Cellular toxicity of expanded RNA repeats: focus on RNA foci (original) (raw)
Abstract
Discrete and punctate nuclear RNA foci are characteristic molecular hallmarks of pathogenesis in myotonic dystrophy type 1 and type 2. Intranuclear RNA inclusions of distinct morphology have also been found in fragile X-associated tremor ataxia syndrome, Huntington's disease-like 2, spinocerebellar ataxias type 8, type 10 and type 31. These neurological diseases are associated with the presence of abnormally long simple repeat expansions in their respective genes whose expression leads to the formation of flawed transcripts with altered metabolisms. Expanded CUG, CCUG, CGG, CAG, AUUCU and UGGAA repeats are associated with the diseases and accumulate in nuclear foci, as demonstrated in variety of cells and tissues of human and model organisms. These repeat RNA foci differ in size, shape, cellular abundance and protein composition and their formation has a negative impact on cellular functions. This review summarizes the efforts of many laboratories over the past 15 years to characterize nuclear RNA foci that are recognized as important triggers in the mutant repeat RNA toxic gain-of-function mechanisms of pathogenesis in neurological disorders.
INTRODUCTION
Expansions of short tandem repeat of tri-, tetra- and pentanucleotides in single genes cause hereditary neurological diseases in humans. The abnormally expanded microsatellites can lead to a variety of effects on genes, including the inhibition of transcription and the loss-of-function of protein products. In several cases, the repeat expansions confer toxicity to the mutant transcripts and to the encoded proteins, both of which are capable of disrupting cell functions, leading to disease.
The discovery that RNA harboring CUG repeat expansion is retained in the cell nucleus, where it colocalizes with some host proteins and forms microscopic ribonuclear inclusions (1,2), had far-reaching consequences for research in the field of neurological disorders. While transcripts are normally destined to deliver the message from the DNA sequence to the cytoplasm for protein expression, abnormally lengthened repeats of the mutant myotonic dystrophy type 1 (DM1) transcript become aberrantly recognized by cellular machinery and pick up an excess of specific proteins, resulting in the nuclear retention of the RNA (1,3,4). In addition to DM1, various microsatellite expansions present in the transcripts of different genes associated with myotonic dystrophy type 2 (DM2), fragile X-associated tremor ataxia syndrome (FXTAS), Huntington's disease-like 2 (HDL2), spinocerebellar ataxias type 8 (SCA8), type 31 (SCA31) and type 10 (SCA10) have also been shown to gain nuclear toxicity (5–10).
Molecular hallmarks of cells expressing expanded repeat RNA are nuclear RNA foci of distinct morphology and abundance. The type of tissue, the expression level of the repeat-containing transcript and the repertoire and abundance of the expressed proteins may influence foci size, shape, colocalization with proteins and intensity of fluorescence when measured by fluorescence in situ hybridization (FISH). Presumably, RNA repeat inclusions are trapped in the nucleus due to their abnormal cargo, which is composed of dozens of molecules of flawed RNA (11) overloaded with proteins (12–15) that non-specifically interact with expanded repeats. Such interactions may lead to robust sequestration of the proteins, as detected for muscleblind 1 (MBNL1) (12), may lead to the recruitment of proteins to RNA foci with limited colocalization, as shown for hnRNP H (16), or may only represent a close association with the RNA inclusions (1,14,17). Recent insight into dynamics of CUG repeat foci has revealed that these are unstable, constantly aggregating and disaggregating structures, and that MBNL1 is directly involved in the stochastic process of foci formation (18). This indicates that CUG repeat foci are formed not just by transcript self-aggregation but the process involves protein immobilization including MBNL1. Whether similar behavior of transcript self-aggregation is part of RNA foci formation by other repeats needs to be resolved. In general, the presence of RNA foci has an adverse effect on host cells, which leads to abnormalities in distinct cellular pathways, including the activation of apoptosis and aberrant alternative splicing (9,19). The muscleblind-like family (MBNL1, MBNL2 and MBNL3), Sam68 and hnRNP K are among the proteins implicated in foci RNA-mediated pathogenesis due to their abnormal interactions with mutant repeats. Whereas MBNL1 is detected in a variety of repeat-formed foci, including CUG, CCUG, CAG and CGG RNA inclusions, Sam68 has only been found in expanded CGG repeat RNA (5,12,20–23), and hnRNP K is associated in situ with the AUUCU mutation of SCA10 (9).
Fluorescence in situ hybridization enables the localization of specific sequences of nucleic acids directly inside cells or tissue (1). Using FISH antisense probes that hybridize to RNA transcripts has been shown to be useful in distinguishing the presence of particular transcripts that harbor simple repeat expansions. In cells expressing such mutant RNA, FISH enables the detection of a variable number of RNA foci scattered throughout the nucleus. By distinguishing mutant and normal alleles, this technique allows both quantitative analysis of the ribonuclear inclusions of repeat expansions and their morphological determination. While DM repeat expansions accumulate as discrete punctate foci, the CAG and CGG expansions form bigger and patchy nuclear aggregates. Presumably the detection of such structures is feasible due to the increased concentration of mutant transcripts in focal inclusions, owing to the altered lability and solubility of expanded repeats and their unspecific interactions with some proteins.
This review emphasizes the involvement of nuclear RNA foci of simple repeat expansions in the pathogenesis of human hereditary neurological diseases. Ribonucleoprotein foci of repeated CUG, CCUG, CGG, CAG, AUUCU and UGGAA motifs present in different human tissues, cultured cells and several model organisms have been characterized in detail providing a complex picture of foci morphology, abundance and molecular composition and their detection method.
RNA FOCI IN MYOTONIC DYSTROPHIES
DM1 and DM2 are adult-onset muscular dystrophies that belong to non-coding repeat expansion disorders. A mutation of the CTG repeat in the 3′UTR of the DMPK gene triggers DM1 (24,25), whereas an abnormal stretch of CCTG repeats in intron 1 of the ZNF9 gene causes DM2 (Fig. 1) (26). Both diseases are autosomal-dominant genetic disorders with multisystemic clinical features. In DM1, these features include progressive weakness and wasting of skeletal and smooth muscles, myotonia, cataracts, cardiac arrhythmias, mild mental retardation and endocrinopathies (reviewed in 27,28). Despite some similarities in the major clinical features between DM1 and DM2, the latter is more benign, and DM2 myopathy is generally not associated with myotonia.
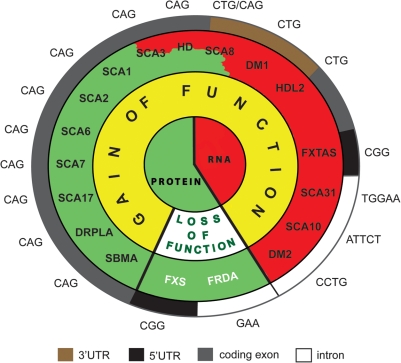
Figure 1.
Simple repeat expansions associated with human neurodegenerative and neuromuscular disorders; mechanisms of pathogenesis mediated by protein loss-of-function (white) and protein and RNA toxic gain-of-function (yellow).
CUG repeat RNA foci in DM1
Normally, the DMPK gene contains 5–37 copies of CTG, but in DM1 patients, the repeat length reaches several kilobases and mild myotonic symptoms occur in carriers of as few as 50–100 copies of CTG (24). The presence of the CTG expansion leads to nuclear retention of the mutant transcript, and on a cellular level, this retention is manifested by the formation of ribonuclear inclusions called RNA foci (1,13,29). CUG foci have been most extensively studied in skeletal muscle, which is the most affected tissue in individuals with DM1 (1,12,20,30). Foci characteristics include abundance, shape and size, fluorescence intensity and colocalization with proteins.
In DM1 biopsies of vastus and derived myoblasts, expansions shorter than 70 copies of CTG are not detected as nuclear RNA foci, whereas sparse inclusions are formed when repeats range from 70 to 100 CTGs (20) (Supplementary Material, Table S1). Studies in MyoD-transformed DM1 fibroblasts found that DMPK transcripts with more than 400 CUGs are completely retained in the nucleus, whereas retention is incomplete when CUG = 150 (4), and undetectable when CUG = 80 (31). However, ectopic expression of a DMPK 3′UTR with as few as 57 CTGs in C2C12 mouse myoblasts was reported to be sufficient for triggering nuclear retention by means of foci formation (3). The CTG repeat length is also a factor in determining the number of foci per nucleus and the fraction of foci-positive nuclei. Botta et al. (32) described that, in DM1 biopsy muscle of the vastus lateralis that expressed 165–430 CUG repeats, RNA foci were present in about 60% of nuclei, and their number ranged from 0 to 5 per nucleus, with an average of 1.18 foci. The number of nuclei without foci decreases with longer CUG mutations, while the number of foci per nucleus increases. In muscle expressing 1250–1900 repeats, only 8% of nuclei have no foci, and up to 18 foci are formed per nucleus, with an average of 2.92 foci per nucleus (Supplementary Material, Table S1).
