Structures of giant icosahedral eukaryotic dsDNA viruses (original) (raw)
. Author manuscript; available in PMC: 2012 Aug 1.
Abstract
In the last twenty years, numerous giant, dsDNA, icosahedral viruses have been discovered and assigned to the nucleocytoplasmic large dsDNA virus (NCLDV) clade. The major capsid proteins of these viruses consist of two consecutive jelly-roll domains, assembled into trimers, with pseudo 6-fold symmetry. The capsomers are assembled into arrays that have either p6 (as in Paramecium bursaria Chlorella virus-1) or p3 symmetry (as in Mimivirus). Most of the NCLDV viruses have a membrane that separates the nucleocapsid from the external capsid.
Keywords: Double-jelly-roll capsid protein, Giant icosahedral DNA viruses, Unique vertices, Assembly
Introduction
Viruses were originally defined as small infectious agents that can pass through the finest known filters [1]. Compared to their host, viruses are very small. For instance, polioviruses or common cold rhinoviruses have a diameter of about 300Å (0.03 μm) whereas a human red blood cell is about 6–8μm in diameter. Larger viruses, such as the oval or brick-shaped poxviruses (2000 to 4000Å in diameter) were discovered later [2]. Large, icosahedral dsDNA viruses that infect eukaryotic cells (e.g. PARAMECIUM BURSARIS CHLORELLA VIRUS 1 (PBCV-1) [3,4*,5–7*], CHILO IRRIDESCENT VIRUS (CIV) [4*,8*], PHAEOCYSTIS POUCHETTI VIRUS (PpV01) [9], MARSEILLE VIRUS [10*]) are now being discovered (Table 1) at exponentially increasing rates in part due to polymerase chain reaction (PCR) and other technological innovations. Most of these nucleocytoplasmic large DNA viruses (NCLDVs) have similarities in their genomes [11,12]. In addition, these viruses replicate their genome in the host's cytoplasm with little help from their host's replication system [11,12]. The NCDLVs will be the primary focus of this review.
Table 1.
List of giant icosahedral eukaryotic dsDNA viruses
| Virus Name | Diameter (Å) | Genome size (kbp) | T number | Fibers | Portal |
|---|---|---|---|---|---|
| Mimivirus | 5000 | 1,181 | 972–1200 | 1200 Å long, densely covering the surface | Starfish shaped |
| CroV | 3000 | 730 | Unknown | Unknown | Unknown |
| Marseillevirus | 2500 | 368 | Unknown | 120Å long, densely covering the surface | Unknown |
| PpV01 | 2200 | 485 | 219 | 3 fibers per trisymmetron | Unknown |
| PBCV-1 | 1900 | 330 | 169 | 3 fibers on 17 of the 20 trisymmetrons | Spike shaped |
| CIV | 1850 | 212 | 147 | On every capsomer | Unknown |
The biggest known virus to date is ACANTHAMOEBA POLYPHAGA MIMIVIRUS (Mimivirus), discovered in 2003 in fresh water [13]. Mimivirus infects single cell amoeba and has an icosahedral capsid with a diameter of about 5,000Å, covered by ~1,250 Å-long closely packed fibers [14]. Mimivirus and other large viruses cannot pass through standard 0.22 μm sized filters that are generally used to separate cellular organisms from virus. Furthermore, Mimivirus's 1.2 Mbp genome is bigger than some small bacteria [15,16]. Another giant virus CroV (Cafeteria roenbergensis virus) was discovered in ocean water infecting single cell microzooplankton. CroV has a 730 kbp genome and has a 3,000Å icosahedral diameter capsid, making it the second largest known virus [17]. Other very large icosahedral NCLDVs, infecting a wide range of eukaryotic cells, are being discovered [18] or rediscovered [19] and their genomes sequenced [10*,15,17], especially from aquatic environments [20].
These large infectious particles are considered to be viruses largely based on their often roughly icosahedral morphologies. The structure of the filtratable infectious agents that had been classified as being “viruses” were shown to have very regular geometric shapes such as rods with helical symmetry and spheres with icosahedral symmetry. These geometries were recognized as being the result of assembling many identical subunits each with the same environment [21,22]. The same rules of symmetry appear to be at least approximately true for many of the larger infectious particles, accounting for them being considered as viruses. However, the increased size of the virions allows the capsid to accommodate a larger genome (Table 1), able to code more functions normally associated with cells. Thus the boundary between a cell able to replicate in a supportive environment and a virus incapable of replicating without a host, has become diffuse [23]. Furthermore, these big viruses are more likely to be degenerative cells rather than advanced replicating molecules as might be the case for the smallest known viruses [24]. A further complication to the definition of viruses has arisen with the discovery of “virophages” for Mimivirus [25**–27] and CroV [28]. These are viruses that utilize other viruses as their host, giving the host virus a role similar to that of a cell. Hence, studies of large viruses are akin to studies of simple cells. Considering the multitude of interactions even in prokaryotic cells, large viruses are ideal for investigating many biological processes at a molecular level.
