Homeodomain and winged-helix transcription factors recruit activated Smads to distinct promoter elements via a common Smad interaction motif (original) (raw)
. 2000 Feb 15;14(4):435–451.
Abstract
We have investigated the regulation of the activin-inducible distal element (DE) of the Xenopus goosecoid promoter. The results show that paired-like homeodomain transcription factors of the Mix family, Mixer and Milk, but not Mix.1, mediate activin/TGF-β-induced transcription through the DE by interacting with the effector domain of Smad2, thereby recruiting active Smad2/Smad4 complexes to the Mixer/Milk-binding site. We identify a short motif in the carboxyl termini of Mixer and Milk, which is demonstrated to be both necessary and sufficient for interaction with the effector domain of Smad2 and is required for mediating activin/TGF-β-induced transcription. This motif is not confined to these homeodomain proteins, but is also present in the Smad2-interacting winged-helix proteins Xenopus Fast-1, human Fast-1, and mouse Fast-2. We demonstrate directly that transcription factors of different DNA-binding specificity recruit activated Smads to distinct promoter elements via a common mechanism. These observations, together with the temporal and spatial expression patterns of Mixer and Milk, lead us to propose a model for mesoendoderm formation in Xenopus in which these homeodomain transcription factor/Smad complexes play a role in initiating and maintaining transcription of target genes in response to endogenous activin-like signals.
Keywords: Activin, homeodomain, Smad, TGF-β, transcriptional regulation, Xenopus
The TGF-β superfamily is a group of secreted signaling molecules produced by cells to influence the behavior of their neighbors, by regulating cell proliferation, survival, adhesion, differentiation, and specification of developmental fate (Massagué 1998). Signals from TGF-β family members are transduced to the nucleus by members of a family of intracellular proteins called Smads (Massagué 1998). Upon ligand stimulation, receptor-activated Smads are phosphorylated directly by specific type I receptor kinases and form activated complexes with a common mediator Smad (Massagué 1998; Howell et al. 1999). These complexes translocate to the nucleus where they regulate transcription of target genes (Whitman 1998). However, Smads bind DNA weakly by themselves (Shi et al. 1998) and are primarily recruited to DNA by other DNA-binding transcription factors (Derynck et al. 1998; Whitman 1998). The prototype is the winged-helix/forkhead protein Fast-1, which forms a complex with activated Smad2/Smad4 in response to activin that is responsible for activin-induced transcription through the activin-responsive element (ARE) of the Xenopus Mix.2 promoter (Chen et al. 1996, 1997). The transcription factors that cooperate with the Smads are likely to be key determinants of cell type specificity of TGF-β signaling, but are mostly still poorly characterized.
The Xenopus embryo provides an excellent system in which to elucidate the basis of specificity in TGF-β signaling pathways. During Xenopus embryogenesis, TGF-β family members act as morphogens, playing key roles in the patterning of different tissues (Green and Smith 1990; Gurdon et al. 1994; Whitman 1998). For example, an activin-like signal, which requires the maternal transcription factor VegT for its production, is produced in the vegetal hemisphere of the embryo and induces mesoderm in the overlying equatorial cells (Harland and Gerhart 1997; Kimelman and Griffin 1998; Zhang et al. 1998). The same signal is also thought to be responsible for specifying endoderm, and it is likely to be composed of more than one type of activin-related molecule (Henry et al. 1996; Clements et al. 1999; Yasuo and Lemaire 1999). Patterning of the mesoderm and endoderm depends on the precise transcriptional responses of cells within the prospective mesoendoderm to this signal. But what determines which genes are induced in response to this activin-like signal in particular cells, and how is their expression maintained? The presence of other cooperating signaling pathways such as Wnt, FGF, and BMP in different regions of the prospective mesoendoderm could obviously play an important role (for review, see Harland and Gerhart 1997; Whitman 1998). In addition, cell type specificity of signaling could be dictated by the presence of different transcription factors that cooperate with Smads. In fact, the existence in Xenopus embryos of multiple transcription factors, which are capable of recruiting activin-activated Smads and have different DNA-binding specificity, has already been proposed, based on the fact that the activin-responsive elements defined in the promoters of differentially expressed mesoendodermal genes share little sequence similarity (for review, see Howell and Hill 1997).
We have studied the regulation of expression of the Xenopus mesoendodermal gene, goosecoid (Blumberg et al. 1991) as a means of understanding how different signaling pathways restrict expression of a gene to a precise domain in the early embryo. Restriction of goosecoid to the dorsal marginal zone of the early gastrula embryo is thought to result from a synergistic interaction between a Wnt-like signal, which is localized to the dorsovegetal region of the embryo, and an activin-like signal, which is active in the vegetal cells and the marginal zone (Watabe et al. 1995; Laurent et al. 1997). The Wnt-like signal acts through a proximal element (PE) in the goosecoid promoter (Watabe et al. 1995; Laurent et al. 1997) and is mediated by the homeodomain protein Xtwin (Laurent et al. 1997). The activin-like signal acts through a distal element (DE) in the promoter (Watabe et al. 1995; Candia et al. 1997). The transcription factors responsible for activin-induced transcription through the DE are unknown but are obviously distinct from Fast-1, as the sequence of the DE bears no resemblance to the ARE from the Mix.2 promoter (Huang et al. 1995; Watabe et al. 1995). Recently it was suggested that a paired-like homeodomain factor might be involved, based on studies of activin-responsive transcription through a paired-like homeodomain-binding site in the zebrafish goosecoid promoter that is related to a sequence within the Xenopus DE (McKendry et al. 1998). This factor has not yet been identified.
Here we identify a DE-binding protein (DEBP) in Xenopus embryos that is synthesized in response to activin and whose binding to the paired-like homeodomain site in the DE correlates well with activin-induced transcription. We demonstrate that DEBP specifically interacts with the effector domain of the activin-activated Smad Smad2, suggesting it acts as a platform for recruiting Smad2. We go on to show that two members of the Xenopus Mix family of paired-like homeodomain transcription factors, Mixer (Henry and Melton 1998) and Milk (Ecochard et al. 1998) are good candidates for endogenous DEBP. We demonstrate that Mixer forms a ligand-dependent transcription factor complex with endogenous Smad2 and Smad4, and prove that this complex is responsible for TGF-β/activin-induced transcription through the DE. We identify and characterize a short Smad interaction motif in Mixer and Milk, which is also present in Xenopus Fast-1, human Fast-1, and mouse Fast-2, which reveals a common mechanism for Smad recruitment to DNA by transcription factors of different DNA-binding specificity. These observations, together with the expression patterns of Mixer and Milk, lead us to a model for mesoendoderm induction in Xenopus in which homeodomain transcription factor/Smad complexes act as direct regulators of activin/TGF-β responsive transcription.
Results
Activin-induced transcription through the DE of the goosecoid promoter is partly dependent on new protein synthesis
The DE in the Xenopus goosecoid promoter is a _cis_-acting element necessary and sufficient to activate transcription in response to activin (Watabe et al. 1995). To understand the molecular mechanism underlying this, we first investigated activin-stimulated transcription through the DE in animal cap assays, and compared it with the transcriptional response of the Mix.2 ARE, which has a completely different sequence and is known to be controlled by the Fast-1/Smad2/Smad4 complex, ARF (Huang et al. 1995; Chen et al. 1996, 1997). Globin reporter genes were used, with four copies of the DE or three copies of the ARE linked to a minimal promoter, and transcription was measured by RNase protection assay and quantitated relative to the activity of a coinjected constitutively-active reference globin gene (Howell and Hill 1997). The minimal promoter was unresponsive to activin (Fig. 1A, left). The reporter driven by four DEs responded strongly to activin, and some of this induction was lost in the presence of the protein synthesis inhibitor cycloheximide (Fig 1A, middle). In contrast, the ARE gave a much higher basal level of transcription, and the activin induction was weaker. As expected, this induction was completely insensitive to cycloheximide (Fig 1A, right), consistent with it being mediated by the maternal transcription factor complex ARF.
Figure 1.
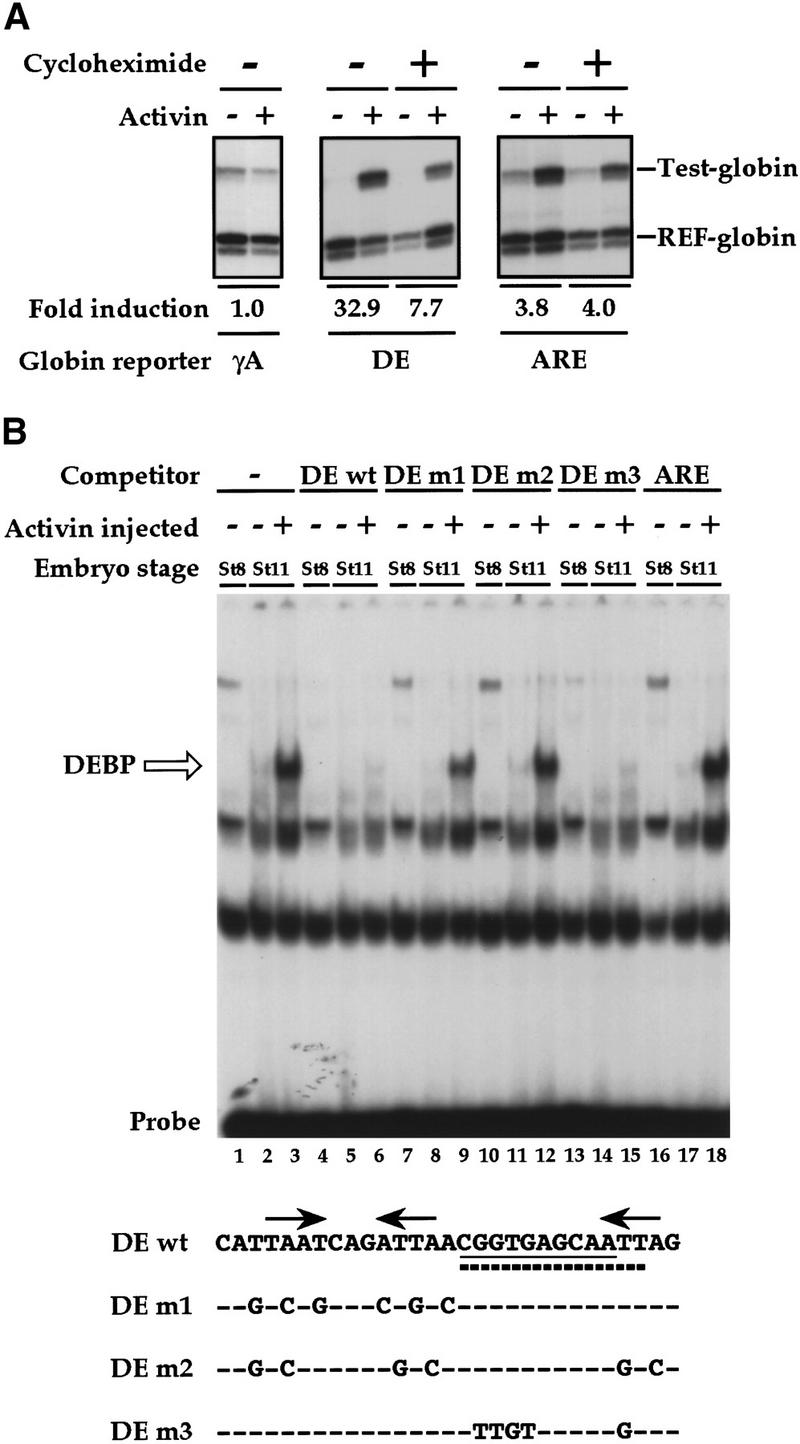
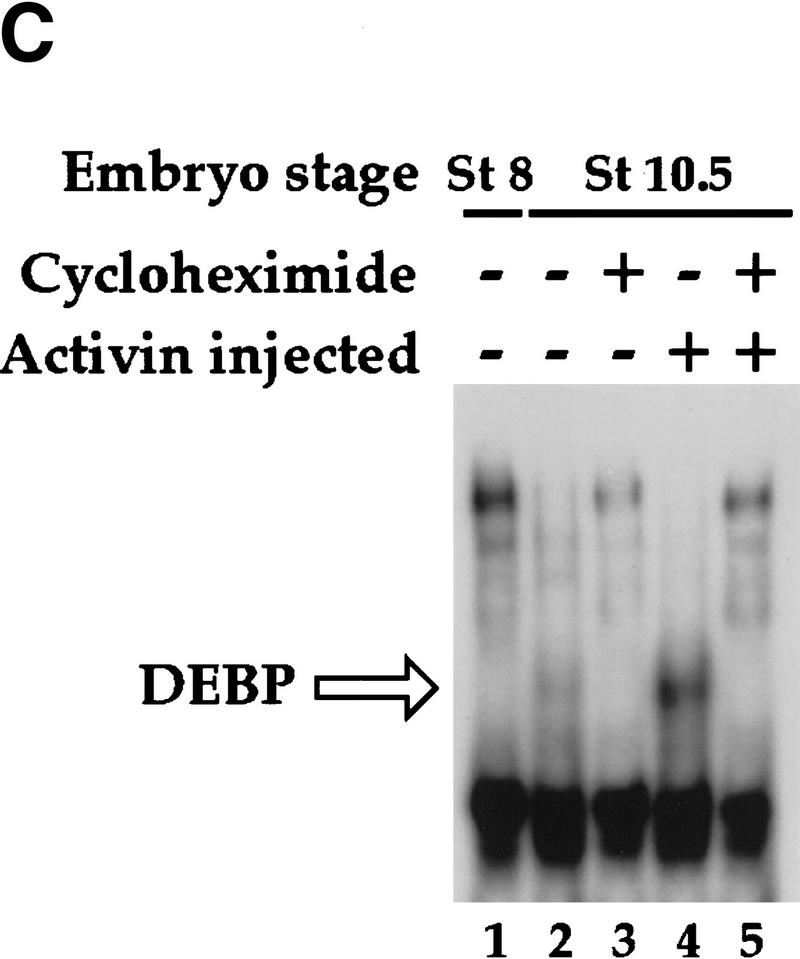
Activin-responsive transcription via the goosecoid DE. (A) Activin-responsive transcription through the DE is partially dependent on new protein synthesis. One-cell embryos were injected with REF–globin internal control together with globin reporters driven by the minimal γ-actin promoter (γA), or by multiple copies of the DE or ARE upstream of the mimimal promoter. Animal caps, cut at stage 8 were cultured for 6 hr with or without activin in the absence or presence of 5 μg/ml cycloheximide. Globin transcripts from reporter genes (Test-globin) or the internal control (REF-globin) were detected by RNase protection (Howell and Hill 1997). Transcriptional activation was calculated as a ratio of the levels of Test-globin to REF-globin. Activin-induced transcription is expressed as fold inductions. (B) An activin-inducible protein (DEBP) binds the goosecoid DE. Whole-cell extracts prepared from stage 8 or stage 11 embryos or stage 11 embryos overexpressing activin, were analyzed by bandshift assay using the single DE probe. Activin-inducible protein DEBP is indicated. Competitor oligonucleotides were used at a 50-fold molar excess over probe where indicated. (Bottom) Sequences of wild-type DE and mutant oligonucleotides indicating only the altered nucleotides. The paired-like homeodomain-binding site comprising two inverted TAAT motifs (Wilson et al. 1993) is denoted by arrows; a third homeodomain core-binding site at the 3′ end is also indicated by an arrow. (Thick dotted line) Sequence reminiscent of a half-site for T-box protein (AGGTGTGAAATT) (Kispert et al. 1995); (underline) almost perfect binding site for the ZFH-1 family of zinc finger homeodomain proteins (AGGTGAGCAA) (Funahashi et al. 1993). (C) Formation of DEBP requires new protein synthesis. Extracts were made from uninjected stage 8 embryos (lane 1), stage 10.5 embryos (lanes 2,3), or stage 10.5 embryos overexpressing activin (lanes 4,5) and analyzed by bandshift using the DE probe. Where indicated, embryos had been preincubated in cycloheximide before stage 8.
