The competence transcription factor of Bacillus subtilis recognizes short A/T-rich sequences arranged in a unique, flexible pattern along the DNA helix (original) (raw)
Abstract
The development of genetic competence in Bacillus subtilis is regulated by a complex signal transduction cascade, which leads to the synthesis of the competence transcription factor (CTF). Previous studies suggested that CTF is encoded by comK. ComK is required for the transcription of comK itself, as well as of the late competence genes encoding the DNA uptake machinery and of genes required for homologous recombination. Here, we used purified ComK to study its role in transcription and to determine the DNA recognition sequence for ComK. In vitro transcription from the comG promoter, which depends on ComK in vivo, was observed on the addition of purified ComK together with Bacillus subtilis RNA polymerase, proving that ComK is CTF. To determine the DNA sequences involved in ComK recognition, footprinting analysis was performed with promoter fragments of the CTF-dependent genes: comC, comE, comF, comG, comK, and addAB. The ComK binding sites determined by DNase I protection experiments were unusually long, with average lengths of ∼65 bp, and displayed only weak sequence similarities. Hydroxy-radical footprinting, performed with the addAB promoter, revealed a unique arrangement of four short A/T-rich sequences. Gel retardation experiments indicated that four molecules of ComK bound the addAB promoter and the dyad symmetrical arrangement of the four A/T-rich sequences implied that ComK functions as a tetramer composed of two dimers each recognizing the motif AAAAN5TTTT. Comparable A/T-rich sequences were identified in all six DNase I footprints and could be used to predict ComK targets in the B. subtilis genome. On the basis of the variability in distance between the ComK–dimer binding sites, ComK-regulated promoters could be divided into three classes, demonstrating a remarkable flexibility in the binding of ComK.The pattern of hydroxy-radical protections suggested that ComK binds at one face of the DNA helix through the minor groove. This inference was strengthened by the observation that minor groove binding drugs inhibited the binding of ComK.
Keywords: Bacillus subtilis, CTF, competence, ComK, footprinting, minor groove
Several prokaryotic species spontaneously develop the ability to take up exogenous DNA, known as competence. The development of genetic competence has been studied most extensively in the Gram-positive bacterium Bacillus subtilis. These studies have revealed the existence of a complex regulatory cascade that senses and interprets environmental conditions and directs development along the competence pathway when these conditions are appropriate. Most of the regulatory proteins that comprise this cascade are also required for other postexponential phase processes. Thus, the competence regulation cascade is integrated in a complex signal transduction network, which not only controls competence, but also sporulation, motility, and the production of degradative enzymes and antibiotics (for review, see Dubnau 1993; Grossman 1995).
Regulation via the competence signal transduction cascade ultimately results in the synthesis of the competence transcription factor (CTF), which activates the transcription of the late competence genes encoding the DNA binding and uptake machinery (comC, comE, comF, comG), as well as genes necessary for recombination (recA, addAB) (Mohan and Dubnau 1990; Van Sinderen et al. 1995; Haijema et al. 1995a,b, 1996). The inference that comK encodes the CTF was derived from (1) extensive epistatic interaction studies (Mohan and Dubnau 1990; Van Sinderen and Venema 1994), (2) the demonstration of transcriptional activation of late competence promoters in a ComK-expressing Escherichia coli strain, and (3) the in vitro demonstration of ComK binding to these promoters (Van Sinderen et al. 1995b). During exponential growth, ComK is kept inactive by the formation of a ternary complex with MecA and the chaperone-like protein, ClpC (Kong and Dubnau 1994; Msadek et al. 1994; Turgay et al. 1997). Toward the end of the exponential growth phase, two quorum-sensing pheromones, ComX and CSF, accumulate in the medium, activating a pathway that results in the phosphorylation of the response regulator ComA (Magnuson et al. 1994; Solomon et al. 1996). ComA∼P activates the transcription of srfA, which encodes the synthetase of the lipopeptide antibiotic, surfactin (Nakano and Zuber 1989; Roggiani and Dubnau 1993). Embedded within the srfA operon is a small gene, comS (D’Souza et al. 1994; Hamoen et al. 1995). The synthesis of ComS in response to the quorum-sensing signal transduction pathway destabilizes the ternary ComK/MecA/ClpC complex in which ComK is held inactive (Turgay et al. 1997). ComS probably acts by binding to MecA (Liu et al. 1996). Because ComK synthesis is positively autoregulated (Van Sinderen and Venema 1994), the release of active ComK from the ternary complex results in the rapid accumulation of CTF. In addition to ComK itself, the DNA binding proteins DegU, AbrB, and SinR are also required for the transcriptional activation of comK (Van Sinderen and Venema 1994; Hahn et al. 1996). For full competence development, repression at the comK promoter caused by the binding of AbrB and CodY must be relieved as well (Hahn et al. 1995; Serror and Sonenshein 1996).
To provide conclusive evidence that ComK is the CTF, we have optimized the purification of ComK and have shown that ComK stimulates transcription in vitro. Despite the availability of a considerable number of ComK-binding promoter sequences, a convincing ComK recognition sequence has escaped definition. This led Van Sinderen et al. (1995b) to propose that ComK, rather than recognizing a specific DNA sequence, might recognize a structural feature of the target DNA such as intrinsic bending. In the present study, we show that intrinsic DNA bending is not involved in the specific recognition of promoter sequences and that instead, ComK binds as a pair of dimers in a sequence specific manner, probably recognizing bases in the minor groove of the DNA helix. The deduced consensus motifs consist of four short A/T-rich stretches organized in a flexible, but unique pattern along the DNA helix.
Results
ComK is sufficient to activate ComK-controlled genes
Previously, we have described the binding of ComK to the promoter regions of the late competence genes which, together with genetic data, strongly suggested that comK encodes CTF (Van Sinderen et al. 1995b). In that study ComK was purified as a fusion to the maltose binding protein (MBP) from E. coli. The cloning of a protease factor-Xa cleavage site between the two protein moieties made it possible to separate ComK from MBP. Our initial attempts to demonstrate ComK-dependent transcription of late competence genes in vitro resulted in failure. Subsequently, we discovered that our ComK preparations were heavily contaminated with chromosomal DNA, which could be removed by DEAE–Sepharose chromatography. The presence of chromosomal DNA offered a plausible explanation for the failure of our in vitro transcription experiments, because this DNA would be expected to compete with added template for RNA polymerase. Although DEAE–Sepharose chromatography readily separated ComK from DNA, pure ComK rapidly precipitated. After screening a variety of conditions, we discovered that the inclusion of phosphate or sulfate as the major anions in our buffers maintained ComK in soluble form.
The activity of DNA-free ComK was tested in an in vitro transcription assay by use of the comG promoter to drive transcription. For this experiment, the comG promoter was cloned in the multiple cloning site of pPAN583, followed by the strong T7 terminator (Predich et al. 1992). The resulting pPAN-G construct was incubated in a transcription buffer containing purified RNA polymerase from B. subtilis. In the absence of ComK, no RNA transcript was visible, whereas in its presence, an abundant transcript of the predicted size was formed (Fig. 1). From these findings, together with our previous results, we conclude that comK encodes CTF.
Figure 1.
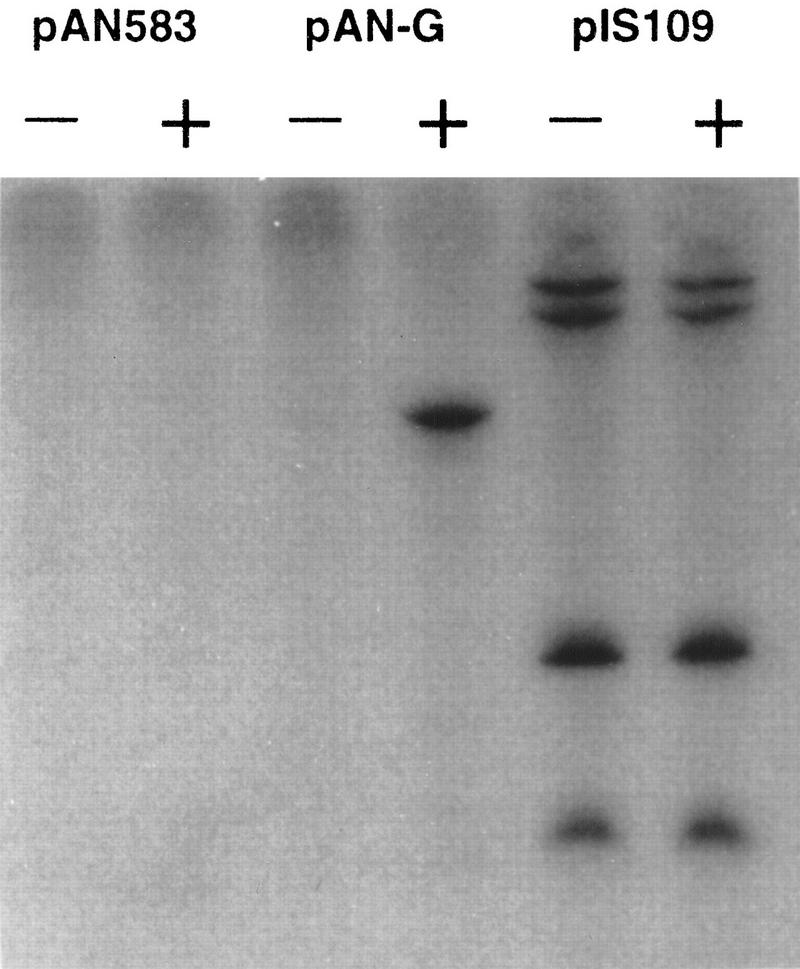
In vitro transcription assays in the absence (−) or presence (+) of ComK. The templates used were plasmid pAN583, plasmid pAN583 containing the comG promoter (pAN-G), and a positive control (pIS109) with transcripts of 142, 184, 455, and 497 nucleotides in length.