The NCLDVgroup of viruses is not a taxonomic family, but is a loosely defined cluster or clade of viruses that include Poxviridae, Asfarviridae, Iridoviridae, Phycodnaviridae [11] as well as the new Mimiviridae families [11–13,15,16,27]. The presence of typical cellular genes within the large genomes of NCLDVs [15,17] as well as their structures [29**–31] have raised debates on the evolutionary relationship between viruses and cellular organisms [23,32–34*,35–38] and the need for a better definition of a virus [39,40]. The name “girus” has been suggested to describe the giant viruses [16,41**]. The existence of genes from different domains of life in the NCLDV genome has given these viruses a reputation as “gene robbers” [42,43] and reignited the debate about whether there is a single tree of life [36,38] that describes the history of how all organisms capable of reproduction have evolved. Some of these giant viruses replicate their large DNA genomes in the host's cytoplasm within a special compartment, called the “virus factory”. The similarity between the “virus factory” and eukaryotic nucleus as well as phylogenetic studies on the viral replication enzymes have led to the hypothesis that these NCLDVs might have had a critical role in the evolution of prokaryotes to eukaryotes [27,34*,44]. Due to the size and complexity of the NCLDVs, structural studies have been a challenge, requiring multiple structural techniques such as X-ray crystallography, scanning electron microscopy (SEM), cryo-electron microscopy (cryo-EM), atomic force microscopy (AFM) and X-ray lasers [14,29**,31,45,46].
The icosahedral capsid and double-jelly-roll
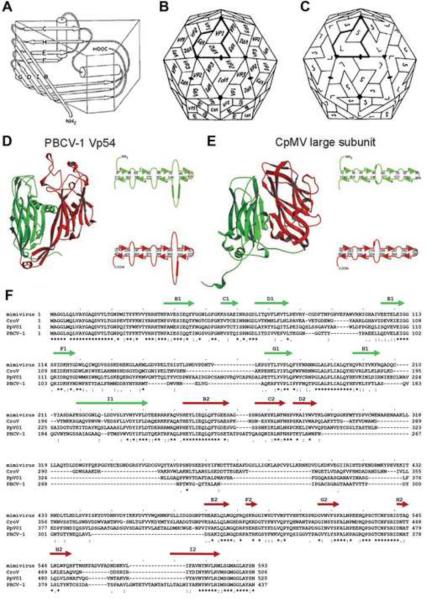
Due to their relatively smaller genome size compared to their host, viruses can accommodate only a limited number of genes that code structural proteins. As mentioned above, this requires that the basic building block is repeated numerous times to form a symmetric capsid. Two of the most common folds utilized for the major capsid protein of viruses are the “jelly-roll” [47] and the “HK97” [48,49] folds. Most NCLDVs use the jelly-roll fold, whereas all known tailed phage structures use the HK97 fold, indicating two major viral lineages in the divergent evolution of their capsids. The “jelly-roll” fold, first found in small RNA plant viruses [50,51] and small RNA animal viruses [52,53], is a β-barrel, wedge-shaped structure composed of eight, anti-parallel, β-strands (B to I) (Figure 1A). The RNA plant viruses have T=3 quasi-symmetry (see below for a description of the triangulation or T number) in which there are 180 subunits, each with a jelly-roll fold, that have quasi-similar environments (Figure 1B). In the picornaviruses such as the rhino- and polioviruses there are three different major capsid proteins (VP1, VP2 and VP3) each with a “jelly-roll” fold, organized exactly as in the T=3 RNA plant viruses (Figure 1B). The VP1 subunits form pentamers around the icosahedral 5-fold axes, whereas the VP2 and VP3 form trimers with quasi-6-fold symmetry around the icosahedral 3-fold axes. The cowpea mosaic plant virus (CpMV) has two of the jelly-rolls (equivalent to VP2 and VP3 in picornavirus) covalently linked together into a single polypeptide chain [54] (Figure 1C, 1E). Similarly, in large dsDNA viruses that utilize a jelly-roll motif for their major capsid protein, including NCLDVs, there are hexagonal and pentagonal capsomers. The hexagonal capsomers are trimers in which the monomer consists of two consecutive double jelly-rolls, giving the capsomers quasi-6-fold symmetry, much as the (VP2–VP3)3 quasi-hexamer of CpMV (Figure 1C). The pentameric capsomers of NCLDVs, like those of CpMV, are located at the 5-fold vertices created from a special vertex single jelly-roll protein [55]. In NCLDVs there are, in general, 12 pentameric capsomers, one at each vertex, and (T-1) quasi-6-fold capsomers, where T is the Casper and Klug triangulation number described below.
Figure 1.
Jelly-roll and double jelly-roll structures of NCLDVs. (A) Cartoon diagram showing the jelly-roll fold of eight anti-parallel β-strands, B to H. From [53 - Hogle JM, Chow M, Filman DJ: Three-dimensional structure of poliovirus at 2.9 Å resolution. Science 1985, 229:1358–1365]. Reprinted with permission from AAAS. (B) Schematic icosahedral arrangement of VP1, VP2 and VP3 in picornaviruses. (C). Schematic icosahedral arrangement of large (VP2 + VP3) and small (VP1) subunits in CpMV. (B and C) adapted from [47]. (D and E) Ribbon diagrams of double jelly-roll structures of PBCV-1 and CpMV, respectively. Length of β-strands (arrows) and loops (curved lines) are shown on the side of each panel in (D) and (E). (D and E) adapted from [5]. (F). Sequence alignment of the major capsid proteins of Mimivirus, CroV, PpV01 and PBCV-1. β-strand regions are assigned based on the PBCV-1 X-ray structure as shown by arrows above the sequence.
Note to publisher concerning Figure 1A: Per AAAS requirements the following statement must be included in any electronic versions, either adjacent to the reprinted AAAS material or in the terms and conditions for use of your electronic products: “Readers may view, browse, and/or download material for temporary copying purposes only, provided these uses are for noncommercial personal purposes. Except as provided by law, this material may not be further reproduced, distributed, transmitted, modified, adapted, performed, displayed, published, or sold in whole or in part, without prior written permission from the publisher.”
