Maternal IL-11Rα function is required for normal decidua and fetoplacental development in mice (original) (raw)
Abstract
In eutherian mammals, implantation and establishment of the chorioallantoic placenta are essential for embryo development and survival. As a maternal response to implantation, uterine stromal cells proliferate, differentiate, and generate the decidua, which encapsulates the conceptus and forms the maternal part of the placenta. Little is known about decidual functions and the molecular interactions that regulate its development and maintenance. Here we show that the receptor for the cytokine interleukin-11 (IL-11Rα) is required specifically for normal establishment of the decidua. Females homozygous for a hypomorphic IL-11Rα allele are fertile and their blastocysts implant and elicit the decidual response. Because of reduced cell proliferation, however, only small deciduae form. Mutant deciduae degenerate progressively, and consequently embryo-derived trophoblast cells generate a network of trophoblast giant cells but fail to form a chorioallantoic placenta, indicating that the decidua is essential for normal fetoplacentation. IL-11Rα is expressed in the decidua as well as in numerous other tissues and cell types, including the ovary and lymphocytes. The differentiation state and proliferative responses of B and T-lymphocytes in mutant females were normal, and wild-type females carrying IL-11Rα mutant ovaries had normal deciduae, suggesting that the decidualization defects do not arise secondarily as a consequence of perturbed IL-11Rα signaling defects in lymphoid organs or in the ovary. Therefore, IL-11Rα signaling at the implantation site appears to be required for decidua development.
Keywords: Decidua, IL-11R, cytokine receptor, chorioallantoic placenta
In eutherian mammals, the establishment of a maternal–fetal interface is a prerequisite for embryonic development and survival. The formation of the maternal–fetal connection begins with implantation and culminates in the generation of the chorioallantoic placenta. The attachment of the embryonic trophoblast to the uterine epithelium elicits the decidual response, apoptosis of the uterine epithelium, recruitment of inflammatory cells, and neovascularization (Cross et al. 1994; Dey 1996; Rinkenberger et al. 1997). As part of the decidual response, uterine stromal (decidual) cells proliferate, differentiate, and form a massively thickened uterine wall (the decidua) that encapsulates the conceptus and generates the implantation chamber. The decidual reaction occurs first at the antimesometrial pole of the implantation chamber, where blastocysts implant. On embryonic day 5 (E5) in the mouse, the primary decidual zone forms around the conceptus, followed by the formation of the secondary decidual zone around the primary decidua on E6 (Huet Hudson et al. 1989). Two days after the formation of the primary decidual zone, at the late egg cylinder stage, the mesometrial decidua forms at the mesometrial pole. Concommitant with the formation of the mesometrial decidua, the egg cylinder begins its expansion into the antimesometrial implantation chamber and cells of the antimesometrial decidua start to die (Welsh and Enders 1985). Primary trophoblast giant cells invade maternal capillaries in the antimesometrial decidua and form maternal blood sinuses surrounding the conceptus. Together with the underlying parietal endoderm they comprise the parietal yolk sac, the earliest placental structure. Later, the chorioallantoic placenta forms at the mesometrial pole of the implantation site tightly connected to the mesometrial decidua and provides the close aposition of maternal and fetal blood vessels.
The molecular interactions that regulate the formation, maintenance, and remodeling of the decidua are not well understood. Implantation and the decidual response depend on ovarian steroid hormones (Psychoyos 1973) and prostaglandins (Lim et al. 1997) and require the maternally produced cytokine leukemia inhibitory factor (LIF) (Stewart et al. 1992). LIF is produced in the uterus specifically before implantation (Stewart et al. 1992), and at later stages of gestation, various other cytokines are expressed in the uterus and placenta (Pollard 1991; Stewart 1994), suggesting that the combinatorial action of systemic and local signals mediated by hormones and cytokines controls implantation and the initial maternal response. Among the cytokines expressed in endometrial and trophoblast cells is interleukin-11 (IL-11), a cytokine with a wide spectrum of biological activities in vitro and in vivo (Du and Williams 1997). The biological effects of IL-11 are mediated by association of the ligand with its receptor (IL-11R) and the signal transducer gp130 (Hilton et al. 1994; Karow et al. 1996). Humans and some mouse strains contain only one IL-11R gene (IL-11Rα), whereas a number of other laboratory mouse strains including 129SvPas and C57BL/6J, which are the strains used in this study, contain a second almost identical IL-11R gene (IL-11Rβ), which is tightly linked to the IL-11Rα gene (Robb et al. 1997) and is co-expressed at low levels with the IL-11Rα gene in many tissues (Bilinski et al. 1996). Consistent with the wide spectrum of IL-11 activities, IL-11R transcripts were detected in numerous tissues and cell types, including the decidua on the ninth day of pregnancy (Neuhaus et al. 1994). Here we report that signals mediated by the IL-11Rα are specifically required for normal female reproduction. Mice homozygous for a disrupted IL-11Rα gene appear phenotypically normal, and homozygous mutant males reproduce. Homozygous mutant females, however, show severe deficiencies in their deciduae, and embryos in homozygous mothers develop abnormal chorioallantoic placentas, resulting in embryonic lethality in the majority of cases.
Results
Targeted mutagenesis of the IL-11Rα gene
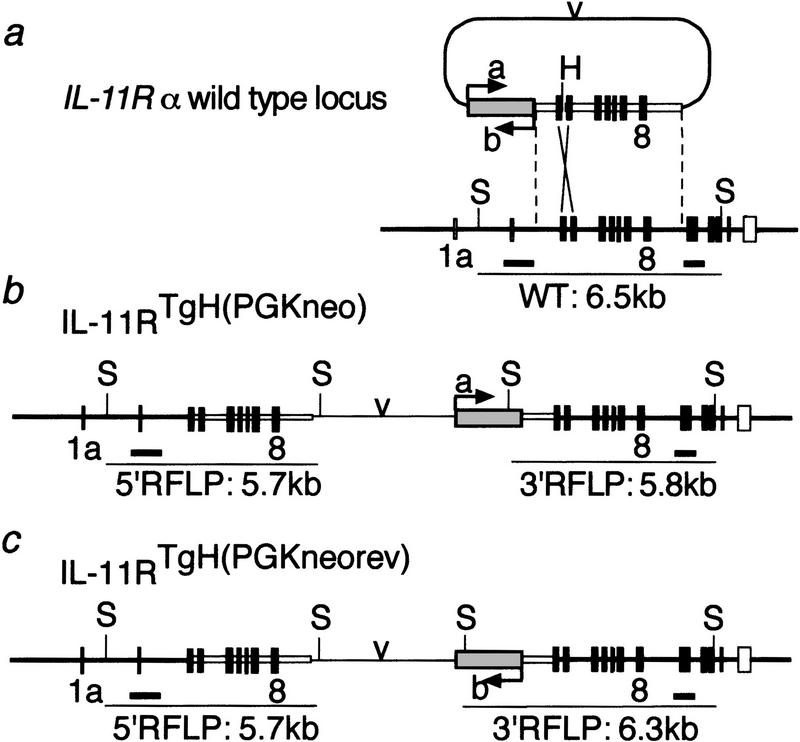
The IL-11Rα gene was disrupted by gene targeting in embryonic stem (ES) cells, such that after homologous recombination vector sequences and the neomycin-resistance (neo) gene driven by the phosphoglycerate kinase (PGK) promoter (PGK–neo) were inserted into intron eight, and a truncated promoterless IL-11Rα gene containing exons 2 to 13 was generated downstream of PGK–neo. Two vectors containing PGK–neo in opposite orientations were used (Fig. 1). In the targeted alleles, transcripts generated from the IL-11Rα promoter do not contain exon 9 encoding the WSXWS box, which is essential for ligand binding (Yawata et al. 1993), and no distinct transcripts should originate from the promoterless IL-11Rα gene portion downstream of the insertion. The mutant IL-11Rα alleles [referred to as IL-11Rα TgH(PGK–neo) and IL-11Rα _TgH(PGK–neo–rev)_] (Fig. 1b,c) were introduced into the mouse germ line, and mutant lines were established on a mixed 129SvPas/C57BL/6J genetic background. Heterozygous mice carrying either allele were phenotypically normal. Intercrosses of heterozygotes gave rise to viable, apparently normal homozygous mutant mice at Mendelian ratios (Table 1A). To address whether the alleles resulted in null mutations, poly(A)+ RNA from kidneys that express readily detectable levels of IL-11Rα transcripts (Bilinski et al. 1996) were analyzed by Northern blot hybridizations. The wild-type IL-11Rα gene gives rise to a 1.9-kb transcript. No wild-type transcript was detected in poly(A)+ RNA from either homozygous mutant. Instead, a larger 2.6-kb transcript not present in wild-type mRNA was detected at low levels in mRNA from IL-11Rα TgH(PGK–neo) mice, and at levels similar to the wild-type transcript in the IL-11Rα TgH(PGK–neo–rev) line (Fig. 2a). Because this transcript was not recognized by a neo probe (Fig. 2b), it most likely was generated by splicing around the PGK–neo insertion. Such a transcript would contain exons 1–8 fused to exons 2–13 and could give rise to a chimeric protein of ∼90 kD. A protein of this approximate molecular mass was detected by Western blot analysis of kidney extracts from IL-11Rα mutant mice with two different anti IL-11R antibodies, but was not found in wild-type extracts (Fig. 2d,e). Both available anti IL-11R antibodies did not allow us to unambiguously identify the wild-type IL-11R protein because of their crossreactivity with multiple proteins. Consistent with the different levels of mutant transcripts, the mutant protein was abundant in IL-11Rα TgH(PGK–neo–rev) extracts, and was present only in low amounts in extracts from IL-11Rα TgH(PGK–neo).