ComK protects extended DNA segments upstream from its target promoters
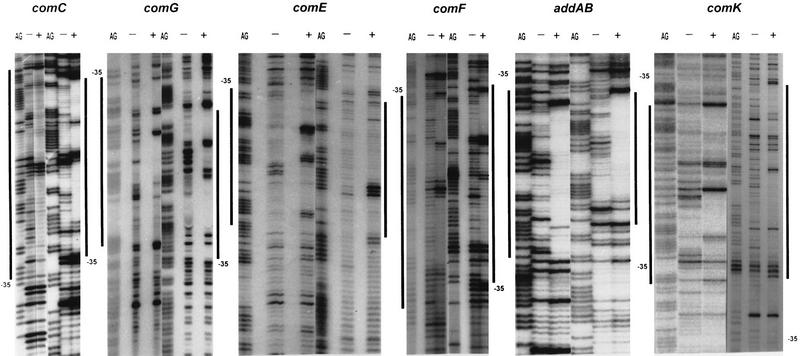
Although a considerable number of ComK-regulated genes are known, no clear DNA binding consensus sequence could be determined. Dubnau (1993) postulated a dyad symmetrical sequence on the basis of a weak similarity between the upstream sequences of comC, comG, comE, and recA. However, this sequence is absent from the upstream regions of more recently characterized ComK-dependent promoters, notably comK and addAB (Van Sinderen et al. 1995; Haijema et al. 1996) . To identify the DNA-binding sequence of ComK, DNase I protection analyses were performed with 32P-labeled PCR fragments containing the comC and comG promoter regions. In the case of comG, binding of ComK resulted in the protection of DNA sequences extending from the −35 consensus to position −110, relative to the start of transcription (Figs. 2 and 3). In addition to the protected regions, several hypersensitive bonds were obvious. As comparison between the comC and comG footprints did not reveal a convincing recognition sequence, additional ComK-regulated promoters were included in the analyses. Figures 2 and 3 show that binding of ComK protected large regions upstream of the comE, comF, comK, and addAB promoters, and again prominent hypersensitive bonds were apparent. The average protected region extended from the −35 sequence to a point ∼65-bp upstream. The proposed CTF recognition sequence was not detectable within the protected regions of the comE, comF, comK, and addAB promoters. Comparison of the six ComK footprints did not reveal an obvious alternative ComK recognition motif.
Figure 2.
DNase I footprinting analysis of ComK binding to the comC, comG, comE, comF, addAB, and comK promoter regions. The left and right halves of each panel represents the footprints of the upper and lower strands, respectively. Footprints obtained in the presence (+) or absence (−) of ComK are flanked by G+A sequence ladders. The protected regions are marked with bars, and the position of the −35 promoter sequence is indicated.
Figure 3.
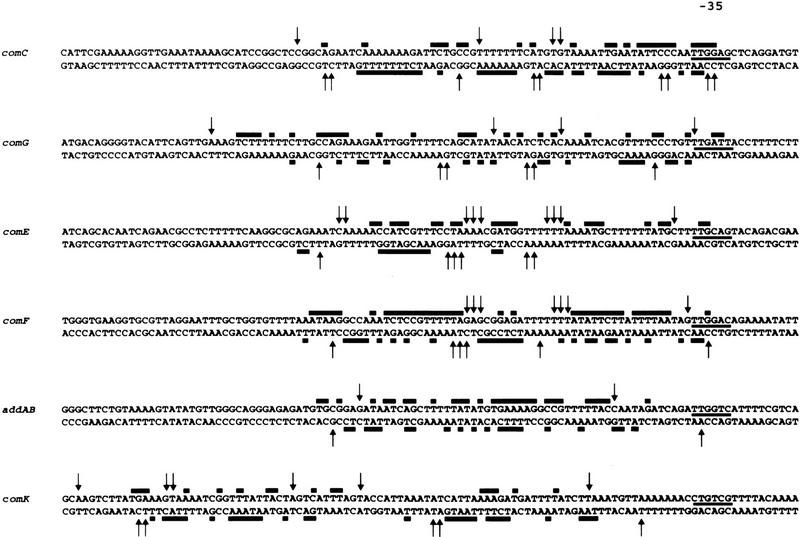
Summary of DNase I footprinting data. Protected bases are marked by solid bars, and hypersensitive sites by arrows. The −35 promoter sequences are underlined.
As noted above, the protected regions are flanked and divided by hypersensitive sites, indicative of local deformation, presumably caused by bending of the helix. Intrinsic bending can be important for protein–DNA recognition, and clear recognition sequences for such regulators may be absent. Because the ComK protected sequences were A/T rich, and short A/T tracks have been shown to bend the DNA helix (Koo et al. 1986; Perez-Martin et al. 1994), we determined whether intrinsic bending in the competence promoters could be involved in the recognition by ComK.
Intrinsic bending is not required for ComK recognition
To determine whether ComK-regulated promoters contain a specific intrinsic bend that could account for the ComK binding specificity, we performed a circular permutation analysis with the plasmid pBend (Kim et al. 1989). For this experiment, the comG promoter was cloned between the tandem multiple cloning sites of pBend. DNA restriction fragments of equal size, carrying the comG sequences at different positions with respect to the fragment termini, were subjected to mobility analysis. The concentration of ComK was such that only a fraction of the DNA fragments were bound to ComK. The results are presented in Figure 4, and show that the unbound DNA fragments migrated at the same rate, indicating that the comG promoter fragment does not contain a detectable intrinsic bend. In contrast, the retardation caused by ComK differed in magnitude among the permuted DNA fragments, implying the existence of an induced bend, for which an apparent curvature of ∼75° was calculated. A comparable experiment with the comF promoter also indicated the absence of intrinsic bending. In this case, the magnitude of the ComK-induced bend was estimated to be ∼65° (results not shown). Although binding of ComK to the competence promoters introduced bending, we may conclude that intrinsic bending of the target DNA was not required for recognition by ComK.
Figure 4.
Gel retardation of permuted DNA fragments containing the comG promoter in the presence of ComK. Restriction enzymes used to release the various fragments from plasmid pBend-G were _Bgl_II (lane 1), _Nhe_I (lane 2), _Eco_RV (lane 3), _Sma_I (lane 4), _Nru_I (lane 5), and _Bam_HI (lane 6). The arrow locates pBend, and retarded and unretarded fragments are marked with + and −, respectively.
Four molecules of ComK bind to its targets
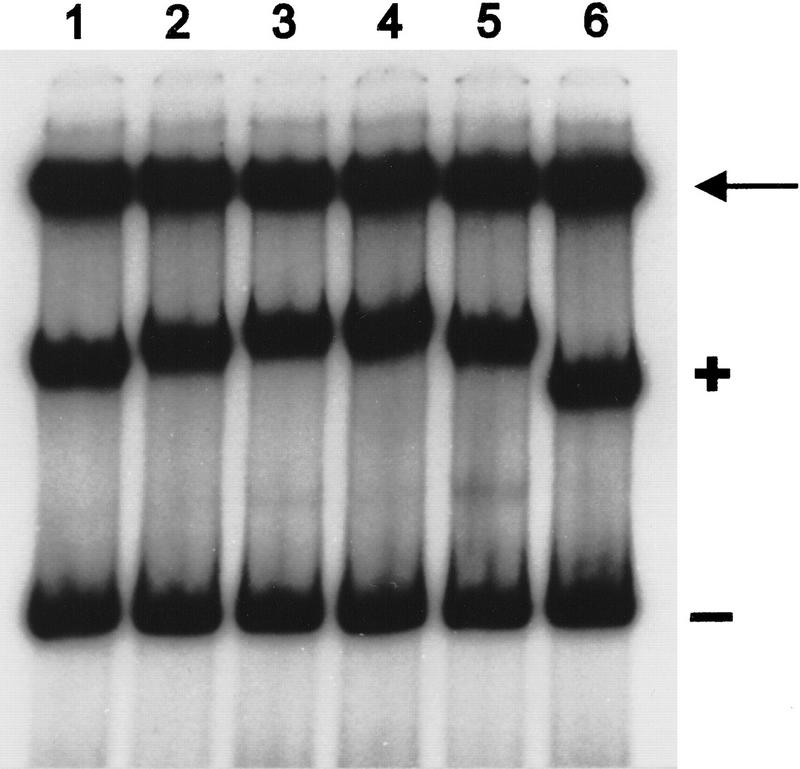
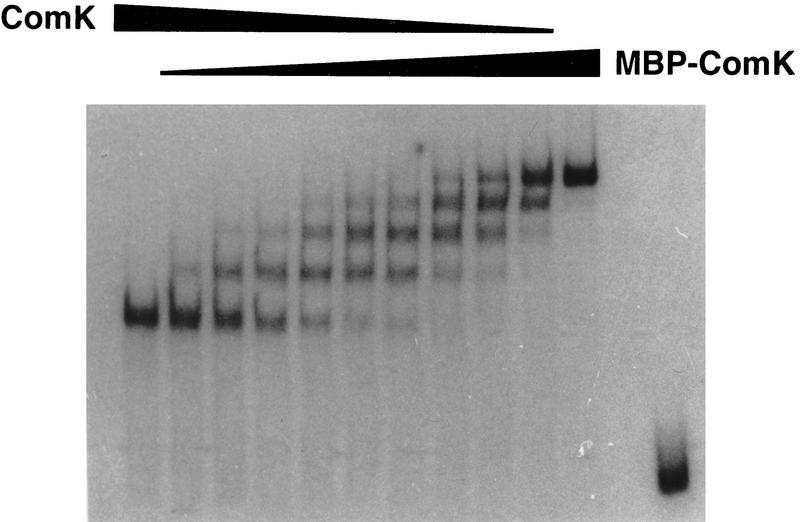
The extension and diversity in footprint lengths between the various promoters hampered the search for conserved sequences involved in ComK recognition. Knowledge about the difference in number of ComK molecules recognizing these promoters, and therefore the number of recognition motifs in each promoter, would facilitate the definition of such motifs. The availability of pure ComK and MBP–ComK fusion protein provided us with a means to determine the number of ComK molecules that bind to ComK-regulated promoters (Lane et al. 1992). From previous studies it was known that, although the MBP moiety of the MBP–ComK fusion protein did not interfere with the binding specificity of the ComK moiety, it caused an increase in retardation because of its additional mass (42 kD; Duplay et al. 1984). Mixtures of the fusion protein with native ComK should, therefore, reveal the stoichiometry of ComK binding in gel retardation experiments. For instance, if one molecule of ComK were to bind to each promoter, a mixture of fusion protein and ComK would result in two retarded DNA fragments: A fast migrating band bound to ComK and a slowly migrating band bound to MBP–ComK. If two molecules of ComK were to bind, an intermediate band would appear. Following this rule, the number of molecules bound to each promoter fragment will always be one more than the number of intermediate bands. In a gel retardation experiment with the comC promoter as probe, incubation with varying mixtures of ComK and MBP–ComK revealed three intermediate bands, indicating that four molecules of ComK bind to the comC promoter (Fig. 5). Although the ComK footprints on the addAB and comK promoters differed substantially in length compared with that obtained with the comC promoter, a comparable titration experiment also revealed three intermediate bands with an addAB or comK promoter fragment (results not shown). Apparently, competence-specific promoters are recognized by four ComK molecules.