The structure of the double-jelly-roll fold was first described for adenovirus [56,57], and later in the bacteriophage PRD1 [58], the algae virus PBCV-1 [5] (Figures 1D, 2A), the archaeal virus Sulfolobus turreted icosahedral virus [59], and the marine bacteriophage PM2 [60]. A double-jelly-roll capsomers has a thickness of about 75 Å, and a diameter of between 74 Å and 85 Å. These viruses, as well as many other NCLDV icosahedral viruses, are sometimes referred to as belonging to the PRD1-adenovirus lineage [61]. Sequence alignment of NCLDV major capsid proteins showed relatively high similarity in the β-strands but often with different numbers of amino acids linking strands, sometimes creating large insertion loops between strands D and E as well as between strands F and G [29**] (Figure 1F). Mostly these insertions form the exterior of these viruses, such as the “towers” in adenovirus.
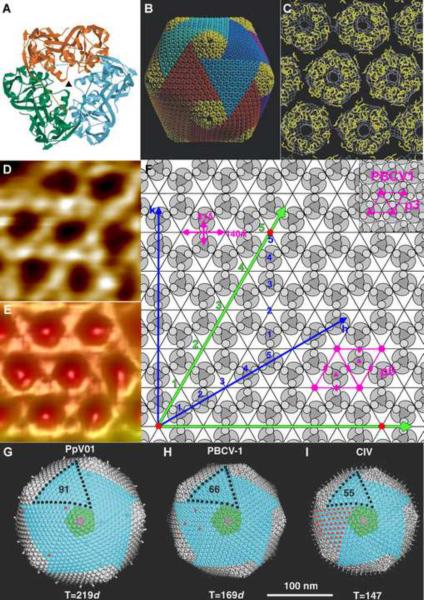
Figure 2.
Capsomer arrangement of icosahedral NCLDVs. (A) The PBCV-1 Vp54 trimer viewed from the inside of the virus with each monomer in a different color. (B) Pseudo-atomic model of the PBCV-1 capsid based on fitting the crystal structure of the Vp54 trimeric major capsid protein into the cryo-EM reconstruction. (C) Enlarged view of the Vp54 Cα backbone (yellow) fitted into the cryo-EM map (white) viewed from outside the virus. (A, B and C) are adapted from [5]. (D) An enlarged atomic force microscopic image of defibered Mimivirus at high magnification shows a honeycomb array of depressions where the capsomers are systematically absent. (E) Cryo-EM reconstruction of Mimivirus on the same scale as (D) (Note: In D and E the white regions are high points and the dark regions are low points on the surface of the virus). (F) Diagram showing the p6 plane group lattice of Mimivirus capsomers as found in C and D. The loose packing of the capsomer in Mimivirus is different to other large dsDNA virus such as PBCV-1 in which the capsomer is close packed with p3 plane group symmetry. The numbers count the potential sites of hexameric capsomers along the h and k axes as described in the text. (D, E and F are adapted from [29**]). (G) PpV01, (H) PBCV-1, and (I) CIV, showing the pentasymmetron in green and one trisymmetron outlined with black dots. Red dots indicate the location of surface fibers. (G, H and I are adapted from [9]).
T number and capsomer arrangement
The simple icosahedral arrangement of protein subunits in viruses was first discussed by Crick and Watson [21]. Their concepts for viral capsid arrangements were later extended by Caspar and Klug [22], using a hexagonal planar lattice to define the position of pentamers in terms of the indices h, k counting capsomers along the axes of the lattice [22] (Figure 2F). The triangulation number, T, is then given by the h, k integers using the equation: T=h2+hk+k2. In the small RNA plant viruses, 60T would be the number of quasi-equivalent subunits. In the more complex large dsDNA viruses, that have double jelly-roll monomers as their major capsid protein, 60T would be the number of quasi-equivalent jelly-roll folds. It has not yet been possible to accurately determine the T number (between 972 and 1200) of Mimivirus due to the low resolution of the cryo-EM reconstruction [29**]. In Mimivirus, neighboring capsomers have a 60° rotation with respect to each other, producing planar arrays that have p3 symmetry, leaving holes in every third position that are as big as a capsomer itself (Figure 2D, 2E, 2F). However, in most other NCLDVs the capsomers are in closely packed hexagonal arrays with p6 plane symmetry (Figure 2C, 2F) [29**]. The above estimate of the T number for Mimivirus assumes that every site on the hexagonal lattice is filled by a capsomer. Therefore, for Mimivirus, the actual number of jelly-rolls per asymmetric unit will be only two thirds of the estimated T number [29**,30*].
Pentasymmetron and trisymmetron
Large, well-organized triangular and pentagonal arrays of capsomers was first observed by Wrigley et al. in a negatively stained Sericesthis iridescent virus (SIV) sample [62]. These arrays were separated by cleavage lines along which the virus came apart after long periods of storage [62]. This suggests that the capsid of large icosahedral NCLDVs is constructed from pre-assembled trigonal and pentagonal patches. The pre-assembled 5-fold (pentasymmetrons) and 3-fold (trisymmetrons) patches (Figure 2B, 2G, 2H, 2I) are separated from each other by cleavage lines, visible in cryo-EM reconstructions [4*–6,8*,9] on the virus surface. The cleavage lines are a consequence of the capsomers having only quasi-6-fold symmetry (Figure 2A), causing the 3-fold symmetric capsomers in neighboring tri- or pentasymmetrons being rotated by 60° relative to each other [6,8*].
