A critical role for TCF-1 in T-lineage specification and differentiation (original) (raw)
. Author manuscript; available in PMC: 2012 Feb 4.
Published in final edited form as: Nature. 2011 Aug 3;476(7358):63–68. doi: 10.1038/nature10279
Abstract
The vertebrate thymus provides an inductive environment for T-cell development. Within the thymus, Notch signals are indispensable for imposing the T-cell fate on multipotential hematopoietic progenitors, but the downstream effectors that impart T-lineage specification and commitment are not well understood. Here we show that transcription factor, T-cell factor 1 (TCF-1), is a critical regulator in T-cell specification. TCF-1 is highly expressed in the earliest thymic progenitors, and its expression is upregulated by Notch signals. Most importantly, when TCF-1 is forcibly expressed in BM progenitors, it drives the development of T-lineage cells in the absence of T-inductive Notch1 signals. Further characterization of these TCF-1-induced cells revealed expression of many T-lineage genes, including T-cell specific transcription factors Gata3, Bcl11b, and components of the T-cell receptor. Our data suggest a model where Notch signals induce TCF-1, and TCF-1 in turn imprints the T-cell fate by upregulating expression of T-cell essential genes.
Within the thymus, Notch1 signals drive development through sequential steps during which alternative lineage potentials are lost and T-lineage specific gene expression (specification) occurs1–4. Notch is necessary for early T-cell development but its downstream effectors remain unclear5–7. We found that HMG box transcription factor, TCF-1, is highly upregulated in early thymic progenitors (ETPs)(Fig. 1a). Indeed, TCF-1 expression is upregulated when progenitors are exposed to Notch1 signals8.
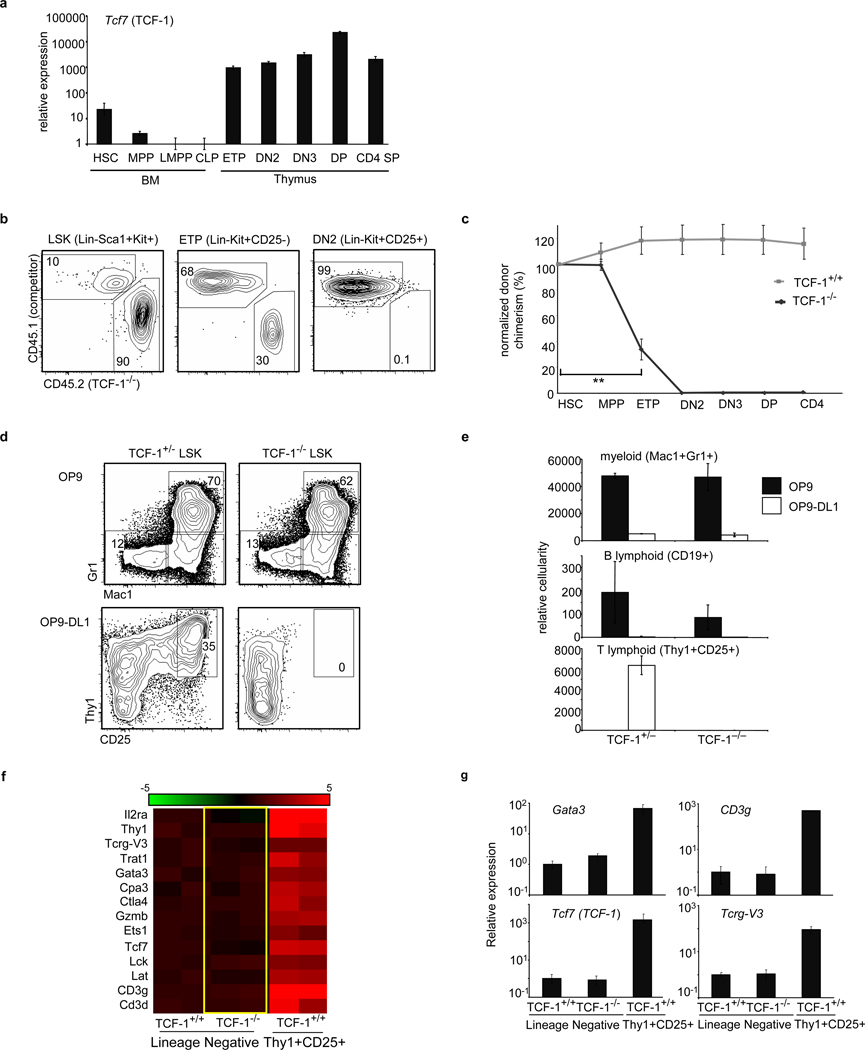
Figure 1. TCF-1 is necessary for early T-lineage development and specification.