Figure 5.
Gel retardation assay by use of 32P-labeled comC promoter fragments incubated with ComK and MBP–ComK mixed in various proportions. (First and penultimate lanes) No MBP–ComK added and no ComK added, respectively. (Last lane) Probe with no protein added.
ComK recognizes a specific pattern of short A/T-rich stretches
Because most recognition sequences are relatively short, the low resolution of DNase I footprints generally presents no obstacle for deducing recognition sequences, but did so in the case of the extended regions protected by ComK binding. To improve the resolution of our analysis, we resorted to hydroxy-radical protection analysis. For this, the addAB promoter was chosen because the DNase I-protected region of this promoter was the smallest of the promoters tested. Binding of ComK to the addAB promoter resulted in a striking periodic pattern of four protected areas of 3–5 nucleotides in length, with a periodicity close to 10 nucleotides (Fig. 6), indicating that ComK molecules bind to one side of the helix (Lane et al. 1992). The protected sequences in the lower and upper strand were within the DNase I-protected regions, and were rich in A/T-nucleotides.
Figure 6.
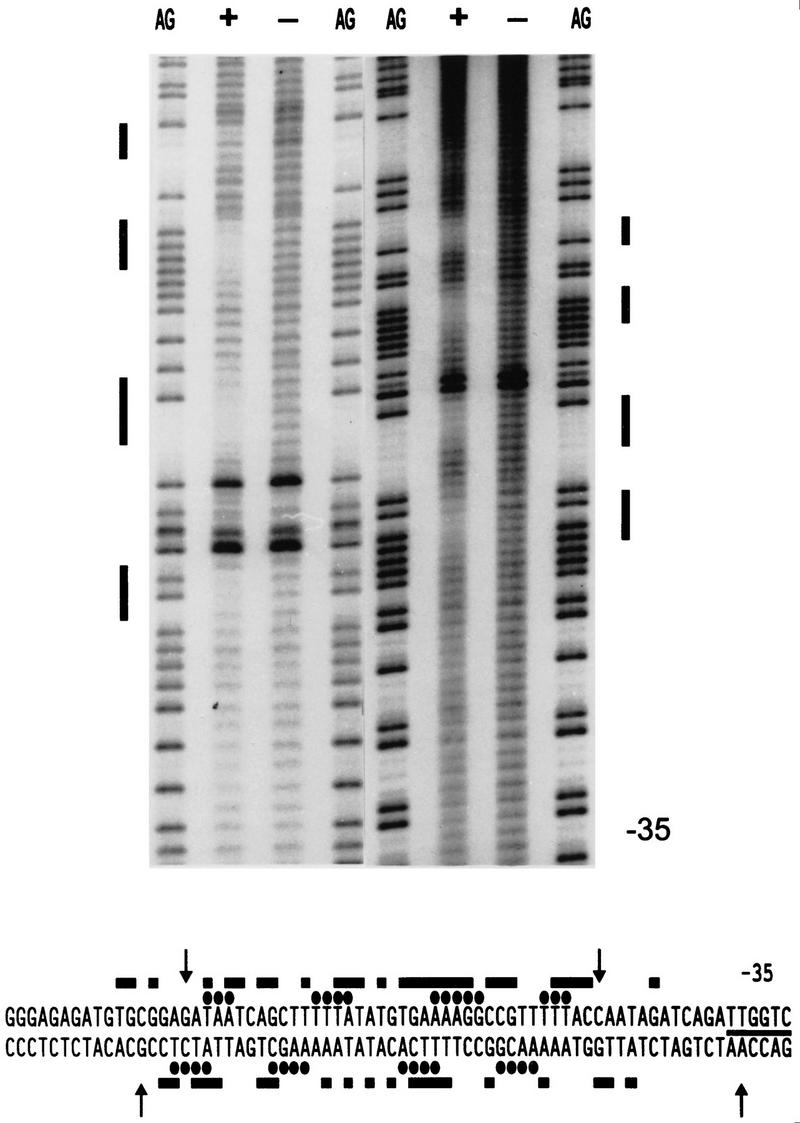
Hydroxy-radical footprinting analysis of ComK binding to the addAB promoter. Footprints obtained in the presence (+) or absence (−) of ComK are flanked by G+A sequence ladders. The left and right halves represent the footprints of the upper and lower strands, respectively. The protected regions are marked with bars and the position of the −35 promoter sequence is indicated. The origin of the hyperactive bands is unknown, but they are not affected by the presence of ComK and do not interfere with the footprint results. In the sequence, hydroxy-radical-protected residues are marked by ellipses, and DNase I protected residues by bars. Arrows indicate hypersensitive sites in the DNase I footprint.
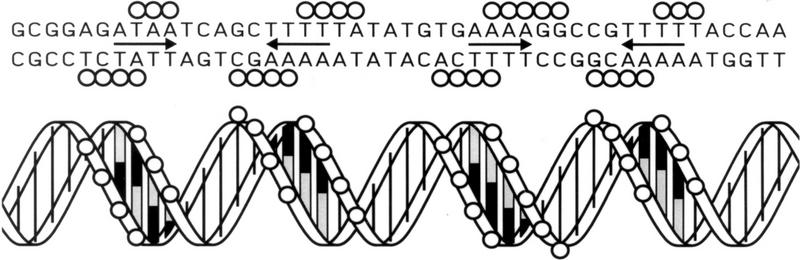
Any model of ComK binding to its target promoters must accommodate the following observations: (1) Four molecules of ComK bind to each competence-promoter; (2) extended sequences are protected from DNase I cleavage; and (3) the protection from hydroxy-radical damage occurs in four periodically distributed stretches of nucleotides. To help interpret these observations, the nucleotide sequences of the addAB, comC, comG, comE, comF, and comK promoter fragments were represented as helical projections (Fig. 7). The four protected regions of the addAB promoter, revealed by the hydroxy-radical footprint, are assumed to correspond to the four molecules of ComK bound to this promoter. Inspection of the hydroxy-radical-protected regions on the helical projection revealed two similar motifs, separated by a single helical turn, exhibiting dyad symmetry, suggesting that dimers of ComK bind to each motif. To examine whether the dimer-specific symmetry was also present in additional ComK-regulated promoters, the DNase I footprints of the other four promoters were searched for the presence of the ComK-dimer recognition pattern noted for addAB. This pattern was found in all of these promoters (Fig. 7). Not only was the sequence similarity within the inferred motifs high, but the localization relative to the −35 sequence was comparable. As compared with addAB, the extra spacing of one helical turn in the comC, comG, comE, and comF promoters and of two helical turns in the comK promoter provided an explanation for the more extended footprints of the latter set of promoters. These results strongly suggest that the deduced ComK recognition pattern is correct and that, in a linear representation, the recognition sequence can be defined as two AAAAN5TTTT tracts (AT box) separated by 8, 18, or 31 nucleotides. The validity of this consensus is supported by the fact that comparable recognition patterns can be identified in all promoter rgions known so far to be under the direct regulation of ComK (see Fig. 12, below, and the Discussion for details).
Figure 7.
