A Mosaic Activating Mutation in AKT1 Associated with the Proteus Syndrome (original) (raw)
. Author manuscript; available in PMC: 2012 Feb 18.
Published in final edited form as: N Engl J Med. 2011 Jul 27;365(7):611–619. doi: 10.1056/NEJMoa1104017
Abstract
BACKGROUND
The Proteus syndrome is characterized by the overgrowth of skin, connective tissue, brain, and other tissues. It has been hypothesized that the syndrome is caused by somatic mosaicism for a mutation that is lethal in the nonmosaic state.
METHODS
We performed exome sequencing of DNA from biopsy samples obtained from patients with the Proteus syndrome and compared the resultant DNA sequences with those of unaffected tissues obtained from the same patients. We confirmed and extended an observed association, using a custom restriction-enzyme assay to analyze the DNA in 158 samples from 29 patients with the Proteus syndrome. We then assayed activation of the AKT protein in affected tissues, using phosphorylation-specific antibodies on Western blots.
RESULTS
Of 29 patients with the Proteus syndrome, 26 had a somatic activating mutation (c.49G→A, p.Glu17Lys) in the oncogene AKT1, encoding the AKT1 kinase, an enzyme known to mediate processes such as cell proliferation and apoptosis. Tissues and cell lines from patients with the Proteus syndrome harbored admixtures of mutant alleles that ranged from 1% to approximately 50%. Mutant cell lines showed greater AKT phosphorylation than did control cell lines. A pair of single-cell clones that were established from the same starting culture and differed with respect to their mutation status had different levels of AKT phosphorylation.
CONCLUSIONS
The Proteus syndrome is caused by a somatic activating mutation in AKT1, proving the hypothesis of somatic mosaicism and implicating activation of the PI3K–AKT pathway in the characteristic clinical findings of overgrowth and tumor susceptibility in this disorder. (Funded by the Intramural Research Program of the National Human Genome Research Institute.)
The proteus syndrome is characterized by patchy or segmental overgrowth and hyperplasia of multiple tissues and organs, along with susceptibility to the development of tumors1,2 (Fig. 1). It is thought that Joseph Merrick, an Englishman who lived in the late 19th century and became the subject of the play and film The Elephant Man, had the Proteus syndrome.
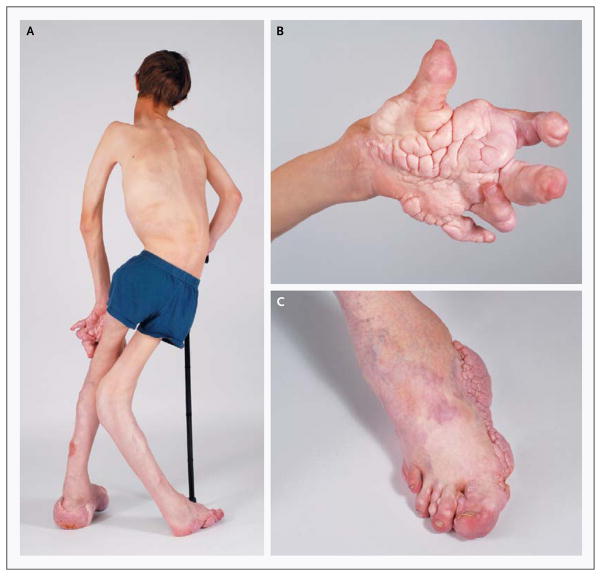
Figure 1. Clinical Manifestations of the Proteus Syndrome in a 12-Year-Old Boy.
Panel A shows severe orthopedic manifestations, including scoliosis, overgrowth with a resultant discrepancy in leg length, and valgus deformity and distortion of the skeleton, in Patient 53. Panels B and C show the characteristic cerebriform connective-tissue nevus and overgrowth and distortion of the hands and feet. Cutaneous vascular anomalies are present on the dorsum of the foot.
This uncommon syndrome (with an incidence of <1 case per 1 million population) has not been reported to recur in a family but has been reported in discordant monozygotic twins.3 These observations support the hypothesis that the Proteus syndrome is caused by a somatic mutation that is lethal when constitutive.4,5 A somatic mutation arises in a somatic cell and is thus present only in that cell and the lineages to which it gives rise, rather than being present in the conceptus and thus constitutively present in every cell of the body.
Some somatic or mosaic disorders, such as the McCune–Albright syndrome, are caused by a single mutation,6 whereas other such disorders (e.g., cancer) are caused by multiple mutations. (A mosaic disorder is one in which cells within the same person have a different genetic composition from one another.) The identification of somatic mutations can be approached by sequencing the exons in the genomes of affected and unaffected tissues from patients with disorders of interest. We used exome sequencing to identify a somatic mutation in patients with the Proteus syndrome.