Wrigley et al. [62], used a modified version of Goldberg's diagram to described the possible ways of building icosahedral particles from different symmetrons. The Goldberg diagram, as also used by Caspar and Klug [22], utilizes a hexagonal lattice to describe the position of pentagonal vertices in forming an icosahedron. Although the relative size of penta- and trisymmetrons can vary to an extent determined by the T number [62,63], in all currently available structures, the pentasymmetrons are always of the same size with 31 pseudo-hexagonal capsomers per pentasymmetron. This size pentasymmetron occurs when h=7 [9,63]. The formation of quasi-equivalence around the 5-fold vertices is a significant problem for the assembly of an icosahedral particle from identical hexagonal capsomers. For the bacteriophage T4 and some other phages there is a special protein, homologous to the major capsid protein, that forms the 5-fold vertex [64]. Apparently formation of the 31 capsomer pentasymmetron cap is sufficient, in general, to take the strain of forming a large capsid.
Other surface features
When Mimivirus was first isolated, the dense, long and Gram-stainable surface fibers led to the misidentification of Mimivirus as a bacterium [13,65]. The fibers are sensitive to proteolysis after treatment with lysozyme [29**,45] suggesting that the surface fibers are cross-linked and protected by peptidoglycan, consistent with the Gram-staining [13]. These fibers might be attached to the center of each capsomer or might be associated with the systematically absent capsomer locations [45]. Possibly Mimivirus uses these fibers to “mimic” bacterial entry into amoeba allowing Mimivirus to be phagocytosed by the amoeba [16]. Some other NCLDVs also have fibers located at the center of all [4*,8*] or some of their capsomers [7*,9]. The function of these fibers in CIV, PBCV and PpV01 are unknown but probably play a role in attaching to the host [7*] as occurs in many bacteriophages [66].
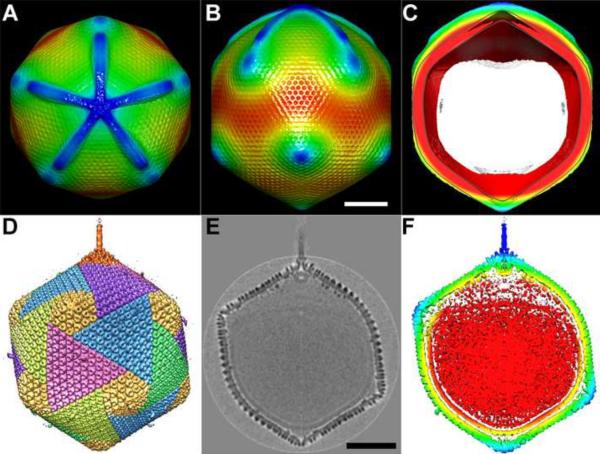
Although the capsids of many viruses have icosahedral symmetry, the internal nucleocapsid is asymmetric as there is only one genome. Unique portals for packaging and delivering genome associated nucleocapsids occur in tailed bacteriophages [64,67,68] as well as in PRD1 [69], PBCV-1 [3] and herpesviruses [70]. Similarly, Mimivirus has a large star-fish-shaped portal on one of its vertices [29**,31]. PBCV-1 also has a unique vertex that is related to genome delivery [7*, X. Zhang et al., unpublished] and modifies the icosahedral symmetry in the vicinity of the special vertex.
Internal Structure
Several common internal structures have been found inside icosahedral NCLDV capsids. Minor capsid proteins beneath the major capsid protein layer, but outside a membrane surrounding the nucleocapsid, are present in many NCLDVs. In PBCV-1 [7*] and CIV [8*], there are fiber-like proteins that link capsomers together, presumably for stability. The nature of the minor capsid proteins in the vicinity of the special vertex is different, probably to initiate virus disassembly when the special vertex has sensed a host [7*].
Both in PBCV-1 and Mimivirus, the unique portal is associated with a pocket, created by the nucleocapsid having a concave depression (Figure 3). In the case of PBCV-1 the pocket contains enzymes which appear to digest part of the host cell wall on entry into the chlorella cells [X. Zhang et al., unpublished]. In the case of Mimivirus the portal is probably associated with the release of the genome into the amoeba cytoplasm to form a virus factory [29**,45].
Figure 3.
The unique vertex/portals in Mimivirus and in PBCV-1. (A, B and C) Top, side and central section views of Mimivirus, respectively. The features in A, B and C are colored based on their radial distance from the center of the virus, with red being being used for features at smaller radial distance, changing to yellow, green and blue to features at larger radial distances from the center. The white area in (C) is where the back of the virus has been removed. The scale bar is 1,000Å long. (D, E and F) Side, grey-scaled central section and color coded central section views of PBCV-1, respectively. The density in (F) is colored radially with red for smaller radii, changing to yellow, green and blue as the radius becomes bigger. Scale bar 500Å long. (A, B and C) are adapted from [29**], (D, E and F) are adapted from [7*].
Conclusion
The number of known and characterized giant, icosahedral, dsDNA eukaryotic viruses both from fresh and salt water environments is increasing rapidly. They have large genomes that contain genes from all three kingdoms of life [15,17]. Many of the NCLDVs have icosahedral or close-to-icosahedral capsid symmetry assembled from quasi-6-fold symmetric capsomers constructed of six jelly-roll domains arranged into penta- and trisymmetrons. The similarly sized pentasymmetron suggests that this is a critical size to relieve the strain around the 5-fold vertices. Mimivirus is different in that it does not have a close packed assembly of its capsomers nor special pentasymmetrons to disperse the strain forces around its vertices. Mimivirus is also unique among known NCLDVs by being covered by a dense surface of fibers and a star-fish-shaped portal [30*]. These differences are probably related to adaptation to different hosts. For instance, the chlorella host of PBCV-1 has a cell wall requiring that only the PBCV-1 genome is inserted. In contrast, amoeba has only a membrane to defend itself, requiring Mimivirus to attract its host to be absorbed by virtue of the amoeba's normal food gathering procedure. More high resolution structural information will not only shed light on how NCLDVs assemble into large and complicated structures, but will probe deeper as to their evolution from simpler viruses or devolution from a higher organisms.
