Translational control of C/EBPα and C/EBPβ isoform expression (original) (raw)
. 2000 Aug 1;14(15):1920–1932.
Abstract
Transcription factors derived from CCAAT/enhancer binding protein (C/EBP)α and C/EBPβ genes control differentiation and proliferation in a number of cell types. Various C/EBP isoforms arise from unique C/EBPβ and C/EBPα mRNAs by differential initiation of translation. These isoforms retain different parts of the amino terminus and therefore display different functions in gene regulation and proliferation control. We show that PKR and mTOR signaling pathways control the ratio of C/EBP isoform expression through the eukaryotic translation initiation factors eIF-2α and eIF-4E, respectively. An evolutionary conserved upstream open reading frame in C/EBPα and C/EBPβ mRNAs is a prerequisite for regulated initiation from the different translation initiation sites and integrates translation factor activity. Deregulated translational control leading to aberrant C/EBPα and C/EBPβ isoform expression or ectopic expression of truncated isoforms disrupts terminal differentiation and induces a transformed phenotype in 3T3-L1 cells. Our results demonstrate that the translational controlled ratio of C/EBPα and C/EBPβ isoform expression determines cell fate.
Keywords: CCAAT, enhancer binding protein, translational control, upstream open reading frame, cellular transformation, 3T3-L1 differentiation, proliferation
Transcription factors of the CCAAT/enhancer binding protein (C/EBP) family have decisive roles during differentiation in a number of cell types, including adipocytes, hepatocytes, enterocytes, keratinocytes, certain cells of the lung, mammary gland, the hematopoietic system, as well as in ovulation (Birkenmeier et al. 1989; Cao et al. 1991; Samuelsson et al. 1991; Lin and Lane 1992; Scott et al. 1992; Chandrasekaran and Gordon 1993; Piontkewitz et al. 1993; Freytag et al. 1994; Müller et al. 1995; Screpanti et al. 1995; Tanaka et al. 1995; Flodby et al. 1996; Pall et al. 1997; Sterneck et al. 1997; Swart et al. 1997; Zhang et al. 1997; Nerlov et al. 1998; Radomska et al. 1998). C/EBPs exert their function by regulating both the expression of tissue-specific genes and cell proliferation (Christy et al. 1989; Kaestner et al. 1990; Park et al. 1990; Umek et al. 1991; Lin et al. 1993; Ness et al. 1993; Buck et al. 1994; Constance et al. 1996; Oelgeschläger et al. 1996; Timchenko et al. 1997; McNagny et al. 1998; Buck et al. 1999; Müller et al. 1999). The importance of C/EBP proteins initially has been demonstrated in tissue culture model systems of adipogenesis and hematopoiesis (Lin and Lane 1992; Freytag et al. 1994; Hu et al. 1995; Müller et al. 1995; Nerlov et al. 1998) and has now been firmly established through analysis of the respective knockout mice (Screpanti et al. 1995; Tanaka et al. 1995; Wang et al. 1995; Flodby et al. 1996; Sterneck et al. 1997; Zhang et al. 1997).
Several C/EBPα and C/EBPβ protein isoforms corresponding to full-length and amino-terminally extended and truncated proteins can be detected in the cell. These isoforms display contrasting functions in gene activation and cell proliferation (Descombes and Schibler 1991; Lin et al. 1993; Ossipow et al. 1993; Buck et al. 1994; Calkhoven et al. 1994, 1997; Freytag et al. 1994; Sears and Sealy 1994; Nerlov et al. 1998; Kowenz-Leutz and Leutz 1999). Changes in the isoform ratio were observed in inductive cellular processes such as acute phase response (An et al. 1996), in liver development and liver regeneration (Diehl et al. 1994; Rana et al. 1995; Timchenko et al. 1998), in mammary glands during lactation (Raught et al. 1995), and in tumorigenic conversion (Raught et al. 1996). These findings suggest that the expression of the C/EBP protein isoforms is regulated and that the ratio of isoforms is important in proliferation and differentiation control.
Initially, differential translation initiation from internal AUG codons via a mechanism called leaky scanning of ribosomes was proposed to be responsible for the generation of truncated cellular C/EBP isoforms (Descombes and Schibler 1991; Lin et al. 1993; Ossipow et al. 1993). Recently, however, it has been suggested that limited proteolytic cleavage accounts for amino-terminally truncated C/EBP isoforms (Baer et al. 1998; Welm et al. 1999). In view of the importance of C/EBP proteins in the determination of cell fate, it is of considerable interest to reveal how C/EBP isoforms are generated and how this process is regulated.
Here we demonstrate that regulated initiation of translation from different sites is the prevailing mechanism for the generation of different protein isoforms from C/EBPα and C/EBPβ mRNAs. The regulated expression of different C/EBP isoforms depends on the integrity of evolutionary conserved upstream open reading frames (uORF) in both C/EBPα and C/EBPβ mRNAs. Signal transduction pathways regulating the function of the translation initiation factors eIF-2 and eIF-4E determine the ratio of C/EBP isoforms. Deregulated expression of truncated C/EBPα and C/EBPβ isoforms interferes with terminal differentiation and induces cell transformation in 3T3-L1 adipocytes. Hence, regulation of translation initiation that determines C/EBP isoform ratio has a crucial role in the control of cell proliferation and differentiation in C/EBPα- and C/EBPβ-expressing cells.
Results
Multiple C/EBPα and C/EBPβ protein isoforms are generated from different translation initiation sites
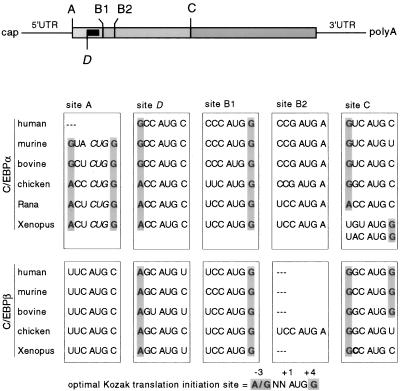
As shown in Figure 1, comparison of C/EBPα and C/EBPβ mRNAs from vertebrates revealed that the distribution and strength of potential translation initiation sites designated A, B1, B2, and C are highly conserved. All known C/EBPβ genes have an upstream translation initiation AUG codon (site A) that may give rise to an extended full-length protein isoform (Descombes and Schibler 1991; Kowenz-Leutz and Leutz 1999). In C/EBPα, an alternative CUG initiation codon is found at the A site, except in human C/EBPα (see Discussion). The most prominent C/EBP full-length translation products initiate at an AUG codon at site B1. In C/EBPα and in chicken C/EBPα a second initiation site, B2, follows a few codons downstream. Amino-terminally truncated C/EBPα and C/EBPβ isoforms may arise from translation initiation at the downstream site, C. Intriguingly, all vertebrate C/EBPα and C/EBPβ mRNAs contain an additional initiation site, D, between sites A and B1, from which a small uORF can be translated (site D, Fig. 1). A salient feature of this uORF is that it is always out of frame with respect to the C/EBP coding frame and terminates a few nucleotides 5′ of site B1.
Figure 1.
Representation of the vertebrate C/EBPα and C/EBPβ mRNA structure and comparison of the potential translation initiation sites. (Top) Shaded areas indicate the C/EBP coding region. The potential translation initiation sites are designated A, B1, B2, and C. The solid box represents the small uORF initiated at site D, which is out of frame with respect to the C/EBP reading frame. (Below) Shading indicates critical nucleotides at position −3 and +4 corresponding to the optimal Kozak translation initiation consensus sequence (bottom).
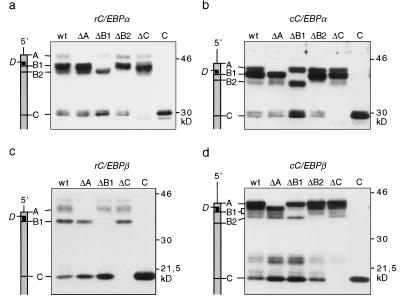
Translation initiation site null (Δ) mutations were introduced in C/EBPα and C/EBPβ cDNAs of rat and chicken to determine whether isoform expression is initiated from multiple translation initiation sites. The resulting expression plasmids were transiently transfected into COS-1 cells, and protein expression was examined by immunoblotting. As shown in Figure 2, mutations of A, B1, B2, or C into noninitiation sites abolished expression of corresponding C/EBPα and C/EBPβ isoforms. In addition, mutations that abrogated expression of the full-length isoforms simultaneously enhanced expression of truncated isoforms. This obvious lack of a precursor—product relationship suggested that C/EBP protein isoforms arise by differential usage of translation initiation sites rather than by limited proteolysis. To unequivocally distinguish between translational vs. proteolytic generation of isoforms, additional artificial initiation sites (X) were introduced, 20–37 codons upstream of site C (Fig. 3). The rationale was that a proteolytic mechanism should process the full-length precursors to truncated isoforms regardless of the presence of site X, whereas translation-based events should give rise to novel proteins that initiate at the X site instead of the wild-type C site. As shown in Figure 3, C/EBP constructs that contain the X site gave rise to larger truncated isoforms at the expense of wild-type truncated isoforms. These results confirm that C/EBP isoforms are generated by a conserved translational mechanism.
Figure 2.
Identification of C/EBPα and C/EBPβ translation initiation sites. Schematic representations of the C/EBP mRNAs with the initiation sites indicated in relation to the protein bands are shown at left. Wild-type cDNA (wt), constructs that lack distinct translation initiation codons (ΔA, ΔB1, ΔB2, ΔC, and ΔD) or an amino-terminal deletion construct (C) were transiently transfected in COS-1 cells. C/EBP protein expression was analyzed by immunoblotting of total cell extracts: (a) rC/EBPα; (b) cC/EBPα; (c) rC/EBPβ; (d) cC/EBPβ.
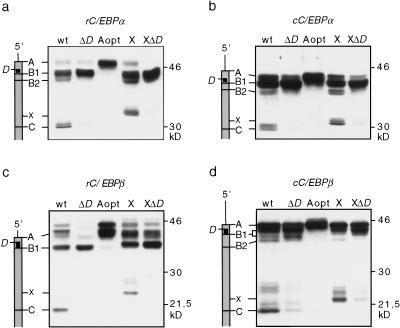
Figure 3.
