Toxin-Antitoxin Systems Influence Biofilm and Persister Cell Formation and the General Stress Response (original) (raw)
Abstract
In many genomes, toxin-antitoxin (TA) systems have been identified; however, their role in cell physiology has been unclear. Here we examine the evidence that TA systems are involved in biofilm formation and persister cell formation and that these systems may be important regulators of the switch from the planktonic to the biofilm lifestyle as a stress response by their control of secondary messenger 3′,5′-cyclic diguanylic acid. Specifically, upon stress, the sequence-specific mRNA interferases MqsR and MazF mediate cell survival. In addition, we propose that TA systems are not redundant, as they may have developed to respond to specific stresses.
INTRODUCTION
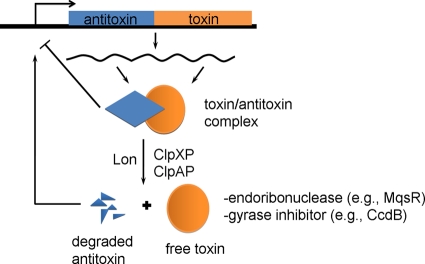
Toxin-antitoxin (TA) systems typically consist of two genes in an operon which encode a stable toxin that disrupts an essential cellular process (e.g., translation via mRNA degradation) and a labile antitoxin (either RNA or a protein) that prevents toxicity (73). RNA antitoxins are known as type I if they inhibit toxin translation as antisense RNA or type III if they inhibit toxin activity; type II antitoxins are proteins that inhibit toxin activity (48). For type II systems (Fig. 1), the antitoxin also acts as a transcriptional repressor and negatively autoregulates the operon by a conserved palindromic motif in the operator region. TA systems were initially discovered in 1983 as plasmid addiction systems on low-copy-number plasmids due to their ability to stabilize plasmids by postsegregational killing (55). TA systems are also ubiquitous as chromosomal elements; for example, of the 126 prokaryotic genomes (16 archaea and 110 bacteria) searched, 671 TA loci were identified (56). Since this report, their prevalence and diversity have increased; for example, in Escherichia coli alone, the number of TA systems has increased from 5 to 37 (71). However, their role in cell physiology is controversial, with nine possible roles identified (51): addictive genomic debris, stabilization of genomic parasites, selfish alleles, gene regulation, growth control, persister cell formation (persister cells are a small fraction of bacteria that demonstrate resistance to antibiotics without genetic change [50]), programmed cell arrest, programmed cell death, and antiphage measures (28, 57). Although they were first thought to be related to cell death, it remains controversial whether TA systems result in cell death (51, 56); hence, the primary role of these systems has been enigmatic. In this review, we present evidence that TA systems regulate genes other than their own operons, mediate the general stress response, and help direct cells toward the formation of biofilm and persister cells.
Fig. 1.
Type II toxin-antitoxin systems. Type II TA systems are typically transcribed in the same operon with the antitoxin gene preceding the toxin gene (although there are exceptions, such as mqsRA). Transcription of the two genes is generally autoregulated by the toxin-antitoxin complex. Proteases Lon, ClpAP, and ClpXP usually degrade labile protein antitoxins and liberate the toxin. Activated toxins function as endoribonucleases or gyrase inhibitors.
TA systems and biofilm formation.
It is well established that bacteria frequently grow in dense, multicellular communities called biofilms (43, 65). Biofilms are formed in aquatic environments by the attachment of bacteria to submerged surfaces, to the air-liquid interface, and to each other. Biofilms attach via appendages such as fimbriae (52) and flagella (33), and microcolonies are formed by the production of microbial products, including polysaccharides (33), glycoproteins (20), and DNA (5). This multicellular behavior is crucial for the disease state since 80% of human bacterial chronic inflammatory and infectious diseases involve biofilms (6). For example, biofilms of E. coli form in the human host in the gastrointestinal tract (9) and in the bladder (3), where uropathogenic E. coli causes urinary tract infections, including both cystitis (bladder infection) and pyelonephritis (kidney infection); these diseases are the most common infections (8 million annual trips to physicians in the United States) and cause enormous financial and health burdens worldwide (23). Understanding pathogenic E. coli infections is important given that there are over 76 million food-related infections annually in the United States (according to the CDC), directly leading to 325,000 hospitalizations, 5,000 deaths, and an economic cost up to $1,426 billion (62).
Biofilms are also important for engineering applications. For example, biofilm formation is deleterious for some fermentations (70), and the economic costs of marine biofouling are estimated be in the billions of dollars (1). In contrast, beneficial biofilms are important for reducing corrosion (30) and for rhizoremediation of chlorinated ethenes (80), as well as holding promise for other applications such as biocatalysis (63). In addition, some groups are beginning to control biofilm formation and dispersal for engineering applications (77).
Hundreds of genes are differentially controlled during the biofilm development process, including stress-associated genes (8, 18, 60, 66). However, early reports indicated that TA systems did not play a role in biofilm formation. For example, in Streptococcus mutans, mutants lacking homologues of the mazF and relE toxin genes had no effect on biofilm formation compared to parental strains (47). The first TA system linked to biofilm formation was the MqsR/MqsA pair of E. coli, since mqsR was induced in a transcriptome study that identified genes that were differentially regulated in biofilm cells (60). The importance of this TA system in biofilm formation was corroborated by the follow-up publication that linked MqsR/MqsA to motility, biofilm formation, and the autoinducer-2 quorum sensing system (26). The mqsR mutation studied in this work likely caused changes in transcription of expression of mqsA (which lies downstream of mqsR) due to the transposon insertion in mqsR (32). The three-dimensional structures show that toxin MqsR is an RNase similar to RelE and YoeB (10) that cleaves mRNA primarily at GCU sites (78) and that antitoxin MqsA binds DNA via its helix-turn-helix motif in its C-terminal domain while binding MqsR at its N-terminal domain (10). These initial results linking MqsR/MqsA to biofilm formation were confirmed recently using 48-h biofilms in which deletion of mqsRA reduced biofilm formation (34) as found in the original report (26). Hence, these studies served to identify a 10th role for TA modules in cell physiology: influencing biofilm formation.
