiation potential. Similarly, Id2-GFP−CD45RAint DCs appear to be immature pDCs. This was further supported by the failure of Id2-GFP−CD45RAint DCs to upregulate CD80 and MHC class II in response to TLR ligands such as LPS or poly I:C and the induction of these activation markers following stimulation with CpG motifs (Supplementary Figure S4). Thus, the in vitro culture contains a combination of early developing and differentiated DCs that may represent the counterparts of blood-derived lymphoid tissue-resident and tissue-derived DCs.
Characteristics of CD103+ DCs in vitro
One property that varies extensively among DC subsets is the ability to take up exogenous antigens and divert these antigens into the MHC class I pathway—a process known as cross-presentation and is thought to be of major importance for the recognition of viral or bacterial antigens when DCs are not directly infected (Carbone and Bevan, 1990). In mice, CD8α+ DCs, and more recently CD103+ DCs, have been identified as the major cross-presenting populations in vivo. Similarly, the CD8α+ DC counterpart in humans, BDCA-3 DCs, also exhibit more efficient cross-presenting ability in vitro (Bachem et al, 2010; Crozat et al, 2010; Jongbloed et al, 2010; Poulin et al, 2010). Despite this, in model systems such as Flt3L bone marrow cultures, the identification of the DC subset(s) that can efficiently and constitutively cross-present exogenous antigens has been more elusive and has limited detailed molecular studies of this cell type. Given our identification of CD103+ DCs in vitro, we tested the ability of CD103−Id2-GFP+CD45RA−Sirp-α+ (pop. 4), CD103−Id2-GFP+CD45RAintSirp-α− (pop. 5; CD8α+ DC equivalents) and CD103+Id2-GFP+ DCs (pop. 6) for their capacity to cross-present cell-associated antigens. Initially, we chose to analyse DCs isolated from day 5 cultures as population 5 reaches its maximal prevalence at this time point. DCs were purified and then co-cultured with OVA-coated bm-1 splenocytes (which cannot present antigen on H-2Kb) and analysed for their capacity to induce proliferation of CFSE-labelled OVA-specific CD8+ T cells (OT-I cells). The presentation of OVA to CFSE-labelled OVA-specific CD4+ T cells (OT-II cells) was used as a control to determine that all populations of DCs could present OVA antigens. Strikingly, strong cross-presentation to CD8+ T cells was only elicited from CD103+Id2-GFP+ stimulators but not from other populations of DCs (Figure 4D). In contrast, all DC populations were able to present exogenous OVA antigen to CD4+ T cells. Similar analysis of DC populations at day 8 of culture showed that CD103+Id2-GFP+CD45RA− DCs remained the only in vitro DC subset capable of cross-presenting cell-associated antigen (Supplementary Figure S5). However, the efficiency of presentation at this time point was reduced (data not shown). Although CD103 was a marker of this discrete subset of DCs, CD103 expression itself was not required for cross-presentation of cell-associated OVA in vivo (Figure 4E) and indeed appeared to be strongly downregulated on activation of DCs (Figure 4F). Thus, unexpectedly, the major in vitro DC that could constitutively cross-present cell-associated antigens were not Sirp-α− DCs (equivalent to CD8α+ DCs) but CD103+ DCs.
Id2-GFP reveals overlapping but non-redundant roles of Irf-8 and Batf3 during CD8α+ DC and CD103+ DC differentiation
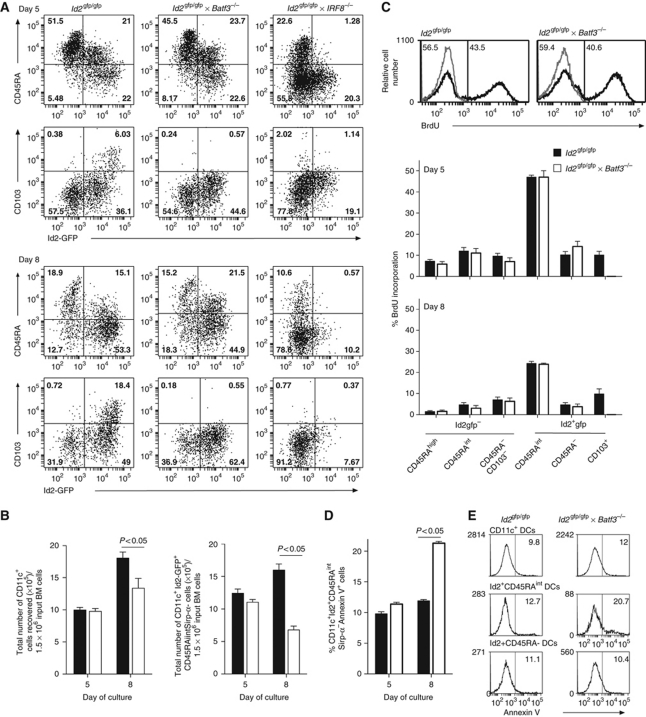
Id2, Irf-8 and Batf3 are transcription factors implicated in the development of CD103+ and CD8α+ DCs (Schiavoni et al, 2004; Hildner et al, 2008; Ginhoux et al, 2009; Edelson et al, 2010). However, precisely where in DC differentiation these transcription factors determine the fate decision between CD103+ and CD8α+ DCs is unknown. First, we analysed the expression pattern of the transcription factors Id2, Irf-8 and Batf3 in the different DC populations in vitro and in vivo (Figure 5). While this demonstrated differential expression of the transcription factors among different DC subsets, the in vitro (Id2-GFP+CD45RA+) and in vivo CD8α+ and CD103+ DCs concordantly expressed high levels of Id2, Irf-8 and Batf3. In an attempt to delineate whether Irf-8 or Batf3 differentially affect particular DC subsets during development, we crossed the _Id2_gfp/gfp mice with _Batf3_−/− or _Irf-8_−/− mice. At days 5 and 8, Flt3L-stimulated bone marrow cultures were analysed. As shown earlier (Figure 4) Id2-GFP marked the differentiating cDC populations, but not pDCs. Removal of Irf-8 resulted in loss of both the Id2-GFP+CD45RAintSirp-α− DCs that give rise to Id2-GFP+Sirp-α− (CD8α+ equivalent) DCs and CD103+Id2-GFP+ DCs, whereas Batf3 was only required for CD103+Id2-GFP+ DCs (Figure 6A). While, _Batf3_−/− cultures generated normal numbers of Id2-GFP+CD45RAintSirp-α− DCs at day 5, a significant reduction in the frequency of these cells was observed at day 8 (Figure 6B). These findings suggest that Id2-GFP+CD45RAintSirp-α− (CD8α