Highlights
- Giant icosahedral dsDNA viruses replicate in the host's cytoplasm.
- Major capsid proteins have a double jelly-roll fold.
- Trimeric capsomers have pseudo-hexagonal symmetry.
- Capsomers assemble into 2D arrays forming tri- and pentasymmetrons.
- Some NCLDVs have special vertices for genome exit and making of virus factories.
- Nucleocapsids inside the capsid are surrounded by a membrane.
Acknowledgements
We are grateful to Matthias Fischer for helpful discussions. We thank Sheryl Kelly for help in preparation of the manuscript. The work was supported by a National Institutes of Health grant award (AI11219) to MGR. CX was supported by funds from the College of Science, University of Texas at El Paso.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
* of special interest
** of outstanding interest
- 1.Levine AJ, Enquist LW. Chapter 1, History of Virology. In: Knipe DM, Howley PM, editors. Fields' Virology. vol 1. Wolters Kluwer Health/Lippincott Williams & Wilkins; 2007. pp. 4–25. 5. [Google Scholar]
- 2.Buller RM, Palumbo GJ. Poxvirus pathogenesis. Microbiol Rev. 1991;55:80–122. doi: 10.1128/mr.55.1.80-122.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Etten JL, Lane LC, Meints RH. Viruses and viruslike particles of eukaryotic algae. Microbiol Rev. 1991;55:586–620. doi: 10.1128/mr.55.4.586-620.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4*.Yan X, Olson NH, Van Etten JL, Bergoin M, Rossmann MG, Baker TS. Structure and assembly of large lipid-containing dsDNA viruses. Nat Struct Biol. 2000;7:101–103. doi: 10.1038/72360. [DOI] [PMC free article] [PubMed] [Google Scholar]; An early paper describing the structures of the tri- and pentasymmetrons in PBCV-1 and CIV. These structures are based on cryo-electron microscopy 3D reconstructions.
- 5.Nandhagopal N, Simpson AA, Gurnon JR, Yan X, Baker TS, Graves MV, Van Etten JL, Rossmann MG. The structure and evolution of the major capsid protein of a large, lipid-containing DNA virus. Proc Natl Acad Sci U S A. 2002;99:14758–14763. doi: 10.1073/pnas.232580699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simpson AA, Nandhagopal N, Van Etten JL, Rossmann MG. Structural analyses of Phycodnaviridae and Iridoviridae. Acta Crystallogr D Biol Crystallogr. 2003;59:2053–2059. doi: 10.1107/s090744490302225x. [DOI] [PubMed] [Google Scholar]
- 7*.Cherrier MV, Kostyuchenko VA, Xiao C, Bowman VD, Battisti AJ, Yan X, Chipman PR, Baker TS, Van Etten JL, Rossmann MG. An icosahedral algal virus has a complex unique vertex decorated by a spike. Proc Natl Acad Sci U S A. 2009;106:11085–11089. doi: 10.1073/pnas.0904716106. [DOI] [PMC free article] [PubMed] [Google Scholar]; Shows that PBCV-1 is not exactly icosahedral. There is a special spike on one of the 12 pentagonal vertices, presumably required for infecting the host. There is also a cavity under the spike which might be filled with enzymes to digest the host's cell wall.
- 8*.Yan X, Yu Z, Zhang P, Battisti AJ, Holdaway HA, Chipman PR, Bajaj C, Bergoin M, Rossmann MG, Baker TS. The capsid proteins of a large, icosahedral dsDNA virus. J Mol Biol. 2009;385:1287–1299. doi: 10.1016/j.jmb.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]; A careful analysis of the cryo-electron microscopy reconstruction of CIV. A model of the major capsid double jelly-roll fold is built based on the crystal structure of the homologous PBCV-1 capsid protein. This structure is fitted to the EM density. Uninterpreted density is interpreted in terms of minor capsid proteins that probably act as the glue to stabilize the viral capsid.
- 9.Yan X, Chipman PR, Castberg T, Bratbak G, Baker TS. The marine algal virus PpV01 has an icosahedral capsid with T=219 quasisymmetry. J Virol. 2005;79:9236–9243. doi: 10.1128/JVI.79.14.9236-9243.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10*.Boyer M, Yutin N, Pagnier I, Barrassi L, Fournous G, Espinosa L, Robert C, Azza S, Sun S, Rossmann MG, et al. Giant Marseillevirus highlights the role of amoebae as a melting pot in emergence of chimeric microorganisms. Proc Natl Acad Sci U S A. 2009;106:21848–21853. doi: 10.1073/pnas.0911354106. [DOI] [PMC free article] [PubMed] [Google Scholar]; Marseillesvirus is one of the newly discovered large dsDNA viruses, many of which use amoeba as their host.