From this experiment we conclude that activin-responsive transcription through the DE has two components: a direct induction mediated by maternal factors, which is insensitive to cycloheximide, and a maintenance phase, which requires new protein synthesis. A similar behavior was recently proposed for an activin-responsive sequence in the zebrafish goosecoid promoter, which shares some similarity with the Xenopus goosecoid DE (McKendry et al. 1998).
The goosecoid DE binds an activin-inducible factor DEBP
We used bandshift assays with a single radiolabeled DE oligonucleotide as a probe to identify DE-binding factors in the embryo that might be responsible for the activin-induced transcription. The DE-binding factor that clearly responds to activin is DEBP (Fig. 1B, lanes 1–3, open arrow). It was absent in extracts prepared from stage 8 embryos, which are transcriptionally inactive (lane 1), present at low levels in extracts from stage 11 embryos in which endogenous activin-like signaling pathways are operating (lane 2; Clements et al. 1999), and highly induced in stage 11 embryos overexpressing activin (lane 3). Binding of this factor to the DE was specific as it was competed by excess homologous unlabeled probe (lanes 4–6).
The region of the DE required for activin-responsive transcription is the paired-like homeodomain-binding site at the 5′ end (arrows, Fig. 1B, bottom; data not shown; Watabe et al. 1995). If DEBP was responsible for the activin-induced transcription, we would expect it to bind to this paired-like homeodomain site. This was addressed by performing competitions with various DE mutants (Fig. 1B). DE m1, which is mutated in the paired-like homeodomain binding site (Watabe et al. 1995), competed very poorly for DEBP binding (lanes 7–9), indicating that indeed this site is required. Consistent with this, DE m2, which is additionally mutated in the core homeodomain binding site at the 3′ end of the DE, did not compete for DEBP binding at all (lanes 10–12). The DE also contains sequences at its 3′ end reminiscent of binding sites for T-box proteins and the ZFH-1 family of zinc finger homeodomain proteins (Fig. 1B). These binding sites are mutated in DE m3, which competed efficiently for DEBP-binding (lanes 13–15), indicating that these sequences are not required. The unrelated ARE did not compete for DEBP binding (lanes 16–18).
Because the activin-responsive transcription through the DE is partly dependent on new protein synthesis, we asked whether the activin-inducible DEBP also required new protein synthesis for its formation. Indeed, preincubation of the embryos with cycloheximide before initiation of zygotic transcription, abolished formation of DEBP either in stage 10.5 embryos or stage 10.5 embryos overexpressing activin, as assayed by bandshift (Fig. 1C).
Thus, we have identified an activin-inducible factor DEBP that binds to the paired-like homeodomain-binding site of the DE. Mutations that inhibit DEBP binding also inhibit activin-responsive transcription through the DE (Watabe et al. 1995), strongly implicating DEBP in the transcriptional activation. DEBP is synthesized in response to activin, suggesting that it mediates the maintenance phase of activin-induced transcription of the DE. However, low levels of maternal DEBP could mediate the cycloheximide-insensitive activin-responsive transcription of the DE (McKendry et al. 1998); see below and Discussion). DEBP has properties very similar to those described for Goosecoid activin element binding protein 1 (GAEBP1), which is an activity shown to bind an activin-responsive paired-like homeodomain-binding site in the zebrafish goosecoid promoter (McKendry et al. 1998). They may correspond to the same factor.
The activin-inducible DEBP interacts specifically with Smad2
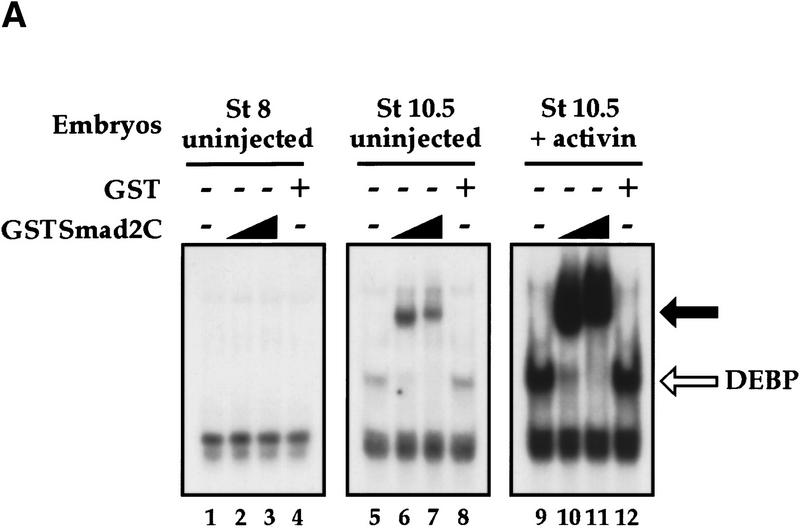
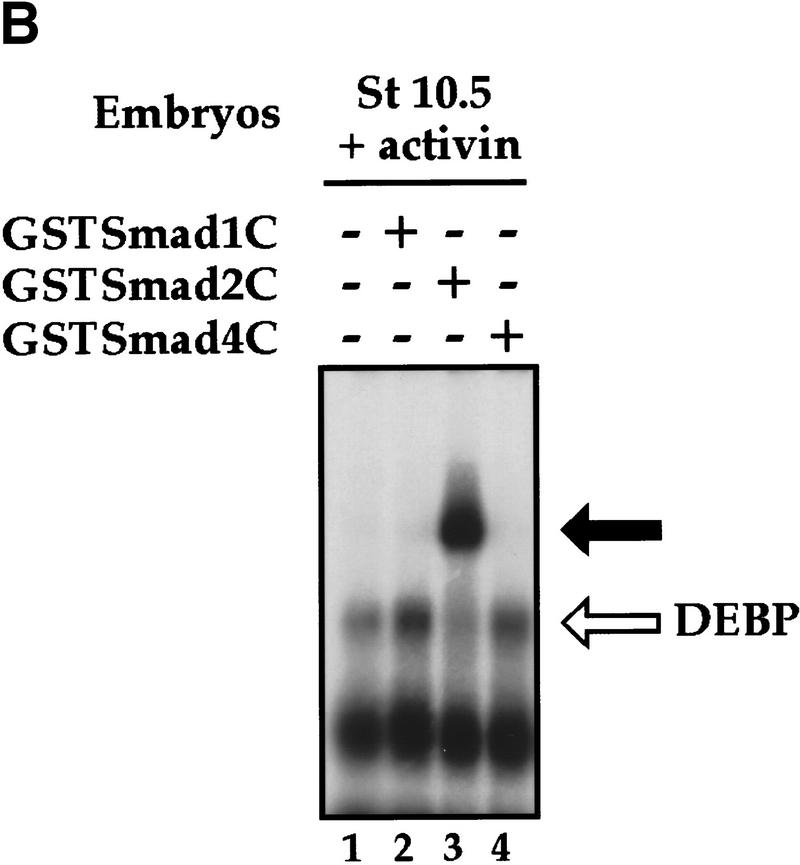
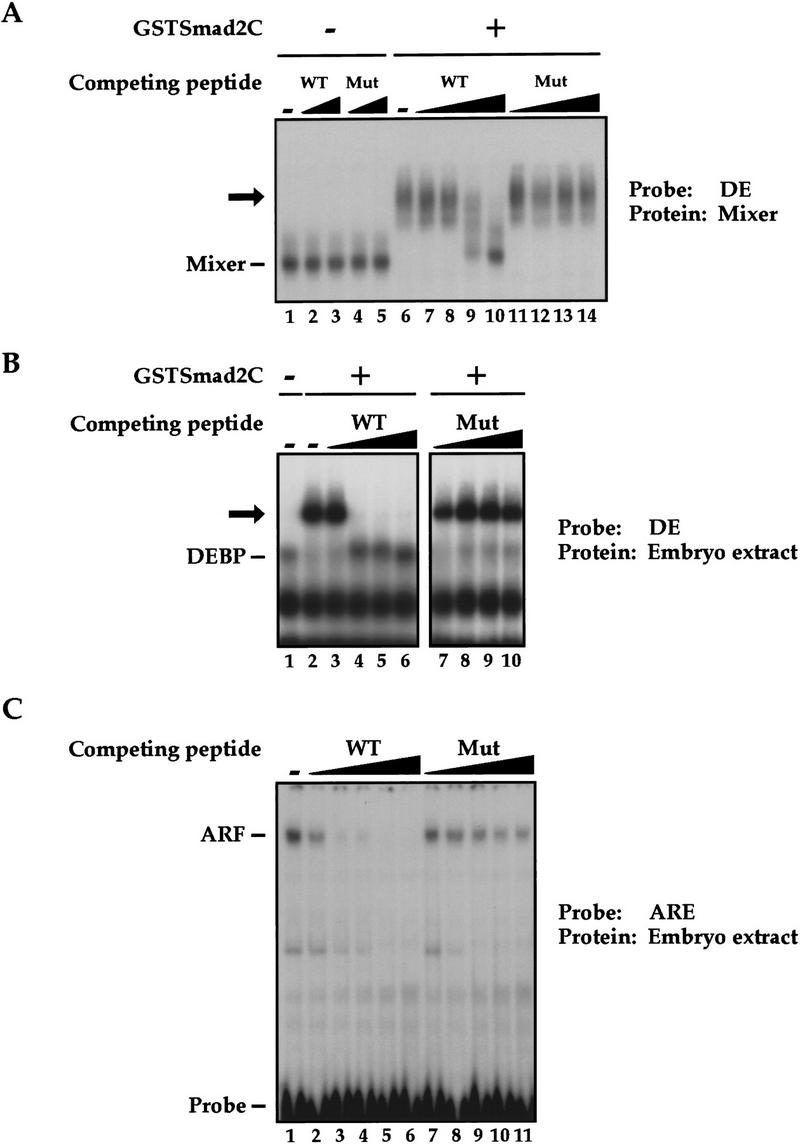
We then investigated how DEBP might function to activate transcription in response to activin. The DEBP complex detected in the bandshift assays does not correspond to a Smad/transcription factor/DNA complex, analogous to the Fast-1/Smad/DNA complex, ARF, as an antibody specific for Smad2 (Nakao et al. 1997) that can supershift ARF (Howell et al. 1999) did not supershift DEBP (data not shown). Another possibility was that DEBP was a transcription factor that recruited activated Smads to DNA. To test this, we investigated whether DEBP could interact with the effector MH2 domain of Smad2 (Smad2C), which is the domain of Smad2 that interacts with Fast-1 in the ARF complex (Chen et al. 1997; Liu et al. 1997). Purified Smad2C, bacterially expressed as a GST fusion protein (GSTSmad2C), bound and stoichiometrically supershifted DEBP generated by the endogenous activin-like signals (Fig. 2A, lanes 5–7) and generated in response to high levels of activin signaling (lanes 9–11). GST alone did not bind DEBP (lanes 8,12). As expected, GSTSmad2C did not bind directly to the DE probe, as seen by the lack of binding activity when added to stage 8 embryo extracts that do not contain DEBP (lanes 1–3). If the ability of DEBP to bind Smad2C reflected a functional interaction involved in activin-induced transcription, we would expect DEBP to interact specifically with activin-activated Smads. We demonstrated this by showing that the effector domain of the BMP-activated Smad1 did not bind DEBP, consistent with the failure of the DE to respond to BMPs (Watabe et al. 1995; Fig. 2B, lane 2). DEBP also did not interact with the effector domain of the common mediator Smad, Smad4 (Fig. 2B, lane 4).