Translation of truncated C/EBPα and C/EBPβ proteins from downstream initiation sites depends on the uORF. Schematic representations of the mRNAs with potential initiation sites indicated in relation to the protein bands are shown at left. COS-1 cells were transiently transfected with C/EBP wild-type (wt) and mutant constructs and analyzed for C/EBP protein expression by immunoblotting. In Δ_D_ mutants, the uORF translation initiation site is mutated to a noninitiation site. In the Aopt mutant, the sequence context of site A is mutated to an optimal Kozak sequence. The X mutants harbor an additional, optimal translation initiation site upstream of site C. (a) rC/EBPα; (b) cC/EBPα; (c) rC/EBPβ; (d) cC/EBPβ.
Previously, it has been suggested that usage of respective initiation sites in chicken C/EBPα (cC/EBPα) may depend on the small uORF between sites A and B1 (Calkhoven et al. 1994). Others suggested that the uORF determines the frequency of initiation from the B1 site only (Lincoln et al. 1998). To resolve whether the uORF directs translation from internal initiation sites, two types of C/EBPα and C/EBPα mutants were constructed and analyzed: (1) mutation of the uORF initiation site (Δ_D_); and (2) optimization of the translation initiation sequence at position A (Aopt), which should prevent leaky scanning of ribosomes to the downstream D site. Figure 3 shows that both types of mutations almost entirely abolished the translation of truncated isoforms from the wild-type C or the X site. In addition, the Δ_D_ mutation reduced translation from the upstream A site, whereas translation from B1 is enhanced [most clearly visible with rat C/EBPβ (rC/EBPβ)]. These results show that the uORF is essential for differential translation initiation from C/EBPα and C/EBPβ mRNAs. To rule out cell-type and species-specific effects, three other cell lines, quail (QT6) fibroblasts, 3T3-L1 pre-adipocytes, and human HeLa cells were examined for expression of C/EBPα and C/EBPβ wild-type and key mutants, Δ_D_, X, and XΔ_D_. Similar results were obtained in all three cell types (data not shown), suggesting that translational control of C/EBP isoform expression mediated by the uORF is conserved during evolution and is not cell type specific.
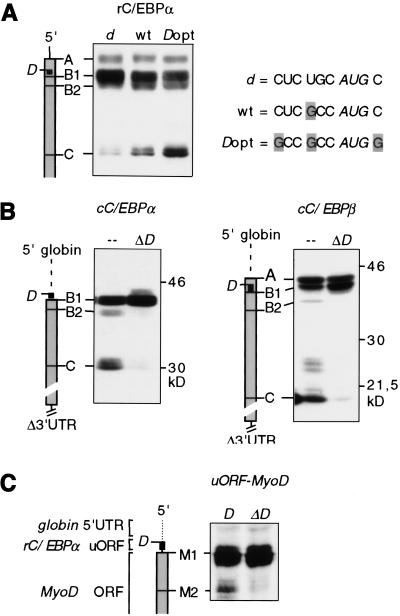
To examine whether the efficiency of the uORF site D selection for translation modulates C/EBP isoform expression we generated mutants of rC/EBPα in which site D, normally in a suboptimal context, was either mutated in a weaker non-Kozak site (d) or placed in stronger optimal Kozak context (_D_opt). Figure 4a shows that the C/EBPα isoform ratio shifts to a relatively more truncated isoform with the increase of site D strength. Hence, more efficient selection of uORF site D shifts the ratio of C/EBP isofom translation to a relatively more truncated isoform.
Figure 4.
The uORF regulates translation from downstream initiation sites. Schematic representations of the mRNAs are shown at left. (a) The C/EBPα isoform ratio is modified by the efficiency of the uORF translation. The wild-type suboptimal sequence context of site D (wt) was placed in an optimal Kozak consensus sequence (D_opt) or in a non-Kozak context (d). Initiation site sequences are shown at right. (b) cC/EBPα and C/EBPβ 5′ UTRs were replaced by a 50-bp sequence of the β-globin leader, and the 3′ UTRs were deleted. The uORF translation initiation site D was left intact (–) or was mutated to a noninitiation site (Δ_D). (c) The Flag-tagged MyoD coding region was placed downstream of a 5′ β-globin leader sequence and the rC/EBPα uORF sequence (D) or the uORF initiation site mutant (Δ_D_) as indicated. All constructs were transiently transfected in COS-1 cells. C/EBP or MyoD protein expression was analyzed by immunoblotting.
Next, we examined whether regulatory elements in the 5′- and/or 3′-untranslated regions (UTRs) would be required in addition to the C/EBP uORF to direct differential initiation of translation. The 5′ and 3′ UTRs were deleted or exchanged against unrelated sequences of the globin-3 gene, as shown in Figure 4b. It is evident that expression of truncated C/EBP isoforms initiated at internal C sites was maintained regardless of the UTRs. Subsequent mutation of the uORF initiation site confirmed that the uORF remained crucial for translation of truncated isoforms (Fig. 4b, Δ_D_).
Finally, we determined whether the C/EBPα uORF could autonomously direct translation initiation to internal sites. To do so, the rC/EBPα uORF was placed in front of the coding sequence of MyoD (uORF–MyoD). As shown in Figure 4c, a truncated MyoD protein was generated in addition to the full-length protein from the uORF–MyoD construct. No such truncated MyoD protein was found when site D was mutated nor with wild-type MyoD (data not shown). Taken together, these results show that the C/EBP uORF is required for translation initiation at multiple sites and that it may function as an autonomous _cis_-regulatory mRNA element.
Translation regulation pathways control C/EBP protein isoform expression
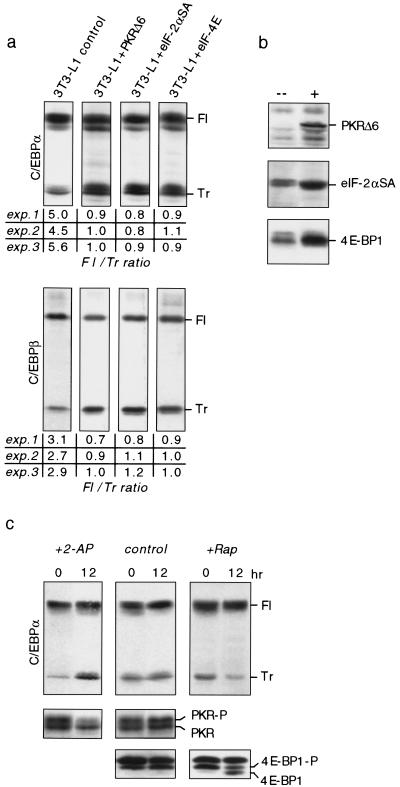
Initiation of translation is affected by a number of pathways that control the activity and level of eukaryotic translation initiation factors (eIFs) (Hershey 1991; Morris 1995). The RNA-dependent protein kinase (PKR) affects translation initiation by phosphorylation-induced inactivation of the rate-limiting eIF-2α, a component of the eIF-2 holocomplex (Meurs et al. 1990). We asked whether interference with PKR or eIF-2 function would modify C/EBPα and C/EBPβ protein isoform ratios. For this purpose we employed the pre-adipocyte 3T3-L1 cell line, which undergoes C/EBPα and C/EBPβ dependent differentiation upon hormone treatment (Yeh et al. 1995b). In these cells the eIF-2 pathway was constitutively activated by retroviral introduction of a kinase-inactive and dominant-negative PKR mutant (PKRΔ6) (Koromilas et al. 1992) or the eIF-2α mutant S52A (Choi et al. 1992), which cannot be phosphorylated and thus resists inactivation by PKR. Transgene-expressing and control (empty vector) 3T3-L1 cultures were subjected to a standard differentiation protocol and analyzed for C/EBPα and C/EBPβ protein isoform expression by immunoblotting. Figure 5a shows that ectopic expression of either PKRΔ6 or eIF-2αS52A, shifts C/EBPα and C/EBPβ isoform expression toward the truncated isoform. A similar shift in C/EBP isoform expression was obtained by treating differentiated 3T3-L1 adipocytes with the PKR inhibitor 2-aminopurine (2-AP) (De Benedetti and Baglioni 1983). As shown in Figure 5c, concomitantly with 2-AP-induced dephosphorylation and inactivation of PKR, expression of truncated C/EBPα isoform was enhanced. These results show that high eIF-2 activity shifts the ratio of C/EBP isoform expression toward a more truncated isoform.
Figure 5.
The ratio of C/EBPα and C/EBPβ protein isoform expression is modulated by translation initiation factor activity. (a) 3T3-L1 cells ectopically expressing PKRΔ6, eIF-2αS52A, eIF-4E, and control cells (empty vector) were induced to undergo adipogenesis and analyzed for expression of endogenous C/EBPα and C/EBPβ isoforms at day 4 of the differentiation protocol by immunoblotting. (Fl) Full-length isoform; (Tr) truncated isoform. Ratios of full-length vs. truncated C/EBP isoforms of three independent experiments are depicted below; immunoblots of experiment 1 are shown. (b) PKRΔ6, eIF-2αSA, and eIF-4E transgene expression were analyzed by immunoblotting in transgene (+) and control (–) cells. (c) 3T3-L1 cells at day 4 of differentiation were treated with 5 mm 2-aminopurine (+2-AP) or 1 μm rapamycin (+Rap) for 12 hr and compared with control cultures for rC/EBPα protein expression. Bottom panels show protein expression of endogenous PKR and 4E-BP1. The respective hyperphosphorylated forms are indicated (-P).