Figure 1.
Targeting vector and structure of the mutant alleles. (a) Structure of the wild-type IL-11Rα locus and targeting vector. Coding exons are indicated by solid boxes, untranslated exons by open boxes. The shaded box represents PGK–neo; (v) the plasmid backbone. (a,b) Two versions of the construct with opposite transcriptional orientation of the neo gene. (b,c) Structure of the IL-11Rα TgH(PGK–neo) and IL-11Rα TgH(PGK–neo–rev) allele, respectively. The probes used for Southern blot hybridizations (thick black horizontal bars) and the DNA fragments indicative for the different alleles (thin horizontal lines) are shown below the maps.
Table 1.
Reproduction in IL-llRα_mutant mice_
| A | Parental genotype | No. of matings | No. of females giving birth | No. of litters | No. of newborns | Average litter size | Genotype of offspring | |||
|---|---|---|---|---|---|---|---|---|---|---|
| male | female | +/+ | +/− | −/− | ||||||
| 1. | nr/+ | nr/+ | 12 | 12 | 12 | 82 | 6.8 | 21 | 42 | 19 |
| 2. | nr/nr | nr/+ | 4 | 4 | 4 | 29 | 7.3 | — | 16 | 13 |
| 3. | nr/+ | nr/nr | 5 | 5 | 9a | 40 | 4.4 | — | 22 | 18 |
| 4. | n/+ | n/+ | 12 | 12 | 12 | 105 | 8.6 | 28 | 55 | 22 |
| 5. | n/n | n/+ | 7 | 7 | 7 | 57 | 8.1 | — | 30 | 27 |
| 6. | n/+ | n/n | 14 | 3 | 5b | 11 | 2.2 | — | 6 | 5 |
| B | Genotype | No. of plugs | No. of pregnant females | No. of implantation sites | Average no. of implantation sites per female |
|---|---|---|---|---|---|
| male | female | ||||
| +/+ | +/+ or n/+ | 19 | 18 | 148 | 8.2 |
| +/+ | n/n | 31 | 26 | 193 | 7.4 |
Figure 2.
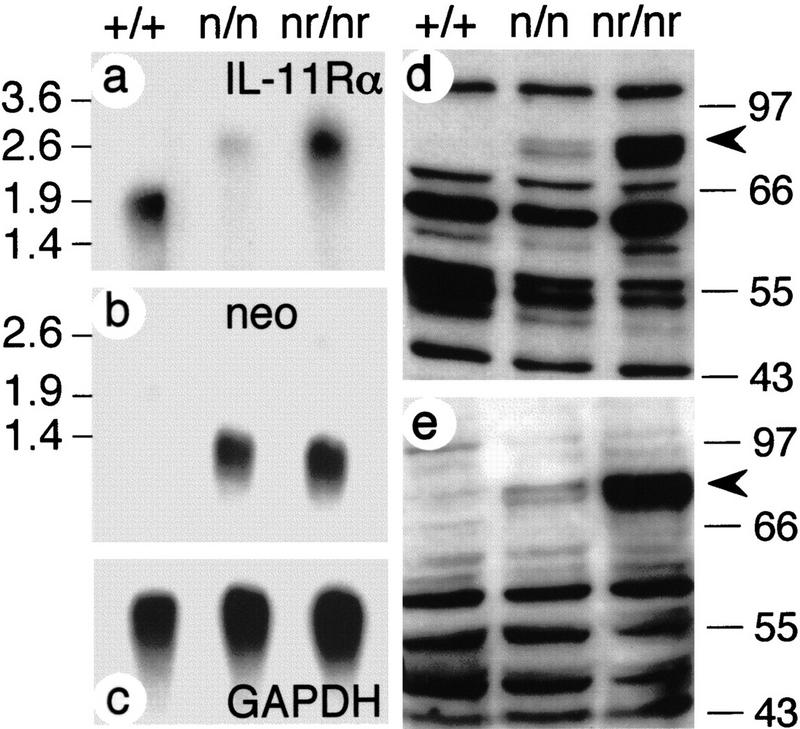
Expression of IL-11Rα in wild-type and mutant mice. (a–c) Northern blot hybridizations of kidney poly(A)+ RNA from wild-type (+/+), homozygous IL-11Rα TgH(PGK–neo) (n/n) and IL-11Rα TgH(PGK–neo–rev) (nr/nr) mice. (d,e) Western blot analysis of kidney extracts with C-20 (d) and N-20 (e) anti-IL-11R antibodies. The abnormal IL-11R protein detected in mutant extracts is indicated by arrowheads. Numbers at the left and right indicate DNA size (kb in a,b) and molecular mass markers (kD in d,e).
Phenotypic analysis of IL-11Rα mutant mice
Mutant males homozygous for either mutant allele were fertile and sired offspring. The fertility of homozygous females carrying either allele was impaired. The five homozygous IL-11Rα TgH(PGK–neo–rev) test mated females all gave rise to litters, but the litter size was reduced in comparison with wild-type females (Table 1A). In contrast, only 3 out of the 14 test-mated IL-11Rα TgH(PGK–neo) females gave rise to litters with only one to three pups. On continued mating with males of proven fertility over several months, only one female gave rise to two additional small litters (Table 1A).