Figure 2.
DEBP specifically interacts with the effector domain of Smad2. (A) Whole-cell extracts prepared from either stage 8 embryos (lanes 1–4), stage 10.5 embryos (lanes 5–8), or stage 10.5 embryos overexpressing activin (lanes 9–12) were analyzed by bandshift using the DE probe. Extracts were mixed with either purified GST protein (100 ng) (lanes 4,8,12) or two concentrations (20 and 100 ng) of purified GSTSmad2C (lanes 2, 3, 6, 7, 10, 11) before addition of probe. (Open arrow) DEBP; (black arrow) GSTSmad2C bound to DEBP. (B) The interaction of DEBP with Smad2C is specific. The assay was as in A using GST fusions of Smad1C, Smad2C, or Smad4C as indicated.
The demonstration that DEBP interacts with the Smad2 effector domain led us to look for an endogenous DEBP/Smad complex. It has proved technically impossible to detect such a complex in extracts made from stage 10.5 Xenopus embryos by bandshift assay (but see below and Discussion).
Paired-like homeodomain proteins Mixer and Milk are good candidates for DEBP
Having established that DEBP is a protein capable of interacting with the Smad2 effector domain, we set out to identify it. DEBP binds the paired-like homeodomain-binding site of the DE, is synthesized in response to activin, and UV cross-linking experiments indicated that it corresponded to a monomer of ∼45–50 kD (data not shown). A group of transcription factors with precisely these properties are the paired-like homeodomain proteins of the Mix family. There are seven known family members: Mix.1 and the highly related Mix.2 (Rosa 1989; Vize 1996), Mixer (Henry and Melton 1998), Milk (Ecochard et al. 1998), also called Bix2 (Tada et al. 1998), and three other Bix genes that are very related to Milk (Tada et al. 1998). They all have molecular masses of ∼44 kD, are first expressed at the mid-to-late blastula stage of Xenopus embryogenesis, and their expression is known to be induced by activin signaling.
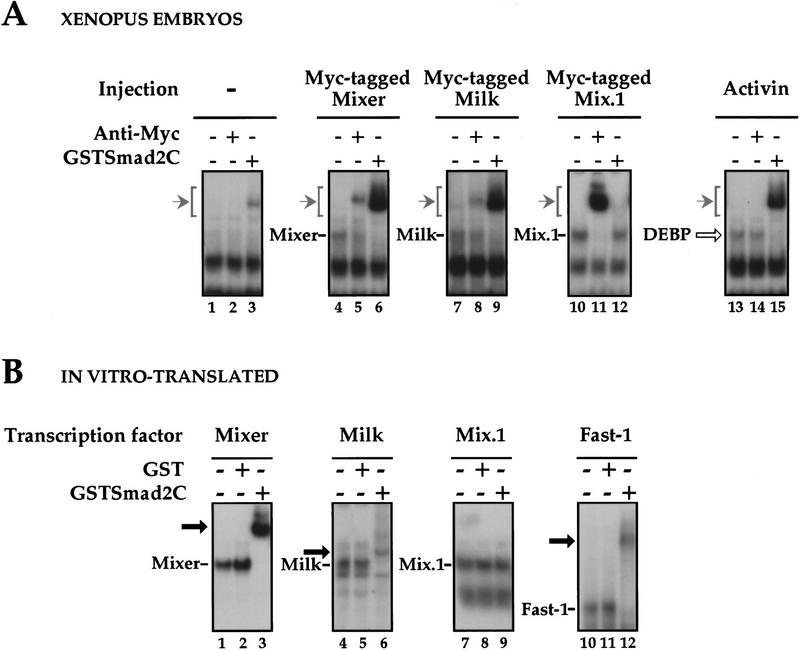
We asked whether three different Mix family members, Mix.1, Mixer, and Milk might be good candidates for DEBP, by investigating whether they shared the same DNA-binding specificity and ability to interact with Smad2C. Expression of myc-tagged Mixer, Milk, or Mix.1, alone in Xenopus embryos gave rise to protein/DNA complexes that comigrated with the activin-induced DEBP (Fig. 3A, cf. lanes 1,4,7,10, with lane 13). These protein/DNA complexes could be supershifted with the anti-myc antibody (lanes 5,8,11) indicating that the myc-tagged proteins are in these complexes. Strikingly, only Mixer and Milk could interact with GSTSmad2C, as shown for endogenous DEBP (cf. lanes 6 and 9 with lanes 3 and 15). Mix.1 could not associate with GSTSmad2C (lane 12).
Figure 3.
Homeodomain proteins, Mixer and Milk, but not Mix.1, interact with Smad2C. (A) Mixer and Milk are good candidates for the endogenous protein DEBP. Whole-cell extracts were prepared from stage 10.5 embryos or embryos injected at the 1-cell stage with mRNA encoding myc-tagged Mixer, Milk, Mix.1, or activin, and DE-binding activity was assayed by bandshift. Anti-myc antibody or purified GSTSmad2C were added where indicated. (Open arrow) DEBP; (gray arrow) supershifted complexes. (B) Interaction of GSTSmad2C with members of the Mix family and Fast-1. In vitro-translated Mixer, Milk, Mix.1, and Fast.1 were assayed by bandshift for their interaction with purified GSTSmad2C or GST using the appropriate radiolabeled DE or ARE probes. Transcription factors complexed with probe are indicated; (black arrow) ternary complex with GSTSmad2C.
An analagous interaction experiment was performed using transcription factors produced in vitro by coupled transcription/translation with identical results (Fig. 3B, lanes 1–9). As shown for DEBP (Fig. 2B), interaction of Mixer and Milk was specific for Smad2C (data not shown). As a control for the supershift bandshift assay we also tested the known Smad2-interacting protein Fast-1 (Chen et al. 1996), which can be supershifted by GSTSmad2C but not by GST alone (Fig. 3B, lanes 10–12).
Thus, using the criteria of DNA-binding specificity and ability to interact with the effector domain of Smad2, Mixer, and Milk are clearly good candidates for endogenous DEBP.
Sequences of Smad2 required for interaction with DEBP, Mixer, Milk, and Fast-1
To investigate whether the abilities of Mixer, Milk, endogenous DEBP, and Fast-1 to interact with Smad2 might reflect a common protein–protein interaction mechanism, we asked whether the same sequences of Smad2 were required in each case. A series of Smad2C deletion mutants were assayed in the supershift bandshift assay described above for binding to these different transcription factors (Table 1). Deletion of the phosphorylation sites in the SSMS motif at the extreme carboxyl terminus of Smad2 had no effect on binding to any of the transcription factors (mutant 198–463). Analysis of further amino- and carboxy-terminal deletions indicated that the integrity of most of the Smad2 MH2 domain was required for binding to the transcription factors (Table 1). Interestingly Mixer behaved identically to the endogenous DEBP in its interaction with Smad2C, whereas Milk behaved like Fast-1 and required additional residues at the carboxy-terminal domain of Smad2C (Table 1, cf. mutants 198-445, 198–440, and 198–426). This suggests that Mixer is a better candidate for DEBP than Milk (see Discussion).
Table 1.
Mapping the transcription factor interaction domain in Smad2
| GST fusions | Endogenous DEBP | Mixer | Milk | Fast-1 |
|---|---|---|---|---|
| Smad2C (198–467)a | + | + | + | + |
| Smad2C (198–463)b | + | + | + | + |
| Smad2C (198–445) | + | + | − | − |
| Smad2C (198–440) | + | + | − | − |
| Smad2C (198–426) | + | + | − | − |
| Smad2C (198–401) | − | − | − | − |
| Smad2C (198–373) | − | − | − | − |
| Smad2C (198–345) | − | − | − | − |
| Smad2C (198–315) | − | − | − | − |
| Smad2C (198–276) | − | − | − | − |
| Smad2C (198–245) | − | − | − | − |
| Smad2C (Δ207–245) | + | + | + | + |
| Smad2C (Δ207–259) | + | + | + | + |
| Smad2C (Δ207–268)c | + | + | + | + |
| Smad2C (Δ207–321)c | − | − | − | − |
| Smad2C (H2 swap)d | − | − | − | − |
The region of Smad2C thought to contact Fast-1 has been elucidated previously and is the α-helix-2 (Chen et al. 1998). We asked whether the same region is required for binding to Mixer, Milk, and endogenous DEBP. A mutant (Smad2C H2 Swap; Chen et al. 1998) was generated in which α-helix2 in Smad2 is replaced with the equivalent region of Smad1 that does not interact with the transcription factors. This mutant failed to bind any of the transcription factors, indicating that the α-helix2 of Smad2 is required for interaction in all cases (Table 1).