What factors can influence the abundance of foci upon expression of DMPK-containing expanded CUG repeats? In cultured cells, the abundance is dependent on the level of expression of the mutant transcript, and myotubes have numerous foci that are larger and brighter than in myoblasts and fibroblasts harboring the same repeat lengths (12,33,34) (Fig. 2). Similarly, MyoD-transformed DM1 fibroblasts contain more CUG RNA foci, which are larger and brighter than in untransformed cells (2,4). Interestingly, in human DM1 biopsies of skeletal muscle, fewer nuclear foci are found than in derived myoblasts and fibroblasts. Numerous reports have described the presence of only 1–3 intense CUG RNA foci in myonuclei from skeletal muscle, while more small foci are found in cultured cells (1,12,20) (Fig. 2). It has been suggested that tissue freezing or improper storage may cause RNA degradation and thus give rise to fewer foci in biopsies, however, Cardani et al. (35) demonstrated that properly stored and thawed muscles have similar numbers of foci.
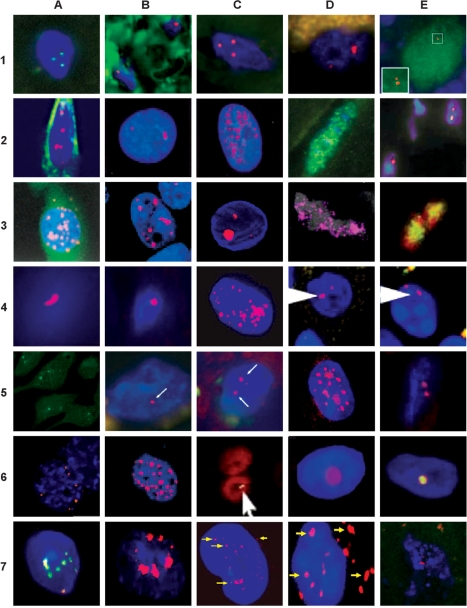
Figure 2.
Representative images of FISH RNA depicting nuclear RNA foci in various tissues and cultured cells expressing CUG, CCUG, CAG, CGG, AUUCU and UGGAA repeat mutations. (1A) Nucleus of human muscle fiber with CUG foci (20) (1B) CUG foci in a gallbladder from a DM1 patient (36) (1C) CUG foci in post-mortem cardiac tissue colocalizing with MBNL2 (29) (1D) CUG inclusions in frontal cortical neurons of a human DM1 brain (13) (1E) small nuclear CUG foci in Purkinje cells from a DM1 brain (13) (2A) ribonuclear CUG foci in DM1 myoblasts (12) (2B) and (2C) CUG nuclear foci in DM1 fibroblasts without MyoD (B) and with MyoD (C) (2) (2D) nuclear CUG foci in skeletal muscle from HSALR-20b DM1 mice (20) (2E) nuclear CUG foci in skeletal muscle from transgenic CTG200 mice (45) (3A) nuclear and cytoplasmic RNA foci in C2C12 cells ectopically expressing 200 CUG from 3′UTR DMPK (37) (3B) nuclear RNA foci in COS7 cells expressing 960 CUG repeats (21) (3C) nuclear CUG foci in the larval muscle of DM1 Drosophila (46) (3D) CUG foci in the nuclei of the body wall muscle cells from DM1 Drosophila (47) (3E) nuclear RNA foci formed by 270 CUG repeats expressed in Drosophila (48) (4A) CCUG nuclear inclusion in human DM2 skeletal muscle (12) (4B) CCUG focus in the left ventricles of an autopsy heart (49) (4C) CCUG foci in human homozygous DM2 myoblasts (10) (4D) CUG foci in Purkinje cells from a human SCA8 cerebellum (7) (4E) CUG foci in Purkinje cells from SCA8 BAC mice (7) (5A) nuclear CUG foci in HEK293 cells expressing an ATXN8OS mutant transcript (50) (5B) CUG nuclear foci in a HDL2 cerebral frontal cortex (6) (5C) CUG foci in HDL2 stratum (6) (5D) CAG nuclear aggregates in human HD fibroblasts (23) (5E) nuclear CAG foci in muscle sections from CAG200 mice (22) (6A) RNA foci in C2C12 cells ectopically expressing 200 CUG colocalizing with MBNL (22) (6B) nuclear foci in COS7 cells expressing 960 CAG repeats (21) (6C) nuclear RNA foci formed by CAG270 expressed in Drosophila (48) (6D) nuclear signal from 5′UTR antisense riboprobe in isolated nuclei of the frontal cortex from post-mortem FXTAS brain (51) (6E) nuclear aggregate of CGG repeat mutation sequesters Sam68 protein in a post-mortem FXTAS brain section of the hippocampal area (5) (7A) nuclear CGG foci in brain sections from mice expressing 98 CGG repeats colocalizing with Sam68 (5) (7B) nuclear RNA aggregates in COS7 cells expressing 100 CGG repeats (5) (7C) nuclear and cytoplasmic AUUCU aggregates in SCA10 human fibroblasts (9) (7D) nuclear and cytoplasmic RNA aggregates in brains from mice expressing expanded AUUCU repeats (9) (7E) nuclear RNA foci in SCA31 Purkinje cells (8).
In DM1 patients, the presence of ribonuclear inclusions of mutant DMPK transcripts has also been shown in non-skeletal muscle tissues, including the smooth muscle (36) and heart (29), and in the central nervous system (brain and spinal cord) (13,19) (Fig. 2). The number and size of RNA foci in the muscle of the gall bladder and the skeletal muscle of DM1 patients have been found to be comparable, and 1–2 foci per nucleus were usually detected in both tissues (36). Another study, performed on paired samples of brain and skeletal muscle from the same patients, showed 3.1-fold more foci in cortical neurons than in muscle (13). The CUG RNA foci were present in over 85% of cortical neurons, and 30% of the cells had more than 1 focus; occasionally, there were up to 15 small foci per nucleus. Except for frontal cortical neurons, foci were also found in the hippocampus, dentate gyrus, thalamus, subcortical substantia nigra, brain stem tegmentum, white matter and corpus callosum (Supplementary Material, Table S1).
CUG RNA foci are nuclear structures, however, the inclusions in dividing cells can also be found in the cytoplasm (37,38). The toxicity of cytoplasmic foci was studied by Dansithong et al. (38) in a transgenic DM1 mouse model designed to express the CTG400 repeat exclusively in the cytoplasm of cardiac cells. The results of this analysis reveal that cellular localization of CUG RNA foci is critical for their deleterious effects, and despite the sequestration of Mbnl1 by the cytoplasmic aggregates and the elevation of Cugbp1 levels, no defects in alternative RNA splicing typical for DM1 were observed in these mice. This result underlines the significance of studies aimed at identifying molecules that increase the transport rate of the flawed transcript outside of the nucleus.
Antisense technology has been employed to disrupt CUG RNA inclusions by targeting the mutant DMPK transcript and preventing its interaction with host cell proteins. Direct administration of morpholino oligonucleotides (39) and 2′-O-methyl-phosphorothioate-modified (CAG)7 AON (antisense oligonucleotides) (40) into DM1 mouse skeletal muscle has been described to be effective in reducing the number of ribonuclear aggregates and in normalizing the effect on aberrant pre-mRNA splicing (Supplementary Material, Table S1). Similar conclusions were derived from work in human DM1 myoblasts with the use of engineered human U7 small nuclear RNAs (hU7-snRNAs) containing a poly-CAG antisense sequence (41). Furthermore, employment of a nuclear-retained hammerhead RNA (ribozyme) designed to cut the 3′UTR of DMPK mRNA (42) and the use of pentamidine, a small molecule that partially targets CUG repeats and releases MBNL1 (43), were reported to result in the destruction of nuclear DM1 foci.