To determine the defect underlying the reproductive failure of homozygous IL-11Rα mutant females, uteri from timed matings of homozygous mutant females with wild-type males (therefore containing embryos carrying one wild-type, functional copy of the IL-11Rα gene) were analyzed. Only 75% (28/37) of the implantation sites in homozygous IL-11Rα TgH(PGK–neo–rev) mutant females analyzed on day 11/12 and day 16/17 of gestation appeared normal, and contained viable, apparently healthy embryos. The remaining implantation sites were small and haemorrhagic, and embryos were completely resorbed as early as on gestational day 11. Only abnormal embryos or resorptions were found in the uteri of eight pregnant IL-11Rα TgH(PGK–neo) females analyzed at gestational days 11 and 12, suggesting that the reduced fertility in homozygous IL-11Rα mutant females is caused by the loss of embryos during postimplantation development before day 11.
To determine the cause of embryonic death, implantation sites from timed matings of the more severely affected homozygous IL-11Rα TgH(PGK–neo) mutant females were analyzed between day 6 and 11 of gestation. Homozygous IL-11Rα TgH(PGK–neo) females contained the same average number of implantation sites as wild-type controls (Table 1B). Their implantation chambers, however, were significantly smaller. On E6, embryos in mutant females were surrounded by a zone of densely packed decidual cells (Fig. 3b), which resembled morphologically the avascular primary decidual zone (Dey 1996). No overt difference in vascularization around the implantation site was found at this stage. On E8, mutant deciduae were morphologically similar to controls except for their reduced size and the presence of blood-filled lacunae in the antimesometrial region (Fig. 3d,f). On E9, the developing mesometrial decidua was smaller in mutant females, and the antimesometrial decidua was discontinuous (Fig. 4c). Subsequently, the mesometrial decidua (decidua basalis) and the antimesometrial decidua (decidua capsularis) degenerated progressively, resulting in the complete absence of a decidua on the eleventh day of gestation in the majority of cases (Fig. 4d,h–j). No indication for increased apoptosis was found by TUNEL labeling (data not shown), however, the labeling index in mutant deciduae as determined by BrdU incorporation was reduced significantly (Figs. 3, h and i, and 5; data not shown).
Figure 3.
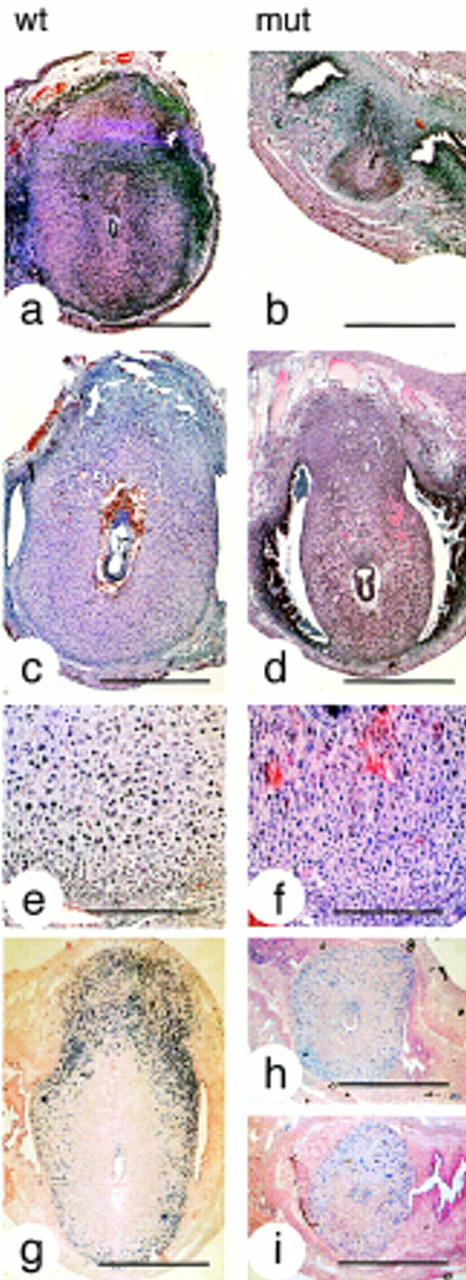
Decidual defects in mutant females. Hematoxilin/eosin-stained paraffin sections of implantation sites in wild-type (a,c,e) and mutant (b,d,f) females, and sections of BrdU-labeled wild-type (g) and mutant (h,i) implantation sites on gestational days 6 (a,b), 7 (g,h,i), and 8 (c,d). In mutant females deciduae were abnormally small, and BrdU incorporation in cells of the secondary and mesometrial decidua reduced (see also Fig. 5). Bars, 1 mm (a–d,g–i) and 250 μm (e,f).
Figure 4.
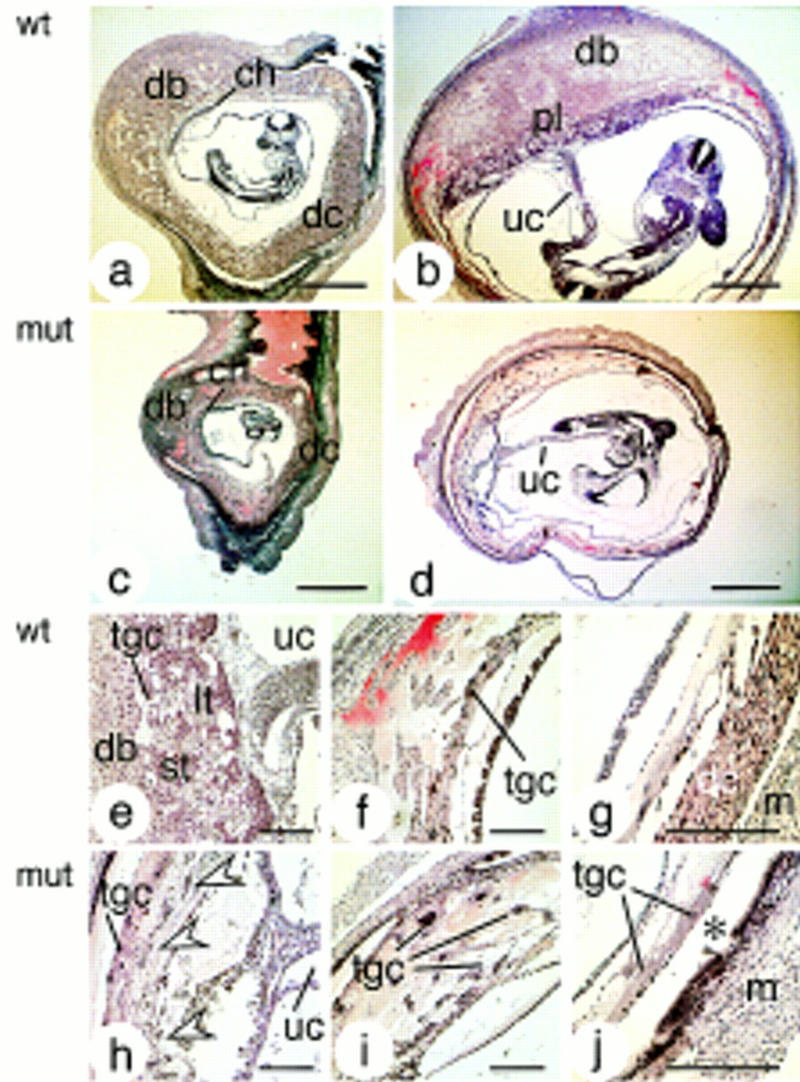
Decidual and placental defects in mutant females. Hematoxilin/eosin-stained paraffin sections of implantation sites in wild-type (a,b,e–g) and mutant (c,d,h–j) females on gestational days 9 (a,c) and 11 (b,d–j). In mutant females, deciduae were abnormally small and progressively degenerated. The chorioallantoic placenta of embryos developing in mutant mothers was severely reduced (d); only scattered remnants resembling spongio and labyrinthine trophoblast were present (arrowheads in h) and trophoblast giant cells filled the space normally occupied by the decidua (i). The asterisk in j indicates the space normally occupied by the decidua capsularis on E11. (ch) Chorion; (dc) decidua capsularis; (db) decidua basalis; (lt) labyrinthine trophoblast; (m) myometrium; (pl) placenta; (st) spongiotrophoblast; (tgc) trophoblast giant cell; (uc) umbilical cord. Bars, 1 mm (a–d) and 250 μm (e–j).