Identification of a Smad interaction motif
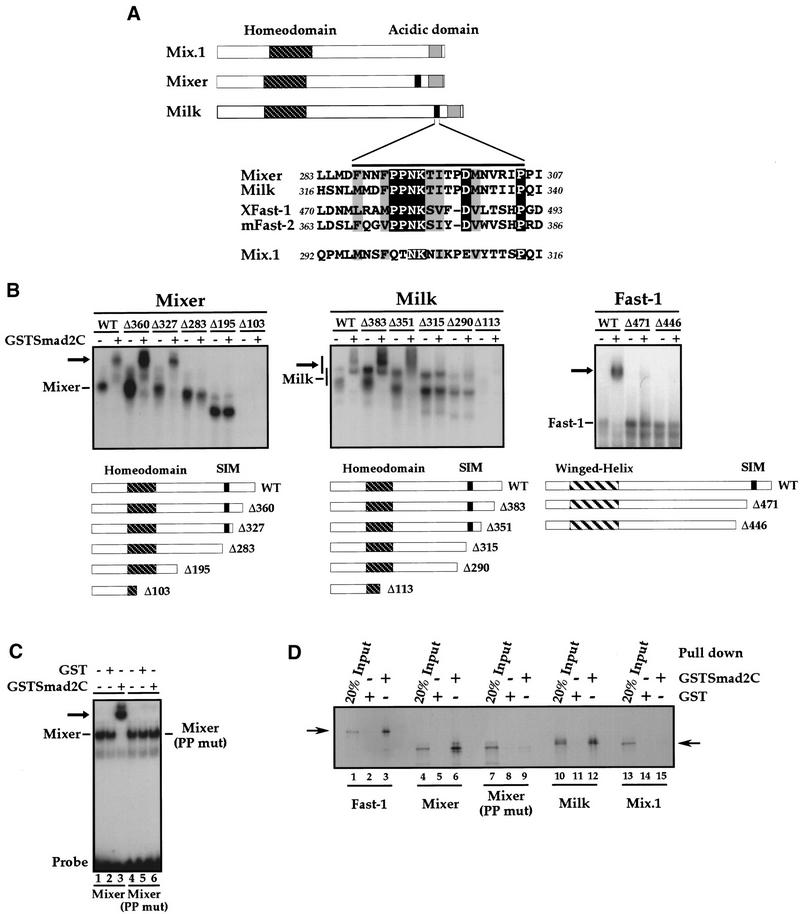
The common property of Smad2 interaction shared by Mixer, Milk, and Fast-1 prompted us to analyze sequence similarities between these transcription factors. Whereas Mixer and Milk belong to the same family of homeodomain transcription factors, Fast-1 belongs to an unrelated family of winged-helix/forkhead transcription factors (Chen et al. 1996; Kaufmann and Knöchel 1996). We identified a short conserved sequence present in the carboxy-terminal region of Mixer, Milk, and Xenopus Fast-1, which was flanked by sequences of no obvious similarity. It is characterized by a completely conserved PPNK core, flanked by other highly conserved residues (Fig. 4A; black line above sequences). Crucially, this sequence has also been conserved in human Fast-1 and mouse Fast-2, which also interact with Smad2 (Labbé et al. 1998; Zhou et al. 1998; Liu et al. 1999; Fig. 4A). Significantly, the PPNK core motif is absent in Mix.1, which does not interact with Smad2.
Figure 4.
Characterization of the SIM. (A) Schematics of Mix.1, Mixer, and Milk, with the conserved homeodomains and a carboxy-terminal acidic domain indicated. (Black box) A region conserved in Milk and Mixer, also present in Xenopus Fast-1 and mouse Fast-2 (expanded below where the black line denotes the boundaries of the conserved sequences). (Black shading) Identical amino acids; (gray shading) similar amino acids. The numbers indicate the positions of these amino acids in the full-length sequences of the individual proteins. (B) Carboxy-terminal deletion mutants of Mixer, Milk, and Fast-1 (schematized below) were produced in vitro and their interaction with GSTSmad2C assayed by bandshift using the DE or ARE probe as appropriate. Complexes of transcription factors and probe are indicated; (black arrow) ternary complex with GSTSmad2C. Smad interaction motif (SIM) and DNA-binding domains are indicated. Note that Milk gives rise to two complexes, both of which shift with GSTSmad2C, which correspond to a dimer of Milk and a higher order complex. (C) Mutation of the prolines in the PPNK core motif abolishes the interaction with Smad2C. Full-length Mixer or a mutant derivative (Mixer PP mut), in which the two prolines in the PPNK-containing motif are mutated to alanines, were produced in vitro and assayed for interaction with GSTSmad2C by bandshift using the DE probe. (D) Interaction of Mixer, Milk, and Fast-1 with Smad2C in solution. 35S-labeled transcription factors as indicated were incubated with Sepharose-bound GST (lanes 2,5,8,11,14) or GSTSmad2C (lanes 3,6,9,12,15) and bound protein was visualized by SDS-PAGE and autoradiography. A fraction of input protein was analyzed for comparison (lanes 1,4,7,10,13).
To address the potential role of this PPNK-containing sequence in Smad2 interaction, a series of carboxy-terminal deletion mutants of Mixer, Milk, and Fast-1 were produced in vitro and assayed by bandshift for their ability to bind the DE and interact with GSTSmad2C. Deletion of the PPNK-containing sequence in the context of either Mixer, Milk, or Fast-1 resulted in the loss of interaction with GSTSmad2C, demonstrating that this sequence is necessary for interaction with Smad2C (Fig. 4B). Larger carboxy-terminal deletions that impinge on the homeodomains of Mixer or Milk completely abolished DNA binding as expected.
The role of the PPNK core motif for Smad2 interaction was investigated in more detail by mutating the two conserved prolines of the PPNK motif to alanine in the context of full-length Mixer [Mixer (PP mut)]. This mutation was sufficient to abolish completely the interaction of Mixer with GSTSmad2C, without affecting its DNA-binding properties (Fig. 4C). Therefore, this short motif is absolutely required for Smad2 interaction.
To prove that the recruitment of Smad2 by these PPNK-containing transcription factors was purely through protein–protein interactions, we established that it could occur in the absence of DNA, using a GST-pull down assay. Mixer, Milk, and Fast-1 interacted efficiently with Sepharose-bound Smad2C, but neither Mixer (PP mut) nor Mix.1, the family member that does not contain the PPNK-containing interaction motif, was able to bind (Fig. 4D).
Thus, we have defined a PPNK-containing sequence present in the carboxy-terminal domain of Mixer and Milk, which is also present in Xenopus Fast-1, human Fast-1, and mouse Fast-2 as a Smad interaction motif (SIM), essential for Smad2 binding.
The SIM is sufficient to bind Smad2
We then investigated whether the SIM was sufficient to interact with Smad2 by testing whether a peptide containing 25 amino acids of Mixer incorporating the SIM (residues 283–307; Fig. 4A) could compete with Mixer for binding to Smad2C. Indeed, wild-type peptide corresponding to ∼10- and 30-fold molar excess over GSTSmad2C was sufficient to inhibit the interaction of Mixer with GSTSmad2C (Fig. 5A, lanes 9,10). The same quantity of the equivalent peptide with the two prolines of the PPNK motif mutated to alanine was ineffective (lanes 13,14). This indicates that the peptide alone is sufficient to bind Smad2C, thus preventing full-length Mixer binding.
Figure 5.
The Smad interaction motif is sufficient to interact with Smad2. (A) A peptide containing the SIM of Mixer competes specifically for interaction of Mixer with Smad2C. In vitro-translated Mixer was incubated with DE probe alone (lane 1) or in the presence of 1 or 10 pmoles of wild-type peptide (lanes 2,3) or mutant peptide (lanes 4,5). GSTSmad2C (20 ng) was included in the reactions in lanes 6–14, with the addition of 0.3, 1, 3, or 10 pmoles of wild-type peptide (lanes 7–10) or mutant peptide (lanes 11–14). Mixer complexed with probe is indicated; (black arrow), ternary complex with GSTSmad2C. (B) A peptide containing the Mixer SIM specifically disrupts the interaction of DEBP with Smad2C. Whole-cell extracts made from activin-injected stage 10.5 embryos were analyzed by bandshift assay with the DE probe in the absence (lane 1) or presence of GSTSmad2C (20 ng; lanes 2–10) with the addition of 5, 15, 30, and 100 pmoles wild-type peptide (lanes 3–6) or mutant peptide (lanes 7–10). DEBP is indicated; (black arrow) ternary complex with GSTSmad2C. (C) A peptide containing the Mixer SIM specifically disrupts the formation of ARF. Whole-cell extracts made from activin-injected stage 8 embryos were analyzed by bandshift assay with the ARE probe in the absence (lane 1) or presence of 10, 30, 60, 100, or 200 pmoles of wild-type peptide (lanes 2–6) or mutant peptide (lanes 7–11). The endogenous ARF complex is indicated.
If, as we have argued, the endogenous DEBP is a Mixer/Milk family member that interacts with Smad2 through the SIM, we would expect that the Mixer SIM peptide should be able to abolish the interaction of DEBP with GSTSmad2C. The experiment shown in Figure 5B clearly demonstrates that this is the case, as the wild-type peptide, but not the mutant, efficiently disrupts the DEBP/Smad2C complex.
To prove that the SIM is a common Smad interaction motif, responsible for recruiting activated Smads in vivo, we investigated whether the Mixer SIM could compete with the Fast-1 SIM for activated endogenous Smad2. Therefore, we determined whether the Mixer SIM-containing peptide would be able to disrupt the formation of the endogenous activin-induced Fast-1/Smad2/Smad4 complex ARF. Wild-type peptide, but not the mutant, is sufficient to inhibit the formation of endogenous Xenopus ARF complex formed in extracts from activin-injected embryos (Fig. 5C). Thus the SIM-containing peptide can bind to endogenous Smad2 and inhibit the interaction of Smad2 with Fast-1. Therefore, the Mixer SIM competes with the Fast-1 SIM for endogenous Smad2, confirming that it is a shared and functional Smad interaction motif.
Mixer recruits an active endogenous Smad complex in vivo
In vivo, activated Smad2 exists as a complex with Smad4 (Massagué 1998). If the interaction of Mixer and Milk with the Smad2 effector domain reflects a physiological function of these proteins, it was critical to prove that they formed stable complexes with ligand-activated Smad2/Smad4 complexes in vivo. NIH-3T3 cells were used for these experiments because they do not normally express Mixer or Milk, and this avoided complications due to the synthesis of these proteins in response to activin in Xenopus embryo explants (Ecochard et al. 1998; Henry and Melton 1998; Tada et al. 1998). Smad2 and Smad4 can be activated in NIH-3T3 cells by TGF-β in the same way as activin activates them in Xenopus embryos (Liu et al. 1997).
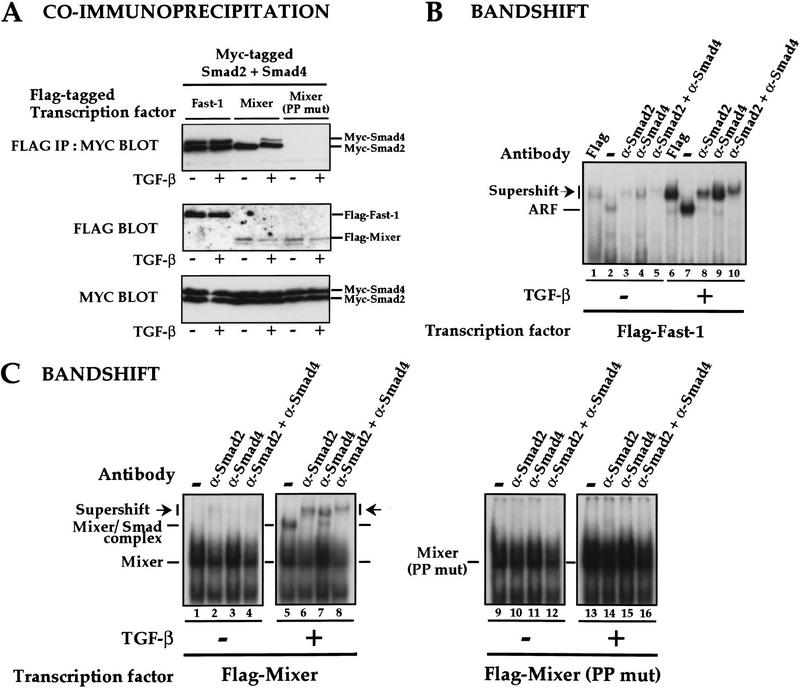
First, we asked whether we could detect ligand-dependent Mixer/Smad complexes in the absence of DNA in a coimmunoprecipitation assay. Flag-tagged transcription factors Mixer, Mixer (PP mut), or Fast-1 (as a control) were immunoprecipitated from cells incubated for 1 hr with or without TGF-β and then Western blotted with anti-myc antibody to detect the presence of coimmunoprecipitating myc-tagged Smads (Fig. 6A). Equal expression of protein was confirmed by Western blotting of whole cell extracts using anti-Flag or anti-myc antibody (Fig. 6A, middle and bottom). In these conditions of overexpressed Smads, Fast-1 interacted constitutively with both Smad2 and Smad4 (Fig. 6A, top; but see below). In the absence of ligand, Mixer interacted with Smad2 only, but after TGF-β stimulation, Mixer coprecipitated with Smad2 in conjunction with a lower level of Smad4 (Fig. 6A, top). Because Mixer does not interact directly with Smad4 (data not shown; see Fig. 2B), we interpret this as Mixer recruiting an activated complex of Smad2 and Smad4 through a direct interaction with Smad2. This is proved below. Consistent with the view that Mixer recruits the Smads through an interaction with Smad2, mutation of the two prolines in the SIM of Mixer (Mixer PP mut) abolished completely the formation of the Mixer/Smad complex in vivo (Fig. 6A, top).
Figure 6.
Mixer and Milk interact with activated Smads in vivo. (A) Mixer forms a ligand-dependent complex with Smad2 and Smad4 in solution. Extracts were prepared from NIH-3T3 cells transfected with myc–Smad2, myc–Smad4, and either Flag–Fast-1, Flag–Mixer, or a Flag-tagged mutant derivative (Mixer PP mut), which had been incubated with or without TGF-β1 (2 ng/ml) for 1 hr. Extracts were assayed either by immunoprecipitation of complexes with anti-Flag antibody followed by Western blotting with anti-Myc antibody (top), or Western blotting the whole extract with anti-Flag antibody (middle), or with anti-Myc antibody (bottom). (B,C) Fast-1 and Mixer form ligand-dependent complexes on DNA with endogenous Smad2 and Smad4. Extracts were prepared from NIH-3T3 cells transfected with Flag-tagged Fast-1, Mixer, or Mixer (PP mut), which had been incubated with or without TGF-β1 (2 ng/ml) for 1 hr. Extracts were analyzed by bandshift assay on the ARE (B) or DE (C) probe. Anti-flag, anti-Smad2, or anti-Smad4 antibodies were included in the binding reactions where indicated. In B ARF and antibody-supershifted ARF are indicated. In C, Mixer or Mixer (PP mut) bound to probe, the Mixer–Smad complex and antibody-supershifted Mixer–Smad complex are indicated.
If ligand-dependent Mixer/Smad2/Smad4 complexes are physiologically relevant, they should form on DNA, and form when the Smads are not overexpressed. Using NIH-3T3 cells expressing a low level of Flag-tagged Mixer, we asked whether a TGF-β inducible Mixer–Smad complex could be detected on the DE in a bandshift assay. This assay relies on endogenous mouse Smads that are not overexpressed. The Smad antibodies were validated by testing their ability to supershift the well-characterized Fast-1/Smad2/Smad4 complex ARF on the ARE probe (Fig. 6B). ARF is strongly ligand-inducible in these conditions when Smads are not overexpressed (lanes 2,7), and clearly contains Flag-tagged Fast-1 and endogenous Smad2 and Smad4 as shown by antibody supershifts (lanes 1–10; Chen et al. 1996, 1997). The results for Mixer are shown in Figure 6C. A Mixer/DNA complex is seen in extracts from cells transfected with low levels of Mixer (lanes 1–8). Strikingly, a strong TGF-β-induced Mixer/Smad complex was detected in extracts made from cells induced with TGF-β for 1 hr (cf. lane 1 with lane 5). We proved that this complex contained endogenous Smad2 and Smad4 by demonstrating that it was quantitatively supershifted with the specific anti-Smad2 and Smad4 antibodies (lanes 6,7). The presence of both Smad2 and Smad4 in the same complex was confirmed by the observation that adding both antibodies together supershifted the complex further than either antibody alone. In addition, we proved that the TGF-β-inducible Mixer/Smad complex contains Mixer as well as the Smads, by showing that no such complex was formed in cells expressing Flag-tagged Mixer (PP mut), which does not interact with Smads (lanes 9–16).
Thus, Mixer forms a strictly ligand-dependent complex with endogenous Smad2 and Smad4 in vivo that binds DNA and requires the integrity of the SIM.
Mixer/Milk–Smad complexes mediate TGF-β-induced transcriptional activation through the DE
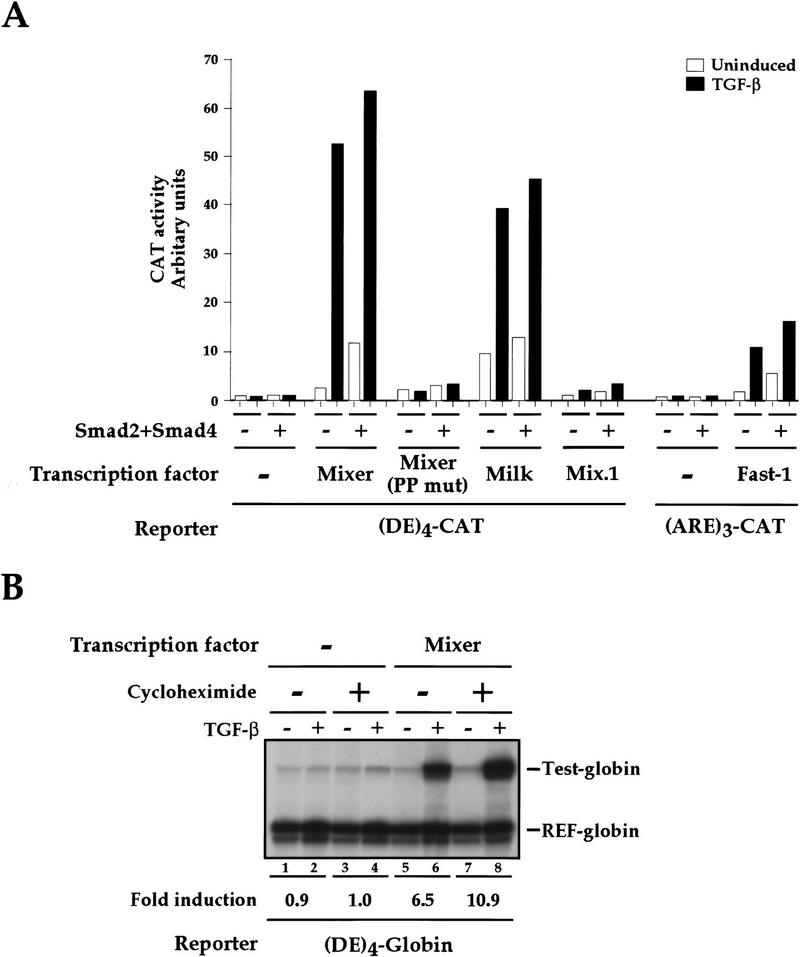
Having demonstrated that Mixer rapidly forms a DNA-binding complex with activated endogenous Smads in response to TGF-β, we investigated whether this complex was transcriptionally active. A DE-driven CAT reporter gene was inactive in NIH-3T3 cells and did not respond to TGF-β induction (Fig. 7A). Cotransfection of Smad2 and Smad4 had no effect, indicating that the Smads could not activate transcription alone. Mixer displayed very little transcriptional activity in the absence of TGF-β, but conferred very strong TGF-β-dependent transcriptional activation on the DE (Fig. 7A). In contrast, the mutant of Mixer that does not bind Smad2 (Mixer PP mut) was completely inactive (Fig. 7A), providing strong evidence that TGF-β induction of transcription via Mixer required recruitment of endogenous Smads. This was corroborated by the observation that overexpression of Smad2 and Smad4 potentiated transcription via Mixer in the absence of TGF-β stimulation. Milk also conferred TGF-β inducibility on the DE. However, Mix.1 was inactive, consistent with the fact that it does not interact with Smad2 (Fig. 7A). These reporter gene assays were performed with four tandem DE elements. Mixer and Milk were also sufficient to confer TGF-β-induced transcription onto a single DE, albeit at a lower level (data not shown). TGF-β-induced transcription mediated by the homeodomain proteins was stronger than that elicited by Fast-1 on the ARE (Fig. 7A; Liu et al. 1997), reflecting what we observe in Xenopus animal cap assays (see Fig. 1A).
Figure 7.
Mixer and Milk mediate TGF-β-dependent transcriptional activation through the DE. (A) NIH-3T3 cells were transfected with the CAT reporters, and plasmids expressing transcription factors, Smad2 and Smad4, as indicated. Cells were cultured with or without TGF-β1 (2 ng/ml) for 8 hr. Cells were harvested and CAT activity measured relative to lacZ activity from the internal control. The data are from a representative experiment, and similar results were obtained in other independent experiments. (B) Mixer mediates TGF-β-dependent transcriptional activation through the DE in the absence of protein synthesis. NIH-3T3 cells were transfected with the (DE)4-globin reporter and REF-globin internal control with or without Mixer expression plasmid. Cells were cultured with or without TGF-β1 (2 ng/ml) for 4 hr in the absence or presence of 50 μg/ml cycloheximide. Globin transcripts from the reporter genes (Test-globin) or the internal control (REF-globin) were detected by RNase protection and quantitated as in Fig. 1.
Finally, we proved that the TGF-β-responsive transcription mediated by Mixer results from the interaction of Mixer with endogenous Smads by demonstrating that it was independent of new protein synthesis. For this experiment we used a globin reporter system and measured globin mRNA levels directly by RNase protection. TGF-β-induced transcription through the DE is absolutely dependent on Mixer (Fig. 7B, lanes 1,2,5,6) and crucially, is not decreased when the protein synthesis inhibitor cycloheximide is added at the same time as TGF-β (lanes 5–8). Thus, when Mixer is present, TGF-β-induced transcription does not require on-going protein synthesis.
Mixer and Milk are expressed appropriately to be endogenous inducers of goosecoid
If, as our data suggest, Mixer and/or Milk are endogenous inducers of goosecoid then we would expect them to be expressed in the same domain as goosecoid. We investigated the spatial expression patterns of Mixer, Milk, and goosecoid by in situ hybridization on stage 10.25 embryos that had been cut in half through the middle of the dorsal lip (Fig. 8A). To aid identification of the prospective mesoendodermal boundary, we also stained embryos for Cerberus mRNA (Fig. 8A; Bouwmeester et al. 1996). goosecoid is expressed in mesoendodermal cells exclusively on the dorsal side, in a domain that includes prospective head mesoderm and dorsal endoderm (Fig. 8A). Both Mixer and Milk are expressed more widely, in a ring, which is clearly seen in these sections as staining on the dorsal and ventral sides (Fig. 8A). Mixer is expressed predominantly in prospective endoderm, but is also detectable in mesodermal cells. It is particularly strong at the mesoendodermal boundary (Fig. 8A; Henry and Melton 1998). Milk is more weakly expressed and the staining is more diffuse than for Mixer. It is clearly expressed in prospective mesoderm and endoderm (Fig. 8A). Thus, goosecoid is expressed in cells on the dorsal side that express Mixer or Milk.
Figure 8.
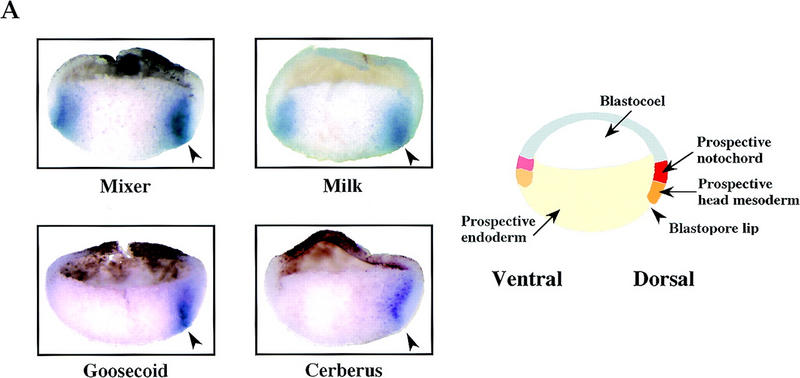
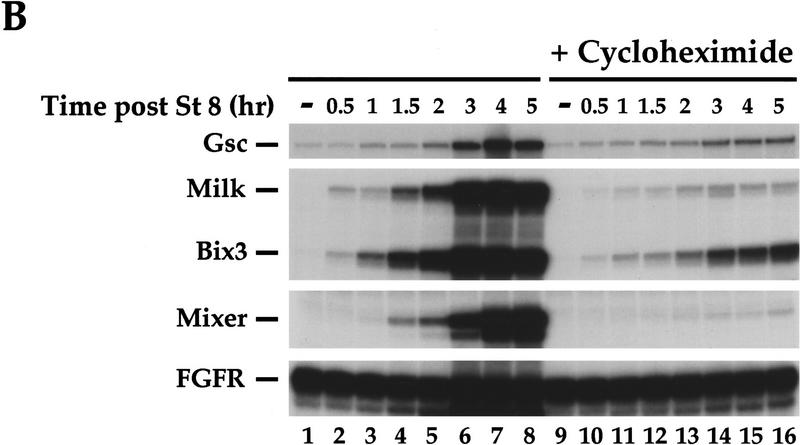
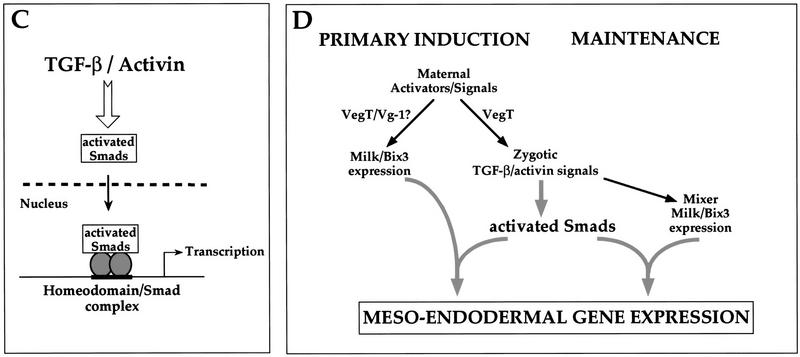
The temporal and spatial expression patterns of Mixer and Milk in Xenopus embryos makes them good candidates for mediating transcription of goosecoid in response to an endogenous activin-like signal. (A) Coexpression of goosecoid with Mixer and Milk at early gastrula stages. Xenopus embryos were fixed at stage 10.25, cut in half through the middle of the dorsal lip, and processed for in situ hybridization with probes against goosecoid, Mixer, Milk, or Cerberus (as a control to visualize the prospective mesoendodermal boundary). (Arrowhead) Blastopore lip. The mRNAs are visualized with a dark blue/purple stain. It is clear that goosecoid is expressed on the dorsal side in prospective mesodermal and endodermal cells expressing Mixer and/or Milk. (Right) Schematic of stage 10.25 embryo cut in the same way as the embryo halves. (Red) Prospective notochord; (orange) prospective head mesoderm; (pink) somitic mesoderm; (light orange) ventral migrating mesoderm; (yellow) prospective endoderm (adapted from Keller 1991). (B) Time course of expression of goosecoid (Gsc), Mixer, Milk, and the FGF receptor (FGFR) assayed by RNase protection. The Milk probe also detects Bix3 (Tada et al. 1998). Embryos were sampled at stage 8 and subsequent times indicated. In lanes 9–16 the embryos had been preincubated with cycloheximide from 30 min before stage 8. (C) A model showing that TGF-β/activin-activated Smads translocate to the nucleus, where they interact with homeodomain transcription factors, Mixer and Milk through the SIM to activate transcription. (D) A model indicating the proposed role of the Mixer/Milk/Bix3-Smad complexes in the formation of mesoderm and endoderm in early Xenopus embryos. (Black arrows) Induction of gene expression; (gray arrows) activation of protein complexes. Milk/Bix3–Smad complexes are involved in the initiation of transcription of mesoendodermal genes and Mixer/Milk/Bix–Smad complexes are involved in the maintenance of gene expression. For discussion, see text.
We also addressed the timing of Mixer and Milk expression relative to goosecoid in Xenopus embryos, using RNase protection assays. The Milk RNase protection probe detects in addition a second transcript, which corresponds to the highly related gene Bix3 (see below; Materials and Methods; Tada et al. 1998). Bix3, like Mixer and Milk binds the goosecoid DE and contains a functional Smad interaction motif (data not shown). Mixer, Milk, and Bix3 are all expressed before the major up-regulation of goosecoid expression at 3 hr after stage 8 (Fig. 8B, lanes 1–8).
Thus, the temporal and spatial expression patterns of Mixer, Milk (and Bix3) are consistent with their being responsible for induction of goosecoid in Xenopus embryos.
The role of Mix family members in mesoendoderm induction
Finally, we addressed the timing of Mixer, Milk, and Bix3 expression relative to the timing of production of the endogenous activin-like signal. In Xenopus embryos the major secreted activin-like signal is zygotic and requires the maternal transcription factor VegT for its production (Kimelman and Griffin 1998; Zhang et al. 1998). Maternal transcription factors, as well as being responsible for producing the activin-like signal that gives rise to the active Smad complexes, might also be responsible for the synthesis of the transcription factors the Smads interact with. Therefore, we tested whether Mixer, Milk, or Bix3 were induced by maternal activators by measuring their transcription in embryos incubated in cycloheximide before stage 8. Inductions mediated by maternal activators are unaffected by this treatment, whereas those mediated by zygotic activators are inhibited.
The expression of Mixer was virtually all abolished by the cycloheximide treatment, indicating that it is solely induced by zygotic activators (Fig. 8B, lanes 1–16; Yasuo and Lemaire 1999). In contrast, Milk and Bix3 were strongly activated in untreated embryos, and some of this activation remained in cycloheximide-treated embryos (particularly evident for Bix3; Fig. 8B; lanes 1–16), suggesting that these genes are weakly induced by maternal activators, and their expression is reinforced by zygotic activators.
Thus, in the embryo, Milk and Bix3 are likely to be the earliest endogenous Mix family partners for Smads to initiate transcription of mesoendodermal genes. In addition, a zygotic signal, probably the endogenous activin-like signal (Clements et al. 1999; Yasuo and Lemaire 1999) induces the synthesis of additional Milk, Bix3, and also Mixer, which likely corresponds to the DEBP that we detect at stage 10.5/11. The complexes this Mixer/Milk/Bix3 form with Smads could be responsible for maintaining the activin-induced transcription of mesoendodermal genes.
Discussion
Mixer and Milk recruit Smads to the goosecoid DE to regulate activin/TGF-β-responsive transcription
Here we have investigated the mechanism of activin-responsive transcription through the DE of the Xenopus goosecoid promoter. We have shown that paired-like homeodomain transcription factors of the Mix family, Mixer and Milk, but not Mix.1, mediate activin/TGF-β-induced transcription through the DE by interacting specifically with the effector domain of Smad2, and thereby recruiting active Smad2/Smad4 complexes to this element (Fig. 8C). We show that Mixer forms a TGF-β-inducible complex with endogenous Smad2 and Smad4 at the goosecoid DE within 1 hour of ligand stimulation, and demonstrate that the Smads are essential for transcriptional regulation mediated by this complex, as the Mixer/Smad complex is at least an order of magnitude more transcriptionally active than Mixer alone.
Our results also reveal that activated Smads are recruited to different promoter elements by a common mechanism. We have identified a short Smad interaction motif (SIM), characterized by the core sequence PPNK, in the carboxy-terminal region of Mixer and Milk, which is both necessary and sufficient for these proteins to interact with the effector domain of Smad2. We demonstrate that it is required for TGF-β-induced transcription mediated by the Mixer/Smad complex. Crucially it is also conserved in the carboxy-terminal regions of the winged-helix/forkhead Smad2-interacting proteins Xenopus Fast-1, human Fast-1, and mouse Fast-2 (Chen et al. 1996, 1997; Labbé et al. 1998; Zhou et al. 1998). Moreover, we demonstrate that the Mixer SIM can disrupt the formation of the endogenous ARF complex and thus is capable of competing with the Fast-1 SIM for its interaction with endogenous activated Smad2. This indicates that transcription factors of completely different DNA-binding specificity recruit activated Smads to distinct promoter elements through the same protein–protein interaction. This finding now explains why activin-responsive elements in the promoters of different Xenopus genes share so little sequence similarity (Howell and Hill 1997). Our characterization of a common Smad interaction motif should now enable identification of more Smad2-interacting proteins involved both in mesoendoderm formation and also in other aspects of TGF-β signaling. In addition, peptides containing this motif have potential as inhibitors of Smad2 activity, and therefore of TGF-β/activin signaling.
Activation of transcription by Smad/transcription factor complexes
The Smads appear to require other transcription factors to recruit them to DNA because they interact with DNA themselves either weakly (Smad3 and Smad4) or not at all (Smad2; Shi et al. 1998; Hill 1999). The interaction of Smads with distinct transcription factors must contribute to cell-type specificity of TGF-β responses, and this is likely to be of particular significance in the patterning of the early Xenopus embryo. The same signaling pathway will activate different genes in distinct regions of the embryo depending on the particular Smad-recruiting transcription factors expressed by the cells in that region. In addition, differential affinities of specific transcription factors for Smads, coupled with the presence or absence of Smad-binding sites on adjacent DNA could allow distinct genes to be activated by different levels of active Smad complexes. This sort of mechanism might underlie the morphogenetic properties of TGF-β family members, whereby different doses of TGF-β ligands elicit different transcriptional responses (Green and Smith 1990). Determination of the relative affinities of the SIMs in Mixer, Milk, Bix proteins, and Fast-1 for Smad complexes will be important to test these ideas.
Regulation of goosecoid
Previous studies of Xenopus goosecoid regulation indicated that there was an activity in the vegetal hemisphere and marginal zone that could regulate activin-induced transcription through the DE (Watabe et al. 1995). This was demonstrated by the observation that a reporter gene driven by six copies of the DE was activated in all blastomeres of the C and D tiers (Watabe et al. 1995). The activity of the DE is not sufficient for regulation of the goosecoid promoter, which in addition requires regulation through the proximal element in the promoter (PE) by a Wnt-like signal operating on the dorsal side of the embryo. The combination of these activities is thought to confine goosecoid expression to the dorsal marginal zone of the early gastrula (Watabe et al. 1995; Laurent et al. 1997).
We now propose that the activin-induced DE transcriptional activator corresponds to a SIM-containing member of the Mixer/Milk family, complexed with activated Smad2/Smad4. Our data from tissue culture cells clearly demonstrate that Mixer forms complexes with endogenous activated Smads in a strictly ligand-dependent manner under physiological conditions. These complexes bind the DE, and are responsible for ligand-induced transcription through the DE. In addition, we have provided compelling evidence to suggest that Mixer/Milk family members could perform this function in the Xenopus embryo. We show that cells that express Mixer/Milk in the embryo are those that receive the endogenous activin-like signal (see Clements et al. 1999; Yasuo and Lemaire 1999, and references therein) and thus will contain activated Smad2/Smad4 complexes. The temporal and spatial expression patterns of Mixer and Milk are appropriate for them to be the endogenous inducers of goosecoid. In addition, we have provided extensive evidence to suggest that the major endogenous factor that binds the goosecoid DE in Xenopus extracts (DEBP) is a member of the Mixer/Milk family: (1) Mixer and Milk have the same DNA-binding specificity as DEBP; (2) Mixer and Milk form complexes at the DE with the same mobility on a bandshift gel as DEBP; (3) DEBP, Mixer, and Milk all interact with Smad2C and require α-helix 2 of Smad2C for the interaction; and (4) The interaction of Mixer and Smad2C is competed by the SIM peptide, as is the interaction between DEBP and Smad2C. Our experiments do not yet allow us to say precisely which member of the Mixer/Milk family corresponds to DEBP, although Mixer appears to be the best candidate. It has so far proved technically impossible to detect the endogenous Mixer/Milk-Smad complex in Xenopus embryos, because the levels of the complex are too low and the complex is too transient and unstable. Therefore, formal proof that these homeodomain proteins function as complexes with Smads in the embryo awaits the development of more sensitive assays.
The original analysis of regulation of the Xenopus goosecoid promoter focused on the combined activites of the DE and PE (Watabe et al. 1995). However, additional elements elsewhere in the promoter may also be involved in confining goosecoid expression to the dorsal mesoendoderm during gastrulation. For instance, a functional Fast-1-binding site was recently identified in the mouse goosecoid promoter, and it is largely conserved in the Xenopus promoter downstream of the DE (Labbé et al. 1998). It will be important to investigate possible functional interactions between Fast-1, Mixer/Milk family members and activated Smads in the context of the full-length Xenopus goosecoid promoter. In addition, overexpression of BMP antagonists weakly induces goosecoid expression throughout the marginal zone in a delayed fashion (Eimon and Harland 1999). This suggests that BMP signaling might normally be required to suppress goosecoid expression in lateral and ventral regions during gastrulation. It will be interesting to investigate the interplay between the BMP and activin-like pathways in the regulation of mesoendodermal genes.
The role of Mixer and Milk in mesoendodermal induction in Xenopus embryos
Recent studies have already implicated Mixer, Milk, and the Bix proteins in endodermal and mesodermal differentiation, based on experiments in which they were overexpressed in prospective ectoderm (animal caps) (Ecochard et al. 1998; Henry and Melton 1998; Tada et al. 1998). However the underlying molecular mechanism was unknown. Our data indicate that Mixer/Milk/Bix have little inherent transcriptional activity, but require bound activated Smads to increase their transcriptional potential, and thus in the embryo, we propose that Mixer/Milk/Bix would activate mesoendodermal genes by cooperating with Smads activated by an endogenous activin-like signal. A prediction would be that the family member Mix.1, which does not interact with Smads, would have a different activity in vivo. In contrast to Mixer and Milk, overexpression of Mix.1 alone in animal caps does not induce endoderm (Ecochard et al. 1998; Henry and Melton 1998; Lemaire et al. 1998; Tada et al. 1998).