equivalent) DCs are generated in the absence of Batf3 and that this transcription factor may have a critical role in the survival and/or maintenance of these DCs. In line with this, in vivo analysis of the _Id2_gfp/gfp × _Batf3_−/− mice revealed a population of DCs that expressed CD8α at low levels and Id2-GFP expression similar to those found in _Id2_gfp/gfp mice (Supplementary Figure S6). To further explore the mechanism by which Batf3 influences cDC development, we measured proliferation (Figure 6C) and cell death (Figure 6D and E) at 5 and 8 days of DC culture. These data showed no difference in the level of incorporation of BrdU between _Id2_gfp/gfp and _Id2_gfp/gfp × _Batf3_−/− DCs on day 5 or 8 of culture (Figure 6C). Intriguingly, most of the proliferative potential at this point lay in the Id2-GFP+CD45RAintSirp-α− population. In the absence of Batf3, the level of death in the Id2-GFP+CD45RAintSirp-α− population was similar to wild-type controls at day 5 of culture; however by day 8, the proportion of cells expressing Annexin V increased ∼2-fold (Figure 6D). This increase appeared to be specific as other DC subsets analysed in the same experiments showed comparable levels of Annexin V staining between _Id2_gfp/gfp and _Id2_gfp/gfp × _Batf3_−/− DCs.
Figure 5.
Quantitative RT–PCR analysis of the transcription factors Id2, Irf-8 and Batf3 in purified _in vitro-_derived and in vivo DC subsets. CD11c+ DCs purified from Flt3L-stimulated bone marrow on day 5 of culture, or isolated directly ex vivo from spleen or bone marrow were analysed. Data show the mean and s.d. relative to HPRT of two biological replicates each assayed in triplicate for each DC population. _In vitro_-derived DC populations correspond to populations 1–6 as outlined in Figure 4.
Figure 6.
Id2, Irf-8 and Batf3 regulate distinct DC subsets in vitro. (A) Flt3L-derived DCs from Id2_gfp/gfp, Id2_gfp/gfp_Irf-8_−/− and _Id2_gfp/gfp_Batf3_−/− mice were analysed for the development of different DC subsets. Cells were stained for their expression of CD11c, CD45RA and CD103 at days 5 and 8 after initiation of cultures. Data show CD11c+ cells and are representative of at least three independent experiments. (B) Total number of CD11c+ and Id2-GFP+CD45RAintCD103− DCs generated per 1.5 × 106 bone marrow input cells from _Id2_gfp/gfp and Id2 gfp/gfp _Batf3_−/− bone marrow at days 5 and 8 in the presence of 100 ng/ml Flt3L. Data show mean±s.e.m. of six individual cultures for each strain at each time point. (C) Histograms (top panels) showing representative profiles of BrdU incorporation in in vitro generated Id2-GFP+CD45RAintCD103− DCs from _Id2_gfp/gfp and _Id2_gfp/gfp_Batf3_−/− bone marrow on day 5 of culture. Bar graphs show BrdU incorporation for each DC subset. Data show the mean±s.e.m. of cultures from bone marrow of 6–9 individual mice for each strain at days 5 and 8. (D, E) Analysis of Annexin V expression on DCs generated as in (C). Cells were stained for surface markers as indicated before staining for Annexin V expression and analysis by flow cytometry. (D) Graph shows expression on days 5 and 8 showing the mean±s.e.m. of cultures from bone marrow of six individual mice pooled from two independent experiments for each strain at days 5 and 8. (E) Histograms show representative profiles of Annexin V staining on day 8 in total CD11c+ DCs, Id2-GFP+CD45RAint and Id2-GFP+CD45RA− DC subsets derived from Id2 gfp or _Id2_gfp/gfp_Batf3_−/− bone marrow. Statistical differences were determined using a one-tailed Student’s _t_-test.
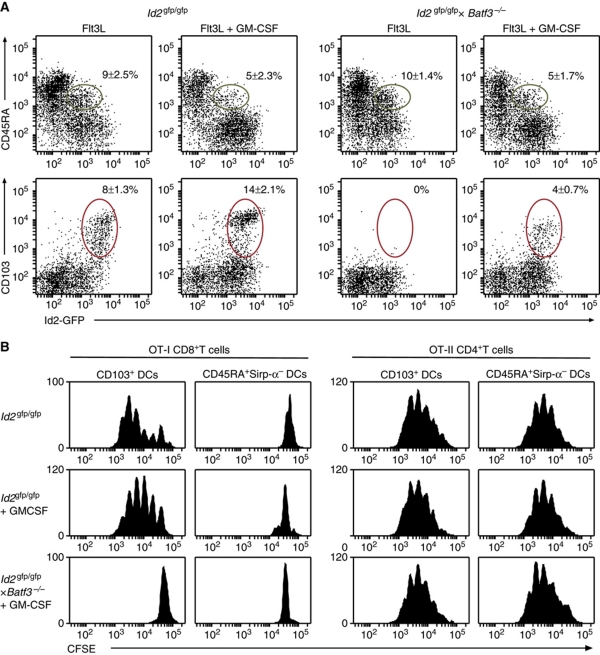
GM-CSF induces CD103 expression in Batf3-deficient DCs but does not restore cross-presentation of cell-associated antigens
Recently, GM-CSF has been implicated in the development of a population of CD103+ DCs (King et al, 2010). To test whether GM-CSF could overcome the failure to develop CD103+ DCs in _Batf3_−/− Flt3L-stimulated cultures, GM-CSF was added to DC cultures 2 days before analysis (Figure 7A). This treatment resulted in an increase in the frequency of CD103+Id2-GFP+ DCs in _Id2_gfp/gfp cultures and the emergence of CD103+Id2-GFP+ DCs from _Id2_gfp/gfp × _Batf3_−/− bone marrow. Next, we tested whether GM-CSF treatment could induce cross-presenting capacity in _Batf3_−/− DCs (Figure 7B). CD103+Id2-GFP+ DCs isolated from _Id2_gfp/gfp cultures exposed to GM-CSF resulted in a two-fold increase in the capacity to amplify OVA-specific CD8+ T cells. In contrast, CD103+Id2-GFP+ DCs isolated from _Id2_gfp/gfp × _Batf3_−/− cultures did not gain the capacity to cross-present cell-associated OVA. Similarly, CD103−Id2-GFP+CD45RAintSirp-α− DCs from exposed to GM-CSF did not gain cross-presenting function regardless of their Batf3 genotype (Figure 7B). Thus, GM-CSF appears to optimize cross-presenting capacity in CD103+Id2-GFP+ DCs but does not overcome defects in cross-presentation in the absence of Batf3 despite the rescue of expression of CD103.