The degeneration of the maternal decidua was accompanied by defects in embryo-derived trophectoderm tissue, perturbed placenta development, and embryonic retardation and death. Most embryos in IL-11Rα TgH(PGK–neo) mutant deciduae appeared phenotypically normal up to E8. They were frequently smaller, however, and developmentally retarded compared with embryos developing in control females (cf. embryos in Figs. 3, c and d, and 4, a–d). Beginning with gestational day 9, mutant deciduae contained an increasing number of dying embryos. On days 9 and 10, ∼50% (7/14 and 7/13, respectively) and on day 11 ∼70% (23/33) of the conceptuses were dead or completely resorbed, and no living embryos were found in three mutant females analyzed after gestational day 12.
Embryos in IL-11Rα mutant mothers formed a chorionic–allantoic connection, but failed to generate a normal chorioallantoic placenta with distinct spongio and labyrinthine trophoblast layers (Fig. 4d,h). Only small dispersed groups of cells resembling spongio- or labyrinthine trophoblast were present (arrowheads in Fig. 4h). A network of trophoblast giant cells filled the space usually occupied by the placenta and decidua, and extended into the area of the missing decidua capsularis, forming large blood-filled sinuses around the embryo (Fig. 4i). The analysis of markers for diploid spongio- and labyrinthine trophoblast (MASH2; Guillemot et al. 1994), spongiotrophoblast (4311; Lescisin et al. 1988), and trophoblast giant cells (PLF; Carney et al. 1993) indicated that these cell types were present in embryos developing in mutant deciduae. Consistent with the histological findings, however, the expression of these markers was altered. PLF expression was high and its expression domain expanded (Fig. 6c,d,i,j), consistent with areas colonized by trophoblast giant cells. In contrast, both MASH2 and 4311 transcripts were present, but their expression domains were small (arrowheads in Fig. 6l,n,p) and hybridization signals were reduced (Fig. 6n,p).
Figure 6.
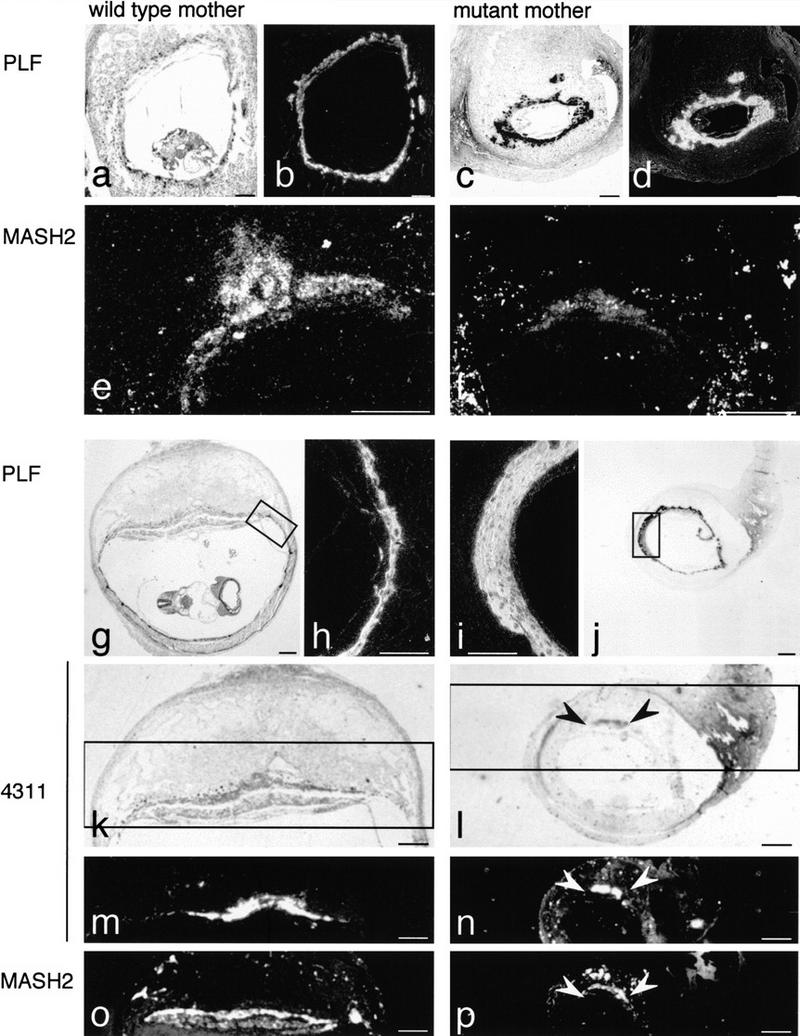
Expression of trophoblast cell markers. Bright-field (a,c,g,j–l) and dark-field (b,d–f,h,i,m–p) microphotographs of paraffin sections of implantation sites from wild-type (a,b,e,g,h,k,m,o) and mutant (c,d, f,i,j,l,n,p) females on E9 (a–f) and 10 (g–p). The expression of MASH2 and 4311, markers for spongio- and labyrinthine trophoblast, was reduced (e,f,m–p), whereas PLF, a marker for trophoblast giant cells, was expressed at high levels in embryos developing in mutant females (b,d,i). (Insets, g and j) The regions of the sections shown at higher magnification in h and i, respectively; (insets, k and l) the regions shown in m and n, respectively, and the regions of neighboring sections (o,p) hybridized to a MASH 2 probe. Arrowheads in (l,n,p) indicate the regions of spongio and labyrinthine trophoblast in placentas of embryos in mutant females. Bars, 500 μm (a–d,e–g,j–p) and 200 μm (h,i).
IL-11Rα is expressed in numerous adult tissues, including the ovary (Bilinski et al. 1996; Robb et al. 1997). In addition, the decidua contains various lymphoid cells (Robertson et al. 1994), whose proliferation and differentiation is stimulated by IL-11 (Du and Williams 1997). This raises the possibility that defective IL-11Rα signaling outside the decidua might contribute to the observed phenotype. To test whether lymphoid cells were affected, the composition and proliferative responses of B and T cells in mutant IL-11Rα TgH(PGK–neo) females were analyzed. Flow fluorimetric analysis of thymocytes or lymph node cells double stained for lymphocyte lineage-specific cell surface (CD3ε, CD4, CD8α, and CD45R) and activation markers (CD69 and CD25) did not reveal qualitative or quantitative differences between IL-11Rα TgH(PGK–neo) and wild-type mice (Fig. 7A). Moreover, proliferative responses of lymph node cells induced by different doses of B or T-cell mitogens were indistinguishable (Fig. 7B). The lack of an immunological defect is consistent with studies of mice carrying a null allele of the IL-11Rα (Nandurkar et al. 1997). To determine whether perturbed IL-11Rα signaling in the ovary might contribute to the decidual defects, potentially by influencing the production of ovarian steroid hormones, ovaries of immunodeficient SCID females were bilaterally replaced by homozygous IL-11Rα TgH(PGK–neo) ovaries. Four females carrying ovary grafts were mated to homozygous mutant males and gave rise to liveborn pups. All newborns analyzed from these matings (n = 37 from seven litters) were homozygous mutant, proving that they were derived from the transplanted ovaries. Implantation sites analyzed in subsequent pregnancies of these females were morphologically indistinguishable from controls and contained normal midgestation homozygous mutant embryos indicating that mutant embryos develop normally in wild-type mothers. Similarly, homozygous mutant blastocysts (n = 12) transferred into a pseudopregnant wild-type female gave rise to normal midgestation embryos surrounded by morphologically normal implantation sites (n = 7).
