The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl (original) (raw)
Abstract
Plant developmental processes are controlled by both endogenous programs and environmental stimuli. As a photomorphogenetic mutant, hy5 of Arabidopsis has been isolated and characterized. Our detailed characterization has revealed that the mutant is deficient in a variety of stimulus responses, including gravitropic response and waving growth of roots, as well as light-dependent hypocotyl elongation. In the roots and hypocotyl, the hy5 mutation also affects greening and specific cell proliferation such as lateral root formation and secondary thickening. Those phenotypes indicate that the HY5 gene is responsible for the regulation of fundamental developmental processes of the plant cell: cell elongation, cell proliferation, and chloroplast development. Molecular cloning of the HY5 gene using a T-DNA-tagged mutant has revealed that the gene encodes a protein with a bZIP motif, one of the motifs found in transcriptional regulators. Nuclear localization of the HY5 protein strongly suggests that the HY5 gene modulates the signal transduction pathways under the HY5-related development by controlling expression of genes downstream of these pathways.
Keywords: Arabidopsis, HY5, stimulus-responses, photomorphogenesis, root development, bZIP protein
Plant development is influenced by many environmental factors. Light, gravity, temperature, touching, and chemical compounds are perceived as stimuli by plants and affect the choice of development programs. Light is one of the most important stimuli to plant development. Seedlings grown in the light or in darkness take quite different morphological strategies, called photomorphogenesis or skotomorphogenesis, respectively. Light-grown seedlings show a short hypocotyl with green and expanded cotyledons. On the other hand, dark-grown, or etiolated seedlings, show a long hypocotyl with yellow and unopened cotyledons. The light-regulated development in young seedlings has been dissected through molecular genetic approaches in Arabidopsis thaliana (for review, see Chory 1993; Deng 1994; McNellis and Deng 1995). A class of hy mutations designated for their long hypocotyl phenotype in the light has defects in light-capturing molecules; the hy1, hy2, phyB (formerly designated hy3), and hy6 mutants have been shown to be deficient in phytochromes, whereas the hy4 mutant is deficient in a blue light receptor (Koornneef et al. 1980; Chory et al. 1989a; Parks and Quail 1991; Ahmad and Cashmore 1993; Reed et al. 1993). Of the six hy loci, the molecular role of the HY5 gene has not been clarified. Another class of mutations, including deetiolated (det) and constitutively photomorphogenic (cop), shows morphology of dark-grown seedlings similar to that of the light-grown seedlings of the wild-type (for review, see Chory 1993; Deng 1994; McNellis and Deng 1995). It is suggested that the DET and the COP gene products work as repressors of photomorphogenesis when grown in darkness, and a number of DET/COP class gene products including DET1 and COP1 are thought to modulate the signal transduction pathways that originate from photoreceptors related to HY1, HY2, PHYB, HY6, and HY4 genes. The DET1 and the COP1 genes have been shown to encode nuclear proteins that are thought to modulate the pathways by repressing light-regulated gene expression (Chory et al. 1989b; Deng et al. 1992; Pepper et al. 1994; von Arnim and Deng 1994). The HY5 gene product also has been regarded as a mediator in the light-dependent signal transduction pathways (Chory 1992; Ang and Deng 1994).
In our study of mechanisms of root morphogenesis and stimulus responses in the root as a model system of plant development, we have examined a series of hy5 mutants and found that these mutants show common interesting phenotypes in the root morphology under the control of environmental stimuli and endogenous programs of development (Okada and Shimura 1994). Recently, extensive genetic studies on root systems of Arabidopsis have been started, and a number of mutants deficient in stimulus response or development of roots have been isolated and characterized (for review, see Okada and Shimura 1992a; Aeschbacher et al. 1994; Dolan and Roberts 1995). Mutants with aberrant root gravitropism, including aux1, dwf, axr1, axr2, and axr4 show resistance to auxin, indicating that those genes work both in the signaling pathway of gravitropism and in auxin-related developmental systems (Wilkins 1966; Juniper 1976; Feldman 1984; Hobbie and Estelle 1995). The AXR1 gene encodes a protein related to ubiquitin-activating enzyme E1, and it has been implied that the ubiquitin pathway may play a role in plant hormone action (Leyser et al. 1993). The AUX1 gene encodes a permease-like protein possibly working in auxin uptake (Bennett et al. 1996). Studies on the developmental steps in root meristem formation have also been progressing, and a number of mutants have been reported to be deficient in specific layers of radial organization, or in cell shape of the root (for review, see Aeschbacher et al. 1994; Dolan and Roberts 1995). The SCARECROW gene, which regulates an asymmetric cell division of the cortex/endodermal initial, encodes a protein with a basic leucine zipper (bZIP)-like domain, and has been indicated as a transcription factor (Di Laurenzio et al. 1996). Although some genes have been cloned, their molecular mechanisms are largely unknown.
Recently, anatomical and genetic studies on lateral roots of Arabidopsis have been started. The lateral roots originate in pericycle cells of the main root. Although the morphology of lateral roots looks similar to that of the main root, the origin and the formation pattern are different from each other (Dolan et al. 1993; Malamy and Benfey 1997). Auxin can induce lateral root initiation (Laskowski et al. 1995). Several mutants, including aux1, axr1, axr4 (Hobbie and Estelle 1995), alf1, alf3, alf4 (Celenza et al. 1995), and sur1 (Boerjan et al. 1995), show abnormality both in lateral root formation and in auxin-related responses.
In this paper we report detailed characterization of the phenotypes of the hy5 mutants, including stimulus responses, lateral root formation, secondary thickening in roots, and photomorphogenesis of young seedlings. We also report the cloning and molecular characterization of the HY5 gene. By integrating the genetic, morphological, and molecular data, we find that the HY5 gene works in the nucleus as a key modulator of signal transduction pathways mediating a wide variety of stimulus responses and developmental processes in the root and hypocotyl.
Results
Phenotypes of hy5 mutants
As shown in Figure 1, A–D, three different alleles of hy5 mutants show similar structural and developmental phenotypes in the hypocotyl and roots of plants on agar plates set in the vertical position. Structural abnormalities were not found in other organs, including leaves, stems, and flowers.
Figure 1.
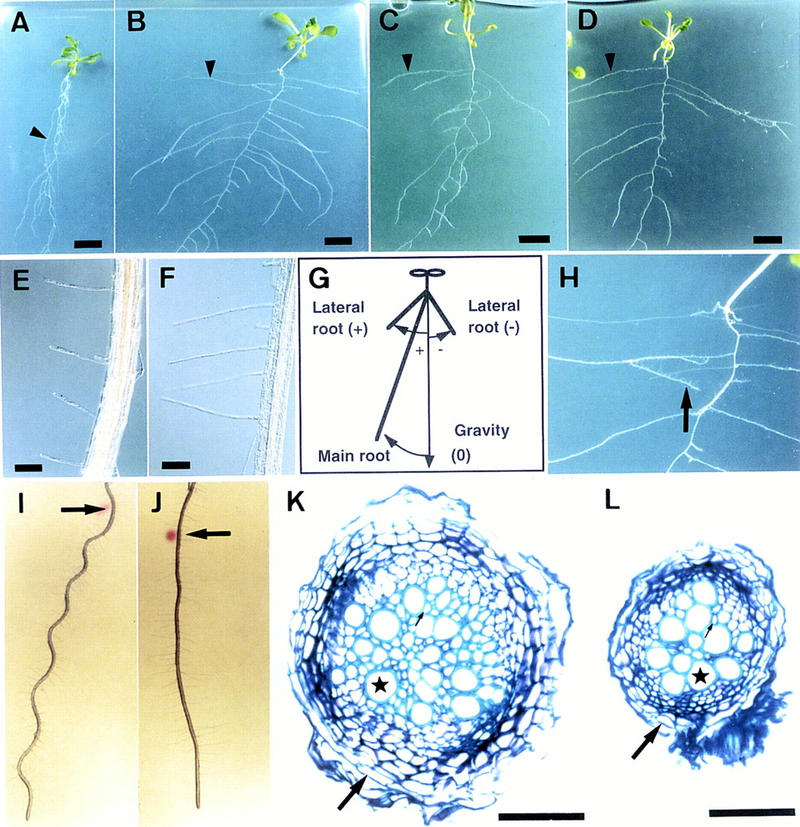
Phenotypes of hy5 mutants. (A_–_D) Plants at 20 DAG grown on vertically positioned agar plates supplemented with sucrose. Wild-type of Landsberg erecta (Ler) ecotype (A), hy5-1 (B), hy5–Ks50 (C), and hy5-215 (D) are shown. The arrowhead in each panel indicates a lateral root. Scale bars, 10 mm. (E,F) Root hairs of wild-type Ler ecotype (E) and hy5-1 (F). The seedlings were grown on agar plates set in a vertical position. Scale bars, 100 μm. (G) A diagram, indicating how to measure angles formed by roots and gravity orientation. Roots of seedlings elongate on an agar plate set in a vertical position. Plus (+) and minus (−) degrees represent a clockwise angle from the gravity orientation and a counterclockwise angle, respectively (see Table 3). (H) A magnified view of Fig. 2B, showing a secondary lateral root of hy5-1 (arrow). (I,J) Wavy pattern of wild-type of Ler ecotype (I) and hy5-1 (J). Seedlings were grown on an agar plate for 3 days set in a vertical position. Then the plate was tilted to 45° (arrows indicate the positions of root tips at the time of the position change), and the seedlings were grown for 3 days. (K,L) Cellular organization in the secondarily thickened roots of wild-type Ler ecotype (K) and hy5-1 (L). Transverse sections were made at the same position of main roots of seedlings at 20 DAG grown on agar plates supplemented with sucrose and stained with toluidine blue (see Materials and Methods). A well-lignified xylem vessel, a peridremal cell, and a fiber element are indicated by a star, a large arrow, and a small arrow, respectively. Scale bars, 50 μm.