METHODS
PATIENTS
The patients who are described here met current clinical criteria for the Proteus syndrome1 and were evaluated at the National Institutes of Health Clinical Center (see Table 1 in the Supplementary Appendix, available with the full text of this article at NEJM.org). All patients and their family members provided written informed consent to participate in this study.
STUDY PROCEDURES
Using standard techniques, we isolated DNA from peripheral blood, tissues, and cell lines from tissues obtained from patients with the Proteus syndrome and from control subjects without the disorder. Tissue samples (or their derivative cell lines) from patients with the Proteus syndrome were labeled “affected” if they were from an area with visible signs of overgrowth or vascular anomaly. “Unaffected” samples were typically obtained by cutaneous punch biopsy of an area that had no signs of overgrowth or vascular anomalies and that was as distant from affected areas as practicable. A designation of “unknown” was assigned to samples that could not be clinically classified (Table 2 in the Supplementary Appendix). Blood samples from patients were categorized as unknown, since no hematologic phenotype of the Proteus syndrome has been identified (other than thrombosis).7 DNA-sequencing libraries were prepared, as described previously.8
We identified genotypes using the most-probable-genotype (MPG) algorithm, in which genotype assignments with an MPG prediction score of 10 or more have been found to provide a balance between sensitivity and accuracy.8 We filtered the sequence data obtained from pairs of affected and unaffected samples to identify candidate variants that were present in the heterozygous state, present in affected samples from patients with the Proteus syndrome but absent or present at lower levels in unaffected samples from these patients (or in the case of the monozygotic twin pair, present in the affected twin and absent in the unaffected twin), or present in a sample from a patient with the Proteus syndrome but absent in both parents of the patient and in the ClinSeq research subjects (currently 401 exomes)9 and the Single Nucleotide Polymorphism Database (dbSNP). When a candidate variant was identified in one affected–unaffected sample pair, the variant was examined in all samples to identify variants that had as many of the preceding attributes as possible.
We carried out follow-up analyses by means of Sanger sequencing and custom restriction-enzyme digestion, using standard methods for polymerase-chain-reaction (PCR) amplification and capillary electrophoresis (primer sequences available on request). Details regarding the sequencing methods, custom restriction-enzyme digestion, and cell-culture methods are provided in the Supplementary Appendix.
For the comparisons of mutation status among the various groups of samples, we dichotomized samples as mutation-positive or mutation-negative, using a threshold of 1% or more for positivity, which was based on the sensitivity of restriction-enzyme digestion. Comparisons were performed with the use of Fisher’s exact test. To assay differences in the activation of AKT1, we compared signals obtained from Western blot luminescence images generated by an antibody that binds AKT family members (AKT1, 2, and 3) only when they are phosphorylated at Ser473, with signals generated by pan-AKT antibody. (Pan-AKT antibody binds all AKT proteins, regardless of phosphorylation status.) We tested for statistically significant differences using a paired t-test.
RESULTS
EXOME SEQUENCING
The initial exome sequencing of DNA from samples obtained from patients with the Proteus syndrome was performed on DNA extracted from cell lines established from surgical and skin-biopsy specimens. We performed exome sequencing of 17 DNA samples from 12 patients. These samples included 11 from 6 patients with the Proteus syndrome, including 4 paired affected–unaffected samples; 1 sample each from 5 parents of patients; and 1 sample from an unaffected identical twin of a patient.6 The average number of sequence reads for each exome was 96,488,818, with 6 of the samples sequenced with one lane each, 10 with two lanes, and 1 with four lanes. We obtained an average of 8,563,387,085 bases per sample. An average of 70.5% of the filtered, aligning reads (i.e., reads that aligned with the reference sequence of the human genome) overlapped the target regions of the Agilent All Exon kit (regions that the kit was designed to amplify, in preparation for sequencing). The average coverage was 87.3% of the 37.6 Mb of targeted DNA regions, with robust determination of both alleles in diploid regions on the basis of an MPG score of 10 or more.