Figure 7.
GM-CSF enhances expression of CD103 but does not restore cross-presenting function in the absence of Batf3. (A) Bone marrow from _Id2_gfp/gfp and _Id2_gfp/gfp × _Batf3_−/− mice was cultured with Flt3L for 5 days. In some cases, GM-CSF was added 2 days before analysis of cultures for the development of different DC subsets. Profiles are gated on CD11c+CD19−NK1.1−CD3− cells. (B) DCs of the indicated genotypes were generated in vitro in response to Flt3L or Flt3L and GM-CSF as described in (A). DCs were flow cytometrically sorted 5 days after initiation of cell culture according to their expression of CD103, CD45RA, Sirp-α and Id2-GFP and analysed for their ability to cross-present cell-associated OVA to CFSE-labelled OVA-specific CD8+ T cells (left panels). The ability of these subsets of present exogenous antigen to CFSE-labelled OVA-specific CD4+ T cells was evaluated as a control (right panels). Data are representative of at least three independent experiments with similar results and show the mean±s.d. (_n_=6–10 per genotype).
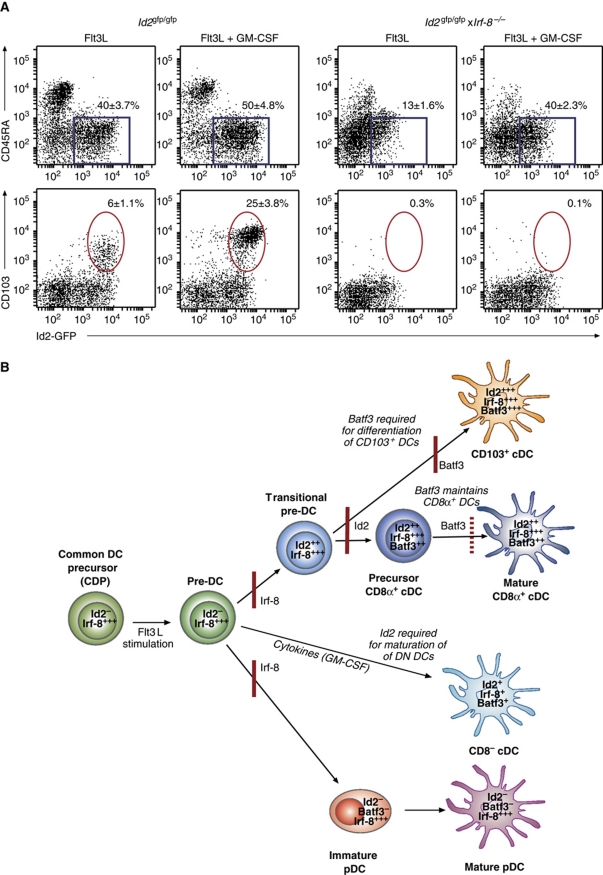
GM-CSF does not amplify CD103+ or Sirp-α− DCs in the absence of Irf-8
Loss of Irf-8 appears to almost completely block CD103+Id2-GFP+ and CD103−Id2-GFP+CD45RAintSirp-α− DC development (Figure 5; Aliberti et al, 2003; Tailor et al, 2008). To determine whether this block in could be overcome by extrinsic signals provided by GM-CSF, _Id2_gfp/gfp × _Irf-8_−/− bone marrow was co-cultured with Flt3L±GM-CSF as described above (Figure 8A). These culture conditions did not induce the generation of either DC subset but Sirp-α+ DCs (Figure 8A; Id2-GFP+CD45RA− DCs) were generated at a similar frequency to _Id2_gfp/gfp cultures. Thus, Irf-8 appears to be a critical regulator CD103+ and CD8α lineages but other Id2-GFP+ cDCs can be induced in response to exogenous growth factors in its absence.
Figure 8.