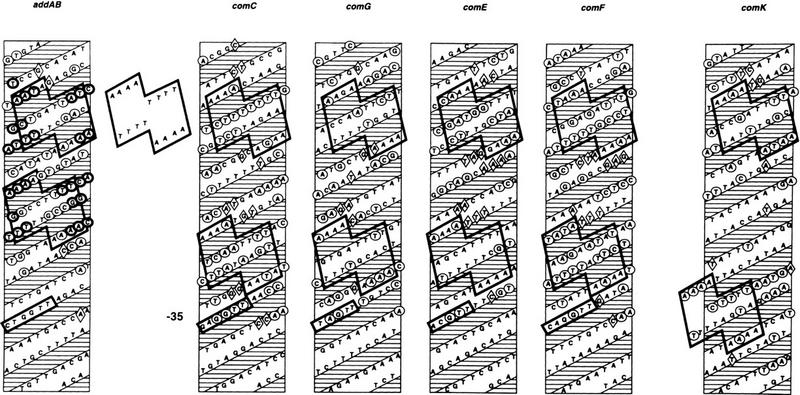
Helical projections summarizing the ComK footprints on the addAB, comC, comG, comE, comF, and comK promoters. Hydroxy-radical protected nucleotides in addAB are represented by heavy contoured circles, DNase I protections are represented by lightly contoured circles, and hypersensitive sites by diamonds. The −35 promoter sequences are boxed. Separately shown is the ComK–dimer binding recognition pattern, which is outlined on each projection.
Figure 12.
Summary of the ComK recognition sequence in the promoter regions of all genes known to be ComK controlled. The ComK dimer binding sites (AT boxes) are shaded and the −35 promoter sequences, if known, are printed in boldface type. cspB is the hypothetical ComK binding site located at the end of the cspB gene.
Deletion of the 4 adenine or 4 thymine tracts in the upstream AT box of the addAB promoter completely abolished ComK binding (results not shown).
ComK binding requires location of the AT boxes at the same face of the DNA helix
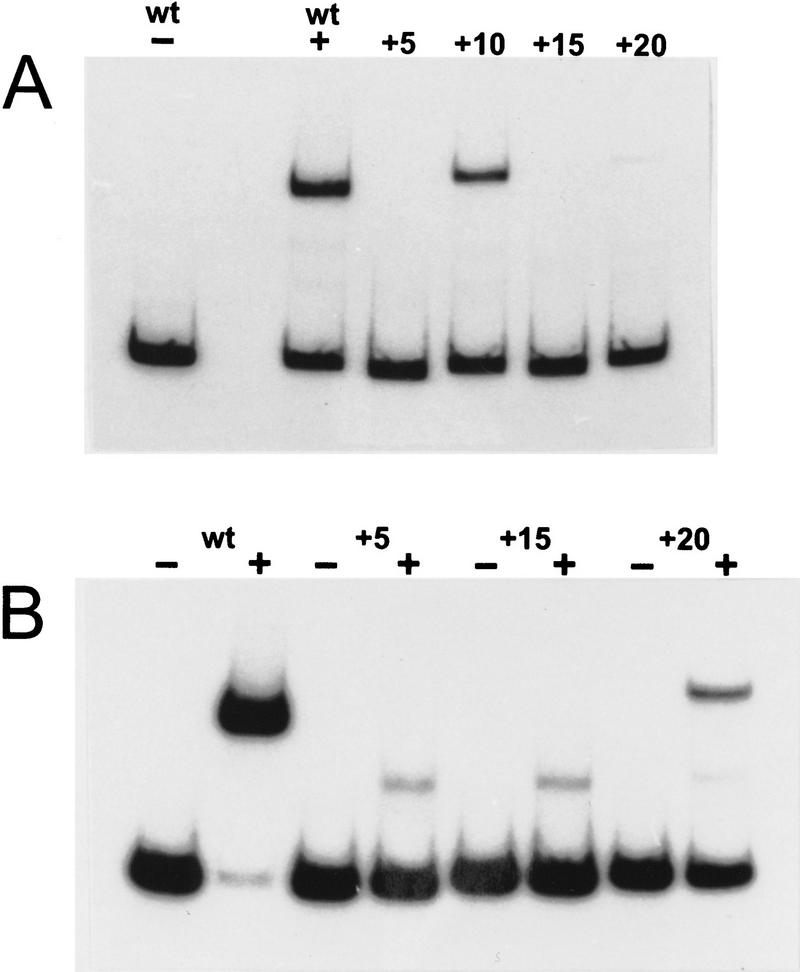
The position of AT boxes in Figure 7 suggests that in ComK-regulated promoters, the distance between the two AT boxes may vary, in a promoter-specific way, provided that the boxes are located at the same face of the DNA helix, such that the ComK dimers are able to form a tetramer. To test this inference, we changed the spacing between the AT boxes of the addAB promoter and examined the effect on ComK binding in a gel retardation experiment. As shown in Figure 8A, an increase in distance between the AT boxes with 5 bp, corresponding to half a helix turn, as a result of which the ComK–dimer binding sites become located at opposite faces of the DNA helix, completely abolished binding of ComK, whereas an increase of 10 bp, corresponding to the distance between AT boxes in the late competence genes, restored ComK binding. Increasing the spacing with one and a half turns again eliminated ComK binding. When the separation between the AT boxes was increased with 20 bp, approaching the distance observed in the comK promoter, hardly any retardation was observed. However, the ComK affinity of the native comK promoter is low compared with that of addAB and the late competence genes, and retardation of the comK promoter fragments requires high ComK concentrations. As indicated in Figure 8B, doubling the ComK concentration resulted in clear retardation. These results show convincingly that binding of ComK requires the location of the two ComK dimers at one face of the DNA helix, and that a separation between the boxes up to three helical turns does not prevent ComK from binding to the boxes. The weak, less retarded bands observed at high ComK concentrations have been shown to correspond to binding of ComK dimers (results not shown). Thus, at high concentrations, ComK can bind weakly to the AT boxes, which is not observed when the two boxes are at the same face of the DNA helix, pointing at strong cooperative binding of the two dimers in the ComK tetramer. The very faint dimer-produced band in the addAB promoter, in which the boxes are separated by 28 nucleotides, may be caused by the distance-imposed constraints for optimal tetramerization.
Figure 8.
Gel retardation of addAB promoter fragments as a function of increasing nucleotide spacing between the AT boxes. The number of base pairs added to the AT box interval of the wild-type addAB promoter (wt) are indicated (+5, +10, +15, and +20, respectively). (A) 0.2 μm ComK was used except for the first lane (−) in which no protein was added; (B) the modified addAB promoter fragments were incubated with 0.4 μm ComK (+).
The inferred ComK recognition pattern has predictive value
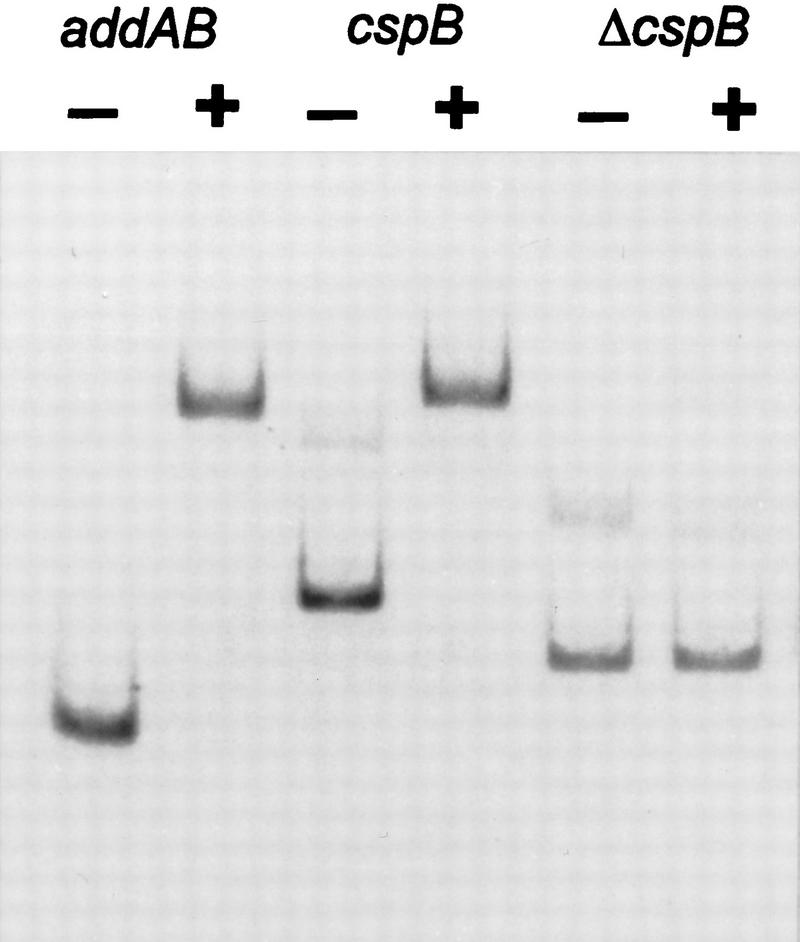
The AT boxes with 8, 18, or 31 nucleotide intervals were screened against the B. subtilis genomic database available at present. Only one fully corresponding sequence emerged, in which the two AT-boxes had a spacing of 8 bases and were located at the terminus of cspB, a gene that encodes a cold shock protein (Fig. 12, below; Schroder et al. 1995). To determine whether this sequence could bind ComK, the terminal 260 bp of cspB were isolated by PCR amplification, and ComK binding was tested by gel retardation (Fig. 9). The addition of ComK to this fragment resulted in complete retardation. This retardation was caused by the ComK recognition sequence, because deletion of the terminally located AT boxes resulted in the loss of retardation of the fragment. It is not known whether the binding of ComK to the terminus of cspB has any biological relevance.