CCUG repeat RNA foci in DM2
In DM2, the expression of an enormous expansion of tetranucleotide repeats in the ZNF9 gene leads to the formation of nuclear CCUG RNA foci as a result of the aberrant degradation of an intronic sequence (26). In skeletal muscle, the length threshold for nuclear retention and foci formation is 100 CCUG repeat copies (44). The inclusions consist exclusively of non-coding CCUG repeat expansions and do not contain other parts of intron 1 of ZNF9 (10). This feature is in contrast to DM1, in which the entire DMPK transcript localizes to CUG foci (1). How does this difference affect foci size? Do larger DM2 mutations reaching up to 11 000 CCTG, and the 8- to 14-fold higher expression level of ZNF9 mRNA than DMPK mRNA in muscle cells affect the abundance of the RNA foci? The results reveal that in human DM1 and DM2 skeletal muscles, the numbers of RNA foci are comparable and range from 0 to 5 per nucleus, however, DM2 inclusions are bigger and more intense (8- to 13-fold) than DM1 inclusions (12) (Supplementary Material, Table S1). The inclusions differ by shape, and whereas DM1 foci are spheroidal, DM2 foci are rod-shaped (Fig. 2). Additionally, the extent of MBNL1 depletion from the nucleoplasm and its accumulation in ribonuclear foci is more extensive in DM2 than in DM1 (30,52).
The differentiation of DM muscle cells is correlated with a decrease in the number of nuclear foci and an increase in their size. DM2 myoblasts from biceps brachii expressing 1000–2500 CCTG repeats have 3–25 foci per nucleus, with an average of 14 foci, while myotubes have fewer but larger and more intense inclusions (35). The presence of cytoplasmic DM2 foci has not been reported in muscle tissue and derived cultured cells (26,35,53).
Protein components of RNA nuclear inclusions in DM
In 2000, Swanson and colleagues (2) presented the first experimental evidence that triplet repeat expansion RNA-binding proteins (EXP), homologous to the Drosophila mbl proteins, bind preferentially to CUG repeat RNA in a length-dependent manner. The authors reported that the EXP protein itself forms nuclear foci in MyoD-transfected DM1 fibroblasts, and that their number per nucleus ranged from 6 to 30, with an average of 15 foci (Supplementary Material, Table S1). The biological significance of the interaction between repeat RNA and the MBNL-family proteins has been manifested through the disruption of alternative splicing, which is a characteristic feature of DM pathogenesis. Muscle wasting and weakness, heart problems and insulin resistance are associated with aberrant alternative pre-mRNA splicing and inappropriate expression of fetal isoforms of CLCN1, INSR and TNNT2 in adult DM tissues (19,54,55).
RNA-binding proteins of the muscleblind-like family, MBNL1, MBNL2 and MBNL3, when either endogenous or exogenously expressed, are sequestered by expanded repeats trapped in nuclear foci. Experimental evidence has revealed the presence of MBNL-positive CUG and CCUG RNA foci in biopsy tissues from skeletal and smooth muscles and in derived cultured cells (12,14,20,30,36,41,52), as well as in cardiac (29) and neuronal cells (13,19) (Supplementary Material, Table S1). Interestingly, as shown in DM2 cultured muscle cells, not all CCUG RNA foci are enriched with muscleblind 1, although each MBNL focus overlaps with the repeat inclusion (35,52).
Cellular RNA-binding proteins may influence the structure of RNA and its trafficking. Several dsRNA-binding proteins and other RNA interacting proteins have been analyzed for their colocalization and sequestration by DM nuclear RNA foci. Among them are CUGBP1, hnRNPs (A1, C1/C2, C, H, F and M), TRBP, PKR, PACT, NF90 and RHA, all of which do not recapitulate the MBNL feature of sequestration by the expanded RNA (2,12–14,29,56,57) (Supplementary Material, Table S1). Some of the proteins are recruited to DM1 foci and colocalize with them to a limited extent. This behavior is also true for the hnRNP H and hnRNP F proteins, which colocalize with CUG foci in human muscle and brain cells, components of the proteosome (20Sα, 11Sγ and 11Sα subunits) recruited to the foci in cortical neurons (13,16) and the Y12, Y14 and 9G8 proteins associated with CUG RNA foci in human myoblasts and fibroblasts (14). For SC-35, a marker of splicing speckles, experimental results showed no overlap with CUG and CCUG RNA foci and only a close association and some random coincidence with the nuclear inclusions (1,14,15,17) (Supplementary Material, Table S1).
CUG RNA foci in DM1 model organisms
In 2000, Thornton and colleagues (58) published the first transgenic DM1 mouse model carrying an untranslated 250 CTG repeat under the human skeletal actin promoter (HSA). These mice develop skeletal muscle myotonia and myopathy and accumulate numerous CUG RNA foci in myonuclei. It was shown that over 50 foci per nucleus can be formed in the mouse muscle, and that the foci are Mbnl1-positive (12,39,40). Other transgenic DM1 mouse models are inducible models that express 960 CTG repeats specifically in the heart (59) or skeletal muscle (60) or 200 CTG in both of these tissues (45). Formation of RNA foci that colocalize with Mbnl1 was reported in all of these models, but the abundance of ribonuclear inclusions depends on the transgene expression levels. Cooper and colleagues (59) showed that, in mouse hearts from the highest-expressing line, a greater fraction of nuclei contained foci than in the moderate-expressing line and that, in the former, multiple foci per nucleus are formed, whereas only single foci per nucleus are usually formed in the latter (Supplementary Material, Table S1). Mahadevan et al. (45) observed the DM1 phenotype of skeletal and cardiac muscles, including aberrant splicing, in both foci-positive DMPK 3′UTR-(CTG)200 expressing mice and littermates who overexpressed (CTG)5 in the absence of detectable foci. Their results raise the question of whether the presence of RNA inclusions is required for the development of DM1 features. In addition, results from transgenic CUGBP1 mouse models (61,62) and knockout Mbnl1 mice (63), both lacking RNA foci but having aberrant splicing, demonstrate that foci formation and aberrant splicing are independent events and that some core features of DM1 do not depend on the presence of either foci or deviated alternative splicing.
In the transgenic DM1 Drosophila models published thus far, RNA foci were formed with CTG ≥ 162 repeats (46,47,64,65) (Supplementary Material, Table S3). In the first fly model, Monckton and colleagues (47) described the presence of dynamic and transient CUG RNA foci with a short half-life, the formation of which is not supported by all cell types. The number and intensity of the inclusions increase throughout the larval stages of development and then decrease during pupation, when the muscle remodels. Interestingly, at the adult stage, despite ubiquitous transgene expression, RNA foci are restricted to abdominal, cranial and pleurosternal muscles and are absent in legs and indirect flight muscles. CUG foci were not found in the nuclei of the salivary gland, brain and peripheral nervous system at any stage in larvae to adults (47). From this work, it was concluded that the expression of expanded CUG repeat RNA is not sufficient to drive foci formation, indicating a requirement for cell-specific factors. The presence of ribonuclear foci was also described in three other DM1 fly models that expressed interrupted 240 and 480 CTG expansions (46,64,65) (Supplementary Material, Table S3). In DM1 Drosophila models, muscleblind type 1 protein colocalizes with CUG foci. The number of RNA inclusions per nucleus decreases in flies co-expressing MBNL1 and increases when CUGBP1 is co-expressed with the CUG repeat mutation (46).
CUG RNA FOCI IN SCA8
SCA 8 is autosomal dominant inherited disease of the central nervous system caused by a CTG/CAG trinucleotide repeat expansion. Bidirectional transcription has been shown in SCA8 because two genes that span the repeat region are expressed in opposite directions (66). The ATXN8 gene on the antisense strand encodes a nearly pure polyglutamine (poly-Q) protein in the CAG direction, and the _ATXN8O_S gene on the sense strand encodes an untranslated CUG expansion RNA. In normal individuals, the length of the CTG repeat is 16–34, whereas it is >74 in affected individuals. SCA8 leads to the impairment of specific nerve fibers carrying messages to and from the brain, resulting in degeneration of the cerebellum with variable involvement of the brainstem and spinal cord.
In 2009, Ranum and colleagues (7) demonstrated that in SCA8 the expression of sense strand repeats is associated with nuclear retention of the transcript, resulting in CUG foci formation as detected in patient post-mortem brains and SCA8 BAC transgenic mice (Fig. 2). CUG ribonuclear inclusions vary in size, number and distribution in different brain regions and among brain samples. In human cerebellar tissue in cases with 109 CTG repeats, the CUG foci are only detected in single molecular layer interneurons. In cases with 400 and 1000 CTG, single foci are found in the nuclei of molecular layer interneurons and the Bergmann glia surrounding the Purkinje cells in the granule cell layer, whereas multiple nuclear foci are detected in the Purkinje cells (Supplementary Material, Table S2). In SCA8 BAC Tg mice expressing 116 CTG repeats, foci have similar distributions in the cerebellar cortex as in human brains with 400 and 1000 repeats (7).