(A) GM-CSF does not induce CD103+ or Sirp-α− DCs but enhances Id2-GFP+Sirp-α+ DC development in the absence of Irf-8. (A) Bone marrow from _Id2_gfp/gfp and _Id2_gfp/gfp × _Irf-8_−/− mice was cultured with Flt3L for 8 days. In some cases, GM-CSF was added 2 days before analysis of cultures for their expression of CD103, CD45RA and Id2-GFP. Dot profiles are gated on CD11c+NK1.1−CD19−CD3ε−PI− cells. (B) The expression of Sirp-α on Id2-GFP+ cells (indicated by the blue box, panel (A)) is shown. The percent of Sirp-α+ and Sirp-α− cells for CD11c+Id2-GFP+ cells is shown on each plot. Data are representative of at least three independent experiments with similar results and show the mean±s.d. (_n_=6–8 per genotype). (B) Model of differentiation of DCs and the requirement for Id2, Irf-8 and Batf3 in this process. Common DC and pre-DCs do not express Id2 and can be activated by Flt3L stimulation. Irf-8 is required for the generation of pDCs, CD8α+ and CD103+ DCs, but not DN DCs which can be induced to differentiate in the absence of both Id2 or Irf-8 when exposed to cytokines such as GM-CSF. Id2 is induced in differentiating cDCs and is required for the generation of CD8α+ and CD103+ DCs, but not pDCs, found in spleen and peripheral LNs. Batf3 is essential for the generation of CD103+ DCs but is dispensable for the development of precursor CD8α+ DCs which depend on Batf3 for their maintenance and full differentiation into mature cells in spleen and peripheral tissues. Precursor cells—CDP and Pre-DCs—are defined as lin−ckitintFlt3+M-CSF+ and lin−CD11cintFlt3+Sirp-α+MHC II−, respectively, as previously defined (Naik et al, 2006; Liu et al, 2009). These precursor populations do not express Id2 suggesting that a transitional cDC stage (transitional pre-DC) occurs in which Id2 expression is switched on but DCs have not yet adopted their mature phenotypes. Mature DCs have been defined as described in Supplementary Table SI. Data describing the role of Id2 are derived from unpublished experiments in which Id2 has been specifically ablated in CD11c-expressing cells using Cre-mediated deletion (JTJ and GTB.; Ginhoux et al, 2009). The presence of precursor CD8α+, but not mature, CD8α+ DCs are evident by their expression of the CD11c+Id2-GFP+CD45RAintSirp-α− phenotype in the absence of Batf3. The red lines represent where loss of a transcription factor blocks further DC development.
Discussion
This study provides new insights into the transcriptional regulation of DC fate decisions and DC subset formation. Our data show that Irf-8 is critical for the development of CD8α+ and CD103+ DCs and places Id2 upstream of Batf3 in regulating differentiation of each of these subsets, respectively (Figure 8B). In addition, the growth factor GM-CSF can induce phenotypic expression of CD103 in DCs but does not induce cross-presenting capacity in these cells.
Despite the known importance of Irf-8, Id2 and Batf3 in DC development, there is little known about precisely how different transcription factors establish the identity of DC subsets in vivo. This is in part attributable to partial overlap between the functions of genes associated with DC differentiation. Furthermore, in vitro approaches that allow the generation of sufficient numbers of DCs to study DC subset gene regulation or differentiation in detail have relatively low resolution. Engineering reporter capacity into a functional Id2 gene has enabled us to begin to investigate in vivo and in vitro differentiation patterns of DC lineages. A recently developed _Id2_del-GFP mouse has been used to trace lung epithelial cell development from progenitors (Rawlins et al, 2009) though this model has not been investigated for other cell types. Analysis of mRNA has provided some insight to the broader expression of Id2 in DCs (Ginhoux et al, 2009); however, this approach lacks the single cell precision of the Id2-GFP reporter and precludes subsequent analysis of function in live cells. In addition, from our studies, crossing the _Id2_gfp mice with strains deficient in various transcription factors has opened the capacity to identify the checkpoints in DC lineage development both in vitro and in vivo that have not been readily resolved using knockout models.
Here, we have shown that Irf-8 and Batf3 differentially regulate DC subset development (Figures 6 and 8B). Batf3 is required for the generation of LN-derived DCs of the CD103+ and CD8α+ lineages (Hildner et al, 2008; Edelson et al, 2010). Although differing in cell surface phenotype, these two cell types in vivo are almost identical with CD103+ DCs being distinguished from LN CD8α+ DCs by their expression of XCR1 and their enhanced capacity to cross-present exogenous antigens. XCR1, a chemokine receptor expressed on murine splenic CD8α+ DCs (Dorner et al, 2009) together with CD103+ DCs, appears also to define the CD11c+CD141+ human homologues of mouse CD8α+ DCs (Bachem et al, 2010; Crozat et al, 2010; Jongbloed et al, 2010; Poulin et al, 2010). The essential function of Batf3 in vitro was restricted to the CD103+ DC subset with substantial reduction in Sirp-α− DCs (CD8α+ equivalents) observed late during culture. This is consistent with earlier observations of reduced Sirp-α− DCs following Flt3L culture (Hildner et al, 2008) but notably when we enumerated the Sirp-α− splenic precursor cells earlier in culture, they were not significantly impaired. This highlights a previously unrecognized role that transcription factors such as Batf3 might have in maintaining DCs, specifically CD8α+ DCs, in the periphery. Interestingly, Flt3L, an important growth factor for DCs, is also required for peripheral DC proliferation (Waskow et al, 2008).
DCs are critical for the collection and processing of antigens for presentation to lymphocytes. Their role as sentinels guarding different tissue environments in the body has led to the development of an intricate network of distinct DC subsets. The regulation of transcription factors that control the differentiation and survival of DCs is essential for DC development. Our work highlights that the developmental hierarchy of DCs is guided by combinatorial activity of transcription factors that drive specific DC subset formation. Intriguingly, transcription factors such as PU.1, and Ikaros appear to have essential roles for the development of virtually all DCs. Ikaros is required for the development of cDCs and pDCs in line with its essential function in normal development of early haematopoietic progenitors (Nichogiannopoulou et al, 1999). PU.1 is intrinsically required for the generation of all DC lineages from early progenitors and regulates the decision-making process through graded expression of Flt3 (Carotta et al, 2010). In contrast, Id2, Batf3 and Irf-8 have non-redundant subset-specific functions that result in the development of distinct DC lineages. Irf-8 is essential for the development of both pDC and CD8α+ DC family members; Id2 guides the subsequent development of cDCs, while Batf3 is not required for the development of CD8α+ DCs, but is necessary for their survival and is essential for the development of CD103+ DCs (Figure 8B).