Figure 7.
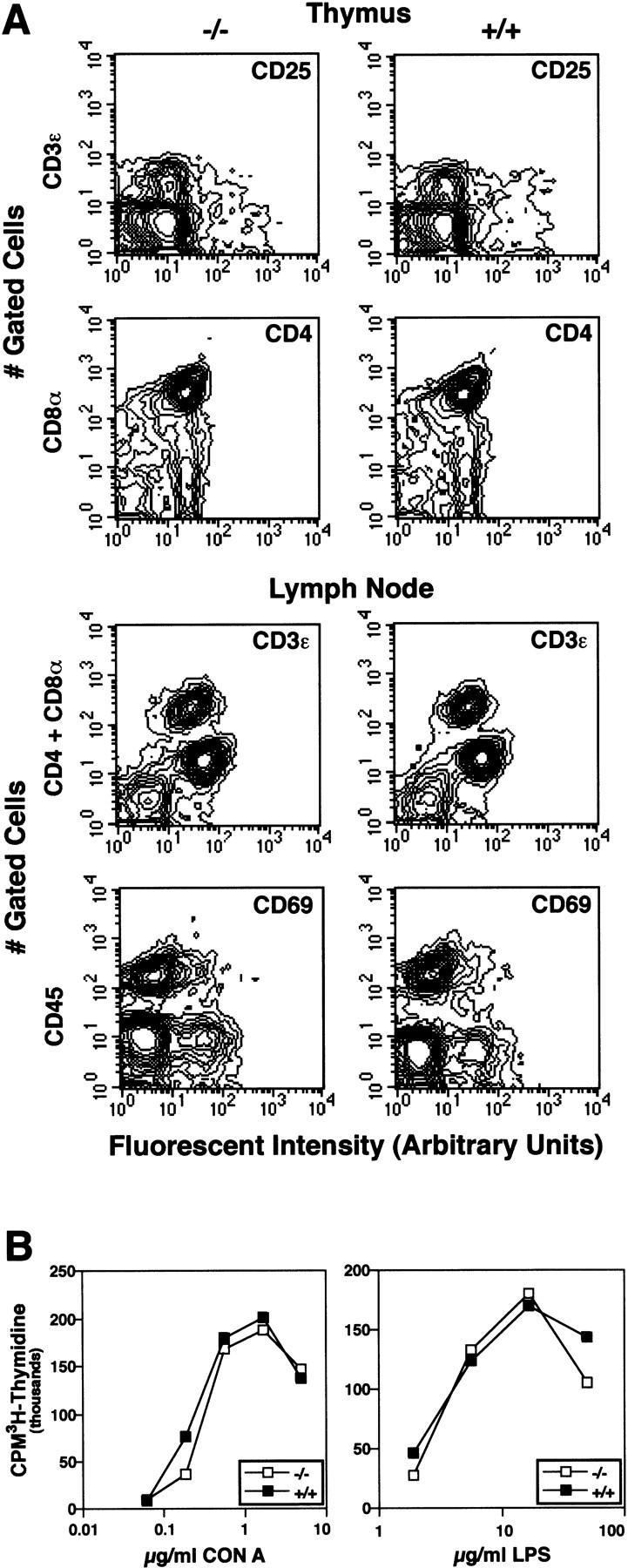
Mutant IL-11Rα TgH(PGK–neo) homozygotes have normal thymus and lymph node cells. Thymocytes and lymphocytes from representative 5-month-old IL-11Rα TgH(PGK–neo) homozygous (−/−) and wild-type mice (+/+). (A) Multiparameter flow cytometric analysis examining expression of the indicated lymphocyte markers. (B) Proliferation of lymph node cells induced by varied doses of the T-cell mitogen CON A or the B cell mitogen LPS. The mean of duplicate or triplicate CPM (counts per minute) of [3H]thymidine incorporation determinations are shown. The mean CPM of lymph node cells without added mitogen was always <7000.
Expression of IL-11Rα and IL-11 in pregnant uteri
To determine the onset and distribution of IL-11R transcripts in pregnant uteri, IL-11R expression was analyzed by RNA in situ hybridization. No expression was detected in uteri prior to implantation. On day 6 of gestation, IL-11R transcripts were present in the peripheral antimesometrial decidua (Fig. 8a). On day 8, IL-11R mRNA was localized in the antimesometrial secondary decidua and in stromal cells underlying the mesometrial luminal epithelium (Fig. 8b; data not shown). Beginning with E9, transcripts were detected throughout the decidua with higher levels in the decidua capsularis (Fig. 8c,d), but were absent from the embryonic part of the placenta (Fig. 8d). Consistent with the low levels of mutant mRNA detected by Northern blot analysis, no IL-11R transcripts were detected in the deciduae of pregnant mutant IL-11Rα TgH(PGK–neo) females (Fig. 8i), also indicating that the highly homologous IL-11Rβ gene (Bilinski et al. 1996; Robb et al. 1997) is not expressed at detectable levels in the decidua. IL-11 is expressed in a partly overlapping and complementary pattern to IL-11Rα. On day 6, IL-11 transcripts were barely detectable in the decidua (Fig. 8e). From the eighth day of development onward, transcripts were found in the antimesometrial decidua partly overlapping with IL-11Rα expression (Fig. 8f,g). In addition, trophoblast cells (arrowheads in Fig. 8g) and the forming chorioallantoic placenta (Fig. 8h) expressed elevated levels of IL-11.
Figure 8.
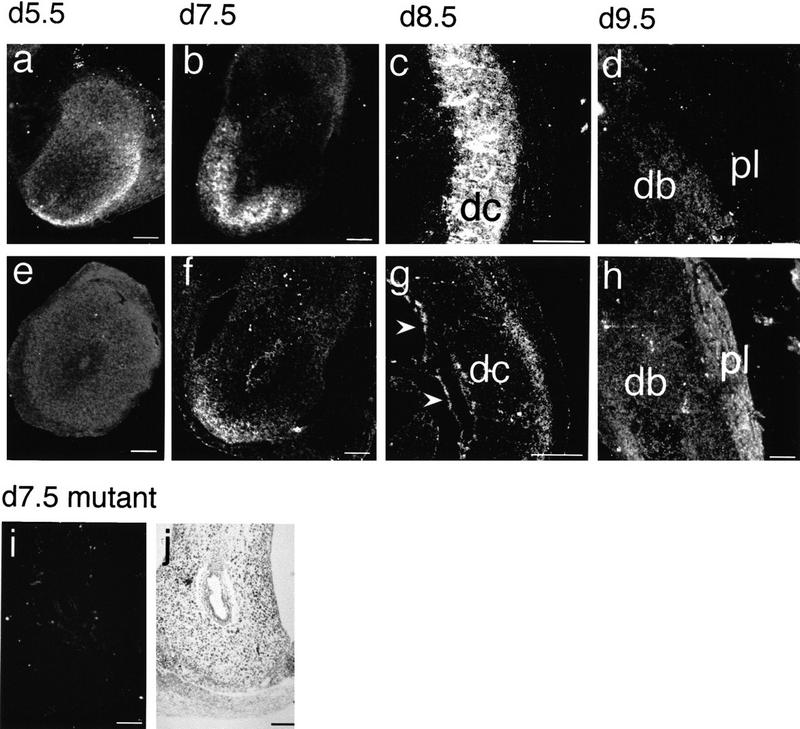
Expression of IL-11Rα and IL-11 in uterine and placental tissues. (a–h) Dark-field microphotographs of paraffin sections of implantation chambers from wild-type females hybridized with a IL-11R (a–d) or IL-11 (e–h) probe. IL-11R transcripts were detected in the secondary decidua at the antimesometrial pole (a,b), and in the decidua capsularis (c) and basalis (d). IL-11 transcripts were detected in the secondary decidua (f), in the peripheral decidua capsularis (g), giant trophoblast cells (arrowheads in g), and in the placenta (h). Dark-field (i) and bright-field (j) microphotographs of paraffin sections of a mutant implantation chamber hybridized with an IL-11R probe. No IL-11R transcripts were detected (i). (dc) Decidua capsularis; (db) decidua basalis; (pl) placenta. Bars, 500 μm.