VARIANT FILTERING
Of the 11 samples from the 6 patients, 7 samples were designated as affected and 4 as unaffected. We identified a total of 265,821 variants that differed from the human reference sequence. Using several filters and VarSifter Next-Gen filtering software to analyze the variants, we identified a variant in AKT1, c.49G→A (predicting a substitution of lysine for glutamine at amino acid 17), in Patient 5, with the use of the affected–unaffected comparison filter. On manual examination, we found that 7 exomes that we analyzed had this variant (2 exomes from 3 patients and 1 exome from a fourth patient). The proportion of mutant sequence reads in these samples ranged from 3.6 to 51% (Table 2 in the Supplementary Appendix). The AKT1 c.49G→A variant was absent in 401 ClinSeq research subjects9 in dbSNP, version 130, and was found in a single read in the 1000 Genomes project.10
SAMPLE SURVEY
To validate the association between AKT1 mutations and the Proteus syndrome, we tested numerous other samples from subjects with the disorder and subjects without the disorder, using a PCR assay and Sanger sequencing. The Sanger sequencing results were generally consistent with the exome sequencing data. We hypothesized that this method was insufficiently sensitive to detect low-level mosaicism. We therefore carried out an assay that was based on restriction-enzyme digestion, the results of which suggested that the mutation was absent in 25 cell lines and 2 fresh tissue samples from subjects who did not have the Proteus syndrome (for details regarding validation, see the Supplementary Appendix). Assays of samples from negative control subjects were repeated throughout the analyses and were consistently negative.
We tested 158 cell lines or uncultured tissue samples from 29 patients with the Proteus syndrome (Fig. 2C, and Table 1 in the Supplementary Appendix). A total of 97 of these samples were classified as affected, and of the affected samples, 75 were positive for the variant. Of 20 samples that were classified as unknown, 11 were positive. Of the 86 samples that tested positive (75 from clinically affected specimens and 11 from clinically unknown specimens), the fraction of mutant DNA in the positive specimens ranged from 1% (our lower limit of detection) to 47%. The mutation-positive specimens included uncultured tissues, which ruled out the hypothesis that the mutation was an artifact of cell culture. Of 41 unaffected samples, 13 were positive for the mutation. DNA that was purified from the peripheral blood of patients with the Proteus syndrome was uniformly negative on Sanger sequencing (data not shown). However, the restriction-enzyme assay showed that two peripheral-blood DNA samples were positive (8 to 9%). Overall, of 29 patients with the Proteus syndrome who were tested, 26 (90%) carried the mutation, as detected in one or more samples.
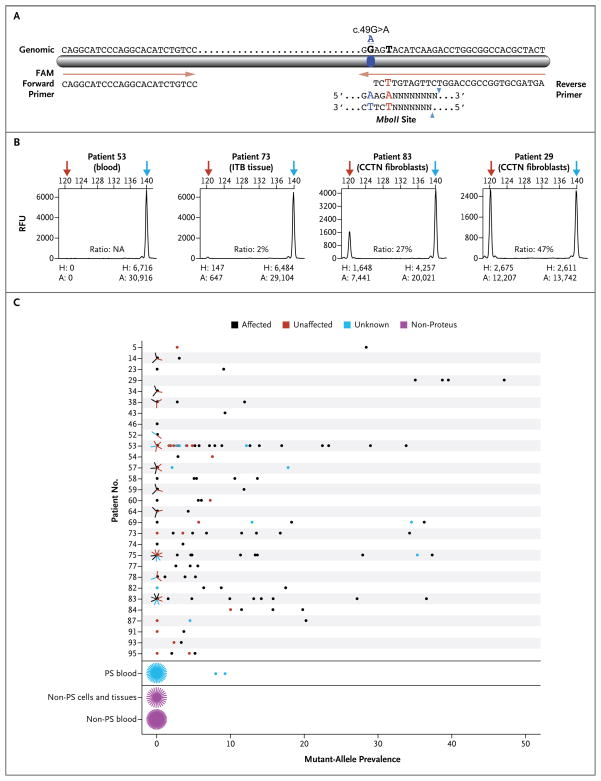
Figure 2. Assay for the Detection of the AKT1 Mutation.
Panel A shows a diagram of the modified polymerase-chain-reaction–restriction-enzyme assay that is used to detect the c.49G→A mutation in AKT1. The assay uses a restriction endonuclease that is derived from an Escherichia coli strain carrying the gene MboII from Moraxella bovis. The genomic sequence is shown above the bar; forward and reverse primers are below. The reverse primer was modified at position 28 (shown in red, chr14:105,246,548), creating an MboII restriction-enzyme site in the presence of the mutation (blue symbol and letters). The MboII consensus site is shown below the reverse primer. Panel B shows electropherograms for four samples from patients with the Proteus syndrome with varying levels of the mutant allele. Red arrows indicate the mutant allele, and blue arrows indicate the wild-type peak. The height (H) and area (A) for each peak are shown below the electropherograms and are expressed in relative fluorescence units (RFU). The numbers at the top of each electropherogram are base pairs. Ratios (provided as percentages) are the areas of the mutant peaks divided by the combined areas of the mutant and wild-type peaks. CCTN denotes cerebriform connective-tissue nevus, and ITB iliotibial band. Panel C shows the results of the MboII assay of samples from patients with the Proteus syndrome and from control subjects. The numbers on the y axis are the individual code numbers for patients for whom cell-line or tissue samples were available for testing. Grouped into single rows at the bottom of the graph are blood samples from 38 patients with the Proteus syndrome (PS), cell and tissue samples from 27 control subjects who did not have the Proteus syndrome, and blood samples from 48 control subjects. Multiple samples with the same value are represented by lines radiating from the data point, with each radiating line representing one sample.