a, TCF-1 gene expression in BM, thymic progenitors and T-cells. Expression is shown relative to 18sRNA and LMPP. b, Mixed BM chimeras were generated using TCF-1−/− BM and wt BM. c, Chimerism of TCF-1−/− cells was normalized to HSC (4 mice/group; 3 independent experiments), **p<0.005. d, TCF-1+/− and TCF-1−/− LSK progenitors were seeded onto OP9 and OP9-DL1 stroma and analyzed for myeloid (Mac1+Gr1+) and T development (Thy1+CD25+). e, Cellularity of d6 cultures, including B (CD19+) is shown. f, TCF-1−/− and TCF-1+/+ LMPPs were seeded onto OP9-DL4 and lineage-negative cells from TCF-1+/+ and TCF-1−/− cultures were harvested at day 4 for gene expression. The right side of panel. f, corresponds to T lineage cells made from normal progenitors at day 4 in culture. Lineage-negative cells from these early cultures retain progenitor activity8. Heat map shows a selection of T-lineage genes enhanced greater than 2 fold from TCF-1+/+ lineage-negative cells and represents the log2 value of normalized signal level. Rows represent two independent samples for each population. g, QRT-PCR validation of selected genes. All error bars, mean +/− s.e.m.
TCF-1 in normal T-lymphopoiesis
TCF-1 deficiency greatly reduces thymic cellularity but does not abrogate T-cell development9–11(Fig. S1). When TCF-1−/− progenitors were assessed in the absence of competition in irradiated mice, small numbers of T-lineage cells developed (Fig. S2a). The related transcription factor LEF-1 can compensate for TCF-112; consistently, TCF-1−/− DN3 cells exhibited elevated LEF-1 expression (Fig. S2d). To more rigorously examine requirements for TCF-1 in early progenitors, we placed TCF-1−/− progenitors in competition with wild-type (wt) cells in mixed BM chimeras. TCF-1−/− progenitors reconstituted BM progenitor populations but were defective in generating ETPs, and downstream thymic populations were almost entirely absent (Fig. 1b,c). These data indicated a dramatic requirement for TCF-1 at very early stages of T-cell development, which was clearly revealed when TCF-1-deficient progenitors were placed in competition with TCF-1-sufficient cells.
To more precisely elucidate the role of TCF-1 in early T-cell development, we used stromal cells expressing Notch ligands (OP9-DL4 or OP9-DL1). In this system, hematopoietic progenitors that respond to Notch signals differentiate into immature Thy1+CD25+ T-lineage cells13,14. Both TCF-1+/− and TCF-1−/− lymphoid-primed multipotent progenitors (LMPPs) generated myeloid and B-lineage cells on control OP9 stroma and these fates were appropriately inhibited when progenitors were signaled through Notch. On OP9-DL1 stroma, however, TCF-1−/− progenitors failed to give rise to T-lineage cells (Fig. 1d,e), even when the survival factor Bcl-xL was ectopically expressed (Fig. S3a). Hence TCF-1 is dispensable for initial Notch1-mediated inhibition of alternative fates but is involved in promoting the T-cell fate.
To better examine the requirement for TCF-1 in promoting T-cell development, we cultured TCF-1−/− and TCF-1+/+ LMPPs on OP9-DL4 for four days and performed global gene expression analysis on TCF-1−/− and TCF-1+/+ lineage-negative precursors as well as TCF-1+/+ Thy1+CD25+ T-lineage cells. We found that TCF-1−/− progenitors failed to upregulate expression of many T-lineage genes (Fig. 1f, g). Both TCF-1+/+ and TCF-1−/− progenitors upregulated expression of Notch target genes Deltex1 and Hes1 (Fig. S4), confirming that TCF-1-deficient progenitors sense Notch signals, but cannot upregulate expression of T-cell genes.
TCF-1 drives early T-cell development
To investigate the possibility that TCF-1 initiates T-lineage gene expression, we ectopically expressed human TCF-1 in LMPPs. T-lineage cells were observed from TCF-1 expressing wt LMPPs on OP9-DL4 stroma, as expected; and ectopic TCF-1 rescued T cell development from TCF-1−/− progenitors (Fig. 2a and S3b). Ectopic TCF-1 and Notch1 signals together enhanced T-cell development (Fig. S5). However, when TCF-1-expressing progenitors were placed on OP9 stromal cells lacking Notch ligands, we also observed the development of T-lineage cells; this population was absent from progenitors transduced with control virus cultured on OP9 stroma (Fig. 2a). Ectopic expression of TCF-1 also efficiently inhibited the development of B-lineage but not myeloid cells (Fig. 2b). However, because Notch signals efficiently inhibited the development of B cells from TCF-1−/− progenitors (Fig. 1e), other mechanisms apart from TCF-1 to enforce lineage commitment must exist.