Further evidence of the role of TA systems in biofilm formation was obtained by studying a strain that had five of the most-studied TA systems deleted, named Δ5; this strain lacks the TA pairs MazF/MazE, RelE/RelB, YoeB/YefM, and YafQ/DinJ and also ChpB. Although the mechanism of toxicity at the molecular level is slightly different, MazF (25), RelE (25), ChpB (25), YoeB (15), and YafQ (59) prevent translation by cleaving RNAs. It was reported that these five deletions had no impact on the stress response of cells (72); however, we reasoned that the TA systems were important for biofilm formation based on our microarray results (60). Upon deletion of these five TA systems, biofilm formation decreased after 8 h and increased after 24 h in rich medium at 37°C (37). Therefore, this work presented additional evidence that TA pairs affect biofilm formation. To determine the mechanism by which these five TA systems affect biofilm formation, transcriptome profiling of biofilm cultures was used to determine that deleting the five TA systems results in the differential expression of a single gene, induction of uncharacterized yjgK. Corroborating the complex phenotype seen upon deleting the TA systems, producing YjgK decreased biofilm formation at 8 h and increased biofilm formation at 24 h. Deleting yjgK also affected biofilm formation in the expected manner: biofilm formation increased at 8 h and decreased at 24 h. Additional transcriptome profiling revealed that YjgK represses fimbria genes at 8 h; hence, YjgK represses (either directly or indirectly) type I fimbriae. In addition, deleting all five toxins and antitoxins reduces dispersal (which explains the increase in biofilm formation at 24 h with the Δ5 strain). These findings were significant because they provided additional evidence for one of the first clear roles for TA systems, regulation of biofilm formation, and showed that antitoxins influence biofilm formation. The results indicating that the TA systems of the Δ5 strain affect biofilm formation were confirmed by a second group, which studied the same systems independently and found that deletion of each system decreased biofilm formation after 8 h and 24 h (42). It was determined that the defect in biofilm formation was mainly a result of decreased cell lysis as a result of deleting the toxin genes mazF and yafQ (42).
TA systems in cryptic prophages have also been found to influence biofilm formation. For example, for the TA pair YpjF-YfjZ (12) in E. coli K-12 from cryptic prophage CP4-57, deletion of toxin YpjF increased biofilm formation 5-fold at 7 h (75, 76). Moreover, the well-studied TA system RelEB is encoded in cryptic Qin prophage, and deletion of relEB decreased biofilm formation 2-fold at 24 h in rich and minimal media (42).
Since biofilm formation often involves quorum sensing (17), the link between TA systems and quorum sensing has been explored. Note that the nutritional environment determines the role that quorum sensing plays in Pseudomonas aeruginosa biofilm formation (68). Along with toxin MqsR (26), toxin MazF has been linked to quorum sensing via the extracellular death factor (Asn-Asn-Trp-Asn-Asn) that increases MazF RNase activity (7). Hence, population sensing is important for activity of this toxin, and this is the first report of a peptide-based quorum sensing system in E. coli.
TA systems and persister cells.
Persister cells are a phenotypic subpopulation of bacteria (maximum of about 1% in the stationary state) (49) that are viable after treatment with lethal concentrations of antibiotics. Persister cells arise primarily in biofilms and in stationary-phase cultures (49). This phenotype was first described in 1944, although there was little investigation of it for 40 years (31). The cells are not drug-resistant mutants, as they revert to wild type upon further culturing (69). Importantly, the phenotype contributes to the tolerance of biofilm bacteria to antibiotics (67), which is responsible for the recalcitrance of human infections (49). Also, this antibiotic resistance occurs in the biofilms of many different genera, including E. coli (where they are best studied), P. aeruginosa, Staphylococcus aureus, Lactobacillus acidophilus, and Gardnerella vaginalis (69).
In terms of the genetic basis of persister formation, the main model is that TA pairs are primarily responsible, as they are used to induce a state of dormancy (31, 49), which enables cells to escape the effects of antibiotics. Only a few TA systems reduce persistence when they are deleted due to the redundancy of these systems (19); however, many toxins increase persistence when they are overexpressed. As with biofilm formation, the first direct evidence that a TA system was related to persister cell formation was found with mqsRA of E. coli; deletion of the mqsRA locus as well as deletion of mqsR alone decreases persister formation and production of MqsR/MqsA increases persistence (39). MqsR relies on Hha and CspD to form persister cells (39); Hha is a putative toxin with antitoxin TomB (24), and CspD is a stress-induced cold shock protein that is a DNA replication inhibitor (79). Furthermore, since the protease Lon degrades CspD primarily during the stationary phase, it has been hypothesized that degradation of CspD may be related to persister cell awakening (i.e., when growth resumes) (46).
Additional findings linking toxin MqsR to persistence include those that mqsR is the most highly induced gene in persister cells compared to nonpersisters (67) and that, in transcriptome studies to probe the impact of kanamycin on cell physiology, mqsRA was found to be one of the most highly induced operons (41). MqsR is so toxic that it is impossible to delete the antitoxin mqsA alone (4, 67); similar results have been seen with other antitoxins, including MazE, ChpS, and YefM (4). Other toxin genes are also highly induced in persister cells, including relE, higB, mazF, yafQ, and yoeB (36, 41, 67). To date, the MqsR/MqsA TA system remains the only TA system that affects persistence upon deletion where the antitoxin is a protein. For biofilm cells, but not for planktonic cells, toxin YafQ has also been shown to increase persistence, and its deactivation decreases persistence (27).