FUNCTIONAL CHARACTERIZATION
We next evaluated the functional consequence of the mutation, which is known to constitutively activate AKT1 through Ser473 and Thr308 phosphorylation.11 Cell lines that were cultured from surgical explants from patients with the Proteus syndrome showed increased Ser473 phosphorylation under conditions of serum starvation, as compared with the control samples (P<0.005) (Fig. 3). We further tested the hypothesis that the mutation underlies the mosaicism of the Proteus syndrome by subcloning cell lines derived by limiting-dilution subcloning from cell lines obtained from patients with the Proteus syndrome. We identified pairs of clones from single-cell lines derived from both affected and unaffected tissues from patients with the Proteus syndrome; these clones were selected such that one was heterozygous for the mutation and the other was negative for the mutation (confirming that the original cell lines were a mixture of both mutation-positive and mutation-negative cells). AKT1 in the mutant clone that was derived from an affected cell line from Patient 93 had increased phosphorylation at both Ser473 and Thr308, as compared with a nonmutant clone derived from the same affected cell line (Fig. 3). Our analyses of two additional single-cell clone pairs, each of which was isolated from a single starting culture, replicated this finding.
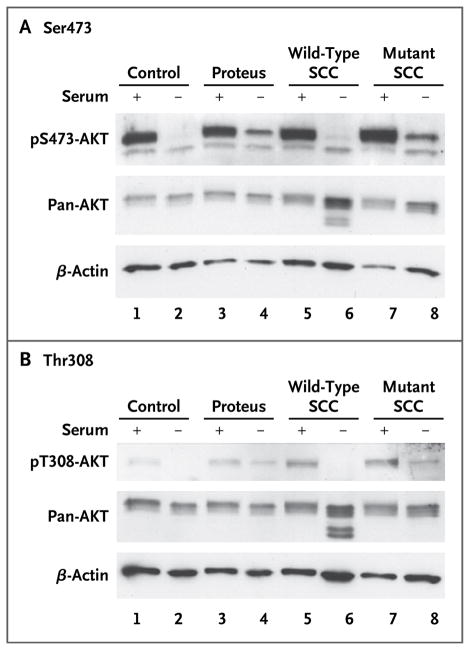
Figure 3. Immunoblot Analyses of AKT Phosphorylation.
To compare signals obtained from Western blot luminescence images, lysates were collected and separated on a 10% TRIS–glycine gel. AKT proteins were visualized with the use of antibodies that recognize phosphorylation at Ser473 (Panel A) or Thr308 (Panel B), and each set of lysates was also hybridized to total AKT (pan-AKT) and _β_-actin antibodies to assess loading variation. Lysates from a control cell line obtained from subjects without the Proteus syndrome are shown in lanes 1 and 2. Lysates from a cell line obtained from a patient with the Proteus syndrome (Patient 83 with cerebriform connective-tissue nevi [CCTN]; mutation level, 37%) are shown in lanes 3 and 4. Lysates from single-cell clones (SCC) isolated from culture established from a CCTN biopsy sample obtained from Patient 93 are shown in lanes 5 through 8, with lysates from a mutation-negative (wild-type) clone shown in lanes 5 and 6 and those from a mutation-positive clone shown in lanes 7 and 8. Lysates in lanes 1, 3, 5, and 7 were grown in serum-containing medium, indicated by a plus sign; those in lanes 2, 4, 6, and 8 were grown in serum-free medium, indicated by a minus sign, for 8 hours before harvesting. Quantitative analyses showed that in cells grown in the absence of factors that can activate the AKT pathway (i.e., in serum-free conditions), the level of phosphorylated AKT protein was higher in mutation-positive cells than in mutation-negative cells (P<0.005).
