Overexpression of Stra13, a novel retinoic acid-inducible gene of the basic helix–loop–helix family, inhibits mesodermal and promotes neuronal differentiation of P19 cells (original) (raw)
Abstract
We report the cDNA cloning of Stra13, a novel retinoic acid (RA)-inducible gene from P19 embryonal carcinoma cells that encodes a basic helix–loop–helix (bHLH) protein that shows the highest sequence similarities to the Drosophila Hairy and Enhancer of split and mouse Hes proteins. Stra13 does not bind to the known consensus motifs (E-box and N-box) for bHLH proteins, but can repress activated transcription (through an α-helix rich domain) in part by interaction with general factors of the basal transcription machinery. During mouse embryogenesis, Stra13 RNA is expressed in the neuroectoderm, and also in a number of mesodermal and endodermal derivatives. Remarkably, overexpression of Stra13 in P19 cells results in neuronal differentiation in monolayer culture, under conditions where wild-type P19 cells only undergo mesodermal/endodermal differentiation. This neuronal differentiation is accompanied by an altered expression of mesodermal and neuronal markers, indicating that Stra13 could be one of the earliest RA target genes whose expression is required for repression of mesodermal/endodermal differentiation and/or induction of neuronal differentiation when P19 cell aggregates are exposed to RA. Our results raise the possibility that Stra13 could be involved as a repressor in a number of decision events occurring during differentiation of various cell lineages.
Keywords: Stra13, RNA, RA, P19 cells, mesodermal, endodermal
Retinoic acid (RA), the active derivative of vitamin A, plays an important role in various physiological processes including vision, reproduction, development, and homeostasis (Chambon 1994; Sporn et al. 1994). Two families of nuclear receptors designated retinoic acid receptors (RARα, RARβ, and RARγ, and their isoforms) and retinoic X receptors (RXRα, RXRβ, and RXRγ, and their isoforms) transduce the RA signal (Chambon 1994, 1996). These receptors are members of the steroid/thyroid superfamily of ligand-activated transcription factors (Gronemeyer and Laudet 1995). RARs bind to, and are activated by, both all-trans retinoic acid (T-RA) and its 9-cis isomer (9-cis RA), whereas RXRs bind and are activated by 9-cis RA only. RAR and RXR heterodimers efficiently activate transcription of target genes by binding to retinoic acid response elements (RAREs), and are believed to transduce the retinoid signal to the transcription machinery and the chromatin template through transcriptional intermediary factors (also called coactivators or mediators) that directly interact with the liganded receptors (Chambon 1996; Glass et al. 1997).
RA regulates the growth and differentiation of many cell types, such as the murine pluripotent P19 embryonal carcinoma (EC) cells. Depending on the concentration of RA and the culture conditions, P19 cells can differentiate into all three embryonic germ layers, that is, endoderm, mesoderm, and ectoderm (McBurney 1993). In monolayer culture, addition of RA induces differentiation into cells with endodermal and mesodermal characteristics (Roguska and Gudas 1985; Mummery et al. 1986). On the other hand, treatment of P19 cell aggregates with RA results primarily, after replating, into neural-like and endodermal-like cells (McBurney et al. 1982). The neuron-like cells that are formed upon aggregation of P19 cells in the presence of RA share several similarities with neurons present in the mammalian nervous system, being postmitotic, containing functional synapses, and expressing a number of neurotransmitters (for review, see Bain et al. 1994). Thus, P19 cells have been used repeatedly as a model system to study neuronal differentiation in vitro.
Since the discovery of the RARs, one major thrust has been to identify RA-responsive genes to elucidate the basis of receptor function in different physiological processes. With the aim of identifying RA target genes involved in the differentiation of P19 cells, we recently reported the isolation of novel RA-responsive genes in a cDNA differential screening of P19 cells treated with RA in monolayers (Bouillet et al. 1995) These genes have been referred to as Stra genes (for stimulated with RA). We report here the molecular cloning and functional characterization of Stra13, which encodes a novel member of the basic helix–loop–helix (bHLH) protein family. We show that Stra13 functions as a repressor of activated transcription and interacts with general factors of the basal transcription machinery. Remarkably, overexpression of Stra13 in P19 cell monolayers induces neuronal differentiation in the presence of RA under conditions where wild-type P19 cells only undergo mesodermal/endodermal differentiation. This ectopic neuronal differentiation in monolayer culture is accompanied by an altered expression of neuronal and mesodermal markers. During mouse development, however, Stra13 expression is not confined to the neuroectoderm, but is also expressed in a number of mesodermal and endodermal derivatives. Taken together, our results indicate that Stra13 expression might be one of the early events occurring during differentiation of P19 cell aggregates into neural cells, and suggest that Stra13 could be involved in the control of differentiation of several cell lineages during mouse development.
Results
Cloning of Stra13 cDNA reveals a novel nuclear bHLH protein
A partial (190-bp) cDNA clone isolated during a differential cDNA screening for RA-induced genes in P19 cell monolayers (Bouillet et al. 1995) was used to screen an oligo(dT)-primed cDNA library prepared from P19 cells treated with RA for 24 hr. Three cDNA clones were obtained, and the longest one (2909 bp) (hereafter called Stra13 cDNA) had a 411 amino acid residue-long open reading frame (45,360 daltons) (Fig. 1). The ATCATGG sequence (nucleotides 214–220) fulfills Kozak’s rule for initiator codons and a typical polyadenylation signal (AATTAAAA) is found 1330-nucleotides downstream from the stop codon.
Figure 1.
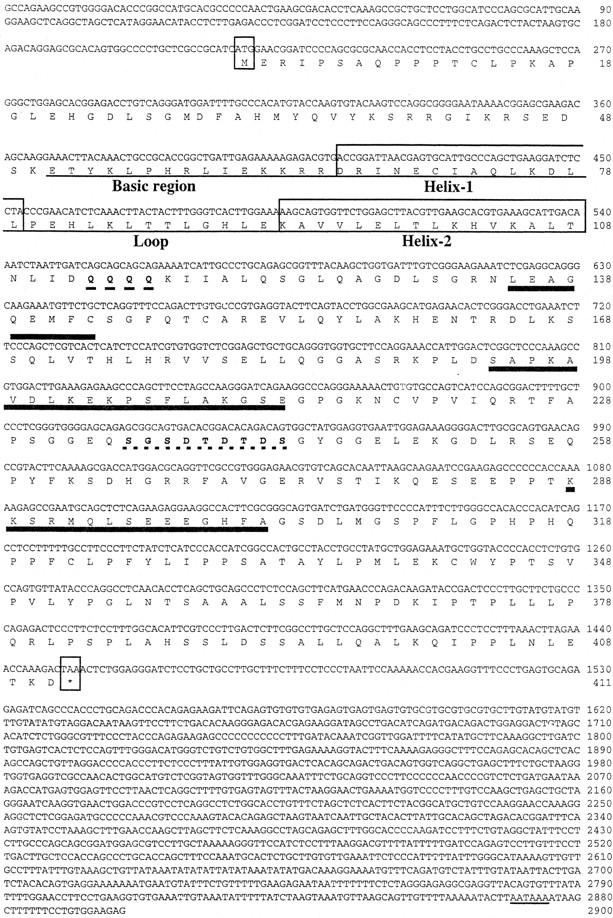
Nucleotide and predicted amino acid sequence of mouse Stra13 cDNA. Numbers on the right correspond to nucleotide or amino acid positions. The first and last codons of the ORF are boxed and the putative polyadenylation signal is underlined. The amino acids corresponding to the putative bHLH regions are underlined and boxed. Amino acids forming putative α-helix structures are underlined with thick lines. Glutamine residues (Q) in the glutamine-rich region and residues of the putative casein kinase 2 motif are in boldface type with a dotted underline.
Protein similarity search revealed a bHLH motif between amino acid residues 51 and 108 (Fig. 1). On the basis of the published alignment of other bHLH proteins (Gradwohl et al. 1996; Ma et al. 1996; and references therein), there is a 20%–48% identity between the bHLH motif of Stra13 and those found in other bHLH proteins. The Stra13 bHLH motif, which contains in the basic region a proline residue also found in Hes, Hairy, and Enhancer of split [E(spl)] bHLH basic regions, is in general more similar to the bHLHs of these proteins than to those of other bHLH proteins (Ishibashi et al. 1993; data not shown). In Stra13, however, this proline residue is located 2 amino acid residues more amino-terminal. Outside the bHLH domain, the Stra13 protein has no obvious similarity with any known proteins; in particular, it does not contain the WRPW motif found in the carboxy-terminal region of Hes/Hairy/E(spl) (Dawson et al. 1995). In addition to the bHLH motif, Stra13 is characterized by four glutamine residues located immediately carboxy-terminal to this motif, and regions between amino acids 135–143, 194–213, and 288–302 (Fig. 1) can fold into α helices according to the Chou-Fasman algorithm for secondary structure prediction. A perfect casein kinase 2 motif (SGSDTDTDS) is located between amino acid residues 235 and 243 (Fig. 1) (Kemp and Pearson 1990). These features distinguish Stra13 from other bHLH proteins.
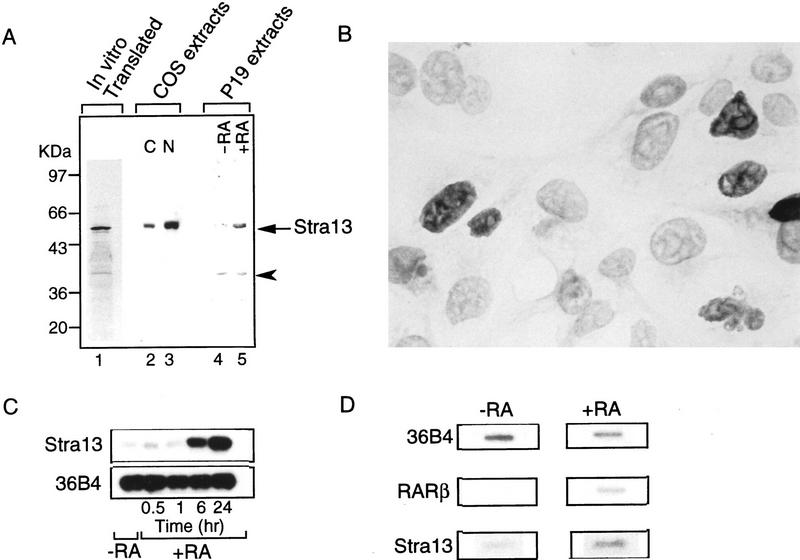
The product of Stra13 mRNA translated in vitro migrated with an apparent molecular weight consistent with the deduced calculated mass of the protein (Fig. 2A, lane 1). The endogenous P19 cell Stra13 protein, or that extracted from COS cells transfected with a Stra13 expression vector, reacted with Stra13 polyclonal antibodies revealing a protein with a size identical to that of the in vitro translated protein (Fig. 2, lanes 2–5). The Stra13 protein was clearly induced in RA-treated P19 cells (Fig. 2, lanes 4,5). Stra13 expressed in transfected COS cells was mostly nuclear as revealed by immunoblotting (Fig. 2A, see lanes 2,3), and this localization was confirmed by immunofluorescence analysis of the same transfected COS cells (Fig. 2B).