Prior to the discovery of mqsRA and persistence, the HipBA TA locus was related to persistence. It was reported that deletion of the hipBA pair caused a 10- to 100-fold repression in persister production under stationary and biofilm culture conditions (36). Unfortunately, this result was recently retracted (19) as the phenotype was due to inadvertent deletion of more than just the TA loci. However, key insights on persistence have been made with HipBA in that this system has been used to demonstrate that once toxin HipA levels reach a threshold, persistence occurs (64).
The second TA system to be related to persistence upon deletion of the toxin is the type I TisAB/IstR-1 system of E. coli, which decreased persistence to ciprofloxacin (19). The TisB toxin (29 amino acids) affects the cell membrane and reduces ATP levels, and IstR-1 is an antisense RNA that acts as the antitoxin by binding the untranslated open reading frame of tisA. This TA locus was used to show that persistence can arise as a result of the SOS response and DNA repair and helps to explain how persister cells arise in exponentially growing cells (but not in the stationary phase).
Quorum sensing has also been related to persistence. A recent study in P. aeruginosa showed that the Las system has a role in persistence through the regulation of RpoS (53). Hence, there may be a close relationship among TA systems, quorum sensing, and biofilm formation.
TA systems and phage abortive infection.
Phage abortive infection is one type of phage resistance, which has been considered altruistic behavior because it favors survival of the cell population following phage infection at the expense of the single cell (14). The seminal report of this behavior for TA systems showed that the type I TA module (RNA-RNA) Hok/Sok from plasmid R1 excludes T4 phage (57). Eight years later, the type II TA module (protein-protein) MazEF was shown to exclude phage P1 (28). Another type II TA module, RnlAB, suppresses T4 propagation by rapidly degrading antitoxin RnlB to release toxin RnlA upon T4 infection (40). The type III TA module (protein-RNA) ToxIN also protects cells against multiple phages (21). It stands to reason that any toxin that reduces host metabolism upon phage attack should cause phage abortive infection, so this behavior should be common for TA systems. Furthermore, this type of altruism should be most important for biofilms, where cells are most likely to be susceptible to phage attack and TA systems should be expressed.
Toxins as global regulators.
Given that toxins that are mRNA interferases degrade mRNA with substrate specificity, they can be viewed as global regulators like Hfq and CsrA that regulate gene expression at a posttranscriptional level (54) by differential mRNA decay. For example, induction of toxin MazF results in the degradation of most mRNA; however, MazF activity also results in the synthesis of a pool of small proteins that are necessary both for toxicity and for survival (2). Critically, some of these enriched small proteins stem from mRNAs that contain MazF cleavage sites, so these mRNAs are protected from cleavage (2). Hence, MazF acts as a global regulatory element (7).
Toxin MqsR also is a global regulator and was in fact the first toxin named based on this property (26). First, deletion of mqsR enriches 76 transcripts in E. coli BW25113 (38) and E. coli MG1655 (26), in which the mqsR coding region was replaced by a kanamycin resistance cassette (4) or in which a transposon insertion was made in mqsR, respectively. These mutations, however, have a polar effect on downstream mqsA, so effects on mRNA levels by both MqsR and MqsA were possible. Therefore, transcriptome studies in which MqsR production alone was used were also assessed, and it was found that MqsR production in the BW25113 wild-type strain causes global changes in the transcriptome profile, with 132 transcripts enriched (38). The global change in the transcriptome profile by MqsR is due to its mRNA interferase activity which has substrate specificity for cellular mRNAs. For example, mRNAs for stress-associated proteins CstA and CspD are reduced upon deleting mqsR and increased by producing MqsR. Other mRNAs for stress-related proteins that are increased by production of MqsR include those for RpoS, ClpP, ClpB, and Dps. Moreover, mRNAs for leader peptides T_naC_, h_isL_, t_rpL_, and p_heL_ are among the most highly enriched mRNAs when MqsR is produced (38). As expected, these four small peptides are among the 14 mRNAs that are do not contain GCU sites (78). Also, 6 of the 14 mRNAs that lack a GCU site (pheL, tnaC, trpL, yciG, ygaQ, and ralR) have genes that are differentially regulated in biofilms (18), which includes three leader peptides. Explorations of these leader peptides and the other 10 proteins in terms of MqsR toxicity and persistence are under way. Preliminary results have confirmed the enrichment of some of these 14 transcripts when MqsR is overproduced and confirmed that they have an effect on cell physiology (T. K. Wood, unpublished data). It is clear that the complexity between mRNA interferase activity and the cellular mRNAs goes beyond the level of the mRNA sequence, since other factors, such as abundance, size, secondary structure, and accessory proteins, also may influence which mRNAs are degraded and which are enriched. Clearly, the cell has devised yet another way to control protein activity: specific mRNA degradation (and protection) via toxin mRNA interferases. Given the prevalence of mRNA interferases, this creates an important and general area for control of cell physiology.
Antitoxins regulate more than their own locus.