DISCUSSION
Our data support the conclusion that the AKT1 c.49G→A variant causes the Proteus syndrome and the mosaicism hypothesis that was advanced more than 20 years ago by Happle.4,5 It has been proposed that a more circumscribed or milder manifestation of the disorder would be associated with a later occurrence of the somatic mutation in an embryo.12 Although we detected the mutation more often in affected tissues than in unaffected tissues, we did not observe an association between the proportion of mutant alleles and the overall clinical severity or specific manifestations of the phenotype. Our data do not suggest a specific stage during development at which the mutation arose in the patients who were included in our analyses.
Although many of the cell cultures derived from the affected tissues carried the mutation, the cells from which we derived these cultures may not have been the cells that caused the tissue to be abnormal. Typical cell-culture conditions support the growth of mesodermal fibroblasts. Since the Proteus phenotype can be manifested in the derivatives of all three germ layers,1,13 it is likely that lineages other than the mesoderm are affected by this mutation. We therefore microdissected sections of skin-biopsy samples (embedded in paraffin blocks) obtained from patients with the Proteus syndrome, purified DNA from the microdissected tissue, and analyzed it on restriction-enzyme digestion. The preliminary data so derived suggest that the prevalence of the mutation is higher in the upper dermis than in the epidermis or lower dermis and that the mutation is absent in glandular tissue (data not shown). Further experiments are necessary to characterize the cells in the many tissues that can be affected in this disorder.
Only 2 of the 38 peripheral-blood DNA samples that were collected from patients with the Proteus syndrome were positive for the mutation, a proportion that is significantly lower than both the proportion of apparently affected samples that were positive (75 of 97, P<0.001) and the proportion of unaffected samples that were positive (13 of 41, P = 0.004). These findings are consistent with a previous study showing that the AKT1 activating mutation is detrimental to hematopoiesis,14 and they suggest that molecular diagnosis of the Proteus syndrome with the use of peripheral-blood DNA may be challenging. Clinical molecular diagnosis on the basis of testing for the mutation in the DNA derived from peripheral blood must therefore await more sensitive methods of detection. With currently available mutation-detection methods, we recommend obtaining biopsy samples for testing.
Samples from three patients with typical Proteus syndrome (Patients 34, 46, and 52) were negative for the mutation. Clinically, we could not distinguish these patients from those with mutation-positive samples. We analyzed only two, one, and three samples, respectively, from these three patients, and we think it is likely that these samples were negative purely by chance. Alternatively, a different activating mutation in AKT1 or a mutation in a different gene may have caused the Proteus syndrome in these patients. However, full sequencing of AKT1 exons and flanking introns in these three patients showed normal sequences (data not shown).
It is interesting that the guanidine residue at position 49 of AKT1 does not include a cytidine–phosphate–guanosine (CpG) dinucleotide and is therefore not predicted to be highly mutable. Moreover, the mutation is present in a mosaic form in patients with the Proteus syndrome. We hypothesize that these two features explain the extreme rarity of this disorder.
The c.49G→A, p.Glu17Lys AKT1 variant is functionally important but uncommon in tumors. According to the Catalogue of Somatic Mutations in Cancer (COSMIC) database,15 this variant has been detected in 116 of 7942 unique cancer samples, including cancers of the breast (72 samples), thyroid (10 samples), urinary tract (9 samples), lung (6 samples), and endometrium (5 samples). It is proposed that constitutive activation of AKT1 through Ser473 and Thr308 phosphorylation underlies the oncogenic mechanism.11 We have found that the up-regulation of AKT1 phosphorylation as a result of a heterozygous mutation in AKT1 occurs in some tissues of patients with the Proteus syndrome and suggest that constitutive activation of the protein underlies the overgrowth and tumor susceptibility in these patients.
Akt1 loss and gain of function have been evaluated extensively in the mouse. The Akt1 null phenotype includes somatic and central-nervous-system growth retardation,16,17 a reduced number and caliber of lymphatic capillaries,18 reduced growth of trabecular bone, reduced formation of endochondral bone,19 dwarfism and reduced ossification of cartilage,20 and platelet dysfunction with prolonged bleeding times.21 Mice with activated forms of Akt1 have also been studied. Fukai et al.20 found that activated Akt1 in vitro stimulated cartilage calcification, a major manifestation of the Proteus syndrome. Mice that were transgenic for an activated (myristolated) form of Akt1 had skin hyperplasia, which is also a manifestation of the Proteus syndrome.22,23 Generally speaking, the mouse phenotype that results from loss of function of Akt1 is the opposite of that of the Proteus syndrome, and the phenotype that results from gain of function is strikingly similar to that of the Proteus syndrome.