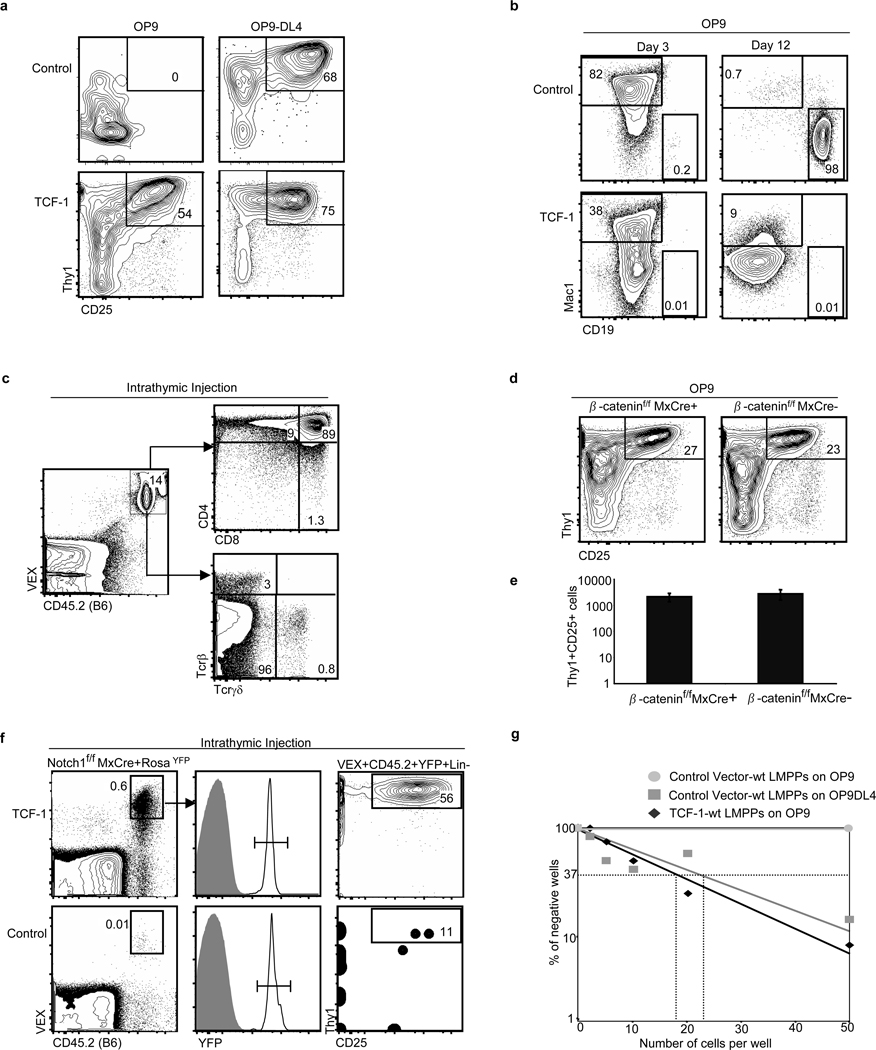
Figure 2. Ectopic expression of TCF-1 elicits T-lineage cells in vitro.
a, Wt LMPPs were transduced with control MSCV-VEX or MSCV containing human TCF-1 (MSCVTCF-1-VEX). Transduced cells were isolated by cell sorting, and seeded onto OP9 or OP9-DL4. Plots are gated on VEX+CD45.2+ Mac1−Gr1− cells, shown on day 12. b, On OP9 stroma, TCF-1-expressing progenitors gave rise to myeloid cells (Mac1+Gr1+), shown on day 3, but TCF-1 inhibited the development of CD19+ B-cells, shown on day 12. c, TCF-1 expressing Thy1+CD25+ cells were isolated from OP9 cultures after 8 days and injected intrathymically into congenic recipients. Shown is 19 days post injection. d, β-cateninf/fMxCre+ and control mice were induced with poly(I:C) and LMPPs were isolated, transduced with MSCV-TCF-1-VEX, and 2000 transduced cells were seeded/well on OP9 stroma and analyzed at day 7 and 12. Plots are gated on VEX+CD45.2+ Mac1−Gr1− cells, shown on day 12. e, Relative cellularity of day 7 cultures. Results represent triplicates +/− SD. f, Notch1f/fMxCre+RosaYFP/+ mice were induced with poly(I:C) and YFP+ LSK progenitors were isolated and transduced with TCF-1 or vector control and intrathymically injected into sublethally irradiated recipients. Shown is day 10 analysis, 2 independent experiments, 4–6 mice per experiment. Frequency of donor-derived TCF-1-expressing Thy1+CD25+ cells compared to control, _p_=0.03. g, Limiting dilution analysis was performed on TCF-1-expressing LMPPs co-cultured with OP9 stroma; this was compared to LMPPs cultured on OP9-DL4 stroma. Frequencies of lineage-competent cells were similar (TCF-1-expressing Thy1+CD25+ lineage on OP9: 1 in 17 [95% confidence interval 1 in 12–26], control Thy1+CD25+ lineage on OP9-DL4: 1 in 23 [95% confidence interval 1 in 15–34].)