Figure 2.
SDS-PAGE of in vitro and in vivo synthesized Stra13 protein and induction of Stra13 expression in P19 cells by RA. (A) (Lane 1) Autoradiography of the product of Stra13 cDNA transcribed and translated in vitro and labeled with [35S] methionine. (Lanes 2,3) Immunological detection of the Stra13 protein (with polyclonal antibodies) in cytoplasmic (C, lane 2) and nuclear (N, lane 3) extracts prepared from COS cells transfected with the pSG5–Stra13 expression vector. (Lanes 4,5) Immunological detection of the endogenous Stra13 protein in nuclear extracts prepared from untreated and treated (1 μm RA, 24 hr) P19 cells, respectively (the arrowhead points to a nonspecific cross-reaction). (B) Immunocytochemistry analysis. The Stra13 expression vector was transfected into COS cells and the expressed protein was revealed by immunocytochemistry with anti-Stra13 polyclonal antibodies at 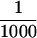 dilution. (C) Time course of Stra13 mRNA expression in P19 cells. P19 cells grown as monolayers were treated with ethanol or 1 μm T-RA. Total RNA was extracted from RA-treated cells after 30 min, 1, 6, and 24 hr, and analyzed by RT–PCR for Stra13 RNA and the invariant 36B4 RNA (Materials and Methods). (D) Nuclear run-on transcription. Nuclei were isolated from ethanol (−RA)- or RA (+RA)-treated P19 cells. The in vitro synthesized radiolabeled RNA was hybridized to an excess of Stra13, RARβ, and 36B4 cDNAs immobilized on nylon membranes, as indicated.
dilution. (C) Time course of Stra13 mRNA expression in P19 cells. P19 cells grown as monolayers were treated with ethanol or 1 μm T-RA. Total RNA was extracted from RA-treated cells after 30 min, 1, 6, and 24 hr, and analyzed by RT–PCR for Stra13 RNA and the invariant 36B4 RNA (Materials and Methods). (D) Nuclear run-on transcription. Nuclei were isolated from ethanol (−RA)- or RA (+RA)-treated P19 cells. The in vitro synthesized radiolabeled RNA was hybridized to an excess of Stra13, RARβ, and 36B4 cDNAs immobilized on nylon membranes, as indicated.
Transcriptional induction of Stra13 mRNA in P19 cells by RA
Stra13 mRNA accumulation was determined in P19 cells cultured as monolayers in the presence of 1 μm RA for increasing time periods by use of a semiquantitative RT–PCR assay. Stra13 transcripts that were present at a low basal level in ethanol (vehicle)-treated P19 cells, increased after a 6-hr period, whereas the same RNA samples did not show any significant variation in the expression of the control invariant 36B4 gene (Fig. 2C).
Nuclear run-on transcription was used to investigate the mechanism responsible for Stra13 mRNA accumulation in RA-treated P19 cells. Nuclei were isolated from P19 cells grown in monolayers after 24 hr of culture in the presence or absence of RA. Transcription of the Stra13 gene was clearly induced in the RA-treated cells (Fig. 2D). The transcription of the RARβ2 gene was analyzed in parallel as a reference gene whose expression is known to be controlled by RA at the transcriptional level (Chambon 1994, 1996). Note that Stra13 mRNA expression was similarly induced by RA treatment in P19 cells growing as aggregates (data not shown).
Stra13 DNA binding and interaction with other bHLH proteins
Most bHLH transcription factors bind to N-box (CANNAG) and/or E-box (CANNTG) DNA motifs either as homodimers or heterodimers (for review, see Anthony-Cahill et al. 1992; Hollenberg et al. 1995). We performed gel electrophoresis mobility shift assays (EMSA) by use of Stra13 protein produced either in vitro (coupled transcription–translation product), in Escherichia coli (purified GST–Stra13 fusion protein), or in transfected COS cells (nuclear extracts), together with several E- or N-boxes (Klambt et al. 1989; Takebayashi et al. 1994; Hollenberg et al. 1995; Gradwohl et al. 1996) as DNA-binding elements. None of the Stra13 protein preparations formed a complex with the elements tested, irrespective of the addition of P19 cell nuclear extracts (data not shown).
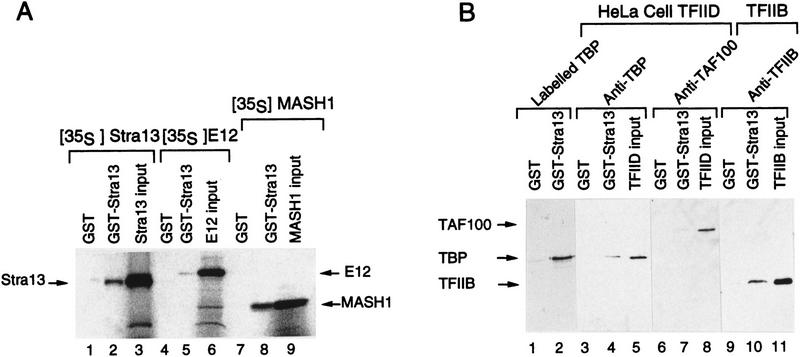
As bHLH proteins are known to bind DNA as dimers, we then tested the ability of Stra13 to homodimerize and to heterodimerize with the bHLH proteins E12 and Mash1 known to form heterodimers (see Gradwohl et al. 1996 for references). GST–Stra13 and GST–E12 were used in pull-down assays with 35S-labeled Stra13 and Mash1 proteins translated in vitro. These assays showed that Stra13 efficiently homodimerized (Fig. 3A, lane 2) and heterodimerized as efficiently as E12 with Mash1 (lane 8; data not shown), whereas it interacted very poorly with E12 (lane 5). However, a Stra13/Mash1 mixture did not bind to an E-box-DNA element (data not shown) known to bind E12/Mash1 heterodimers (see Gradwohl et al. 1996).
Figure 3.
Dimerization of Stra13 and interaction with other bHLH proteins. (A) Thirty microliters of glutathione–Sepharose preloaded with either GST (control) or GST–Stra13 protein (10 μg) were incubated with 5 μl of 35S-labeled Stra13, E12, or MASH1 (Gradwohl et al. 1996) protein translated in vitro (input lanes). Mixtures were incubated at 4°C for 2 hr in a total volume of 100 μl binding buffer (5 mm Tris-HCl at pH 8.0, 100 mm NaCl, 0.3 mm DTT, 10 mm MgCl2, 10% glycerol, 0.1% NP40), washed with the same buffer containing 250 mm KCl, and resolved by 12% SDS-PAGE as described in Jacq et al. (1994). 35S-Labeled Stra13 (lanes 1,2,3), E12 (lanes 4,5,6), and MASH1 (lanes 7,8,9) were revealed by autoradiography. (B) Interactions between Stra13 and general transcription factors. Glutathione–Sepharose beads preloaded as above with GST or GST–Stra13 proteins were incubated with 5 μl of 35S-labeled in vitro translated TBP (Caron et al. 1993), 10 μg of immunopurified HeLa cell TFIID complex (Brou et al. 1993), or 10 μg of recombinant TFIIB (Moncollin et al. 1992). 35S-Labeled TBP was detected by autoradiography (lanes 1,2); TBP, TAF100, and TFIIB were revealed by Western blotting (lanes 3–11) with cognate monoclonal antibodies (Jacq et al. 1994). TFIID and TFIIB input lanes contained 1 μg of protein.
Transcriptional activity of Stra13
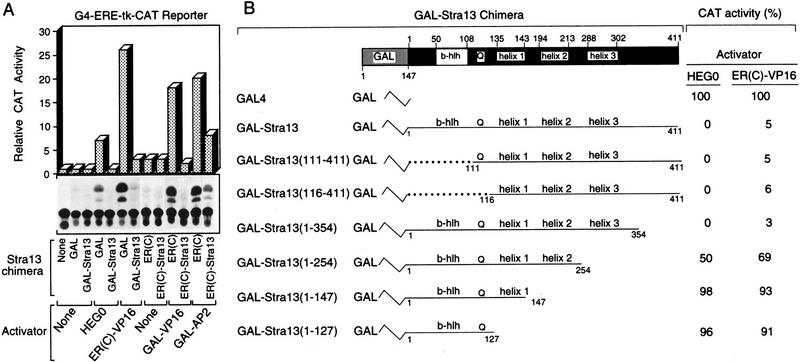
Most bHLH proteins act either as positive or negative regulators of transcription (Benezra et al. 1990). The transcriptional activity of the Stra13 protein was investigated in transfected COS cells expressing chimeric proteins [GAL–Stra13 and ER(C)–Stra13] containing either the amino-terminal 147 amino acids of the yeast transactivator GAL4 [harboring the GAL4 DNA binding domain, dimerization domain, and nuclear localization signal; (Carey et al. 1989)] or the DNA-binding domain (DBD) of the estrogen receptor (ER) [Region C, amino acids 174 to 258; ER(C); Tora et al. 1989]. These chimeric proteins were coexpressed with a reporter gene containing a DNA-binding element (17-mer tetramer) for GAL4 (G4), followed by an estrogen response element (ERE) located upstream of the thymidine kinase (tk) promoter region and the CAT gene (G4–ERE–tk–CAT). This reporter gene is responsive to ER (HEG0) and activators fused to either the ER DBD [e.g., ER(C)–VP16] or the GAL4 DBD [e.g., GAL–AP2 (May et al. 1996) or GAL–VP16 (Tora et al. 1989)]. Under these conditions, the expression of the reporter gene was not stimulated by GAL–Stra13 or ER(C)–Stra13 (Fig. 4A), thus indicating that Stra13 is unlikely to be a transcriptional activator.
Figure 4.