Several groups have reported that patients with the Proteus syndrome had PTEN mutations.24–26 Other investigators have argued that persons with PTEN mutations were clinically distinct from those with the Proteus syndrome and that persons with bona fide Proteus syndrome did not have PTEN mutations.27–31 Among patients with segmental overgrowth disorders, there is clinical overlap between those with somatic PTEN mutations — now designated as the segmental overgrowth, lipomatosis, arteriovenous malformation, and epidermal nevus (SOLAMEN) syndrome,29 or type 2 segmental Cowden syndrome (T2SCS)30 — and those with the Proteus syndrome. AKT1 is activated by loss-of-function mutations in PTEN,32 which explains why patients with such mutations (those with the SOLAMEN syndrome) and patients with activating mutations in AKT1 (those with the Proteus syndrome) have overlapping but distinct clinical manifestations. The Proteus and SOLAMEN syndromes may be members of a larger family of disorders related to dysfunction in the PI3K–AKT pathway. We hypothesize that multiple disorders are caused by mutated genes encoding proteins in this pathway.
The importance of somatic genetic variation is widely appreciated in oncology, in which a broad range of variations contributes to the pathogenesis of cancer. In contrast, the Proteus syndrome is caused by a de novo somatic mutation in a single gene. Several disorders share this attribute, with the prototype being the McCune–Albright syndrome, caused by a somatic mutation in GNAS.6 The SOLAMEN syndrome29 and autosomal dominant polycystic kidney disease33 are caused by two mutations, each of which affects an allelic copy of the same gene. One of these mutations is inherited in the germline. Somatic acquisition of the second mutation results in disease. Germline or inherited disorders are termed simple or complex on the basis of whether they can be attributed to a single gene variant or multiple gene variants, respectively. We suggest that mosaic disorders are analogous to inherited disorders in that some of them (e.g., the Proteus and McCune–Albright syndromes) are caused by a single variant and others (e.g., many cancers) arise only after the accumulation of many somatic mutations.
Supplementary Material
Supplement1
Acknowledgments
Supported by the Intramural Research Program of the National Human Genome Research Institute. The Proteus Syndrome Foundations in the United States and the United Kingdom contributed financial support for the sequencing.
We thank the patients and their families for their participation in this study; Danielle Brinckman, Steve Crane, Stefanie Dugan, Flavia Facio, Jennifer Sloan, and numerous fellows and genetic counseling trainees for their assistance in clinical research; Julie Nadel and Timothy Huber for their laboratory support; the staff of the National Institutes of Health Intramural Sequencing Center for sequence generation and analysis; Julia Fekecs for graphics support; Larry Singh for advice on statistics; M. Michael Cohen, Jr., for assistance in the development of the clinical diagnostic criteria; Scott Vacha for early research on the Proteus syndrome; and Paul Meltzer and Barbara Biesecker for their critical review of a previous version of the manuscript.
APPENDIX
The authors’ affiliations are as follows: the National Human Genome Research Institute (M.J.L., J.C.S., J.K.T., J.J.J., E.M.F., K.P., J.T., J.L.C., C.B., G.G., D.N., K.O., P.L.S., J.C.M, L.G.B.), National Institute of Dental and Craniofacial Research (N.C., S.A.K., P.G.R.), National Cancer Institute (D.J.S.), and the Department of Dermatology, Uniformed Services University of the Health Sciences (T.N.D.) — all in Bethesda; and National Institutes of Health Intramural Sequencing Center, Rockville (J.C.M., L.G.B.) — all in Maryland; the Department of Pediatrics, Division of Genetics, Medical College of Wisconsin, Milwaukee (D.B.); the Division of Orthopaedics and Sports Medicine (L.B., L.L.T.) and the Department of Surgery (K.N.), Children’s National Medical Center, Washington, DC; the Department of Pediatrics and Pediatric Neurology, Georg August University, Göttingen, Germany (K.B.); the Royal National Orthopaedic Hospital, Stanmore, United Kingdom (P.C., R.T.); the Division of Genetics, Children’s Hospital of Philadelphia, Philadelphia (M.A.D., E.H.Z.); Greenwood Genetics Center, Greenwood, SC (D.B.E.); the Department of Genetics and Developmental Biology, University of Connecticut, West Hartford (R.M.G.); the Ear Institute, Palm Desert, CA (B.M.K.); the Department of Pediatrics, Division of Genetics, University of Iowa, Iowa City (K.M.K-N.); the Department of Otolaryngology, Indiana University–Purdue University Indianapolis, Indianapolis (R.T.M.); the Department of Pediatric Surgery, Rocky Mountain Hospital for Children, Denver (S.R.); the Department of Pediatric Plastic and Craniofacial Surgery, Children’s Mercy Hospitals and Clinics; Kansas City, MO (V.S.); the Department of Plastic Surgery, Children’s Hospital Boston, Boston (J.U.); the Department of Pediatric Orthopaedics, Dana Children’s Hospital, Tel Aviv Medical Center, Tel Aviv University, Tel Aviv, Israel (S.W.); Proteus Syndrome Foundation, Colorado Springs, CO (K.H.); and Proteus Syndrome Foundation UK, Bexhill-on-Sea, United Kingdom (T.W.-N.).