- 11.Iyer LM, Aravind L, Koonin EV. Common origin of four diverse families of large eukaryotic DNA viruses. J Virol. 2001;75:11720–11734. doi: 10.1128/JVI.75.23.11720-11734.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iyer LM, Balaji S, Koonin EV, Aravind L. Evolutionary genomics of nucleo-cytoplasmic large DNA viruses. Virus Res. 2006;117:156–184. doi: 10.1016/j.virusres.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 13.La Scola B, Audic S, Robert C, Jungang L, de Lamballerie X, Drancourt M, Birtles R, Claverie JM, Raoult D. A giant virus in amoebae. Science. 2003;299:2033. doi: 10.1126/science.1081867. [DOI] [PubMed] [Google Scholar]
- 14.Xiao C, Chipman PR, Battisti AJ, Bowman VD, Renesto P, Raoult D, Rossmann MG. Cryo-electron microscopy of the giant Mimivirus. J Mol Biol. 2005;353:493–496. doi: 10.1016/j.jmb.2005.08.060. [DOI] [PubMed] [Google Scholar]
- 15.Raoult D, Audic S, Robert C, Abergel C, Renesto P, Ogata H, La Scola B, Suzan M, Claverie JM. The 1.2-megabase genome sequence of Mimivirus. Science. 2004;306:1344–1350. doi: 10.1126/science.1101485. [DOI] [PubMed] [Google Scholar]
- 16.Claverie JM, Ogata H, Audic S, Abergel C, Suhre K, Fournier PE. Mimivirus and the emerging concept of “giant” virus. Virus Res. 2006;117:133–144. doi: 10.1016/j.virusres.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 17.Fischer MG, Allen MJ, Wilson WH, Suttle CA. Giant virus with a remarkable complement of genes infects marine zooplankton. Proc Natl Acad Sci U S A. 2010;107:19508–19513. doi: 10.1073/pnas.1007615107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Matos AP, Caeiro MF, Papp T, Matos BA, Correia AC, Marschang RE. New viruses from Lacerta monticola (Serra da Estrela, Portugal): further evidence for a new group of nucleo-cytoplasmic large deoxyriboviruses. Microsc Microanal. 2011;17:101–108. doi: 10.1017/S143192761009433X. [DOI] [PubMed] [Google Scholar]
- 19.Claverie JM, Grzela R, Lartigue A, Bernadac A, Nitsche S, Vacelet J, Ogata H, Abergel C. Mimivirus and Mimiviridae: giant viruses with an increasing number of potential hosts, including corals and sponges. J Invertebr Pathol. 2009;101:172–180. doi: 10.1016/j.jip.2009.03.011. [DOI] [PubMed] [Google Scholar]
- 20.Ghedin E, Claverie JM. Mimivirus relatives in the Sargasso sea. Virol J. 2005;2:62. doi: 10.1186/1743-422X-2-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crick FH, Watson JD. Structure of small viruses. Nature. 1956;177:473–475. doi: 10.1038/177473a0. [DOI] [PubMed] [Google Scholar]
- 22.Caspar DL, Klug A. Physical principles in the construction of regular viruses. Cold Spring Harb Symp Quant Biol. 1962;27:1–24. doi: 10.1101/sqb.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- 23.Forterre P. Manipulation of cellular syntheses and the nature of viruses: The virocell concept. Comptes Rendus Chimie. 2011;14:392–399. [Google Scholar]
- 24.Hendrix RW, Lawrence JG, Hatfull GF, Casjens S. The origins and ongoing evolution of viruses. Trends Microbiol. 2000;8:504–508. doi: 10.1016/s0966-842x(00)01863-1. [DOI] [PubMed] [Google Scholar]
- 25**.La Scola B, Desnues C, Pagnier I, Robert C, Barrassi L, Fournous G, Merchat M, Suzan-Monti M, Forterre P, Koonin E, et al. The virophage as a unique parasite of the giant mimivirus. Nature. 2008;455:100–104. doi: 10.1038/nature07218. [DOI] [PubMed] [Google Scholar]; This paper records the discovery of the first virophage called “Sputnik” Sputnik uses Mimivirus as a host, which in turn requires amoeba for its reproduction.
- 26.Sun S, La Scola B, Bowman VD, Ryan CM, Whitelegge JP, Raoult D, Rossmann MG. Structural studies of the Sputnik virophage. J Virol. 2010;84:894–897. doi: 10.1128/JVI.01957-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Claverie JM, Abergel C. Mimivirus and its virophage. Annu Rev Genet. 2009;43:49–66. doi: 10.1146/annurev-genet-102108-134255. [DOI] [PubMed] [Google Scholar]
- 28.Fischer MG, Suttle CA. A virophage at the origin of large DNA transposons. Science. 2011;332:231–234. doi: 10.1126/science.1199412. [DOI] [PubMed] [Google Scholar]
- 29**.Xiao C, Kuznetsov YG, Sun S, Hafenstein SL, Kostyuchenko VA, Chipman PR, Suzan-Monti M, Raoult D, McPherson A, Rossmann MG. Structural studies of the giant mimivirus. PLoS Biol. 2009;7:e1000092. doi: 10.1371/journal.pbio.1000092. [DOI] [PMC free article] [PubMed] [Google Scholar]; The most detailed account of the 3D structure of Mimivirus is given in this paper. The work depends on both cryo-electron microscopy and atomic force microscopy.
- 30*.Klose T, Kuznetsov YG, Xiao C, Sun S, McPherson A, Rossmann MG. The three-dimensional structure of Mimivirus. Intervirology. 2010;53:268–273. doi: 10.1159/000312911. [DOI] [PMC free article] [PubMed] [Google Scholar]; This is a review of the structural information on Mimivirus.