Repression of activated transcription by Stra13. (A) Transcriptional repression activity of Stra13. One microgram of G4–ERE–tk-CAT reporter gene (see text for details) was transfected into COS cells either alone, or with the following expression vectors as indicated: GAL4 DNA-binding domain (1–147) (GAL), estrogen receptor (ER) DNA-binding domain [ER(C)], GAL–Stra13, or ER(C)–Stra13 (1 μg each); ER(HEG0), ER(C)–VP16 (Tora et al. 1989), GAL–AP2 (May et al. 1996) (200 ng each); GAL–VP16 (1 ng). When HEG0 was used, estrogen was added to the culture medium at a final concentration of 10−7 m. Cells were harvested 48 hr after transfection, and the extracts were assayed for CAT activity that was plotted in a graph. (B) Schematic representation of deletion mutants of the GAL–Stra13 fusion protein and their corresponding repressive activities with the G4–ERE–tk–CAT reporter. The CAT activity of G4–ERE–tk–CAT upon COS cell cotransfection with vectors (1 μg) encoding the various GAL–Stra13 deletion mutants and either HEG0 or ER(C)–VP16 vector (200 ng) was expressed relative to the activity obtained with the unfused GAL4 DBD, which was taken as 100%.
To determine whether Stra13 could be a repressor, the effect of either GAL–Stra13 or ER(C)–Stra13 was tested on the same reporter gene activated by either the ER (HEG0), ER(C)–VP16, GAL–VP16, or GAL–AP2 [when ER (HEG0) was used as a transactivator, estradiol (E2, 10−7 m) was added to the medium]. GAL–Stra13 very efficiently repressed transcription activated by ER or ER(C)–VP16 (Fig. 4A). This repression was dependent on GAL–Stra13 amounts (data not shown) and reached >90% when the ratio between the transfected vectors encoding GAL–Stra13 and either ER (HEG0) or ER(C)–VP16 was 5:1. Similarly, ER(C)–Stra13 repressed by ∼60% transcription activated by GAL–AP2. In contrast, Stra13 not fused to either the GAL4 DBD or the ER DBD had no effect on activated transcription, irrespective of the transactivator tested (data not shown).
To map the Stra13 domain responsible for this repressive activity, we generated deletion mutants of the GAL–Stra13 chimeric protein (Fig. 4B). Similar effects were observed on transcription activated by either the ER (HEG0) or ER(C)–VP16. GAL–Stra13(111–411) that lacks the bHLH motif and GAL–Stra13(116–411) which, in addition to the bHLH domain, lacks the poly(Q) region repressed as efficiently as GAL–Stra13. Deletion from the carboxy-terminal region of GAL–Stra13 [GAL–Stra13(1–354)] showed that amino acid residues 354–411 were dispensible for the repressing activity. GAL–Stra13(1–254), from which helix 3 was deleted, lost ∼50% of the repressing activity, whereas further deletion of either helix 2 or helices 1 and 2 [GAL–Stra13(1–147) or GAL–Stra13(1–127), respectively] abolished the repression. These results demonstrate that the Stra13 region located between amino acids 147–354, which is rich in putative α-helix structures, is required for the repressing activity of Stra13.
Transcriptional activation and repression of class II genes transcribed by RNA polymerase II may involve interactions between activators/repressors bound to regulatory elements and the general transcription factors (GTFs) that assemble on the TATA box and/or the intiator element (Tjian and Maniatis 1994). Therefore, we investigated whether Stra13 could interact with either TATA-binding protein (TBP), TFIID, or TFIIB. Interestingly, a GST–Stra13 fusion protein interacted with in vitro-translated TBP (Fig. 3B, cf. lanes 1 and 2). The purified TFIID complex also interacted with GST–Stra13 (presumably through TBP), as shown by the presence of TBP (lanes 3–5) and TAFII100 (lanes 6–8) in pull-down assays with GST–Stra13. GST–Stra13 also interacted with bacterially expressed recombinant TFIIB (lanes 9–11), but no interaction could be detected with another general transcription factor, TFIIH (data not shown).
Neuronal differentiation of P19 cells constitutively expressing Stra13
To investigate the possible role of Stra13 protein in cell differentiation and proliferation, we electroporated P19 EC cells with either the parental expression vector pSG5 or Stra13 cDNA cloned in pSG5, together with a vector expressing the selectable neomycin-resistance gene (Metzger et al. 1995). Resistant clones were isolated after 10 days of selection with geneticin. The presence of pSG5 or pSG5–Stra13 cDNA in these clones was determined by Southern blotting, and the expression of Stra13 transcript and protein was determined by RT–PCR and Western blotting (data not shown). Clones that had integrated the Stra13 cDNA fell into three groups, depending on the level of constitutive expression of Stra13 (either low, medium, or high levels). For proliferation and differentiation studies, we analyzed only clones expressing medium levels of Stra13 transcription and protein (identical results were obtained with three independent medium expressing clones), as these levels were similar to those induced in RA-treated P19 cells (see Fig. 2A,C; data not shown). Clones expressing higher amounts of the protein died after the second passage in culture and low expressing clones did not undergo efficient neuronal differentiation in monolayers in the presence of RA and in aggregates in the absence of RA (data not shown). Clones that had integrated the pSG5 vector served as control.
In the absence of RA, proliferation of clones constitutively expressing medium amounts of Stra13 (“Stra13” cells) was consistently two- to threefold higher than that of control cells. However, RA treatment had the same antiproliferative effect on “Stra13” cells as on control wild-type P19 cells (data not shown).
Depending upon the treatment and culture conditions used, P19 cells can differentiate into derivatives of all three germ layers, that is, endoderm, mesoderm, and ectoderm (Bain et al. 1994). RA treatment of P19 cells growing as monolayers induces the formation of endodermal and mesodermal derivatives, whereas RA treatment of aggregated P19 cells results in their differentiation into cells that resemble neurons, glia, and fibroblast-like cells (Mummery et al. 1986). When grown as monolayers in the absence of RA, “Stra13” cells (Fig. 5A) were morphologically altered when compared with untreated wild-type P19 cells (Fig. 5D) and immunostaining revealed that they had lost the embryonic cell surface antigen SSEA1 characteristic of wild-type P19 cells (Fig. 6A–C). The expression of SSEA1, which was high in undifferentiated wild-type P19 cells (Fig. 6A) and markedly down-regulated in the presence of RA (Fig. 6B), was severely reduced in “Stra13” cells even in the absence of RA (Fig. 6C). Interestingly, when monolayers of “Stra13” cells were treated with 1 μm RA, compact clusters of cells started to form by the second day of treatment (Fig. 5B), whereas similar clusters did not appear in RA-treated wild-type P19 cells (Fig. 5E). Strikingly, on the fifth day of treatment, individual “Stra13” cells exhibited neurite-like structures projecting from the clusters (Fig. 5C), whereas wild-type P19 cells treated in parallel with RA exclusively differentiated into mesodermal-like cells as expected (Fig. 5F).
Figure 5.
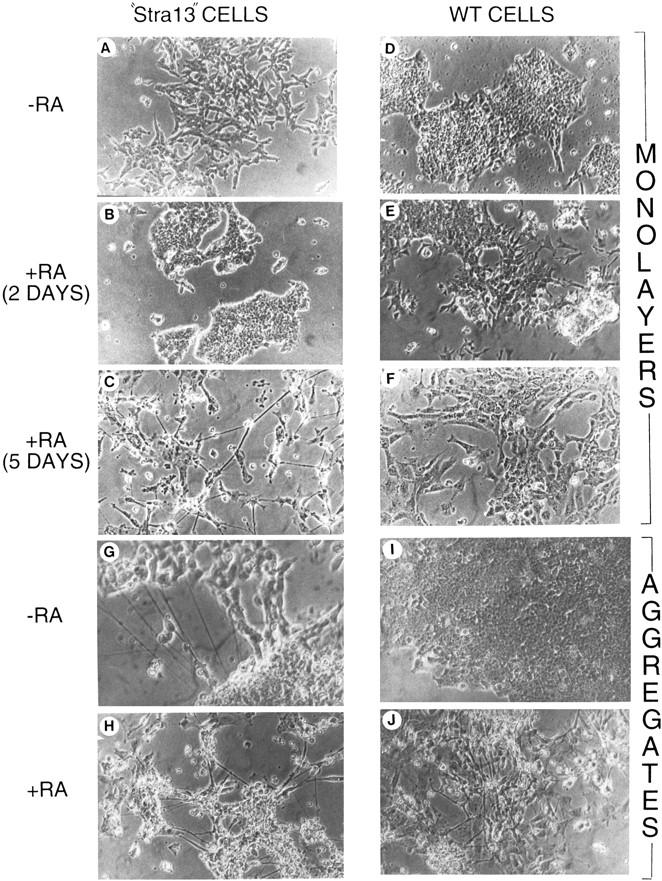
Morphological differentiation of Stra13-expressing and wild-type P19 cells cultured as monolayers or aggregates. (A–F) Phase-contrast micrography of P19 cells constitutively expressing medium levels of Stra13 (A–C) and pSG5-electroporated control cells (D–F) grown as monolayers in the absence (A,D) or after two (B,E) and 5 days culture (C,F) in the presence of 1 μm T-RA. (G–J) Phase-contrast micrography of cells 5 days after plating aggregates formed by Stra13-expressing P19 cells and control cells either in the absence (G,I) or in the presence of 1 μm T-RA (H,J), respectively.
Figure 6.