Figure 9.
Gel retardation assay with a 32P-labeled addAB promoter fragment, a PCR fragment containing the ComK recognition sequence at the end of the cspB gene, and the same fragment from which the ComK recognition sequence was removed (Δ_cspB_). The ComK (+) concentration was 0.3 μm in all cases.
Minor groove binding drugs interfere with ComK binding
The symmetrical location of the hydroxy-radical protected stretches at the AT boxes of addAB is indicative of ComK dimers recognizing these sequences. Because the upper and lower strand protections, at the center of the AT box dyad symmetries, are offset from each other by 6.5 and 5.5 nucleotides, the ComK monomers appear to make contact across the major groove of DNA (Tullius et al. 1987). As illustrated in Figure 10, this implies that ComK binds to the minor groove of the addAB promoter. To examine whether ComK binding is sensitive toward minor groove binding drugs, gel retardation assays were performed in the presence of increasing concentrations of actinomycin D, chromomycin A3, and distamycin A (Fig. 11). Although there is a clear difference in sensitivity, with actinomycin D being the least and chromomycin A3 the most potent inhibitor, it is apparent that all drugs affect ComK binding in the micromolar range, supporting the notion that ComK interacts with the DNA through the minor groove (Copenhaver et al. 1994; Quinn et al. 1996).
Figure 10.
Location of the hydroxy-radical-protected sugar residues on the addAB promoter backbone. Hydroxy-radical-protected residues are marked by open circles, and the dyad symmetry in the two AT boxes is indicated with arrows. Shaded and black bars in the DNA helix represent adenine and thymine residues, respectively.
Figure 11.
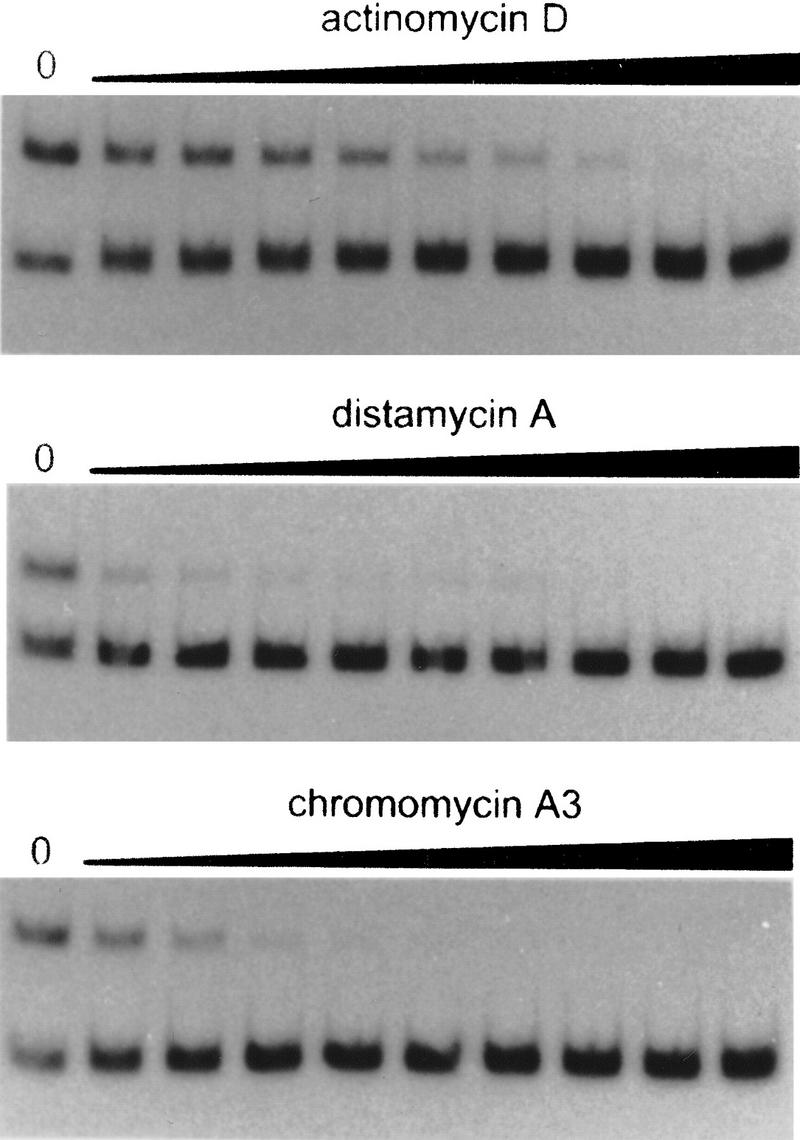
Competition of minor groove binding drugs with ComK binding to the addAB promoter. Actinomycin D and distamycin A concentrations were increased in twofold increments from 0.4 to 100 μm, chromomycin A3 concentrations were increased in twofold increments from 0.04 to 10 μm. Left lanes present ComK-induced retardation of the addAB probe in the absence of drugs.
Discussion
Extensive epistatic interaction studies have assigned a central role for comK in the regulation of competence in B. subtilis (Hahn et al. 1994; Van Sinderen and Verema 1994). ComK activates and binds specifically to the promoters of the late competence genes and genes required for recombination, and it was therefore postulated that comK encodes the CTF (Haijema et al. 1995, 1996; Van Sinderen et al. 1995b). In this study we confirm this hypothesis by showing a specific stimulation of comG transcription, using purified ComK, in vitro.
To advance our understanding of the interaction of ComK with its target sites, we undertook a footprinting analysis. On the basis of DNase I protection analysis with six different ComK-regulated promoters, the ComK binding sites were localized directly upstream of the −35 promoter sequences. Comparison of the six footprints did not reveal a ComK recognition sequence. The comG and comC promoters exhibited no intrinsic bend, thus rendering it unlikely that bending-induced distortion of the helix was involved in recognition by ComK. Hydroxy-radical protection analysis was carried out on the addAB promoter, revealing four protected stretches of 4–5 nucleotides within the extended DNase I-protected region. This corresponded with the finding that four molecules of ComK bound to the addAB promoter fragment.
This correspondence, together with the existence of two sets of dyad symmetries in the hydroxy-radical protected nucleotide sequence, strongly suggested that two dimers of ComK associate with the region upstream of the addAB promoter, and that the binding motif is AAAAN5TTTT (AT box). Strong support for this conclusion was provided by the presence of similar sequences in the DNase I footprints of the other ComK regulated promoters examined (Fig. 7), by the observation that ComK binding required the two AT boxes to be in phase with the helical turn, and ultimately, by the observation that ComK bound to a DNA fragment carrying this motif, which was identified by a search of the known B. subtilis genome sequences.
ComK binds as two dimers to its target, and it is very likely that ComK functions as a tetramer. First, all ComK-dependent promoter regions identified so far contain two ComK–dimer binding sites (AT boxes). Second, deletion of a single dimer binding site from the comC and comK promoters destroys competence and ComK binding in vitro (Mohan and Dubnau 1990; Van Sinderen et al. 1994). Third, in gel retardation experiments, the mobility of the target DNA invariably corresponds to the binding of four molecules; less retarded bands that might be caused by binding of a single dimer were never observed in unmodified promoters. Consistent with this, protection of one AT box has never been observed in footprinting experiments. Fourth, location of the ComK–dimer binding sites on opposite faces of the DNA helix completely eliminated ComK binding. Fifth, for specificity, the binding of ComK tetramers is essential, as the incidence of single AT boxes in the B. subtilis genome is high. We conclude that at least in association with its targets, ComK probably acquires a tetrameric configuration, but with a remarkable recognition flexibility, because, depending on the ComK target, one to three helical turns can be accommodated between the dimer binding sites. This type of flexibility is, to our knowledge, unique for tetrameric transcription factors and may be made possible by the ability of ComK to distort the DNA helix, as confirmed by the induced bending obtained with the comF and comG promoters.
Figure 12 illustrates three classes of promoters on the basis of the separation of the ComK dimer binding sites. In the first class (addAB, recA, dinA, nucA, and cspB), the dimer binding sites are separated by 8 nucleotides. Repetition of the AT boxes occurs at an interval of 21 nucleotides, placing both AT boxes on the same face of the DNA helix, assuming 10.5 nucleotides per helical turn (Lane et al. 1992). RecA expression is induced during the development of competence and this induction is under the control of ComK. Haijema et al. (1996) have shown that ComK contacts a large region upstream of the recA promoter. Within this region, two ComK dimer binding motifs are located with a spacing similar to that in addAB. Another DNA repair gene, dinA, is competence stimulated as well. Gel retardation experiments indicate that ComK binds to the region upstream from the dinA promoter (J. Sun and R. Yasbin, pers. comm.), and it is likely that two dimers of ComK bind to this sequence. The same applies to the promoter-upstream region of nucA, which is regulated by ComK in vivo, and is known to bind ComK (Van Sinderen et al. 1995a).
The second class of ComK-recognition motifs is present in the promoters of the late competence genes; comC, comG, comE, and comF. In this class, the dimer binding sites are separated by an additional helical turn. Repetition of the two AT boxes occurs at an interval of 31 nucleotides, corresponding to three helical turns, again locating the two boxes on the same face of the DNA helix. Deletion analysis of the comC promoter indicated that removal of the upstream AT box abolished comC expression and reduced ComK binding in vitro (Mohan and Dubnau 1990). Thus, for optimal competence expression, both AT boxes were necessary.