Recently, an unexpected role for GM-CSF in promoting differentiation of CD103+ cells was uncovered (King et al, 2010). GM-CSF is a growth factor that promotes differentiation of myeloid cells but also influences the homeostatic development of DCs (Hamilton and Anderson, 2004; Kingston et al, 2009). We found that GM-CSF in combination with Flt3L stimulation could induce the expression of CD103 on the surface of DCs. However, while this enhanced the efficiency of cross-presentation in CD103+ DC, it failed to induce cross-presenting capacity in Id2-GFP+Sirp-α− DCs from wild-type mice or DCs derived from _Batf3_−/− bone marrow. This suggests that extrinsic factors such as GM-CSF may modulate DC differentiation and phenotype during development and enhance antigen presentation to promote T-cell responses (King et al, 2010). Furthermore, it indicates that while _Batf3_−/− mice do possess CD8α+ DCs, the major defect may lie in their intrinsically impaired ability to cross-present antigens combined with the failure of specific DC subsets to differentiate and survive.
In conclusion, we describe an approach that allowed the prospective identification of individual DC populations using the Id2-GFP reporter mouse but which now can be more broadly applied to other mouse strains. Translation of these analyses to our highly defined in vitro system now enables us to investigate the collaborative transcription program of DC differentiation and the plasticity of DC subsets responding to extrinsic signals such as cytokines and inflammatory stimuli. Furthermore, it is now feasible to generate murine DC subsets, which have equivalent human counterparts, in sufficient numbers to permit detailed molecular analysis of their behaviour at steady state and in response to pathogen stimuli. This will shed light on the role of DC network composed of multiple DC subsets with distinct functions required to maintain tolerance or drive immunity.
Materials and methods
Construction of Id2gfp/gfp reporter mice
The Id2 targeting construct used the pKW11 vector consisting of a splice acceptor, stop codons in all reading frames, an IRES, eGFP cDNA, an SV40 polyadenylation signal and a PGK-Neo r gene. Genomic DNA containing LoxP flanked Id2 exons 1–2, containing the entire Id2 coding region was cloned in front of the pKW11 insert. Homology arms of 5794 bp (5′) and 6161 bp (3′) were amplified from an _Id2_-containing BAC and cloned into the final targeting vector. The linear targeting vector was introduced into the Id2 locus by homologous recombination in C57BL/6 ES cells. The targeting vector was inserted so that the 5′ LoxP sites were introduced into the 5′ untranslated region of Id2 at position −71 bp compared with the Id2 start codon. Neomycin-resistant clones were screened by Southern hybridization using 5′ (digested with _Xba_I, giving wild type 8819 kb and Id2 fl-gfp 7255 kb) and 3′ (digested with _Sac_I, giving wild type 12 578 kb and Id2 fl-gfp 8916 kb) probes. Targeted ES cell clones were injected into BALB/c blastocysts to obtain chimeric founders. Germ-line transmission was achieved with two clones, resulting in the generation of two lines. Founders for the reporter lines lacked the 5′ Lox P site and were designated as Id2-reporter (_Id2_gfp/gfp). PCR genotyping was performed using the primer combination: a: 5′-TGCCTATGTGGTAAGTCAAGCGG-3′, b: 5′-GCGGAATTCATTTAATCACCCA-3′, c: 5′-CTCCAAGCTCAAGGAACTGG-3′. Primer combination a/b/c gave PCR fragments of 688 bp (a/c wild type) and 959 bp (a/_b Id2_gfp).
Mice
_Id2_gfp/gfp, _Id2_gfp/+, C57BL/6, B6.CH-2bm-1 (bm1), OT-I (Hogquist et al, 1994), OT-II (Barnden et al, 1998), _Batf3_−/− (Hildner et al, 2008), _Irf-8_−/− (Holtschke et al, 1996) and _Itgae_−/− (Schon et al, 2000) mice were used at 6–8 weeks. All mice were bred and maintained under specific pathogen-free conditions at the Walter and Eliza Hall Institute animal breeding facility according to institute guidelines.
Bone marrow cultures
Bone marrow cells were extracted, and erythrocytes were removed by brief exposure to 0.168 M NH4Cl. Cells were cultured at a density of 1.5 × 106 cells/ml in mouse osmolarity RPMI 1640 medium with 10% (vol/vol) fetal bovine serum containing mouse Flt3L (100 ng/ml, Peprotech) at 37°C in 10% CO2 (Naik et al, 2005). In some cases, GM-CSF (200 ng/ml, R&D Systems) was added to FLT3L cultures 2 days before analysis.
Isolation of DC and DC precursors from spleen, thymus, LN and bone marrow
Spleens or LNs were chopped, then were digested for 20 min at 25°C with collagenase and DNase then treated for 5 min with EDTA to disrupt T cell–DC complexes (Vremec et al, 2000; Henri et al, 2001). Light-density cells were isolated from cell suspensions using Nycodenz density (spleen 1.076 g/cm3, LN 1.082 g/cm3 and thymus 1.086 g/cm3) and were centrifuged at 1700 g for 10 min at 4°C. The light-density cells were collected, counted and washed, then depleted of non-DC lineages before staining for analysis (Henri et al, 2001). Precursor DCs were isolated from BM and spleen as previously described (Liu and Nussenzweig, 2010).
Flow cytometric staining
DC was stained with varying combinations of mAbs to CD11c (HL3), CD45RA (14.8), CD11b (M1/70), CD24 (M1/69), CD4 (GK1.5), CD8 (53-6.7), signal regulatory protein (SIRP)-α (p84), CD103 (M290), CD24 (M1/69), MHC II (M5/114) and Ly5.2 (104). Cells not of the DC lineage and dead cells were excluded by staining for CD19 (ID3), NK1.1 (NKR.PIC), CD3ε (17A2) and propidium iodide. Some samples were stained with anti-Annexin V (BD Biosciences) according to the manufacturer's instructions. Cell sorting and analysis were performed on a FACSVantageSE DiVa, LSRII or FACScan instruments (BD Instruments).
OVA-coated spleen cells
To prepare cell-associated OVA, bm1 spleen cells were γ-irradiated (1000 rads), washed and incubated with 10 mg/ml OVA in RPMI 1640 medium for 10 min at 37°C then washed (Li et al, 2001).