The hy5 mutation enhances cell elongation in hypocotyl and root hairs
When germinated in the light, hypocotyls in hy5 mutants are longer than those in the wild type (Table 1). The longitudinal length of epidermal cells was ∼1.9 times that of the wild type, indicating that the longer hypocotyl of the mutants largely depends on the higher degree of cell elongation. It is well known that the length of the hypocotyl is increased when seeds are germinated in darkness, namely, elongation of hypocotyl cells is repressed by light. Interestingly, the hypocotyl length of the mutant was identical to that of the wild type when both plants were grown in darkness (Table 1). This suggests that the hy5 mutation abolishes the light-dependent repression of cell elongation and does not simply promote cell elongation independent of light stimulus.
Table 1.
Length of hypocotyl and epidermal cells of the hypocotyl
| Allele | Hypocotyl lengtha (mm ± s.e.) | Epidermal cell lengthb (μm ± s.e.) | |
|---|---|---|---|
| light | dark | ||
| Wild type (Ler) | 2.06 ± 0.01 | 12.42 ± 0.03 | 156 ± 14 |
| hy5-1 | 5.38 ± 0.02 (2.6) | 12.73 ± 0.03 (1.0) | 294 ± 25 (1.9) |
Excessive cell elongation was also observed in root hairs of the hy5 mutant. As shown in Table 2 and Figure 1, E and F, the root hair length of the mutant was 1.5 times greater than that of the wild type, whereas the length of root-hair-bearing epidermal cells was less affected by the hy5 mutation.
Table 2.
Length of root hairs and root hair-bearing epidermal cells
| Allele | Root haira (μm ± s.e.) | Root-hair-bearing epidermal cellb (μm ± s.e.) |
|---|---|---|
| Wild type (Ler) | 293 ± 23 | 183 ± 8 |
| hy5-1 | 441 ± 22 (1.5) | 221 ± 16 (1.2) |
The hy5 mutation alters the gravitropic response and touching response in roots
One of the clear characters of hy5 mutants grown on agar plates was the widely spread lateral roots (Fig. 1A–D, arrowheads). Compared with that of wild-type plants, the direction of lateral root growth was nearly horizontal rather than downward, indicating alteration of the gravitropic response. To examine the degree of the gravitropism, we measured the angles between the lateral roots and the orientation of gravity. The angles of lateral roots of the mutant were larger than those of the wild type (Table 3). In addition, secondary lateral roots grew and changed their direction of elongation to the horizontal (Fig. 1H). These results suggest that lateral roots of the hy5 mutant lack normal gravitropism and may obtain a new type, called diageotropism (Darwin 1882). The main root of the hy5 mutant also showed a reduced gravitropic response. As shown in Figure 1A, the main root of the wild type elongated with a slight slant to the viewer’s left (clockwise). The angle of slant of hy5-1 was two times greater than that of the wild type (Table 3), indicating that the main root of the hy5 mutant shows reduced gravitropism and/or increased circumnutation (Simmons et al. 1995). However, the reduced gravitropic response of the main root of the mutant was also shown by changing the position of the agar plate, after which the main root of wild-type Arabidopsis forms a hairpin loop. hy5-1 showed a larger arc than the wild type because of a slow response to the position shift (data not shown). These results suggest that the gravitropic response of the main root is reduced in the hy5 mutant.
Table 3.
Angles (degree ± s.e. ) formed by roots and gravity orientation plus root length (cm ± s.e. )
| Allele | Main roots | Lateral roots (+) | Lateral roots (−) | |||
|---|---|---|---|---|---|---|
| angle | length | angle | length | angle | length | |
| Wild type (Ler) | 16.2 ± 1.0 | 3.93 ± 0.18 | 24.5 ± 2.8 | 1.03 ± 0.10 | −9.7 ± 1.9 | 0.95 ± 0.08 |
| hy5-1 | 32.8 ± 1.3 (2.0) | 3.72 ± 0.14 (0.95) | 81.5 ± 3.3 (3.3) | 1.91 ± 0.15 (1.85) | −62.4 ± 8.1 (6.4) | 2.08 ± 0.10 (2.19) |
When seedlings of wild-type Arabidopsis are incubated on 1.5% agar plates set at an angle of 45° from the vertical position, the roots bend along the line of gravity but cannot penetrate the agar surface, as they perceive the obstacle-touching stimulus (Okada and Shimura 1990; Simmons et al. 1995). On the angled plates, roots of the wild type grew in a wavy pattern (Fig. 1I); however, roots of hy5-1 grew straight and did not show the wavy pattern (Fig. 1J). This indicates that the hy5 mutation also gives rise to a deficiency in the obstacle-touching response of roots.
The hy5 mutation enhances the initiation and elongation of lateral roots
In young seedlings of Arabidopsis, lateral roots originate from pericycle cells that swell and divide periclinally into two cell layers, and the divisions that follow form the lateral root meristem (Dolan et al. 1993; Laskowski et al. 1995; Malamy and Benfey 1997). To examine the frequency of initiation of lateral root formation, we counted the number of lateral root primordia formed in the main root by observation with Nomarski microscopy. Two to three lateral root primordia per seedling were detected in wild-type and hy5 mutants of both ecotypes at 2 days after germination (DAG) (Table 4). At 4 DAG, the number of lateral root primordia were not increased in the Landsberg erecta (Ler) wild type but were increased in hy5-1. The number of primordia were increased in the Wassilewskija (Ws) wild type at 4 DAG, but the increase was two times greater in the hy5–Ks50 mutant than in Ws (Table 4). Growth rates of the main root of the hy5 mutants were about the same as those of the wild-type plants (Table 4). The elongation of lateral roots was also enhanced in the hy5 mutant. Table 3 shows the length of main roots and lateral roots of Ler wild-type and hy5-1. The lateral roots of hy5-1 were about twice as long as those of the wild type, whereas the length of the main root of hy5-1 was about the same as that of the wild type. These results indicate that the hy5 mutation enhances the initiation of lateral root primordia and their growth.
Table 4.
Number of lateral root primordia and length of main roots
| Allele | Seedlings at 2 DAG | Seedlings at 4 DAG | ||
|---|---|---|---|---|
| number of lateral root primordiaa | main root length (cm) | number of lateral root primordiaa | main root length (cm) | |
| Wild type (Ler) | 2.0 | 0.90 | 1.3 | 1.53 |
| hy5-1 | 2.0 (1.00) | 0.66 (0.73) | 3.7 (2.85) | 1.18 (0.77) |
| Wild type (Ws) | 2.7 | 1.22 | 4.3 | 2.11 |
| hy5–Ks50 | 3.0 (1.11) | 1.30 (1.07) | 8.3 (1.93) | 1.97 (0.93) |
The hy5 mutation reduces the secondary thickening of the root and hypocotyl
Roots of Arabidopsis grow thicker through successive cell proliferation, termed secondary thickening. The secondary cell growth occurs acropetally. As shown in Figure 1K, the main root of a typical wild-type plant at 20 DAG was 0.2 mm in diameter at ∼0.5 cm below the root–hypocotyl joint. The cell walls of many oval-shaped xylem vessels were widely lignified, and angular fiber elements were present in the spaces among the vessels (Fig. 1K, small arrow). Thick peridermal cells surrounded the root (Fig. 1K, large arrow). Compared with that of the wild type, the main root of the hy5 mutant at the same age showed a decreased amount of secondary cell proliferation. As shown in Figure 1L, the number of lignified oval-shaped xylem vessels was decreased to ∼60% of the wild-type number. In addition, angular fiber elements were hardly developed, and the size of peridermal cells was smaller than that of the wild-type ones.
The hypocotyl of the Arabidopsis plant also undergoes secondary thickening. At 20 DAG, the hypocotyl of the wild-type plant was ∼0.5 mm in diameter. The hy5 mutant showed reduced secondary thickening of the hypocotyl, with a hypocotyl diameter of ∼50% of that in the wild type at the same stage. The number of lignified xylem vessels and fiber elements was reduced in the hypocotyl of the mutant as well (data not shown).
The hy5 mutation reduces greening of the hypocotyl and root
In the hypocotyl of light-grown Arabidopsis seedlings, chloroplasts are well developed. As shown in Figure 2A, the morphology and distribution of chloroplasts of the hy5 mutant were indistinguishable from those of wild type only at the upper part of the hypocotyl. However, a reduced number of chloroplasts, which were pale green, were observed at the middle part of the mutant. In the lower part of the hypocotyl, the number of chloroplasts was reduced in wild-type plants; however, the hy5 mutant showed only a few small and transparent plastids.