Discussion
We have generated two mutant alleles of the IL-11Rα gene, which affect female reproduction and indicate a specific requirement for IL-11Rα-mediated signals for decidua formation and maintenance. No normal IL-11Rα mRNA was detected in either homozygous mutant, suggesting that no wild-type IL-11Rα protein is present in these mice. Different levels of a mutant IL-11Rα protein, however, were found in both mutants. The mutant protein likely retains activity as an IL-11 receptor, as the mutant phenotype is much milder in IL-11Rα TgH(PGK–neo–rev) mice that express this protein at higher levels. Therefore, IL-11Rα TgH(PGK–neo) and IL-11Rα TgH(PGK–neo–rev) most likely are two hypomorphic mutations, and IL-11Rα TgH(PGK–neo) represents the stronger allele that with rare exceptions reduces IL-11Rα activity below the threshold required for normal decidua development.
Female sterility attributable to defective decidualization has been reported recently for mice carrying a null allele of the IL-11Rα gene (Robb et al. 1998). Homozygous females carrying the null allele did not give rise to any offspring, and no embryos survived past gestational day 11, but, as IL-11Rα TgH(PGK–neo) mice, IL-11Rα null mice did not display defects in any other tissue (Nandurkar et al. 1997). This suggests that the IL-11Rα TgH(PGK–neo) phenotype is very similar to, but somewhat milder than, the null phenotype. In addition, the mutant phenotype caused by the IL-11Rα TgH(PGK–neo) allele showed greater variability than the null phenotype. A few embryos in homozygous IL-11Rα TgH(PGK–neo) mutant mothers even developed to term, suggesting that in rare cases the level and/or residual activity of the mutant protein in homozygous IL-11Rα TgH(PGK–neo) females is sufficient for decidualization and embryonic survival. This interpretation is supported further by the similar but weaker phenotype of the IL-11Rα TgH(PGK–neo–rev) allele, which expresses larger amounts of the mutant protein. Because our analysis was done on a genetic background nonsegregating for the IL-11Rβ gene, the presence or absence of IL-11Rβ cannot account for the observed variability. Because the mutation was analyzed on a mixed 129SvPas/C57BL/6 genetic background, however, segregating modifier loci may contribute to the variable expressivity of the mutation.
The normal number of implantation sites in homozygous mutant females (see Table 1) indicates that mutant females produce normal numbers of oocytes, that these oocytes can be fertilized, and that preimplantation development and implantation are not compromised by impaired maternal IL-11Rα signaling. Implanted embryos were surrounded by cells that morphologically resembled primary decidua cells, suggesting that the initial decidual reaction occurs in mutant females. From E6 onward, deciduae in mutant mothers were significantly smaller than in controls. BrdU incorporation in the peripheral antimesometrial (secondary) decidual zone and in the mesometrial decidua, both areas of high mitotic activity (O’Shea et al. 1983), was reduced. This strongly suggests that the small deciduae are caused by decreased cell proliferation, and that IL-11Rα signaling stimulates decidual cell division early during postimplantation development. At present, however, we cannot distinguish whether cell proliferation is reduced in all decidual cells, or whether only a subset of decidual cells is affected. Because the decidua contains polyploid cells in its antimesometrial portion at this time of gestation (O’Shea et al. 1983), the reduced BrdU incorporation could also indicate reduced endo-reduplication of cells. The degeneration of the antimesometrial decidua is a physiological process that during normal pregnancy leads to the complete decay of the decidua capsularis late in gestation (Welsh and Enders 1985). Therefore, the early loss of the antimesometrial decidua in homozygous IL-11Rα TgH(PGK–neo) females might not be attributable to a specific requirement for IL-11R signaling for decidua cell survival, but might merely reflect the degeneration of too few decidual cells at a normal rate. The mesometrial decidua, however, normally persists during pregnancy and might specifically require IL-11R signaling for survival.
The spatial and temporal distribution of IL-11R transcripts in uteri of pregnant females correlates with the observed decidual phenotype. The results of the ovary transplantations demonstrate that perturbed IL-11Rα function in the ovary does not cause the decidual defects. Similarly, the normal composition and proliferative responses of B and T cells in mutant IL-11Rα TgH(PGK–neo) females suggest that defects in the lymphoid system are unlikely to contribute to the observed decidual degeneration, which is supported further by the normal reproductive capacity of genetically immunodeficient mice (Croy and Chapeau 1990; Koller et al. 1990). The normal development of homozygous mutant embryos in wild-type females carrying homozygous mutant ovary transplants, as well as the normal development of homozygous mutant blastocysts after transfer into a pseudopregnant wild-type female indicates that the trophectodermal defects are not caused by the loss of IL-11Rα function in the embryo. Heterozygous embryos carry one copy of the functional IL-11Rα gene, and, as wild-type embryos, they develop normally in wild-type females. When they develop in homozygous mutant mothers, however, they have severe defects in their trophoblasts, further supporting that IL-11R signaling in maternal tissues is critical. Taken together, our data strongly suggest that abnormal decidua development and decidua degeneration, as well as the defects in the embryo-derived trophoblast, are likely to be solely consequences of defective IL-11Rα signaling in the decidua.
Embryos in mutant deciduae appeared developmentally or growth retarded already on E8, well before the chorioallantoic placenta is established, and many embryos died before placentation, suggesting that the decidua has a critical role in promoting and sustaining early postimplantation development. Massive embryonic death was observed beginning with gestational days 9–10, a time when the chorioallantoic placenta is established and begins to function as the primary means of nutrient, gas, and waste exchange (Cross et al. 1994; Rossant 1995; Rinkenberger et al. 1997). The failure of establishing a proper chorioallantoic placenta therefore appears to be the major cause for the demise of the embryos after E9.
The overlapping expression of IL-11Rα and IL-11 in the decidua suggests paracrine or autocrine functions of IL-11. The complementary expression of IL-11Rα in maternal tissue and IL-11 in the trophoblast and placenta suggests that IL-11, in addition to its multiple known hematopoietic and non-hematopoietic effects (Du and Williams 1997), might be a fetal signal that is involved in an ongoing cross-talk between feto-placental unit and uterus, and required for normal decidua development. Conversely, the severe deficiencies in the embryo-derived spongio and labyrinthine trophoblast suggest that decidua-derived signals are essential for fetoplacental development, and the decidua may have a trophic role that is specifically required for spongio and labyrinthine trophoblast cells.
In summary, our results demonstrate that signals mediated by the IL-11Rα are essential for the normal formation of the decidua and its maintenance, and these signals are likely to act directly in uterine tissues. Therefore, like LIF, which is required for implantation (Stewart et al. 1992), IL-11 appears to be an essential component of a intrauterine cytokine network (Pollard 1991) that is required for embryo implantation, decidua formation, and the establishment of the maternal–fetal interface during pregnancy.