Antigen presentation to naive T cells in vitro
In all, 2–2.5 × 104 purified DC subsets were washed and resuspended in 200 μl mouse tonicity complete RPMI 1640 medium containing 2 × 105 OVA-coated irradiated bm1 splenocytes and 1 × 105 CFSE-labelled OT-I or OT-II T cells. After 60 h culture, T cells were stained for CD8α or CD4 and proliferation was analysed by flow cytometry as previously described (Belz et al, 2002).
Proliferation of DCs
At 5 and 7 days after the initiation of Flt3L-stimulated bone marrow cultures from _Id2_gfp/gfp and _Id2_gfp/gfp × _Batf3_−/− mice, cells were pulsed with BrdU (3 mg/ml) for 4 h and DC subsets identified as previously described by staining for CD11c, CD45RA and CD103. Cells were fixed with 2% paraformaldehyde and permeabilized 0.1% Tween-20 for 72 h, treated with 100 μg/ml DNase (Sigma) for 2 h at 37°C and stained with anti-BrdU mAb (Invitrogen) for 40 min then analysed by flow cytometry.
Real-time PCR analysis
Total RNA was prepared from flow cytometrically purified DCs using an RNeasy kit (Qiagen). cDNA was synthesized from total RNA with random hexamers and SuperScript III reverse transcriptase (Invitrogen) or oligo dT using Thermoscript (Invitrogen) RT–PCR system. Real-time PCR of Hprt, GFP and Id2 was performed using the QuantiTect SYBR Green PCR kit (Qiagen) or GoTaq qPCR Master Mix (Promega) as per manufacturers’ instructions combined with both the forward and reverse primers for the gene of interest and were measured using the ABI7900HT (Applied Biosystems). Primers for Id2 were obtained from Applied Biosystems Taqman Gene Expression Assays (Probe Mm_00711781_m1) or as described below. Primer sequences are as follows:
Irf8:
5′-CAG GAG GTG GAT GCT TCC ATC-3′5′-GCA CAG CGT AAC CTC GTC TTC-3′;
Batf3:
5′-CAG AGC CCC AAG GAC GATG-3′5′-GCA CAA AGT TCA TAG GAC ACA GC-3′;
Id2:
5′-ATG AAA GCC TTC AGT CCG GTG-3′5′-AGC AGA CTC ATC GGG TCGT-3′;
Hprt:
5′-GGG GGC TAT AAG TTC TTT GC-3′5′-TCC AAC ACT TCG AGA GGT CC-3′;
GFP:
5′-AGT CCG CCC TGA GCA AAG A-3′5′-TCA CGA ACT CCA GCA GGA CC-3′.
Analyses were performed in triplicate and the mean normalized expression calculated using the Q-Gene application with Hprt serving as a reference gene.
Supplementary Material
Supplementary Information
Review Process File
Acknowledgments
We thank Ken Murphy for providing the _Batf3_−/− mice; Frank Battye, Vicki Lapatis and David Baloyan for cell sorting; Elaine Major, Tara Carle and Liana Mackiewicz for expert animal care; and Lorraine O’Reilly and Sarah Oraki for technical advice. This work was supported by grants and fellowships from the National Health and Medical Research Council of Australia (GTB, SLN, LW and GKS), the Australian Research Council Future Fellowship (AK), Swiss National Science Foundation (FM), the Sylvia and Charles Viertel Foundation and the Howard Hughes Medical Institute (GTB) and the Pfizer Australia Research Fellowship (SLN).
Author contributions: JTJ, FM, AD, SC, AX, MJC, AMM, AK, LW, SN and GTB designed and performed research; JTJ, JH, CL and GKS analysed gene expression data; GTB and SN prepared an initial draft based on a systematic review of published literature, and discussed the draft with JTJ, FM, AD, SC, AX, MJC, AMM, AK, LW and GKS.
Footnotes
The authors declare that they have no conflict of interest.
References
- Aliberti J, Schulz O, Pennington DJ, Tsujimura H, Reis e Sousa C, Ozato K, Sher A (2003) Essential role for ICSBP in the in vivo development of murine CD8alpha + dendritic cells. Blood 101: 305–310 [DOI] [PubMed] [Google Scholar]
- Allan RS, Smith CM, Belz GT, van Lint AL, Wakim LM, Heath WR, Carbone FR (2003) Epidermal viral immunity induced by CD8alpha+ dendritic cells but not by Langerhans cells. Science 301: 1925–1928 [DOI] [PubMed] [Google Scholar]
- Allenspach EJ, Lemos MP, Porrett PM, Turka LA, Laufer TM (2008) Migratory and lymphoid-resident dendritic cells cooperate to efficiently prime naive CD4 T cells. Immunity 29: 795–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachem A, Guttler S, Hartung E, Ebstein F, Schaefer M, Tannert A, Salama A, Movassaghi K, Opitz C, Mages HW, Henn V, Kloetzel PM, Gurka S, Kroczek RA (2010) Superior antigen cross-presentation and XCR1 expression define human CD11c+CD141+ cells as homologues of mouse CD8+ dendritic cells. J Exp Med 207: 1273–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnden MJ, Allison J, Heath WR, Carbone FR (1998) Defective TCR expression in transgenic mice constructed using cDNA-based alpha- and beta-chain genes under the control of heterologous regulatory elements. Immunol Cell Biol 76: 34–40 [DOI] [PubMed] [Google Scholar]
- Belz GT, Behrens GM, Smith CM, Miller JF, Jones C, Lejon K, Fathman CG, Mueller SN, Shortman K, Carbone FR, Heath WR (2002) The CD8alpha(+) dendritic cell is responsible for inducing peripheral self- tolerance to tissue-associated antigens. J Exp Med 196: 1099–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belz GT, Smith CM, Eichner D, Shortman K, Karupiah G, Carbone FR, Heath WR (2004) Cutting edge: conventional CD8 alpha+ dendritic cells are generally involved in priming CTL immunity to viruses. J Immunol 172: 1996–2000 [DOI] [PubMed] [Google Scholar]