The third class of ComK-regulated promoters contains a single member, comK itself. ComK expression is subject to positive autoregulation and it has been shown that ComK binds upstream of the comK −35 promoter sequence. As shown in Figure 12, two AT boxes are present. Interestingly, in this case, the repetition of the two boxes occurs at an interval of 44 nucleotides, corresponding to four helical turns. Deletion of the upstream dimer binding site of the comK promoter prevented the development of competence (Van Sinderen et al. 1994). Thus, interaction of two dimers to form a ComK tetramer is apparently required to produce functionally active ComK.
The three classes of ComK-dependent promoters may reflect regulatory differences. For instance, the comK promoter (class III) is negatively regulated by AbrB and CodY binding, and requires the DNA binding proteins AbrB, SinR, and DegU for maximal activity in vivo. Recent results (L. Hamoen, A. Van Werkhoven, D. Dubnau, and G. Venema, unpubl.) have shown that DegU can bind to sequences upstream from the comK −35 site, and that DegU and ComK bind synergistically to the comK promoter region. Perhaps DegU facilitates the distortion of DNA necessary to permit a ComK tetramer to bind simultaneously with high affinity to both AT boxes. The late competence proteins (class II) appear to be regulated solely by ComK. Class I promoters include some (addAB and possibly nucA) which may be regulated solely by ComK as well as other promoters, like recA (Haijema et al. 1996), which are subject to more complex regulation. In spite of these differences, in all of the promoters presented in Figure 12, ComK functions as a transcription activator. The close proximity of the ComK binding sites to the −35 promoter sequences suggests an association with RNA polymerase, probably to recruit the latter for stable promoter binding (Ptashne and Gann 1997).
In the hydroxy-radical footprint of the addAB promoter, the minima of the protected patches at the center of the AT-box dyad symmetries are offset from each other by 5.5 and 6.5 bp. According to Tullius et al. (1987) an offset of 3 bp results when dimers contact each other across the minor groove because the backbone positions of B-DNA, which are closest to each other across a minor groove, are 3 bp apart in the sequence. For binding across a major groove this distance would be 7 bp. This rule applies to dimers in which monomers are positioned exactly at the same face of the DNA helix. Although the ComK dimers are located precisely at the same face of the helix, the two monomers that comprise a dimer are not. As can be seen in Figure 10, the monomers approach each other toward the center of the AT-box dyad symmetry, resulting in an offset slightly smaller than 7 bp. A more qualitative indication for protein contact across the minor groove is that the central protected domains will be offset to the 3′ direction, relative to the center of a dyad symmetrical recognition sequence. If the subunits contact each other across a major groove, the central protections would be offset toward the 5′ direction, as is the case for ComK. Protein contact across a major groove suggests that the monomers bind to the minor groove of DNA. Inhibition of ComK binding because of the presence of minor groove binding drugs provides support for the implied minor groove binding of ComK.
Van Sinderen et al. (1995b) have pointed to the existence of a limited degree of amino acid sequence similarity between the carboxy-terminal domain of ComK and the DNA binding domain present in the so-called HMG boxes of the eukaryotic regulatory proteins SRY and TCF-1, the human testis determining factor and T cell factor 1, respectively (Ferrari et al. 1992; Waterman et al. 1991). The HMG family of proteins are minor groove binding proteins that induce DNA bending and that recognize motifs rich in A/T residues (Geierstanger et al. 1994; Grosschedl et al. 1994). Sensitivity of HMG box containing proteins toward minor groove binding drugs has been used to distinguish them from the more usual type of transcriptional regulators, those which bind through the major groove of DNA (Copenhaver et al. 1994; Quinn et al. 1996). Thus, in addition to limited amino acid similarity, the mechanism of base recognition by the prokaryotic ComK may resemble that of the eukaryotic HMG class of proteins.
Materials and methods
DNA manipulations and materials
All molecular cloning procedures were carried out by use of standard techniques (Sambrook et al. 1989). Restriction endonucleases were obtained from either Boehringer Mannheim Biochemicals or New England Biolabs. Labeled nucleotides were from Amersham. Media for growth of B. subtilis and E. coli have been described previously (Venema et al. 1965; Sambrook et al. 1989). B. subtilis chromosomal DNA was purified as described by Venema et al. (1965).
PCR amplifications
PCR reactions were carried out as described by Innes and Gelfand (1990) by use of Vent DNA polymerase (New England Biolabs) and B. subtilis 8G5 chromosomal DNA as template. For the various gel retardation experiments and footprinting analyses, the sets of primers listed in Table 1 were used in the PCR reactions: Primers C1 and C2 for the comC promoter region, primers E1 and E2 for the comE promoter region, primers F1 and F2 for the comF promoter region, primers G1 and G2 for the comG promoter region, primers K1 and K2 for the comK promoter region, and primers AB1 and AB2 for the addAB promoter region. The cspB fragment containing a terminally located ComK binding site was isolated by use of primers CB1 and CB2. The cspB fragment lacking the ComK binding site was isolated by use of primers CB1 and CB3.
Table 1.
Nucleotide sequence of the primers used for PCR amplification
| Primer | Sequence |
|---|---|
| C1 | 5‘-TCTGAATTCCGAAAAGGTTGAAATAAAAG-3‘ |
| C2 | 5‘-ATTGGATCCTGGTATGGATTGTAG-3‘ |
| E1 | 5‘-CCGGAATTCACGAAGAAATGCTTAAG-3‘ |
| E2 | 5‘-CGCGGATCCTTTCATGTGTAGACGAG-3‘ |
| F1 | 5‘-CCGGAATTCGGTGAAGGTGCGTTAGG-3‘ |
| F2 | 5‘-CGCGGATCCGCCTCCTTTCGAAACAG-3‘ |
| G1 | 5‘-CTAGAATTCATGGTCACCATGTCTCGC-3‘ |
| G2 | 5‘-CGCGGATCCCCTCTCCTTTCAACGC-3‘ |
| K1 | 5‘-CCGGAATTCAGAATCCCCCCAATGCC-3‘ |
| K2 | 5‘-CGCGGATCCCTAATAATCTATCATCTG-3‘ |
| AB1 | 5‘-TTCTGTAAAAGTATATGTTG-3‘ |
| AB2 | 5‘-CCCTCTCTTTTTATCTCTC-3‘ |
| CB1 | 5‘-CTGCAACTGCATTTATTGC-3‘ |
| CB2 | 5‘-TTATTAGTTTGCAATATCC-3‘ |
| CB3 | 5‘-CTGCAGTTTTTATTTTGACA-3‘ |
Purification of ComK
ComK was purified as a MBP–ComK fusion protein on an amylose resin (New England Biolabs) column and separated from MBP by cleavage with protease Factor Xa as described previously (Van Sinderen et al. 1995b). After cleavage was complete, Factor Xa was inactivated by the addition of 1 mm PMSF. To separate ComK from MBP and DNA, the protein mixture was loaded onto a DEAE–Sepharose column (Pharmacia) equilibrated with 20 mm Tris-HCl at pH 8, 1 mm EDTA, and 0.5 mm dithiotreitol. MBP and ComK were sequentially eluted with a 0–50 mm Na2SO4 gradient and then with a 0–1 m KCl gradient (containing 50 mm Na2SO4). Fractions were collected, and the Na2SO4 concentration increased to 100 mm to prevent precipitation of ComK. The ComK containing fractions were checked for the absence of contaminating DNA by ethidium bromide-stained agarose–gel electrophoresis, aliquoted, and stored at −70°C. Purification and cleavage of MBP–ComK were followed by SDS-PAGE.
Gel retardation analysis
Gel retardation with ComK and MBP–ComK was carried out essentially as described (Van Sinderen et al. 1995b). The PCR-generated DNA probes were end labeled with T4 polynucleotide kinase by use of [γ-32P]ATP. The purified proteins and probes were premixed on ice in binding buffer [20 mm Tris-HCl at pH 8, 100 mm KCl, 5 mm MgCl2, 0.5 mm dithiotreitol, 0.05 mg/ml poly[(I-C)] and 0.05 mg/ml BSA]. After 20 min incubation at room temperature, one-fourth volume of 50% glycerol solution was added and the samples were loaded on a nondenaturing 4% polyacrylamide gel. Gels were run in TAE buffer (40 mm Tris-acetate at pH 8, 2 mm EDTA) at 100 V, dried, and autoradiographed. To study the inhibition with minor groove binding drugs, addAB probe was incubated with the indicated concentrations of actinomycin D, chromomycin A3, or distamycin A, for 30 min prior to incubation with ComK.
In vitro transcription assays
In vitro transcription experiments were performed essentially as described by Predich et al. (1992). The DNA template, pAN-G, was constructed by cloning a PCR fragment containing the comG promoter between the _Sma_I and _Bam_HI sites of the pAN583 vector (Predich et al. 1992). The comG promoter fragment was isolated by PCR with the primers G3 (5′-CTTTTCAGTGATGCCGG-3′) and G2 (Table 1), and subsequent digestion with _Eco_RV and _Bam_HI. The transcription reactions were performed in the binding buffer described for the gel retardation experiments, except for the omission of poly(I-C). DNA templates, purified B. subtilis RNA polymerase, and purified ComK were incubated for 15 min at 37°C in a final volume of 20 μl, before the addition of 3 μl of a nucleotide mix (1 mm ATP, 1 mm UTP, 1 mm GTP, 0.5 μl [α-32P]CTP). After 1 min incubation at room temperature, 2 μl of 0.3% heparin was added to the mixture and incubation resumed for another 10 min at 37°C. After the addition of 2μl of 1 mm CTP, incubation was continued for 10 min before terminating the reaction by the addition of 18 μl of formamide containing 0.05% bromophenol blue and 0.05% xylene blue. After heating for 3 min at 90°C, the samples were loaded on an 8% polyacrylamide–urea gel and run at 300 V. Gels were subjected to autoradiography immediately after electrophoresis without prior drying.