- 31.Zauberman N, Mutsafi Y, Halevy DB, Shimoni E, Klein E, Xiao C, Sun S, Minsky A. Distinct DNA exit and packaging portals in the virus Acanthamoeba polyphaga mimivirus. PLoS Biol. 2008;6:e114. doi: 10.1371/journal.pbio.0060114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Claverie JM, Ogata H. Ten good reasons not to exclude giruses from the evolutionary picture. Nat Rev Microbiol. 2009;7:615. doi: 10.1038/nrmicro2108-c3. author reply 615. [DOI] [PubMed] [Google Scholar]
- 33.Hegde NR, Maddur MS, Kaveri SV, Bayry J. Reasons to include viruses in the tree of life. Nat Rev Microbiol. 2009;7:615. doi: 10.1038/nrmicro2108-c1. author reply 615. [DOI] [PubMed] [Google Scholar]
- 34*.Koonin EV, Senkevich TG, Dolja VV. Compelling reasons why viruses are relevant for the origin of cells. Nat Rev Microbiol. 2009;7:615. doi: 10.1038/nrmicro2108-c5. author reply 615. [DOI] [PubMed] [Google Scholar]; A discussion is given on what defines a virus and of the co-evolution of viruses with their hosts.
- 35.Ludmir EB, Enquist LW. Viral genomes are part of the phylogenetic tree of life. Nat Rev Microbiol. 2009;7:615. doi: 10.1038/nrmicro2108-c4. author reply 615. [DOI] [PubMed] [Google Scholar]
- 36.Moreira D, Lopez-Garcia P. Ten reasons to exclude viruses from the tree of life. Nat Rev Microbiol. 2009;7:306–311. doi: 10.1038/nrmicro2108. [DOI] [PubMed] [Google Scholar]
- 37.Navas-Castillo J. Six comments on the ten reasons for the demotion of viruses. Nat Rev Microbiol. 2009;7:615. doi: 10.1038/nrmicro2108-c2. author reply 615. [DOI] [PubMed] [Google Scholar]
- 38.Raoult D. There is no such thing as a tree of life (and of course viruses are out!) Nat Rev Microbiol. 2009;7:615. doi: 10.1038/nrmicro2108-c6. author reply 615. [DOI] [PubMed] [Google Scholar]
- 39.Raoult D, Forterre P. Redefining viruses: lessons from Mimivirus. Nature Reviews Microbiology. 2008;6:315–319. doi: 10.1038/nrmicro1858. [DOI] [PubMed] [Google Scholar]
- 40.Claverie JM, Abergel C. Mimivirus: the emerging paradox of quasi-autonomous viruses. Trends Genet. 2010;26:431–437. doi: 10.1016/j.tig.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 41**.Van Etten JL, Lane LC, Dunigan DD. DNA viruses: the really big ones (giruses) Annu Rev Microbiol. 2010;64:83–99. doi: 10.1146/annurev.micro.112408.134338. [DOI] [PMC free article] [PubMed] [Google Scholar]; This is a review of the properties and characteristics of PBCV-1 by the major expert, J. van Etten, and his colleagues.
- 42.Filee J, Pouget N, Chandler M. Phylogenetic evidence for extensive lateral acquisition of cellular genes by Nucleocytoplasmic large DNA viruses. BMC Evol Biol. 2008;8:320. doi: 10.1186/1471-2148-8-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moreira D, Brochier-Armanet C. Giant viruses, giant chimeras: the multiple evolutionary histories of Mimivirus genes. BMC Evol Biol. 2008;8:12. doi: 10.1186/1471-2148-8-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Forterre P. The origin of viruses and their possible roles in major evolutionary transitions. Virus Research. 2006;117:5–16. doi: 10.1016/j.virusres.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 45.Kuznetsov YG, Xiao C, Sun S, Raoult D, Rossmann M, McPherson A. Atomic force microscopy investigation of the giant mimivirus. Virology. 2010;404:127–137. doi: 10.1016/j.virol.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 46.Seibert MM, Ekeberg T, Maia FR, Svenda M, Andreasson J, Jonsson O, Odic D, Iwan B, Rocker A, Westphal D, et al. Single mimivirus particles intercepted and imaged with an X-ray laser. Nature. 2011;470:78–81. doi: 10.1038/nature09748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rossmann MG, Johnson JE. Icosahedral RNA virus structure. Annu Rev Biochem. 1989;58:533–573. doi: 10.1146/annurev.bi.58.070189.002533. [DOI] [PubMed] [Google Scholar]
- 48.Wikoff WR, Liljas L, Duda RL, Tsuruta H, Hendrix RW, Johnson JE. Topologically linked protein rings in the bacteriophage HK97 capsid. Science. 2000;289:2129–2133. doi: 10.1126/science.289.5487.2129. [DOI] [PubMed] [Google Scholar]
- 49.Helgstrand C, Wikoff WR, Duda RL, Hendrix RW, Johnson JE, Liljas L. The refined structure of a protein catenane: the HK97 bacteriophage capsid at 3.44 Å resolution. J Mol Biol. 2003;334:885–899. doi: 10.1016/j.jmb.2003.09.035. [DOI] [PubMed] [Google Scholar]
- 50.Harrison SC, Olson AJ, Schutt CE, Winkler FK, Bricogne G. Tomato bushy stunt virus at 2.9 Å resolution. Nature. 1978;276:368–373. doi: 10.1038/276368a0. [DOI] [PubMed] [Google Scholar]
- 51.Abad-Zapatero C, Abdel-Meguid SS, Johnson JE, Leslie AG, Rayment I, Rossmann MG, Suck D, Tsukihara T. Structure of southern bean mosaic virus at 2.8 Å resolution. Nature. 1980;286:33–39. doi: 10.1038/286033a0. [DOI] [PubMed] [Google Scholar]