In cerebellar sections of human and mouse SCA8 samples, CUG RNA foci co-localize with MBNL1 in molecular layer interneurons. Interestingly, in Purkinje cells, where MBNL1 is predominantly expressed in the cytoplasm, the nuclear RNA foci are MBNL1-negative (7), indicating that expression of the SCA8 mutation does not trigger the intracellular translocation of MBNL1 and its accumulation in the nucleus where it could amplify the toxicity of CUG RNA foci.
CUG RNA FOCI IN HDL2
Nuclear RNA inclusions resembling those in DM1 brains are detected in HDL2 brains but with relatively short CUG repeat expansions. HDL2 is an autosomal dominant and adult-onset neurodegenerative disease caused by a CTG repeat expansion located in the variably spliced exon 2A of the JPH3 gene (Fig. 1). However, most recent results from BAC transgenic mouse models of HDL2 have provided some evidence to consider the disease also a polyglutamine disorder with bidirectional transcription similar to what is found in SCA8 (67). The repeat length in healthy individuals ranges from 6 to 28 triplets, whereas HDL2 symptoms develop with 40–59 repeats. Pathological features of HDL2 include cortical and basal ganglia degeneration and a loss of medium-size neurons in the striatum in a dorsal-to-ventral gradient.
In the frontal cortex and striatum of HDL2 patient brains, untranslatable JPH3 transcripts with CUG repeat expansions are found in ribonuclear inclusions (Fig. 2). The RNA foci include, in addition to CUG repeats, other parts of the JPH3 message with sequestered MBNL1. Margolis and colleagues (6) reported that an abundance of foci in post-mortem HDL2 brains correlates with CTG repeat length, and shorter repeats give rise to fewer riboinclusions. In the frontal cortex in cases with 51–55 CTG, about 30% of neurons contain foci; most of the neurons have more than one focus, with as many as 13 foci per nucleus. However, in the striatum, which is the most severely affected brain region in HDL2, about 15–20% of neurons have 5–20 foci, which appear brighter than those in the cortex (6). In dentate nuclei of the cerebellum and hippocampus, foci are found only occasionally, in <5% of neurons (Supplementary Material, Table S2).
In HEK293 and HT22 cells, overexpression of truncated JPH3 with exon 2A harboring untranslated 53 CTG repeats was shown to be toxic to the cells and led to the formation of RNA inclusions. Nuclear CUG inclusions are also found in brains of BAC transgenic mice of HDL2 where some of the RNA foci colocalize with nuclear inclusions positive for ubiquitin and immunoreactive with polyQ antibodies (67). In all these cells, CUG foci colocalize with both endogenous and exogenous MBNL1. The presence of MBNL1 is also detected in foci of the frontal cortex of HDL2 patients, however, alternative splicing of APP and MAPT genes, although somehow deviated, is less pronounced than in DM1 (13). Together, these results indicate that expanded CUG repeats in the size range of the HDL2 mutation can be toxic to various mammalian cells (6).
CGG RNA FOCI IN FXTAS
FXTAS is a common genetic disease of the central nervous system that results from an expansion of CGG repeats from 6–54 copies to 55–200 repeats located in the 5′UTR of the fragile X syndrome gene FMR1 (Fig. 1). All individuals with FXTAS are carriers of a pre-mutation of the FMR1, and the full mutation of this gene with more than 200 repeats results in fragile X mental retardation syndrome (FXS) (68). Carriers of the pre-mutation may have some features of the FXS phenotype, including progressive cerebellar tremor and ataxia, cognitive impairment, mild parkinsonian symptoms and brain atrophy (69).
In FXTAS brains, the expression of CGG repeat expansion is associated with the formation of ubiquitin-positive intranuclear inclusions (70). It was therefore asked whether the elevated levels of FMR1 mRNA that are found in pre-mutation carriers have a cumulative toxic effect, leading to the formation of RNA inclusions. In 2004, Hagerman and colleagues (51) identified the presence of FMR1 mRNA within the intranuclear inclusions isolated from the frontal cortex of FXTAS post-mortem human brains (Fig. 2). A single-spot signal of enlarged size was detectable in the nuclei with each of three antisense riboprobes targeting either coding or non-coding (5′UTR and 3′UTR) portions of the FMR1 message (Supplementary Material, Table S2). The inclusions, however, were found only in a subgroup (6–11%) of the analyzed nuclei. These results provided some evidence to indicate that FXTAS is an RNA-mediated disease. However, a later report by Sellier et al. (5) demonstrated that an RNA gain-of-function mechanism and spliceopathy are also features of FXTAS pathogenesis. As shown, expanded FMR1 repeats of pre-mutation lengths are retained in the nuclei of FXTAS post-mortem brains and in the brains of transgenic mice that express 98 CGG repeats. Additionally, ectopic expression of FXTAS repeats in a variety of cells results in the formation of enlarged aggregates, which are dynamic and increase over time (Fig. 2). Interestingly, while PC12, COS7 and SKOV3 cells can support the formation of CGG inclusions, they are not formed in HeLa, HEK293, Neuro-2a and SK-N-MC cells.
In cells expressing expanded CGG repeat RNA, the formation of enlarged foci is associated with the recruitment variety of RNA-binding proteins, including Sam68, hnRNP G and MBNL1. Surprisingly, although MBNL1 is employed as a foci constituent, its free-pool functional level is not exhausted, and there is no aberrant splicing alteration of MBNL1-sensitive genes in expanded CGG repeat-expressing cells. Nevertheless, spliceopathy typical for DM1 has been observed in cells expressing the FXTAS mutation, where a subset of Sam68-sensitive genes is aberrantly spliced (5).
NUCLEAR CAG RNA FOCI
Expansions of CAG repeats are associated with a group of dominantly inherited neurological disorders known as polyglutamine diseases represented by spinocerebellar ataxias type 1, 2, 3, 6, 7 and 17, Huntington's disease (HD), dentatorubral-pallidoluysian atrophy and spinal and bulbar muscular atrophy (Fig. 1). Causative CAG mutations are located in the coding sequences of single unrelated genes and their translation gives rise to polyglutamine-rich proteins. These disorders are characterized by variable degrees of degeneration in the cerebellum, spinocerebellar tracts and brain stem neurons (reviewed in 71).
In CAG repeat expansion diseases, the presence of coding mutation may confer toxicity to the mutant transcripts and to encoded poly-Q proteins. However, the most studied mechanism of pathogenesis is centered on the aberrant ability of mutant proteins to attract cellular proteins, such as ubiquitin, HSP70, proteosome proteins and transcription factors, which leads to the formation of nuclear and cytoplasmic inclusion bodies (reviewed in 71). A role for mutant CAG repeat RNA in poly-Q disorders has been increasingly recognized over the past few years, with experimental evidence revealing its toxic capacity. The results of the studies in human (23) and primate cells (21) and in transgenic fly (48), worm (72) and mouse (22) models unequivocally demonstrate that expanded CAG RNA exhibits some toxic features and therefore could be considered an auxiliary toxic agent (reviewed in 73).
In human HD and SCA3 fibroblasts, endogenous expression of HTT and ATXN3 mutant transcripts has recently been correlated with the formation of enlarged nuclear aggregates that are composed of CAG repeat-containing RNA and MBNL1 protein (23) (A. Mykowska et al., manuscript in preparation). Similar phenomena were observed in primate cells ectopically expressing large and interrupted CAG960 repeat RNA (21). COSM6 cells were found to support the formation of nuclear MBNL1-positive CAG RNA foci with an average number of 13 per nucleus; for comparison, there were 11 RNA inclusions per cell when CUG960 repeats were expressed in COSM6 cells (Supplementary Material, Table S2).