We further investigated the TCF-1 mediated generation of Thy1+CD25+ cells on OP9 stroma. These cells appeared early and expanded in number over time. They expressed surface markers of Double-negative (DN) 2 and DN3 pro-T cell stages. A different retroviral vector that expresses TCF-1 at lower levels failed to generate Thy1+CD25+ cells, indicating a threshold level of TCF-1 expression is necessary. The generation of Thy1+CD25+cells was unaffected by inhibitors of Notch signaling (Fig. S6). When injected intrathymically, these cells completed T-cell differentiation, reconstituting both αβ and γδ T-cell lineages (Fig. 2c).
TCF-1 can function with β-catenin to mediate canonical Wnt signaling; however, deletion of β-catenin does not affect T-cell development15,16. Consistently, the generation of Thy1+CD25+ cells was unaffected by deletion of β-catenin (Fig 2d,e). Furthermore, ectopic expression of a small molecule inhibitor of β and γ-catenin, ICAT17, had no effect on the generation of TCF-1 expressing Thy1+CD25+ cells, demonstrating that TCF-1 is not acting as an effector of canonical Wnt signaling in early T cell development (Fig. S7). Ectopic expression of TCF-1 in long term-HSCs but not in myeloerythroid progenitors resulted in development of T-lineage cells on OP9 stroma, indicating that TCF-1-directed T-lineage development is not restricted to lymphoid-biased progenitors (Fig. S8a), and requires factors absent from committed myeloerythroid progenitors (Fig. S8b). These results indicate TCF-1 is sufficient to induce the development of primitive hematopoietic progenitors into cells phenotypically and functionally resembling early T-cell precursors.
We studied the effects of ectopic expression of TCF-1 in vivo. When TCF-1-expressing progenitors were injected intravenously into irradiated mice, we did not observe T-cell leukemia, unlike forced expression of intracellular Notch1 (ICN1) (Fig. S9)18. These data signify that key gene targets of ICN1 that control growth and oncogenesis are not similarly triggered by TCF-1. We next intrathymically injected TCF-1-expressing or control vector-expressing progenitors from Notch1f/fMxCre+RosaYFP/+ mice that had been induced with poly(I:C). TCF-1-expressing progenitors lacking Notch1 gave rise to DN2/3-like Thy1+CD25+ cells whereas control progenitors lacking Notch1 developed into B-lineage cells (Fig. 2d and S10). Hence forced expression of TCF-1 can drive early T-cell development in the absence of Notch1 signals in the thymus.
To investigate the frequency of TCF-1-expressing LMPPs able to give rise to T-lineage cells, we performed limiting dilution analysis with TCF-1-expressing LMPPs on OP9 stromal cells and vector-control expressing LMPPs on OP9-DL4. The frequencies of T-lineage cells developing in these cultures were similar (Fig. 2e). Thus, ectopic TCF-1 generates phenotypic T-cell precursors with frequencies comparable to Notch.
TCF-1 directs T-lineage specification
To understand whether TCF-1 is sufficient to direct a program of T-lineage specific gene expression, we performed global gene expression analysis on TCF-1-expressing Thy1+CD25+ T-lineage cells that developed on OP9 stroma. We found upregulated expression of many T-cell genes, including transcription factors Gata3 and Bcl11b, and T-cell structural genes including components of the T cell receptor (Fig. 3a). Established direct Notch1 gene targets such as Ptcra and Deltex119 failed to be upregulated, confirming that these T-lineage cells arose independently of Notch1 signals (Fig. 3b). QRT-PCR confirmed expression of key T-lineage genes, including Gata3, Bcl11b, CD3g, Lat, Lck, and endogenous Tcf7 (TCF-1) (Fig. 3b). At the time-points examined (d10–d14), expression of some genes in adult TCF-1-expressing Thy1+CD25+ cells was lower than levels in DN3 thymocytes. Fetal liver progenitors exhibit accelerated differentiation in vitro14; consistently, TCF-1-expressing Thy1+CD25+ cells from fetal liver expressed T-cell genes at levels comparable to DN3 thymocytes by day 10 in culture (Fig. S11). However, some genes such as endogenous Tcf7 and CD3g never reached DN3 levels, suggesting additional regulatory inputs. These data indicate ectopic expression of TCF-1 drives expression of many T-cell lineage-specific genes.