Footnotes
The authors’ affiliations are listed in the Appendix.
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
References
- 1.Biesecker L. The challenges of Proteus syndrome: diagnosis and management. Eur J Hum Genet. 2006;14:1151–7. doi: 10.1038/sj.ejhg.5201638. [DOI] [PubMed] [Google Scholar]
- 2.Idem. The multifaceted challenges of Proteus syndrome. JAMA. 2001;285:2240–3. doi: 10.1001/jama.285.17.2240. [DOI] [PubMed] [Google Scholar]
- 3.Brockmann K, Happle R, Oeffner F, Konig A. Monozygotic twins discordant for Proteus syndrome. Am J Med Genet A. 2008;146A:2122–5. doi: 10.1002/ajmg.a.32417. [DOI] [PubMed] [Google Scholar]
- 4.Happle R. Lethal genes surviving by mosaicism: a possible explanation for sporadic birth defects involving the skin. J Am Acad Dermatol. 1987;16:899–906. doi: 10.1016/s0190-9622(87)80249-9. [DOI] [PubMed] [Google Scholar]
- 5.Idem. Cutaneous manifestation of lethal genes. Hum Genet. 1986;72:280. doi: 10.1007/BF00291899. [DOI] [PubMed] [Google Scholar]
- 6.Weinstein LS, Shenker A, Gejman PV, Merino MJ, Friedman E, Speigel AM. Activating mutations of the stimulatory G protein in the McCune–Albright syndrome. N Engl J Med. 1991;325:1688–95. doi: 10.1056/NEJM199112123252403. [Erratum, N Engl J Med 1992;326:1648.] [DOI] [PubMed] [Google Scholar]
- 7.Slavotinek AM, Vacha SJ, Peters KF, Biesecker LG. Sudden death caused by pulmonary thromboembolism in Proteus syndrome. Clin Genet. 2000;58:386–9. doi: 10.1034/j.1399-0004.2000.580509.x. [DOI] [PubMed] [Google Scholar]
- 8.Teer JK, Bonnycastle LL, Chines PS, et al. Systematic comparison of three genomic enrichment methods for massively parallel DNA sequencing. Genome Res. 2010;20:1420–31. doi: 10.1101/gr.106716.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Biesecker LG, Mullikin JC, Facio FM, et al. The ClinSeq Project: piloting large-scale genome sequencing for research in genomic medicine. Genome Res. 2009;19:1665–74. doi: 10.1101/gr.092841.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Durbin RM, Abecasis GR, Altshuler DL, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–73. doi: 10.1038/nature09534. [Erratum, Nature 2011;473:544.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carpten JD, Faber AL, Horn C, et al. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature. 2007;448:439–44. doi: 10.1038/nature05933. [DOI] [PubMed] [Google Scholar]
- 12.Happle R. The McCune-Albright syndrome: a lethal gene surviving by mosaicism. Clin Genet. 1986;29:321–4. doi: 10.1111/j.1399-0004.1986.tb01261.x. [DOI] [PubMed] [Google Scholar]
- 13.Biesecker LG. Proteus syndrome. In: Cassidy SB, Allanson JE, editors. Management of genetic syndromes. 2. Hoboken, NJ: John Wiley; 2005. pp. 449–56. [Google Scholar]
- 14.Kharas MG, Okabe R, Ganis JJ, et al. Constitutively active AKT depletes hematopoietic stem cells and induces leukemia in mice. Blood. 2010;115:1406–15. doi: 10.1182/blood-2009-06-229443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forbes SA, Bindal N, Bamford S, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945–D950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen WS, Xu PZ, Gottlob K, et al. Growth retardation and increased apoptosis in mice with homozygous disruption of the Akt1 gene. Genes Dev. 2001;15:2203–8. doi: 10.1101/gad.913901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cho H, Thorvaldsen JL, Chu Q, Feng F, Birnbaum MJ. Akt1/PKBalpha is required for normal growth but dispensable for maintenance of glucose homeostasis in mice. J Biol Chem. 2001;276:38349–52. doi: 10.1074/jbc.C100462200. [DOI] [PubMed] [Google Scholar]