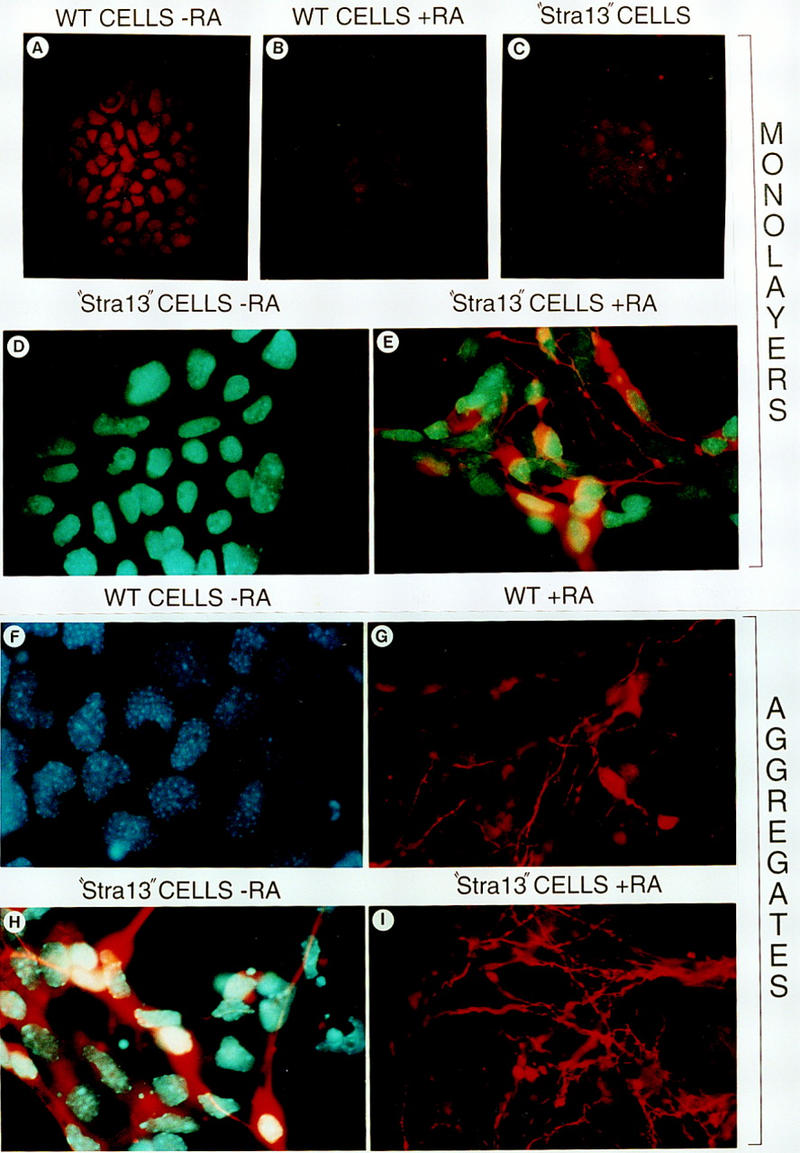
Immunofluorescence detection of SSEA-1 and βIII-tubulin expression in control and Stra13-expressing P19 cells. (A–C) Immunofluorescence detection (red color) of SSEA-1 expression in P19 control cells in the absence of T-RA (A) and after a 4 day treatment with T-RA (B), and in Stra13-expressing P19 cells cultured as monolayers in the absence of T-RA (C). (D,E) Anti-βIII-tubulin immunofluorescence (red color) of Stra13-expressing cells cultured in the absence (D) and presence (E) of T-RA for 5 days, (F–I) βIII-tubulin expression 5 days after plating cell aggregates formed by control (wild-type, F,G) and Stra13-expressing, (H,I) P19 cells untreated (F,H) or treated (G,I) with 1 μm T-RA.
To support the neuronal-like phenotype of the RA-treated “Stra13” cells, we stained them with anti-βIII-tubulin antibodies, a major microtubule protein that can be used as a marker for neuronal differentiation (Banerjee et al. 1990). Irrespective of RA treatment, no expression of βIII-tubulin was ever detected in monolayers of wild-type P19 cells (data not shown) or in RA-untreated “Stra13” cells (Fig. 6D). In contrast, a strong signal was detected in “Stra13” cells treated with RA, and the labeling was along the developing neurite-like outgrowth (Fig. 6E).
As aggregates of wild-type P19 cells are known to differentiate into neuronal-like cells upon RA treatment, we then grew “Stra13” and wild-type P19 cells in aggregates for four days in the presence and absence of 1 μm RA, and plated them on normal cell culture plates to differentiate. All “Stra13” clones tested formed aggregates as did control cells (data not shown). Five days after plating, cells derived from aggregates of RA-untreated wild-type cells grew as undifferentiated P19 cells (Fig. 5I) and did not express β-III tubulin (Fig. 6F). In contrast, even in the absence of RA treatment, cells derived from “Stra13” aggregates looked partially differentiated with neuronal-like structures (Fig. 5G), and the expression of βIII-tubulin was strong even in cells that did not extend neurite-like filaments (Fig. 6H). The aggregates formed by either “Stra13” or wild-type cells in the presence of RA differentiated similarly into neuron-like cells (Fig. 5H,J, respectively), and a strong βIII-tubulin labeling was detected (Fig. 6G,I).
These results clearly demonstrate that “Stra13” cells differentiate into neuronal-like cells when treated with RA in monolayer culture or after aggregation in the absence of RA and, therefore, that Stra13 overexpression promotes neuronal differentiation of P19 cells.
Ectopic expression of Stra13 leads to an altered expression of neuronal and mesodermal markers
The constitutive expression of Stra13 resulted in neuronal differentiation in monolayer culture (see above), under conditions where wild-type P19 cells differentiate only into mesodermal and endodermal derivatives. As these results suggest that the mesodermal fate might be repressed in Stra13 overexpressing cells, the expression of several markers of neuronal and mesodermal differentiation was analyzed by semiquantitative RT–PCR in wild-type P19 and “Stra13” cells. This analysis was performed in monolayer culture upon treatment with 100 nm T-RA for 72 hr. Cells in aggregates, treated or not with T-RA (100 nm), were analyzed 4 days after plating (Fig. 7).
Figure 7.
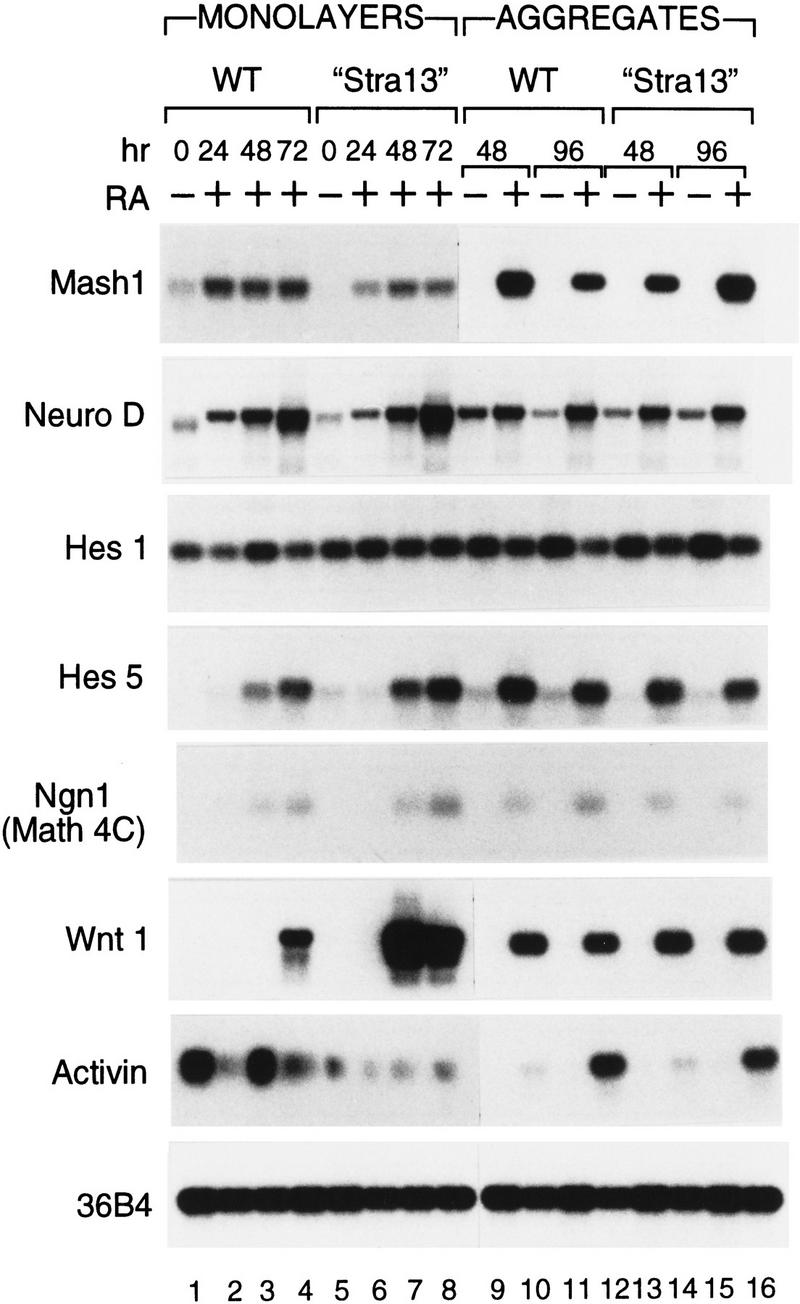
Constitutive expression of Stra13 in P19 cells results in an altered expression of neuronal and mesodermal markers. Wild-type P19 cells and “Stra13” cells were grown as monolayers or aggregates in the absence (−) or presence (+) of T-RA (100 nm). Total RNA was extracted from monolayers of wild-type P19 and “Stra13” cells after 24, 48, and 72 hr of RA treatment. RNA from aggregates of wild-type P19 and “Stra13” cells was isolated 2 and 4 days after plating cells. The transcripts for each gene were analyzed by semiquantitative RT–PCR, and the amount of RNA in each reaction was normalized with transcripts of the 36B4 gene (Krowczynska et al. 1989) that is unresponsive to retinoid treatment.
In contrast to wild-type P19 cells, in which Mash1 is expressed at a low level in the absence of T-RA, Mash1 RNA was not detectable in the absence of RA in “Stra13” cells in monolayer culture (Fig. 7, lanes 1 and 5, respectively), indicating that Stra13 overexpression abrogates Mash1 expression. However, Mash1 expression was induced with T-RA in “Stra13” cells, although to a lesser level than in wild-type cells. In agreement with previous studies (Johnson et al. 1992; Smolich and Papkoff 1994), Mash1 was expressed to a high level in aggregates from wild-type cells cultured in the presence of T-RA, at days 2 and 4 after plating (lanes 10,12). Under the same conditions, however, “Stra13” cells exhibited a lower level of Mash1 expression at day 2 relative to wild-type cells (lane 14), although the expression was induced to a higher level in “Stra13” cells after 4 days (lane 16). In contrast to the change in Mash1 expression, constitutive expression of Stra13 did not result in any significant change in the expression of Neuro D, Hes1, Hes5, and Ngn1 (Math4C) relative to wild-type cells, irrespective of the cell culture conditions (monolayers or aggregates).
Interestingly, the expression of activin, a mesodermal marker (Smith et al. 1990), was strongly reduced in “Stra13” cells in monolayer culture compared with wild-type cells (cf. lanes 1–4 with lanes 5–8), indicating a suppression of the mesodermal differentiation program that normally occurs under these conditions. As expected when compared with wild-type cells, there was no significant alteration in the expression of activin in “Stra13” cells when cultured as aggregates, which results in neuronal differentiation. Interestingly, the expression of Wnt1 which is normally associated with neuronal differentiation of P19 cells (St. Arnaud et al. 1989; Schuuring et al. 1989) was very strongly induced in RA-treated “Stra13” cells in monolayer culture (cf. lanes 7 and 8 with lanes 2–4), indicating that Stra13 overexpression can stimulate the expression of a neuronal marker. No significant difference in Wnt1 expression was observed between aggregates from “Stra13” cells and wild-type cells.
Expression pattern of Stra13 in adult and developing mouse tissues
The presence of Stra13 transcripts in mouse embryo and adult tissues was investigated by RT–PCR analysis. Stra13 transcripts increased steadily during mouse embryogenesis from 9.5 to 12.5 days postcoitum (dpc) and were detected in a number of adult mouse tissues (e.g., brain, liver, female genital tract, lung, kidney, spleen, and heart) (data not shown).
The developmental expression pattern of Stra13 was also investigated by in situ hybridization (ISH) on cryosections of mouse embryos from 8.5 to 17.5 dpc. No specific signal was detected in the embryo proper at 8.5 dpc, although strong signals were seen in the decidual tissue and in trophoblast giant cells surrounding the yolk sac (data not shown). By 9.5 dpc, Stra13 expression was detected in the fifth rhombomere of the developing hindbrain and more weakly in some mesenchymal areas, especially at the level of the heart (Fig. 8A). Labeling became apparent in the prevertebral condensations and the outflow tract of the heart by 10.5 dpc (data not shown). Embryos at 11.5 and 12.5 dpc showed specific labeling of the ventricular layer of the neuroepithelium, both at the level of the brain (Fig. 8C) and spinal cord (Fig. 8B–D). Labeling was more intense in the basal (ventral) region of the spinal cord ventricular layer (open arrow in Fig. 8D). Stra13 was also expressed in a subset of the cells of the spinal cord ventral horns (filled arrow in Fig. 8D). In the developing eye, at 11.5 dpc, strong expression was seen in the optic stalk and prospective retinal pigment epithelium, whereas the lens and the retrolenticular mesenchyme were weakly labeled (Fig. 8B; data not shown). There was also strong expression at the level of the outflow tract of the heart and interventricular septum, and weak expression in other regions of the ventricular myocardium (Fig. 8B,C). The prevertebrae and rib anlagen were strongly labeled at 11.5–12.5 dpc (Fig. 8B,D), as well as the mesonephric tubules (not shown).
Figure 8.
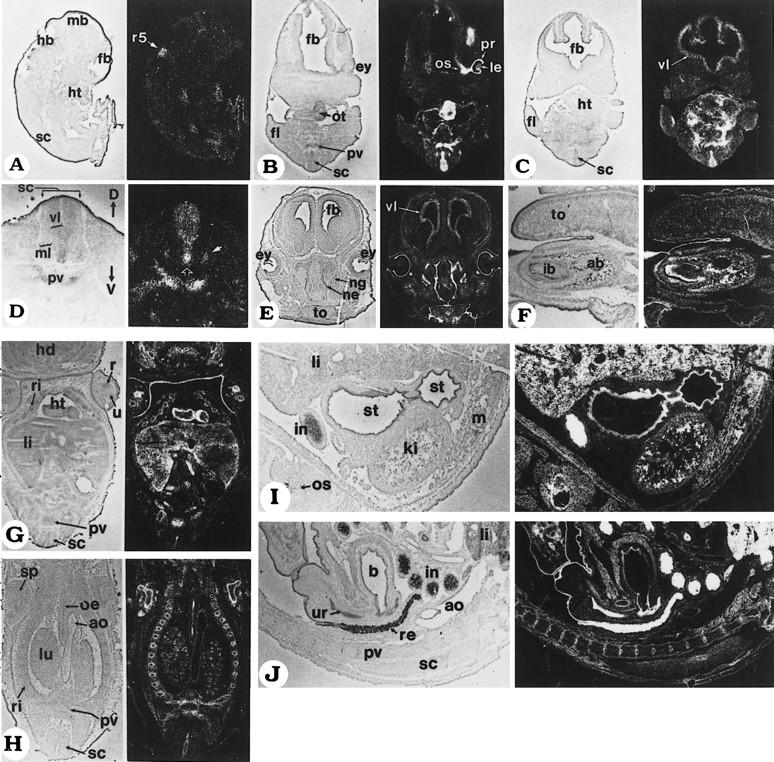
In situ analysis of Stra13 transcript expression during mouse development. Mouse embryo sections were hybridized to a 35S-labeled Stra13 antisense riboprobe; all panels are composed of a bright-field view (left) and a dark-field view (right) of the same section, where signal grain appears as white dots. (A) Sagittal section of a 9.5 dpc embryo. (B,C) Two serial coronal sections through the forebrain, heart, and forelimbs of an 11.5 dpc embryo. The section in B crosses one of the eyes, as well as the heart outflow tract. (D) Transverse section of the cervical spinal cord of a 12.5 dpc embryo. The open arrow indicates enhanced labeling in the ventral region of the ventricular layer and the filled arrow points to the signal in the ventral horn. (E) Coronal section of the head of a 15.5 dpc fetus. (F) Sagittal section at the level of the lower incisor bud of a 17.5 dpc fetus. (G,H) Two coronal sections of a 13.5 dpc fetus. G is at the level of the liver and the apex of the heart ventricles. H is a more dorsal section crossing the lungs, rib cage, and scapula. (I) Sagittal section through the stomach and kidney of a 17.5 dpc fetus. (J) Mid-sagittal section through the rectum, urinary bladder, and urethra of a 17.5 dpc fetus. Abbreviations: (ab) alveolar bone; (ao) aorta; (b) urinary bladder; (D) dorsal; (ey) eye; (fb) forebrain; (fl) forelimb; (hb) hindbrain; (hd) head; (ht) heart; (ib) incisor bud; (in) intestine; (ki) kidney; (le) lens; (li) liver; (lu) lung; (m) muscle; (mb) midbrain; (ml) mantle layer; (ne) nasal epithelium; (ng) nasal gland; (oe) oesophagus; (os) optic stalk; (ot) outflow tract of the heart; (pr) prospective retinal pigment epithelium; (pv) prevertebra; (r) radius; (r5) rhombomere 5; (re) rectum; (ri) rib; (sc) spinal cord; (sp) scapula; (st) stomach; (to) tongue; (u) ulna; (ur) urethra; (V) ventral; (vl) ventricular layer.
From 13.5 to 17.5 dpc, Stra13 expression persisted in the ventricular layer of the brain (Fig. 8E) and spinal cord (Fig. 8G,H,J). Specific signals were detected in the retinal pigment epithelium and developing eyelids (Fig. 8E), the nasal epithelium and serous glands (Fig. 8E), the vibrissae (not shown), and the epithelium of the mouth cavity and of the tooth buds (Fig. 8F). In the heart, there was strong labeling of the interventricular septum (not shown) and of the external layer of the ventricular myocardium (Fig. 8G). Strong signals were seen in the developing thymus and adrenal glands (not shown). Labeling was also detected in the small bronchi of the lungs (Fig. 8H), the liver parenchyma (Fig. 8G,I), the kidney tubules (Fig. 8I), the epithelium of the esophagus (Fig. 8H), stomach (Fig. 8I), and intestine (Fig. 8I,J; note that labeling was particularly intense in the intestine epithelium at 17.5 dpc). The developing urinary bladder and urethra were labeled from 15.5 to 17.5 dpc (Fig. 8J). Labeling was detected around the cartilaginous bone models at 13.5 dpc (Fig. 8G,H: ribs, scapula, and limb cartilages) and later within the ossification centers (Fig. 8F: alveolar bone). Finally, Stra13 expression was detected in developing muscles by 17.5 dpc (Fig. 8F: tongue; and 8I: back and body wall muscles).
Discussion
Stra13 is a novel member of the bHLH protein family
We report here the identification of a novel gene, Stra13, and the functional characterization of the encoded protein, a novel member of the bHLH family. The bHLH domain is located between amino acid residues 50–108 and, within this domain, exhibits maximum identity (48%) with HES-1. The basic region of Stra13 contains a proline residue that is characteristic of the Hes, Hairy, and E(spl) group of bHLH factors (Ish-Horowicz and Pinichin 1987; Ishibashi et al. 1993; Hollenberg et al. 1995). The Stra13 proline residue, however, is located two amino acid residues more amino-terminal than in Hes, Hairy, and E(spl).
Another unique feature of Stra13 is the presence of a stretch of four glutamine residues just carboxy-terminal to the bHLH domain. In general, in other bHLH proteins, polyglutamine or polyproline stretches are located at distances of at least 70 amino acids from the carboxy-terminal or amino-terminal end of the bHLH motif (Sasai et al. 1992). Besides the bHLH motif, Stra13 has no other structural motif(s), such as the WRPW motif found in the carboxy-terminal region of Hes, Hairy, and E(spl) (Dawson et al. 1995), or the leucine zipper motif found in the myc family (Kato and Dang 1992), which are conserved between members of the same group or class of bHLH proteins. These features distinguish Stra13 from other bHLH proteins, and Stra13 may, therefore, represent the first member of a new class of bHLH factors.
Stra13 is a putative transcription repressor when fused to a DNA-binding domain
Most bHLH factors bind to consensus motifs, the E-box (Blackwell and Weintraub 1990) or the N-box (Akazawa et al. 1992; Sasai et al. 1992). Stra13, however, is unable to bind to either the E- or the N-box. bHLH proteins are characterized by the presence of two conserved amino acid residues in their basic region, which are determinants for specific sequence recognition (Hollenberg et al. 1995). These residues, which correspond to histidine 57 and arginine 58 in the basic region of Stra13, are not identical to those found in any other bHLH protein. These residues, as well as the proline residue present in the basic region of Stra13, might affect its DNA-binding specificity. It has been reported that the introduction of a proline residue in MyoD at the same position as in Stra13, abrogates its binding to the E-box (Davis et al. 1990). Furthermore, the bHLH domains of Hes-1 and Hes-5 also contain a proline residue that prevents these factors from binding the E-box, but not the N-box, indicating that the presence of a proline residue in the bHLH domain might change its DNA-binding specificity (Akazawa et al. 1992; Sasai et al. 1992). As the position of the proline residue found in the basic region of Hes, Hairy, and E(spl) is not the same as in Stra13, the shift in the position of the proline residue in Stra13 could lead to a further change in DNA-binding specificity.
With the exception of c-myc (Amati et al. 1993), which heterodimerizes with a specific partner, most bHLH proteins either homodimerize or heterodimerize with E12 or E47, and then bind DNA. Stra13 that efficiently homodimerizes, can also heterodimerize with Mash1 in vitro, but not efficiently with E12 or E47. Thus, the failure of Stra13 to efficiently bind DNA on its own or in the presence of Mash1, E12, and E47 might reflect a need for heterodimerization with an as yet unidentified partner and/or its binding to a nonconsensus DNA-binding site.
In transient transfection assays, Stra13 fused to a DNA-binding domain is a potent transcriptional repressor which inhibits transcription activated by the ER, and by other activators such as VP16 and AP2. GAL–Stra13 and ER(C)–Stra13 fusion proteins repress transcription in a dose-dependent manner, and this repression requires DNA-binding, as Stra13 not fused to a DNA-binding domain failed to inhibit activated transcription. Analyses of bHLH transcriptional repressors, such as Hairy and E(Spl), have revealed that three conserved regions involved in protein–protein interactions (bHLH; Orange, a helix located 20–30 amino acids carboxy-terminal to the bHLH motif; and WRPW), are required for the repressing functions of these proteins (Dawson et al. 1995). Stra13 does not contain a WRPW motif, and CAT assays with GAL–Stra13 mutants show that the bHLH motif is not required for its repressing activity either. Instead, the α-helix rich region that resides between amino acids 147–354 (Fig. 4) appears to be the main domain involved in Stra13 repressing activity.
Although the precise mechanism through which Stra13 acts as a repressor remains to be analyzed, the in vitro interactions between Stra13 and either TBP or TFIIB support the idea that Stra13 might repress transcription by interacting with general factors of the basal transcription machinery. In this respect, we note that coexpression of TFIIB can partially reverse the repressing activity of Stra13 (M. Boudjelal and P. Chambon, unpubl.; the effect of TBP overexpression on Stra13-mediated repression could not be studied, as TBP itself was inhibitory for transcription activated by the ER). Regardless of the mechanism by which Stra13 represses activated transcription, these results provide the first direct evidence for a biochemical interaction between a bHLH protein and components of the basal transcription machinery, which results in an inhibition of its activity.
Constitutive expression of Stra13 represses the RA-induced mesodermal/endodermal differentiation of P19 cell monolayers and promotes neuronal differentiation
Mesodermal and endodermal differentiation of P19 cell in monolayers requires RA, whereas cell aggregation, in addition to RA, is required to trigger neuronal differentiation (Mummery et al. 1986). When grown as monolayers, cells constitutively overexpressing Stra13 (“Stra13”) are morphologically altered in the absence of RA treatment and do not express SSEA-1, a marker of undifferentiated EC cells (Solter and Knowles 1978). Thus, Stra13 overexpression appears to induce some differentiation events. It is, however, remarkable that “Stra13” cells, when treated with RA in monolayer culture, differentiate into neuronal-like cells (Fig. 6,7) and express βIII-tubulin, a marker of neuronal differentiation, under conditions where wild-type P19 cells undergo mesodermal and endodermal differentiation (Mummery et al. 1986). In this respect, it should be stressed that wild-type P19 cells in monolayer culture never undergo neuronal differentiation, even though Stra13 is expressed in wild-type P19 cell monolayers at a low level, and is induced to a higher level in cells treated with RA for 24 hr. These results suggest that the endogenous level of Stra13 in P19 cell monolayers is insufficient to drive them into the neuronal differentiation pathway, and that the induction of Stra13 expression by RA probably occurs too late, well after genes presumably required for mesodermal/endodermal differentiation have been induced, thus committing the cells to a non-neuronal fate. In contrast, it appears that some aggregation-induced events and RA-induced levels of Stra13 can act synergistically in P19 cells grown in aggregates to promote neuronal differentiation. Thus, ectopically overexpressed Stra13 in P19 cell monolayers may, in some way, bypass the requirement for these aggregation-induced events.
The high level of constitutive Stra13 expression in “Stra13” cells, therefore, may repress the expression of genes required for initiating the mesodermal/endodermal differentiation program, and neuronal differentiation of these cells upon RA treatment would occur as a default mechanism (Hemmati-Brivanlou and Melton 1997). The lowered expression of activin, a mesodermal marker, as well as the increased expression of the neuronal marker Wnt1, in “Stra13” cells, is consistent with this hypothesis. Neuronal differentiation of P19 cells may not occur normally in RA-treated monolayer culture because of the relatively late induction of neurogenic genes (Wnt1, Mash1, NeuroD, etc.), thus allowing genes involved in formation of mesoderm and endoderm to drive the cells into a distinct pathway of differentiation.
Unlike monolayers of “Stra13” cells, aggregates of “Stra13” cells can undergo a certain degree of neuronal differentiation in the absence of RA. Thus, as in monolayers, overexpression of Stra13 in aggregates might prime the cells towards a neuronal differentiation pathway, and partially trigger some events normally induced by RA in aggregates of wild-type P19 cells. RA has been shown to affect patterning and neurogenesis within the neural plate in vivo (Papalopulu and Kinter 1996; Maden and Holder 1997). The observation that Stra13 is transcriptionally up-regulated by RA and appears to be crucial for neuronal differentiation strongly suggests that Stra13 is likely to be one of the earliest RA target genes required for mediating the neurogenic effects of RA in P19 cells aggregates. In this respect, note that in contrast with the ability of overexpressed Stra13 to promote neuronal differentiation in P19 cell aggregates, overexpression of Mash1 does not result in any neuronal differentiation of P19 cell aggregates in the absence of RA (Johnson et al. 1992). Furthermore, Mash1 (Xhash1) overexpression has no, or limited, effect on neurogenesis in Xenopus. Two recently identified neural bHLH factors, NeuroD (Lee et al. 1995) and Neurogenin (Ma et al. 1996), however, have been shown to be sufficient to induce neuronal differentiation in vivo, and these two genes appear to function in a unidirectional cascade with Neurogenin being upstream of NeuroD (Ma et al. 1996). The kinetics of expression of these proteins in “Stra13” P19 cells remains to be investigated.
Stra13 could be involved in the control of differentiation of several cell lineages during mouse development
The expression of Stra13 is tissue-specific as early as 9.5 dpc, and Stra13 transcripts are detected in certain fetal tissues up to 17.5 dpc. Importantly, unlike most neural bHLH factors, Stra13 expression is not confined to neuroectoderm, but is also expressed in a number of mesodermal and endodermal (gastrointestinal epithelia, bronchi, liver parenchyma) derivatives. Furthermore, in the neuroectoderm, Stra13 expression occurs rather late, and is more related to the phase of differentiation/proliferation, rather than to that of induction of the neuroectoderm. The major Stra13 expression domains correspond to various epithelia, in particular the neuroepithelium, as well as to developing cartilages and muscles, including specific regions of the developing heart (see Results). Interestingly, the expression pattern of Stra13 overlaps in part with that of Hes-1 at various stages of embryogenesis. Hes-1, as Stra13, is expressed in various embryonal epithelia, including the embryonal neuroepithelial cells, as well as in mesoderm-derived tissues such as developing muscle (Sasai et al. 1992; Ishibashi et al. 1993, 1994).
Both Hes-1 and Mash1 have been shown to play essential roles in neurogenesis. Mash1 is a positive regulator of neurogenesis, required for differentiation of olfactory and autonomic neurons (Guillemot and Joyner 1993). Hes-1, however, antagonizes the activity of Mash1 (Sasai et al. 1992; Ishibashi et al. 1995). The observation that overexpression of Stra13 in P19 cells (see above) represses the expression of Mash1, indicates that under certain conditions, Stra13 may directly or indirectly negatively control the expression of Mash1. However, no significant change in the expression of Hes-1 was seen in “Stra13” cells, indicating that the down-regulation of Mash1 expression in these cells is not caused by an activation of Hes-1 expression by Stra13. It is also important to emphasize that although Stra13 most closely resembles Hes-1 in its amino acid sequence, expression domains (see above) and repressing function, the overexpression of Stra13 and Hes-1 can have opposing effects on neuronal differentiation. Constitutive Hes-1 overexpression has been reported to inhibit neuronal differentiation in vivo (Ishibashi et al. 1994), whereas the present study shows that overexpression of Stra13 can promote neuronal differentiation.
Taken together, our functional characterization of Stra13 as a transcriptional repressor, combined with its ability to repress mesodermal and endodermal differentiation and to induce neuronal differentiation in P19 cells, strongly suggests that Stra13 in concert with other bHLH proteins may play a critical role during development. Both Hes-1 and Stra13, which are expressed in multiple tissues of different origin at various stages of development, could be involved as negative regulators in the spatiotemporal control of differentiation of multiple cell lineages. Genetic analysis is clearly required to elucidate the actual roles played by Stra13 in this control.
Materials and methods
Plasmid constructs
Full-length Stra13 cDNA was cloned into the _Eco_RV site of the expression vector pSG5 to yield pSG5–STRA13. cDNA fragments used to construct GAL–Stra13 mutants were generated by PCR and cloned in either _Bam_HI- or _Bam_HI/_Sac_I-digested pG4M vector (Tora et al. 1989). ER(C)–Stra13 was constructed as follows: Two _Bss_HI sites at position 758 and 1073 of the human ER cDNA were generated by PCR (Tora et al. 1989); the _Bss_HI fragment coding for the estrogen receptor DNA-binding domain [ER(C)] was cloned into the Stra13-coding sequence of _Bss_HI-digested pSG5–Stra13, resulting in a vector expressing the ER(C)–Stra13 protein that lacks the first six amino acid residues of Stra13. For bacterial expression of Stra13, a _Nde_I site was created at position 217 of its cDNA by PCR. The _Nde_I–_Nde_I fragment (nucleotide 217 to 1782) was ligated into the _Nde_I site of the pET15b expression vector containing six amino-terminal histidine codons (Novagen). To construct the GST–Stra13 fusion expression vector, _Bam_HI and _Bgl_II sites were generated at positions 240 and 1465 of Stra13 cDNA, respectively. This fragment was cloned into _Bam_HI-digested PGEX-2TK expression vector (Pharmacia) that was used to produce GST–Stra13 fusion protein in E. coli following the manufacturer’s instructions (Pharmacia). All constructs were verified by automated sequencing.
Cell culture and retinoic acid treatment
P19 cells were cultured in Dulbecco’s modified Eagle medium (DMEM) supplemented with 5% fetal calf serum (McBurney 1993). In monolayer culture, cells were plated at a density of 106/75 cm2 flask in 20 ml of medium. For aggregation, cells were grown at a density of 5 × 105 in 60-mm bacterial grade petri dishes. After 4 days, the aggregates were transfered into tissue culture dishes as described (McPherson and McBurney 1995) and cultured for an additional 2–5 days. For differentiation, T-RA was added to the medium at a final concentration of 1 μm. Control cells were treated with 0.1% ethanol (vehicle).
Transfection assays
Transfections were carried out with the calcium phosphate coprecipitation method after seeding 2 × 106 COS cells in 9-cm dishes for 24 hr. DNA was made up to a total of 15 μg with Bluescript M13+ (Stratagene). One microgram of G4–ERE–tk–CAT (Tora et al. 1989) reporter plasmid was transfected either alone, or together, with one or two expression vectors as indicated in the legend to Figure 4. To determine the efficiency of each transfection, cells were cotransfected with 1 μg of pCH110 β-galactosidase expression vector. After 24 hr, cells were washed with DMEM, and fresh medium containing 10−7 m estradiol was added when ER was used. Cells were cultured for an additional 24 hr, pelleted, washed, resuspended in 100 mm Tris-HCl at pH 7.5, and assayed for CAT activity with samples containing equivalent amounts of β-galactosidase activity.
Production of recombinant Stra13, Stra13 polyclonal antibodies, and in vitro-translated Stra13 protein
One to two μg of the Stra13 pET15b construct (see above) was transfected into E. coli BL21 (DE3). Rabbit polyclonal antibodies were generated against the purified His–Stra13 protein expressed in E. coli (Jacq et al. 1994). Antibodies were used directly or after purification on a sulfolink column (Pharmacia). For in vitro protein synthesis, full-length Stra13 cDNA cloned in pBSK− was linearized by _Xba_I and the RNA was transcribed and translated with a TNT reticulocyte lysate system according to the manufacturer’s instructions (Promega).
In vitro protein interaction
Interaction assays between proteins were performed as described (Le Douarin et al. 1995). Immunopurified TFIID complex from HeLa cells and recombinant TFIIB were gift from L. Tora (IGBMC). TBP, TAF100, and TFIIB were detected on Western blot with cognate antibodies as described (Jacq et al. 1994).
Western blotting and immunofluorescence
For Western blot analysis, proteins were transferred to nitrocellulose membranes (Towbin et al. 1979). After blocking in 5% nonfat dry milk in PBS for 1 hr, the nitrocellulose sheets were incubated with 1000-fold diluted anti-Stra13 polyclonal antibodies for 2 hr at room temperature, and then with anti-rabbit IgG conjugated with horseradish peroxidase followed by detection with the ECL kit (Amersham). Monoclonal anti-βIII-tubulin antibodies were purchased from Sigma and immunofluorescence analysis was as described (Le Douarin et al. 1995).
RNA extraction and RT–PCR
Total RNA preparation, semi-quantitative RT–PCR, and Southern blotting were performed as described (Bouillet et al. 1995). The sequences of the PCR primers used in Figures 2C and 7 are available upon request. RT–PCR blots were probed with the cognate 32P-labeled cDNAs and autoradiography was quantified with a PhosphorImager system (BAS2000, Fuji).
In situ hybridization (ISH)
Stra13 cDNA cloned in pBluescriptSK− (Stratagene) was used in T7 polymerase in vitro transcription reactions including 35S-labeled CTP (Amersham) to produce antisense riboprobes. Probe length was reduced by partial alkaline hydrolysis with NaCO3 (pH 10.2). ISH on cryosections was carried out as described (Décimo et al. 1995).
P19 cells constitutively expressing Stra13 protein (“Stra13” cells)
Monolayer P19 cells cultured (5 × 106), suspended in 500 μl of phosphate-buffered saline, were electroporated with 10 μg of purified DNA [mixture of 9 μg of pSG5 or pSG5–Stra13 and 1 μg of PGK–neo (Metzger et al. 1995)] with a Bio-Rad Gene Pulser set at 200 V and 950 μF. Cells were plated at a density of 6 × 105 per 100-mm culture plates, incubated for 24 hr, and then subjected to neomycin selection (Geneticin, G418 sulfate, 400 μg/ml) for 10 days. Individual colonies were isolated and analyzed by Southern blotting as described (Lufkin et al. 1991).
Acknowledgments
We are most grateful to François Guillemot for his suggestions and critical reading of the manuscript and thank the members of his laboratory for discussions, advice, and the generous gift of cDNA probes and primers for Mash1, NeuroD, Hes1, Hes5, and Math4C. We thank Allan Bradley for the activin cDNA; Jan Kitajewski and Marion Wassef for Wnt1 cDNA; Lazlo Tora for GAL fusion plasmids, TFIID complex purified from HeLa cells, anti-TAF100, and anti-TBP antibodies; Jean-Marc Egly for anti-TFIIB antibodies and TFIIB expression plasmids. We also thank Andrée Krust for useful discussions, and Jean-Luc Plassat, Nathalie Chartoire, and Steve Roth for technical assistance, as well as the staff of the sequencing, oligonucleotide, antibody production, and cell culture facilities. We are grateful to C. Werle, S. Metz, B. Boulay, J.-M. Lafontaine, J.-L. Vonesch, and the secretarial staff for help with preparation of the manuscript. This work was supported by funds from the CNRS, INSERM, the Collège de France, the Centre Hospitalier Universitaire Régional, the Association pour la Recherche sur la Cancer (ARC), the Fondation pour la Recherche Médicale (FRM), the Human Frontier Science Program, and Bristol-Myers-Squibb. M.B. and S.M. were supported by funds from the Université Louis Pasteur and INSERM, respectively; R.T. was a postdoctoral fellow from the CNRS, the FRM, and the ULP.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Note
The nucleotide sequence described in this paper has been submitted to the GenBank data library under accession no. AF010305.
Footnotes
E-MAIL igbmc@igbmc.u-strasbg.fr; FAX (33) 88 65 32 03.
References
- Akazawa C, Sasai Y, Nakanishi S, Kagemaya R. Molecular characterization of a rat negative regulator with a basic helix-loop-helix structure predominantly expressed in the developing nervous system. J Biol Chem. 1992;267:21879–21885. [PubMed] [Google Scholar]
- Amati B, Brooks MW, Levy N, Littlewood TD, Evan GI. Oncogenic activity of the c-Myc protein requires dimerization with Max. Cell. 1993;72:233–245. doi: 10.1016/0092-8674(93)90663-b. [DOI] [PubMed] [Google Scholar]
- Anthony-Cahill SJ, Benfield PA, Fairman R, Wasserman ZR, Brenner SL, Stafford WF, Altenbach C, Hubbell WL, de Grado WF. Molecular characterization of helix-loop-helix peptides. Science. 1992;255:979–983. doi: 10.1126/science.1312255. [DOI] [PubMed] [Google Scholar]
- Bain G, Ray WJ, Yao M, Gottlieb DI. From embryonal carcinoma cells to neurons: The P19 pathway. BioEssays. 1994;16:343–348. doi: 10.1002/bies.950160509. [DOI] [PubMed] [Google Scholar]
- Banerjee A, Roach MC, Trcka P, Luduena RF. Increased microtubule assembly in bovine brain tubulin lacking the type III isotype of β-tubulin. J Biol Chem. 1990;265:1794–1799. [PubMed] [Google Scholar]
- Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H. The protein Id: A negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61:49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- Blackwell TK, Weintraub H. Differences and similarities in DNA-binding preferences of MyoD and E2A protein complexes revealed by binding site selection. Science. 1990;250:1104–1110. doi: 10.1126/science.2174572. [DOI] [PubMed] [Google Scholar]
- Bouillet P, Oulad-Abdelghani M, Vicaire S, Garnier JM, Schuhbaur B, Dollé P, Chambon P. Efficient cloning of cDNAs of retinoic acid-responsive genes in P19 embryonal carcinoma cells and characterization of a novel mouse gene, Stra1 (Mouse LERK-2/Eplg2) Dev Biol. 1995;170:420–433. doi: 10.1006/dbio.1995.1226. [DOI] [PubMed] [Google Scholar]
- Brou C, Chaudhary S, Davidson I, Lutz Y, Wu J, Egly J-M, Tora L, Chambon P. Distinct TFIID complexes mediate the effect of different transcriptional activators. EMBO J. 1993;12:489–499. doi: 10.1002/j.1460-2075.1993.tb05681.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey M, Kakidani H, Leatherwood J, Mostashari F, Ptashne M. An amino-terminal fragment of GAL4 binds DNA as a dimer. J Mol Biol. 1989;209:423–432. doi: 10.1016/0022-2836(89)90007-7. [DOI] [PubMed] [Google Scholar]
- Caron C, Rousset R, Beraud C, Moncollin V, Egly J-M, Jalinot P. Functional and biochemical interaction of the HTLV-I Tax1 transactivator with TBP. EMBO J. 1993;12:4269–4278. doi: 10.1002/j.1460-2075.1993.tb06111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambon P. The retinoid signaling pathway: Molecular and genetic analyses. Semin Cell Biol. 1994;5:115–125. doi: 10.1006/scel.1994.1015. [DOI] [PubMed] [Google Scholar]
- ————— A decade of molecular biology of retinoic acid receptors. FASEB J. 1996;10:940–954. [PubMed] [Google Scholar]
- Davis RL, Cheng PF, Lassar AB, Weintraub H. The MyoD DNA binding domain contains a recognition code for muscle-specific gene activation. Cell. 1990;60:733–746. doi: 10.1016/0092-8674(90)90088-v. [DOI] [PubMed] [Google Scholar]
- Dawson SR, Turner DL, Weintraub H, Parkhurst SM. Specificity for the hairy/enhancer of split basic helix-loop-helix (bHLH) proteins maps outside the bHLH domain and suggests two separable modes of transcriptional repression. Mol Cell Biol. 1995;15:6923–6931. doi: 10.1128/mcb.15.12.6923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decimo D, Georges-Labouesse E, Dollé P. In situ hybridization of nucleic acid probes to cellular RNA. In: Hames BD, Higgins S, editors. Gene probes 2, a practical approach. Oxford, UK: Oxford University Press; 1995. pp. 183–210. [Google Scholar]
- Glass CK, Rose DW, Rosenfeld MG. Nuclear receptor coactivators. Curr Opin Cell Biol. 1997;9:222–232. doi: 10.1016/s0955-0674(97)80066-x. [DOI] [PubMed] [Google Scholar]
- Gradwohl G, Fode C, Guillemot F. Restricted expression of a novel murine atonal-related bHLH protein in undifferentiated neural precursors. Dev Biol. 1996;180:227–241. doi: 10.1006/dbio.1996.0297. [DOI] [PubMed] [Google Scholar]
- Gronemeyer H, Laudet V. Transcription factors 3: Nuclear receptors. Protein Profile. 1995;2:1173–1236. [PubMed] [Google Scholar]
- Guillemot F, Joyner AL. Dynamic expression of the murine Achaete-Scute homologue Mash-1 in the developing nervous system. Mech Dev. 1993;42:171–185. doi: 10.1016/0925-4773(93)90006-j. [DOI] [PubMed] [Google Scholar]
- Hemmati-Brivanlou A, Melton D. Vertebrate embryonic cells will become nerve cells unless told otherwise. Cell. 1997;88:13–17. doi: 10.1016/s0092-8674(00)81853-x. [DOI] [PubMed] [Google Scholar]
- Hollenberg SM, Sternglanz R, Cheng PF, Weintraub H. Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid system. Mol Cell Biol. 1995;15:3813–3822. doi: 10.1128/mcb.15.7.3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ish-Horowicz D, Pichin SM. Pattern abnormalities induced by ectopic expression of the Drosophila gene hairy are associated with repression of ftz transcription. Cell. 1987;51:405–415. doi: 10.1016/0092-8674(87)90636-2. [DOI] [PubMed] [Google Scholar]
- Ishibashi M, Sasai Y, Nakanishi S, Kageyama R. Molecular characterization of HES-2, a mammalian helix-loop-helix factor structurally related to Drosophila hairy and Enhancer of split. Eur J Biochem. 1993;215:645–652. doi: 10.1111/j.1432-1033.1993.tb18075.x. [DOI] [PubMed] [Google Scholar]
- Ishibashi M, Moriyoshi K, Sasai Y, Shiota K, Nakanishi S, Kageyama R. Persistent expression of helix-loop-helix factor HES-1 prevents mammalian neural differentiation in the central nervous system. EMBO J. 1994;13:1799–1805. doi: 10.1002/j.1460-2075.1994.tb06448.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi M, Ang SL, Shiota K, Nakanishi S, Kageyama R, Guillemot F. Targeted disruption of mammalian hairy and enhancer of split homolog-1 (HES-1) leads to up-regulation of neural helix–loop–helix factors, premature neurogenesis, and severe neural tube defects. Genes & Dev. 1995;9:3136–3148. doi: 10.1101/gad.9.24.3136. [DOI] [PubMed] [Google Scholar]
- Jacq X, Brou C, Lutz Y, Davidson I, Chambon P, Tora L. Human TAFII30 is present in a distinct TFIID complex and is required for transcriptional activation by the estrogen receptor. Cell. 1994;79:107–117. doi: 10.1016/0092-8674(94)90404-9. [DOI] [PubMed] [Google Scholar]
- Johnson JE, Zimmerman K, Saito T, Anderson DJ. Induction and repression of mammalian achaete-scute homologue (MASH) gene expression during neuronal differentiation of P19 embryonal carcinoma cells. Development. 1992;114:75–87. doi: 10.1242/dev.114.1.75. [DOI] [PubMed] [Google Scholar]
- Kato GJ, Dang CV. Function of the c-Myc oncoprotein. FASEB J. 1992;6:3065–3072. doi: 10.1096/fasebj.6.12.1521738. [DOI] [PubMed] [Google Scholar]
- Kemp BE, Pearson RB. Protein kinase recognition sequence motifs. Trends Biochem Sci. 1990;15:342–346. doi: 10.1016/0968-0004(90)90073-k. [DOI] [PubMed] [Google Scholar]
- Klambt C, Knust E, Tietze K, Campos-Ortega JA. Closely related transcripts encoded by the neurogenic gene complex enhancer of split of Drosophila melanogaster. EMBO J. 1989;8:203–210. doi: 10.1002/j.1460-2075.1989.tb03365.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krowczynska AM, Coutts M, Makrides S, Brawerman G. The mouse homologue of the human acidic ribosomal phosphoprotein PO: A highly conserved polypeptide that is under translational control. Nucleic Acids Res. 1989;17:6408. doi: 10.1093/nar/17.15.6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeDouarin B, Zechel C, Garnier J-M, Lutz Y, Tora L, Pierrat B, Heery D, Gronemeyer H, Chambon P, Losson R. The N-terminal part of TIF-1, a putative mediator of the ligand-dependent activation function (AF-2) of nuclear receptors, is fused to B-raf in the oncogenic protein T18. EMBO J. 1995;14:2020–2033. doi: 10.1002/j.1460-2075.1995.tb07194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JE, Hollenberg SM, Snider L, Turner DL, Lipnick N, Weintraub H. Conversion of Xenopus ectoderm into neurons by NeuroD, a basic helix-loop-helix protein. Science. 1995;268:836–844. doi: 10.1126/science.7754368. [DOI] [PubMed] [Google Scholar]
- Lufkin T, Dierich A, Le Meur M, Mark M, Chambon P. Disruption of the Hox-1.6 homeobox gene results in defects in a region corresponding to its rostral domain of expression. Cell. 1991;66:1105–1119. doi: 10.1016/0092-8674(91)90034-v. [DOI] [PubMed] [Google Scholar]
- Ma Q, Kintner C, Anderson DJ. Identification of neurogenin, a vertebrate neuronal determination gene. Cell. 1996;87:43–52. doi: 10.1016/s0092-8674(00)81321-5. [DOI] [PubMed] [Google Scholar]
- MacPherson PA, McBurney MW. P19 embryonal carcinoma cells: A source of cultured neurons amenable to genetic manipulation. Methods: Companion Methods Enzymol. 1995;7:238–252. [Google Scholar]
- Maden M, Holder N. Retinoic acid and development of the central nervous system. BioEssays. 1992;14:431–438. doi: 10.1002/bies.950140702. [DOI] [PubMed] [Google Scholar]
- May M, Mengus G, Lavigne A-C, Chambon P, Davidson I. Human TAF(II28) promotes transcriptional stimulation by activation function 2 of the retinoid X receptors. EMBO J. 1996;15:3093–3104. [PMC free article] [PubMed] [Google Scholar]
- McBurney MW. P19 embryonal carcinoma cells. Int J Dev Biol. 1993;37:135–140. [PubMed] [Google Scholar]
- McBurney MW, Jones-Villeneuve EMV, Edwards MKS, Anderson PJ. Control of muscle and neuronal differentiation in a cultured embryonal carcinoma cell line. Nature. 1982;299:165–167. doi: 10.1038/299165a0. [DOI] [PubMed] [Google Scholar]
- Metzger D, Clifford J, Chiba H, Chambon P. Conditional site-specific recombination in mammalian cells using a ligand-dependent chimeric Cre recombinase. Proc Natl Acad Sci. 1995;92:6991–6995. doi: 10.1073/pnas.92.15.6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moncollin V, Schaeffer L, Chalut C, Egly J-M. Expression in Escherichia coli: Purification and properties of the recombinant human general transcription factor rTFIIB. Protein Expres Purif. 1992;3:374–379. doi: 10.1016/s1046-5928(05)80038-5. [DOI] [PubMed] [Google Scholar]
- Mummery CL, Feijen A, Moolenaar WH, van den Brink CE, de Laat SW. Establishment of a differentiated mesodermal line from P19 EC cells expressing functional PDGF and EGF receptors. Exp Cell Res. 1986;165:229–242. doi: 10.1016/0014-4827(86)90547-1. [DOI] [PubMed] [Google Scholar]
- Papalopulu N, Kintner C. A posteriorising factor, retinoic acid, reveals that anteroposterior patterning controls the timing of neuronal differentiation in Xenopus neuroectoderm. Development. 1996;122:3409–3418. doi: 10.1242/dev.122.11.3409. [DOI] [PubMed] [Google Scholar]
- Roguska MA, Gudas LJ. An increase in prolyl-4-hydroxylase activity occurs during the retinoic acid-induced differentiation of mouse teratocarcinoma stel cell lines F9 and P19. J Biol Chem. 1985;260:13893–13896. [PubMed] [Google Scholar]
- Sasai Y, Kageyama R, Tagawa Y, Shigemoto R, Nakanishi G. Two mammalian helix-loop-helix factors structurally related to Drosophila Hairy and Enhancer of split. Genes & Dev. 1992;6:2620–2634. doi: 10.1101/gad.6.12b.2620. [DOI] [PubMed] [Google Scholar]
- Schuuring E, van Deemter L, Roelink H, Nusse R. Transient expression of the proto-oncogene int-1 during differentiation of P19 embryonal carcinoma cells. Mol Cell Biol. 1989;9:1357–1361. doi: 10.1128/mcb.9.3.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JC, Price BMJ, Van Nimmmen K, Huylebroeck D. Identification of a potent Xenopus mesoderm-inducing factor as a homolog of activin A. Nature. 1990;345:729–731. doi: 10.1038/345729a0. [DOI] [PubMed] [Google Scholar]
- Smolich BD, Papkoff J. Regulated expression of Wnt family members during neuroectodermal differentiation of P19 embryonal carcinoma cells: Overexpression of Wnt-1 perturbs normal differentiation-specific properties. Dev Biol. 1994;166:300–310. doi: 10.1006/dbio.1994.1316. [DOI] [PubMed] [Google Scholar]
- Solter D, Knowles BB. Monoclonal antibody defining a stage-specific mouse embryonic antigen (SSEA-1) Proc Natl Acad Sci. 1978;75:5565–5569. doi: 10.1073/pnas.75.11.5565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporn MB, Roberts AB, Goodman DS. The retinoids: Biology, chemistry and medicine. New York, NY: Raven Press; 1994. [Google Scholar]
- St. Arnaud R, Craig J, McBurney MW, Papkoff J. The int-1 proto-oncogene is transcriptionally activated during neuroectodermal differentiation of P19 mouse embryonal carcinoma cells. Oncogene. 1989;4:1077–1080. [PubMed] [Google Scholar]
- Takebayashi K, Sasai Y, Watanabe T, Nakanishi S, Kageyama S. Structure, chromosomal locus, and promoter analysis of the gene encoding the mouse helix-loop-helix factor HES-1. Negative autoregulation through the multiple N box elements. J Biol Chem. 1994;269:5150–5156. [PubMed] [Google Scholar]
- Tjian R, Maniatis T. Transcriptional activation: A complex puzzle with few easy pieces. Cell. 1994;77:5–8. doi: 10.1016/0092-8674(94)90227-5. [DOI] [PubMed] [Google Scholar]
- Tora L, White JH, Brou C, Tasset D, Webster N, Sheer E, Chambon P. The human estrogen receptor has two independent nonacidic transcriptional activation functions. Cell. 1989;59:477–487. doi: 10.1016/0092-8674(89)90031-7. [DOI] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]