- Carbone FR, Bevan MJ (1990) Class I-restricted processing and presentation of exogenous cell-associated antigen in vivo. J Exp Med 171: 377–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carotta S, Dakic A, D’Amico A, Pang SH, Greig KT, Nutt SL, Wu L (2010) The transcription factor PU.1 controls dendritic cell development and Flt3 cytokine receptor expression in a dose-dependent manner. Immunity 32: 628–641 [DOI] [PubMed] [Google Scholar]
- Cisse B, Caton ML, Lehner M, Maeda T, Scheu S, Locksley R, Holmberg D, Zweier C, den Hollander NS, Kant SG, Holter W, Rauch A, Zhuang Y, Reizis B (2008) Transcription factor E2-2 is an essential and specific regulator of plasmacytoid dendritic cell development. Cell 135: 37–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crozat K, Guiton R, Contreras V, Feuillet V, Dutertre CA, Ventre E, Vu Manh TP, Baranek T, Storset AK, Marvel J, Boudinot P, Hosmalin A, Schwartz-Cornil I, Dalod M (2010) The XC chemokine receptor 1 is a conserved selective marker of mammalian cells homologous to mouse CD8alpha+ dendritic cells. J Exp Med 207: 1283–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Haan JM, Bevan MJ (2002) Constitutive versus activation-dependent cross-presentation of immune complexes by CD8(+) and CD8(-) dendritic cells in vivo. J Exp Med 196: 817–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorner BG, Dorner MB, Zhou X, Opitz C, Mora A, Guttler S, Hutloff A, Mages HW, Ranke K, Schaefer M, Jack RS, Henn V, Kroczek RA (2009) Selective expression of the chemokine receptor XCR1 on cross-presenting dendritic cells determines cooperation with CD8+ T cells. Immunity 31: 823–833 [DOI] [PubMed] [Google Scholar]
- Dzionek A, Fuchs A, Schmidt P, Cremer S, Zysk M, Miltenyi S, Buck DW, Schmitz J (2000) BDCA-2, BDCA-3, and BDCA-4: three markers for distinct subsets of dendritic cells in human peripheral blood. J Immunol 165: 6037–6046 [DOI] [PubMed] [Google Scholar]
- Edelson BT, Kc W, Juang R, Kohyama M, Benoit LA, Klekotka PA, Moon C, Albring JC, Ise W, Michael DG, Bhattacharya D, Stappenbeck TS, Holtzman MJ, Sung SS, Murphy TL, Hildner K, Murphy KM (2010) Peripheral CD103+ dendritic cells form a unified subset developmentally related to CD8alpha+ conventional dendritic cells. J Exp Med 207: 823–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissmann F, Manz MG, Jung S, Sieweke MH, Merad M, Ley K (2010) Development of monocytes, macrophages, and dendritic cells. Science 327: 656–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux F, Liu K, Helft J, Bogunovic M, Greter M, Hashimoto D, Price J, Yin N, Bromberg J, Lira SA, Stanley ER, Nussenzweig M, Merad M (2009) The origin and development of nonlymphoid tissue CD103+ DCs. J Exp Med 206: 3115–3130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacker C, Kirsch RD, Ju X-S, Heironymous T, Gust TC, Kuhl C, Jorgas T, Kurz SM, Rose-John S, Yokota Y, Zenke M (2003) Transcription profiling identifies Id2 function in dendritic cell development. Nat Immunol 4: 380–386 [DOI] [PubMed] [Google Scholar]
- Hamilton JA, Anderson GP (2004) GM-CSF biology. Growth Factors 22: 225–231 [DOI] [PubMed] [Google Scholar]
- Henri S, Vremec D, Kamath A, Waithman J, Williams S, Benoist C, Burnham K, Saeland S, Handman E, Shortman K (2001) The dendritic cell populations of mouse lymph nodes. J Immunol 167: 741–748 [DOI] [PubMed] [Google Scholar]
- Hildner K, Edelson BT, Purtha WE, Diamond M, Matsushita H, Kohyama M, Calderon B, Schraml BU, Unanue ER, Diamond MS, Schreiber RD, Murphy TL, Murphy KM (2008) Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science 322: 1097–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogquist KA, Jameson SC, Heath WR, Howard JL, Bevan MJ, Carbone FR (1994) T cell receptor antagonist peptides induce positive selection. Cell 76: 17–27 [DOI] [PubMed] [Google Scholar]
- Holtschke T, Lohler J, Kanno Y, Fehr T, Giese N, Rosenbauer F, Lou J, Knobeloch KP, Gabriele L, Waring JF, Bachmann MF, Zinkernagel RM, Morse HC III, Ozato K, Horak I (1996) Immunodeficiency and chronic myelogenous leukemia-like syndrome in mice with a targeted mutation of the ICSBP gene. Cell 87: 307–317 [DOI] [PubMed] [Google Scholar]
- Honda K, Mizutani T, Taniguchi T (2004) Negative regulation of IFN-alpha/beta signaling by IFN regulatory factor 2 for homeostatic development of dendritic cells. Proc Natl Acad Sci USA 101: 2416–2421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa E, Hida S, Omatsu Y, Shimoyama S, Takahara K, Miyagawa S, Inaba K, Taki S (2004) Defective development of splenic and epidermal CD4+ dendritic cells in mice deficient for IFN regulatory factor-2. Proc Natl Acad Sci USA 101: 3909–3914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongbloed SL, Kassianos AJ, McDonald KJ, Clark GJ, Ju X, Angel CE, Chen CJ, Dunbar PR, Wadley RB, Jeet V, Vulink AJ, Hart DN, Radford KJ (2010) Human CD141+ (BDCA-3)+ dendritic cells (DCs) represent a unique myeloid DC subset that cross-presents necrotic cell antigens. J Exp Med 207: 1247–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King IL, Kroenke MA, Segal BM (2010) GM-CSF-dependent, CD103+ dermal dendritic cells play a critical role in Th effector cell differentiation after subcutaneous immunization. J Exp Med 207: 953–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingston D, Schmid MA, Onai N, Obata-Onai A, Baumjohann D, Manz MG (2009) The concerted action of GM-CSF and Flt3-ligand on in vivo dendritic cell homeostasis. Blood 114: 835–843 [DOI] [PubMed] [Google Scholar]