- 52.Rossmann MG, Arnold E, Erickson JW, Frankenberger EA, Griffith JP, Hecht HJ, Johnson JE, Kamer G, Luo M, Mosser AG, et al. Structure of a human common cold virus and functional relationship to other picornaviruses. Nature. 1985;317:145–153. doi: 10.1038/317145a0. [DOI] [PubMed] [Google Scholar]
- 53.Hogle JM, Chow M, Filman DJ. Three-dimensional structure of poliovirus at 2.9 Å resolution. Science. 1985;229:1358–1365. doi: 10.1126/science.2994218. [DOI] [PubMed] [Google Scholar]
- 54.Lin T, Chen Z, Usha R, Stauffacher CV, Dai JB, Schmidt T, Johnson JE. The refined crystal structure of cowpea mosaic virus at 2.8 Å resolution. Virology. 1999;265:20–34. doi: 10.1006/viro.1999.0038. [DOI] [PubMed] [Google Scholar]
- 55.Liu H, Wu L, Zhou ZH. Model of the trimeric fiber and its interactions with the pentameric penton base of human adenovirus by cryo-electron microscopy. J Mol Biol. 2011;406:764–774. doi: 10.1016/j.jmb.2010.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Athappilly FK, Murali R, Rux JJ, Cai Z, Burnett RM. The refined crystal structure of hexon, the major coat protein of adenovirus type 2, at 2.9 Å resolution. J Mol Biol. 1994;242:430–455. doi: 10.1006/jmbi.1994.1593. [DOI] [PubMed] [Google Scholar]
- 57.Burnett RM, Grutter MG, White JL. The structure of the adenovirus capsid. I. An envelope model of hexon at 6 Å resolution. J Mol Biol. 1985;185:105–123. doi: 10.1016/0022-2836(85)90186-x. [DOI] [PubMed] [Google Scholar]
- 58.Stewart PL, Ghosh S, Bamford DH, Burnett RM. Crystallization of the major coat protein of PRD1, a bacteriophage with an internal membrane. J Mol Biol. 1993;230:349–352. doi: 10.1006/jmbi.1993.1148. [DOI] [PubMed] [Google Scholar]
- 59.Khayat R, Tang L, Larson ET, Lawrence CM, Young M, Johnson JE. Structure of an archaeal virus capsid protein reveals a common ancestry to eukaryotic and bacterial viruses. Proc Natl Acad Sci U S A. 2005;102:18944–18949. doi: 10.1073/pnas.0506383102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huiskonen JT, Kivela HM, Bamford DH, Butcher SJ. The PM2 virion has a novel organization with an internal membrane and pentameric receptor binding spikes. Nat Struct Mol Biol. 2004;11:850–856. doi: 10.1038/nsmb807. [DOI] [PubMed] [Google Scholar]
- 61.Benson SD, Bamford JK, Bamford DH, Burnett RM. Does common architecture reveal a viral lineage spanning all three domains of life? Mol Cell. 2004;16:673–685. doi: 10.1016/j.molcel.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 62.Wrigley NG. An electron microscope study of the structure of Sericesthis iridescent virus. J Gen Virol. 1969;5:123–134. doi: 10.1099/0022-1317-5-1-123. [DOI] [PubMed] [Google Scholar]
- 63.Sinkovits RS, Baker TS. A tale of two symmetrons: rules for construction of icosahedral capsids from trisymmetrons and pentasymmetrons. J Struct Biol. 2010;170:109–116. doi: 10.1016/j.jsb.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fokine A, Leiman PG, Shneider MM, Ahvazi B, Boeshans KM, Steven AC, Black LW, Mesyanzhinov VV, Rossmann MG. Structural and functional similarities between the capsid proteins of bacteriophages T4 and HK97 point to a common ancestry. Proc Natl Acad Sci U S A. 2005;102:7163–7168. doi: 10.1073/pnas.0502164102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Raoult D, La Scola B, Birtles R. The discovery and characterization of mimivirus, the largest known virus and putative pneumonia agent. Clinical Infectious Diseases. 2007;45:95–102. doi: 10.1086/518608. [DOI] [PubMed] [Google Scholar]
- 66.Leiman PG, Kanamaru S, Mesyanzhinov VV, Arisaka F, Rossmann MG. Structure and morphogenesis of bacteriophage T4. Cell Mol Life Sci. 2003;60:2356–2370. doi: 10.1007/s00018-003-3072-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Morais MC, Choi KH, Koti JS, Chipman PR, Anderson DL, Rossmann MG. Conservation of the capsid structure in tailed dsDNA bacteriophages: the pseudoatomic structure of phi29. Mol Cell. 2005;18:149–159. doi: 10.1016/j.molcel.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 68.Jiang W, Baker ML, Jakana J, Weigele PR, King J, Chiu W. Backbone structure of the infectious epsilon15 virus capsid revealed by electron cryomicroscopy. Nature. 2008;451:1130–1134. doi: 10.1038/nature06665. [DOI] [PubMed] [Google Scholar]
- 69.Gowen B, Bamford JK, Bamford DH, Fuller SD. The tailless icosahedral membrane virus PRD1 localizes the proteins involved in genome packaging and injection at a unique vertex. J Virol. 2003;77:7863–7871. doi: 10.1128/JVI.77.14.7863-7871.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Baker ML, Jiang W, Rixon FJ, Chiu W. Common ancestry of herpesviruses and tailed DNA bacteriophages. J Virol. 2005;79:14967–14970. doi: 10.1128/JVI.79.23.14967-14970.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]