Based on our discovery that MqsR/MqsA affect many aspects of cell physiology, including motility (26), we hypothesized that MqsA, through its DNA-binding motif, likely regulates more than its own synthesis (10, 38). Hence, not only does MqsR affect cell physiology by degrading nearly all the cellular mRNA (thereby inducing dormancy) but also antitoxin MqsA affects cell dormancy by regulating other cellular systems, including other toxins. To investigate this possibility, a systems biology approach was utilized so that all promoters in the genome could be explored. Using three sets of transcriptome studies and two nickel-enrichment DNA binding microarrays coupled with cell survival studies in which MqsR was overproduced in isogenic mutants, we identified that MqsR/MqsA are related to cspD (38). Quantitative real-time PCR showed that (i) MqsA represses cspD (encoding a DNA replication inhibitor), (ii) MqsR overproduction increases cspD mRNA, (iii) stress induces cspD, and (iv) stress fails to induce cspD when both MqsR/MqsA are produced or when mqsRA is deleted. Electrophoretic mobility shift assays show that the MqsR/MqsA complex binds the promoter of cspD. In addition, proteases Lon and ClpXP are necessary for MqsR toxicity (38). Together, these results indicate that antitoxin MqsA represses cspD, which may be derepressed by titrating MqsA with MqsR or by degrading MqsA via stress conditions through proteases Lon and ClpPX. Therefore, MqsR/MqsA are the first TA pair shown to regulate more than their own synthesis (38); this creates a new paradigm where antitoxins of TA systems may be viewed as regulators.
TA systems regulate the GSR.
As with cspD, antitoxin MqsA also helps mediate the general stress response (GSR). The GSR in bacteria is accompanied by a significantly reduced growth rate, and it appears that TA systems are the means by which growth is slowed (56). The GSR allows cells to survive long periods of starvation and different environmental stresses. Importantly, the GSR has been shown to be a modulated switch rather than an on/off-type switch and thus is a reversible state. The _rpoS_-encoded sigma factor σS is the master regulator of the GSR (29) in Gram-negative bacteria, such as in E. coli. It is thus probable that the increased dormancy in biofilms and the dramatically reduced growth rates of persister cells are the major reasons for the reduced susceptibility of biofilms to antibiotics (13).
The GSR has been directly implicated in chronic infections due to biofilm formation (22); thus, a detailed molecular and functional understanding of GSR regulation in biofilms will provide essential insights for potential routes for novel and potent therapeutic interventions in biofilm-dependent infections. Indeed, quorum-sensing GSR signaling pathways are activated in chronic infections in cystic fibrosis patients (22).
Critically, TA pairs are implicated in the regulation of the GSR via RpoS as demonstrated by work with MqsR/MqsA. Antitoxin MqsA, much as it does for cspD, directly represses the transcription of the master regulator of stress, RpoS (74). MqsA recognizes _mqsRA-_like palindromes (11) in several promoter sequences like those of rpoS and csgD; mutation of the palindrome in the rpoS promoter abolishes the binding of MqsA (74). Upon oxidative stress, MqsA is degraded by the protease Lon (74) with the result that rpoS is derepressed. Conversely, production of MqsA represses rpoS and reduces concentrations of the second messenger 3′,5′-cyclic diguanylic acid (c-di-GMP) due to repression of diguanylate cyclases that are controlled by RpoS. For example, MqsA indirectly represses adrA (which encodes a diguanylate cyclase related to cellulose production), which is positively regulated by RpoS (45), and reduces c-di-GMP levels by inhibiting other genes that encode diguanylate cyclases (e.g., ydaM, yegE, and yedQ) all in a manner opposite from how RpoS regulates these genes. In addition, csgD, which encodes the regulator for curli and cellulose, which is activated by RpoS (58), is repressed by MqsA (74). The result of repressing these RpoS-regulated genes by MqsA leads to increased motility and a reduction in the cell adhesins curli and cellulose as well as a reduction in biofilm formation (74). Hence, degradation of MqsA and activation of MqsR reduce motility (74), and this is why this TA system was first linked to motility (26). Further evidence that MqsA blocks the GSR is that repression of rpoS by MqsA leads to an 850-fold reduction in oxidative stress resistance via repression of catalase activity (RpoS is a positive regulator of catalase activity via katG and katE [44]). Therefore, four of the expected phenotypes related to the GSR are controlled by MqsA. Hence, one way that external stress alters gene regulation is via toxin-antitoxin systems (74) and their regulation of rpoS transcription; this is one of the first clear mechanisms of how external stress is propagated in terms of gene regulation and creates a vital new role for TA systems.
TA systems as the genetic basis of biofilm formation.
External stress also alters gene regulation and the GSR and leads to a switch from the planktonic state (high motility) to the biofilm state (low motility) (58). During stress, MqsA is degraded, which in turn activates MqsR and RpoS (74). Activation of RpoS leads to reduced motility and increased production of cell adhesions (74); this results in increased biofilm formation (74). Hence, MqsA regulates biofilm formation by regulating RpoS. This also serves to explain why persister cells are seen primarily in biofilms, since it is in this state that toxin MqsR is activated (39). It appears that there is a spectrum of MqsR activity, with some cells using MqsR to redirect cellular metabolism to RpoS-induced genes (by degrading mRNA of transcripts from exponential growth), whereas other cells (a small percentage) utilize MqsR to make the cells dormant, i.e., to create persister cells (74).
Significance of redundant TA systems.