Materials and methods
Generation of mice carrying a germ-line mutation in the IL-11Rα gene
The targeting vector was constructed by cloning a genomic 129SvPas-derived 4.5-kb _Bam_HI–_Xba_I fragment containing exons 2–8 of the IL-11Rα gene (Bilinski et al. 1996) into the Bluescript pBSIIKS+ vector (Stratagene) containing a PGK–neo cassette either in the same or opposite transcriptional orientation as the IL-11Rα gene. This vector was linearized with _Hin_dIII and electroporated into D3 ES cells (Doetschman et al. 1985). Correctly targeted clones were identified and verified by Southern blot analysis (Ausubel et al. 1994) using external probes from the 5′ and 3′ region of the targeted area, and were injected into C57BL/6J blastocysts to obtain germ-line transmission. Germ-line chimeras were bred to C57BL/6J mice, and mutant lines maintained by brother sister matings.
RNA isolation and Northern blot analysis
Total RNA was isolated according to standard protocols (Ausubel et al. 1994), poly(A)+ RNA using Oligotex dT cellulose (Quiagen) according to the manufacturer’s instructions. For Northern blot analysis 2–5 μg of poly(A)+ RNA were electrophoresed and transferred to nylon membranes as described (Lehrach et al. 1977; Sambrook et al. 1989). Filters were hybridized as described (Neuhaus et al. 1994).
In situ hybridization
Tissues were dissected, washed twice in PBS, and fixed in 4% paraformaldehyde in PBS at 4°C overnight. In situ hybridization on tissue sections was carried out essentially as described (Wilkinson et al. 1990) with [33P]UTP instead of [35S]UTP. After autoradiography using Kodak NTB2, emulsion sections were stained with hematoxilin or toluene blue and embedded as described (Ausubel et al. 1994).
Western blot analysis
Proteins were separated by SDS-PAGE using 10% polyacrylamide gels in a Bio-Rad Mini Protean gel chamber and blotted onto Nitrocellulose filters in a Bio-Rad Mini Trans Blot chamber according to the manufacturer’s instructions. Proteins were detected using C-20 and N-20 anti-IL-11R antibodies (Santa Cruz Biochemicals) using the ECL detection system (Amersham).
Histological analysis
Tissues were dissected, washed twice in PBS, fixed in Bouin’s or 4% paraformaldehyde in PBS at 4°C overnight, and processed as described (Ausubel et al. 1994). Eight-micrometer sections were cut using a Leica 1512 microtome, stained with hematoxilin/eosin and photographed with a Leica WILD or Leica MRXE microscope.
Labeling index analysis
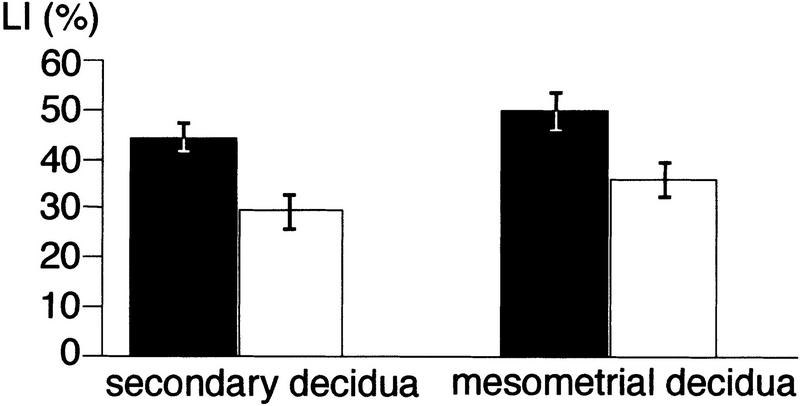
Females were sacrificed 1 hr after intraperitoneal injection of 100 μg of BrdU/gram bodyweight, and tissues processed as described above. BrdU incorporation was detected immunohistochemically using rat anti-BrdU antibodies (Harlan, Sera-Lab) and biotinylated secondary antibodies (Jackson Immuno Research). Bound antibodies were visualized after incubation with strepavidin-conjugated β-galactosidase by X-gal staining as described (Gossler and Zachgo 1993). Labeling indices were calculated from the numbers of labeled and unlabeled cells that were counted in several standardized areas (9500 μm2) of different sections in the mesometrial and secondary decidua.
Analysis of lymphocytes
Thymocyte or lymph node cell surface differentiation and activation markers were evaluated by multi-parameter flow cytometry using conventional procedures (Christianson et al. 1997) and the following antibodies: 145-2C11 anti-CD3ε-FITC; 56-6.72 anti-CD8α-PE; GK1.5 anti-CD4-CY3; 53-6.7 anti-CD8α-PE; PC61 anti-CD25/IL2R-FITC, clone RA3-6B2 CD45R/B220-PE, clone H1.2F3 anti-CD69-biotin/SA, obtained from Pharmingen, Inc., San Diego, CA, or prepared by The Jackson Laboratory Flow Cytometry Service. Lymph node cell proliferative responses were induced by varied doses of concanavalin A (CON A) and lipopolysaccharide (LPS) in microtiter cultures with each well seeded with 2.5 × 105 nucleated cells in 0.2 ml of Dulbecco’s modified Eeagle medium. Cultures were labeled after 48 hr of incubation with [3H]thymidine, harvested 12 hr later, and 3H incorporation was determined.
Figure 5.
Labeling indices in wild-type and mutant secondary and mesometrial deciduae on gestational day 8. Labeling indices were calculated from 10–17 counted areas from sections of two wild-type and mutant BrdU-labeled deciduae, respectively, and analyzed statistically by single factor ANOVA. P values for the comparison of labeling indices in the secondary and mesometrial decidua are 2.5 × 10−5 and 1.8 × 10−5, respectively. (Solid bars) Wild type; (open bars) mutant.
Acknowledgments
We thank Drs. Patsy Nishina and John Schimenti for critical comments, and Janet Rossant and Jay Cross for helpful discussions and probes. This work was supported by the Deutsche Forschungsgemeinschaft (DFG grant Go449/3-1) and by the Mallinckrodt Jr. Foundation, and facilitated by the Cancer Center Core facilities of The Jackson Laboratory.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL ago@aretha.jax.org; FAX 0211 811 2279.