Footprinting analyses
The DNase I footprint was performed following the description supplied with the Sure Track Footprinting Kit (Pharmacia). The DNA probes were obtained by PCR amplification with primers that were end labeled by use of [γ-32P]ATP. Binding reactions were performed as for the gel retardation experiments, in a total volume of 40 μl. DNA fragments containing ∼30,000 cpm were added to each reaction mixture. After 20 min incubation at room temperature, 10 μl of 10 mm CaCl2 and 5 Units of DNase I were added, and after 1 min, the reactions were terminated with 140 μl of stop solution (192 mm Na-acetate, 32 mm EDTA, 0.14% SDS, 64 μg/ml yeast tRNA). The samples were extracted with phenol–chloroform and ethanol precipitated. The precipitates were resuspended in 3 μl loading buffer. Analysis of the DNase I products was carried out by electrophoresis on a 6% polyacrylamide–urea gel. Maxam–Gilbert G+A reactions were run with each experiment to locate sequence positions and protected regions (Sambrook et al. 1989).
Hydroxy-radical footprints were performed as described by Tullius and Dombroski (1986) with the modifications described by O’Halloran et al. (1989). Binding reaction conditions were as for the gel retardation experiments, except that glycerol was omitted. DNA fragments, containing ∼30,000 cpm, were added to each 50-μl reaction mixture. After 20 min at room temperature, 1 μl of 5.6 mm (NH4)2Fe(SO2)2 ⋅ 6 H2O was mixed with 1 μl of 11.2 mm EDTA and added to the incubation mixture, followed by the addition of 2 μl of 3.36% H2O2 and 2 μl of 112 mm Na-ascorbate. Reactions were incubated for 1 min at room temperature and terminated with 44 μl of stop solution (10 μl of 3 m Na-acetate, 6 μg of yeast tRNA, and 10 μl of 320 mm thiourea) and the subsequent addition of 300 μl of ethanol. Samples were extracted with phenol–chloroform and analyzed as described for the DNase I footprint experiments.
Circular permutation analysis
Circular permutation analysis was carried out essentially as described by Kim et al. (1989). The comG and comF promoter regions were generated by PCR amplification with the primer couples G1/G2 and F1/F2, respectively. After digestion with _Bam_HI and _Eco_RI (located in the primers) and filling in the ends with Klenow polymerase, the promoter fragments were cloned in the _Sal_I site, blunt ended by Klenow polymerase. In this manner the comG and comF promoter fragments were placed in the multiple cloning site of pBend2, resulting in the plasmids pBend-G and pBend-F. Various DNA fragments containing the promoter regions were isolated by digesting plasmid pBend-G and pBend-F with the appropriate restriction enzymes, as described in Figure 4. The fragments were end labeled by T4-polynucleotide kinase and [γ-32P]ATP, after alkaline phosphatase treatment. The increase in retardation as a result of the binding of ComK was observed as in a standard gel retardation experiment. The bending angle (α) was calculated according to the following equation; (mm/me) = cosα/2, where mm = mobility of the complex with protein bound at the center of DNA and me = mobility of the complex with protein at the end of the DNA (Kim et al. 1989).
Mutational analysis of the addAB promoter region
The 4 adenine or 4 thymine tracts in the upstream AT box of the addAB promoter were replaced following a PCR-based mutagenesis, by use of primers RB and F1A (minus the 4 adenine tract) and RB and F1T (minus the 4 thymine tract), respectively (Table 2), and B. subtilis 8G5 chromosomal DNA as template. The _Sac_I and _Nae_I restriction sites thus created were used to verify the mutations. As positive control, a wild-type addAB promoter fragment was amplified with primers RB and Fwt (Table 2).
Table 2.
Primers used for mutational analysis of the addAB promoter
| Primer | Sequence | Restriction site |
|---|---|---|
| IIE | 5‘-CCCG_GAATTC_TGTAAAAGTATATGTGG-3‘ | _Eco_RI |
| RB | 5‘-GCGC_GGATCC_TCTCTATATTTTATCGAATG-3‘ | _Bam_HI |
| AP + 5 | 5‘-AACGGCCTTTTCA_GATAT_ _C_ATATAAAAAGCTGA-3‘ | _Eco_RV |
| AP + 10 | 5‘-CGGCCTTTTCAGATAT_GGTAC_ _C_ATATAAAAAGC-3‘ | _Kpn_I |
| AP + 15 | 5‘-CGGCCTTTTCAGATAT_GTCGAC_GTACCATATA-3‘ | _Sal_I |
| AP + 20 | 5‘-AACGGCCTTTTCA_GCTG_ATATGT_CTA_GACGTA-3‘ | _Xba_I |
| _Pvu_II | ||
| Fwt | 5‘-CCCGG_AATTC_GCAGGGAGAGATGTGCGGA-3‘ | _Eco_RI |
| F1A | 5‘-CCCGG_AATTC_GCAGGGAGAGATGTGCGGAG_GAGCTC_AGCTTTTTATATGTGAA-3‘ | _Eco_RI |
| _Sac_I | ||
| F1T | 5‘-CCCG_GAATTC_GCAGGGAGAGATGTGCGGAGATAATCA_GCCGGC_TATATGTGAAAAGGCCGTT-3‘ | _Eco_RI _Nae_I |
The addAB promoter region was amplified by PCR with the primers IIE and RB (Table 2). After digestion with _Eco_RI and _Bam_HI, the 128-bp fragment was cloned into _Eco_RI–_Bam_HI-digested pBluescript (SK+). The resulting plasmid (pAP8) was used as the template for PCR-based mutagenesis. Insertions between the two AT boxes in the addAB promoter were made with the two-step megaprimer PCR method as described by Datta (1995). The mutant primers used to create insertions of 5, 10, 15, and 20 bp, respectively, are listed in Table 2. Each mutant fragment was used as the new template for additional insertions. All fragments were cloned into pBluescript (SK+), yielding pAP13, pAP18, pAP23, and pAP28, respectively. Primers used for the insertions were designed in such a way that new restriction sites were created, which were used to verify the mutations. PCR-generated DNA fragments were end labeled with T4-polynucleotide kinase and [γ-32P]ATP, and used in gel retardation experiments.
Acknowledgments
We dedicate this paper to the memory of Gopalan Nair, whose helpfulness and warmth continue to inspire those who knew him. We are indebted to Liyun Kong for constructing the MBP–ComK fusion plasmid, Gopalan Nair and Issar Smith for purified B. subtilis RNA polymerase and for valuable comments, Charles Lovett for suggesting the minor groove binding of ComK, Ji Sun and Ronald Yasbin for discussing their results before publication, Bert Jan Haijema for helpful discussions, and Henk Mulder for preparing the figures. This work was supported by the Netherlands Organization of Scientific Research (NWO) under the auspices of the Netherlands Foundation for Chemical Research (SON) and by National Institutes of Health grant AI10311.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL G.VENEMA@BIOL.RUG.NL; FAX (050) 3632348.