Work in transgenic model organisms bearing untranslated and pathogenic-length CAG repeats revealed the formation of intranuclear RNA inclusions and the occurrence of various abnormalities in the eye, nervous system and muscle, which are not observed in control littermates. In Drosophila, the toxic effect of CAG RNA was repeat-length dependent, and the loss of neuronal integrity detectable with 100 CAG repeats was elevated with 250 CAG repeats (48). Interestingly, overexpression of MblA caused enhanced neurotoxicity, correlated with an increased level of non-coding RNA. Nuclear foci of CAG250 RNA, which were detected in a limited number of cells, were smaller than the robust foci of CUG repeat RNA observed in these flies (Fig. 2). In mouse (22) and Caenorhabditis elegans (72) models, the expression of CAG expansions was correlated with pathogenic changes in muscle structure and function, shortened life span and decreased brood size. In mouse muscle, 200 CAG-containing transcripts accumulated in punctate nuclear RNA foci that sequester MBNL1. Similarly, in C2C12 myoblasts, CAG RNA inclusions are formed after the expression of either 200 or 58 repeats. The abundance of these foci is repeat-length dependent, and twice as few inclusions are formed with 58 CAG repeats than with 200 repeats (3.4 and 6.7 foci/nucleus, respectively) (22). The repeat length, however, does not influence the proportion of foci-positive nuclei. In the CAG58 line, 13.6% of nuclei are foci-positive compared to 17.4% of nuclei in the CAG200 line. No difference in foci number per nucleus was found when comparing CAG200- and CUG200-expressing cells (Supplementary Material, Table S2). In C. elegans, expression of either CAG125 or CUG125 repeats in muscle results in the formation of nuclear RNA foci that are of similar size. Both types of foci recruit C. elegans muscleblind protein (CeMBL), and ∼92% of CAG RNA foci and 60% of CUG foci are enriched with CeMBL. In expanded CAG worms, the overexpression of CeMBL partially reverses the repeat-triggered pathogenesis, as does knock-down of repeat expression with siRNA (72). Thus, toxicity of CAG repeat RNA seems to be expressed by compromising CeMBL function, which is in agreement with results obtained in a variety of cells that express mutant CUG repeat RNA.
RNA FOCI IN SCA10 AND SCA31
SCA10 is an autosomal dominant neurodegenerative disease of the central nervous system with progressive impairment of specific nerve fibers leading to the degradation of cerebellum. The disease-causing mutation is an enormous expansion of a tandem 5-base ATTCT unit present in intron 9 of the ATXN10 gene (reviewed in 74). In normal individuals, the repeat ranges from 10 to 29 and increases up to 4500 in SCA10 patients.
The pathomechanism of SCA10 has been associated with the expression of expanded AUUCU repeat RNA, and the mutant ATXN10 transcript was shown to be the principal molecule capable of triggering neuronal death in SCA10 (9). The mutant RNA expression is associated with the activation of apoptosis and the functional inactivation of the hnRNP K protein, which interacts in situ with AUUCU repeat expansions. The intronic AUUCU repeat mutation is properly spliced out but becomes resistant to degradation and is deposited as numerous aggregates in nuclei and cytoplasm in SCA10 cells. Their presence was shown in SCA10 human fibroblasts and in cells ectopically expressing untranslated AUUCU repeat expansions (9) (Fig. 2). Additionally, SCA10-like aggregates were detected in the brain of transgenic mice expressing ∼500 AUUCU repeats from the β-globin intron sequence, and their abundance was higher in 6-month-old mice than in 3-month-old mice (Supplementary Material, Table S2). Collectively, these results show that the vast SCA10 intronic repeat mutation develops a resistance to degradation and forms insoluble nuclear aggregates similar to the DM2 repeat mutation.
SCA31 is an adult-onset autosomal-dominant neurodegenerative disorder with progressive cerebellar ataxia mainly affecting Purkinje cells. This disease is associated with a pentanucleotide (TGGAA)n repeat expansion, ranging from 2.5 to 3.8 kb in length and spanning two genes, TK2 and BEAN, which are transcribed in opposite directions. Intranuclear RNA foci bearing sense transcripts of BEAN that contains the (UAAAAUAGAA)n repeat were detected in SCA31 Purkinje cells in which about 30–50% of nuclei were foci-positive (Fig. 2). RNA foci were not detected with a probe for the antisense (UUCUAUUUUA)n repeat corresponding to the TK2 transcript (Supplementary Material, Table S2) (8).
CONCLUDING REMARKS AND FUTURE PERSPECTIVES
This review article brings together comprehensive information regarding nuclear foci formed by transcripts of different mutant genes that contain simple sequence repeat expansions and are implicated in the pathogenesis of human hereditary neurological diseases (Fig. 1). It appears that nuclear RNA foci are formed by transcripts harboring different types of repeated motifs, including CUG, CCUG, CGG, CAG, AUUCU and UGGAA of different lengths and expression levels (Fig. 2). We provided detailed characteristics of these foci present in different human tissues and cultured cells and in several model organisms. The foci characteristics include their detection method, morphology, abundance and molecular composition (Supplementary Material, Tables S1, S2 and S3). The picture that emerges from this analysis shows the current state of research on ribonuclear foci, reveals areas of more and less developed research, and allows the pinpointing of some issues that need to be resolved.
It is known that development and progression of RNA-mediated pathology involve expression of expanded repeats (5–7,21,22,75) and the presence of mutant transcript is required to drive RNA foci formation (9,10,37,58). These microscopic RNA structures are found in all cases in which pathology develops and thus are linked to the degeneration process. However, this is still a matter of debate whether RNA foci are a cause of pathology or they are epiphenomena which accompany other symptoms of pathogenesis. Our present knowledge speaking in favor of toxicity of nuclear RNA foci is mostly based on their propensity to affect essential alternative splicing factors causing their sequestration, and compromising their functionality in pre-mRNA processing (2,20,43). One might speculate, however, that the real situation is more complex and the RNA foci might be considered a sink for at least temporary immobilization of many other proteins. One could also imagine putative toxic interactions of mutant transcripts with cellular proteins that occur outside of foci and are triggered by soluble fraction of transcripts. In such scenarios, complexity and variability of symptoms associated with RNA repeat-mediated mechanisms could be explained by differences in protein-binding properties of different repeats and heterogeneity of protein environment in different tissues.
For almost two decades, detection of nuclear repeat expansions has been conducted with FISH, as initially described by Singer and colleagues (1) with some minor modifications. Antisense probes used in FISH varied in repeat length, type and number of fluorescent labels and the presence of backbone modifications, such as PNA or LNA, and were used to enhance the binding efficiency and sensitivity of transcript detection. However, these differences in detection method are unlikely to be responsible for the foci diversity presented here, and most of these varieties must have biological sources. In general, brighter foci detected by FISH are indicative of a higher number of repeats because a greater number of probe molecules hybridize to a target, thereby yielding a strong and clearly distinguishable signal. For example, this phenomenon occurs in DM1 and DM2, which are caused by several hundreds or thousands of repeated units present in the transcripts.
As shown in Supplementary Material, Tables S1 and S2, RNA foci associated with DM1 pathogenesis are the best characterized. The studies on DM2, SCA8 or FXTAS foci are less advanced, and the characterization of nuclear RNA inclusions in cells with CAG repeat expansions that contribute to the pathogenesis of polyglutamine diseases has just begun. Distinct features of the different repeat RNA foci observed in various tissues include their abundance and characteristic morphology (Fig. 2). While DM expansions accumulate as discrete punctate inclusions with sizes and numbers that are somehow related to the expression levels of the mutant transcript, the CAG expansions seem to aggregate as bigger splotches, similar to what is found in expanded CGG expressing cells. The repeat RNA sequence, its secondary structure and the host cell protein environment might modulate nuclear foci morphology. Importantly, the majority of transcripts containing different types of repeats colocalize robustly with MBNL1. It is conceivable that other proteins that have been detected to overlap with foci to some extent or to closely juxtapose with them represent transient and secondary protein–protein interactions. The functions of such proteins do not seem to be severely compromised, as with MBNL1, but their activity is somehow modified, as recently reported in FXTAS (5). It is of importance to resolve the relationship of repeat RNA foci with known nuclear bodies, such as splicing speckles and paraspeckles, which look similar to the RNA foci described for CGG and CAG repeat expansions under the microscope. This association was previously investigated for DM1 and DM2 mutations (1,14,15,17,47).
Therefore, studies in a broader range of cells and tissues combined with careful foci characterization will be essential to explain issues such as cellular factors, which are necessary for foci formation, foci morphology, abundance and their molecular composition. This issue applies to all diseases in which RNA toxicity is involved and to different tissues in which mutant RNA is expressed, not just to tissues in which pathology primarily develops. In the scope of this analysis is also the importance of careful RNA sequencing studies, given the recent discovery that antisense transcription is more ubiquitous than originally thought and since the presence of antisense transcripts has been demonstrated for many genes harboring trinucleotide repeat expansions associated with human neurological disorders (reviewed in 76). In two of them, SCA8 and HDL2, the CAG antisense transcripts seem to be additional sources of cellular toxicity (66,67). A comprehensive analysis of these factors may contribute to a better understanding of the complexity of diseases in tissues affected by pathological processes.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
FUNDING
This work was supported by European Regional Development Fund within Innovative Economy Programme (POIG.01.03.01-00-098/08 to W.J.K.) and Ministry of Science and Higher Education (N N401 572140 to M.W.). Funding to pay the Open Access publication charges for this article was provided by European Regional Development Fund within Innovative Economy Programme (POIG.01.03.01-00-098/08 to W.J.K.).