The FKBP12–rapamycin-associated protein (FRAP)/mammalian target of rapamycin (mTOR) (Brown et al. 1994) enhances the level of accessible eIF-4E (Lawrence and Abraham 1997): mTOR phosphorylates and inhibits phosphatase PP2A, which keeps the inhibitory 4E-binding protein 1 (4E-BP1, also called PHAS1) in an active unphosphorylated state (Lin et al. 1994; Pause et al. 1994; Brunn et al. 1997; Peterson et al. 1999). We generated 3T3-L1 cells that overexpressed eIF-4E by retroviral transfer and subjected them to the standard differentiation protocol. As shown in Figure 5a, enhanced levels of eIF-4E shift C/EBPα isoform expression toward the truncated isoform. On the other hand, when mTOR was inhibited by rapamycin, expression of the truncated C/EBPα isoform was reduced concomitantly with the dephosphorylation of 4E-BP1 (Fig. 5c). Similar results were obtained with C/EBPβ (data not shown). In conclusion, two rate-limiting translation initiation factors control the ratio of C/EBP isoforms: At high eIF-2 and eIF-4E activity relatively more truncated C/EBP isoforms are expressed, whereas at lower eIF activity expression of the full-length isoforms dominates.
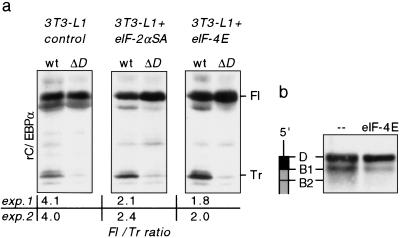
To investigate the role of the uORF in conjunction with eIF activity for the regulation of C/EBPα and C/EBPβ isoform expression, we transiently transfected undifferentiated 3T3-L1 control cells and 3T3-L1 cells expressing eIF-2αSA or eIF-4E transgenes with C/EBPα wild-type or the corresponding mutated uORF (Δ_D_) expression constructs. Figure 6a shows that the expression of truncated C/EBP isoforms depends on the uORF under conditions of enhanced eIF activity as well. eIF-4E has been implicated in the selection of upstream initiation sites in bicistronic messengers (Tahara et al. 1991). We examined the effect of increased eIF-4E levels on translation initiation from the uORF site D. To do so we fused the uORF of C/EBPα in frame to the C/EBPα coding sequence and tested the effect of eIF-4E on translation initiation from D and B1. As shown in Figure 6b, overexpression of eIF-4E enhanced translation initiation from the uORF initiation site D at the expense of initiation from site B1. These results indicate that the uORF is crucial for the modulation of C/EBPα isoform ratio through eIF activity.
Figure 6.
The uORF is essential for the modulation of C/EBP isoform expression through eIF activity in 3T3-L1 cells. (a) Undifferentiated 3T3-L1 control cells and cells expressing eIF-2αS52A or eIF-4E were transiently transfected with wild-type rC/EBPα expression constructs (wt) or constructs with mutated uORF initiation site (Δ_D_). Cell extracts were analyzed for rC/EBPα expression by immunoblotting. (Fl) Full-length isoform; (Tr) truncated isoform. Ratios of full-length vs. truncated C/EBP isoforms of two independent experiments are depicted below; immunoblots of experiment 1 are shown. (b) A construct expressing a uORF–rC/EBPα fusion protein was cotransfected with an eIF-4E expression construct or empty vector (–) in COS-1 cells. uORF–rC/EBPα fusion protein expression was analyzed by immunoblotting of total cell extracts. A schematic representation of the mRNA is shown at left.
Enhanced expression of the truncated C/EBPα or C/EBPβ isoform alters proliferation and differentiation in 3T3-L1 cells
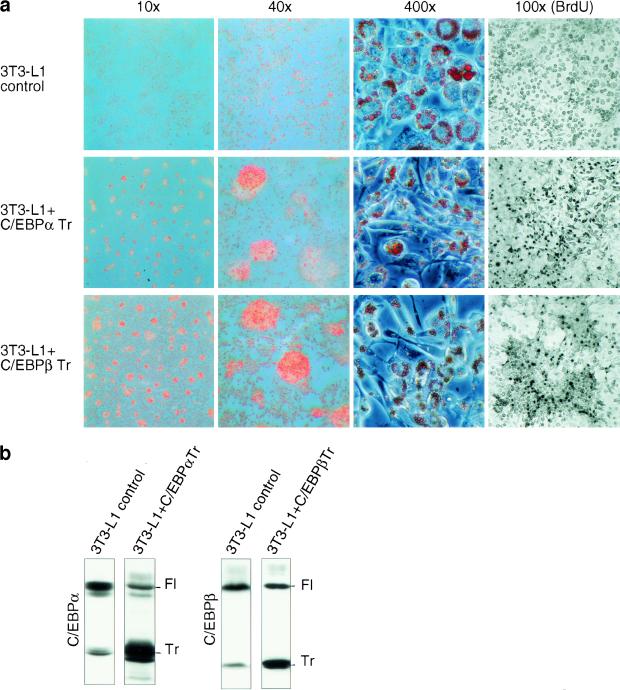
Terminal differentiated 3T3-L1 adipocytes form monolayers of contact-inhibited fat cells that store large amounts of lipids. We noticed that interference with translational control pathways altered the differentiation program of 3T3-L1 cells. Increased eIF activity reduced contact inhibition and resulted in foci formation in differentiating cultures (data not shown). To determine whether deregulated expression of C/EBP isoforms might account for the altered growth properties, we stably introduced truncated C/EBPα or C/EBPβ isoforms by retroviral gene transfer and induced adipogenesis by the standard protocol. As shown in Figure 7, sustained ectopic expression of truncated C/EBPα or truncated C/EBPβ isoform gave rise to a heterogeneous population of predominantly small and spindle-shaped cells that were poorly differentiated. The cells displayed reduced adherence to the culture dish, formed foci, and continued to multiply during differentiation, as shown by BrdU incorporation. Thus, up-regulation of truncated C/EBP isoforms prevents proliferation arrest and contact inhibition, interferes with terminal adipogenic differentiation, and induces a transformed phenotype. Apparently, proper translational regulation of C/EBP isoform expression is a prerequisite for terminal fat cell differentiation and proliferation arrest.
Figure 7.
Constitutive ectopic expression of truncated C/EBPα or C/EBPβ induces cellular transformation in differentiating 3T3-L1 cells. (a) 3T3-L1 control cells and 3T3-L1 ectopically expressing truncated rC/EBPα or truncated rC/EBPβ were induced to undergo adipogenesis. At day 8 of differentiation cells were fixed and stained with Oil Red O (lipid staining). Microscopic pictures of different magnifications (as indicated) are shown. In parallel experiments the cells were incubated for 8 hr with BrdU, fixed and stained by immunohistochemistry (right). (b) Ectopic and endogenous expression of C/EBP protein isoforms was analyzed by immunoblotting. (Fl) Full-length isoform; (Tr) truncated isoform.
Discussion
Aberrant translational control of C/EBPα and C/EBPβ isoform expression disrupts differentiation and induces cellular transformation
In this paper we show that the availability and activity of translation initiation factors determine the translation of C/EBPα and C/EBPβ mRNAs into different transcription factor isoforms. The individual C/EBPα and C/EBPβ protein isoforms have different biological activities that determine cell fate. Full-length C/EBPα and C/EBPβ isoforms, translated from the B1 site, are transcriptional activators and induce differentiation and cell-cycle arrest in various cell types (Ness et al. 1993; Ossipow et al. 1993; Buck et al. 1994; Freytag et al. 1994; Müller et al. 1995; Wu et al. 1995; Nerlov et al. 1998). Recently, it was shown that the extended C/EBPβ isoform, generated from the A site, may recruit the chromatin remodeling SWI/SNF complex to activate a different set of target genes in comparison to full-length C/EBPβ (Kowenz-Leutz and Leutz 1999). Truncated isoforms, initiated at the C site, display little if any transactivation and permit, or even induce, the cell cycle to proceed (Lin et al. 1993). Furthermore, the truncated isoforms, even at substoichiometric levels, may counteract the functions of full-length isoform levels (Descombes and Schibler 1991; Ossipow et al. 1993; Raught et al. 1995; Calkhoven et al. 1997). Thus, the biological effect evoked by C/EBPα and C/EBPβ proteins will depend strongly on the ratio of their isoforms. Full-length C/EBP isoforms prevail in differentiating cells both in vivo and in tissue culture, reflecting proliferation arrest and expression of tissue-specific C/EBP target genes. Enhanced expression of the truncated isoform results in a transformed phenotype of 3T3-L1 adipocytes. These cells lost contact inhibition and continued to proliferate during differentiation, which suggests that down-regulation of truncated C/EBPα isoforms is essential for cessation of proliferation and terminal differentiation. Constitutive activation of eIF-2 or eIF-4E, which deregulated C/EBPα and C/EBPβ isoform expression, induced a similar transformed phenotype in 3T3-L1 adipocytes.
Deregulated eIF activity has been shown to transform fibroblasts (Koromilas et al. 1992; Donze et al. 1995). Although deregulated eIF activity will affect translation of many genes, it is tempting to speculate that inadequate adjustment of C/EBP isoform ratios contributes to neoplastic conversion in tissues that express C/EBPα and/or C/EBPβ. A correlation has been found between enhanced eIF-2 activity and enhanced truncated C/EBPβ expression in mammary epithelial cancer cells (Raught et al. 1996). In addition, up-regulation of eIF-4E is an early event in colon cancer cells known to express C/EBPs (Rosenwald et al. 1999). Also, the myxoid liposarcoma-specific chromosomal rearrangement of the CHOP gene, TLS–CHOP, which is required for oncogenic transformation, interferes with C/EBP function (Zinszner et al. 1994).
Differential initiation of translation generates different C/EBP isoforms
Initially, truncated C/EBPα and C/EBPβ isoforms were proposed to arise by a ribosomal scanning mechanism to alternative translation initiation sites in their mRNAs (Descombes and Schibler 1991; Lin et al. 1993; Ossipow et al. 1993; An et al. 1996). In contrast, limited proteolysis recently has been suggested to account for the generation of truncated isoforms (Baer et al. 1998; Welm et al. 1999). We showed by a mutagenesis approach that different C/EBPα and C/EBPβ protein isoforms originate from all evolutionary conserved translation initiation sites (termed A, B1, B2, and C; Fig. 1). Elimination of upstream initiation sites increases expression of truncated C/EBP isoforms. Moreover, introduction of novel initiation sites (X) between B and C sites results in production of novel isoforms initiated at the X rather than at the C site. Shift of translation initiation to downstream initiation sites once upstream sites were removed, and substitution of the wild-type truncated products (C) by alternative products once an extra initiation codon (X) was introduced argue strongly in favor of a translational mechanism in the generation of truncated C/EBP isoforms. Although we cannot rule out that limited proteolysis of C/EBPs might occur under specific conditions (Welm et al. 1999), our results are incompatible with precursor–product relationships between full-length and truncated proteins and consistent with observations reported by Sears and Sealy (1994), who also failed to find a precursor–product relationship by pulse-chase labeling. Additional experiments, using bicistronic contructs also ruled out the possibility that truncated proteins are generated by internal ribosomal entry sites in C/EBP mRNAs (data not shown). We conclude that translational control is the prevailing mechanism of C/EBPα and C/EBPβ isoform expression in vertebrates.