Figure 2.
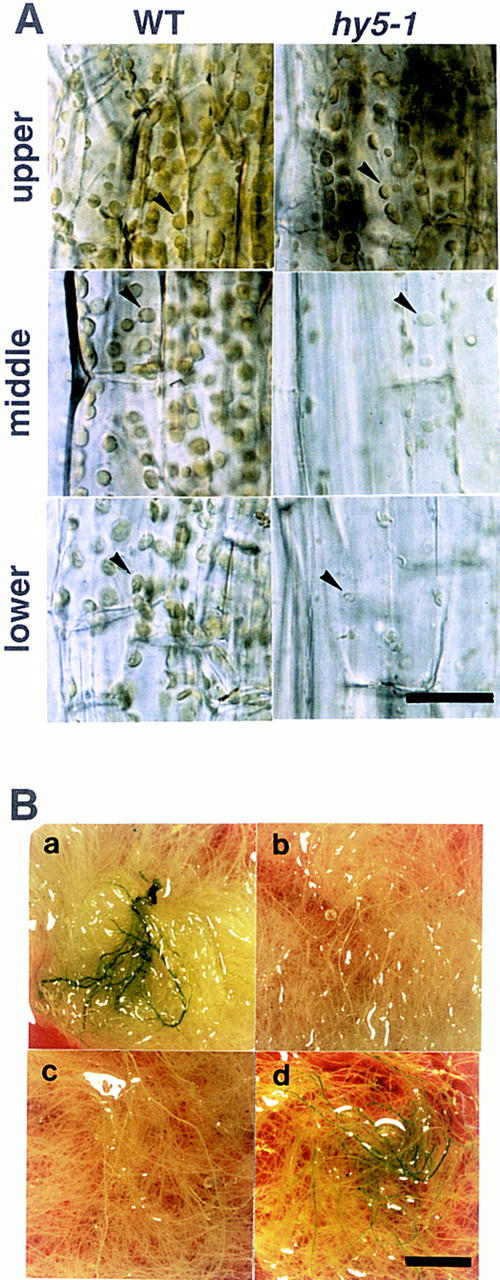
Greening in the hypocotyl and roots. (A) Chloroplasts in the hypocotyl of wild-type Ler ecotype (left) and hy5-1 (right) plants at 20 DAG grown on agar plates supplemented with sucrose. Upper, middle, and lower parts of the hypocotyl are shown in respectively labeled panels. Scale bar, 25 μm. (B) Roots of plants at 30 DAG grown in a liquid medium. Leaves and the hypocotyl are removed from the plants. Wild-type of Ler ecotype (a), hy5-1 (b), hy5–Ks50 (c), and cop1-6 (d) are shown. Scale bar, 10 mm.
Roots of wild-type plants turn green when grown under the light. As shown in Figure 2B, the roots of wild-type plants were green (panel a); whereas those of hy5 mutants (panels b,c) remained white after having been cultured for 30 days under light. It is known that chloroplast development is enhanced in roots of seedlings of cop1 mutants (Deng and Quail 1992). Liquid-cultured roots of cop1-6 showed greening (Fig. 2B, panel d). The chlorophyll content in roots of the wild-type plant (Ws) was 1.1 μg of chlorophyll/gram of fresh roots (See Materials and Methods). Chlorophyll was not detected in roots of hy5-1 or hy5–Ks50. Roots of cop1-6 contained ∼20-fold the amount of chlorophyll of the wild type. Thus, the hy5 mutation has an inhibitory effect on the greening of roots, which is opposite that of the cop1 mutation.
Molecular characterization of the HY5 gene
Molecular cloning of the HY5 gene
To understand the molecular mechanism of the HY5 gene, we cloned the gene using the hy5–Ks50 line. The genomic sequence around the T-DNA insertion site is shown in Figure 3A. The longest cDNA clone was 842 bp, which is close to the estimated mRNA size, 0.9 kb, by Northern blot analysis (data not shown). The gene, which was divided into four exons, covered a 1.5-kb genomic region. To confirm that the genomic region included the HY5 gene, we introduced a 4.5-kb _Pst_I-digested fragment (one of the _Pst_I sites is shown in Fig. 3A) into hy5–Ks50 mutants. The transformants showed the same hypocotyl length as found in the wild type in white light, normal growth pattern in the root, and normal secondary thickening and greening of the root and hypocotyl (Fig. 3C). The complemented phenotype of the mutant thus indicated that the introduced genomic fragment included the whole region of the HY5 gene.
Figure 3.
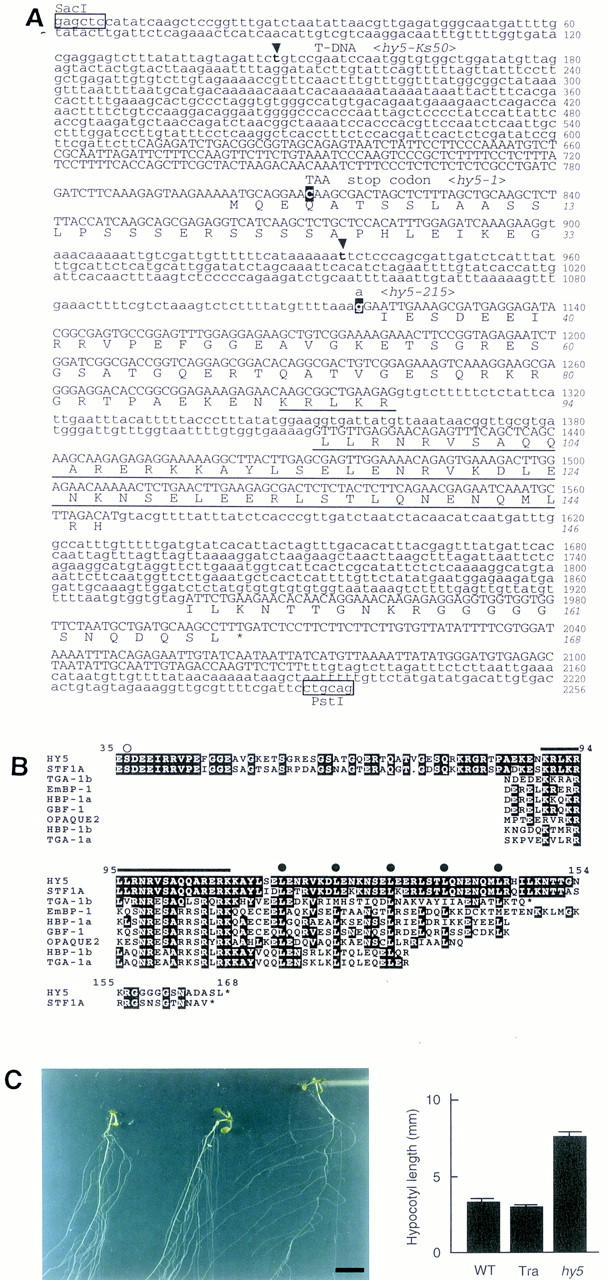
Sequence analysis of the HY5 gene. (A) Genomic DNA sequence and deduced amino acid sequence of HY5. The genomic sequence from _Sac_I to _Pst_I in the HY5 locus of the Ws wild type is shown. The first G in the _Sac_I site is designated as number 1. Numbers in italics indicate the amino acid sequence of the HY5 protein. Exons are shown in uppercase letters. The bZIP domain is underlined. In hy5–Ks50, sequences between t-147 and t-935 nucleotides (in boldface at arrowheads) are deleted and a T-DNA concatemer is inserted. In hy5-1, the nucleotide C-811 (white letter) is replaced by T, which results in a stop of translation. In hy5-215, the nucleotide g-1117 (white letter), which is the last nucleotide in the first intron, is replaced by an a. The HY5 cDNA sequence has been deposited in DDBJ/GenBank/EMBL databases (accession no. AB005295). (B) Amino acid comparison of plant bZIP proteins. The deduced HY5 amino acid sequence is compared with the sequences of eight related proteins in plants: STF1A from soybean (GenBank accession no. L28003, J.-C. Hong, pers. comm.), TGA-1b and TGA-1a from tobacco (Katagiri et al. 1989), EmBP-1 from wheat (Guiltinan et al. 1990), HBP-1a and HBP-1b from wheat (Tabata et al. 1989, 1991), GBF-1 from Arabidopsis (Schindler et al. 1992a), and OPAQUE2 from maize (Hartings et al. 1989; Schmidt et al. 1990). White letters represent residues identical to the HY5 sequence. A serine residue that is predicted to be phosphorylated by CKII is indicated (○). The basic region (from K-90 to K-109) is overlined. (•) The heptad repeat of leucines in the leucine zipper region. Asterisks (*) indicate the carboxyl-terminus of a given protein. (C) Complementation of hy5–Ks50 with a genomic sequence including the HY5 gene. (Left) Plants at 20 DAG grown in the light on a vertically positioned agar plate supplemented with sucrose. The plants are wild-type of Ws ecotype, hy50–Ks50 transformed with the genomic sequence, and hy5–Ks50 (from left to right). Leaves were removed from the plants. Scale bar, 10 mm. (Right) Bar graph of hypocotyl lengths of seedlings at 7 DAG grown in the light. The bars represent wild-type of Ws ecotype, hy5–Ks50 transformed with the genomic sequence, and hy5-Ks50, respectively. Numbers of the measured samples are 18 (wild-type) or 40 (the transformant and hy5–Ks50).