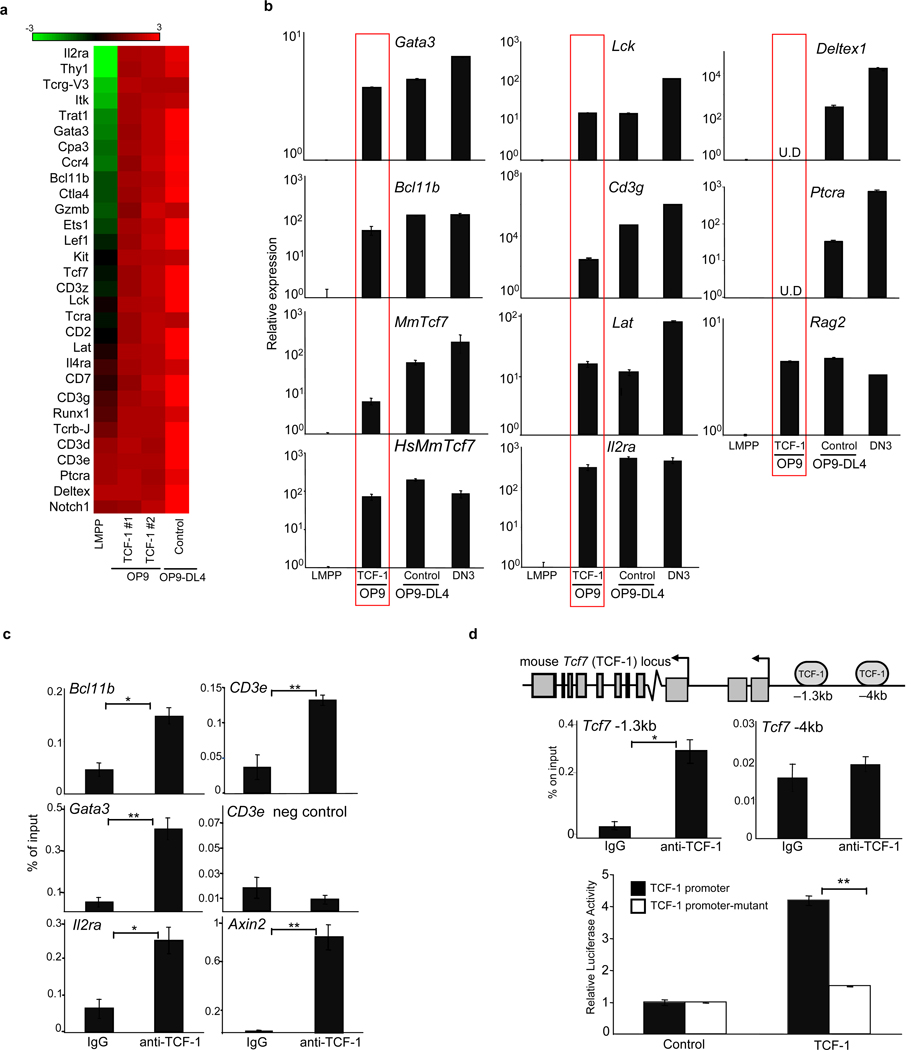
Figure 3. TCF-1 upregulates expression of T-lineage specific genes.
a, Microarray-based analysis of gene expression in TCF-1-expressing Thy1+CD25+ T cells on OP9, control Thy1+CD25+ on OP9-DL4, and LMPPs. Shown are selected T-lineage genes upregulated greater than 2-fold in TCF-1-expressing T-lineage cells. Scale represents the log2 value of normalized signal level. b, QRT-PCR validation of selected genes normalizing to GAPDH and LMPP. U.D=undetectable. c, ChIP on DN thymocytes utilizing TCF-1 or IgG antibodies. QRT-PCR was performed with primers flanking putative TCF-1-binding sites. Axin2 is a positive control and CD3ε negative control refers to region lacking TCF-1 binding sites. d, ChIP as described in (c). e, TCF-1 enhances TCF-1 promoter activity. 293T cells were cotransfected with the pGL3 vector containing the TCF-1 promoter and −1.3kb TCF-1 binding site or a mutated TCF-1 binding site, and with either empty vector or MSCV-TCF-1. Luciferase activity is shown relative to Renilla and normalized to empty vector. All bars are means ± s.e.m of triplicate samples. *p<0.05, **p<0.005.