- 18.Zhou F, Chang Z, Zhang L, et al. Akt/Protein kinase B is required for lymphatic network formation, remodeling, and valve development. Am J Pathol. 2010;177:2124–33. doi: 10.2353/ajpath.2010.091301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vandoorne K, Magland J, Plaks V, et al. Bone vascularization and trabecular bone formation are mediated by PKB alpha/Akt1 in a gene-dosage-dependent manner: in vivo and ex vivo MRI. Magn Reson Med. 2010;64:54–64. doi: 10.1002/mrm.22395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fukai A, Kawamura N, Saito T, et al. Akt1 in murine chondrocytes controls cartilage calcification during endochondral ossification under physiologic and pathologic conditions. Arthritis Rheum. 2010;62:826–36. doi: 10.1002/art.27296. [DOI] [PubMed] [Google Scholar]
- 21.Chen J, De S, Damron DS, Chen WS, Hay N, Byzova TV. Impaired platelet responses to thrombin and collagen in AKT-1-deficient mice. Blood. 2004;104:1703–10. doi: 10.1182/blood-2003-10-3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Segrelles C, Lu J, Hammann B, et al. Deregulated activity of Akt in epithelial basal cells induces spontaneous tumors and heightened sensitivity to skin carcinogenesis. Cancer Res. 2007;67:10879–88. doi: 10.1158/0008-5472.CAN-07-2564. [DOI] [PubMed] [Google Scholar]
- 23.Segrelles C, Moral M, Lorz C, et al. Constitutively active Akt induces ectodermal defects and impaired bone morphogenetic protein signaling. Mol Biol Cell. 2008;19:137–49. doi: 10.1091/mbc.E07-08-0764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith JM, Kirk EP, Theodosopoulos G, et al. Germline mutation of the tumour suppressor PTEN in Proteus syndrome. J Med Genet. 2002;39:937–40. doi: 10.1136/jmg.39.12.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou X, Hampel H, Thiele H, et al. Association of germline mutation in the PTEN tumour suppressor gene and Proteus and Proteus-like syndromes. Lancet. 2001;358:210–1. doi: 10.1016/s0140-6736(01)05412-5. [DOI] [PubMed] [Google Scholar]
- 26.Zhou XP, Marsh DJ, Hampel H, Mulliken JB, Gimm O, Eng C. Germline and germline mosaic PTEN mutations associated with a Proteus-like syndrome of hemi-hypertrophy, lower limb asymmetry, arteriovenous malformations and lipomatosis. Hum Mol Genet. 2000;9:765–8. doi: 10.1093/hmg/9.5.765. [DOI] [PubMed] [Google Scholar]
- 27.Barker K, Martinez A, Wang R, et al. PTEN mutations are uncommon in Proteus syndrome. J Med Genet. 2001;38:480–1. doi: 10.1136/jmg.38.7.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Biesecker LG, Rosenberg MJ, Vacha S, Turner JT, Cohen MM., Jr PTEN mutations and Proteus syndrome. Lancet. 2001;358:2079–80. doi: 10.1016/S0140-6736(01)07109-4. [DOI] [PubMed] [Google Scholar]
- 29.Caux F, Plauchu H, Chibon F, et al. Segmental overgrowth, lipomatosis, arteriovenous malformation and epidermal nevus (SOLAMEN) syndrome is related to mosaic PTEN nullizygosity. Eur J Hum Genet. 2007;15:767–73. doi: 10.1038/sj.ejhg.5201823. [DOI] [PubMed] [Google Scholar]
- 30.Happle R. Type 2 segmental Cowden disease vs. Proteus syndrome. Br J Dermatol. 2007;156:1089–90. doi: 10.1111/j.1365-2133.2007.07818.x. [DOI] [PubMed] [Google Scholar]
- 31.Thiffault I, Schwartz CE, Der Kaloustian V, Foulkes WD. Mutation analysis of the tumor suppressor PTEN and the glypican 3 (GPC3) gene in patients diagnosed with Proteus syndrome. Am J Med Genet A. 2004;130A:123–7. doi: 10.1002/ajmg.a.30335. [DOI] [PubMed] [Google Scholar]
- 32.Cantley LC, Neel BG. New insights into tumor suppression: PTEN suppresses tumor formation by restraining the phosphoinositide 3-kinase/AKT pathway. Proc Natl Acad Sci U S A. 1999;96:4240–5. doi: 10.1073/pnas.96.8.4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brasier JL, Henske EP. Loss of the polycystic kidney disease (PKD1) region of chromosome 16p13 in renal cyst cells supports a loss-of-function model for cyst pathogenesis. J Clin Invest. 1997;99:194–9. doi: 10.1172/JCI119147. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplement1