Ctf7p is essential for sister chromatid cohesion and links mitotic chromosome structure to the DNA replication machinery (original) (raw)
Abstract
CTF7 (chromosome transmission fidelity) gene in budding yeast encodes an essential protein that is required for high-fidelity chromosome transmission and contains regions of identity conserved from yeast to man. ctf7 mutant cells arrested prior to anaphase onset contain separated sister chromatids. Thus, Ctf7p is essential for cohesion. Cohesion is established during S phase and then maintained until mitosis. However, Ctf7p activity is required only during S phase, suggesting that Ctf7p functions in the establishment of cohesion. In addition, ctf7 genetically interacts with DNA metabolism mutations pol30 (PCNA) and ctf18 (an RF-C like protein) and ctf7 temperature sensitivity and chromosome loss are rescued by high levels of POL30. These findings provide the first evidence that links the establishment of sister chromatid cohesion to the DNA replication machinery and suggest that the assembly of cohesion (and possibly condensation) complexes are coupled to PCNA-dependent DNA replication. The analysis of Ctf7p also reveals an important connection between sister chromatid cohesion, spindle integrity and the spindle assemby checkpoint.
Keywords: CTF7, chromosome loss, BUB3, MAD2, POL30, chromatid cohesion, condensation
Cohesion between replicated sister chromatids is fundamental to cell viability. Cohesion tethers replicated sister chromatids together along their entire lengths from S phase until anaphase. Chromatid cohesion also sterically constrains sister kinetochores so that they face away from each other, promoting proper chromosome attachment to microtubules nucleated from opposing spindle poles (Rieder and Salmon 1994; Hyman and Sorger 1995). On attachment of kinetochores to spindle microtubules, each sister chromatid experiences microtubule-dependent poleward forces. These poleward forces are resisted by cohesion so that sister chromatids do not separate but instead come under tension. Observations made in meiotic cells have led to the proposal that tension is monitored by the cell to ensure that each sister chromatid is properly associated with microtubules. Only after all of the chromosomes are placed under tension does the cell signal for the onset of anaphase (Nicklas 1997). Thus, chromatid cohesion may also be a critical link in the mechanism that regulates anaphase onset. At anaphase, cohesion complexes are inactivated and the sister chromatids segregate from one another and move toward their spindle poles.
Despite the essential role of chromatid cohesion in mitosis and cell division, only recently has progress been made in elucidating the molecular basis of this form of chromosome metabolism. In budding yeast, genetic and biochemical analyses have identified a complex that contains two regulators of sister chromatid cohesion, Pds1p and Esp1p (Cohen-Fix et al. 1996; Yamamoto et al. 1996 a,b; Ciosk et al. 1998). Pds1p is an inhibitor of anaphase onset and is degraded at the metaphase-to-anaphase transition by a ubiquitin-dependent process involving the anaphase promoting complex (APC). Pds1p proteolysis is thought to free Esp1p, which in turn promotes the inactivation of chromatid cohesion. The Pds1p–Esp1p complex is functionally related to the fission yeast Cut1p–Cut2p complex and potential mammalian homologs of Esp1p have been identified (Funabiki et al. 1996). In addition to these regulatory proteins, several candidate cohesion proteins have been identified: Mcd1p/Scc1p, Scc2p, Smc1p, and Smc3p from budding yeast (Guacci et al. 1997; Michaelis et al. 1997) and Mis4p from fission yeast (Furuya et al. 1998). Smc1p and Smc3p are members of the recently discovered gene family required for the structural maintenance of chromosomes (SMC) and are related by protein motifs first identified in Smc1p (Strunnikov et al. 1993). Inactivation of Mcd1p, Scc2p, Smc1p, Smc3p, or Mis4p causes precocious sister separation in M phase. Consistent with Mcd1p’s role in the maintenance of cohesion, Mcd1p associates with chromosomes from S phase through M phase. Mcd1p both exists in a complex with Smc1p and requires Smc1p to associate with chromatin (Guacci et al. 1997; Michaelis et al. 1997). Xenopus homologs of Mcd1p, Smc1p, and Smc3p, along with two other proteins, form a cohesin complex that is required in interphase to maintain cohesion in the subsequent mitosis (Losada et al. 1998). These similarities between yeast and vertebrate proteins suggest that the mechanisms of chromatid cohesion is likely to be conserved in all eukayotes.
In budding yeast, the cohesion factor Mcd1p is also required for chromosome condensation (Guacci et al. 1997), revealing an important, but poorly understood, link between cohesion and condensation. Chromosome condensation ensures both disentanglement of sisters and compaction of the chromosome arms away from the cytokinetic furrow (Koshland and Strunnikov 1996). Interestingly, two other SMC family members, Smc2p and Smc4p, are required for condensation and appear conserved across evolution (Chuang et al. 1994; Hirano and Mitchison 1994; Saitoh et al. 1994; Saka et al. 1994; Strunnikov et al. 1995; Jessberger et al. 1996; Sutani and Yanagida 1997). In Xenopus, Smc2p (XCAP-E) and Smc4p (XCAP-C) homologs combine with three additional proteins to form a condensin complex that associates with chromosomes (Hirano et al. 1997). Biochemical analysis of Xenopus extracts have revealed that cohesin and condensin complexes function independent of one another, indicating that the processes of cohesion and condensation are uncoupled in this cell system (Losada et al. 1998).
Although recent advances have been made in the identification of proteins required for cohesion and condensation, little is known about how cohesion proteins are assembled onto chromatin and, once assembled, how those complexes are regulated. For instance, temporal analysis of cohesion in human cells and genetic analysis of cohesion in yeast have shown that cohesion is established during S phase (Selig et al. 1992; Guacci et al. 1997; Michaelis et al. 1997; Furuya et al. 1998; Uhlmann and Nasmyth 1998). Yet, specialized factors that assemble cohesion complexes or load those complexes onto chromatin remain to be discovered. Another fundamental issue pertains to the regulation of chromatid cohesion. In early mitotic vertebrate cells, sister chromatids are paired along their entire length. In metaphase-arrested cells, however, the chromosome arms are no longer paired, whereas the centromeres can remain paired for an extended period of time (Sumner 1991). A similar spatial and temporal regulation is observed in meiotic cells: Cohesion is lost from the chromosome arms during meiosis I to resolve recombinant intermediates, but cohesion is retained at the centromeres until meiosis II. In budding yeast, mcd1 mutant cells are defective in cohesion at both the centromere and arms (Guacci et al. 1997; Michaelis et al. 1997). These results suggest that although a common cohesion mechanism exists for the entire chromosome, cohesion along the chromosome can be differentially regulated. Finally, Losada et al. (1998) recently reported that the Mcd1–Smc1–Smc3 protein complex in Xenopus dissociates from chromatin early in mitosis, suggesting that the process of cohesion may be more mechanistically complicated than anticipated and that other factors involved in cohesion have yet to be identified.
Previously, a collection of 136 chromosome transmission fidelity (ctf) mutants was identified in Saccharomyces cerevisiae on the basis of the increased rate of loss of a nonessential chromosome fragment (Spencer et al. 1990). ctf mutants that allowed both transcription to proceed through a centromere and the stable propagation of a dicentric chromosome (in vivo assays for kinetochore integrity) proved to contain mutations in genes subsequently shown to encode kinetochore components that bind the yeast centromere (Lechner and Carbon 1991; Doheny et al. 1993; Goh and Kilmartin 1993; Jiang et al. 1993). However, the vast majority of ctf mutants do not appear to correspond to integral kinetochore components and therefore constitute a powerful resource to identify proteins involved in novel processes crucial for proper chromosome transmission. In this paper, we report that CTF7 encodes an essential nuclear protein that is required for cohesion. The analysis of CTF7 also establishes a novel connection between cohesion and DNA replication.
Results
Ctf7p is an essential nuclear protein and contains domains that are highly conserved
In this study, we characterized ctf7 mutants, one complementation group of the ctf collection that exhibit elevated loss of a nonessential chromosome fragment (Spencer et al. 1990). The CTF7 gene was cloned on the basis of complementation of the chromosome loss phenotype and confirmed by integrative mapping (Materials and Methods). CTF7 was shown to encode an essential gene by both tetrad analyses of a heterozygous CTF7/ctf7Δ strain and plasmid shuffle methods (see Materials and Methods). CTF7 is identical to the unannotated ORF YFR027W on chromosome VI (Murakami 1995) and encodes a 281 amino acid protein with a predicted molecular mass of 31.8 kD (Fig. 1A). A search of the expressed sequence tag database (dBEST) revealed that portions of Ctf7p contained strong homologies to several human and mouse EST sequences (Fig. 1B). Similar homology was found for an ORF of unknown function in fission yeast. These results show that Ctf7p, an essential yeast protein, contains a motif that is conserved across kingdoms and suggests that Ctf7p-related proteins may play a role important for chromosome transmission in all eukaryotic cells. Overexpressed HA-tagged Ctf7p localized to the nucleus (HA-tagged Ctf7p under the control of the endogenous CTF7 promoter was undetectable; Materials and Methods), consistent with Ctf7p’s essential role in chromosome transmission.
Figure 1.
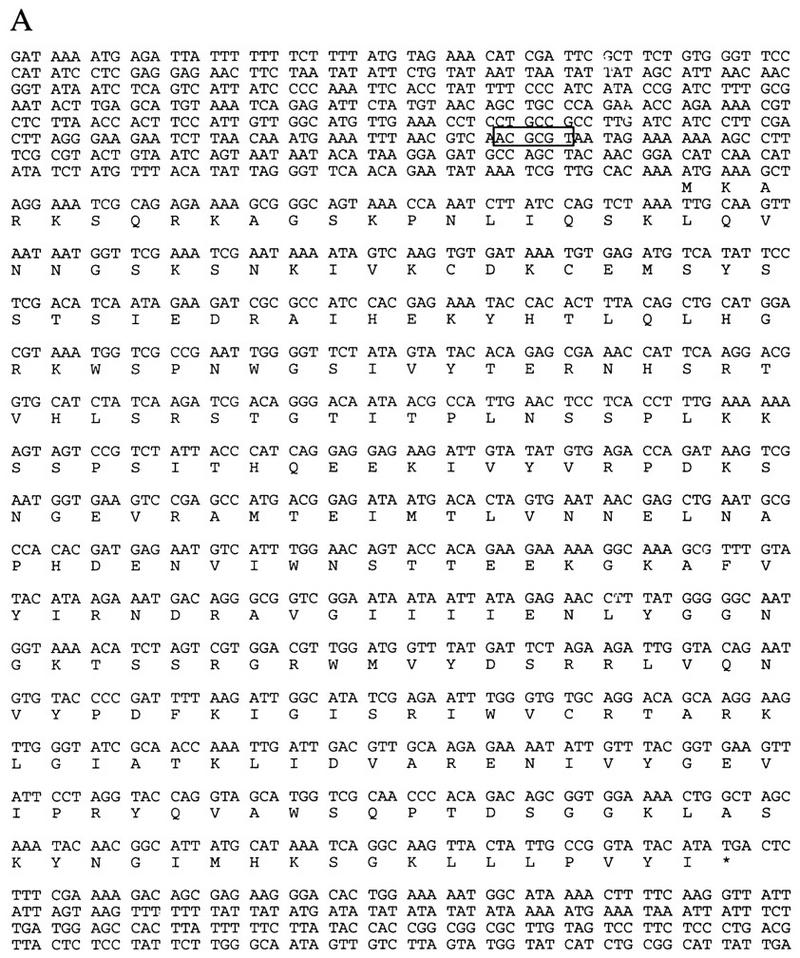
(A) Nucleotide and amino acid sequences of CTF7. The _Mlu_I cell cycle box in the 5′ untranslated region, which directs the expression of many genes required during S phase, is boxed (Johnston and Lowndes 1992; Toyn et al. 1995). (B) Ctf7p homology to fission yeast, mouse and human ESTs are shown. Identical residues are indicated by placement of the amino acid above the sequence, conserved residues are indicated with a plus. The carboxy-terminal region of Ctf7p (211–267 amino acids) is 49%, 40%, and 41% identical and 70%, 63%, and 60% similar to fission yeast, mouse, and human ESTs, respectively.
ctf7 mutants delay in mitosis with a 2C DNA content, separated DNA masses, and partially elongated spindles
To investigate the function of Ctf7p in vivo, three temperature-sensitive alleles of CTF7 were obtained by a PCR-based mutagenesis strategy and integrated into the genome (see Materials and Methods). At the permissive temperature, all three temperature-sensitive alleles, ctf7-201, ctf7-202, and ctf7-203, exhibited increased chromosome fragment loss (Fig. 2A), recapitulating the original Ctf phenotype. At the restrictive temperature, ctf7 mutant strains rapidly lost viability compared with wild type, indicating that all three alleles caused an irreversible defect (Fig. 2B).
Figure 2.
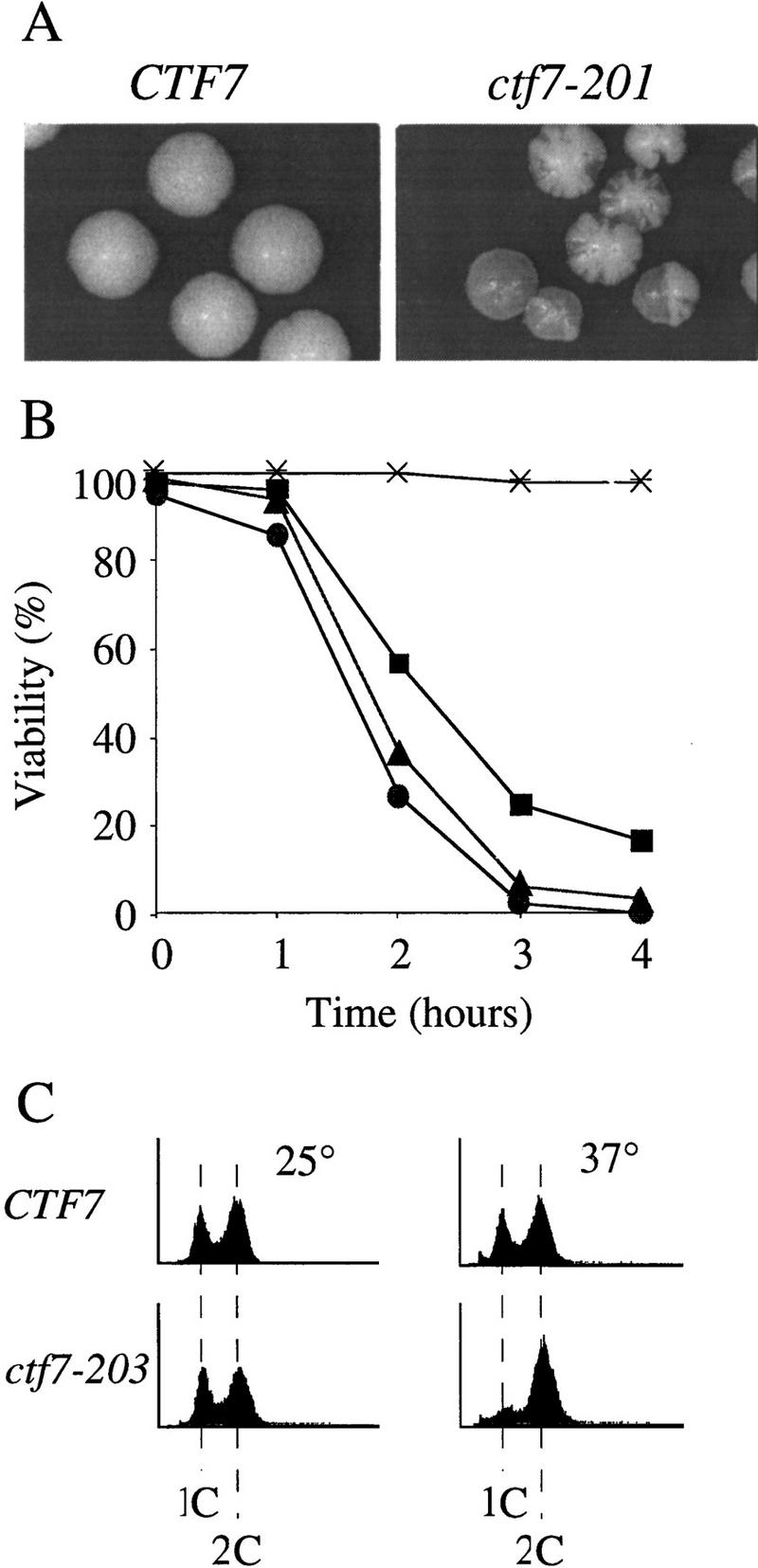
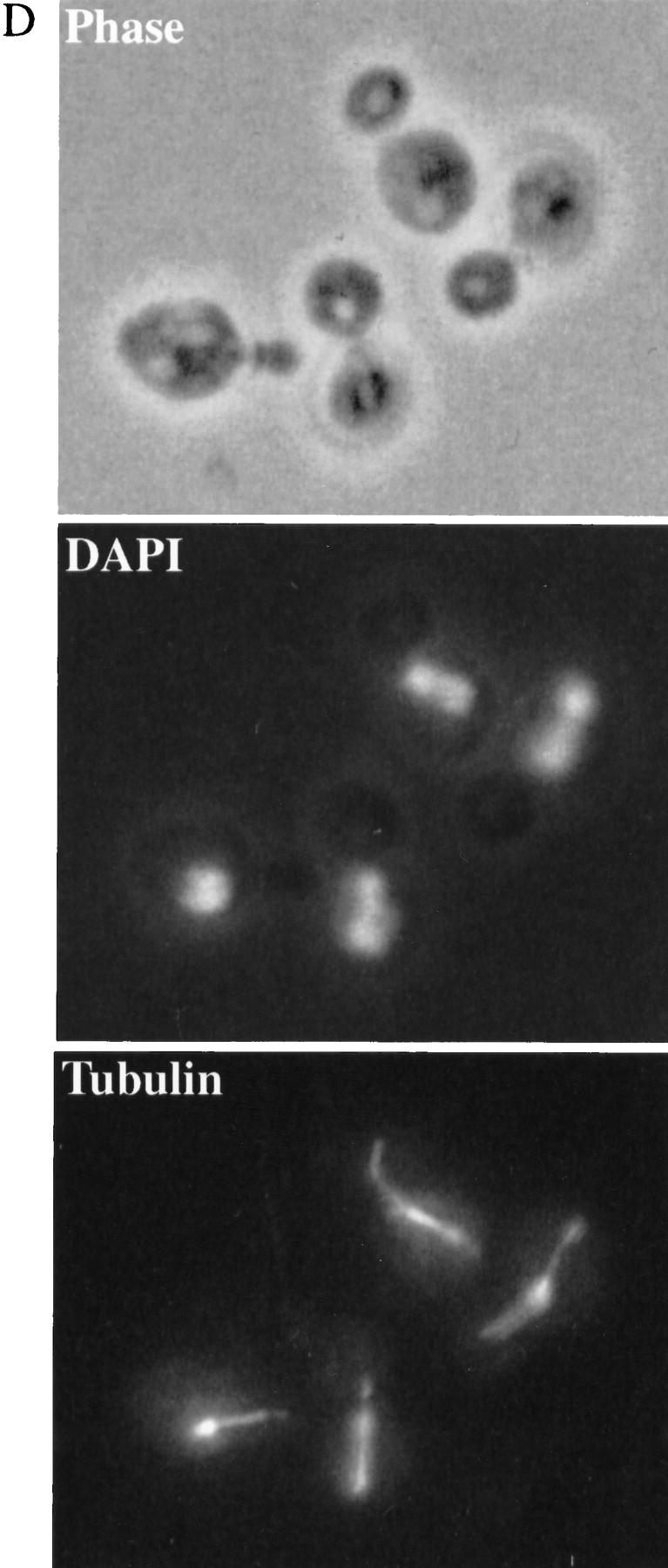
Phenotypes of ctf7 conditional mutants. (A) ctf7 cells (ctf7-201 shown) exhibit elevated chromosome loss over wild type. Faithful maintenance of a reporter chromosome fragment yields white colonies. Loss of the reporter chromosome results in red sectored colonies. (B) ctf7 mutants are temperature-sensitive lethal. Log-phase cultures were shifted from 25°C to 37°C and assayed for cell viability. (●) ctf7-201; (█) ctf7-202; (▴) ctf7-203; (*) wild type. (C) ctf7 mutants delay in mitosis. The DNA content of log phase cultures grown at 25°C or 37°C for 3 hr were assessed by flow cytometry (ctf7-203 shown). (D) Micrographs of ctf7 mutant cell (Phase), DNA (DAPI), and microtubule (Tubulin) morphologies after 3 hr growth at 37°C.
To ascertain the terminal phenotype of the ctf7 mutant strains, log phase cultures of each mutant were shifted from 25°C to 37°C and processed to assess DNA content and cell morphology. At 25°C, both wild-type and mutant strains exhibited similar DNA contents, as expected for cycling cells. However, after 3 hr growth at 37°C, a significant increase in the number of cells with a 2C DNA content was observed in ctf7 mutant strains compared with CTF7 control cells (Fig. 2C). Analyses of cell morphologies from this time point showed that ∼51% of ctf7 mutant cells were large budded and of these, 63% contained separating or bi-lobed DNA masses (Fig. 2D; Table 1A). The bilobed DNA masses often remained entirely within the mother cell and occasionally contained unequal amounts of DNA. Spindles from these cells were also atypical in that they were entirely contained within the mother cell (Fig. 2D) and appeared longer than preanaphase spindles. To quantify this last observation, log phase cultures of CTF7, ctf7-203, and ctf13-30 (a kinetochore mutant that arrests prior to anaphase onset) were shifted to 35°C for 3 hr, fixed, and immunostained for tubulin to visualize the spindle. Measurements of spindle lengths revealed that ctf7 mutant cells with bilobed DNA masses contained partially elongated spindles of 3.7 μm (±0.4, n = 7) in length; ∼2.5 times longer than the 1.4 μm (±0.3, n = 8) preanaphase spindles in ctf13-30 mutants but roughly half as long as the 6.9 μm (±0.9, n = 11) elongated spindles in anaphase CTF7 wild-type cells. The increased chromosome loss, large-budded cell morphology, 2C DNA content, abnormal spindle morphology, and unequal chromatin distribution observed in ctf7 mutation strains indicate that loss of Ctf7p function leads to a defect in mitosis and a mitotic delay.
Table 1.
ctf7 morphologies at 37°C
The mitotic delay in ctf7 cells requires the kinetochore/spindle assembly checkpoint
Mitotic delays typically arise by activation of either the DNA damage or kinetochore/spindle assembly checkpoint pathways. To determine whether ctf7 cells required either of these checkpoint mechanisms to delay in mitosis, we constructed strains containing ctf7 in combination with either mutations in the DNA damage checkpoint pathway (rad9Δ and mec1-1) (Weinert and Hartwell 1988, 1990; Weinert et al. 1994) or mutations in the kinetochore/spindle assembly checkpoint pathway (mad2-1 and bub3Δ) (Hoyt et al. 1991; Li and Murray 1991). Log phase cultures of the resulting strains were shifted to 37°C for 3 hr and their DNA content measured by flow cytometry. Neither rad9Δ nor mec1-1 mutations abrogated the ctf7-203 mediated 2C DNA accumulation (Fig. 3, data not shown). In contrast, both ctf7-203 mad2-1 and ctf7-203 bub3Δ strains exhibited a complete loss of the 2C DNA accumulation at the restrictive temperature (Fig. 3; data not shown). Consistent with this result, the ctf7-203 mad2-1 double mutant strain also failed to accumulate partially elongated spindles or bilobed DNA masses, but instead proceeded through mitosis as evidenced by the reappearance of G1-like (unbudded) cells (Table 1B). These results show that the mitotic delay in ctf7 cells requires a functional kinetochore/spindle assembly checkpoint but not a functional DNA damage checkpoint.
Figure 3.
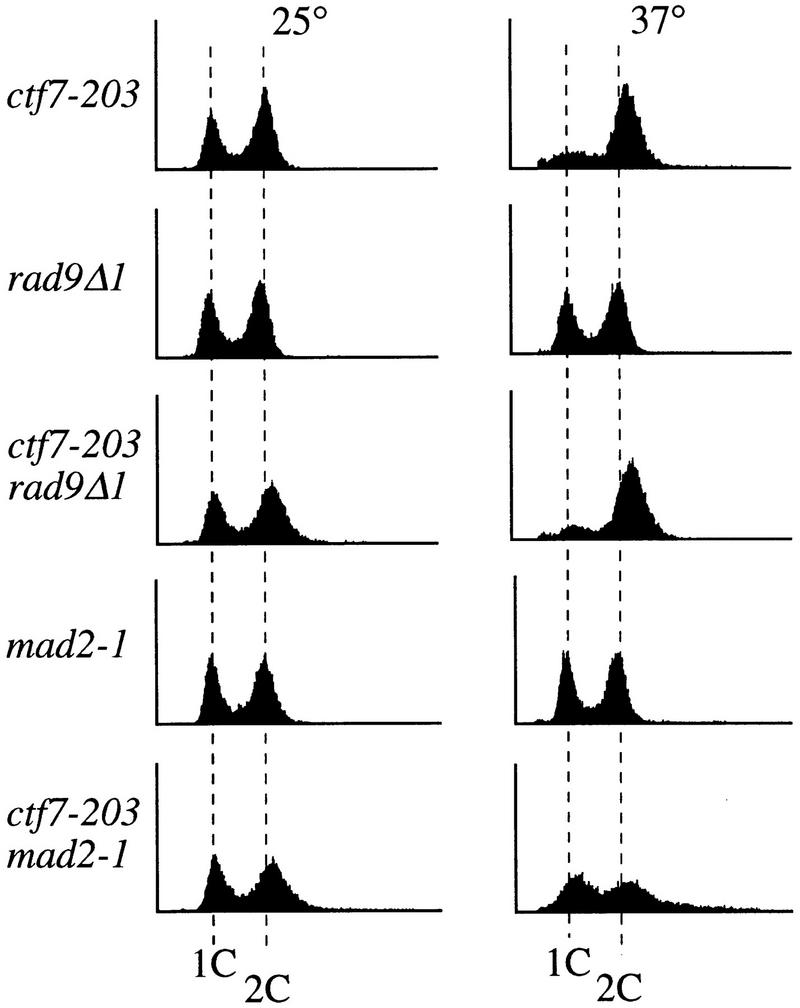
The mitotic delay in ctf7 cells requires the kinetochore/spindle checkpoint pathway. Flow cytometry analysis of DNA content of log phase ctf7, checkpoint mutant, and double-mutant strains grown for 3 hr at 25°C (left) or 37°C (right).
ctf7 cells delay in metaphase prior to anaphase onset
Typically, an activated kinetochore/spindle assembly checkpoint causes cells to arrest prior to anaphase onset and those cells contain short spindles and a single DNA mass. However, the _MAD/BUB_-dependent mitotic delay in ctf7 mutant cells lead to an accumulation of cells that contained partially elongated spindles and separated DNA masses. This unusual phenotype could represent an aberrant metaphase. An alternative possibility was that ctf7 cells were delayed in midanaphase, suggesting a novel postmetaphase role for Mad/Bub checkpoint proteins. To determine whether the mitotic delay occurred prior to anaphase onset, we assessed the levels of Pds1p in wild-type and ctf7 mutant cells. Pds1p is an inhibitor of anaphase onset and is degraded at the metaphase-to-anaphase transition (Cohen-Fix et al. 1996; Yamamoto et al. 1996a,b).
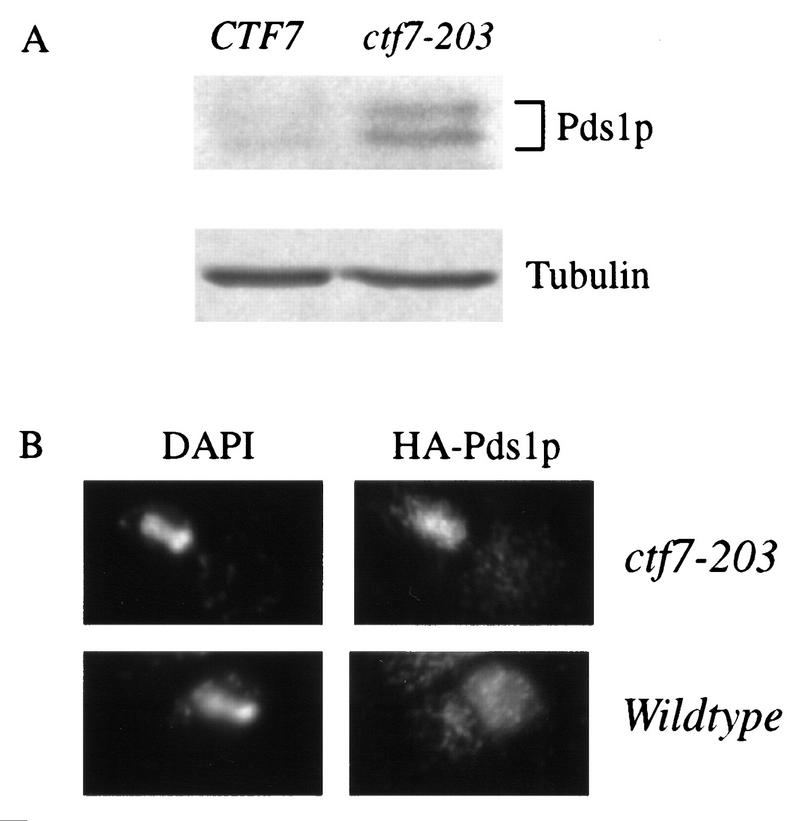
Wild-type and ctf7 mutant cells were transformed with a plasmid directing the expression of HA–Pds1p (Cohen-Fix et al. 1996). The level of HA–Pds1p was analyzed after synchronized G1 cells were released into rich medium and grown for 3 hr at 37°C. Western blot analysis revealed that wild-type cells had low levels of HA–Pds1p because most cells had already completed mitosis (Fig. 4A). In contrast, ctf7 cells contained high HA–Pds1p levels, indicating that ctf7 cells were delayed prior to anaphase onset. To confirm that ctf7 cells were delayed in metaphase, HA–Pds1p was visualized by immunofluorescence in wild-type and ctf7 cells treated as above. As expected, wild-type cells with separating DNA masses did not contain HA–Pds1p (Cohen-Fix et al. 1996). In contrast to wild-type cells, a significant population of ctf7 cells with separating DNA masses contained high levels of HA–Pds1p (Fig. 4B). Thus, these ctf7 cells were delayed prior to anaphase onset. It was not surprising that some ctf7 cells with separating DNA masses did not contain detectable HA–Pds1p levels, because ctf7 cells are only delayed in the cell cycle and eventually exit mitosis. These results reveal that partial spindle elongation and DNA separation occurs prior to anaphase onset in ctf7 mutant cells.
Figure 4.
ctf7 cells are delayed prior to anaphase onset. (A). The Western blot shows HA–Pds1p levels in wild-type and ctf7 cells that were synchronized in G1 and then released into medium and grown at 37°C for 3 hr. Tubulin levels verified that equal protein loads were present in each lane. (B) Micrographs show the localization of DNA (DAPI) and HA–Pds1p (HA–Pds1p) in ctf7 mutant and wild-type cells treated as in A.
Ctf7p is required for sister chromatid cohesion
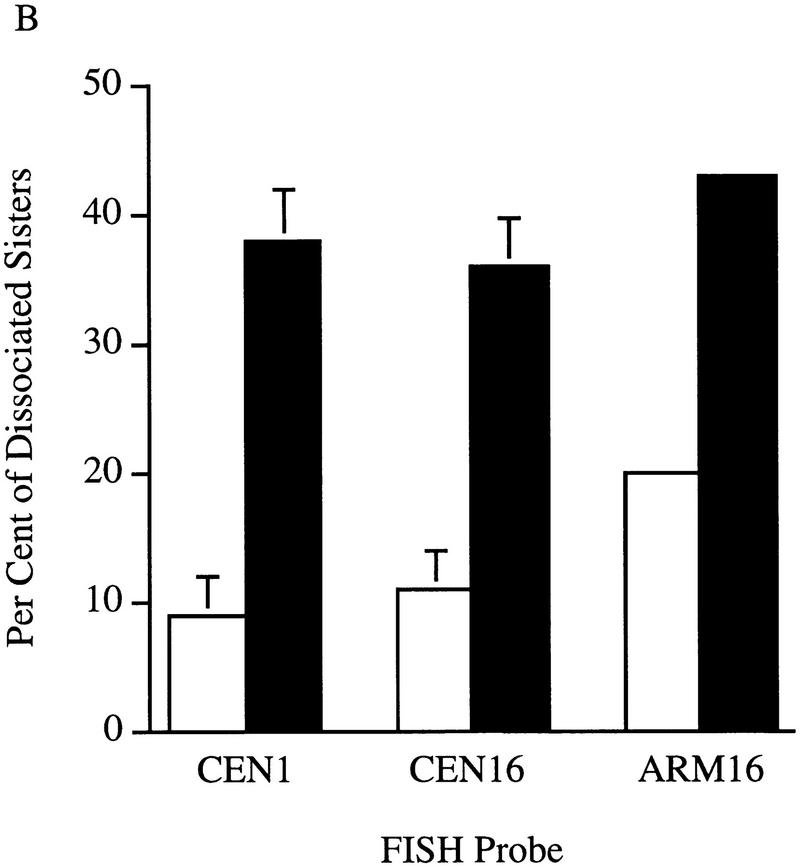
How can cells delayed prior to anaphase onset have separated DNA masses? One possibility was that defects in sister chromosome cohesion allowed sister chromatids to separate prematurely, resulting in the movement of disjoined sisters toward the spindle poles. To test this model, wild-type and ctf7-203 cells were synchronized in G1 with α-factor, released into medium that contained nocodazole, and grown for 3 hr at 37°C. Nocodazole blocks both microtubule assembly and anaphase onset such that sister chromatids typically remain closely apposed for extended periods of time. FISH has been used previously to demonstrate the pairing of sister chromatids in M-phase yeast cells and can further assess the pairing at different loci such as the centromere and chromosome arms (Guacci et al. 1994). Therefore, we used FISH to ask if sister chromatid cohesion was normal in ctf7 mutant cells. In wild-type cells arrested in mitosis, only ∼10% of the DNA masses contained two FISH signals when using a CEN16 proximal DNA probe (Fig. 5A,B). In contrast, ∼40% of the DNA masses from ctf7 cells contained two FISH signals, often with sister centromeres residing at opposite sides of the nuclear masses (Fig. 5A,B). The ctf7 defect affected other chromosomes, ∼40% of DNA masses in ctf7 cells contained two FISH signals with a CEN1 proximal probe (Fig. 5B). Finally, a similar loss of cohesion was obtained by use of centromere-distal (arm) probes to chromosome XVI, indicating that Ctf7p was required for cohesion along the entire length of the chromosome (Fig. 5B). These findings reveal that Ctf7p is essential for sister chromosome cohesion and supports previous findings that cohesion is mediated by a mechanism common throughout the chromosome (Guacci et al. 1997).
Figure 5.
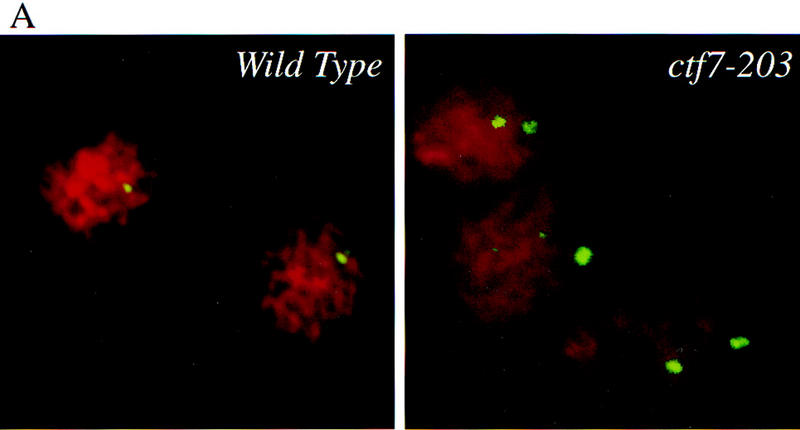
Ctf7p is required for sister chromatid cohesion. (A) Micrographs of wild-type and ctf7 mutant cells grown for 3 hr at 37°C in medium containing nocodazole and processed for FISH. A single FISH signal represents paired sister chromatids, two FISH signals represents disjoined sister chromatids. (red) Propidium iodide staining DNA mass; (green) hybridization to CEN16 probe. (B) Quantification of single (nondisjoined chromosomes) vs. double FISH signals (disjoined chromosomes) within propidium iodide staining DNA masses. Synchronized G1 wild-type and ctf7 cells contained an equal percentage of single FISH signals (data not shown), indicating that the increase in two FISH signals observed for ctf7 mutant cells was not due to pre-existing aneuploid.
Ctf7p is required during S phase
Sister chromatid cohesion is apparently established during S phase and maintained until anaphase onset. To assess the role of Ctf7p in the establishment and maintenance of cohesion, we used reciprocal temperature-shift experiments in ctf7 cells to map that portion of the cell cycle that requires Ctf7p function (Fig. 6). A shift-up experiment marks the earliest point in the cell cycle when Ctf7p can be inactivated without reducing cell viability (i.e., when Ctf7p function is no longer essential). ctf7-203 cells were synchronized in G1 with α-factor and then released into medium at 23°C to allow the cells to proceed through the cell cycle. Starting at the release from α-factor (t = 0), aliquots were shifted to 37°C at 20 min intervals. At the time of the shift, aliquots were also harvested for flow cytometry and immunofluorescence analyses to position cells within the cell cycle (see Materials and Methods). At the end of the experiment, all samples were plated at 23°C to assess viability (Materials and Methods). Cultures exhibited low viability when shifted to 37°C any time during the first 60 min (after α-factor release but prior to DNA replication) (Fig. 6A,B). Cultures exhibited high viability when shifted to 37°C after 80 min of growth at 23°C (Fig. 6A). The increase in viability correlated with progression of DNA replication: Maximum viability was realized only after the bulk of DNA replication was completed (80 min) (Fig. 6B). Furthermore, maximal cell viability preceded anaphase onset (spindle elongation) by >20 min (Fig. 6A). These results suggest that Ctf7p is required during S phase but not during mitosis.
Figure 6.
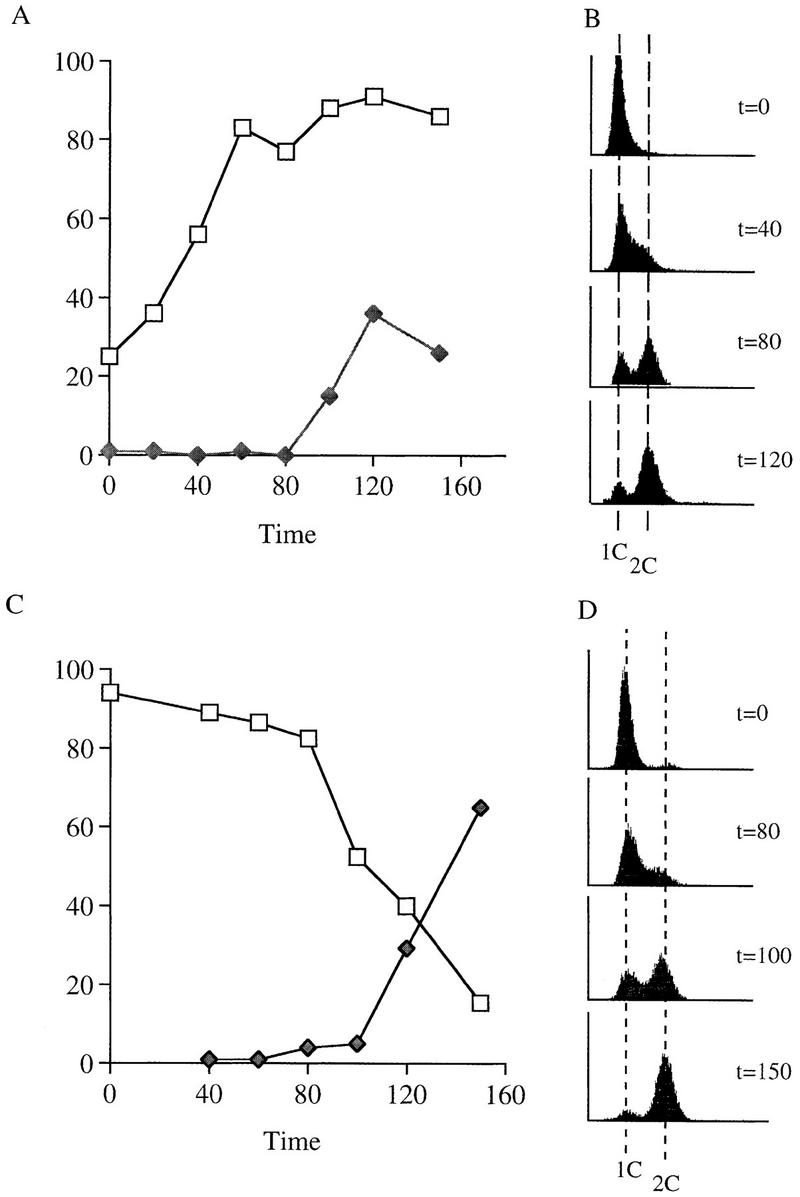
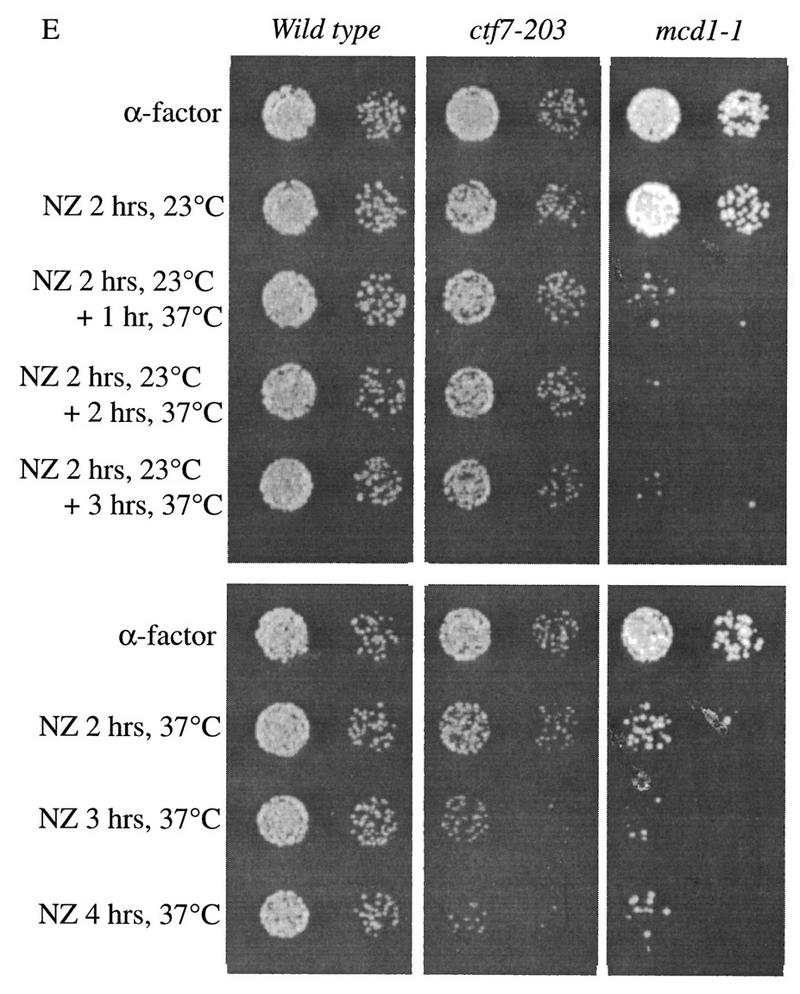
Ctf7p is required only during S phase. (A,B) Shift-up experiment: G1 ctf7-203 cells were released from α-factor and shifted from 23°C to 37°C at 20–30 min increments. (C,D) Shift-down experiment: G1 ctf7-203 cells were released from α-factor into 37°C medium and shifted down to 25°C at 20–30 min increments. Wild-type strains retain high viability throughout either procedures (data not shown). For each time point, an aliquot was assayed for spindle elongation/DNA mass separation (♦), viability (□), and DNA content (flow cytometry profile). (E, top) Viability of G1 synchronized wild-type, ctf7-203, and mcd1-1 cells released into medium containing nocodazole and permitted to arrest in mitosis at 23°C prior to shifting to 37°C. (E, bottom) Viability of G1 synchronized wild-type, ctf7-203, and mcd1-1 cells released into medium containing nocodazole at 37°C. A 10-fold dilution for each culture is shown.
A shift-down experiment marks the latest point in the cell cycle when Ctf7p must be active to produce viable cells. ctf7-203 cultures were synchronized in G1 with α-factor at 23°C, then released into 37°C rich medium in the absence of α-factor to allows cells to proceed through the cell cycle. Aliquots were then shifted down to 23°C at 20 min intervals and also harvested for flow cytometry and immunofluoresence analyses as described above. On release from α-factor (t = 0), cells retained high viability for 80 min of growth at 37°C prior to shifting down to 23°C (Fig. 6C). Decreased cell viability (starting at t = 80) correlated with entry into DNA replication (Fig. 6C,D) and a further decline in cell viability coincided with progression through S phase. Thus, Ctf7p must be active as cells enter S phase to obtain viable cells. Activation later in the cell cycle resulted in cell death. A further finding from this experiment was that ctf7 mutants progressed through S phase with kinetics similar to wild-type cells (data not shown). Thus, the lesion incurred during S phase in ctf7 mutant cells is not manifested as a defect in DNA replication. The combination of results from the shift-up and shift-down experiments in ctf7 cells suggest that Ctf7p function is essential during S phase when cohesion is established but not in mitosis when cohesion is maintained.
We decided to use a second method to test whether Ctf7p is required during M phase and S phase. Mcd1p appears to be required during both M phase and S phase and was included as an indicator for loss of cohesion (Guacci et al. 1997). To test whether Ctf7p was required during mitosis, wild-type, ctf7-203, and mcd1-1 mutant cells synchronized in G1 were released into medium containing nocodazole and allowed to proceed from G1 into mitosis at the permissive temperature of 23°C. Aliquots of the cultures were then shifted to 37°C at 1 hr intervals. After incubation at 37°C, the cultures were plated and allowed to grow for 2 days at 23°C to assess viability (Fig. 6E). Wild-type cells were not adversely affected by this procedure and maintained high viability. As expected, mcd1-1 cells exhibited a dramatic decrease in viability within the first hour of incubation at 37°C. In contrast, ctf7-203 cells retained high viability even after 3 hr incubation at 37°C. Thus, Ctf7p is not required during mitosis when sister chromatid cohesion is maintained. However, Ctf7p is required during S phase when cohesion is established. ctf7-203 and mcd1-1 mutant cells both exhibited decreased viability when synchronized cells were allowed to proceed from G1 in to mitosis at 37°C. The combination of these results suggest that Ctf7p plays a role in cohesion distinct from Mcd1p.
The high viability of ctf7-203 cells allowed to proceed into mitosis before shifting them to the restrictive temperature implied that Ctf7p was not required for the maintenance of cohesion. To demonstrate this unambiguously, we performed FISH analysis on wild-type and ctf7-203 cells. As before, G1 synchronized cells were released into medium containing nocodazole and allowed to proceed into mitosis at the permissive temperature. After 3 hr at 23°C, the cultures were shifted to 37°C and held at this restrictive temperature for 2 hr. Aliquots of the cultures were then processed for FISH analysis by use of both CEN1 and CEN16 proximal probes. Wild-type cells retained sister chromatid cohesion following this procedure, as only 22% (CEN1) and 23% (CEN16) of the DNA masses contained two FISH signals. ctf7-203 cells also retained sister chromatid cohesion, 26% (CEN1) and 27% (CEN16) of the DNA masses contained two FISH signals (data not shown). The slight increase in the number of DNA masses that contained two FISH signals observed for both strains was most likely due to the prolonged mitotic arrest and exposure to nocodazole. Thus, by use of both criteria of viability and localization of sister centromeres, Ctf7p is not required for the maintenance of sister chromatid cohesion.
Genetic interactions between CTF7 and the DNA replication machinery
Typically, high copy suppressors are proteins that either interact with the target protein or function in a parallel pathway. To gain further insight into the role of Ctf7p in cohesion, high-copy suppressors of the temperature sensitive ctf7 phenotype were identified. Three plasmids were obtained that enabled ctf7 mutant cells to grow at 34°C but not at 37°C. The three plasmids contained overlapping DNA sequences, suggesting suppression was due to a single gene. ctf7 temperature sensitivity was suppressed by a minimal DNA sequence defined by _Bgl_II to _Stu_I restriction sites (Fig. 7A). Sequence analysis revealed that this 1.7-kb fragment contained POL30, which encodes the DNA replication processivity factor proliferating cell nuclear antigen (PCNA) (Bauer and Burgers 1990; Burgers 1991). A frame-shift mutation in the POL30 ORF rendered the resulting plasmid incapable of rescuing ctf7-203 temperature sensitivity, indicating that POL30 is a high-copy suppressor of ctf7-203. Overexpression of POL30 also rescued the temperature sensitivity of ctf7-201 but failed to rescue a ctf7Δ1 deletion strain (not shown).
Figure 7.
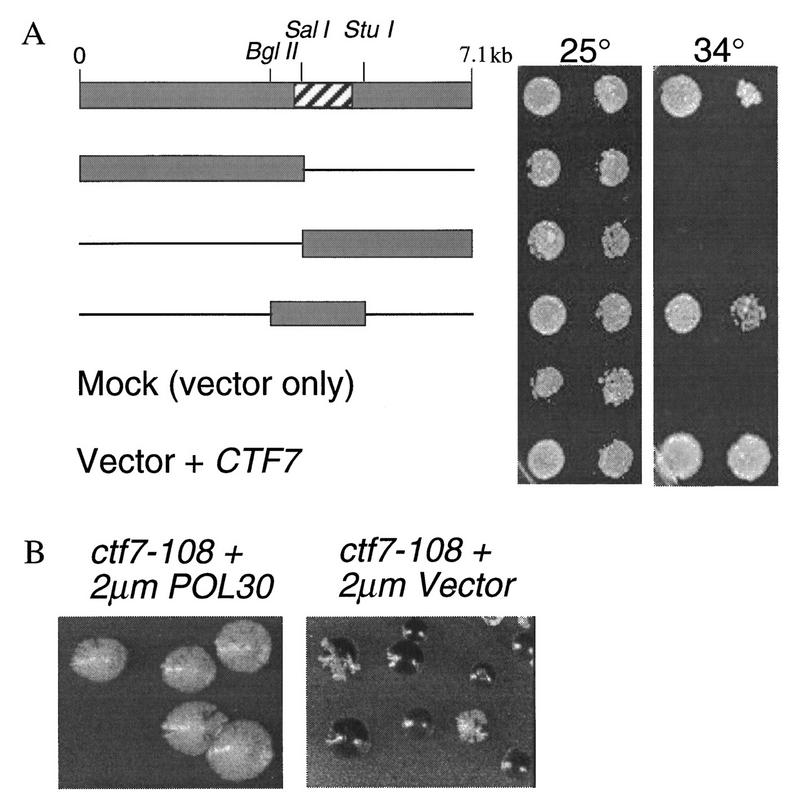
POL30 is an overexpression suppressor of ctf7 mutant phenotypes. (A) A 7.1-kb insert was identified that suppressed ctf7 temperature sensitivity at 34°C. Neither vector alone (Mock) nor vector containing insert DNA sequences to either the left or right of the _Sal_l digestion site rescued the ctf7 mutant at 34°C. A 1.7-kb fragment flanked by _Bgl_ll and _Stu_l restriction sites rescued ctf7 mutant temperature sensitivity but not to the same level of growth as CTF7 (Vector + CTF7). The hatched box in starting clone denotes POL30 OFR. (B) ctf7 mutations containing reporter chromosomes were transformed with 2 μm vector ± POL30 and grown on medium that allowed for chromosome fragment loss but retention of the 2 μm plasmid (ctf7-108 is a non-temperature-sensitive strain obtained from the original ctf collection).
On the basis of the finding that POL30 is a high-copy suppressor of ctf7 temperature sensitivity, we asked whether POL30 could also rescue ctf7 chromosome loss. To test this possibility, POL30 was overexpressed in strains containing either ctf7 temperature-sensitive alleles (ctf7-201 and ctf7-203) or a non-temperature-sensitive allele (ctf7-108) that was obtained in the original ctf collection (Spencer et al. 1990). Identical results were obtained in each case: Overexpression of POL30 decreased the chromosome loss associated with all ctf7 mutations tested (Fig. 7B).
To both test whether the genetic interaction between CTF7 and POL30 reflected an important link between the establishment of sister chromatid cohesion and DNA replication machinery, and to further identify gene products that might require CTF7 function, we assayed for synthetic lethal interactions. A strain harboring the ctf7-203 allele was mated to strains containing either checkpoint, kinetochore, DNA metabolism, or other chromosome loss mutants. The inability to recover double-mutant spores was scored as a synthetic lethal interaction (Table 2). Synthetic lethal interactions were observed for ctf7-203 in combination with either ctf18Δ1 or pol30-104. CTF18 encodes an RF-C-like protein and RF-C complexes are known to load PCNA onto double-stranded DNA (Burgers 1988; Kouprina et al. 1994; Ayyagari et al. 1995; Amin and Holm 1996). In contrast to these findings, no synthetic lethal interactions were observed when ctf7-203 was combined with checkpoint mutants (rad9Δ, mec1-1, mad2-1, or bub3Δ), kinetochore mutants (ctf13-30, ctf14-42, skp1-4, or skp1-3), or other mutants involved in DNA replication (ctf4-25, an allele of the Polα-binding protein, and cdc6/ctf10-103, an allele of the ORC interacting protein). Of 12 of the mutant strains that we tested (Table 3), synthetic lethality was only observed for ctf7 in combination with pol30 or ctf18 mutations. The specificity of these genetic interactions is substantiated by the related functions of PCNA and Ctf18p and by the finding that POl30 is a high-copy suppressor of ctf7 phenotypes. These genetic interactions link Ctf7p with proteins that are required during S phase, consistent with our mapping experiments, and together suggest that the establishment of sister chromatid cohesion is facilitated by components of the DNA replication machinery.
Table 2.
ctf7-203 is synthetically lethal with cft18Δ1 and pol30-104
| | | Observed | Expected | | ------------------------------------------------ | ----------------------------- | -------- | | pol30–104mms–LEU2 × ctf7Δ∷HIS3 ctf7-203ts:LEU2 | 61% viable (68 0f 112 spores) | | | wild type | 32+ | 28 | | pol30 | 28 | 28 | | ctf7 | 8 | 14 | | pol30 ctf7 | 0 | 14 | | ctf18Δ∷URA3 × ctf7Δ∷HIS3 ctf7-203ts:LEU2 | 55% viable (57 of 104 spores) | | | wild type | 30 | 26 | | ctf18Δ | 13 | 26 | | ctf7 | 14 | 13 | | ctf18Δ ctf7 | 0 | 13 |
Table 3.
Strains used in this study
| Strains | Genotypes | References |
|---|---|---|
| YBS2 | MATa/MATα ura3-52/ura3-52 lys2-801/lys2-801 ade2-101/ade2-101 trpl-Δ63/trp1-Δ63 his3-Δ200/his3-Δ200 leu2-Δ/leu2-Δ1 ctf7Δ1∷HIS3/CTF7 CFIII (CEN3.L) TRP1 SUP11 | this study |
| YBS3 | MATa ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 CTF7:HIS3 | |
| YBS5 | MATa ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 CTF7∷HIS3 CFIII (CEN3.L) TRP1 SUP11 pRS316_CTF7_ | |
| YBS204 | MATa ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 ctf7Δ1∷HIS3 CTF7:LEU2 rad9Δ1∷TRP1 | |
| YBS205 | MATa ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 ctf7Δ1∷HIS3 ctf7-203:LEU2 rad9Δ1∷TRP1 | |
| YBS213 | MATa ura3-52 lys2-801 ade2-101 trp1-Δ1,63 his3-Δ200 leu2-Δ1 ctf7Δ1∷HIS3 CTF7:LEU2 mad2-1 | |
| YBS214 | MATa ura3-52 lys2-801 ade2-101 trp1-Δ1,63 his3-Δ200 leu2-Δ1 ctf7Δ1∷HIS3 ctf7-203:LEU2 mad2-1 | |
| YBS504 | MATa ura3-52 lys2-801 ade2-101 his3-Δ200 leu2-Δ1 ctf7Δ1∷HIS3 CTF7:LEU2 bub3Δ1∷LEU2 | |
| YBS568 | MATa ura3-52 lys2-801 ade2-101 his3-Δ200 leu2-Δ1 ctf7Δ1∷HIS3 ctf7-203:LEU2 bub3Δ1∷LEU2 | |
| YBS255 | MATa ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 ctf7Δ1∷HIS3 CTF7:LEU2 | |
| YBS514 | MATα ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 ctf7Δ1∷HIS3 ctf7-203:LEU2 | |
| YBS268 | MATα ura3 trp1 his3 leu2 ctf7Δ1∷HIS3 ctf7-203:LEU2 mec1-1:HIS3 | |
| YBS271 | MATα ura3 trp1 his3 leu2 ctf7Δ1∷HIS3 CTF7–LEU2 mec1-1:HIS3 | |
| YCTF108 | MATa ura3-52 lys2-801 ade2-101 trp1-Δ1 leu2-Δ1 ctf7-108 CFIII (CEN3.L) URA3 SUP11 | Spencer et al. (1990) |
| YCTF103 | MATa ura3-52 lys2-801 ade2-101 trp1-Δ1 leu2-Δ1 ctf10-103 CFIII (CEN3.L) URA3 SUP11 | |
| YCTF42 | MATα ura3-52 lys2-801 ade2-101 his3-Δ200 leu2-Δ1 ctf14-42 CFIII (CEN3.L) URA3 SUP11 | |
| YCTF30 | MATα ura3-52 lys2-801 ade2-101 his3-Δ200 leu2-Δ1 ctf13-30 CFIII (CEN3.L) URA3 SUP11 | |
| YCTF25 | MATα ura3-52 lys2-801 ade2-101 his3-Δ200 leu2-Δ1 cf4-25 CFIII (CEN3.L) URA3 SUP11 | |
| YCTF10 | MATα ura3-52 lys2-801 ade2-101 his3-Δ200 leu2-Δ1 ctf7-10 CFIII (CEN3.L) URA3 SUP11 | |
| YPH1182 | MATa ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 skp1-4:LEU2 | Connelly and Heiter (1996) |
| YPH1188 | MATa ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 skp1-3:LEU2 | |
| YCH2165 | MATa leu2-3,112 ura3-52 POL30:LEU2 | Amin and Holm (1996) |
| YCH2161 | MATa leu2-3,112 ura3-52 pol30–104:LEU2 | |
| YJE106 | MATa ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 ctf18Δ1:LEU2 | Kouprina et al. (1994) |
| YJE188 | MATa ura3-52 lys2-801 ade2-101 trp1-Δ1 ctf18Δ1∷URA3 | |
| DLY285 | MATa ura3 trp1 his3 leu2 trp1 mec1-1–HIS3 | Paulovich (1995) |
| YCB624 | MATa ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 rad9Δ1∷TRP1 | Brachmann (1995) |
| YFS451 | MATa ura3-52 lys2-801 ade2-101 trp1-Δ1,63 his3-Δ200 leu2-Δ1 mad2-1 CFIII (CEN3.L) URA3 SUP11 | Pangilinan (1996) |
| YFP74 | MATα ura3-52 lys2-801 ade2-101 his3-Δ200 leu2-Δ1 bub3Δ1∷LEU2 | |
| VG955-7D | MATa mcd1-1 trp1 leu2 bar1 gal1 | Guacci et al. (1997) |
Discussion
Ctf7p, a novel link between sister chromatid cohesion, S phase, and the DNA replication machinery
In this study, we have identified Ctf7p as a protein required for sister chromatid cohesion. HA–Ctf7p is a nuclear protein and mutations in CTF7 result in elevated chromosome loss, consistent with Ctf7p’s role in chromosome segregation (this paper; Spencer et al. 1990). We discovered further that Ctf7p is required for cohesion at both centromeres and chromosome arms, as found for the cohesion factor Mcd1p (this paper; Guacci et al. 1997; Michaelis et al. 1997). Independent analysis has also shown that Ctf7p (called Eco1p) is required for cohesion (A. Toth, R. Ciosk, F. Uhlmann, M. Galova, A. Schleiffer, K. Nasmyth, pers. comm.). Therefore, although there is evidence from vertebrate cell chromosomes that cohesion is differentially regulated along the length of the chromosome (Sumner 1991), Ctf7p and Mcd1p appear to function in aspects of cohesion that are common to all regions of the chromosome.
Cell viability and sister chromatid cohesion are abolished when ctf7 cells are placed at the restrictive temperature during S phase but not during mitosis. These findings demonstrate that Ctf7p, unlike Mcd1p, functions specifically in the establishment of cohesion (this paper; Guacci et al. 1997; Michaelis et al. 1997). An independent allele of ctf7 (eco1-1) is also defective in the establishment but not the maintenance of cohesion (A. Toth, R. Ciosk, F. Uhlmann, M. Galova, A. Schleiffer, K. Nasmyth, pers. comm.), indicating that the inability to reveal a function for Ctf7p in the maintenance of cohesion is not an allele-specific effect. The model in which Ctf7p functions in the establishment of cohesion during S phase is further supported by the genetic interactions observed between mutations in CTF7 and the DNA metabolism genes POL30 and CTF18 (Kouprina et al. 1994; Amin et al. 1996). POL30 was the only gene found in a genome-wide screen that, at high copy, suppressed ctf7 temperature sensitivity and chromosome loss. In addition, pol30-104 and ctf18Δ1 are synthetically lethal with ctf7-203. The significance of these genetic interactions is augmented by their specificity; no synthetic lethal interactions were observed for ctf7 in combination with mutations that affect either the initiation of DNA replication, kinetochore function, or DNA damage or spindle assembly checkpoint pathways. The genetic interactions between CTF7, POL30, and CTF18 do not appear to reflect a general response of ctf7 mutant strains to alterations in S-phase progression. For instance, both ctf18Δ1 and pol30-104 are cold-sensitive mutations that, at elevated temperatures, exhibit normal S-phase kinetics (Kouprina et al. 1994; Amin and Holm 1996). In addition, ctf7 mutant strain temperature sensitivity was not suppressed by levels of hydroxyurea that slow DNA elongation (R.V. Skibbens and D. Koshland, unpubl.). Taken together, these observations suggest that Ctf7p functions during S phase for the establishment of cohesion and reveals an important link between DNA replication and sister chromatid cohesion.
The connection between CTF7 and POL30 is particularly intriguing. PCNA forms doughnut-like sliding clamps that are continuously loaded behind DNA polymerases at the replication fork (Krishna et al. 1994). Others have noted that PCNA both appears to be landing pads for a variety of proteins not directly related to DNA replication and may play a role in chromatin remodeling (Kelman 1997). In combination with our results, these data suggest that the loading of cohesin complexes onto chromatin is coupled with PCNA-dependent DNA replication. This model is attractive because it places cohesin complexes proximal to newly replicated sister chromatids to ensure the pairing of sister chromatids but preclude the pairing of nonsister chromatids. Loading of cohesin complexes onto chromatin can be uncoupled from replication, as has been observed in Xenopus extracts and budding yeast cells, but at least in yeast this association fails to generate functional sister chromatid cohesion (Losada et al. 1998; Uhlmann and Nasmyth 1998).
In budding yeast, Mcd1p is required for both cohesion and condensation along chromosome arms, providing the first link between these two processes (Guacci et al. 1997). Given this precedent, we found that Ctf7p was also required for condensation at the rDNA locus, although the defect in ctf7 mutants was not as pronounced as that observed in mcd1 mutants (R.V. Skibbens and D. Koshland, unpubl.). Concomitant with our study, Trf4 protein has also been shown to be required for both cohesion and condensation (Castano et al. 1996; I. Castano and M. Christman, pers. comm.). Thus, their study of Trf4p and our study of _ctf7_p further establishes that yeast cohesin and condensin complexes share either common components, regulators, or assembly factors. In contrast, Losada et al. (1998) reported that depleting cohesin components from Xenopus extracts did not adversely affect chromosome condensation. In addition, Xenopus 13S condensin and 14S cohesin complexes were found to associate with chromatin independent of one another, indicating a separation of function (Losada et al. 1998). It will be important to pursue whether the coupling/uncoupling of cohesion and condensation reflect differences in organisms, cell systems (somatic versus embryonic) or segregation cycles (mitotic versus meiotic). Such differences have been well documented and what once appeared as conflicting data has historically proven to be of tremendous scientific value.
Cohesion, checkpoints, and spindle integrity
Our results showed that ctf7 mutant cells exhibited a mitotic delay that required the kinetochore/spindle assembly checkpoint. Consistent with the preanaphase delay typically imposed by this checkpoint, ctf7 cells contained high levels of the anaphase inhibitor Pds1p. On the other hand, ctf7 cells contained elongated spindles and separated DNA masses, uncharacteristic of preanaphase cells. This aberrant metaphase delay raises three issues that further reveal important functions of sister chromatid cohesion.
First, how does the ctf7 mutation disrupt spindle structure? In preanaphase budding yeast cells, spindle pole bodies are mechanically linked by a few interdigitating polar microtubules and by many kinetochore microtubules that are indirectly connected by virtue of their attachment to paired sister chromatids (O’Toole et al. 1997). We propose that the dissolution of sister chromatid cohesion complexes weakens this linkage and profoundly affects spindle integrity. Abnormal spindles have been reported for other mutants defective in cohesion such as smc1 and mcd1 (Strunnikov et al. 1993; Guacci et al. 1997; Michaelis et al. 1997). Thus, sister chromatid cohesion appears to function as part of the mechanical linkage that is required to maintain spindle integrity.
The second question raised by our observations is why do ctf7 mutants activate the kinetochore/spindle assembly checkpoint and delay in mitosis? To date, the kinetochore/spindle damage checkpoint pathway has been implicated in monitoring defects in centromere, kinetochore, or spindle function and responds by delaying cells in mitosis (Skibbens and Hieter 1998). Currently, we cannot rule out a model whereby ctf7 cells contain both defective kinetochores and sister chromatid cohesion complexes. However, it is not necessary to invoke a kinetochore defect to explain the checkpoint-mediated delay in ctf7 cells. Interestingly, other cohesion mutants also exhibit a mitotic delay, although it is unknown whether this delay is dependent on a spindle assembly checkpoint (Strunnikov et al. 1993; Guacci et al. 1997; Michaelis et al. 1997). On the basis of our evidence, we envision cohesion defects to activate this checkpoint by altering the state of tension exerted on chromosomes. Chromosomes attached to microtubules experience poleward forces that place the chromosome under tension. Evidence from meiotic cell systems revealed that tension exerted on chromosomes is critical for signaling anaphase onset (Nicklas 1997): A decrease or loss of tension generates a mitotic delay. One predicted effect for the dissolution of chromatid cohesion is to decrease the level of tension exerted on chromosomes by spindle microtubules and activate the spindle assembly checkpoint (Wells and Murray 1996). This model suggests that sister chromatid cohesion is also a fundamental component of the mechanism that maintains tension on chromosomes.
Typically, checkpoint mechanisms maintain cell viability by delaying the cell cycle and allowing cells time to repair damage or correct mistakes. In mutations that induce such damage, the additional loss of checkpoint function often results in synthetic lethality. Therefore, a third question is why is ctf7 not synthetically lethal with mutations in the kinetochore/spindle assembly checkpoint? The loss of the checkpoint has little impact on ctf7 cells for two reasons. First, one component of the checkpoint mechanism is to block sister separation. However, ctf7 cells are defective in sister chromatid cohesion and subvert this component of checkpoint function. Second, sister separation is an irreversible event. Thus, whether the cell delays or not in response to checkpoint function, cells with precociously separated sisters are beyond rescue. Our results are analogous to those reported over a decade ago, double-strand breaks generated in G1 haploid cells resulted in a _RAD9_-mediated cell cycle delay at G2/M. Despite this delay, the cells were inviable (Weinert and Hartwell 1988).
Materials and methods
Yeast strains and medium
Medium used for growth and sporulation was described previously (Rose et al. 1990). Yeast transformations were performed as described (Ito et al. 1983; Schiestl and Gietz 1989). S288C-derived Saccharomyces cerevisiae yeast strains (YPH) and plasmids (pRS) were described previously (Sikorski and Hieter 1989; Doheny et al. 1993; Connelly and Hieter 1996). Other strains are listed in Table 3. To establish genetic interactions, two independent double heterozygous diploids were obtained, sporulated, and haploid spores containing both mutations identified. The nonessential reporter chromosome of ∼150 kb contains the short arm and centromere of chromosome III for stable propagation and SUP11 to suppress the formation of the red byproduct of the adenine biosynthetic pathway in ade2 mutations (Spencer et al. 1990).
Cloning and molecular characterization of CTF7
A region of DNA that rescued the chromosome fragment-loss phenotype in a ctf7 mutant strain (ctf7-108) from the original ctf collection (Spencer et al. 1990) was identified from a CEN/LEU2 library and subcloned following procedures and reagents described previously (Gerring et al. 1990). This 2.1-kb region was shown to encode the chromosome transmission fidelity locus of CTF7 in the following manner. The 2.1-kb fragment was cloned into pRS303, linearized within the CTF7 ORF with _Xba_I and integrated into the genome of YPH250. His+ transformants were shown to contain _HIS3_-marked CTF7 integrated at the proper locus by Southern analysis on the basis of the known restriction map of the region. The resulting strain (YBS3) was mated with a strain from the original collection that contains ctf7-10 (Spencer et al. 1990) and sporulated. His+ spores (22 of 22) exhibited wild-type levels of chromosome loss, whereas all 14 His− spores tested sectored dramatically when grown on nonselective medium. Database searches of the yeast genome indicated that CTF7 is contained on Chromosome VI. This chromosome assignment was confirmed by hybridization to yeast genomic clone contigs (L. Riles and M. Olson, pers. comm.).
The CTF7 ORF was replaced with the selectable marker HIS3 by a PCR strategy described previously (Baudin et al. 1993). The deletion DNA fragments were generated by PCR by the primers 5′-AGTAATAATACATAAGGAGATGCCAGCTACAACGGACATCAACATATATCTAGATTGTACTGAGAGTGCACC-3′ and 5′-ACCTTGAAAAGTTTTATGCCATTTTTCCAGTGTCCCTTCTCGCTGTCTTTCTGTGCGGTATTTCACACCG-3′. The wild-type diploid strain YPH987 was transformed with PCR product and His3+ colonies identified to obtain a heterozygous CTF7/ctf7Δ deletion strain (YBS2). CTF7 deletion was confirmed by Southern blot analysis. Tetrad analysis yielded 2+:2− spore death with no recovery of His+ spores. To confirm that CTF7 encodes an essential protein, the diploid YBS2 (ctf7Δ::HIS3/CTF7) was transformed with pRS316–CTF7 and sporulated. All His+ Ura+ spores tested were inviable on medium plates containing 5-FOA (Boeke et al. 1987).
Generation, integration and suppression of conditional ctf7 alleles
PCR-based mutagenesis of pRS316–CTF7 was performed as described (Kassenbrock et al. 1993) with minor modifications. YBS5 (ctf7Δ covered by pRS316–CTF7) was cotransformed with pooled PCR product and Mlu_I–_Nde_I-gapped pRS315–_CTF7 (this digestion removes the entire CTF7 ORF but leaves 5′ and 3′ ends homologous to the CTF7 genomic locus). Of 12,500 Leu+ transformants screened, 11 scored positive as temperature-sensitive nonpetite strains by virtue of their inability to grow on 5-FOA plates at 37°C and growth on glycerol plates at 25°C. Plasmids containing putative mutated CTF7 sequences were recovered from those strains. YBS5 was then transformed with the rescued plasmids to confirm plasmid-dependent temperature sensitivity. Three of the eleven plasmids that exhibited the lowest reversion rate and conferred the tightest temperature sensitivity were chosen for integration into the genome. The 2.1-kb insert was cloned into pRS304 and the resulting plasmid linearized with Hpa_I to target integration to the LEU2 locus in YBS5. After passage on 5-FOA, His+ Leu+ Ura− transformants were temperature sensitive for growth. The temperature-sensitive phenotype was rescued on retransformation with pRS316–_CTF7. Hence, these mutations are recessive.
For overexpression suppression, ctf7-203 was transformed with a 2 μm of URA3 library (Connelly et al. 1996). Of 24,900 transformants, 26 plasmids were obtained that, on retransformation, rescued growth at 34°C. Twenty-three of the twenty-six plasmids contained the CTF7 ORF. The remaining three were found to contain overlapping sequences and one plasmid was subcloned to identify the suppressor ORF.
Morphological and growth analyses
Flow cytometry, Western blot analysis, and indirect immunofluorescence (IIF) were performed as described previously (Gerring et al. 1990; Cohen-Fix et al. 1996) with minor modifications. Viability was determined as described previously (Morrow et al. 1995) with minor modifications. Briefly, cells were plated after experimental procedures onto rich YPD medium plates and grown at the permissive temperature for 18 hr. The plates were then assessed microscopically for the formation of microcolonies. Typically, microcolonies were rescored after an additional 24 hr. For execution experiments, the incidence of anaphase onset was based on either spindle elongation or separation of DNA masses. In shift-down experiments, the percent of anaphase cells rose dramatically because of the cell cycle delay observed in ctf7 mutants, consistent with the accumulation of a 2C DNA content. For localization of Ctf7p, the entire CTF7 ORF was placed in-frame with the encoded HA epitope tag contained on pAD5 (Neiman et al. 1993). YBS5 was transformed with this plasmid and pRS316-CTF7 shuffled out from the resulting strain by use of medium plates containing 5-Fluororotic acid, FOA (Boeke et al. 1987) prior to immunolocalization. Immunostaining was performed by use of the anti-HA antibody 12CA5 (BabCo) in combination with fluorescein- or Cy3-conjugated goat anti-mouse antibodies (Jackson ImmunoResearch) to visualize epitope-tagged Ctf7p and Pds1p (Cohen-Fix et al. 1996). Not all nuclei stained with 12CA5, indicating that protein levels varied considerably when constructs are carried on a 2 μm plasmid. YBS5 similarly transformed with the pAD5 vector only (containing no CTF7 insert) exhibited no nuclear staining, nor was a nuclear signal observed with secondary antibody alone. YOL1/34 (Serotec) and fluorescein- or rhodamine-conjugated goat anti-rat antibodies (Cappel and Boehringer Mannhein) were used to visualize microtubules. DNA masses were visualized with DAPI (Sigma). For Western blot analysis of HA–Pds1, cell densities were normalized by optical density. Equal loads of cell extracts were confirmed by Western blot analysis of tubulin levels.
Reagents used to perform FISH analysis and detailed procedures are described previously (Guacci et al. 1994, 1997).
Acknowledgments
We thank A. Toth, R. Ciosk, F. Uhlmann, M. Galova, A. Schleiffer, and K. Nasmyth for sharing unpublished results. We also thank Drs. C.-M. Fan, J. Boeke, C. Holm, A.W. Murray, P. Megee, V. Guacci, S. Laloraya, B. Lavoie, Z. Feng, and O. Cohen-Fix for their critical reading of this manuscript. We also thank F. Spencer and members of the Koshland, Hieter, Murray, and T. Kelly laboratories for helpful discussions. In addition, we thank members of the Hieter, Koshland, and Boeke laboratories, especially C. Connelly, M. Basrai, P. Megee, E. Hogan, O. Cohen-Fix, V. Guacci, and M. Kenna, for their support, guidance, and use of reagents throughout the course of this project. We also thank N. Amin, C. Holm, F. Spencer and Hieter and Boeke laboratory members for generously sharing strains and J. Flook for his expertise in Flow cytometry. This work was supported by National Institutes of Health grants GM17770 to R.V.S, GM1718 to D.K., who is also a Howard Hughes investigator, and CA16519 to P.H.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL skibbens@mail1.ciwemb.edu; FAX (410) 243-6311
References
- Amin NS, Holm C. In vivo analysis reveals that the interdomain region of the yeast proliferating cell nuclear antigen is important for DNA replication and DNA repair. Genetics. 1996;144:479–493. doi: 10.1093/genetics/144.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayyagari R, Impellizzeri KJ, Yoder BL, Gary SL, Burgers PM. A mutational analysis of the yeast proliferating cell nuclear antigen indicates distinct roles in DNA replication and DNA repair. Mol Cell Biol. 1995;15:4420–4429. doi: 10.1128/mcb.15.8.4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer GA, Burgers PM. Molecular cloning, structure and expression of the yeast proliferating cell nuclear antigen gene. Nucleic Acids Res. 1990;18:261–265. doi: 10.1093/nar/18.2.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke JD, Trueheart J, Natsoulis G, Fink GR. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- Brachmann CB, Sherman JM, Devine SE, Cameron EE, Pillus L, Boeke JD. The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression, and chromosome stability. Genes & Dev. 1995;9:2888–2902. doi: 10.1101/gad.9.23.2888. [DOI] [PubMed] [Google Scholar]
- Burgers PM. Mammalian cyclin/PCNA (DNA polymerase delta auxiliary protein) stimulates processive DNA synthesis by yeast DNA polymerase III. Nucleic Acids Res. 1988;16:6297–6307. doi: 10.1093/nar/16.14.6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Saccharomyces cerevisiae replication factor C. II. Formation and activity of complexes with the proliferating cell nuclear antigen and with DNA polymerases delta and epsilon. J Biol Chem. 1991;266:22698–22706. [PubMed] [Google Scholar]
- Castano IB, Brzoska PM, Sadoff BU, Chen H, Christman MF. Mitotic chromosome condensation in the rDNA requires TRF4 and DNA topoisomerase I in Saccharomyces cerevisiae. Genes & Dev. 1996;10:2564–2576. doi: 10.1101/gad.10.20.2564. [DOI] [PubMed] [Google Scholar]
- Chuang P-T, Albertson DG, Meyer BJ. DPY-27: A chromosome condensation protein homolog that regulates C. elegans dosage compensation through association with the X chromosome. Cell. 1994;79:459–474. doi: 10.1016/0092-8674(94)90255-0. [DOI] [PubMed] [Google Scholar]
- Ciosk R, Zachariae W, Michaelis C, Shevchenko A, Mann M, Nasmyth K. An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell. 1998;93:1067–1076. doi: 10.1016/s0092-8674(00)81211-8. [DOI] [PubMed] [Google Scholar]
- Cohen-Fix O, Peters JM, Kirschner MW, Koshland D. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes & Dev. 1996;10:3081–3093. doi: 10.1101/gad.10.24.3081. [DOI] [PubMed] [Google Scholar]
- Connelly C, Hieter P. Budding yeast SKP1 encodes an evolutionarily conserved kinetochore protein required for cell cycle progression. Cell. 1996;86:275–285. doi: 10.1016/S0092-8674(00)80099-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doheny KF, Sorger PK, Hyman AA, Tugendreich S, Spencer F, Hieter P. Identification of essential components of the S. cerevisiae kinetochore. Cell. 1993;73:761–774. doi: 10.1016/0092-8674(93)90255-O. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabiki H, Kumada K, Yanagida M. Fission yeast Cut1 and Cut2 are essential for sister chromatid separation, concentrate along the metaphase spindle and form large complexes. EMBO J. 1996;15:6617–6628. [PMC free article] [PubMed] [Google Scholar]
- Furuya K, Takahashi K, Yanagida M. Faithful anaphase is ensured by Mis4, a sister chromatid cohesion molecule required in S phase and not destroyed in G1 phase. Genes & Dev. 1998;12:3408–3418. doi: 10.1101/gad.12.21.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerring SL, Spencer F, Hieter P. The CHL1 (CTF1) gene product of Saccharomyces cerevisiae is important for chromosome transmission and normal cell cycle progression in G2/M. EMBO J. 1990;9:4347–4358. doi: 10.1002/j.1460-2075.1990.tb07884.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh PY, Kilmartin JV. NDC10: A gene involved in chromosome segregation in Saccharomyces cerevisiae. J Cell Biol. 1993;121:503–512. doi: 10.1083/jcb.121.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guacci V, Hogan E, Koshland D. Chromosome condensation and sister chromatid pairing in budding yeast. J Cell Biol. 1994;125:517–530. doi: 10.1083/jcb.125.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guacci V, Koshland D, Strunnikov A. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell. 1997;91:47–57. doi: 10.1016/s0092-8674(01)80008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T, Mitchison TJ. A heterodimeric coiled-coil protein required for mitotic chromosome condensation in vitro. Cell. 1994;79:449–458. doi: 10.1016/0092-8674(94)90254-2. [DOI] [PubMed] [Google Scholar]
- Hirano T, Kobayashi R, Hirano M. Condensins, chromosome condensation protein complexes containing XCAP-C, XCAP-E and a Xenopus homolog of the Drosophila Barren protein. Cell. 1997;89:511–521. doi: 10.1016/s0092-8674(00)80233-0. [DOI] [PubMed] [Google Scholar]
- Hoyt MA, Totis L, Roberts BT. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell. 1991;66:507–517. doi: 10.1016/0092-8674(81)90014-3. [DOI] [PubMed] [Google Scholar]
- Hyman AA, Sorger PK. Structure and function of kinetochores in budding yeast. Annu Rev Cell Dev Biol. 1995;11:471–495. doi: 10.1146/annurev.cb.11.110195.002351. [DOI] [PubMed] [Google Scholar]
- Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessberger R, Riwar B, Baechtold H, Akhmedov A. SMC proteins constitute two subunits of the mammalian recombination complex RC-1. EMBO J. 1996;15:4061–4068. [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Lechner J, Carbon J. Isolation and characterization of a gene (CBF2) specifying a protein component of the budding yeast kinetochore. J Cell Biol. 1993;121:513–519. doi: 10.1083/jcb.121.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston LH, Lowndes NF. Cell cycle control of DNA synthesis in budding yeast. Nucleic Acids Res. 1992;20:2403–2410. doi: 10.1093/nar/20.10.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassenbrock CK, Cao W, Douglas MG. Genetic and biochemical characterization of ISP6, a small mitochondrial outer membrane protein associated with the protein translocation complex. EMBO J. 1993;12:3023–3034. doi: 10.1002/j.1460-2075.1993.tb05971.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelman Z. PCNA: Structure, functions and interactions. Oncogene. 1997;14:629–640. doi: 10.1038/sj.onc.1200886. [DOI] [PubMed] [Google Scholar]
- Koshland D, Strunnikov A. Mitotic chromosome condensation. Annu Rev Cell Dev Biol. 1996;12:305–333. doi: 10.1146/annurev.cellbio.12.1.305. [DOI] [PubMed] [Google Scholar]
- Kouprina N, Kroll E, Kirillov A, Bannikov V, Zakharyev V, Larionov V. CHL12, a gene essential for the fidelity of chromosome transmission in the yeast Saccharomyces cerevisiae. Genetics. 1994;138:1067–1079. doi: 10.1093/genetics/138.4.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna TS, Kong XP, Gary S, Burgers PM, Kuriyan J. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell. 1994;79:1233–1243. doi: 10.1016/0092-8674(94)90014-0. [DOI] [PubMed] [Google Scholar]
- Lechner J, Carbon J. A 240 kd multisubunit protein complex, CBF3, is a major component of the budding yeast centromere. Cell. 1991;64:717–725. doi: 10.1016/0092-8674(91)90501-o. [DOI] [PubMed] [Google Scholar]
- Li R, Murray AW. Feedback control of mitosis in budding yeast. Cell. 1991;66:519–531. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- Losada A, Hirano M, Hirano T. Identification of Xenopus SMC protein complexes required for sister chromatid cohesion. Genes & Dev. 1998;12:1986–1997. doi: 10.1101/gad.12.13.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis C, Ciosk R, Nasmyth K. Cohesins: Chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- Morrow DM, Tagle DA, Shiloh Y, Collins FS, Hieter P. TEL1, an S. cerevisiae homolog of the human gene mutated in ataxia telangiectasia, is functionally related to the yeast checkpoint gene MEC1. Cell. 1995;82:831–840. doi: 10.1016/0092-8674(95)90480-8. [DOI] [PubMed] [Google Scholar]
- Murakami Y, Naitou M, Hagiwara H, Shibata T, Ozawa M, Sasanuma S-I, Tsuchiya Y, Soeda E, Yokoyama K, Yamazaki M, Tashiro H, Eki T. Analysis of the nucleotide sequence of chromosome VI from saccharomyces cervisiae. Nature Genet. 1995;10:261–268. doi: 10.1038/ng0795-261. [DOI] [PubMed] [Google Scholar]
- Neiman AM, Stevenson BJ, Xu HP, Sprague GF, Jr, Herskowitz I, Wigler M, Marcus S. Functional homology of protein kinases required for sexual differentiation in Schizosaccharomyces pombe and Saccharomyces cerevisiae suggests a conserved signal transduction module in eukaryotic organisms. Mol Biol Cell. 1993;4:107–120. doi: 10.1091/mbc.4.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklas RB. How cells get the right chromosomes. Science. 1997;275:632–637. doi: 10.1126/science.275.5300.632. [DOI] [PubMed] [Google Scholar]
- O’Toole ET, Mastronarde DN, Giddings TH, Jr, Winey M, Burke DJ, McIntosh JR. Three-dimensional analysis and ultrastructural design of mitotic spindles from the cdc20 mutant of Saccharomyces cerevisiae. Mol Biol Cell. 1997;8:1–11. doi: 10.1091/mbc.8.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pangilinan F, Spencer F. Abnormal kinetochore structure activates the spindle assembly checkpoint in budding yeast. Mol Biol Cell. 1996;7:1195–1208. doi: 10.1091/mbc.7.8.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulovich AG, Hartwell LH. A checkpoint regulates the rate of progression through S phase in S. cerevisiae in response to DNA damage. Cell. 1995;82:841–847. doi: 10.1016/0092-8674(95)90481-6. [DOI] [PubMed] [Google Scholar]
- Rieder CL, Salmon ED. Motile kinetochores and polar ejection forces dictate chromosome position. J Cell Biol. 1994;124:223–233. doi: 10.1083/jcb.124.3.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P. Methods in yeast genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- Saitoh N, Goldberg IG, Wood ER, Earnshaw WC. ScII: An abundant chromosome scaffold protein is a member of a family of putative ATPases with an unusual predicted tertiary structure. J Cell Biol. 1994;127:303–318. doi: 10.1083/jcb.127.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saka Y, Sutani T, Yamashita Y, Saitoh S, Takeuchi M, Nakaseko Y, Yanagida M. Fission yeast cut3 and cut14, members of a ubiquitous protein family, are required for chromosome condensation and segregation in mitosis. EMBO J. 1994;13:4938–4952. doi: 10.1002/j.1460-2075.1994.tb06821.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiestl RH, Gietz RD. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- Selig S, Okumura K, Ward DC, Cedar H. Delineation of DNA replication time zones by fluorescence in situ hybridization. EMBO J. 1992;11:1217–1225. doi: 10.1002/j.1460-2075.1992.tb05162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibbens, R.V. and P. Hieter. 1998. Kinetochores and the checkpoint mechanism that monitors for defects in the chromosome segregation machinery. Annu. Rev. Genet. (in press). [DOI] [PubMed]
- Spencer F, Gerring SL, Connelly C, Hieter P. Mitotic chromosome transmission fidelity mutants in Saccharomyces cerevisiae. Genetics. 1990;124:237–249. doi: 10.1093/genetics/124.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunnikov AV, Larionov VL, Koshland D. SMC1: An essential yeast gene encoding a putative head-rod-tail protein is required for nuclear division and defines a new ubiquitous protein family. J Cell Biol. 1993;123:1635–1648. doi: 10.1083/jcb.123.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunnikov AV, Hogan E, Koshland D. SMC2, a Saccharomyces cerevisiae gene essential for chromosome segregation and condensation, defines a subgroup within the SMC family. Genes & Dev. 1995;9:587–599. doi: 10.1101/gad.9.5.587. [DOI] [PubMed] [Google Scholar]
- Sumner AT. Scanning electron microscopy of mammalian chromosomes from prophase to telophase. Chromosoma. 1991;100:410–418. doi: 10.1007/BF00337519. [DOI] [PubMed] [Google Scholar]
- Sutani T, Yanagida M. DNA renaturation activity of the SMC complex implicated in chromosome condensation. Nature. 1997;388:798–801. doi: 10.1038/42062. [DOI] [PubMed] [Google Scholar]
- Toyn JH, Johnson AL, Johnston LH. Segregation of unreplicated chromosomes in Saccharomyces cerevisiae reveals a novel G1/M checkpoint. Mol Cell Biol. 1995;15:5312–5321. doi: 10.1128/mcb.15.10.5312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann F, Nasmyth K. Cohesion between sister chromatids must be established during DNA replication. Curr Biol. 1998;8:1095–1101. doi: 10.1016/s0960-9822(98)70463-4. [DOI] [PubMed] [Google Scholar]
- Weinert TA, Hartwell LH. The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science. 1988;241:317–322. doi: 10.1126/science.3291120. [DOI] [PubMed] [Google Scholar]
- ————— Characterization of RAD9 of Saccharomyces cerevisiae and evidence that its function acts posttranslationally in cell cycle arrest after DNA damage. Mol Cell Biol. 1990;10:6554–6564. doi: 10.1128/mcb.10.12.6554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert TA, Kiser GL, Hartwell LH. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes & Dev. 1994;8:652–665. doi: 10.1101/gad.8.6.652. [DOI] [PubMed] [Google Scholar]
- Wells WA, Murray AW. Aberrantly segregating centromeres activate the spindle assembly checkpoint in budding yeast. J Cell Biol. 1996;133:75–84. doi: 10.1083/jcb.133.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Guacci V, Koshland D. Pds1p is required for faithful execution of anaphase in the yeast, Saccharomyces cerevisiae. J Cell Biol. 1996a;133:85–97. doi: 10.1083/jcb.133.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Pds1p, an inhibitor of anaphase in budding yeast, plays a critical role in the APC and checkpoint pathway(s) J Cell Biol. 1996b;133:99–110. doi: 10.1083/jcb.133.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]