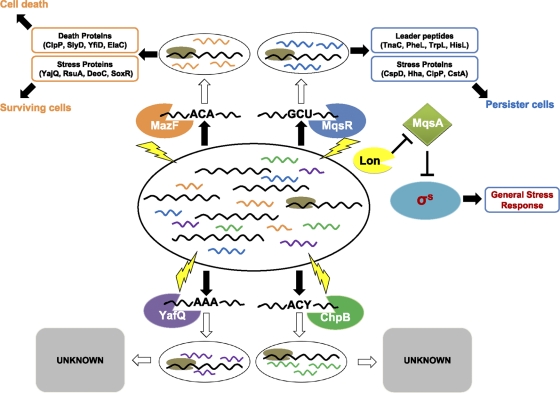
Although speculative, given the importance of the MqsR/MqsA TA system for the oxidative stress response of E. coli and given that there are at least 37 TA systems in this species (71) as well as redundant TA systems in many if not most bacteria (56) (e.g., Mycobacterium tuberculosis has at least 88 TA systems [7]), it is tempting to speculate that the reason for the redundancy is that each TA system allows the cell to respond to a specific stress or group of stresses in a highly regulated, elegant fashion. Hence, there may be at least six RNases in E. coli (MqsR, MazF, RelE, ChpB, YafQ, and YoeB), with each degrading a distinct group of mRNAs based on a specific stress. The evidence for this hypothesis is that the stresses to induce each TA system are different, although there is some overlap (Table 1); the overlap is not surprising given that one important class of environmental stresses, antibiotics, works through a common reactive oxygen species mechanism (41). Second, MqsR, MazF, YafQ, and ChpB cleave mRNA at the GCU (78), ACA (81), AAA (59), and ACY (Y = A or G) (25) sites, respectively (Fig. 2 ); therefore, their functions are not entirely redundant. Moreover, upon antibiotic stress, MazF degrades most mRNAs; however, specific proteins are produced from nondegraded mRNAs even when they contain the ACA degradation site with some of the pool of newly synthesized proteins used for toxicity and some used for cell survival (2). Similarly, for MqsR, upon oxidative stress, once MqsA is rapidly degraded, the released MqsR cleaves mRNAs specifically and enriches mRNAs that encode DNA replication inhibitor CspD and the TnaC, TrpL, HisL, and PheL leader peptides. Cell death has been regarded as an important feature of biofilms and under other stress conditions (61). The exact role of the toxic peptides and the leader peptides in mediating toxicity and surviving after stress is under investigation. Other stress proteins, RpoS, ClpP, ClpB, and CstA, are also induced by active MqsR and thus help cells to cope with stress and survive. Therefore, upon a specific stress, the role of each TA system may be to reduce growth and direct metabolism toward a new set of mRNAs (that are primarily not cleaved) as well as to create persister cells for a small subpopulation of cells.
Table 1.
Summary of stress conditions that induce toxin-antitoxin systems
| Stress condition | Induced TA system(s) (reference[s]) |
|---|---|
| Biofilm | MqsRA (60), YoeB/YefM (60) |
| Amino acid starvation | YafNO (16), HigBA (16), MqsRA (16) |
| Oxidative stress (H2O2) | MqsRA (38) |
| Ampicillin (100 μg/ml) | MqsRA (41, 67) |
| Chloramphenicol (30 μg/ml) | YafNO (16), HigBA (16), MqsRA (16) |
| Mitomycin C or SOS response | YafNO (16), YafQ/DinJ (59), TisB/IstR-1 (19), SymE/SymR (35) |
| Kanamycin (5 μg/ml) | MqsRA (41), YoeB/YefM (41), RelEB (41), HigBA (41), MazFE (41) |
| Ciprofloxacin (0.1 μg/ml) | TisB/IstR-1 (19) |
Fig. 2.
Schematic of how toxicity is mediated by mRNA interferases MqsR, MazF, YafQ, and ChpB. Toxin-antitoxin systems are induced by various environmental stresses (indicated by lightning bolts) (Table 1) which serve to induce proteases such as Lon that degrade antitoxins. Upon degradation of the antitoxin, the free toxin cleaves most cellular mRNAs in a sequence-specific manner. This leads to reduced production of large proteins (encoded by mRNAs with cleavage sites shown in black) and an enrichment of small proteins (encoded by mRNAs that lack cleavage sites and shown in orange for MazF, blue for MqsR, green for ChpB, and purple for YafQ) that are death proteins or stress proteins (which help the cell cope with the stress). Cellular mRNAs that contain cleavage sites for each toxin but remain uncleaved are marked with an accessory protein (in brown); these mRNAs may also be protected by secondary structure. The roles of YafQ and ChpB in determining the fate of the cells need to be investigated further.
Conclusions.
As outlined here, TA systems appear to be integral regulators of cellular activity as they clearly can impact motility, biofilm formation, quorum sensing, and persistence; hence, they are far more than genomic debris. Furthermore, one of the most important TA systems is the MqsR/MqsA locus of E. coli, which is the first TA system to be related to biofilm formation, quorum sensing, persistence, and global regulation. It remains to be investigated whether this interesting TA system of E. coli is as important in other bacteria, since MqsR sequences are present in 40 of 914 genomes (34) and in many genera, including Yersinia pseudotuberculosis, Yersinia pestis, Bordetella bronchiseptica, and Pseudomonas fluorescens (38). Based on the insights gleaned from MqsR/MqsA and other TA systems, it seems that the primary function of TA systems is to mediate the response of the cell to external stress by initiating programmed cell arrest, persister cell formation, and biofilm formation.
ACKNOWLEDGMENT
This work was supported by the National Institutes of Health (R01 GM089999).
Biographies

Xiaoxue Wang was a postdoctoral fellow in Thomas K. Wood's lab at Texas A&M University (2008 to 2011). She has been exploring the roles of cryptic prophage genes and toxin-antitoxin systems in the stress response of E. coli. She is also interested in horizontal gene transfer in generating diversity in biofilms. She obtained her Ph.D. in genetics from Texas A&M University in 2006 by studying the quantitative genetics of fish and obtained her B.E. from the Ocean University of China in 2000. Her first postdoctoral research position was at the Haskin Shellfish Research Laboratory at Rutgers University (2006 to 2007). Funded by the Recruitment Program of Global Experts program, she is starting her academic career as a research professor in the Chinese Academy of Sciences Key Laboratory of Marine Bio-Resource Sustainable Utilization at the South China Sea Institute of Oceanology and plans to study biofilm formation under various marine environmental conditions and with potential manipulative strategies.