References
- Ausubel FM, Brent R, Kingston RE, Moore D, Seidman JG, Smith JA, Struhl K, editors. Current protocols in molecular biology. New York, NY: John Wiley & Sons; 1994. [Google Scholar]
- Bilinski P, Hall MA, Neuhaus H, Gissel C, Heath JK, Gossler A. Two differentially expressed IL-11 receptor genes in the mouse genome. Biochem J. 1996;320:359–363. doi: 10.1042/bj3200359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carney EW, Prideaux V, Lye SJ, Rossant J. Progressive expression of trophoblast-specific genes during formation of mouse trophoblast giant cells in vitro. Mol Reprod. 1993;34:357–368. doi: 10.1002/mrd.1080340403. [DOI] [PubMed] [Google Scholar]
- Christianson G, Brooks W, Vekasi S, Manolfi E, Niles J, Roopenian S, Roths J, Rothlein R, Roopenian D. β2 microglobulin-deficient mice are protected from hypergammaglobulinemia and have defective antibody responses because of increased IgG catabolism. J Immunol. 1997;159:4780–4792. [PubMed] [Google Scholar]
- Cross JC, Werb Z, Fisher SJ. Implantation and the placenta: Key pieces of the development puzzle. Science. 1994;266:1508–1518. doi: 10.1126/science.7985020. [DOI] [PubMed] [Google Scholar]
- Croy BA, Chapeau C. Evaluation of the pregnancy immunotrophism hypothesis by assessment of the reproductive performance of young adult mice of genotype scid/scid.bg/bg. J Reprod Fertil. 1990;88:231–239. doi: 10.1530/jrf.0.0880231. [DOI] [PubMed] [Google Scholar]
- Dey SK. Implantation. In: Adashi EY, Rock JA, Rosenwaks Z, editors. Reproductive endocrinology, surgery, and technology. Philadelphia, PA: Lippincott-Raven; 1996. pp. 421–434. [Google Scholar]
- Doetschman TC, Eistetter H, Katz M, Schmidt W, Kemler R. The in vitro development of blastocyst-derived embryonic stem cell lines: Formation of visceral yolk sac, blood islands and myocardium. J Embryol Exp Morphol. 1985;87:27–45. [PubMed] [Google Scholar]
- Du X, Williams DA. Interleukin-11: Review of molecular, cell biology, and clinical use. Blood. 1997;89:3897–3908. [PubMed] [Google Scholar]
- Gossler A, Zachgo J. Gene and enhancer trap screens in ES cell chimaeras. In: Joyner A, editor. Gene targeting: A practical approach. Oxford, U.K: Oxford University Press; 1993. pp. 181–227. [Google Scholar]
- Guillemot F, Nagy A, Auerbach A, Rossant J, Joyner AL. Essential role of Mash-2 in extraembryonic development. Nature. 1994;371:333–336. doi: 10.1038/371333a0. [DOI] [PubMed] [Google Scholar]
- Hilton DJ, Hilton AA, Raicevic A, Rakar S, Harrison-Smith M, Gough NM, Begley CG, Metcalf D, Nicola NA, Willson TA. Cloning of a murine IL-11 receptor alpha-chain; requirement for gp130 for high affinity binding and signal transduction. EMBO J. 1994;13:4765–4775. doi: 10.1002/j.1460-2075.1994.tb06802.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huet Hudson YM, Andrews GK, Dey SK. Cell type-specific localization of c-myc protein in the mouse uterus: Modulation by steroid hormones and analysis of the periimplantation period. Endocrinology. 1989;125:1683–1690. doi: 10.1210/endo-125-3-1683. [DOI] [PubMed] [Google Scholar]
- Karow, J., K.R. Hudson, M.A. Hall, A.B. Vernallis, A. Gossler, and J.K. Heath. 1996. Mediation of Interleukin-11 dependant biological responses by a soluble form of the Interleukin-11 receptor. Biochem. J. (in press). [DOI] [PMC free article] [PubMed]
- Koller BH, Marrack P, Kappler JW, Smithies O. Normal development of mice deficient in β2M, MHC class I proteins, and CD8+ T cells. Science. 1990;248:1227–1230. doi: 10.1126/science.2112266. [DOI] [PubMed] [Google Scholar]
- Lehrach H, Diamond D, Wozney JM, Boedtker H. RNA molecular weight determinations by gel electrophoresis under denaturing conditions, a critical reexamination. Biochemistry. 1977;16:4743–4751. doi: 10.1021/bi00640a033. [DOI] [PubMed] [Google Scholar]
- Lescisin KR, Varmuza S, Rossant J. Isolation and characterization of a novel trophoblast-specific cDNA in the mouse. Genes & Dev. 1988;2:1639–1646. doi: 10.1101/gad.2.12a.1639. [DOI] [PubMed] [Google Scholar]
- Lim H, Paria BP, Das SK, Dinchuk JE, Langenbach R, Trzaskos JM, Dey SK. Multiple female reproductive failures in cyclooxygenase 2-deficient mice. Cell. 1997;91:197–208. doi: 10.1016/s0092-8674(00)80402-x. [DOI] [PubMed] [Google Scholar]
- Nandurkar HH, Robb L, Tarlinton D, Barnett L, Köntgen F, Begley CG. Adult mice with targeted mutation of the interleukin-11 receptor (IL11Ra) display normal hematopoiesis. Blood. 1997;90:2148–2159. [PubMed] [Google Scholar]
- Neuhaus H, Bettenhausen B, Bilinski P, Simon-Chazottes D, Guénet J-L, Gossler A. Etl2, a novel putative type-I-cytokine receptor expressed during mouse embryogenesis at high levels in skin and cells with skeletogenic potential. Dev Biol. 1994;166:531–542. doi: 10.1006/dbio.1994.1335. [DOI] [PubMed] [Google Scholar]
- O’Shea JD, Kleinfeld RG, Morrow HA. Ultrastructure of decidualization in the pseudopregnant rat. Am J Anat. 1983;166:271–298. doi: 10.1002/aja.1001660304. [DOI] [PubMed] [Google Scholar]
- Pollard JW. Lymphohematopoietic cytokines in the female reproductive tract. Curr Opin Immunol. 1991;3:772–777. doi: 10.1016/0952-7915(91)90112-e. [DOI] [PubMed] [Google Scholar]
- Psychoyos A. Hormonal control of ovoimplantation. Vitam Horm. 1973;31:201–256. doi: 10.1016/s0083-6729(08)60999-1. [DOI] [PubMed] [Google Scholar]
- Rinkenberger JL, Cross JC, Werb Z. Molecular genetics of implantation in the mouse. Dev Genet. 1997;21:6–20. doi: 10.1002/(SICI)1520-6408(1997)21:1<6::AID-DVG2>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Robb L, Hilton DJ, Brook-Carter PT, Begley CG. Identification of a second murine interleukin-11 receptor α-chain gene (IL11Ra2) with a restricted pattern of expression. Genomics. 1997;40:387–394. doi: 10.1006/geno.1996.4579. [DOI] [PubMed] [Google Scholar]
- Robb L, Li R, Hartley L, Nandukar H, Koentgen F, Begley G. Infertility in female mice lacking the receptor for interleukin 11 is due to a defective uterine response to implantation. Nature Med. 1998;4:303–308. doi: 10.1038/nm0398-303. [DOI] [PubMed] [Google Scholar]
- Robertson SA, Seamark RF, Guilbert LJ, Wegmann TG. The role of cytokines in gestation. Crit Rev Immunol. 1994;14:239–292. doi: 10.1615/critrevimmunol.v14.i3-4.30. [DOI] [PubMed] [Google Scholar]
- Rossant J. Development of the extraembryonic lineages. Dev Biol. 1995;6:237–247. [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Stewart CL. The role of leukemia inhibitory factor (LIF) and other cytokines in regulating implantation in mammals. Ann NY Acad Sci. 1994;734:157–165. doi: 10.1111/j.1749-6632.1994.tb21743.x. [DOI] [PubMed] [Google Scholar]
- Stewart CL, Kaspar P, Brunet LJ, Bhatt H, Gadi I, Kontgen F, Abbondanzo SJ. Blastocyst implantation depends on maternal expression of leukaemia inhibitory factor. Nature. 1992;359:76–79. doi: 10.1038/359076a0. [DOI] [PubMed] [Google Scholar]
- Welsh AO, Enders AC. Light and electron microscopic examination of the mature decidual cells of the rat with emphasis on the antimesometrial decidua and its degeneration. Am J Anat. 1985;172:1–29. doi: 10.1002/aja.1001720102. [DOI] [PubMed] [Google Scholar]
- Wilkinson DG, Bhatt S, Herrmann BG. Expression pattern of the mouse T gene and its role in mesoderm formation. Nature. 1990;343:657–659. doi: 10.1038/343657a0. [DOI] [PubMed] [Google Scholar]
- Yawata H, Yasukawa K, Natsuka S, Murakami M, Yamasaki K, Hibi M, Taga T, Kishimoto T. Structure-function analysis of human IL-6 receptor: Dissociation of amino acid residues required for IL-6-binding and for IL-6 signal transduction through gp130. EMBO J. 1993;12:1705–1712. doi: 10.1002/j.1460-2075.1993.tb05815.x. [DOI] [PMC free article] [PubMed] [Google Scholar]