Our interaction data, together with the expression patterns of these homeodomain proteins allow us to propose how these proteins might play a role in mesoendodermal formation in the Xenopus embryo (Fig. 8D). The major activin-like mesoendoderm-inducing activity that would activate Smad2 and Smad4 is zygotic, and requires the maternal transcription factor VegT for its production (Kimelman and Griffin 1998; Zhang et al. 1998). This activity is likely to be composed of several different activin-related molecules, including derrière, Xnr1, Xnr2, Xnr4, and activin βB (Clements et al. 1999; Yasuo and Lemaire 1999). Our experiments indicate that Milk and Bix3 are also induced (weakly) in Xenopus embryos by a maternal activator (Fig. 8D). This could be VegT itself, as the Bix genes have been shown to be VegT targets (Tada et al. 1998). Alternatively, the maternal activator could be the signaling pathway activated by the maternal activin-related protein Vg-1, as the Bix genes are also known to be directly induced by activin (Ecochard et al. 1998; Tada et al. 1998). Thus, low levels of Milk and Bix3 would be available in the embryo to bind the Smad2/Smad4 complexes activated by the zygotic activin-like ligand, thereby initiating transcription of downstream genes like goosecoid (Fig. 8D). In addition, there may be low levels of ubiquitously maternally expressed Milk/Bix genes that could account for the cycloheximide-insensitive activin-induced transcription of the DE seen in the animal caps in Figure 1A. Milk, Bix3, and also Mixer are themselves induced by the zygotic activin-like signaling pathway (Ecochard et al. 1998; Henry and Melton 1998; Tada et al. 1998; see also Clements et al. 1999; Yasuo and Lemaire 1999). We propose that these proteins would be involved in maintaining transcription in response to the zygotic activin-like ligand through their formation of transcriptionally active complexes with activated Smads (Fig. 8D). Determining precisely which mesoendodermal genes are regulated in this way by which Mixer/Milk family members presents a challenge for the future.
Materials and methods
Plasmids constructs
Mix.1 (Rosa 1989), Milk (Ecochard et al. 1998), and Mixer (Henry and Melton 1998) were isolated by PCR from a stage 11 Xenopus cDNA library and their coding sequences and that of Fast-1 (Chen et al. 1997) were subcloned into pFTX5 (Howell and Hill 1997) and EF–Flag. Human Smad4 and Xenopus Smad2 were subcloned in EF–Myc. EF–Flag, and EF–myc were derivatives of EF–Plink (Hill et al. 1995). Prolines 291 and 292 of Mixer were mutated to alanines by PCR. In DE4–CAT and ARE3–CAT four copies of the goosecoid DE or three copies of the Mix.2 ARE are upstream of the minimal γ-actin promoter driving CAT. In the globin versions, human β-globin replaced CAT (Howell and Hill 1997). REF-globin was as described (Howell and Hill 1997). In GSTSmad2C amino acids 198–467 of XSmad2 were subcloned into pGEX–KG (Poon et al. 1993). GSTSmad1C contained amino acids 172–468 of XSmad1, and GSTSmad4C contained amino acids 226–552 of hSmad4. Deletions of GSTSmad2C were made using standard methods and named according to the positions of the deletion such that GSTSmad2C(198–245) lacks sequence after codon 245, whereas GSTSmad2C (Δ207–245) lacks sequence between codons 208 and 244. α-Helix 2 of Smad2 was replaced by α-helix 2 of Smad1 in GSTSmad2C using PCR. All constructs were verified by sequencing.
Oligonucleotides
- CTAGCCATTAATCAGATTAACGGTGAGCAATTAGA (DE-top).
- CCGACTAGTATCTGCTGCCCTAAAATGTGTATTCCATGGAAATG (ARE top).
- CCGGCTAGCTAGGGAGAGAAGGGCAGACATTTCCATGGAATAC (ARE bot).
- CTAGCCAGTCAGCAGCTGACCGGTGAGCAATTAGA (DE m1 top).
- CTAGCCAGTCATCAGAGTCACGGTGAGCAAGTCGA (DE m2 top).
- CTAGCCATTAATCAGATTAACTTGTAGCAAGTAGA (DE m3 top).
GST fusion protein purification, GST “pull-downs,” and in vitro transcription/translation
Expression of GST fusion proteins, SDS-PAGE, and in vitro-coupled transcription/translation in reticulocyte lysate (Promega) were performed using standard methods. Mixer, Milk, and Fast-1 carboxy-terminal deletion mutants were synthesized in vitro using linear templates generated by restriction enzyme digestion. For “pull-down” experiments, 35S-labeled transcription factors were mixed with GST or GST–Smad2C Sepharose beads for 2 hr at 4°C in 20 mm Tris-HCl (pH 7.5), 20% glycerol, 1 mm EDTA, 5 mm MgCl2, 0.1% NP-40, and 220 mm NaCl. The beads were washed three times with five-bead volumes of binding buffer, and the protein remaining bound to the beads was analyzed by SDS-PAGE, followed by autoradiography.
Embryo manipulations, RNase protection assays, and in situ hybridizations
The production, maintenance, and manipulation of Xenopus embryos was previously described (Howell and Hill 1997). mRNA for microinjection was generated in vitro (Howell and Hill 1997) and injected at the one-cell stage: 200 pg of activin βA mRNA per embryo, 1.5 ng of mRNA encoding myc-tagged Mix.1, Mixer, or Milk. When embryos were treated with cycloheximide, it was added at 20 μg/ml in 0.1× NAM 30 min before stage 8. RNA isolation and RNase protection assays were performed as described (Howell and Hill 1997). The antisense probes were as follows: human β-globin (Howell and Hill 1997); goosecoid (Blumberg et al. 1991); Xenopus FGF receptor, protecting amino acids 539–580; Mixer, amino acids 173–237; Milk, amino acids 143–226. The Milk probe also detects a smaller product, which we have identified as the highly _Milk-_related gene Bix3 (Tada et al. 1998). In situ hybridizations were carried out on embryo halves essentially as described (Harland 1991), using probes against goosecoid, Mixer, and Milk, which were identical to those used in the RNase protections, and Cerberus, which protects 9 nucleotides of 5′ UTR and the region encompassing amino acids 1–95.
Transfections, transcriptional assays, and immunoprecipitation
NIH-3T3 cells were transfected using lipofectamine (GIBCO BRL). The following amounts of plasmids were used per 6-cm dish for transcriptional assays: 0.5 μg CAT reporters, 0.2 μg of transcription factors, 0.3 μg of EF–Smad2 and EF–Smad4 and 0.5 μg of EF–LacZ as an internal control for transfection efficiency, as indicated in the Figure 7 legend. For globin transcriptional assays, 1 μg of globin reporter, 0.45 μg of REF–globin, and 0.2 μg of Mixer were transfected. For immunoprecipitations, two 6-cm plates were transfected with 0.6 μg of each plasmid. For the bandshift assay, one 6-cm plate was transfected with 1.2 μg of transcription factor. The amounts of DNA transfected was kept constant by adding control plasmid EF–plink as appropriate. After transfection, cells were maintained 18 hr in DMEM containing 10% FBS, before induction by TGF-β1 (2 ng/ml, Calbiochem) for times indicated in the Figure 6 and 7 legends.
For transcription assays, cells were lysed in 200 μl of 20 mm Tris-HCl (pH 7.5), 150 mm NaCl, 1 mm EDTA, and 0.5 % NP-40. CAT assays were performed exactly as previously described (Hill et al. 1993). β-Galactosidase assays were performed using CDGP (Calbiochem) as a substrate and quantitated spectrophotometrically. RNA was extracted from NIH-3T3 cells for the globin assays as described (Hill et al. 1995) and the RNase protection assays were as above.
For immunoprecipitations, cells were lysed in 100 μl of buffer containing 20 mm Tris-HCl (pH 7.4), 150 mm NaCl, 1 mm EDTA, 1 mm EGTA, 5 mm NaF, 10 mm β-glycerophosphate, 10% glycerol, 1% Triton, and protease inhibitors (10 μg/ml leupeptin, E-64, aprotinin, 20 μg/ml pepstatin, 0.5 mm benzamidine, and 0.4 mm Pefabloc SC). Flag-tagged transcription factors were immunoprecipitated with anti-Flag M2 affinity gel (Sigma) for 2 hr at 4°C, and washed three times with lysis buffer. Immunoprecipitates were separated by SDS–15% polyacrylamide gel electrophoresis and Western blotted with anti-myc antibody 9E10.
Bandshift assays and peptides
Bandshift probes corresponding to the ARE (oligonucleotides 2 and 3) and DE (oligonucleotide 1 and its complement) were labeled with [α32P]dATP and [α32P]dCTP by PCR. Competitions were performed with double-stranded oligonucleotides: DE m1, DE m2, DE m3, or ARE (produced by annealing and filling in oligonucleotides 2 and 3). Whole-cell Xenopus embryo extracts were prepared by homogenizing embryos in buffer (10 μl per embryo) containing 200 mm KCl, 50 mm Tris-HCl (pH 7.4), 10% glycerol, 25 mm β-glycerophosphate, 1 mm EGTA, 1 mm EDTA, 2 mm DTT, and protease inhibitors as above. Lysates were cleared by repeated centrifugation. Binding reactions were performed with 30 μg of protein extract incubated with 0.2 ng of DE probe in 20 μl of buffer containing 140 mm KCl, 8 mm MgCl2, 12.5 mm β-glycerophosphate, 1 mm EGTA, 1 mm EDTA, 1 mm NaF, and 0.5 μg poly[d(I-C)], 2 mm DTT, and protease inhibitors for 20 min at room temperature. Twenty nanograms of purified GST fusion proteins were used to study interactions with GSTSmad2C. Extracts for the ARF bandshift in Figure 5C were prepared from activin-injected stage 8 embryos and the bandshift conditions were as described (Huang et al. 1995). For in vitro translated Mix family members, bandshift conditions were as described (Wilson et al. 1993). For in vitro translated Fast-1, the final buffer concentrations were 8 mm HEPES (pH 7.6), 90 mm KCl, 5 mm MgCl2, 4 mm β-glycerophosphate, 40 μm EDTA, 40 μm Spermidine, 2 μg poly[d(I-C)], and 5% glycerol. Extracts from NIH-3T3 cells transfected with Flag-tagged transcription factors were prepared as described (Marais et al. 1993) and final bandshift conditions were 14 μg total protein in 10 mm HEPES (pH 7.5), 15% glycerol, 210 mm KCl, 5.5 mm MgCl2, 0.2% Triton, 5 mm EGTA, 2.5 mm EDTA, 2 μg of poly[d(I-C)], 0.25 mm DTT, and protease inhibitors with 0.2 ng of labeled probe. Specific antibodies (1 μl) anti-Smad2 (Nakao et al. 1997), anti-Smad4 (B8; Santa Cruz), or anti-Flag were added to the binding reactions. In all cases electrophoresis was in 5% polyacrylamide gels/0.5× TBE containing 2.5% glycerol.
The wild-type SIM-containing peptide used in Figure 5 was Biotin.Aminohexanoicacid-RQIKIWFQNRRMKWKKLLMDFNNFPPNKTITPDMNVRIPPI. The first 16 amino acids are from the helix 3 of Antennapedia, which allows internalization of these peptides into live cells (Derossi et al. 1998); the last 25 amino acids are codons 283–307 of Mixer. The mutant was the same except that the two prolines at positions 25 and 26 were alanines. Peptides were included in the binding reactions at the concentrations given in the Figure legend.
Acknowledgments
We thank Rik Derynck, Jonathan Graff, Michael King, Tim Mohun, Jim Smith, and Richard Treisman for plasmids and libraries and the NHPP for human recombinant activin A [lot 15365-36(1)]. We are grateful to Les Dale, David Ish-Horowicz, Helen McNeill, Tim Mohun, Francisco Nicolás, Malcolm Parker, Christophe Pierreux, and Richard Treisman for useful discussions and comments on the manuscript. The early part of this work was performed at the Ludwig Institute for Cancer Research, University College London School of Medicine Branch where C.S.H. held a Royal Society University Research Fellowship. This work was also supported by the Imperial Cancer Research Fund and the Société Française d'Hypertension Artérielle through a fellowship to S.G.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL c.hill@icrf.icnet.uk; FAX 44 171 269 3093.