Supplementary Material
Supplementary Data
REFERENCES
- 1.Taneja K.L., McCurrach M., Schalling M., Housman D., Singer R.H. Foci of trinucleotide repeat transcripts in nuclei of myotonic dystrophy cells and tissues. J. Cell Biol. 1995;128:995–1002. doi: 10.1083/jcb.128.6.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miller J.W., Urbinati C.R., Teng-Umnuay P., Stenberg M.G., Byrne B.J., Thornton C.A., Swanson M.S. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. EMBO J. 2000;19:4439–4448. doi: 10.1093/emboj/19.17.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amack J.D., Paguio A.P., Mahadevan M.S. Cis and trans effects of the myotonic dystrophy (DM) mutation in a cell culture model. Hum. Mol. Genet. 1999;8:1975–1984. doi: 10.1093/hmg/8.11.1975. [DOI] [PubMed] [Google Scholar]
- 4.Davis B.M., McCurrach M.E., Taneja K.L., Singer R.H., Housman D.E. Expansion of a CUG trinucleotide repeat in the 3′ untranslated region of myotonic dystrophy protein kinase transcripts results in nuclear retention of transcripts. Proc. Natl Acad. Sci. USA. 1997;94:7388–7393. doi: 10.1073/pnas.94.14.7388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sellier C., Rau F., Liu Y., Tassone F., Hukema R.K., Gattoni R., Schneider A., Richard S., Willemsen R., Elliott D.J., et al. Sam68 sequestration and partial loss of function are associated with splicing alterations in FXTAS patients. EMBO J. 2010;29:1248–1261. doi: 10.1038/emboj.2010.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rudnicki D.D., Holmes S.E., Lin M.W., Thornton C.A., Ross C.A., Margolis R.L. Huntington's disease–like 2 is associated with CUG repeat-containing RNA foci. Ann. Neurol. 2007;61:272–282. doi: 10.1002/ana.21081. [DOI] [PubMed] [Google Scholar]
- 7.Daughters R.S., Tuttle D.L., Gao W., Ikeda Y., Moseley M.L., Ebner T.J., Swanson M.S., Ranum L.P. RNA gain-of-function in spinocerebellar ataxia type 8. PLoS Genet. 2009;5:e1000600. doi: 10.1371/journal.pgen.1000600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sato N., Amino T., Kobayashi K., Asakawa S., Ishiguro T., Tsunemi T., Takahashi M., Matsuura T., Flanigan K.M., Iwasaki S., et al. Spinocerebellar ataxia type 31 is associated with ‘inserted’ penta-nucleotide repeats containing (TGGAA)n. Am. J. Hum. Genet. 2009;85:544–557. doi: 10.1016/j.ajhg.2009.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.White M.C., Gao R., Xu W., Mandal S.M., Lim J.G., Hazra T.K., Wakamiya M., Edwards S.F., Raskin S., Teive H.A., et al. Inactivation of hnRNP K by expanded intronic AUUCU repeat induces apoptosis via translocation of PKCdelta to mitochondria in spinocerebellar ataxia 10. PLoS Genet. 2010;6:e1000984. doi: 10.1371/journal.pgen.1000984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Margolis J.M., Schoser B.G., Moseley M.L., Day J.W., Ranum L.P. DM2 intronic expansions: evidence for CCUG accumulation without flanking sequence or effects on ZNF9 mRNA processing or protein expression. Hum. Mol. Genet. 2006;15:1808–1815. doi: 10.1093/hmg/ddl103. [DOI] [PubMed] [Google Scholar]
- 11.Taneja K.L. Localization of trinucleotide repeat sequences in myotonic dystrophy cells using a single fluorochrome-labeled PNA probe. Biotechniques. 1998;24:472–476. doi: 10.2144/98243rr02. [DOI] [PubMed] [Google Scholar]
- 12.Mankodi A., Teng-Umnuay P., Krym M., Henderson D., Swanson M., Thornton C.A. Ribonuclear inclusions in skeletal muscle in myotonic dystrophy types 1 and 2. Ann. Neurol. 2003;54:760–768. doi: 10.1002/ana.10763. [DOI] [PubMed] [Google Scholar]
- 13.Jiang H., Mankodi A., Swanson M.S., Moxley R.T., Thornton C.A. Myotonic dystrophy type 1 is associated with nuclear foci of mutant RNA, sequestration of muscleblind proteins and deregulated alternative splicing in neurons. Hum. Mol. Genet. 2004;13:3079–3088. doi: 10.1093/hmg/ddh327. [DOI] [PubMed] [Google Scholar]
- 14.Holt I., Mittal S., Furling D., Butler-Browne G.S., Brook J.D., Morris G.E. Defective mRNA in myotonic dystrophy accumulates at the periphery of nuclear splicing speckles. Genes Cells. 2007;12:1035–1048. doi: 10.1111/j.1365-2443.2007.01112.x. [DOI] [PubMed] [Google Scholar]
- 15.Perdoni F., Malatesta M., Cardani R., Giagnacovo M., Mancinelli E., Meola G., Pellicciari C. RNA/MBNL1-containing foci in myoblast nuclei from patients affected by myotonic dystrophy type 2: an immunocytochemical study. Eur. J. Histochem. 2009;53:151–158. doi: 10.4081/ejh.2009.e18. [DOI] [PubMed] [Google Scholar]
- 16.Kim D.H., Langlois M.A., Lee K.B., Riggs A.D., Puymirat J., Rossi J.J. HnRNP H inhibits nuclear export of mRNA containing expanded CUG repeats and a distal branch point sequence. Nucleic Acids Res. 2005;33:3866–3874. doi: 10.1093/nar/gki698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith K.P., Byron M., Johnson C., Xing Y., Lawrence J.B. Defining early steps in mRNA transport: mutant mRNA in myotonic dystrophy type I is blocked at entry into SC-35 domains. J. Cell Biol. 2007;178:951–964. doi: 10.1083/jcb.200706048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Querido E., Gallardo F., Beaudoin M., Menard C., Chartrand P. Stochastic and reversible aggregation of mRNA with expanded CUG-triplet repeats. J. Cell Sci. 2011;124:1703–1714. doi: 10.1242/jcs.073270. [DOI] [PubMed] [Google Scholar]
- 19.Wheeler T.M., Lueck J.D., Swanson M.S., Dirksen R.T., Thornton C.A. Correction of ClC-1 splicing eliminates chloride channelopathy and myotonia in mouse models of myotonic dystrophy. J. Clin. Invest. 2007;117:3952–3957. doi: 10.1172/JCI33355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mankodi A., Urbinati C.R., Yuan Q.P., Moxley R.T., Sansone V., Krym M., Henderson D., Schalling M., Swanson M.S., Thornton C.A. Muscleblind localizes to nuclear foci of aberrant RNA in myotonic dystrophy types 1 and 2. Hum. Mol. Genet. 2001;10:2165–2170. doi: 10.1093/hmg/10.19.2165. [DOI] [PubMed] [Google Scholar]
- 21.Ho T.H., Savkur R.S., Poulos M.G., Mancini M.A., Swanson M.S., Cooper T.A. Colocalization of muscleblind with RNA foci is separable from mis-regulation of alternative splicing in myotonic dystrophy. J. Cell Sci. 2005;118:2923–2933. doi: 10.1242/jcs.02404. [DOI] [PubMed] [Google Scholar]
- 22.Hsu R.J., Hsiao K.M., Lin M.J., Li C.Y., Wang L.C., Chen L.K., Pan H. Long tract of untranslated CAG repeats is deleterious in transgenic mice. PLoS One. 2011;6:e16417. doi: 10.1371/journal.pone.0016417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Mezer M., Wojciechowska M., Napierala M., Sobczak K., Krzyzosiak W.J. Mutant CAG repeats of Huntingtin transcript fold into hairpins, form nuclear foci and are targets for RNA interference. Nucleic Acids Res. 2011;39:3852–3863. doi: 10.1093/nar/gkq1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brook J.D., McCurrach M.E., Harley H.G., Buckler A.J., Church D., Aburatani H., Hunter K., Stanton V.P., Thirion J.P., Hudson T., et al. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell. 1992;68:799–808. doi: 10.1016/0092-8674(92)90154-5. [DOI] [PubMed] [Google Scholar]