The conservation of translation initiation site distribution and sequence context further supports the experimental data. The sequence context of initiation codons determines the fidelity and frequency of their selection, as has been described by Kozak (1989). Only two deviations from the conserved initiation sites were found. First, the A site in human C/EBPα is apparently absent. However, an immunoreactive band corresponding to an extended isoform was detected in extracts of the human HL-60 leukemia cell line, indicating that translation may be initiated from a functionally equivalent A site (a GUG codon; C.F. Calkhoven, unpubl.). The second exception is that a B2 site is present in all C/EBPα isoforms examined and in cC/EBPβ but not in mammalian and Xenopus laevis C/EBPβ.
The C/EBP uORF mediates differential translation initiation
A small uORF is located between the A and the B sites in all vertebrate C/EBPα and C/EBPβ mRNAs. The uORF is out of frame with respect to the C/EBP reading frame and terminates just upstream of the major initiation site B1. Fusion of the uORF to the C/EBP reading frame revealed that site D is selected for translation. Our data show that the uORF has a crucial role in the regulation of isoform expression. Three lines of evidence show that the uORF is paramount for translation initiation at downstream initiation sites. First, different types of mutations that disrupt the function of the uORF concomitantly abrogate translation initiation at downstream sites. Second, the efficiency of translation from site C is proportional to the efficiency of site D selection. Third, the uORF mediates translation from downstream initiation sites in a different mRNA context (MyoD transcript) and thus displays an autonomous function.
The observation that removal of the uORF initiation codon abolished initiation at the C site (or X site) and optimization of the uORF site D enhances initiation at C indicates that uORF translation is required for its function. This implies that translation reinitiation rather than leaky scanning is the mechanism of downstream initiation at C (or X). Translation from site B1 is not dependent on the uORF. In contrast, it is inversely regulated to the strength of the D site. This indicates that translation from B1 mainly results from leaky scanning over the wild-type D site (or the weak mutant site d). However, the optimization of the D site (_D_opt), which does not allow leaky scanning of ribosomes (data not shown), diminishes but does not completely abolish translation from B1 (as one would expect with leaky scanning as the only mechanism). Hence, immediate reinitiation after uORF translation seems to occur as well. Taken together, translation from site C appears to depend strictly on uORF translation, whereas initiation at site B1 can occur by either leaky scanning or by immediate reinitiation.
Removal of the D site also revealed that the uORF is involved in regulation of initiation at site A. We presume that under certain cellular conditions translation of the uORF may cause cueing of ribosomes and thus increase the probability of initiation at the weak initiation site A. Although several details need to be solved, the data show that a conserved uORF in vertebrate C/EBPα and C/EBPβ mRNAs directs the translation from multiple translation sites.
Translational control and cell fate
Kinases PKR and mTOR, implicated in C/EBP isoform translation, are involved in many cellular processes. A major function of PKR is to mediate translational repression after virus infection and interferon signaling (Wek 1994). However, regulating PKR activity also appears to be essential for normal cell proliferation and differentiation. Others showed that both the inactivation of PKR in quiescent cells or ectopic expression of the PKR phosphorylation-defective eIF-2αS52A mutant induces proliferation and even transformation (Koromilas et al. 1992; Meurs et al. 1993; Barber et al. 1994; Donze et al. 1995). We also observed loss of contact inhibition and persistent proliferation in differentiating 3T3-L1 cells expressing dominant-negative PKRΔ6 or eIF-2αS52A (data not shown). Augmentation of rate-limiting eIF-2α enhanced expression of truncated C/EBP forms in an uORF-dependent manner. A paradigm of eIF-2 function and translational control is provided by the yeast transcription factor GCN4. In yeast, high levels of eIF-2 mediate multiple translation reinitiation events after initial uORF translation (Mueller and Hinnebusch 1986; Hinnebusch 1994). In a similar fashion, elevated eIF-2 levels might enhance translation reinitiation at C/EBP C sites after translation of the uORF.
The FRAP/mTOR kinases are involved in the regulation of several cellular processes, including cell proliferation, transcriptional response to nutrients, and mRNA translation (Brown et al. 1994; Zheng and Schreiber 1997; Peterson and Schreiber 1998; Beck and Hall 1999; Cardenas et al. 1999; Dennis et al. 1999; Hardwick et al. 1999; Kuruvilla and Schreiber 1999). Our data show that inhibition of FRAP/mTOR function by rapamycin reduced expression of the truncated C/EBP isoforms, whereas overexpression of eIF-4E enhanced expression of the truncated isoforms in an uORF-dependent manner. Others have shown that eIF-4E is implicated in selection of upstream initiation sites in bicistronic messengers (Tahara et al. 1991). Accordingly, we observed enhanced translation initiation from uORF site D upon overexpression of eIF-4E (Fig. 6b). As the translation of the uORF determines reinitiation at the downstream site C (Fig. 4a), it is presumably eIF-4E-mediated enhanced _D_-site usage that leads to increased expression of truncated C/EBP isoforms. Our data are also in accordance with that of Yeh et al. (1995a), who showed that treatment of 3T3-L1 cells with rapamycin at the onset of the differentiation program inhibits their clonal expansion, a prerequisite for 3T3-L1 differentiation. It is possible that the observed repression of truncated C/EBP isoforms is responsible for failure of clonal expansion.
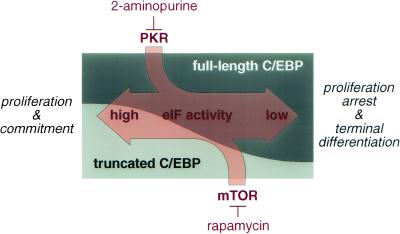
Taken together, pathways that modulate the activity of eIF-2 and eIF-4E alter the ratio of C/EBP isoforms in conjunction with their uORF. According to the working model shown in Figure 8, pathways that activate translation (e.g., by various growth factors) concomitantly enhance expression of truncated C/EBP isoforms, which in turn supports proliferation. In contrast, pathways that decrease eIF functions will preferentially shift C/EBP expression to full-length isoforms and thus support growth arrest and differentiation. We suggest that the C/EBP uORF acts as a mediator for translation factor activity to adjust the ratio of C/EBP isoforms accordingly and so determine cell fate.
Figure 8.
Translation initiation factor activity determines cell fate through modulation of the C/EBP isoform ratio.
It is noteworthy that in addition to C/EBPs, a number of other key regulatory proteins involved in proliferation and differentiation are regulated at the translational level. Among these proteins are the CDK inhibitor p27 (Hengst and Reed 1996), cyclin D1 (Rousseau et al. 1996), CLN3 (Polymenis and Schmidt 1997), thrombopoietin (Ghilardi et al. 1998), PDGF2 (Bernstein et al. 1995), BCL-2 (Harigai et al. 1996), AdoMetDC (Hill and Morris 1993), and c-Myc (Hann et al. 1992). Fewer than 10% of vertebrate mRNAs have upstream initiation codons and/or uORFs. However, two-thirds of the transcripts encoding growth regulatory proteins (growth factors, cytokines, oncogenes, etc.) have such features (Kozak 1987, 1991). An attractive possibility, therefore, is that the expression of mRNAs encoding critical proteins that determine cell fate are restricted in their translation to permissive eIF activities that are under environmental control. Only in a specific window of eIF activity such proteins are generated or their composition adjusted to allow proliferation or differentiation. It is evident that such a safeguard, which controls entry into the cell cycle, independently of transcriptional regulation, is also prone to tumorigenic conversion.