References
- Copenhaver GP, Putnam CD, Denton ML, Pikaard CS. The RNA-polymerase I transcription factor UBF is a sequence-tolerant HMG-box protein that can recognize structured nucleic acids. Nucleic Acids Res. 1994;22:2651–2657. doi: 10.1093/nar/22.13.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta AK. Efficient amplification using ’megaprimer’ by asymmetric polymerase chain reaction. Nucleic Acids Res. 1995;23:4530–4531. doi: 10.1093/nar/23.21.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza C, Nakano MM, Zuber P. Identification of comS, a gene of the srfA operon that regulates the establishment of genetic competence in Bacillus subtilis. Proc Natl Acad Sci. 1994;91:9397–9401. doi: 10.1073/pnas.91.20.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau D. Genetic exchange and homologous recombination. In: Sonenshein AL, Hoch JA, Losick R, editors. Bacillus subtilis and other Gram-positive bacteria. Washington, D.C.: American Society for Microbiology; 1993. pp. 555–584. [Google Scholar]
- Duplay P, Bedouelle H, Fowler A, Zabin I, Saurin W, Hofnung M. Sequences of the malE gene and of its product, the maltose-binding protein of Escherichia coli K12. J Biol Chem. 1984;259:10606–10613. [PubMed] [Google Scholar]
- Ferrari S, Harley VR, Pontiggia A, Goodfellow PN, Lovell-Badge R, Bianchi M. SRY, like HMG1, recognizes sharp angles in DNA. EMBO J. 1992;11:4497–4506. doi: 10.1002/j.1460-2075.1992.tb05551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geierstanger BH, Volkman BF, Kremer W, Wemmer DE. Short peptide fragments derived from HMG-I/Y proteins bind specifically to the minor groove of DNA. Biochemistry. 1994;33:5347–5355. doi: 10.1021/bi00183a043. [DOI] [PubMed] [Google Scholar]
- Grosschedl R, Giese K, Pagel J. HMG domain proteins: Architectural elements in the assembly of nucleoprotein structures. Trends Genet. 1994;10:94–100. doi: 10.1016/0168-9525(94)90232-1. [DOI] [PubMed] [Google Scholar]
- Grossman AD. Genetic networks controlling the initiation of sporulation and the development of genetic competence in Bacillus subtilis. Annu Rev Genet. 1995;29:477–508. doi: 10.1146/annurev.ge.29.120195.002401. [DOI] [PubMed] [Google Scholar]
- Hahn J, Kong L, Dubnau D. The regulation of competence transcription factor synthesis constitutes a critical control point in the regulation of competence in Bacillus subtilis. J Bacteriol. 1994;176:5733–5761. doi: 10.1128/jb.176.18.5753-5761.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn J, Roggiani M, Dubnau D. The major role of SpoOA in genetic competence is to downregulate abrB, an essential competence gene. J Bacteriol. 1995;177:3601–3605. doi: 10.1128/jb.177.12.3601-3605.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn J, Luttinger A, Dubnau D. Regulatory inputs for the synthesis of ComK, the competence transcription factor of Bacillus subtilis. Mol Microbiol. 1996;21:763–775. doi: 10.1046/j.1365-2958.1996.371407.x. [DOI] [PubMed] [Google Scholar]
- Haijema BJ, Hamoen LW, Kooistra J, Venema G, Van Sinderen D. Expression of the ATP-dependent deoxyribonuclease of Bacillus subtilis is under competence-mediated control. Mol Microbiol. 1995;15:203–211. doi: 10.1111/j.1365-2958.1995.tb02235.x. [DOI] [PubMed] [Google Scholar]
- Haijema BJ, Van Sinderen D, Winterling K, Kooistra J, Venema G, Hamoen LW. Regulated expression of the dinR and recA genes during competence development and SOS induction in Bacillus subtilis. Mol Microbiol. 1996;22:75–85. doi: 10.1111/j.1365-2958.1996.tb02657.x. [DOI] [PubMed] [Google Scholar]
- Hamoen LW, Eshuis H, Jongbloed J, Venema G, Van Sinderen D. A small gene, designated comS, located within the coding region of the fourth amino acid-activation domain of srfA, is required for competence development in Bacillus subtilis. Mol Microbiol. 1995;15:55–63. doi: 10.1111/j.1365-2958.1995.tb02220.x. [DOI] [PubMed] [Google Scholar]
- Innes MA, Gelfand DH. Optimization of PCRs. In: Innes MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: A guide to methods and applications. San Diego, CA: Academic Press; 1990. pp. 3–12. [Google Scholar]
- Kim J, Zwieb C, Wu C, Adhya S. Bending of DNA by gene-regulatory proteins: Construcion and use of a DNA bending vector. Gene. 1989;85:15–23. doi: 10.1016/0378-1119(89)90459-9. [DOI] [PubMed] [Google Scholar]
- Kong L, Dubnau D. Regulation of competence-specific gene expression by Mec-mediated protein-protein interaction in Bacillus subtilis. Proc Natl Acad Sci. 1994;91:5793–5797. doi: 10.1073/pnas.91.13.5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo H, Wu H, Crothers DM. DNA bending at adenine-thymine tracts. Nature. 1986;320:501–506. doi: 10.1038/320501a0. [DOI] [PubMed] [Google Scholar]
- Lane D, Prentki P, Chandler M. Use of gel retardation to analyze protein-nucleic acid interactions. Microbiol Rev. 1992;56:509–528. doi: 10.1128/mr.56.4.509-528.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, L., M.M. Nakano, O.H. Lee, and P. Zuber. 1996. Presented at the 12th International Spores Conference, Madison, WI.
- Magnuson R, Solomon J, Grossman AD. Biochemical and genetic characterization of a competence pheromone from B. subtilis. Cell. 1994;77:207–216. doi: 10.1016/0092-8674(94)90313-1. [DOI] [PubMed] [Google Scholar]
- Mohan S, Dubnau D. Transcriptional regulation of comC: Evidence for a competence-specific factor in Bacillus subtilis. J Bacteriol. 1990;172:4064–4071. doi: 10.1128/jb.172.7.4064-4071.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Msadek T, Kunst F, Rapoport G. MecB of Bacillus subtilis is a pleiotropic regulator of the ClpC ATPase family, controlling competence gene expression and survival at high temperature. Proc Natl Acad Sci. 1994;91:5788–5792. doi: 10.1073/pnas.91.13.5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano MM, Zuber P. Cloning and characterization of srfB, a regulatory gene involved in surfactin production and competence in Bacillus subtilis. J Bacteriol. 1989;171:5347–5353. doi: 10.1128/jb.171.10.5347-5353.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Halloran TV, Frantz B, Shin MK, Ralston DM, Wright JG. The MerR heavy metal receptor mediates positive activation in a topologically novel transcription complex. Cell. 1989;56:119–129. doi: 10.1016/0092-8674(89)90990-2. [DOI] [PubMed] [Google Scholar]
- Perez-Martin J, Rojo F, de Lorenzo V. Promoters responsive to DNA bending: A common theme in prokaryotic gene expression. Microbiol Rev. 1994;58:268–290. doi: 10.1128/mr.58.2.268-290.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Predich M, Nair G, Smith I. Bacillus subtilis early genes kinA, spo0F and spo0A are transcribed by the RNA polymerase containing σH. J Bacteriol. 1992;174:2771–2778. doi: 10.1128/jb.174.9.2771-2778.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- Quinn J, Fyrberg AM, Ganster RW, Schmidt MC, Peterson CL. DNA-binding properties of the yeast SWI/SNF complex. Nature. 1996;379:844–847. doi: 10.1038/379844a0. [DOI] [PubMed] [Google Scholar]
- Roggiani M, Dubnau D. ComA, a phosphorylated response regulator protein of Bacillus subtilis, binds to the promoter region of srfA. J Bacteriol. 1993;175:3182–3187. doi: 10.1128/jb.175.10.3182-3187.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schroder K, Graumann P, Schnuchel A, Holak TA, Marahiel MA. Mutational analysis of the putative nucleic acid-binding surface of the cold-shock domain, CspB, revealed an essential role of aromatic and basic residues in binding of single-stranded DNA containing the Y-box motif. Mol Microbiol. 1995;16:699–708. doi: 10.1111/j.1365-2958.1995.tb02431.x. [DOI] [PubMed] [Google Scholar]
- Serror P, Sonenshein AL. CodY is required for nutritional repression of Bacillus subtilis genetic competence. J Bacteriol. 1996;178:5910–5915. doi: 10.1128/jb.178.20.5910-5915.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon JM, Lazazzera BA, Grossman AD. Purifaction and characterization of an extracellular peptide factor that affects two different developmental pathways in Bacillus subtilis. Genes & Dev. 1996;10:2014–2024. doi: 10.1101/gad.10.16.2014. [DOI] [PubMed] [Google Scholar]
- Tullius TD, Dombroski BA. Hydroxyl radical “footprinting”: High-resolution information about DNA-protein contacts and application to λ repressor and Cro protein. Proc Natl Acad Sci. 1986;83:5469–5473. doi: 10.1073/pnas.83.15.5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullius TD, Dombroski BA, Churchill MEA, Kam L. Hydroxyl radical footprinting: A high-resolution method for mapping protein-DNA contacts. Methods Enzymol. 1987;155:537–558. doi: 10.1016/0076-6879(87)55035-2. [DOI] [PubMed] [Google Scholar]
- Turgay K, Hamoen LW, Venema G, Dubnau D. Biochemical characterization of a molecular switch involving the heat shock protein ClpC, which controls the activity of ComK, the competence transcription factor of Bacillus subtilis. Genes & Dev. 1997;11:119–128. doi: 10.1101/gad.11.1.119. [DOI] [PubMed] [Google Scholar]
- Van Sinderen D, Venema G. ComK acts as an autoregulatory control switch in the signal transduction route to competence in Bacillus subtilis. J Bacteriol. 1994;176:5762–5770. doi: 10.1128/jb.176.18.5762-5770.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Sinderen D, Ten Berge A, Hayema BJ, Hamoen L, Venema G. Molecular cloning and sequence of comK, a gene required for genetic competence in Bacillus subtilis. Mol Microbiol. 1994;11:695–703. doi: 10.1111/j.1365-2958.1994.tb00347.x. [DOI] [PubMed] [Google Scholar]
- Van Sinderen D, Kiewiet R, Venema G. Differential expression of two closely related deoxyribonuclease genes, nucA and nucB, in Bacillus subtilis. Mol Microbiol. 1995a;15:213–223. doi: 10.1111/j.1365-2958.1995.tb02236.x. [DOI] [PubMed] [Google Scholar]
- Van Sinderen D, Luttinger A, Kong L, Dubnau D, Venema G, Hamoen L. ComK encodes the competence transcription factor, the key regulatory protein for competence development in Bacillus subtilis. Mol Microbiol. 1995b;15:455–462. doi: 10.1111/j.1365-2958.1995.tb02259.x. [DOI] [PubMed] [Google Scholar]
- Venema G, Pritchard RH, Venema-Schroder T. Fate of transforming deoxyribonucleic acid in Bacillus subtilis. J Bacteriol. 1965;89:1250–1255. doi: 10.1128/jb.89.5.1250-1255.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman ML, Fisher WH, Jones KA. A thymus-specific member of the HMG protein family regulates the human T Cell receptor Ca enhancer. Genes & Dev. 1991;5:656–669. doi: 10.1101/gad.5.4.656. [DOI] [PubMed] [Google Scholar]