- 25.Mahadevan M., Tsilfidis C., Sabourin L., Shutler G., Amemiya C., Jansen G., Neville C., Narang M., Barcelo J., O'Hoy K., et al. Myotonic dystrophy mutation: an unstable CTG repeat in the 3′ untranslated region of the gene. Science. 1992;255:1253–1255. doi: 10.1126/science.1546325. [DOI] [PubMed] [Google Scholar]
- 26.Liquori C.L., Ricker K., Moseley M.L., Jacobsen J.F., Kress W., Naylor S.L., Day J.W., Ranum L.P. Myotonic dystrophy type 2 caused by a CCTG expansion in intron 1 of ZNF9. Science. 2001;293:864–867. doi: 10.1126/science.1062125. [DOI] [PubMed] [Google Scholar]
- 27.Wheeler T.M., Thornton C.A. Myotonic dystrophy: RNA-mediated muscle disease. Curr. Opin. Neurol. 2007;20:572–576. doi: 10.1097/WCO.0b013e3282ef6064. [DOI] [PubMed] [Google Scholar]
- 28.Osborne R.J., Thornton C.A. RNA-dominant diseases. Hum. Mol. Genet. 2006;15(Spec no. 2):R162–169. doi: 10.1093/hmg/ddl181. [DOI] [PubMed] [Google Scholar]
- 29.Mankodi A., Lin X., Blaxall B.C., Swanson M.S., Thornton C.A. Nuclear RNA foci in the heart in myotonic dystrophy. Circ. Res. 2005;97:1152–1155. doi: 10.1161/01.RES.0000193598.89753.e3. [DOI] [PubMed] [Google Scholar]
- 30.Lin X., Miller J.W., Mankodi A., Kanadia R.N., Yuan Y., Moxley R.T., Swanson M.S., Thornton C.A. Failure of MBNL1-dependent post-natal splicing transitions in myotonic dystrophy. Hum. Mol. Genet. 2006;15:2087–2097. doi: 10.1093/hmg/ddl132. [DOI] [PubMed] [Google Scholar]
- 31.Hamshere M.G., Newman E.E., Alwazzan M., Athwal B.S., Brook J.D. Transcriptional abnormality in myotonic dystrophy affects DMPK but not neighboring genes. Proc. Natl Acad. Sci. USA. 1997;94:7394–7399. doi: 10.1073/pnas.94.14.7394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Botta A., Rinaldi F., Catalli C., Vergani L., Bonifazi E., Romeo V., Loro E., Viola A., Angelini C., Novelli G. The CTG repeat expansion size correlates with the splicing defects observed in muscles from myotonic dystrophy type 1 patients. J. Med. Genet. 2008;45:639–646. doi: 10.1136/jmg.2008.058909. [DOI] [PubMed] [Google Scholar]
- 33.Furling D., Coiffier L., Mouly V., Barbet J.P., St Guily J.L., Taneja K., Gourdon G., Junien C., Butler-Browne G.S. Defective satellite cells in congenital myotonic dystrophy. Hum. Mol. Genet. 2001;10:2079–2087. doi: 10.1093/hmg/10.19.2079. [DOI] [PubMed] [Google Scholar]
- 34.Holt I., Jacquemin V., Fardaei M., Sewry C.A., Butler-Browne G.S., Furling D., Brook J.D., Morris G.E. Muscleblind-like proteins: similarities and differences in normal and myotonic dystrophy muscle. Am. J. Pathol. 2009;174:216–227. doi: 10.2353/ajpath.2009.080520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cardani R., Mancinelli E., Giagnacovo M., Sansone V., Meola G. Ribonuclear inclusions as biomarker of myotonic dystrophy type 2, even in improperly frozen or defrozen skeletal muscle biopsies. Eur. J. Histochem. 2009;53:107–111. doi: 10.4081/ejh.2009.e13. [DOI] [PubMed] [Google Scholar]
- 36.Cardani R., Mancinelli E., Saino G., Bonavina L., Meola G. A putative role of ribonuclear inclusions and MBNL1 in the impairment of gallbladder smooth muscle contractility with cholelithiasis in myotonic dystrophy type 1. Neuromuscul. Disord. 2008;18:641–645. doi: 10.1016/j.nmd.2008.06.366. [DOI] [PubMed] [Google Scholar]
- 37.Amack J.D., Mahadevan M.S. The myotonic dystrophy expanded CUG repeat tract is necessary but not sufficient to disrupt C2C12 myoblast differentiation. Hum. Mol. Genet. 2001;10:1879–1887. doi: 10.1093/hmg/10.18.1879. [DOI] [PubMed] [Google Scholar]
- 38.Dansithong W., Wolf C.M., Sarkar P., Paul S., Chiang A., Holt I., Morris G.E., Branco D., Sherwood M.C., Comai L., et al. Cytoplasmic CUG RNA foci are insufficient to elicit key DM1 features. PLoS One. 2008;3:e3968. doi: 10.1371/journal.pone.0003968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wheeler T.M., Sobczak K., Lueck J.D., Osborne R.J., Lin X., Dirksen R.T., Thornton C.A. Reversal of RNA dominance by displacement of protein sequestered on triplet repeat RNA. Science. 2009;325:336–339. doi: 10.1126/science.1173110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mulders S.A., van den Broek W.J., Wheeler T.M., Croes H.J., van Kuik-Romeijn P., de Kimpe S.J., Furling D., Platenburg G.J., Gourdon G., Thornton C.A., et al. Triplet-repeat oligonucleotide-mediated reversal of RNA toxicity in myotonic dystrophy. Proc. Natl Acad. Sci. USA. 2009;106:13915–13920. doi: 10.1073/pnas.0905780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Francois V., Klein A.F., Beley C., Jollet A., Lemercier C., Garcia L., Furling D. Selective silencing of mutated mRNAs in DM1 by using modified hU7-snRNAs. Nat. Struct. Mol. Biol. 2011;18:85–87. doi: 10.1038/nsmb.1958. [DOI] [PubMed] [Google Scholar]
- 42.Langlois M.A., Lee N.S., Rossi J.J., Puymirat J. Hammerhead ribozyme-mediated destruction of nuclear foci in myotonic dystrophy myoblasts. Mol. Ther. 2003;7:670–680. doi: 10.1016/s1525-0016(03)00068-6. [DOI] [PubMed] [Google Scholar]
- 43.Warf M.B., Nakamori M., Matthys C.M., Thornton C.A., Berglund J.A. Pentamidine reverses the splicing defects associated with myotonic dystrophy. Proc. Natl Acad. Sci. USA. 2009;106:18551–18556. doi: 10.1073/pnas.0903234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lucchiari S., Pagliarani S., Corti S., Mancinelli E., Servida M., Fruguglietti E., Sansone V., Moggio M., Bresolin N., Comi G.P., et al. Colocalization of ribonuclear inclusions with muscle blind like-proteins in a family with myotonic dystrophy type 2 associated with a short CCTG expansion. J. Neurol. Sci. 2008;275:159–163. doi: 10.1016/j.jns.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 45.Mahadevan M.S., Yadava R.S., Yu Q., Balijepalli S., Frenzel-McCardell C.D., Bourne T.D., Phillips L.H. Reversible model of RNA toxicity and cardiac conduction defects in myotonic dystrophy. Nat. Genet. 2006;38:1066–1070. doi: 10.1038/ng1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Haro M., Al-Ramahi I., De Gouyon B., Ukani L., Rosa A., Faustino N.A., Ashizawa T., Cooper T.A., Botas J. MBNL1 and CUGBP1 modify expanded CUG-induced toxicity in a Drosophila model of myotonic dystrophy type 1. Hum. Mol. Genet. 2006;15:2138–2145. doi: 10.1093/hmg/ddl137. [DOI] [PubMed] [Google Scholar]
- 47.Houseley J.M., Wang Z., Brock G.J., Soloway J., Artero R., Perez-Alonso M., O'Dell K.M., Monckton D.G. Myotonic dystrophy associated expanded CUG repeat muscleblind positive ribonuclear foci are not toxic to Drosophila. Hum. Mol. Genet. 2005;14:873–883. doi: 10.1093/hmg/ddi080. [DOI] [PubMed] [Google Scholar]
- 48.Li L.B., Yu Z., Teng X., Bonini N.M. RNA toxicity is a component of ataxin-3 degeneration in Drosophila. Nature. 2008;453:1107–1111. doi: 10.1038/nature06909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schoser B.G., Ricker K., Schneider-Gold C., Hengstenberg C., Durre J., Bultmann B., Kress W., Day J.W., Ranum L.P. Sudden cardiac death in myotonic dystrophy type 2. Neurology. 2004;63:2402–2404. doi: 10.1212/01.wnl.0000147335.10783.e4. [DOI] [PubMed] [Google Scholar]