Materials and methods
DNA constructs
C/EBP constructs
All mutations were generated by site-directed mutagenesis following the method of Kunkel (Ausubel et al. 1993; Kunkel et al. 1991). cC/EBPα mutants were generated on the cC/EBPαwt–pSG5 template (Calkhoven et al. 1994) with oligonucleotide primers ΔA (accctgg → ccccggg), 5′-ccgccccgtccgaccccgggtttgccggagccc-3′; ΔB1 (ttcatgg → ttcctcg), 5′-ggctgtaggtgcttcctcgagcaagccaacttc-3′; ΔB2 (ccgatga → cggggag), 5′-cccggcccccggggagcagcggccagcaccacc-3′; ΔC (ggcatgc → gggatcc), 5′-ttccacgggatccacggggcc-3′; ΔD (accatgc → aggatcc), 5′-ccgggcccttcaggatccccggcaggctg-3′; Aopt (cgaaccctgg → gccaccatgg), 5′-cgcgccccgtcgccaccatggattgccggagc-3′; and X (gacctct → gccatgg), 5′-cgagttcctggccgccatggtccagcacagcaagc-3′. The C mutant was generated by introducing an _Eco_RI site with primer 5′-ggggatttcgaattccacggcatg-3′ and subsequent removal of the Eco_RI fragment containing the 5′ UTR and amino-terminal C/EBP sequences. For the cC/EBPα–5′ β-globin/Δ3′ UTR and corresponding Δ_D mutants a _Hin_dIII site was introduced upstream of site D with primer 5′-gcagagccgccgcaagcttgtccgaaccctgg-3′, and the _Kpn_I–_Hin_dIII 5′ UTR fragment was exchanged for the _Kpn_I–_Hin_dIII fragment of pBAT (Annweiler et al. 1991) containing the β-globin leader sequence. The 3′ UTR was removed by _Apa_I–_Bam_HI digestion. A cC/EBPβ (NF-M) _Eco_RI cDNA fragment (Katz et al. 1993) was cloned into pSG5, and mutants were generated on the cC/EBPβwt–pSG5 template with the primers ΔA (ttcatgc → tgaattc), 5′-ccccctttgcttgaattcaacgcctggtgg-3′; ΔB1 (tccatgg → tccattgaa), 5′-cgcctttaaatccattgaagtggctaatttctattacgaggcgg-3′; ΔB2 (tccatga → tctagaa), 5′-gggccgctctagaaccgaacttaccgtagg-3′; ΔC (ggcatgt → ggaattc), 5′-ggaccggggggaattccctcgccctacggc-3′; ΔD (agcatgc → agcaagc), 5′-ggcctgggacgcagcttgcctccccattcagcc-3′; Aopt (gctttcatgc → gccaccatgg), 5′-gcatccccctttgccaccatggaacgcctggtggcc-3′; _D_opt (cgcagcatgc → gccaccatgg), 5′-ggtggcctgggcgccaccatggctccccattcagcc-3′; and X (gagaccctgg → gccaccatgg), 5′-ggagccggtcttcgccaccatggactcttgcaaagg-3′. The C mutant was generated by introducing an _Eco_RI site with primer 5′-ccgtaaggaagaattcggagcggggccagg-3′ and subsequent removal of the Eco_RI fragment containing the 5′ UTR and amino-terminal cC/EBPβ sequences. For cC/EBPβ–5′ β-globin/Δ3′ UTR and corresponding Δ_D mutants a _Hin_dIII site was introduced upstream of site D with primer 5′-ccgtcttctcctccaagcttccccctttgc-3′, and the _Kpn_I–_Hin_dIII 5′ UTR fragment was exchanged for the _Kpn_I–_Hin_dIII fragment of pBAT (Annweiler et al. 1991) containing the β-globin leader sequence. The 3′ UTR was removed by _Bam_HI–_Eco_RI digestion after introduction of a _Bam_HI site downstream of the C/EBPβ stop codon with primer 5′-cgctgctgaccccggatccggccgcgc-3′. A rC/EBPα wild-type clone containing the cDNA sequence was generated by cloning a 590-bp _Eco_RI–_Not_I PCR fragment (primers, 5′-ccggaattccattcgcgacccaaagctgcg-3′ and 5′-cgcggatccgatctggaactgcaagtgaggg-3′) from genomic DNA containing the 5′UTR, together with the _Not_I–_Bam_HI rC/EBPα cDNA fragment (Landschulz et al. 1988) into pSG5, which was used as template for further mutagenesis with the primers ΔA (gtactgg → cggatcc), 5′-gggcgagttgggcggatccgtgggcggcgg-3′; ΔB1 (cccatgg → cccatcg), 5′-ctctaactcccccatcgagtcggccgac-3′; ΔB2 (ccgatga → cggatcc), 5′-cggccccggatccgcagccacctcc-3′; ΔC (gtcatgt → gtgaatt), 5′-ggcggtgcggtgaattccgcgggggcgcacgg-3′; ΔD (gccatgc → gggatcc), 5′-ccgaggctcgggatcccgggagaactctaactccc-3′; Aopt (ggggtactgg → gccgccatgg), 5′-gggcgagttgccgccatgggtgggcggcgg-3′; d (gccatgc → tgcatgc), 5′-ccgccgaggctctgcatgccgggagaactc-3′; _D_opt (ctcgccatgc → gccgccatgg), 5′-gctggaggccgtcgacggccgccatggcgggagaactctaactcc-3′; X (gccgacctct → gccgccatgg), 5′-cgagttcctggccgccatggtccagcacagccggc-3′; and uORF+C/EBPα fusion (taactc → aactc), 5′-ccatgccgggagagctcaactcccccatgg-3′. The C mutant was generated by cloning a 725-bp _Eco_RI–_Bam_HI PCR fragment (primers 5′-gcgaattcatgtccgcgggggcgcacggacc-3′ and 5′-gcggatcctcacgcgcagttgcccatggccttgacc-3′) into pcDNA3. A rat C/EBPβwt–pSG5 vector was generated by cloning the IL-6–DBP Eco_RI cDNA fragment (Poli et al. 1990) into pSG5 and used as template for further mutagenesis with the primers ΔA (ttcatgc → tgaattc), 5′-ggccccgcgtgaattcaccgcctgctggcc-3′; ΔB1 (cccatgg → cccattg), 5′-gcctttagacccattgaagtggccaacttc-3′; ΔC (gccatgg → gcgatcg), 5′-cgacgcgcccgcgatcgcggccggcttccc-3′; Δ_D (agcatgc → agaattc), 5′-ggcctgggacgcagaattcctcccgccgcc-3′; Aopt (gcgttcatgc → gccaccatgg), 5′-gggccccgccaccatggcccgcctgctggc-3′; and X (gccgcactca → gccgccatgg), 5′-gcctcccgccgccatggaggccgagccggg-3′. The C mutant was generated by removal of the Eco_RI fragment containing the 5′ UTR and amino-terminal C/EBP sequences from the rC/EBPβX–pSG5 construct. The MyoD–uORF–pSG5 [gccatgccgggagaactctaa (uORF) ctcccccatgg (MyoD)] and MyoD–ΔuORF–pSG5 [ggg atc (Δ_D uORF) ccgggagaactctaactcccccatgg (MyoD)] vectors were generated by combined cloning of a Kpn_I–_Nco_I fragment containing the β-globin leader and rC/EBPα–uORF or –Δ_D sequences (from the cC/EBPα–5′ β-globin/Δ3′ UTR mutants), together with a _Nco_I–_Xba_I fragment containing amino-terminal Flag-tagged human MyoD coding region into pSG5.
pBabe–puro retroviral constructs
rC/EBPαTr–pBabe–puro was generated by cloning the _Eco_RI–_Bam_HI fragment from rC/EBPαC–pSG5 into pBabe–puro (Morgenstern and Land 1990). rC/EBPβpBabe–puro was generated by cloning the _Eco_RI fragment from rC/EBPβC–pSG5 into pBabe–puro. A human eIF-2α _Eco_RI–_Hin_dIII 1.6-kb fragment was cloned from SP65-2a (Ernst et al. 1987) into pSG5. eIF-2αwt–pSG5 was used for creating the S52A mutant (Choi et al. 1992) with the primer 5′-cttagtgaattggccagaaggcgtatccg-3′. The _Bam_HI eIF-2αS52A fragment from eIF-2αS52A–pSG5 was cloned into pBabe–puro. Human PKR (Meurs et al. 1990) was cloned by PCR from HeLa cells using the primers 5′-gggaatcaacatccacacttccg-3′ and 5′-gggagactgtgtcattgcactcc-3′, tagged with _Bam_HI sites and cloned into pSG5. The dominant-negative PKRΔ6 mutant (Koromilas et al. 1992) was generated on PKR–pSG5 with primer 5′-ggtcaaagactaagtgcttctgtgataaagggaccttgg-3′, and the _Bam_HI fragment was cloned into pBabe–puro. Human eIF-4E (Rychlik et al. 1987) was cloned by PCR from HeLa cells using the primers 5′-gattcagatcgatctaagatgg-3′ and 5′-cctatgagaatactcagaagg-3′, tagged with _Bam_HI sites, and cloned into pBabe–puro.
Cells and tissue culture
COS-1 cells (ATCC, CRL-1650) were propagated in DMEM, F12, and 5% FCS (GIBCO); Hela cells (ATCC, CCL-2) in DMEM and 10% FCS; 3T3-L1 cells (ATCC, CL-173) in DMEM and 10% FCS (Seromed); QT6 cells (ATCC, CRL 1708) in DMEM, 8% FCS, and 2% heat-inactivated chicken serum; and Phoenix E cells (G.P. Nolan, Stanford University School of Medicine, Stanford, CA; ATCC, SD 3444) in DMEM and 10% FCS in a humidified atmosphere with 5% CO2 at 37°C. Induction of adipogenetic differentiation in 3T3-L1 cells was induced in 2-day confluent cultures (designated day 0) with 2 days of incubation in medium containing 10 μg/ml insulin (Sigma), 1 μm dexamethasone (Sigma), and 0.5 mm 3-isobutyl-1-methylxanthine (Sigma) (days 1–2), followed by incubation in 10 μg/ml insulin with medium exchange every second day (days 3–8) (Yeh et al. 1995b). Medium for pBabe–puro-infected 3T3-L1 cells contained an additional 0.5 μg/ml puromycin (Sigma). Rapamycin (Calbiochem) was used in a concentration of 1 μm, and 2-amunopurine (Calbiochem) was used in a concentration of 5 mm.
Oil-Red-O staining
3T3-L1 cells were washed with PBS, fixed with 4% paraformaldehyde overnight at 4°C, stained with Oil-Red-O solution for 5 min [2:3, 0.3% (wt/vol) Oil Red O (Sigma) in isopropanol and water before filtering], and analyzed by bright-field microscopy.
BrdU labeling
3T3-L1 cells were labeled with BrdU for 8 hr following the manufacturer's protocol (BrdU Labeling and Detection Kit II, Boehringer Mannheim).
Retroviral methods
The ecotropic-packaging cell line Phoenix E was transiently transfected with the calcium phosphate–DNA precipitation method, and infectious virus was harvested after 48 hr. 3T3-L1 target cells (5 × 105) were infected as described in Pear et al. (1993) and selected for puromycin (2 μg/ml) resistance.
Transfections
COS-1 cells were transfected with 5 μg of pSG5-based expression vector using DEAE–dextran/chloroquine as described by Gonzalez and Joly (1995). HeLa and 3T3-L1 cells were transfected with 0.75 μg of pSG5-based expression vector using GenePORTER (Gene Therapy Systems, Inc) following the protocol of the manufacturer in six-well culture trays. Transfected cells were harvested 24 hr after transfection. QT6 cells were transfected with 5 μg of pSG5-based expression vector using the calcium phosphate–DNA precipitation method as described in Ausubel et al. (1993).