Thomas K. Wood is the O'Connor Endowed Chair and Professor in the Department of Chemical Engineering at Texas A&M University. He was formerly the Northeast Utilities Endowed Chair in Environmental Engineering at the University of Connecticut (1998 to 2005) and started his teaching career at the University of California, Irvine, in 1991. He obtained his Ph.D. in chemical engineering from North Carolina State University in 1991 by studying heterologous protein production and obtained his B.S. from the University of Kentucky in 1985. His current research pursuits include understanding the genetic basis of biofilm formation to prevent disease and to utilize biofilms for beneficial biotransformations, including remediation, green chemistry, and energy production. He also uses systems biology approaches to understand cell resistance, specifically discerning the role of toxin-antitoxin systems and cryptic prophage in resistance. He also has utilized protein engineering to control biofilm formation as well as for bioremediation and green chemistry.
Footnotes
▿
Published ahead of print on 17 June 2011.
REFERENCES
- 1.Aldred N., Phang I. Y., Conlan S. L., Clare A. S., Vancso G. J. 2008. The effects of a serine protease, Alcalase, on the adhesives of barnacle cyprids (Balanus amphitrite). Biofouling 24:97–107 [DOI] [PubMed] [Google Scholar]
- 2.Amitai S., Kolodkin-Gal I., Hananya-Meltabashi M., Sacher A., Engelberg-Kulka H. 2009. Escherichia coli MazF leads to the simultaneous selective synthesis of both “death proteins” and “survival proteins.” PLoS Genet. 5:e1000390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson G. G., et al. 2003. Intracellular bacterial biofilm-like pods in urinary tract infections. Science 301:105–107 [DOI] [PubMed] [Google Scholar]
- 4.Baba T., et al. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barken K. B., et al. 2008. Roles of type IV pili, flagellum-mediated motility and extracellular DNA in the formation of mature multicellular structures in Pseudomonas aeruginosa biofilms. Environ. Microbiol. 10:2331–2343 [DOI] [PubMed] [Google Scholar]
- 6.Barraud N., et al. 2009. Nitric oxide-mediated dispersal in single- and multi-species biofilms of clinically and industrially relevant microorganisms. Microb. Biotechnol. 2:370–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belitsky M., et al. 2011. The Escherichia coli extracellular death factor EDF induces the endoribonucleolytic activities of the toxins MazF and ChpBK. Mol. Cell 41:625–635 [DOI] [PubMed] [Google Scholar]
- 8.Beloin C., et al. 2004. Global impact of mature biofilm lifestyle on Escherichia coli K-12 gene expression. Mol. Microbiol. 51:659–674 [DOI] [PubMed] [Google Scholar]
- 9.Bollinger R. R., Barbas A. S., Bush E. L., Lin S. S., Parker W. 2007. Biofilms in the normal human large bowel: fact rather than fiction. Gut 56:1481–1482 [PMC free article] [PubMed] [Google Scholar]
- 10.Brown B. L., et al. 2009. Three dimensional structure of the MqsR:MqsA complex: a novel toxin:antitoxin pair comprised of a toxin homologous to RelE and an antitoxin with unique properties. PLoS Pathog. 5:e1000706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown B. L., Wood T. K., Peti W., Page R. 2011. Structure of the Escherichia coli antitoxin MqsA (YgiT/b3021) bound to its gene promoter reveals extensive domain rearrangements and the specificity of transcriptional regulation. J. Biol. Chem. 286:2285–2296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown J. M., Shaw K. J. 2003. A novel family of Escherichia coli toxin-antitoxin gene pairs. J. Bacteriol. 185:6600–6608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown M. R., Collier P. J., Gilbert P. 1990. Influence of growth rate on susceptibility to antimicrobial agents: modification of the cell envelope and batch and continuous culture studies. Antimicrob. Agents Chemother. 34:1623–1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chopin M.-C., Chopin A., Bidnenko E. 2005. Phage abortive infection in lactococci: variations on a theme. Curr. Opin. Microbiol. 8:473–479 [DOI] [PubMed] [Google Scholar]
- 15.Christensen S. K., et al. 2004. Overproduction of the Lon protease triggers inhibition of translation in Escherichia coli: involvement of the _yefM_-yoeB toxin-antitoxin system. Mol. Microbiol. 51:1705–1717 [DOI] [PubMed] [Google Scholar]
- 16.Christensen-Dalsgaard M., Jørgensen M. G., Gerdes K. 2010. Three new RelE-homologous mRNA interferases of Escherichia coli differentially induced by environmental stresses. Mol. Microbiol. 75:333–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davies D. G., et al. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295–298 [DOI] [PubMed] [Google Scholar]
- 18.Domka J., Lee J., Bansal T., Wood T. K. 2007. Temporal gene-expression in Escherichia coli K-12 biofilms. Environ. Microbiol. 9:332–346 [DOI] [PubMed] [Google Scholar]
- 19.Dörr T., Vulić M., Lewis K. 2010. Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLoS Biol. 8:e1000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elvers K. T., Lappin-Scott H. M. 2000. Biofilms and biofouling, 2nd ed., vol. 1. Academic Press, San Diego, CA [Google Scholar]
- 21.Fineran P. C., et al. 2009. The phage abortive infection system, ToxIN, functions as a protein-RNA toxin-antitoxin pair. Proc. Natl. Acad. Sci. U. S. A. 106:894–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Foley I., Marsh P., Wellington E. M., Smith A. W., Brown M. R. 1999. General stress response master regulator rpoS is expressed in human infection: a possible role in chronicity. J. Antimicrob. Chemother. 43:164–165 [DOI] [PubMed] [Google Scholar]
- 23.Foxman B. 2002. Epidemiology of urinary tract infections: incidence, morbidity, and economic costs. Am. J. Med. 113:5–13 [DOI] [PubMed] [Google Scholar]