References
- Blumberg B, Wright CV, De Robertis EM, Cho KW. Organizer-specific homeobox genes in Xenopus laevis embryos. Science. 1991;253:194–196. doi: 10.1126/science.1677215. [DOI] [PubMed] [Google Scholar]
- Bouwmeester T, Kim S, Sasai Y, Lu B, De Robertis EM. Cerberus is a head-inducing secreted factor expressed in the anterior endoderm of Spemann's organizer. Nature. 1996;382:595–601. doi: 10.1038/382595a0. [DOI] [PubMed] [Google Scholar]
- Candia AF, Watabe T, Hawley SH, Onichtchouk D, Zhang Y, Derynck R, Niehrs C, Cho KW. Cellular interpretation of multiple TGF-β signals: Intracellular antagonism between activin/BVg1 and BMP-2/4 signaling mediated by Smads. Development. 1997;124:4467–4480. doi: 10.1242/dev.124.22.4467. [DOI] [PubMed] [Google Scholar]
- Chen X, Rubock MJ, Whitman M. A transcriptional partner for MAD proteins in TGF-β signalling. Nature. 1996;383:691–696. doi: 10.1038/383691a0. [DOI] [PubMed] [Google Scholar]
- Chen X, Weisberg E, Fridmacher V, Watanabe M, Naco G, Whitman M. Smad4 and FAST-1 in the assembly of activin-responsive factor. Nature. 1997;389:85–89. doi: 10.1038/38008. [DOI] [PubMed] [Google Scholar]
- Chen YG, Hata A, Lo RS, Wotton D, Shi Y, Pavletich N, Massagué J. Determinants of specificity in TGF-β signal transduction. Genes & Dev. 1998;12:2144–2152. doi: 10.1101/gad.12.14.2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements D, Friday RV, Woodland HR. Mode of action of VegT in mesoderm and endoderm formation. Development. 1999;126:4903–4911. doi: 10.1242/dev.126.21.4903. [DOI] [PubMed] [Google Scholar]
- Derossi D, Chassaing G, Prochiantz A. Trojan peptides: The penetratin system for intracellular delivery. Trends Cell Biol. 1998;8:84–87. [PubMed] [Google Scholar]
- Derynck R, Zhang Y, Feng XH. Smads: Transcriptional activators of TGF-β responses. Cell. 1998;95:737–740. doi: 10.1016/s0092-8674(00)81696-7. [DOI] [PubMed] [Google Scholar]
- Ecochard V, Cayrol C, Rey S, Foulquier F, Caillol D, Lemaire P, Duprat AM. A novel Xenopus mix-like gene milk involved in the control of the endomesodermal fates. Development. 1998;125:2577–2585. doi: 10.1242/dev.125.14.2577. [DOI] [PubMed] [Google Scholar]
- Eimon PM, Harland RM. Xenopus embryos, BMP heterodimers are not required for mesoderm induction, but BMP activity is necessary for dorsal/ventral patterning. Dev Biol. 1999;216:29–40. doi: 10.1006/dbio.1999.9496. [DOI] [PubMed] [Google Scholar]
- Funahashi J, Sekido R, Murai K, Kamachi Y, Kondoh H. δ-Crystallin enhancer binding protein δEF1 is a zinc finger-homeodomain protein implicated in postgastrulation embryogenesis. Development. 1993;119:433–446. doi: 10.1242/dev.119.2.433. [DOI] [PubMed] [Google Scholar]
- Green JB, Smith JC. Graded changes in dose of a Xenopus activin A homologue elicit stepwise transitions in embryonic cell fate. Nature. 1990;347:391–394. doi: 10.1038/347391a0. [DOI] [PubMed] [Google Scholar]
- Gurdon JB, Harger P, Mitchell A, Lemaire P. Activin signalling and response to a morphogen gradient. Nature. 1994;371:487–492. doi: 10.1038/371487a0. [DOI] [PubMed] [Google Scholar]
- Harland RM. In situ hybridization: An improved whole-mount method for Xenopus embryos. Methods Cell Biol. 1991;36:685–695. doi: 10.1016/s0091-679x(08)60307-6. [DOI] [PubMed] [Google Scholar]
- Harland R, Gerhart J. Formation and function of Spemann's organizer. Annu Rev Cell Dev Biol. 1997;13:611–667. doi: 10.1146/annurev.cellbio.13.1.611. [DOI] [PubMed] [Google Scholar]
- Henry GL, Melton DA. Mixer, a homeobox gene required for endoderm development. Science. 1998;281:91–96. doi: 10.1126/science.281.5373.91. [DOI] [PubMed] [Google Scholar]
- Henry GL, Brivanlou IH, Kessler DS, Hemmati-Brivanlou A, Melton DA. TGF-β signals and a pre-pattern in Xenopus laevis endodermal development. Development. 1996;122:1007–1015. doi: 10.1242/dev.122.3.1007. [DOI] [PubMed] [Google Scholar]
- Hill CS. The Smads. Int J Biochem Cell Biol. 1999;31:1249–1254. doi: 10.1016/s1357-2725(99)00093-x. [DOI] [PubMed] [Google Scholar]
- Hill CS, Marais R, John S, Wynne J, Dalton S, Treisman R. Functional analysis of a growth factor-responsive transcription factor complex. Cell. 1993;73:395–406. doi: 10.1016/0092-8674(93)90238-l. [DOI] [PubMed] [Google Scholar]
- Hill CS, Wynne J, Treisman R. The Rho family GTPases RhoA, Rac1, and CDC42Hs regulate transcriptional activation by SRF. Cell. 1995;81:1159–1170. doi: 10.1016/s0092-8674(05)80020-0. [DOI] [PubMed] [Google Scholar]
- Howell M, Hill CS. XSmad2 directly activates the activin-inducible, dorsal mesoderm gene XFKH1 in Xenopus embryos. EMBO J. 1997;16:7411–7421. doi: 10.1093/emboj/16.24.7411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell M, Itoh F, Pierreux CE, Valgeirsdottir S, Itoh S, ten Dijke P, Hill CS. Xenopus Smad4β is the co-Smad component of developmentally regulated transcription factor complexes responsible for induction of early mesodermal genes. Dev Biol. 1999;214:354–369. doi: 10.1006/dbio.1999.9430. [DOI] [PubMed] [Google Scholar]
- Huang H-C, Murtaugh LC, Vize PD, Whitman M. Identification of a potential regulator of early transcriptional responses to mesoderm inducers in the frog embryo. EMBO J. 1995;14:5965–5973. doi: 10.1002/j.1460-2075.1995.tb00285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann E, Knöchel W. Five years on the wings of forkhead. Mech Dev. 1996;57:3–20. doi: 10.1016/0925-4773(96)00539-4. [DOI] [PubMed] [Google Scholar]
- Keller R. Early embryonic development of Xenopus laevis. Methods Cell Biol. 1991;36:61–113. doi: 10.1016/s0091-679x(08)60273-3. [DOI] [PubMed] [Google Scholar]
- Kimelman D, Griffin KJ. Mesoderm induction: A postmodern view. Cell. 1998;94:419–421. doi: 10.1016/s0092-8674(00)81582-2. [DOI] [PubMed] [Google Scholar]
- Kispert A, Koschorz B, Herrmann BG. The T protein encoded by Brachyury is a tissue-specific transcription factor. EMBO J. 1995;14:4763–4772. doi: 10.1002/j.1460-2075.1995.tb00158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbé E, Silvestri C, Hoodless PA, Wrana JL, Attisano L. Smad2 and Smad3 positively and negatively regulate TGF β-dependent transcription through the forkhead DNA-binding protein FAST2. Mol Cell. 1998;2:109–120. doi: 10.1016/s1097-2765(00)80119-7. [DOI] [PubMed] [Google Scholar]
- Laurent MN, Blitz IL, Hashimoto C, Rothbacher U, Cho KW. The Xenopus homeobox gene twin mediates Wnt induction of goosecoid in establishment of Spemann's organizer. Development. 1997;124:4905–4916. doi: 10.1242/dev.124.23.4905. [DOI] [PubMed] [Google Scholar]
- Lemaire P, Darras S, Caillol D, Kodjabachian L. A role for the vegetally expressed Xenopus gene Mix.1 in endoderm formation and in the restriction of mesoderm to the marginal zone. Development. 1998;125:2371–2380. doi: 10.1242/dev.125.13.2371. [DOI] [PubMed] [Google Scholar]
- Liu B, Dou CL, Prabhu L, Lai E. FAST-2 is a mammalian winged-helix protein which mediates transforming growth factor β signals. Mol Cell Biol. 1999;19:424–430. doi: 10.1128/mcb.19.1.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Pouponnot C, Massagué J. Dual role of the Smad4/DPC4 tumor suppressor in TGFβ-inducible transcriptional complexes. Genes & Dev. 1997;11:3157–3167. doi: 10.1101/gad.11.23.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marais R, Wynne J, Treisman R. The SRF accessory protein Elk-1 contains a growth factor-regulated transcriptional activation domain. Cell. 1993;73:381–393. doi: 10.1016/0092-8674(93)90237-k. [DOI] [PubMed] [Google Scholar]
- Massagué J. TGF-β signal transduction. Annu Rev Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- McKendry R, Harland RM, Stachel SE. Activin-induced factors maintain goosecoid transcription through a paired homeodomain binding site. Dev Biol. 1998;204:172–186. doi: 10.1006/dbio.1998.9065. [DOI] [PubMed] [Google Scholar]
- Nakao A, Imamura T, Souchelnytskyi S, Kawabata M, Ishisaki A, Oeda E, Tamaki K, Hanai J, Heldin CH, Miyazono K, et al. TGF-β receptor-mediated signalling through Smad2, Smad3 and Smad4. EMBO J. 1997;16:5353–5362. doi: 10.1093/emboj/16.17.5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon RY, Yamashita K, Adamczewski JP, Hunt T, Shuttleworth J. The cdc2-related protein p40MO15 is the catalytic subunit of a protein kinase that can activate p33cdk2 and p34cdc2. EMBO J. 1993;12:3123–3132. doi: 10.1002/j.1460-2075.1993.tb05981.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa FM. Mix.1, a homeobox mRNA inducible by mesoderm inducers, is expressed mostly in the presumptive endodermal cells of Xenopus embryos. Cell. 1989;57:965–974. doi: 10.1016/0092-8674(89)90335-8. [DOI] [PubMed] [Google Scholar]
- Shi Y, Wang YF, Jayaraman L, Yang H, Massagué J, Pavletich NP. Crystal structure of a Smad MH1 domain bound to DNA: Insights on DNA binding in TGF-β signaling. Cell. 1998;94:585–594. doi: 10.1016/s0092-8674(00)81600-1. [DOI] [PubMed] [Google Scholar]
- Tada M, Casey ES, Fairclough L, Smith JC. Bix1, a direct target of Xenopus T-box genes, causes formation of ventral mesoderm and endoderm. Development. 1998;125:3997–4006. doi: 10.1242/dev.125.20.3997. [DOI] [PubMed] [Google Scholar]
- Vize PD. DNA sequences mediating the transcriptional response of the Mix.2 homeobox gene to mesoderm induction. Dev Biol. 1996;177:226–231. doi: 10.1006/dbio.1996.0158. [DOI] [PubMed] [Google Scholar]
- Watabe T, Kim S, Candia A, Rothbacher U, Hashimoto C, Inoue K, Cho KW. Molecular mechanisms of Spemann's organizer formation: Conserved growth factor synergy between Xenopus and mouse. Genes & Dev. 1995;9:3038–3050. doi: 10.1101/gad.9.24.3038. [DOI] [PubMed] [Google Scholar]
- Whitman M. Smads and early developmental signaling by the TGFβ superfamily. Genes & Dev. 1998;12:2445–2462. doi: 10.1101/gad.12.16.2445. [DOI] [PubMed] [Google Scholar]
- Wilson D, Sheng G, Lecuit T, Dostatni N, Desplan C. Cooperative dimerization of paired class homeo domains on DNA. Genes & Dev. 1993;7:2120–2134. doi: 10.1101/gad.7.11.2120. [DOI] [PubMed] [Google Scholar]
- Yasuo H, Lemaire P. A two-step model for the fate determination of presumptive endodermal blastomeres in Xenopus embryos. Curr Biol. 1999;9:869–879. doi: 10.1016/s0960-9822(99)80391-1. [DOI] [PubMed] [Google Scholar]
- Zhang J, Houston DW, King ML, Payne C, Wylie C, Heasman J. The role of maternal VegT in establishing the primary germ layers in Xenopus embryos. Cell. 1998;94:515–524. doi: 10.1016/s0092-8674(00)81592-5. [DOI] [PubMed] [Google Scholar]
- Zhou S, Zawel L, Lengauer C, Kinzler KW, Vogelstein B. Characterization of human FAST-1, a TGF β and activin signal transducer. Mol Cell. 1998;2:121–127. doi: 10.1016/s1097-2765(00)80120-3. [DOI] [PubMed] [Google Scholar]