The HY5 locus encodes a protein with a bZIP motif
The longest open reading frame (ORF) in the HY5 cDNA clones encoded a protein of 18.5 kD composed of 168 amino acid residues (Fig. 3A). A comparison of the amino acid sequence with available databases revealed the carboxy-terminal half of the HY5 protein to be homologous to a DNA-binding and dimerization domain of bZIP-class proteins (Fig. 3B). The sequences at the other regions were not found in other bZIP proteins except STF1A, which was isolated from soybean (Y.-H. Cheong and J.-C. Hong, pers. comm.). STF1A protein showed high similarity to HY5 in the amino- and carboxy-terminal regions, including the bZIP domain (Fig. 3B). The amino acid sequence in the basic region was completely identical between HY5 and STF1A proteins. The two proteins contained five heptad repeats of leucine, and only three amino acid residues are different in the leucine zipper region. In addition, there was a casein kinase II (CKII) phosphorylation site (S/TXXE/D, rich in acidic amino acids) (Pearson and Kemp 1991) in a highly conserved region from E-35 to E-49 of the HY5 protein (Fig. 3B). Genomic Southern analysis using HY5 cDNA as the probe showed a single band corresponding to the HY5 gene under high stringency conditions and several weak bands under low stringency conditions, indicating that there are no genes highly homologous to the HY5 gene in the genome of Arabidopsis (data not shown).
Molecular lesions in the three hy5 mutant genes
Molecular lesions in three hy5 mutant alleles, hy5–Ks50, hy5-1, and hy5-215, were determined by sequencing the hy5 loci. In the genome of hy5–Ks50, the insertion of a T-DNA concatemer was accompanied by a deletion of a 790-bp region including the first exon and 5′ upstream region of the HY5 gene. In the genome of hy5-1, the fourth codon (CAA = Q) was substituted for a stop codon (=TAA). In the genome of hy5-215, the splicing acceptor site of the first intron (=G) (Padgett et al. 1986) was replaced by A, suggesting that this mutation causes aberrant RNA processing. As shown in Figure 4A, HY5 mRNA did not accumulate in the three mutants. Therefore, all three mutants may have no functional HY5 gene product and can be considered to be null alleles.
Figure 4.
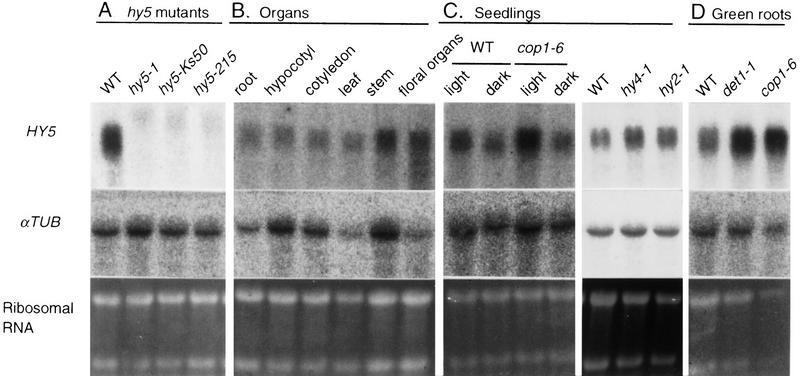
Northern blot analysis of HY5 mRNA. An aliquot of the total RNA (10 μg/lane) was subjected to agarose gel electrophoresis, transferred to nylon membranes, and hybridized with the probe of HY5 cDNA, as described in Materials and Methods. The same membranes were subsequently hybridized with the probe of the α_TUBULIN_ (αTUB) coding region of Arabidopsis (Kopczak et al. 1992). rRNA was detected by staining gels with ethidium bromide. (A) Total RNA was extracted from roots of plants at 30 DAG grown in a liquid medium. (B) The total RNA was extracted from root, hypocotyl, and cotyledon tissues isolated from light-grown seedlings of wild-type (Ler) at 3 DAG on agar plates supplemented with sucrose. Leaf, stem, and floral organs were isolated from mature wild-type plants grown in soil. Samples of the floral organs contained different tissues of the inflorescent meristem, floral buds, and mature flowers. (C) Total RNA was extracted from light- and dark-grown seedlings of wild-type Ler ecotype and the cop1-6 mutant or from light-grown seedlings of the hy2 and the hy4 mutants. All seedlings were grown on agar plates supplemented with sucrose for 3 days. (D) Total RNA was extracted from roots of plants at 30 DAG of each genotype grown in a liquid medium.
Expression patterns of the HY5 gene
In the wild-type plant, HY5 mRNA was accumulated in all tissues examined: root, hypocotyl, cotyledon, leaf, stem, and floral organs (Fig. 4B). To examine the light dependency of HY5 expression, we examined accumulation levels of HY5 mRNA in seedlings of wild-type, cop1-6 grown in the light and in darkness, and hy2-1 and hy4-1 grown in the light. It is known that cop1-6 mutants show photomorphogenesis in darkness (McNellis et al. 1994). As shown in Figure 4C, HY5 mRNA accumulated in wild-type seedlings to an extent approximately two times greater in the light than in darkness, indicating that the expression of the HY5 gene is induced by light. In cop1-6, the level of HY5 mRNA was higher than that in the wild type, and the enhanced accumulation in the light was also observed. The accumulation levels of HY5 mRNA in light- and dark-grown seedlings of cop1-6 were ∼2 and 1.6 times, respectively, as high as those of wild-type seedlings. This indicates that the accumulation of HY5 mRNA in seedlings is modulated by the COP1 gene. However, the accumulation levels of HY5 mRNA in light-grown hy2-1 and hy4-1 seedlings were about the same as that in wild type (Fig. 4C). The two mutants show a long hypocotyl, like the hy5 mutant, in white light because the hy2 and hy4 mutants are defective in the phytochrome (Koornneef et al. 1980; Parks and Quail 1991) and in a blue-light receptor named CRY1 (Ahmad and Cashmore 1993; Lin et al. 1995), respectively. These results suggest that the induction of the HY5 gene by light may require photoreceptors other than the phytochrome and CRY1. In roots of plants at 30 DAG grown in a liquid medium, cop1-6 and det1-1 accumulated approximately five and three times higher levels of HY5 mRNA, respectively, than the wild type (Fig. 4D), indicating that both COP1 and DET1 genes repress the expression of the HY5 gene in roots of wild-type.
The HY5 protein is localized in the nucleus
To confirm the possibility that the HY5 protein works as a transcription regulatory factor, we examined the subcellular localization of the HY5 protein. The HY5 protein was stained with anti-HY5 antiserum. In protoplasts prepared from roots of wild-type seedlings, signals of anti-HY5 staining overlapped with the DAPI staining signals, indicating that the HY5 protein is localized in the nucleus (Fig. 5, top panels). In protoplasts prepared from hy5–Ks50, however, signals of background level were detected in the cytoplasm (Fig. 5, bottom panels). It will be necessary to examine whether the basic region of the HY5 protein works as a nuclear localization signal, because the sequence of the basic region of the HY5 protein is different from those of several plant bZIP proteins, including TGA-1a, TGA-1b, and OPAQUE2, which have been shown to act as a nuclear localization signal (for review, see Foster et al. 1994).
Figure 5.
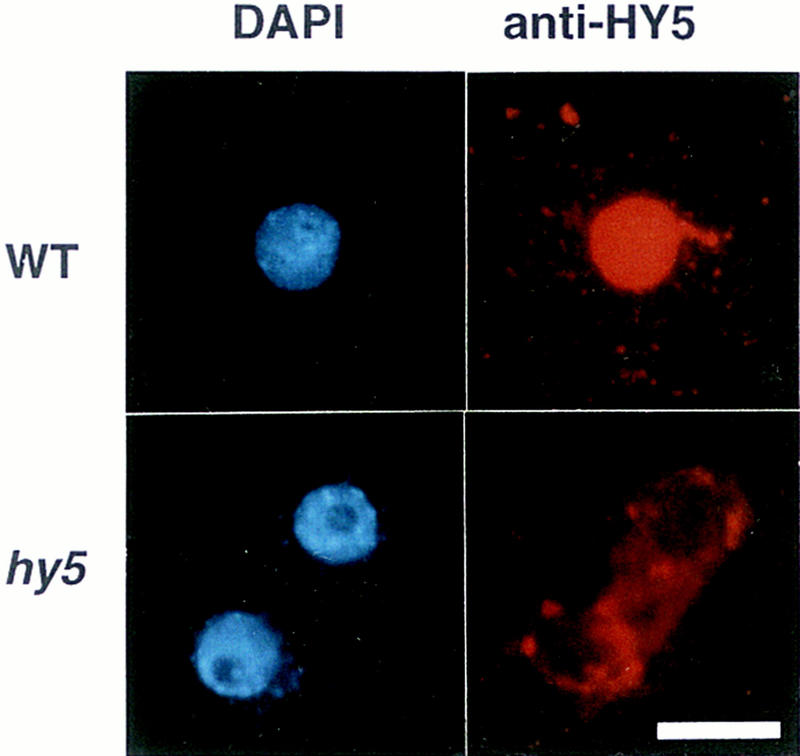
Subcellular localization of the HY5 protein. Protoplasts were prepared from roots of seedlings at 7 DAG grown on agar plates supplemented with sucrose in the light. The top and bottom panels show the wild-type Ws ecotype and hy5-Ks50, respectively. The left and right panels show DAPI staining of the nuclei and Texas Red imaging of the same cells detected with anti-HY5 antiserum, respectively. Scale bar, 10 μm.