- Li M, Davey GM, Sutherland RM, Kurts C, Lew AM, Hirst C, Carbone FR, Heath WR (2001) Cell-associated ovalbumin is cross-presented much more efficiently than soluble ovalbumin in vivo. J Immunol 166: 6099–6103 [DOI] [PubMed] [Google Scholar]
- Liu K, Nussenzweig MC (2010) Origin and development of dendritic cells. Immunol Rev 234: 45–54 [DOI] [PubMed] [Google Scholar]
- Liu K, Victora GD, Schwickert TA, Guermonprez P, Meredith MM, Yao K, Chu FF, Randolph GJ, Rudensky AY, Nussenzweig M (2009) In vivo analysis of dendritic cell development and homeostasis. Science 324: 392–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merad M, Manz MG (2009) Dendritic cell homeostasis. Blood 113: 3418–3427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount AM, Smith CM, Kupresanin F, Stoermer K, Heath WR, Belz GT (2008) Multiple dendritic cell populations activate CD4+ T cells after viral stimulation. PLoS One 3: e1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik SH, Metcalf D, van Nieuwenhuijze A, Wicks I, Wu L, O’Keeffe M, Shortman K (2006) Intrasplenic steady-state dendritic cell precursors that are distinct from monocytes. Nat Immunol 7: 663–671 [DOI] [PubMed] [Google Scholar]
- Naik SH, Proietto AI, Wilson NS, Dakic A, Schnorrer P, Fuchsberger M, Lahoud MH, O’Keeffe M, Shao QX, Chen WF, Villadangos JA, Shortman K, Wu L (2005) Cutting edge: generation of splenic CD8+ and CD8- dendritic cell equivalents in Fms-like tyrosine kinase 3 ligand bone marrow cultures. J Immunol 174: 6592–6597 [DOI] [PubMed] [Google Scholar]
- Nichogiannopoulou A, Trevisan M, Neben S, Friedrich C, Georgopoulos K (1999) Defects in hemopoietic stem cell activity in Ikaros mutant mice. J Exp Med 190: 1201–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palucka K, Banchereau J, Mellman I (2010) Designing vaccines based on biology of human dendritic cell subsets. Immunity 33: 464–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulin LF, Salio M, Griessinger E, Anjos-Afonso F, Craciun L, Chen JL, Keller AM, Joffre O, Zelenay S, Nye E, Le Moine A, Faure F, Donckier V, Sancho D, Cerundolo V, Bonnet D, Reis ESC (2010) Characterization of human DNGR-1+ BDCA3+ leukocytes as putative equi. J Exp Med 207: 1261–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathinam C, Geffers R, Yucel R, Buer J, Welte K, Moroy T, Klein C (2005) The transcriptional repressor Gfi1 controls STAT3-dependent dendritic cell development and function. Immunity 22: 717–728 [DOI] [PubMed] [Google Scholar]
- Rawlins EL, Clark CP, Xue Y, Hogan BL (2009) The Id2+ distal tip lung epithelium contains individual multipotent embryonic progenitor cells. Development 136: 3741–3745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiavoni G, Mattei F, Borghi P, Sestili P, Venditti M, Morse HC III, Belardelli F, Gabriele L (2004) ICSBP is critically involved in the normal development and trafficking of Langerhans cells and dermal dendritic cells. Blood 103: 2221–2228 [DOI] [PubMed] [Google Scholar]
- Schiavoni G, Mattei F, Sestili P, Borghi P, Venditti M, Morse HC III, Belardelli F, Gabriele L (2002) ICSBP is essential for the development of mouse type I interferon-producing cells and for the generation and activation of CD8alpha(+) dendritic cells. J Exp Med 196: 1415–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schon MP, Schon M, Warren HB, Donohue JP, Parker CM (2000) Cutaneous inflammatory disorder in integrin alphaE (CD103)-deficient mice. J Immunol 165: 6583–6589 [DOI] [PubMed] [Google Scholar]
- Steinman RM, Idoyaga J (2010) Features of the dendritic cell lineage. Immunol Rev 234: 5–17 [DOI] [PubMed] [Google Scholar]
- Suzuki S, Honma K, Matsuyama T, Suzuki K, Toriyama K, Akitoyo I, Yamamoto K, Suematsu T, Nakamura M, Yui K, Kumatori A (2004) Critical roles of interferon regulatory factor 4 in CD11bhighCD8alpha- dendritic cell development. Proc Natl Acad Sci USA 101: 8981–8986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tailor P, Tamura T, Morse HC III, Ozato K (2008) The BXH2 mutation in IRF8 differentially impairs dendritic cell subset development in the mouse. Blood 111: 1942–1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura T, Tailor P, Yamaoka K, Kong HJ, Tsujimura H, O’Shea JJ, Singh H, Ozato K (2005) IFN regulatory factor-4 and -8 govern dendritic cell subset development and their functional diversity. J Immunol 174: 2573–2581 [DOI] [PubMed] [Google Scholar]
- Vremec D, Pooley J, Hochrein H, Wu L, Shortman K (2000) CD4 and CD8 expression by dendritic cell subtypes in mouse thymus and spleen. J Immunol 164: 2978–2986 [DOI] [PubMed] [Google Scholar]
- Waskow C, Liu K, Darrasse-Jeze G, Guermonprez P, Ginhoux F, Merad M, Shengelia T, Yao K, Nussenzweig M (2008) The receptor tyrosine kinase Flt3 is required for dendritic cell development in peripheral lymphoid tissues. Nat Immunol 9: 676–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Liu YJ (2007) Development of dendritic-cell lineages. Immunity 26: 741–750 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Review Process File