Analysis of T-lineage genes upregulated upon ectopic TCF-1 expression revealed many to contain evolutionarily conserved TCF-1 binding sites, suggesting a role for TCF-1 in directly regulating these genes. To validate these putative TCF-1 binding sites we performed chromatin immunoprecipitation assay (ChIP) on CD4−CD8− (DN) thymocytes with an antibody against TCF-1. We found TCF-1 was enriched at Gata3, Bcl11b, Il2ra, Cd3ε and TCF-1 itself (Fig. 3C). In addition, T-lineage genes were already upregulated in TCF-1-expressing Lin−Sca1+Kit+ (LSK) progenitors (Fig. S12). Indeed, TCF-1 was initially cloned as a factor enriched at the CD3ε enhancer20 and TCF-1 has also been shown to regulate Gata3 in Th2 cells21. Gata3 is required in ETPs22, which may explain the paucity of ETPs from TCF-1−/− progenitors. Bcl11b is critical for maintenance of T-lineage commitment, as deletion of Bcl11b in committed T-cells results in developmental arrest or diversion to the NK lineage23–25.
Regulation of TCF-1
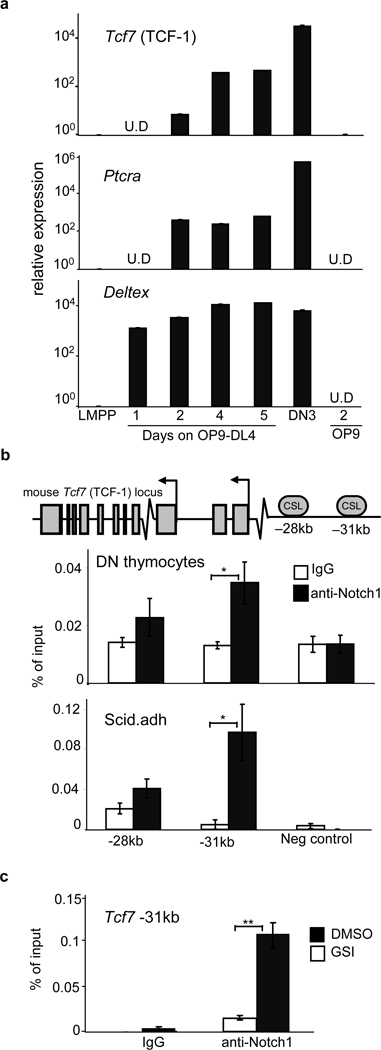
To examine how TCF-1 expression is initially upregulated by Notch signals, we cultured LMPPs on OP9-DL4. We found upregulated TCF-1 expression within 2 days that continued to rise over time, as expected8 (Fig. 4a). ChIP revealed enrichment of Notch1 at a conserved −31kb CSL binding site in DN thymocytes and in “DN3-like” Scid.adh cells (Fig. 4b); this binding was greatly decreased when Notch1 signals were blocked in vitro (Fig. 4c). The −31kb CSL binding site was also active in a reporter assay (Fig S13). These data indicate Notch1 regulates TCF-1 expression.
Figure 4. TCF-1 is expressed in the earliest T cell progenitors and is downstream of Notch1.
a, LMPPs from wt BM were seeded onto OP9-DL4 and Mac1−Gr1− cells were harvested over a five-day period. Relative gene expression of Tcf7 and the Notch target genes Ptcra and Deltex is shown after normalizing to 18sRNA and LMPP. b, TCF-1 locus with conserved putative CSL binding sites. ChIP on DN thymocytes utilizing Notch1 or control IgG antibodies. QRT-PCR was performed with primers flanking putative CSL-binding sites. Shown is the relative percentage of input DNA. c, Scid.adh cells were treated with 1um GSI or DMSO for 6 hours in culture. Cells were subjected to ChIP analysis as in (b). Shown is the relative percentage of input DNA in GSI or DMSO treated cultures. All bars are means ± s.e.m of triplicate samples, *p<0.05, **p<0.005.
Although TCF-1 is initially expressed downstream of Notch1 signals, TCF-1 may also regulate its own expression. TCF-1 binds to the TCF-1 locus (Fig. 3d), and ectopic expression of human TCF-1 is sufficient to induce mouse TCF-1 gene expression (Fig. 3b). Consistently, we found that TCF-1 activates a reporter containing the TCF-1 promoter; mutation of the TCF-1 binding site decreased activation (Fig. 3d). Positive autoregulation may be one mechanism by which TCF-1 remains highly expressed after Notch1 signals cease after the β-selection checkpoint26,27, contributing to the stability of T-cell specific gene expression
Conclusions
In B-cells, a network of transcription factors composed of E47, EBF1, FoxO1 and Pax5 drives B-lineage gene expression28. For T-cells, similar factors were previously unknown; the present study implicates TCF-1 in this role. Our results suggest a model in which TCF-1 is induced by Notch signals in ETPs, and subsequently TCF-1 drives T-cell lineage specification. Among the genes induced by TCF-1 are components of the TCR, as well as T-cell essential transcription factors Gata3 and Bcl11b. TCF-1 likely plays a role in inhibiting the B-cell fate early in T-cell development, although redundant mechanisms to inhibit B-cell development from ETPs must exist29. Additional functions for TCF-1 in T-cell development and function remain to be explored in future work. The present study establishes TCF-1 as a critical regulator that is not only essential for normal T-cell development but is sufficient to establish many components of T-cell identity.