Western blot analysis
Cells were lysed rapidly in 0.5 m NaOH, neutralized by adding 0.5 m HCl, or directly lysed in RIPA buffer (150 mm NaCl, 50 mm Tris-HCl at pH 7.5, 1% NP-40, 0.1% SDS), supplemented with SDS loading buffer, sonicated, and boiled. The proteins were separated on a 12.5% SDS–polyacrylamide gel and electroblotted on PVDF membrane (Immobilon-P, Millipore). Western blot analysis was performed, as described in Calkhoven et al. (1994), followed by luminescent detection according to the manufacturer's protocol (Amersham Life Technologies, ECL system). The following antisera were used: 1:1500 cC/EBPα (Calkhoven et al. 1994) and 1:3000 cC/EBPβ/NF-M (Katz et al. 1993); 0.5 μg/ml rC/EBPα (14AA), rC/EBPβ (C-19), PKR (M-515), PKR (K-17), 4E-BP1 (R-113), and eIF-2α (C-20); 1:2000 anti-goat immunoglobulin HRP (Sc-2020) (all from Santa Cruz Biotechnology Inc.); 0.5 μg/ml eIF-4E (E27620) (Transduction Laboratories); 10 μg/ml anti-Flag M2 (IB13026) (Eastman Kodak Company); 1:2000 anti-mouse immunoglobulin HRP (NA931) and 1:5000 anti-rabbit immunoglobulin HRP (NA934) (Amersham Life Technologies). Protein bands were quantified using the FUJIFILM Science Lab/Image Gauge computer program.
Acknowledgments
We offer special thanks to Marion Bengs for extensive technical assistance. We thank Dr. Garry P. Nolan for providing the Phoenix E cells, Dr. Adri A.M. Thomas for providing an eIF-2α clone, Dr. Steven L. McKnight for providing a rC/EBPα clone, Valeria Poli for providing an IL-6DBP clone, Dr. Barbara Winter for providing a MyoD clone, Dr. Stéphane Ansieau for the Flag-tagged MyoD construct, and Dr. Dipak Ramji for X. laevis C/EBPβ sequence information. This research is supported by the Deutsche Forschungsgemeinschaft by grants to A.L. (LE77072-2/LE770/3-1). C.F.C was supported by a Marie Curie /TMR fellowship (ERBFMBICT961254).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL aleutz@mdc-berlin.de; FAX 49-(0)-30-9406-3298.
E-MAIL calkhov@mdc-berlin.de; FAX 49 (0) 30 9406 3298.
References
- An MR, Hsieh CC, Reisner PD, Rabek JP, Scott SG, Kuninger DT, Papaconstantinou J. Evidence for posttranscriptional regulation of C/EBPα and C/EBPβ isoform expression during the lipopolysaccharide-mediated acute-phase response. Mol Cell Biol. 1996;16:2295–2306. doi: 10.1128/mcb.16.5.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annweiler A, Hipskind RA, Wirth T. A strategy for efficient in vitro translation of cDNAs using the rabbit β-globin leader sequence. Nucleic Acids Res. 1991;19:3750. doi: 10.1093/nar/19.13.3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. New York, NY: Green Publishing Associates/John Wiley & Sons; 1993. [Google Scholar]
- Baer M, Williams SC, Dillner A, Schwartz RC, Johnson PF. Autocrine signals control CCAAT/enhancer binding protein beta expression, localization, and activity in macrophages. Blood. 1998;92:4353–4365. [PubMed] [Google Scholar]
- Barber GN, Thompson S, Lee TG, Strom T, Jagus R, Darveau A, Katze MG. The 58-kilodalton inhibitor of the interferon-induced double-stranded RNA-activated protein kinase is a tetratricopeptide repeat protein with oncogenic properties. Proc Natl Acad Sci. 1994;91:4278–4282. doi: 10.1073/pnas.91.10.4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck T, Hall MN. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature. 1999;402:689–692. doi: 10.1038/45287. [DOI] [PubMed] [Google Scholar]
- Bernstein J, Shefler I, Elroy-Stein O. The translational repression mediated by the platelet-derived growth factor 2/c-sis mRNA leader is relieved during megakaryocytic differentiation. J Biol Chem. 1995;270:10559–10565. doi: 10.1074/jbc.270.18.10559. [DOI] [PubMed] [Google Scholar]
- Birkenmeier EH, Gwynn B, Howard S, Jerry J, Gordon JI, Landschulz WH, McKnight SL. Tissue-specific expression, developmental regulation, and genetic mapping of the gene encoding CCAAT/enhancer binding protein. Genes & Dev. 1989;3:1146–1156. doi: 10.1101/gad.3.8.1146. [DOI] [PubMed] [Google Scholar]
- Brown EJ, Albers MW, Shin TB, Ichikawa K, Keith CT, Lane WS, Schreiber SL. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature. 1994;369:756–758. doi: 10.1038/369756a0. [DOI] [PubMed] [Google Scholar]
- Brunn GJ, Hudson CC, Sekulic A, Williams JM, Hosoi H, Houghton PJ, Lawrence JC, Jr, Abraham RT. Phosphorylation of the translational repressor PHAS-1 by the mammalian target of rapamycin. Science. 1997;277:99–101. doi: 10.1126/science.277.5322.99. [DOI] [PubMed] [Google Scholar]
- Buck M, Turler H, Chojkier M. LAP (NF-IL-6), a tissue-specific transcriptional activator, is an inhibitor of hepatoma cell proliferation. EMBO J. 1994;13:851–860. doi: 10.1002/j.1460-2075.1994.tb06328.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck M, Poli V, van der Geer P, Chojkier M, Hunter T. Phosphorylation of rat serine 105 or mouse threonine 217 in C/EBPβ is required for hepatocyte proliferation induced by TGF alpha. Mol Cell. 1999;4:1087–1092. doi: 10.1016/s1097-2765(00)80237-3. [DOI] [PubMed] [Google Scholar]
- Calkhoven CF, Bouwman PR, Snippe L, Ab G. Translation start site multiplicity of the CCAAT/enhancer binding protein alpha mRNA is dictated by a small 5′ open reading frame. Nucleic Acids Res. 1994;22:5540–5547. doi: 10.1093/nar/22.25.5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calkhoven CF, Snippe L, Ab G. Differential stimulation by CCAAT/ enhancer-binding protein alpha isoforms of the estrogen-activated promoter of the very-low-density apolipoiprotein II gene. Eur J Biochem. 1997;249:113–120. doi: 10.1111/j.1432-1033.1997.t01-2-00113.x. [DOI] [PubMed] [Google Scholar]
- Cao Z, Umek RM, McKnight SL. Regulated expression of three C/EBP isoforms during adipose conversion of 3T3-L1 cells. Genes & Dev. 1991;5:1538–1552. doi: 10.1101/gad.5.9.1538. [DOI] [PubMed] [Google Scholar]
- Cardenas ME, Cutler NS, Lorenz MC, Di Como CJ, Heitman J. The TOR signaling cascade regulates gene expression in response to nutrients. Genes & Dev. 1999;13:3271–3279. doi: 10.1101/gad.13.24.3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekaran C, Gordon JI. Cell lineage-specific and differentiation-dependent patterns of CCAAT/enhancer binding protein alpha expression in the gut epithelium of normal and transgenic mice. Proc Natl Acad Sci. 1993;90:8871–8875. doi: 10.1073/pnas.90.19.8871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SY, Scherer BJ, Schnier J, Davies MV, Kaufman RJ, Hershey JWB. Stimulation of protein synthesis in COS cells transfected with variants of the alpha- subunits of initiation factor eIF-2. J Biol Chem. 1992;267:286–293. [PubMed] [Google Scholar]
- Christy RJ, Yang VW, Ntambi JM, Geiman DE, Landschulz WH, Friedman AD, Nakabeppu Y, Kelly TJ, Lane MD. Differentiation-induced gene expression in 3T3-L1 preadipocytes: CCAAT/enhancer binding protein interacts with and activates the promoters of two adipocyte-specific genes. Genes & Dev. 1989;3:1323–1335. doi: 10.1101/gad.3.9.1323. [DOI] [PubMed] [Google Scholar]
- Constance CM, Morgan JIT, Umek RM. C/EBPα regulation of the growth-arrest-associated gene gadd45. Mol Cell Biol. 1996;16:3878–3883. doi: 10.1128/mcb.16.7.3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Benedetti A, Baglioni C. Phosphorylation of initiation factor eIF-2 alpha, binding of mRNA to 48S complexes, and its reutilization in initiation of protein synthesis. J Biol Chem. 1983;258:14556–14562. [PubMed] [Google Scholar]
- Dennis PB, Fumagalli S, Thomas G. Target of rapamycin (TOR): Balancing the opposing forces of protein synthesis and degradation. Curr Opin Genet Dev. 1999;9:49–54. doi: 10.1016/s0959-437x(99)80007-0. [DOI] [PubMed] [Google Scholar]
- Descombes P, Schibler U. A liver-enriched transcriptional activator protein, LAP, and a transcriptional inhibitory protein, LIP, are translated from the same mRNA. Cell. 1991;67:569–579. doi: 10.1016/0092-8674(91)90531-3. [DOI] [PubMed] [Google Scholar]
- Diehl AM, Michaelson P, Yang SQ. Selective induction of CCAAT/enhancer binding protein isoforms occurs during rat liver development. Gastroenterology. 1994;106:1625–1637. doi: 10.1016/0016-5085(94)90420-0. [DOI] [PubMed] [Google Scholar]
- Donze O, Jagus R, Koromilas AE, Hershey JW, Sonenberg N. Abrogation of translation initiation factor eIF-2 phosphorylation causes malignant transformation of NIH 3T3 cells. EMBO J. 1995;14:3828–3834. doi: 10.1002/j.1460-2075.1995.tb00052.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst H, Duncan RF, Hershey JW. Cloning and sequencing of complementary DNAs encoding the alpha-subunit of translational initiation factor eIF-2. Characterization of the protein and its messenger RNA. J Biol Chem. 1987;262:1206–1212. [PubMed] [Google Scholar]
- Flodby P, Barlow C, Kyleford H, Ährlund-Richter L, Xanthopoulos G. Increased hepatic cell proliferation and lung abnormalities in mice deficient in CCAAT/enhancer binding protein alpha. J Biol Chem. 1996;271:24753–24760. doi: 10.1074/jbc.271.40.24753. [DOI] [PubMed] [Google Scholar]