- 24.Garcia-Contreras R., Zhang X. S., Kim Y., Wood T. K. 2008. Protein translation and cell death: the role of rare tRNAs in biofilm formation and in activating dormant phage killer genes. PLoS One 3:e2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gerdes K., Christensen S. K., Løbner-Olesen A. 2005. Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 3:371–382 [DOI] [PubMed] [Google Scholar]
- 26.González Barrios A. F., et al. 2006. Autoinducer 2 controls biofilm formation in Escherichia coli through a novel motility quorum-sensing regulator (MqsR, B3022). J. Bacteriol. 188:305–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harrison J. J., et al. 2009. The chromosomal toxin gene yafQ is a determinant of multidrug tolerance for Escherichia coli growing in a biofilm. Antimicrob. Agents Chemother. 53:2253–2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hazan R., Engelberg-Kulka H. 2004. Escherichia coli _mazEF_-mediated cell death as a defense mechanism that inhibits the spread of phage P1. Mol. Genet. Genomics 272:227–234 [DOI] [PubMed] [Google Scholar]
- 29.Hengge-Aronis R. 1996. Back to log phase: σS as a global regulator in the osmotic control of gene expression in Escherichia coli. Mol. Microbiol. 21:887–893 [DOI] [PubMed] [Google Scholar]
- 30.Jayaraman A., Mansfeld F. B., Wood T. K. 1999. Inhibiting sulfate-reducing bacteria in biofilms by expressing the antimicrobial peptides indolicidin and bactenecin. J. Ind. Microbiol. Biotechnol. 22:167–175 [DOI] [PubMed] [Google Scholar]
- 31.Jayaraman R. 2008. Bacterial persistence: some new insights into an old phenomenon. J. Biosci. 33:795–805 [DOI] [PubMed] [Google Scholar]
- 32.Kang Y., et al. 2004. Systematic mutagenesis of the Escherichia coli genome. J. Bacteriol. 186:4921–4930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karatan E., Watnick P. 2009. Signals, regulatory networks, and materials that build and break bacterial biofilms. Microbiol. Mol. Biol. Rev. 73:310–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kasari V., Kurg K., Margus T., Tenson T., Kaldalu N. 2010. The Escherichia coli mqsR and ygiT genes encode a new toxin-antitoxin pair. J. Bacteriol. 192:2908–2919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kawano M., Aravind L., Storz G. 2007. An antisense RNA controls synthesis of an SOS-induced toxin evolved from an antitoxin. Mol. Microbiol. 64:738–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Keren I., Shah D., Spoering A., Kaldalu N., Lewis K. 2004. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J. Bacteriol. 186:8172–8180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim Y., Wang X., Ma Q., Zhang X.-S., Wood T. K. 2009. Toxin-antitoxin systems in Escherichia coli influence biofilm formation through YjgK (TabA) and fimbriae. J. Bacteriol. 191:1258–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim Y., et al. 2010. Escherichia coli toxin/antitoxin pair MqsR/MqsA regulate toxin CspD. Environ. Microbiol. 12:1105–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim Y., Wood T. K. 2010. Toxins Hha and CspD and small RNA regulator Hfq are involved in persister cell formation through MqsR in Escherichia coli. Biochem. Biophys. Res. Commun. 391:209–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koga M., Otsuka Y., Lemire S., Yonesaki T. 2011. Escherichia coli rnlA and rnlB compose a novel toxin-antitoxin system. Genetics 187:123–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kohanski M. A., Dwyer D. J., Hayete B., Lawrence C. A., Collins J. J. 2007. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130:797–810 [DOI] [PubMed] [Google Scholar]
- 42.Kolodkin-Gal I., Verdiger R., Shlosberg-Fedida A., Engelberg-Kulka H. 2009. A differential effect of E. coli toxin-antitoxin systems on cell death in liquid media and biofilm formation. PLoS One 4:e6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kolter R., Losick R. 1998. One for all and all for one. Science 280:226–227 [DOI] [PubMed] [Google Scholar]
- 44.Lacour S., Landini P. 2004. σS-dependent gene expression at the onset of stationary phase in Escherichia coli: function of σS-dependent genes and identification of their promoter sequences. J. Bacteriol. 186:7186–7195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Landini P. 2009. Cross-talk mechanisms in biofilm formation and responses to environmental and physiological stress in Escherichia coli. Res. Microbiol. 160:259–266 [DOI] [PubMed] [Google Scholar]
- 46.Langklotz S., Narberhaus F. 2011. The Escherichia coli replication inhibitor CspD is subject to growth-regulated degradation by the Lon protease. Mol. Microbiol. 80:1313–1325 [DOI] [PubMed] [Google Scholar]
- 47.Lemos J. A. C., Brown J. T. A., Abranches J., Burne R. A. 2005. Characteristics of Streptococcus mutans strains lacking the MazEF and RelBE toxin-antitoxin modules. FEMS Microbiol. Lett. 253:251–257 [DOI] [PubMed] [Google Scholar]
- 48.Leplae R., et al. 21 March 2011. Diversity of bacterial type II toxin-antitoxin systems: a comprehensive search and functional analysis of novel families. Nucleic Acids Res. [Epub ahead of print.] doi:10.1093/nar/gkr131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lewis K. 2008. Multidrug tolerance of biofilms and persister cells, p. 107–131 _In_Romeo T. (ed.), Bacterial biofilms. Springer-Verlag, Berlin, Germany: [DOI] [PubMed] [Google Scholar]
- 50.Lewis K. 2007. Persister cells, dormancy and infectious disease. Nat. Rev. Microbiol. 5:48–56 [DOI] [PubMed] [Google Scholar]