Methods Summary
Mice
Mice were males or females, age 5–18 weeks. C57BL/6 (CD45.2) and B6-Ly5.2 (CD45.1) mice were purchased from the NCI animal facility. Other mice used were _Tcf7_−/− (TCF-1−/− ΔVII) mice9, Notch1f/fMxCre+RosaYFP/+ mice30, and β-cateninf/fMxCre+/− mice31. All live animal experiments were performed according to protocols approved by the Office of Regulatory Affairs of the University of Pennsylvania in accordance with guidelines set forth by NIH.
Intravenous transfers and Intrathymic Injections
Chimeric mice were generated by intravenously injecting T-depleted TCF-1+/+ or TCF-1−/− BM (CD45.2) that was mixed with wt T-depleted BM (CD45.1) at 1:1 or 2:1 ratios into lethally (900 rad) irradiated mice (CD45.1). Mice were analyzed after 12–14 weeks for donor chimerism. Notch1f/fMxCre+RosaYFP/+ LSK progenitors were transduced with TCF-1 or control virus; 24 hours later 2×104 cells were intrathymically injected into sublethally (650rad) irradiated mice (CD45.1). Mice were analyzed 10–16 days later. For intrathymic injections of TCF-1 expressing Thy1+CD25+ cells, cells were isolated by cell sorting from day 8 cultures and 3×105 cells were injected into sublethally irradiated mice and analyzed for thymic reconstitution 1–3 weeks later.
OP9 and OP9-DL cell culture
OP9-GFP (OP9), OP9-DL1, and OP9-DL4 cells were provided by Dr. J.C. Zuniga-Pflucker (University of Toronto, Canada) and used as described13.
Supplementary Material
1
2
3
4
5
6
7
Abbreviations used in this paper
BM
bone marrow
CLP
common lymphoid progenitor
ELP
early lymphoid progenitor
ETP
early thymic progenitor
HSC
hematopoietic stem cell
LSK
lineage marker-negative Sca1+ Kit+
LMPP
lymphoid-primed multipotent progenitor
NK
natural killer
Footnotes
References
- 1.Schwarz BA, et al. Selective thymus settling regulated by cytokine and chemokine receptors. J Immunol. 2007;178(4):2008–2017. doi: 10.4049/jimmunol.178.4.2008. [DOI] [PubMed] [Google Scholar]
- 2.Spangrude GJ, Scollay R. Differentiation of hematopoietic stem cells in irradiated mouse thymic lobes. Kinetics and phenotype of progeny. J Immunol. 1990;145(11):3661–3668. [PubMed] [Google Scholar]
- 3.Doulatov S, et al. Revised map of the human progenitor hierarchy shows the origin of macrophages and dendritic cells in early lymphoid development. Nat Immunol. 11(7):585–593. doi: 10.1038/ni.1889. [DOI] [PubMed] [Google Scholar]
- 4.Rothenberg EV, Zhang J, Li L. Multilayered specification of the T-cell lineage fate. Immunol Rev. 238(1):150–168. doi: 10.1111/j.1600-065X.2010.00964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pui JC, et al. Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity. 1999;11(3):299–308. doi: 10.1016/s1074-7613(00)80105-3. [DOI] [PubMed] [Google Scholar]
- 6.Radtke F, et al. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity. 1999;10(5):547–558. doi: 10.1016/s1074-7613(00)80054-0. [DOI] [PubMed] [Google Scholar]
- 7.Sambandam A, et al. Notch signaling controls the generation and differentiation of early T lineage progenitors. Nat Immunol. 2005;6(7):663–670. doi: 10.1038/ni1216. [DOI] [PubMed] [Google Scholar]
- 8.Taghon TN, David ES, Zuniga-Pflucker JC, Rothenberg EV. Delayed, asynchronous, and reversible T-lineage specification induced by Notch/Delta signaling. Genes Dev. 2005;19(8):965–978. doi: 10.1101/gad.1298305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Verbeek S, et al. An HMG-box-containing T-cell factor required for thymocyte differentiation. Nature. 1995;374(6517):70–74. doi: 10.1038/374070a0. [DOI] [PubMed] [Google Scholar]
- 10.Schilham MW, et al. Critical involvement of Tcf-1 in expansion of thymocytes. J Immunol. 1998;161(8):3984–3991. [PubMed] [Google Scholar]
- 11.Goux D, et al. Cooperating pre-T-cell receptor and TCF-1-dependent signals ensure thymocyte survival. Blood. 2005;106(5):1726–1733. doi: 10.1182/blood-2005-01-0337. [DOI] [PubMed] [Google Scholar]
- 12.Okamura RM, et al. Redundant regulation of T cell differentiation and TCRalpha gene expression by the transcription factors LEF-1 and TCF-1. Immunity. 1998;8(1):11–20. doi: 10.1016/s1074-7613(00)80454-9. [DOI] [PubMed] [Google Scholar]