Discussion
The HY5 gene was shown earlier to be a positive regulator of photomorphogenesis of young seedlings (Koornneef et al. 1980; Chory 1992; Ang and Deng 1994). Detailed characterization of the mutant phenotype has revealed that the hy5 mutant is defective in various aspects of morphogenesis and stimulus responses in the hypocotyl and root. The pleiotropic phenotypes can be classified under three criteria: whether the phenotype is mainly related to cell elongation, to cell proliferation, or to chloroplast development (Table 5).
Table 5.
Classification of phenotypes of the hy5 mutant
| Group | Phenotype | Physical stimulus related to the phenotype |
|---|---|---|
| Cell elongation | enhanced hypocotyl elongation | light |
| enhanced root hair elongation | touching | |
| aberrant gravitropic response of roots | gravity | |
| no waving growth of roots | touching | |
| Cell proliferation | enhanced lateral root elongation | none |
| enhanced lateral root initiation | none | |
| reduced secondary thickening | none | |
| Chloroplast development | reduced greening of root and hypocotyl | light |
The first group includes four phenotypes. Gravitropic and touching responses of roots are placed in the first group because these responses are thought to be a consequence of regulation of cell elongation (Wilkins 1966; Okada and Shimura 1994). It has been shown that the length of root hairs is affected by the touching stimulus (Okada and Shimura 1994). Although the type of stimulus is different, the four phenotypes appear to be based on aberrant cell elongation. Therefore, we propose that one of the molecular functions of the HY5 gene is negative regulation at some common step(s) of the intracellular signal transduction pathways that mediate signals triggered by different physical stimuli for cell elongation.
The second group includes three phenotypes: lateral root elongation, lateral root initiation, and secondary thickening. The lateral root elongation appears to be related to cell elongation. However, the length of mature cells in the lateral roots of the hy5 mutant was about the same as that of the wild type, indicating that the enhanced lateral root elongation of the mutant may be caused by enhanced cell proliferation in the meristem. The hy5 mutation also promoted the initiation of lateral roots, suggesting that the differentiation from pericycle cells to a lateral root primordium may be enhanced in the mutant. Because the structure of lateral root primordia of the mutant looked similar to that of the wild type, the HY5 gene is likely to control the initiation by repressing the cell proliferation in wild-type plants. In the secondary thickening of root and hypocotyl, the HY5 gene is likely to promote or maintain the meristimatic activity in the vascular cambial cells that proliferate and differentiate into xylem and fiber elements. Thus, the HY5 gene controls cell proliferation positively in the secondary thickening, and negatively in the lateral root formation. HY5 may regulate cell division in a tissue-specific manner. Interestingly, the three phenotypes in the second group are not directly related to environmental stimuli; however, they are possibly stimulated by endogenous nutrition conditions or balance of phytohormones.
The third group includes a phenotype of the deficiency of greening of root and hypocotyl. The HY5 gene is likely to promote chloroplast development in the root and hypocotyl, possibly stimulated by light, as in cotyledons and leaves. Because the chloroplast development in roots of wild-type plants is usually accompanied by secondary thickening, the phenotype of reduced greening in the hy5 mutant may be dependent on the reduced secondary thickening in roots. It is worthy to note that the hy5 mutation also gives rise to a reduced gravitropic response, longer root hairs, and enhanced lateral root formation in darkness (data not shown). This indicates that the HY5 gene is likely to play roles in the root growth and the gravitropic response independent of the light signal.
Our molecular analysis has revealed that the HY5 gene encodes a bZIP protein localized in the nucleus. Many bZIP proteins have been isolated from various plant species. In many cases, the bZIP proteins bind to DNA containing an ACGT core sequence. Although the ACGT sequence is known as a core motif for _cis_-acting elements in promoters of various stimulus-responsive genes in plants, the physiological roles of bZIP class _trans_-acting factors have been demonstrated only in some cases (for review, see Foster et al. 1994; Menkens et at. 1995). Many of the plant bZIP proteins contain extra domains such as a proline-rich region, acidic region, or glutamine-rich region, in addition to the bZIP domain. A proline-rich region of the Arabidopsis GBF1 protein was shown to have the activity of transcriptional activation (Schindler et al. 1992b). Highly acidic regions are a common feature of transcriptional activators in eukaryotes (Cress and Triezenberg 1991). The HY5 protein has no obvious domains except the bZIP domain. The lack of potential transcriptional activation domains implies that the HY5 protein may not work as a transcriptional activator. The amino acid sequence of this protein from E-35 to the last amino acid residue shares 70% identity with the STF1A protein of soybean, and the basic region of the HY5 protein is completely identical to that of the STF1A protein. The STF1A protein preferentially binds to DNA sequences containing a TGACGT core, and it weakly binds to DNA sequences containing a G box (Y.-H. Cheong and J.-C. Hong, pers. comm.). A crystallographic study on GCN4 (a yeast bZIP protein)/DNA complex clearly showed that the basic region interacts with the target DNA (Ellenberger et al. 1992). Moreover, the amino acid sequence of the basic region in bZIP proteins was shown to specify the target nucleotide sequence (Suckow et al. 1993). Therefore, the complete identity of basic regions between the HY5 protein and the STF1A protein strongly suggests that the HY5 protein likely binds to the same DNA sequences as the STF1A protein. In addition, a region from E-35 to E-49 of the HY5 protein is highly homologous to that of the STF1A protein (Fig. 3B). This region contains a consensus sequence for phosphorylation by CKII. Because phosphorylation of an Arabidopsis bZIP protein, GBF1, by CKII was shown to stimulate its DNA-binding activity (Klimczak et al. 1992), phosphorylation at the putative CKII phosphorylation sites of the HY5 and STF1A proteins may enhance their DNA-binding activity.
As discussed above, the HY5 protein is likely to have roles as a transcriptional regulatory factor in a wide variety of stimulus responses and developmental processes in the hypocotyl and root. As for the molecular mechanism governing HY5 protein function, studies on photomorphogenesis of young seedlings have provided useful information because molecular genetic data on the HY5 gene and other genes have been accumulated (for review, see Chory 1993; Deng 1994; McNellis and Deng 1995). In the mechanism of hypocotyl elongation of light-grown seedlings, it has been proposed that the HY5 protein works as a positive regulator downstream of photoreceptors in the signaling pathway (Koornneef et al. 1980; Chory 1992; Ang and Deng 1994). In addition to the HY5 gene, the DET1 and COP1 genes also work downstream of photoreceptors in light-signaling pathways, however, as negative regulators (Chory et al. 1989b; Deng et al. 1991; Chory 1992; Ang and Deng 1994). Both DET1 and COP1 proteins have been shown to be localized in the nucleus, and they negatively regulate expression of light-induced genes in dark-grown seedlings (Pepper et al. 1994; von Arnim and Deng 1994). Because the HY5 protein is also localized in the nucleus and likely acts as a transcriptional regulatory factor, these three proteins may be associated with a common transcription complex that regulates the expression of light-induced genes. It has been reported that several light-regulated genes contain the ACGT sequence as a light-responsive _cis_-acting element in their promoters (Batschauer et al. 1994), suggesting that HY5 may bind directly to those promoters. Interestingly, HY5 mRNA was accumulated at a higher level than wild type in roots of both det1 and cop1 mutants. Thus, the amount of the HY5 gene product is likely to be modulated by the DET1 and COP1 genes. Because the det1 and cop1 mutants show excess chloroplast development in their roots—just the reverse effect of the hy5 mutant (Chory and Peto; Deng and Quail 1992)—the DET1 and COP1 genes may repress chloroplast development in the root of the wild type by repressing the expression of the HY5 gene.
As discussed above, hy5 is a unique mutant because it shows pleiotropic effects on a variety of stimulus responses and on the development of both root and hypocotyl. As a similar mutant with a wide range of phenotypes, the diagiotropica (dgt) mutant was isolated from the tomato (Zobel 1974). The dgt mutant shows diageotropism in both shoots and roots and, interestingly, also shows morphological defects, that is, it lacks large secondary xylem vessels in the stem and shows a loss of lateral roots and an open hypocotyl hook. Although not all of the phenotypes of the dgt mutant coincide with those of the hy5 mutant, the resemblance of phenotypes between the two mutants suggests that they affect common signaling pathways in the tomato and in Arabidopsis. Interestingly, the dgt mutant is also known to be insensitive to auxin (Kelly and Bradford 1986). It is known that auxin mediates gravitropism and lateral root formation (Wilkins 1966; Juniper 1976; Boerjan et al. 1995; Laskowski et al. 1995). In addition, auxin is likely to be involved in cambium proliferation and secondary thickening (Wareing and Phillips 1981). As summarized in Table 5, the HY5 gene is involved not only in the light response but also in gravitropic and touching responses and the process of secondary thickening, in which auxin plays an important role. Therefore, it could be argued that the HY5 protein may be involved in the signaling pathway mediated by auxin and may regulate expression of auxin-induced genes. It has been shown that the promoter of the soybean GH3 gene contains TGACGT elements that confer the auxin inducibility (Liu et at. 1994). Further investigation of molecular mechanisms underlying how the HY5 protein perceives stimulus-induced signals and how the HY5 protein controls the expression of the target genes will unravel the role of the HY5 gene in the development and signaling pathways in plants.
Materials and methods
Plant materials and growth conditions
Mutant lines hy2-1, hy4-1, and hy5-1 were obtained from the Arabidopsis Biological Resource Center (The Ohio State University, Columbus, OH). The ecotype of the mutants is Ler. The cop1-6, det1-1, and hy5-215 (Columbia background) were provided by Y. Komeda (Hokkaido University, Sapporo, Japan), J. Chory (The Salk Institute, San Diego, CA), and X.-W. Deng (Yale University, New Haven, CT), respectively.
Plants were grown on agar plates under conditions described previously (Okada and Shimura 1992b). In several experiments, the agar medium was supplemented with 20 grams/liter of sucrose. For the liquid culture of plants, sterilized seeds were germinated and cultured with gentle shaking (60 rpm) in a liquid medium containing 1× Gamborg’s B5 medium salt mixture (Nihon-Seiyaku, Co., Ltd. Tokyo, Japan) and 20 grams/liter of sucrose, adjusted to pH 5.7 with KOH, at 22°C under white light of 50–100 μE/m2 per sec.
Isolation of hy5-Ks50
hy5–Ks50 was isolated from T-DNA insertion lines that were produced by the in planta transformation method developed by Chang et al. (1994) and modified by T. Ohsumi and R.F. Whittier (pers. comm.). An Agrobacterium tumefaciens strain, C58C1::rif, carrying an intermediate type Ti plasmid vector, pGV3850::hyg, was kindly provided by Mitui Plant Biotechnology Institute and was used to infect Ws wild-type plants (T0 plants). The seeds of T0 plants were harvested and sown on a selection medium containing 1× Gamborg’s B5 salt mixture [pH 5.7 with KOH, 1% sucrose, 10 mg/liter of hygromycin B (Wako Pure Chemical Industries, Ltd., Osaka, Japan)], and 8 grams/liter of agar. Seedlings showing the hygromycin resistance (T1 plants) were transplanted to soil, and the seeds (T2 seeds) were harvested from each T1 plant. Mutants were screened from the T2 lines. A recessive mutant line, Ks50, was isolated and showed the long hypocotyl phenotype. That line was subjected to an allelism test with hy1-1, hy2-1, phyB-1, hy4-1, and hy5-1 (Koornneef et al. 1980) and was shown to be allelic to the hy5 mutant (data not shown). We therefore designated Ks50 as hy5–Ks50.
Count of lateral root number
Two DAG and four DAG seedlings were fixed in 4% paraformaldehyde in PBS and inspected under an Olympus Provia AX70 microscope.
Sectioning and microscopy
Secondarily thickened roots were fixed for 12 hr in 4% paraformaldehyde in PBS. Fixed samples were dehydrated in a series of 50% (ice-cold), 90%, 100%, 100% ethanol (30 min in each step), an ethanol-_tert_-butanol series, 3:1, 1:1, 1:3, then through two changes of 100% _tert_-butanol (20 min in each step). The samples were passed through paraffin (solidifying point 51°C–53°C; Merck, Darmstadt, Germany)–_tert_-butanol of 1:1 and 100% paraffin at 55°C for 1 hr in each step, and embedded in 100% paraffin. Eight-micrometer sections were made on Erma 02631A microtome (Erma, Tokyo, Japan). Sections were stained in 0.05% toluidine blue (Kodak) and inspected under an Olympus Provia AX70 microscope.
Measurement of chlorophyll content
Roots of plants at 30 DAG grown in liquid medium were homogenized in 80% acetone in a Potter–Elvehjem-type glass homogenizer. The homogenate was centrifuged, and absorbance spectrum of the supernatant was measured. Two plants of each allele were measured. Total chlorophyll contents were calculated according to the following equation (Arnon 1949): total chlorophyll (μg/ml) = 8.02(_A_663−_A_710) + 20.2(_A_645−_A_710).
Molecular analysis
Standard protocols were used for enzyme reactions, DNA blotting, and RNA blotting (Sambrook et al. 1989). DNA and RNA were isolated from plants by the methods described by Ausubel et al. (1987). In Southern analysis, plaque, and colony screening, the blotting was done onto nylon membranes (Hybond-N, Amersham), and hybridization was performed with a solution [5× SSC, 1% blocking reagent (Boehringer Mannheim), 0.1% _N_-lauroylsarcosine, 0.02% SDS] at 65°C containing a probe, and washing was performed under stringent conditions (1× SSC, 0.1% SDS) at 65°C. Probes were labeled with [32P]dCTP (Amersham) by use of a random primer DNA labeling kit (Takara Shuzo, Ohtsu, Japan). Autoradiographs were scanned and analyzed with a Bio-Imaging Analyzer (BAS 1000 or 2000, Fuji). In Northern analysis, hybridization was performed with a solution [6× SSC, 5× Denhardt’s solution, 0.5% SDS, 0.1 mg/ml of denatured salmon sperm DNA, 50% formamide] at 42°C containing a probe, and washing was performed under stringent conditions (0.1× SSC, 0.1% SDS) at 65°C. DNA sequencing was carried out by Applied Biosystems automated sequencers (model 373A) using dye primers or dye terminators as recommended by the manufacturer.
Isolation of the HY5 gene
Linkage between the hy5 phenotype and the inserted T-DNA in the hy5–Ks50 mutant was examined in 11 plants of the T2 generation. Because one of the three T-DNA insertion loci cosegregated with the hy5 locus, we isolated a genomic DNA fragment flanking the T-DNA using the TAIL PCR method (Liu et al. 1995). The PCR reactions were carried out in a Perkin Elmer Cetus thermal cycler (model 9600). We tried several sets of primers specific for the left and right borders of the T-DNA, and a 1.1-kb PCR fragment containing a genomic sequence flanking the left border of the T-DNA was amplified by use of a set of primers: an arbitrary degenerate primer, 5′-NTCGA(G/C)T(A/T)T(G/C)G(A/T)GTT-3′ (15-mer); a specific primer for the primary reaction, 5′-CACATCATCTCATTGATGCTTGGT-3′ (24-mer); and a specific primer for the secondary reaction, 5′-GTGTTATTAAGTTGTCTAAGCGTC-3′ (24-mer). Because the 1.1-kb PCR fragment showed restriction fragment length polymorphism (RFLP) between Ws (4.5 kb) and Ler (12 kb) ecotypes when digested with _Pst_I, 165 mutants in the F2 generation [hy5-1 (Ler) × Ws wild-type parents] were examined for the RFLP. Because all of them showed the Ler-type band pattern, we proved that the PCR fragment was tightly linked to the HY5 locus. We screened a genomic library prepared from Arabidopsis DNA (Ws wild type) that was partially digested with _Sau_3AI and ligated into the Lambda DASH II vector (Stratagene), using the 1.1-kb PCR fragment as the probe. Three independent clones were isolated and subcloned into the pBluescript II vector (Stratagene). A 0.7-kb genomic fragment including the bZIP region was used for screening a library constructed from poly(A)+ RNA prepared from light-grown seedlings of Arabidopsis (Ler). Three independent cDNA clones were isolated and analyzed.
Complementation test
A 4.5-kb genomic fragment, containing the HY5 gene, was ligated into a binary vector, pARK5mcs (gift from Meiji-Seika, Kaisha, Ltd., Tokyo, Japan), which carries the bialaphos herbicide resistance marker gene for plant selection. The plasmid was introduced into A. tumefaciens strain C58::pGV2260 by electroporation using a Gene Pulsar (Bio-Rad). Transformation of hy5–Ks50 was performed by a vacuum transformation procedure (Bechtold et al. 1993). Seeds of T0 plants were harvested and sown on selection agar medium containing 1× Gamborg’s B5 salt mixture, 5 mg/liter of bialaphos (gift from Meiji-Seika), and 8 grams/liter of agar, adjusted to pH 5.7 with KOH. Seedlings surviving in the selection agar medium were transplanted to soil and grown to obtain their seeds.
Sequence analysis of three hy5 mutants
Genomic fragments of hy5–Ks50 including a junction of T-DNA border and the genome were amplified by PCR, and the fragments were ligated into the pBluescript II vector. Total DNA was extracted from plants of hy5-1 and hy5-215, digested with _Xho_I, and separated by gel electrophoresis. Because HY5 cDNA was included in a 3.4-kb _Xho_I fragment, minilibraries were constructed by extracting DNA fragments of ∼3.4 kb from the gel and ligating them into the pBluescript II vector. Then the minilibraries were screened with the HY5 cDNA used as a probe. Sequences of those subcloned hy5 genes of the mutants were determined.
Immunofluorescence detection in protoplasts
The glutathione _S_-transferase (GST)–HY5 fusion protein was purified from Escherichia coli by use of a GST gene fusion system (Pharmacia). Rabbits were immunized with the GST–HY5 fusion protein. Protoplast preparation and immunological detection of HY5 protein were made by basically following the procedures of Matsui et al. (1995). We used the enzyme solution containing 1% cellulase RS (Yakult Pharmaceutical Ind. Co. Ltd., Tokyo, Japan), 0.25% pectolyase Y23 (Seishin Corporation, Tokyo, Japan), 0.4 m mannitol, and 10 mm 2-(_N_-monopholino) ethanesulfonic acid (pH 5.7). As the primary antibody, anti-GST–HY5 antiserum of a 1:100 dilution was used. As the secondary antibody, Texas Red-conjugated goat antibody against rabbit IgG (Organon Teknika, West Chester, PA) of a concentration of 5 μg/ml was used. Cells were inspected under an Olympus Provia AX70 microscope.
Acknowledgments
This work was started in 1993 when the authors were at Division 1 of Gene Expression and Regulation, National Institute for Basic Biology, Okazaki 444, Japan. We are grateful to members of our laboratory, especially A. Kawai, for production of transgenic plant lines; and to S. Sawa, T. Wada, T. Ito, S. Ishiguro, H. Ito, and A. Tanaka for technical assistance and helpful discussions on this work. We thank T. Oosumi, Y.-G. Liu, and R. F. Whittier for providing an Agrobacterium strain, information on the in planta transformation procedure, and permission to use the TAIL PCR method before publication; T. Meshi for discussion about bZIP proteins; X.-W. Deng for providing a mutant line and for helpful discussions on photomorphogenesis; J.-C. Hong for information on STF1 genes; and J. Chory and Y. Komeda for providing mutant lines. T.O. was the recipient of a fellowship from Research Fellowships of the Japan Society for the Promotion of Science for Young Scientists. This work was supported by a Grant-in-Aid for Scientific Research (A) (no. 08408031) and a Grant-in-Aid for Scientific Research on Priority Areas (no. 06278103) from the Japanese Ministry of Education, Science, Culture, and Sports and by funds from the Joint Studies Program for Advanced Studies from the Science and Technology Agency of Japan.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL kiyo@ok-lab.bot.kyoto-u.ac.jp; FAX +81-75-753-4257.
References
- Aeschbacher RA, Schiefelbein JW, Benfey PN. The genetic and molecular basis of root development. Annu Rev Plant Physiol Plant Mol Biol. 1994;45:25–45. [Google Scholar]
- Ahmad M, Cashmore AR. HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature. 1993;366:162–166. doi: 10.1038/366162a0. [DOI] [PubMed] [Google Scholar]
- Ang L-H, Deng X-W. Regulatory hierarchy of photomorphogenic loci: allele-specific and light-dependent interaction between the HY5 and COP1 loci. Plant Cell. 1994;6:613–628. doi: 10.1105/tpc.6.5.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnon DI. Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol. 1949;24:1–15. doi: 10.1104/pp.24.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current protocols in molecular biology. New York, NY: Greene/Wiley; 1987. [Google Scholar]
- Batschauer A, Gilmartin PM, Nagy F, Schäfer E. The molecular biology of photoregulated genes. In: Kendrick R E, Kronenberg G H M, editors. Photomorphogenesis in plants. 2nd ed. Dordrecht, Netherlands: Kluwer Academic Publishers; 1994. pp. 559–599. [Google Scholar]
- Bechtold N, Ellis J, Pelletier G. In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. CR Acad Sci Ser III Sci Vi. 1993;316:1194–1199. [Google Scholar]
- Bennett MJ, Marchant A, Green HG, May ST, Ward SP, Millner PA, Walker AR, Schulz B, Feldmann KA. Arabidopsis AUX1 gene: A permease-like regulator of root gravitropism. Science. 1996;273:948–950. doi: 10.1126/science.273.5277.948. [DOI] [PubMed] [Google Scholar]
- Boerjan W, Cervera M-T, Delarue M, Beeckman T, Dewitte W, Bellini C, Caboche M, Van Onckelen H, Van Montagu M, Inzé D. superroot, a recessive mutation in Arabidopsis, confers auxin overproduction. Plant Cell. 1995;7:1405–1419. doi: 10.1105/tpc.7.9.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celenza JL, Jr, Grisafi PL, Fink GR. A pathway for lateral root formation in Arabidopsis thaliana. Genes & Dev. 1995;9:2131–2142. doi: 10.1101/gad.9.17.2131. [DOI] [PubMed] [Google Scholar]
- Chang SS, Park SK, Kim BJ, Kang BJ, Kim DU, Nam HG. Stable genetic transformation of Arabidopsis thaliana by Agrobacterium inoculation in planta. Plant J. 1994;5:551–558. [Google Scholar]
- Chory J. A genetic model for light-regulated seedling development in Arabidopsis. Development. 1992;115:337–354. [Google Scholar]
- Chory J. Out of darkness: Mutants reveal pathways controlling light-regulated development in plants. Trends Genet. 1993;9:167–172. doi: 10.1016/0168-9525(93)90163-c. [DOI] [PubMed] [Google Scholar]
- Chory J, Peto CA. Mutations in the DET1 gene affect cell-type-specific expression of light-regulated genes and chloroplast development in Arabidopsis. Proc Natl Acad Sci. 1990;87:8776–8780. doi: 10.1073/pnas.87.22.8776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chory J, Peto CA, Ashbaugh M, Saganich R, Pratt L, Ausubel F. Different roles for phytochrome in etiolated and green plants deduced from characterization of Arabidopsis thaliana mutants. Plant Cell. 1989a;1:867–880. doi: 10.1105/tpc.1.9.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chory J, Peto CA, Feinbaum R, Pratt L, Ausubel F. Arabidopsis thaliana mutant that develops as a light-grown plant in the absence of light. Cell. 1989b;58:991–999. doi: 10.1016/0092-8674(89)90950-1. [DOI] [PubMed] [Google Scholar]
- Cress WD, Triezenberg SJ. Critical structural elements of the VP16 transcriptional activation domain. Science. 1991;251:87–90. doi: 10.1126/science.1846049. [DOI] [PubMed] [Google Scholar]
- Darwin C. The power of movement in plants. London, UK: John Murray; 1882. [Google Scholar]
- Deng X-W. Fresh view of light signal transduction in plants. Cell. 1994;76:423–426. doi: 10.1016/0092-8674(94)90107-4. [DOI] [PubMed] [Google Scholar]
- Deng X-W, Quail PH. Genetic and phenotypic characterization of cop1 mutants of Arabidopsis thaliana. Plant J. 1992;2:83–95. [Google Scholar]
- Deng X-W, Casper T, Quail PH. COP1, a regulatory locus involved in the light-controlled development and gene expression in Arabidopsis. Genes & Dev. 1991;5:1172–1182. doi: 10.1101/gad.5.7.1172. [DOI] [PubMed] [Google Scholar]
- Deng X-W, Matsui M, Wei N, Wagner D, Chu A M, Feldmann K A, Quail P H. COP1, an Arabidopsis regulatory gene, encodes a protein with both a zinc-binding motif and a Gβ homologous domain. Cell. 1992;71:791–801. doi: 10.1016/0092-8674(92)90555-q. [DOI] [PubMed] [Google Scholar]
- Di Laurenzio L, Wysocka-Diller J, Malamy J, Pysh L, Helariutta Y, Freshour G, Hahn MG, Feldmann KA, Benfey PN. The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell. 1996;86:423–433. doi: 10.1016/s0092-8674(00)80115-4. [DOI] [PubMed] [Google Scholar]
- Dolan L, Roberts K. Plant development: Pulled up by the roots. Curr Opin Genet Dev. 1995;5:432–438. doi: 10.1016/0959-437x(95)90045-i. [DOI] [PubMed] [Google Scholar]
- Dolan L, Janmaat K, Willemsen V, Linstead P, Poethig S, Roberts K, Scheres B. Cellular organisation of the Arabidopsis thaliana root. Development. 1993;119:71–84. doi: 10.1242/dev.119.1.71. [DOI] [PubMed] [Google Scholar]
- Ellenberger TE, Brandl CJ, Struhl K, Harrison SC. The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted α helices: crystal structure of the protein-DNA complex. Cell. 1992;71:1223–1237. doi: 10.1016/s0092-8674(05)80070-4. [DOI] [PubMed] [Google Scholar]
- Feldman LJ. Regulation of root development. Annu Rev Plant Physiol. 1984;35:223–242. doi: 10.1146/annurev.pp.35.060184.001255. [DOI] [PubMed] [Google Scholar]
- Foster R, Izawa T, Chua N-H. Plant bZIP proteins gather at ACGT elements. FASEB J. 1994;8:192–200. doi: 10.1096/fasebj.8.2.8119490. [DOI] [PubMed] [Google Scholar]
- Guiltinan MJ, Marcotte WRJ, Quatrano RS. A plant leucine zipper protein that recognizes an abcisic acid response element. Science. 1990;250:267–271. doi: 10.1126/science.2145628. [DOI] [PubMed] [Google Scholar]
- Hartings H, Maddaloni M, Lazzaroni N, Di Fonzo N, Motto M, Salamini F, Thompson R. The O2 gene which regulates zein deposition in maize endosperm encodes a protein with structural homologies to transcriptional activators. EMBO J. 1989;8:2795–2801. doi: 10.1002/j.1460-2075.1989.tb08425.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbie L, Estelle M. The axr4 auxin-resistant mutants of Arabidopsis thaliana define a gene important for root gravitropism and lateral root initiation. Plant J. 1995;7:211–220. doi: 10.1046/j.1365-313x.1995.7020211.x. [DOI] [PubMed] [Google Scholar]
- Juniper BE. Geotropism. Annu Rev Plant Physiol. 1976;27:385–406. [Google Scholar]
- Katagiri F, Lam E, Chua N-H. Two tobacco DNA-binding proteins with homology to the nuclear factor CREB. Nature. 1989;340:727–730. doi: 10.1038/340727a0. [DOI] [PubMed] [Google Scholar]
- Kelly MO, Bradford KJ. Insensitivity of the Diageotropica tomato mutant to auxin. Plant Physiol. 1986;82:713–717. doi: 10.1104/pp.82.3.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimczak LJ, Schindler U, Cashmore AR. DNA binding activity of the Arabidopsis G-box binding factor GBF1 is stimulated by phosphorylation by Casein Kinase II from broccoli. Plant Cell. 1992;4:87–98. doi: 10.1105/tpc.4.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Rolff E, Spruit CJP. Genetic control of light-inhibited hypocotyl elongation in Arabidopsis thaliana (L.) Heynh. Z Pflanzenphysiol. 1980;100:147–160. [Google Scholar]
- Kopczak SD, Haas NA, Hussey PJ, Silflow CD, Snustad DP. The small genome of Arabidopsis contains at least six expressed α-tubulin genes. Plant Cell. 1992;4:539–547. doi: 10.1105/tpc.4.5.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski MJ, Williams ME, Nusbaum HC, Sussex IM. Formation of lateral root meristems is a two-stage process. Development. 1995;121:3303–3310. doi: 10.1242/dev.121.10.3303. [DOI] [PubMed] [Google Scholar]
- Leyser HMO, Lincoln CA, Timpte C, Lammer D, Turner J, Estelle M. Arabidopsis auxin-resistance gene AXR1 encodes a protein related to ubiquitin-activating enzyme E1. Nature. 1993;364:161–164. doi: 10.1038/364161a0. [DOI] [PubMed] [Google Scholar]
- Lin C, Robertson DE, Ahmad M, Raibekas AA, Jorns MS, Dutton PL, Cashmore AR. Association of flavin adenin dinucleotide with the Arabidopsis blue light receptor CRY1. Science. 1995;269:968–970. doi: 10.1126/science.7638620. [DOI] [PubMed] [Google Scholar]
- Liu Y-G, Mitsukawa N, Oosumi T, Whittier RF. Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 1995;8:457–463. doi: 10.1046/j.1365-313x.1995.08030457.x. [DOI] [PubMed] [Google Scholar]
- Liu Z-B, Ulmasov T, Shi X, Hagen G, Guilfoyle TJ. Soybean GH3 promoter contains multiple auxin-inducible elements. Plant Cell. 1994;6:645–675. doi: 10.1105/tpc.6.5.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy JE, Benfey PN. Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development. 1997;124:33–44. doi: 10.1242/dev.124.1.33. [DOI] [PubMed] [Google Scholar]
- Matsui M, Stoop CD, von Arnim AG, Wei N, Deng X-W. Arabidopsis COP1 protein specifically interacts in vitro with a cytoskeleton-associated protein, CIP1. Proc Natl Acad Sci. 1995;92:4239–4243. doi: 10.1073/pnas.92.10.4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNellis TW, Deng X-W. Light control of seedling morphogenetic pattern. Plant Cell. 1995;7:1749–1761. doi: 10.1105/tpc.7.11.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNellis TW, von Arnim AG, Araki T, Komeda Y, Miséra S, Deng X-W. Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional roles for the multiple protein domains. Plant Cell. 1994;6:487–500. doi: 10.1105/tpc.6.4.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menkens AE, Schindler U, Cashmore AR. The G-box: A ubiquitous regulatory DNA element in plants bound by the GBF family of bZIP proteins. Trends Biochem Sci. 1995;20:506–510. doi: 10.1016/s0968-0004(00)89118-5. [DOI] [PubMed] [Google Scholar]
- Okada K, Shimura Y. Reversible root tip rotation in Arabidopsis seedlings induced by obstacle-touching stimulus. Science. 1990;250:274–276. doi: 10.1126/science.250.4978.274. [DOI] [PubMed] [Google Scholar]
- ————— Aspects of recent developments in mutational studies of plant signaling pathways. Cell. 1992a;70:369–372. doi: 10.1016/0092-8674(92)90159-a. [DOI] [PubMed] [Google Scholar]
- ————— Mutational analysis of root gravitropism and phototropism of Arabidopsis thaliana seedlings. Aust J Plant Physiol. 1992b;19:439–448. [Google Scholar]
- ————— . Modulation of root growth by physical stimuli. In: Meyerowitz EM, Somerville CR, editors. Arabidopsis. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. pp. 665–684. [Google Scholar]
- Padgett RA, Grabowski PJ, Konarska MM, Seiler S, Sharp PA. Splicing of messenger RNA precursors. Annu Rev Biochem. 1986;55:1119–1150. doi: 10.1146/annurev.bi.55.070186.005351. [DOI] [PubMed] [Google Scholar]
- Parks BM, Quail PH. Phytochrome-deficient hy1 and hy2 long hypocotyl mutants of Arabidopsis are defective in phytochrome chromophore biosynthesis. Plant Cell. 1991;3:1177–1186. doi: 10.1105/tpc.3.11.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson RB, Kemp BE. Protein phosphorylation site sequences and consensus specificity motifs: Tabulations. Methods Enzymol. 1991;200:62–81. doi: 10.1016/0076-6879(91)00127-i. [DOI] [PubMed] [Google Scholar]
- Pepper A, Delaney T, Washburn T, Poole D, Chory J. DET1, a negative regulator of light-mediated development and gene expression in Arabidopsis, encodes a novel nuclear-localized protein. Cell. 1994;78:109–116. doi: 10.1016/0092-8674(94)90577-0. [DOI] [PubMed] [Google Scholar]
- Reed JW, Nagpal P, Poole DS, Furuya M, Chory J. Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell. 1993;5:147–157. doi: 10.1105/tpc.5.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- Schindler U, Menkens AE, Beckmann H, Ecker JR, Cashmore AR. Heterodimerization between light-regulated and ubiquitoulsy expressed Arabidopsis GBF bZIP proteins. EMBO J. 1992a;11:1261–1273. doi: 10.1002/j.1460-2075.1992.tb05170.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler U, Terzaghi W, Beckmann H, Kadesch T, Cashmore AR. DNA binding site preferences and transcriptional activation properties of the Arabidopsis transcription factor GBF1. EMBO J. 1992b;11:1275–1289. doi: 10.1002/j.1460-2075.1992.tb05171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt RJ, Burr FA, Aukerman MJ, Burr B. Maize regulatory gene opaque-2 encodes a protein with a “leucine-zipper” motif that binds to zein DNA. Proc Nat Acad Sci. 1990;87:46–50. doi: 10.1073/pnas.87.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons C, Söll D, Migliaccio F. Circumnutation and gravitropism cause root waving in Arabidopsis thaliana. J Exp Bot. 1995;46:143–150. [Google Scholar]
- Suckow M, von Wilcken-Bergmann B, Müller-Hill B. Identification of three residues in the basic regions of the bZIP proteins GCN4, C/EBP and TAF-1 that are involved in specific DNA binding. EMBO J. 1993;12:1193–1200. doi: 10.1002/j.1460-2075.1993.tb05760.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata T, Takase H, Takayama S, Mikami K, Nakatsuka A, Kawata H, Nakayama T, Iwabuchi M. A protein that binds to a cis-acting element of wheat histone genes has a leucine zipper motif. Science. 1989;245:965–967. doi: 10.1126/science.2772648. [DOI] [PubMed] [Google Scholar]
- Tabata T, Nakayama T, Mikami K, Iwabuchi M. HBP-1a and HBP-1b: Leucine zipper-type transcription factors of wheat. EMBO J. 1991;10:1459–1467. doi: 10.1002/j.1460-2075.1991.tb07666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Arnim AG, Deng X-W. Light inactivation of Arabidopsis photomorphogenic repressor COP1 involves a cell-specific regulation of its nucleocytoplasmic partitioning. Cell. 1994;79:1035–1045. doi: 10.1016/0092-8674(94)90034-5. [DOI] [PubMed] [Google Scholar]
- Wareing PF, Phillips IDJ. Growth and differentiation in plants. 3rd ed. Oxford, UK: Pergamon Press; 1981. [Google Scholar]
- Wilkins MB. Geotropism. Annu Rev Plant Physiol. 1966;17:379–408. [Google Scholar]
- Zobel RW. Control of morphogenesis in the ethylene-requiring tomato mutant, diageotropica. Can J Bot. 1974;52:735–741. [Google Scholar]