- Freytag SO, Paielli DL, Gilbert JD. Ectopic expression of the CCAAT/enhancer-binding protein a promotes the adipogenic program in a variety of mouse fibroblastic cells. Genes & Dev. 1994;8:1654–1663. doi: 10.1101/gad.8.14.1654. [DOI] [PubMed] [Google Scholar]
- Ghilardi N, Wiestner A, Skoda RC. Thrombopoietin production is inhibited by a translational mechanism. Blood. 1998;92:4023–4030. [PubMed] [Google Scholar]
- Gonzalez AL, Joly E. A simple procedure to increase efficiency of DEAE-dextran transfection of COS cells. Trends Genet. 1995;11:216–217. doi: 10.1016/s0168-9525(00)89051-4. [DOI] [PubMed] [Google Scholar]
- Hann SR, Sloan-Brown K, Spotts GD. Translational activation of the non-AUG-initiated c-myc 1 protein at high cell densities due to methionine deprivation. Genes & Dev. 1992;6:1229–1240. doi: 10.1101/gad.6.7.1229. [DOI] [PubMed] [Google Scholar]
- Hardwick JS, Kuruvilla FG, Tong JK, Shamji AF, Schreiber SL. Rapamycin-modulated transcription defines the subset of nutrient-sensitive signaling pathways directly controlled by the Tor proteins. Proc Natl Acad Sci. 1999;96:14866–14870. doi: 10.1073/pnas.96.26.14866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harigai M, Miyashita T, Hanada M, Reed JC. A cis-acting element in the BCL-2 gene controls expression through translational mechanisms. Oncogene. 1996;12:1369–1374. [PubMed] [Google Scholar]
- Hengst L, Reed SI. Translational control of p27kip1 accumulation during the cell cycle. Science. 1996;271:1861–1864. doi: 10.1126/science.271.5257.1861. [DOI] [PubMed] [Google Scholar]
- Hershey JW. Translational control in mammalian cells. Annu Rev Biochem. 1991;60:717–755. doi: 10.1146/annurev.bi.60.070191.003441. [DOI] [PubMed] [Google Scholar]
- Hill JR, Morris DR. Cell-specific translational regulation of S-adenosylmethionine decarboxylase mRNA. Dependence on translation and coding capacity of the cis-acting upstream open reading frame. J Biol Chem. 1993;268:726–731. [PubMed] [Google Scholar]
- Hinnebusch AG. Translational control of GCN4: An in vivo barometer of initiation-factor activity. Trends Biochem Sci. 1994;19:409–414. doi: 10.1016/0968-0004(94)90089-2. [DOI] [PubMed] [Google Scholar]
- Hu E, Tontonoz P, Spiegelman BM. Transdifferentiation of myoblasts by the adipogenic transcription factors PPARγ and C/EBPα. Proc Natl Acad Sci. 1995;92:9856–9860. doi: 10.1073/pnas.92.21.9856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaestner KH, Christy RJ, Lane MD. Mouse insulin-responsive glucose transporter gene: Characterization of the gene and trans-activation by the CCAAT/enhancer binding protein. Proc Natl Acad Sci. 1990;87:251–255. doi: 10.1073/pnas.87.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz S, Kowenz LE, Müller C, Meese K, Ness SA, Leutz A. The NF-M transcription factor is related to C/EBPβ and plays a role in signal transduction, differentiation and leukemogenesis of avian myelomonocytic cells. EMBO J. 1993;12:1321–1332. doi: 10.1002/j.1460-2075.1993.tb05777.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koromilas AE, Roy S, Barber GN, Katze MG, Sonenberg N. Malignant transformation by a mutant of the IFN-inducible dsRNA-dependent protein kinase. Science. 1992;257:1685–1689. doi: 10.1126/science.1382315. [DOI] [PubMed] [Google Scholar]
- Kowenz-Leutz E, Leutz A. A C/EBP beta isoform recruits the SWI/SNF complex to activate myeloid genes. Mol Cell. 1999;4:735–743. doi: 10.1016/s1097-2765(00)80384-6. [DOI] [PubMed] [Google Scholar]
- Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— The scanning model for translation: An update. J Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— An analysis of vertebrate mRNA sequences: Intimations of translational control. J Cell Biol. 1991;115:887–903. doi: 10.1083/jcb.115.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel TA, Bebenek K, McClary J. Efficient site-directed mutagenesis using uracil-containing DNA. Methods Enzymol. 1991;204:125–139. doi: 10.1016/0076-6879(91)04008-c. [DOI] [PubMed] [Google Scholar]
- Kuruvilla FG, Schreiber SL. The PIK-related kinases intercept conventional signaling pathways. Chem Biol. 1999;6:R129–R136. doi: 10.1016/S1074-5521(99)80070-2. [DOI] [PubMed] [Google Scholar]
- Landschulz WH, Johnson PF, Adashi EY, Graves BJ, McKnight SL. Isolation of a recombinant copy of the gene encoding C/EBP. Genes & Dev. 1988;2:786–800. doi: 10.1101/gad.2.7.786. [DOI] [PubMed] [Google Scholar]
- Lawrence JCJ, Abraham RT. PHAS/4E-BPs as regulators of mRNA translation and cell proliferation. Trends Biochem Sci. 1997;22:345–349. doi: 10.1016/s0968-0004(97)01101-8. [DOI] [PubMed] [Google Scholar]
- Lin F-T, Lane MD. Antisense CCAAT/enhancer-binding protein RNA suppresses coordinate gene expression and triglyceride accumulation during differentiation of 3T3-L1 preadipocytes. Genes & Dev. 1992;6:533–544. doi: 10.1101/gad.6.4.533. [DOI] [PubMed] [Google Scholar]
- Lin FT, MacDougald OA, Diehl AM, Lane MD. A 30-kDa alternative translation product of the CCAAT/enhancer binding protein alpha message: Transcriptional activator lacking antimitotic activity. Proc Natl Acad Sci. 1993;90:9606–9610. doi: 10.1073/pnas.90.20.9606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin TA, Kong X, Haystead TA, Pause A, Belsham G, Sonenberg N, Lawrence JCJ. PHAS-I as a link between mitogen-activated protein kinase and translation initiation. Science. 1994;266:653–656. doi: 10.1126/science.7939721. [DOI] [PubMed] [Google Scholar]
- Lincoln AJ, Monczak Y, Williams SC, Johnson PF. Inhibition of CCAAT/enhancer-binding protein alpha and beta translation by upstream open reading frames. J Biol Chem. 1998;273:9552–9560. doi: 10.1074/jbc.273.16.9552. [DOI] [PubMed] [Google Scholar]
- McNagny KM, Sieweke MH, Doderlein G, Graf T, Nerlov C. Regulation of eosinophil-specific gene expression by a C/EBP-Ets complex and GATA-1. EMBO J. 1998;17:3669–3680. doi: 10.1093/emboj/17.13.3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meurs E, Chong K, Galabru J, Thomas NSB, Kerr IM, Williams BRG, Hovanessian AG. Molecular cloning and characterization of the human double-stranded RNA-activated protein kinase induced by interferon. Cell. 1990;62:379–390. doi: 10.1016/0092-8674(90)90374-n. [DOI] [PubMed] [Google Scholar]
- Meurs EF, Galabru J, Barber GN, Katze MG, Hovanessian AG. Tumor suppressor function of the interferon-induced double-stranded RNA-activated protein kinase. Proc Natl Acad Sci. 1993;90:232–236. doi: 10.1073/pnas.90.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgenstern JP, Land H. Advanced mammalian gene transfer: High titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris DR. Growth control of translation in mammalian cells. Prog Nucleic Acid Res Mol Biol. 1995;51:339–363. doi: 10.1016/s0079-6603(08)60883-1. [DOI] [PubMed] [Google Scholar]
- Mueller PP, Hinnebusch AG. Multiple upstream AUG codons mediate translational control of GCN4. Cell. 1986;45:201–207. doi: 10.1016/0092-8674(86)90384-3. [DOI] [PubMed] [Google Scholar]
- Müller C, Kowenz-Leutz E, Grieser-Ade S, Graf T, Leutz A. NF-M (chicken C/EBPb) induces eosinophilic differentiation and apoptosis in a hematopoietic progenitor cell line. EMBO J. 1995;14:6127–6135. doi: 10.1002/j.1460-2075.1995.tb00303.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller C, Alunni-Fabbroni M, Kowenz-Leutz E, Mo X, Tommasino M, Leutz A. Separation of C/EBPα-mediated proliferation arrest and differentiation pathways. Proc Natl Acad Sci. 1999;96:7276–7281. doi: 10.1073/pnas.96.13.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nerlov C, McNagny KM, Doderlein G, Kowenz-Leutz E, Graf T. Distinct C/EBP functions are required for eosinophil lineage commitment and maturation. Genes & Dev. 1998;12:2413–2423. doi: 10.1101/gad.12.15.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ness SA, Kowenz LE, Casini T, Graf T, Leutz A. Myb and NF-M: Combinatorial activators of myeloid genes in heterologous cell types. Genes & Dev. 1993;7:749–759. doi: 10.1101/gad.7.5.749. [DOI] [PubMed] [Google Scholar]
- Oelgeschläger M, Nuchprayoon I, Lüscher B, Friedman AD. C/EBP, c-Myb, and PU.1 cooperate to regulate the neutrophil elastase promotor. Mol Cell Biol. 1996;16:4717–4725. doi: 10.1128/mcb.16.9.4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossipow V, Descombes P, Schibler U. CCAAT/enhancer-binding protein mRNA is translated into multiple proteins with different transcription activation potentials. Proc Natl Acad Sci. 1993;90:8219–8223. doi: 10.1073/pnas.90.17.8219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pall M, Hellberg P, Brannstrom M, Mikuni M, Peterson CM, Sundfeldt K, Norden B, Hedin L, Enerback S. The transcription factor C/EBP-β and its role in ovarian function; evidence for direct involvement in the ovulatory process. EMBO J. 1997;16:5273–5279. doi: 10.1093/emboj/16.17.5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park EA, Roesler WJ, Liu J, Klemm DJ, Gurney AL, Thatcher JD, Shuman J, Friedman A, Hanson RW. The role of the CCAAT/enhancer-binding protein in the transcriptional regulation of the gene for phosphoenolpyruvate carboxykinase (GTP) Mol Cell Biol. 1990;10:6264–6272. doi: 10.1128/mcb.10.12.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pause A, Belsham GJ, Gingras AC, Donze O, Lin TA, Lawrence JC, Jr, Sonenberg N. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature. 1994;371:762–767. doi: 10.1038/371762a0. [DOI] [PubMed] [Google Scholar]