- 51.Magnuson R. D. 2007. Hypothetical functions of toxin-antitoxin systems. J. Bacteriol. 189:6089–6092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Müller C. M., et al. 2009. Type 1 fimbriae, a colonization factor of uropathogenic Escherichia coli, are controlled by the metabolic sensor CRP-cAMP. PLoS Pathog. 5:e1000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murakami K., et al. 2005. Role for rpoS gene of Pseudomonas aeruginosa in antibiotic tolerance. FEMS Microbiol. Lett. 242:161–167 [DOI] [PubMed] [Google Scholar]
- 54.Nogueira T., Springer M. 2000. Post-transcriptional control by global regulators of gene expression in bacteria. Curr. Opin. Microbiol. 3:154–158 [DOI] [PubMed] [Google Scholar]
- 55.Ogura T., Hiraga S. 1983. Mini-F plasmid genes that couple host cell division to plasmid proliferation. Proc. Natl. Acad. Sci. U. S. A. 80:4784–4788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pandey D. P., Gerdes K. 2005. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 33:966–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pecota D. C., Wood T. K. 1996. Exclusion of T4 phage by the hok/sok killer locus from plasmid R1. J. Bacteriol. 178:2044–2050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pesavento C., et al. 2008. Inverse regulatory coordination of motility and curli-mediated adhesion in Escherichia coli. Genes Dev. 22:2434–2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Prysak M. H., et al. 2009. Bacterial toxin YafQ is an endoribonuclease that associates with the ribosome and blocks translation elongation through sequence-specific and frame-dependent mRNA cleavage. Mol. Microbiol. 71:1071–1087 [DOI] [PubMed] [Google Scholar]
- 60.Ren D., Bedzyk L. A., Thomas S. M., Ye R. W., Wood T. K. 2004. Gene expression in Escherichia coli biofilms. Appl. Microbiol. Biotechnol. 64:515–524 [DOI] [PubMed] [Google Scholar]
- 61.Rice K. C., Bayles K. W. 2008. Molecular control of bacterial death and lysis. Microbiol. Mol. Biol. Rev. 72:85–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Roberts T. 2007. WTP estimates of the societal costs of U.S. food-borne illness. Am. J. Agric. Econ. 89:1183–1188 [Google Scholar]
- 63.Rosche B., Li X. Z., Hauer B., Schmid A., Buehler K. 2009. Microbial biofilms: a concept for industrial catalysis? Trends Biotechnol. 27:636–643 [DOI] [PubMed] [Google Scholar]
- 64.Rotem E., et al. 2010. Regulation of phenotypic variability by a threshold-based mechanism underlies bacterial persistence. Proc. Natl. Acad. Sci. U. S. A. 107:12541–12546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sauer K., Camper A. K. 2001. Characterization of phenotypic changes in Pseudomonas putida in response to surface-associated growth. J. Bacteriol. 183:6579–6589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schembri M. A., Kjærgaard K., Klemm P. 2003. Global gene expression in Escherichia coli biofilms. Mol. Microbiol. 48:253–267 [DOI] [PubMed] [Google Scholar]
- 67.Shah D., et al. 2006. Persisters: a distinct physiological state of E. coli. BMC Microbiol. 6:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shrout J. D., et al. 2006. The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol. Microbiol. 62:1264–1277 [DOI] [PubMed] [Google Scholar]
- 69.Singh R., Ray P., Das A., Sharma M. 2009. Role of persisters and small colony variants in antibiotic resistance of planktonic and biofilm-associated Staphylococcus aureus: an in vitro study. J. Med. Microbiol. 5:1067–1073 [DOI] [PubMed] [Google Scholar]
- 70.Sung B. H., et al. 2006. Development of a biofilm production-deficient Escherichia coli strain as a host for biotechnological applications. Appl. Environ. Microbiol. 72:3336–3342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tan Q., Awano N., Inouye M. 2011. YeeV is an Escherichia coli toxin that inhibits cell division by targeting the cytoskeleton proteins, FtsZ and MreB. Mol. Microbiol. 79:109–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tsilibaris V., Maenhaut-Michel G., Mine N., Van Melderen L. 2007. What is the benefit to Escherichia coli of having multiple toxin-antitoxin systems in its genome? J. Bacteriol. 189:6101–6108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Van Melderen L., Saavedra De Bast M. 2009. Bacterial toxin-antitoxin systems: more than selfish entities? PLoS Genet. 5:e1000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang X., et al. 2011. Antitoxin MqsA mediates the bacterial general stress response. Nat. Chem. Biol. 7:359–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang X., et al. 2010. Cryptic prophages help bacteria cope with adverse environments. Nat. Commun. 1:147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang X., Kim Y., Wood T. K. 2009. Control and benefits of CP4-57 prophage excision in Escherichia coli biofilms. ISME J. 3:1164–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wood T. K., Hong S. H., Ma Q. 2011. Engineering biofilm formation and dispersal. Trends Biotechnol. 29:87–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yamaguchi Y., Park J.-H., Inouye M. 2009. MqsR, a crucial regulator for quorum sensing and biofilm formation, is a GCU-specific mRNA interferase in Escherichia coli. J. Biol. Chem. 284:28746–28753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yamanaka K., Zheng W., Crooke E., Wang Y.-H., Inouye M. 2001. CspD, a novel DNA replication inhibitor induced during the stationary phase in Escherichia coli. Mol. Microbiol. 39:1572–1584 [DOI] [PubMed] [Google Scholar]
- 80.Yee D. C., Maynard J. A., Wood T. K. 1998. Rhizoremediation of trichloroethylene by a recombinant, root-colonizing Pseudomonas fluorescens strain expressing toluene _ortho_-monooxygenase constitutively. Appl. Environ. Microbiol. 64:112–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang Y., et al. 2003. MazF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli. Mol. Cell 12:913–923 [DOI] [PubMed] [Google Scholar]