- 13.Schmitt TM, Zuniga-Pflucker JC. T-cell development, doing it in a dish. Immunol Rev. 2006;209:95–102. doi: 10.1111/j.0105-2896.2006.00353.x. [DOI] [PubMed] [Google Scholar]
- 14.Huang J, et al. Propensity of adult lymphoid progenitors to progress to DN2/3 stage thymocytes with Notch receptor ligation. J Immunol. 2005;175(8):4858–4865. doi: 10.4049/jimmunol.175.8.4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cobas M, et al. Beta-catenin is dispensable for hematopoiesis and lymphopoiesis. J Exp Med. 2004;199(2):221–229. doi: 10.1084/jem.20031615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jeannet G, et al. Long-term, multilineage hematopoiesis occurs in the combined absence of beta-catenin and gamma-catenin. Blood. 2008;111(1):142–149. doi: 10.1182/blood-2007-07-102558. [DOI] [PubMed] [Google Scholar]
- 17.Tago K, et al. Inhibition of Wnt signaling by ICAT, a novel beta-catenin-interacting protein. Genes Dev. 2000;14(14):1741–1749. [PMC free article] [PubMed] [Google Scholar]
- 18.Pear WS, et al. Exclusive development of T cell neoplasms in mice transplanted with bone marrow expressing activated Notch alleles. J Exp Med. 1996;183(5):2283–2291. [Google Scholar]
- 19.Deftos ML, et al. Notch1 signaling promotes the maturation of CD4 and CD8 SP thymocytes. Immunity. 2000;13(1):73–84. doi: 10.1016/s1074-7613(00)00009-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van de Wetering M, Oosterwegel M, Dooijes D, Clevers H. Identification and cloning of TCF-1, a T lymphocyte-specific transcription factor containing a sequence-specific HMG box. EMBO J. 1991;10(1):123–132. doi: 10.1002/j.1460-2075.1991.tb07928.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu Q, et al. T cell factor 1 initiates the T helper type 2 fate by inducing the transcription factor GATA-3 and repressing interferon-gamma. Nat Immunol. 2009;10(9):992–999. doi: 10.1038/ni.1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hosoya T, et al. GATA-3 is required for early T lineage progenitor development. J Exp Med. 2009;206(13):2987–3000. doi: 10.1084/jem.20090934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ikawa T, et al. An essential developmental checkpoint for production of the T cell lineage. Science. 329(5987):93–96. doi: 10.1126/science.1188995. [DOI] [PubMed] [Google Scholar]
- 24.Li L, Leid M, Rothenberg EV. An early T cell lineage commitment checkpoint dependent on the transcription factor Bcl11b. Science. 329(5987):89–93. doi: 10.1126/science.1188989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li P, et al. Reprogramming of T cells to natural killer-like cells upon Bcl11b deletion. Science. 329(5987):85–89. doi: 10.1126/science.1188063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taghon T, et al. Developmental and molecular characterization of emerging beta- and gammadelta-selected pre-T cells in the adult mouse thymus. Immunity. 2006;24(1):53–64. doi: 10.1016/j.immuni.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 27.Yashiro-Ohtani Y, et al. Pre-TCR signaling inactivates Notch1 transcription by antagonizing E2A. Genes Dev. 2009;23(14):1665–1676. doi: 10.1101/gad.1793709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin YC, et al. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat Immunol. 11(7):635–643. doi: 10.1038/ni.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wendorff AA, et al. Hes1 Is a Critical but Context-Dependent Mediator of Canonical Notch Signaling in Lymphocyte Development and Transformation. Immunity. 33(5):671–684. doi: 10.1016/j.immuni.2010.11.014. [DOI] [PubMed] [Google Scholar]
- 30.Liu Z, et al. Notch1 loss of heterozygosity causes vascular tumors and lethal hemorrhage in mice. J Clin Invest. 121(2):800–808. doi: 10.1172/JCI43114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brault V, et al. Inactivation of the beta-catenin gene by Wnt1-Cre-mediated deletion results in dramatic brain malformation and failure of craniofacial development. Development. 2001;128(8):1253–1264. doi: 10.1242/dev.128.8.1253. [DOI] [PubMed] [Google Scholar]
- 32.Huang J, et al. Pivotal role for glycogen synthase kinase-3 in hematopoietic stem cell homeostasis in mice. J Clin Invest. 2009;119(12):3519–3529. doi: 10.1172/JCI40572. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2
3
4
5
6
7