- Pear WS, Nolan GP, Scott ML, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson RT, Schreiber SL. Translation control: Connecting mitogens and the ribosome. Curr Biol. 1998;8:R248–R250. doi: 10.1016/s0960-9822(98)70152-6. [DOI] [PubMed] [Google Scholar]
- Peterson RT, Desai BN, Hardwick JS, Schreiber SL. Protein phosphatase 2A interacts with the 70-kDa S6 kinase and is activated by inhibition of FKBP12-rapamycinassociated protein. Proc Natl Acad Sci. 1999;96:4438–4442. doi: 10.1073/pnas.96.8.4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piontkewitz Y, Enerback S, Hedin L. Expression and hormonal regulation of the CCAAT enhancer binding protein-alpha during differentiation of rat ovarian follicles. Endocrinology. 1993;133:2327–2333. doi: 10.1210/endo.133.5.8404685. [DOI] [PubMed] [Google Scholar]
- Poli V, Mancini FP, Cortese R. IL-6DBP, a nuclear protein involved in interleukin-6 signal transduction, defines a new family of leucine zipper proteins related to C/EBP. Cell. 1990;63:643–653. doi: 10.1016/0092-8674(90)90459-r. [DOI] [PubMed] [Google Scholar]
- Polymenis M, Schmidt V. Coupling of cell division to cell growth by translational control of the G1 cyclin CLN3 in yeast. Genes & Dev. 1997;11:2522–2531. doi: 10.1101/gad.11.19.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radomska HS, Huettner CS, Zhang P, Cheng T, Scadden DT, Tenen DG. CCAAT/enhancer binding protein alpha is a regulatory switch sufficient for induction of granulocytic development from bipotential myeloid progenitors. Mol Cell Biol. 1998;18:4301–4314. doi: 10.1128/mcb.18.7.4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana B, Xie Y, Mischoulon D, Bucher NL, Farmer SR. The DNA binding activity of C/EBP transcription factor is regulated in the G1 phase of the hepatocyte cell cycle. J Biol Chem. 1995;270:18123–18132. doi: 10.1074/jbc.270.30.18123. [DOI] [PubMed] [Google Scholar]
- Raught B, Liao WS, Rosen JM. Developmentally and hormonally regulated CCAAT/enhancer-binding protein isoforms influence β-casein gene expression. Mol Endocrinol. 1995;9:1223–1232. doi: 10.1210/mend.9.9.7491114. [DOI] [PubMed] [Google Scholar]
- Raught B, Gingras AC, James A, Medina D, Sonenberg N, Rosen JM. Expression of a translationally regulated, dominant-negative CCAAT/enhancer-binding protein beta isoform and up-regulation of the eukaryotic translation initiation factor 2α are correlated with neoplastic transformation of mammary epithelial cells. Cancer Res. 1996;56:4382–4386. [PubMed] [Google Scholar]
- Rosenwald IB, Chen JJ, Wang S, Savas L, London IM, Pullman J. Upregulation of protein synthesis initiation factor eIF-4E is an early event during colon carcinogenesis. Oncogene. 1999;18:2507–2517. doi: 10.1038/sj.onc.1202563. [DOI] [PubMed] [Google Scholar]
- Rousseau D, Kaspar R, Rosenwald IB, Gehrke L, Sonenberg N. Translation initiation of ornithine decarboxilase and nucleocytoplasmic transport of cyclin D1 mRNA are increased in cells overexpressing eukaryotic initiation factor 4E. Proc Natl Acad Sci. 1996;93:1065–1070. doi: 10.1073/pnas.93.3.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rychlik W, Domier LL, Gardner PR, Hellmann GM, Rhoads RE. Amino acid sequence of the mRNA cap-binding protein from human tissues (published erratum appears in Proc. Natl. Acad. Sci. 1992, 89: 1148) Proc Natl Acad Sci. 1987;84:945–949. doi: 10.1073/pnas.84.4.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuelsson L, Stromberg K, Vikman K, Bjursell G, Enerback S. The CCAAT/enhancer binding protein and its role in adipocyte differentiation: Evidence for direct involvement in terminal adipocyte development. EMBO J. 1991;10:3787–3793. doi: 10.1002/j.1460-2075.1991.tb04948.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott LM, Civin CI, Rorth P, Friedman AD. A novel temporal expression pattern of three C/EBP family members in differentiating myelomonocytic cells. Blood. 1992;80:1725–1735. [PubMed] [Google Scholar]
- Screpanti I, Romani L, Musiani P, Modesti A, Fattori E, Lazzaro D, Sellitto C, Scarpa S, Bellavia D, Lattanzio G, et al. Lymphoproliferative disorder and imbalanced T-helper response in C/EBP beta-deficient mice. EMBO J. 1995;14:1932–1941. doi: 10.1002/j.1460-2075.1995.tb07185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears RC, Sealy L. Multiple forms of C/EBP beta bind the EFII enhancer sequence in the Rous sarcoma virus long terminal repeat. Mol Cell Biol. 1994;14:4855–4871. doi: 10.1128/mcb.14.7.4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterneck E, Tessarollo L, Johnson PF. An essential role for C/EBPβ in female reproduction. Genes & Dev. 1997;11:2153–2162. doi: 10.1101/gad.11.17.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swart GW, van Groningen JJ, van Ruissen F, Bergers M, Schalkwijk J. Transcription factor C/EBPalpha: Novel sites of expression and cloning of the human gene. Biol Chem. 1997;378:373–379. doi: 10.1515/bchm.1997.378.5.373. [DOI] [PubMed] [Google Scholar]
- Tahara SM, Dietlin TA, Dever TE, Merrick WC, Worrilow LM. Effect of eukaryotic initiation factor 4F on AUG selection in a bicistronic mRNA. J Biol Chem. 1991;266:3594–3601. [PubMed] [Google Scholar]
- Tanaka T, Akira S, Yoneda Y, Shirafuji N, Fujiwara H, Suematsu S, Yoshida N, Kishimoto T. Targeted disruption of the NF-IL6 gene discloses its essential role in bacteria killing and tumor cytotoxity by macrophages. Cell. 1995;80:353–361. doi: 10.1016/0092-8674(95)90418-2. [DOI] [PubMed] [Google Scholar]
- Timchenko NA, Harris TE, Wilde M, Bilyeu TA, Burgess-Beusse BL, Finegold MJ, Darlington GJ. CCAAT/enhancer binding protein alpha regulates p21 protein and hepatocyte proliferation in newborn mice. Mol Cell Biol. 1997;17:7353–7361. doi: 10.1128/mcb.17.12.7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timchenko NA, Wilde M, Kosai KI, Heydari A, Bilyeu TA, Finegold MJ, Mohamedali K, Richardson A, Darlington GJ. Regenerating livers of old rats contain high levels of C/EBPalpha that correlate with altered expression of cell cycle associated proteins. Nucleic Acids Res. 1998;26:3293–3299. doi: 10.1093/nar/26.13.3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umek RM, Friedman AD, McKnight SL. CCAAT-enhancer binding protein: A component of a differentiation switch. Science. 1991;251:288–292. doi: 10.1126/science.1987644. [DOI] [PubMed] [Google Scholar]
- Wang ND, Finegold MJ, Bradley A, Ou CN, Abdelsayed SV, Wilde MD, Taylor LR, Wilson DR, Darlington GJ. Impaired energy homeostasis in C/EBPα knockout mice. Science. 1995;269:1108–1112. doi: 10.1126/science.7652557. [DOI] [PubMed] [Google Scholar]
- Wek RC. eIF-2 kinases: Regulators of general and gene-specific translation initiation. Trends Biochem Sci. 1994;19:491–496. doi: 10.1016/0968-0004(94)90136-8. [DOI] [PubMed] [Google Scholar]
- Welm AL, Timchenko NA, Darlington GJ. C/EBPalpha regulates generation of C/EBPbeta isoforms through activation of specific proteolytic cleavage. Mol Cell Biol. 1999;19:1695–1704. doi: 10.1128/mcb.19.3.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Xie Y, Bucher NL, Farmer SR. Conditional ectopic expression of C/EBPβ in NIH-3T3 cells induces PPARγ and stimulates adipogenesis. Genes & Dev. 1995;9:2350–2363. doi: 10.1101/gad.9.19.2350. [DOI] [PubMed] [Google Scholar]
- Yeh WC, Bierer BE, McKnight SL. Rapamycin inhibits clonal expansion and adipogenic differentiation of 3T3-L1 cells. Proc Natl Acad Sci. 1995a;92:11086–11090. doi: 10.1073/pnas.92.24.11086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh WC, Cao Z, Classon M, McKnight SL. Cascade regulation of terminal adipocyte differentiation by three members of the C/EBP family of leucine zipper proteins. Genes & Dev. 1995b;9:168–181. doi: 10.1101/gad.9.2.168. [DOI] [PubMed] [Google Scholar]
- Zhang DE, Zhang P, Wang ND, Hetherington CJ, Darlington GJ, Tenen DG. Absence of granulocyte colony-stimulating factor signaling and neutrophil development in CCAAT enhancer binding protein alpha-deficient mice. Proc Natl Acad Sci. 1997;94:569–574. doi: 10.1073/pnas.94.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng XF, Schreiber SL. Target of rapamycin proteins and their kinase activities are required for meiosis. Proc Natl Acad Sci. 1997;94:3070–3075. doi: 10.1073/pnas.94.7.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinszner H, Albalat R, Ron D. A novel effector domain from the RNA-binding protein TLS or EWS is required for oncogenic transformation by CHOP. Genes & Dev. 1994;8:2513–2526. doi: 10.1101/gad.8.21.2513. [DOI] [PubMed] [Google Scholar]