- 50.Chen I.C., Lin H.Y., Lee G.C., Kao S.H., Chen C.M., Wu Y.R., Hsieh-Li H.M., Su M.T., Lee-Chen G.J. Spinocerebellar ataxia type 8 larger triplet expansion alters histone modification and induces RNA foci. BMC Mol. Biol. 2009;10:1–8. doi: 10.1186/1471-2199-10-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tassone F., Iwahashi C., Hagerman P.J. FMR1 RNA within the intranuclear inclusions of fragile X-associated tremor/ataxia syndrome (FXTAS) RNA Biol. 2004;1:103–105. doi: 10.4161/rna.1.2.1035. [DOI] [PubMed] [Google Scholar]
- 52.Cardani R., Mancinelli E., Rotondo G., Sansone V., Meola G. Muscleblind-like protein 1 nuclear sequestration is a molecular pathology marker of DM1 and DM2. Eur. J. Histochem. 2006;50:177–182. [PubMed] [Google Scholar]
- 53.Cardani R., Mancinelli E., Sansone V., Rotondo G., Meola G. Biomolecular identification of (CCTG)n mutation in myotonic dystrophy type 2 (DM2) by FISH on muscle biopsy. Eur. J. Histochem. 2004;48:437–442. doi: 10.4081/918. [DOI] [PubMed] [Google Scholar]
- 54.Savkur R.S., Philips A.V., Cooper T.A. Aberrant regulation of insulin receptor alternative splicing is associated with insulin resistance in myotonic dystrophy. Nat. Genet. 2001;29:40–47. doi: 10.1038/ng704. [DOI] [PubMed] [Google Scholar]
- 55.Philips A.V., Timchenko L.T., Cooper T.A. Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science. 1998;280:737–741. doi: 10.1126/science.280.5364.737. [DOI] [PubMed] [Google Scholar]
- 56.Fardaei M., Larkin K., Brook J.D., Hamshere M.G. In vivo co-localisation of MBNL protein with DMPK expanded-repeat transcripts. Nucleic Acids Res. 2001;29:2766–2771. doi: 10.1093/nar/29.13.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fardaei M., Rogers M.T., Thorpe H.M., Larkin K., Hamshere M.G., Harper P.S., Brook J.D. Three proteins, MBNL, MBLL and MBXL, co-localize in vivo with nuclear foci of expanded-repeat transcripts in DM1 and DM2 cells. Hum. Mol. Genet. 2002;11:805–814. doi: 10.1093/hmg/11.7.805. [DOI] [PubMed] [Google Scholar]
- 58.Mankodi A., Logigian E., Callahan L., McClain C., White R., Henderson D., Krym M., Thornton C.A. Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science. 2000;289:1769–1773. doi: 10.1126/science.289.5485.1769. [DOI] [PubMed] [Google Scholar]
- 59.Wang G.S., Kearney D.L., De Biasi M., Taffet G., Cooper T.A. Elevation of RNA-binding protein CUGBP1 is an early event in an inducible heart-specific mouse model of myotonic dystrophy. J. Clin. Invest. 2007;117:2802–2811. doi: 10.1172/JCI32308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Orengo J.P., Chambon P., Metzger D., Mosier D.R., Snipes G.J., Cooper T.A. Expanded CTG repeats within the DMPK 3′ UTR causes severe skeletal muscle wasting in an inducible mouse model for myotonic dystrophy. Proc. Natl Acad. Sci. USA. 2008;105:2646–2651. doi: 10.1073/pnas.0708519105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Timchenko N.A., Patel R., Iakova P., Cai Z.J., Quan L., Timchenko L.T. Overexpression of CUG triplet repeat-binding protein, CUGBP1, in mice inhibits myogenesis. J. Biol. Chem. 2004;279:13129–13139. doi: 10.1074/jbc.M312923200. [DOI] [PubMed] [Google Scholar]
- 62.Ho T.H., Bundman D., Armstrong D.L., Cooper T.A. Transgenic mice expressing CUG-BP1 reproduce splicing mis-regulation observed in myotonic dystrophy. Hum. Mol. Genet. 2005;14:1539–1547. doi: 10.1093/hmg/ddi162. [DOI] [PubMed] [Google Scholar]
- 63.Kanadia R.N., Johnstone K.A., Mankodi A., Lungu C., Thornton C.A., Esson D., Timmers A.M., Hauswirth W.W., Swanson M.S. A muscleblind knockout model for myotonic dystrophy. Science. 2003;302:1978–1980. doi: 10.1126/science.1088583. [DOI] [PubMed] [Google Scholar]
- 64.Le Mee G., Ezzeddine N., Capri M., Ait-Ahmed O. Repeat length and RNA expression level are not primary determinants in CUG expansion toxicity in Drosophila models. PLoS One. 2008;3:e1466. doi: 10.1371/journal.pone.0001466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Garcia-Lopez A., Monferrer L., Garcia-Alcover I., Vicente-Crespo M., Alvarez-Abril M.C., Artero R.D. Genetic and chemical modifiers of a CUG toxicity model in Drosophila. PLoS One. 2008;3:e1595. doi: 10.1371/journal.pone.0001595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moseley M.L., Zu T., Ikeda Y., Gao W., Mosemiller A.K., Daughters R.S., Chen G., Weatherspoon M.R., Clark H.B., Ebner T.J., et al. Bidirectional expression of CUG and CAG expansion transcripts and intranuclear polyglutamine inclusions in spinocerebellar ataxia type 8. Nat. Genet. 2006;38:758–769. doi: 10.1038/ng1827. [DOI] [PubMed] [Google Scholar]
- 67.Wilburn B., Rudnicki D.D., Zhao J., Weitz T.M., Cheng Y., Gu X., Greiner E., Park C.S., Wang N., Sopher B.L., et al. An antisense CAG repeat transcript at JPH3 locus mediates expanded polyglutamine protein toxicity in Huntington's disease-like 2 mice. Neuron. 2011;70:427–440. doi: 10.1016/j.neuron.2011.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schneider A., Hagerman R.J., Hessl D. Fragile X syndrome—from genes to cognition. Dev. Disabil. Res. Rev. 2009;15:333–342. doi: 10.1002/ddrr.80. [DOI] [PubMed] [Google Scholar]
- 69.Hagerman R.J., Leehey M., Heinrichs W., Tassone F., Wilson R., Hills J., Grigsby J., Gage B., Hagerman P.J. Intention tremor, parkinsonism, and generalized brain atrophy in male carriers of fragile X. Neurology. 2001;57:127–130. doi: 10.1212/wnl.57.1.127. [DOI] [PubMed] [Google Scholar]
- 70.Greco C.M., Hagerman R.J., Tassone F., Chudley A.E., Del Bigio M.R., Jacquemont S., Leehey M., Hagerman P.J. Neuronal intranuclear inclusions in a new cerebellar tremor/ataxia syndrome among fragile X carriers. Brain. 2002;125:1760–1771. doi: 10.1093/brain/awf184. [DOI] [PubMed] [Google Scholar]
- 71.Orr H.T., Zoghbi H.Y. Trinucleotide repeat disorders. Ann. Rev. Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- 72.Wang L.C., Chen K.Y., Pan H., Wu C.C., Chen P.H., Liao Y.T., Li C., Huang M.L., Hsiao K.M. Muscleblind participates in RNA toxicity of expanded CAG and CUG repeats in Caenorhabditis elegans. Cell. Mol. Life Sci. 2011;68:1255–1267. doi: 10.1007/s00018-010-0522-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wojciechowska M., Krzyzosiak W.J. CAG repeat RNA as an auxiliary toxic agent in polyglutamine disorders. RNA Biol. 2011;8:1–7. doi: 10.4161/rna.8.4.15397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lin X., Ashizawa T. Recent progress in spinocerebellar ataxia type-10 (SCA10) Cerebellum. 2005;4:37–42. doi: 10.1080/14734220510007897. [DOI] [PubMed] [Google Scholar]
- 75.Amack J.D., Reagan S.R., Mahadevan M.S. Mutant DMPK 3′-UTR transcripts disrupt C2C12 myogenic differentiation by compromising MyoD. J. Cell Biol. 2002;159:419–429. doi: 10.1083/jcb.200206020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Batra R., Charizanis K., Swanson M.S. Partners in crime: bidirectional transcription in unstable microsatellite disease. Hum. Mol. Genet. 2010;19:R77–82. doi: 10.1093/hmg/ddq132. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data