Phosphorylation-dependent regulation of cyclin D1 nuclear export and cyclin D1–dependent cellular transformation (original) (raw)
Abstract
GSK-3β-dependent phosphorylation of cyclin D1 at Thr-286 promotes the nuclear-to-cytoplasmic redistribution of cyclin D1 during S phase of the cell cycle, but how phosphorylation regulates redistribution has not been resolved. For example, phosphorylation of nuclear cyclin D1 could increase its rate of nuclear export relative to nuclear import; alternatively, phosphorylation of cytoplasmic cyclin D1 by GSK-3β could inhibit nuclear import. Here, we report that GSK-3β-dependent phosphorylation promotes cyclin D1 nuclear export by facilitating the association of cyclin D1 with the nuclear exportin CRM1. D1-T286A, a cyclin D1 mutant that cannot be phosphorylated by GSK-3β, remains nuclear throughout the cell cycle, a consequence of its reduced binding to CRM1. Constitutive overexpression of the nuclear cyclin D1-T286A in murine fibroblasts results in cellular transformation and promotes tumor growth in immune compromised mice. Thus, removal of cyclin D1 from the nucleus during S phase appears essential for regulated cell division.
Keywords: cyclin D1, GSK-3β, CRM1, tumorigenesis, nuclear export
During G1 phase, the D-type cyclins (D1, D2, D3) accumulate and assemble with either cyclin-dependent kinase 4 or 6 (CDK4 or CDK6) in response to mitogenic growth factors. The active cyclin D holoenzyme promotes G1 progression by inactivating the growth-suppressive properties of the retinoblastoma protein (Rb) through site-specific phosphorylation and by virtue of its ability to titrate CDK inhibitors such as p27Kip1 and p21Cip1 (Weinberg 1995; Sherr and Roberts 1999). Titration of p27Kip1 and p21Cip1, in turn, facilitates activation of the cyclin E/CDK2 complex and subsequent entry and progression through the DNA synthetic (S) phase of the cell cycle. Although p27Kip1 and p21Cip1 are effective inhibitors of cyclin E/CDK2 and cyclin A/CDK2 complexes, recent evidence demonstrates that they promote the assembly of cyclin D/CDK complexes (LaBaer et al. 1997) and are found in catalytically active cyclin D/CDK complexes in vivo (Cheng et al. 1999).
Extracellular mitogens promote cellular proliferation via receptor-mediated signaling circuitry that ultimately converges on the cell cycle machine, and the D-type cyclins function as critical sensors of these signals. For example, receptor-dependent activation of Ras promotes the accumulation of active cyclin D1/CDK4 complexes via at least two pathways. First, activated Ras promotes transcription of the cyclin D1 gene through a kinase cascade involving Raf1, mitogen-activated protein kinase kinase (MEK), and the extracellular signal-regulated protein kinases (Albanese et al. 1995; Aktas et al. 1997; Lavoie et al. 1996; Winston et al. 1996; Kerkhoff and Rapp 1997; Weber et al. 1997). Assembly and activation of the cyclin D1/CDK4 holoenzyme are also augmented by this same pathway (Cheng et al. 1998). Second, the rate of cyclin D1 proteasomal degradation is mediated by GSK-3β-dependent phosphorylation of a single threonine residue (Thr-286) near the C terminus of cyclin D1 (Diehl et al. 1998). Mitogens inactivate GSK-3β via a pathway involving Ras, phosphatidylinositol 3-kinase, and protein kinase B/Akt (Rodriguez-Viciana et al. 1994, 1997; Franke et al. 1995, 1997; Klinghoffer et al. 1996; Kauffmann-Zeh et al. 1997; Vanhaesebroeck et al. 1997). Maximal accumulation of the active cyclin D1/CDK4 holoenzyme requires activation of both Ras-dependent signaling pathways.
Alterations in the subcellular distribution of cyclin D1 during the cell cycle may also regulate cyclin D1/CDK4 function. Cyclin D1 accumulates in the nucleus throughout G1 phase, but it relocalizes to the cytoplasm during the remainder of interphase (Baldin et al. 1993; Diehl et al. 1998). Like its proteolysis, the cell cycle–specific redistribution of cyclin D1 correlates with its phosphorylation on Thr-286 by GSK-3β (Diehl et al. 1998). Overexpression of GSK-3β redirects nuclear cyclin D1 to the cytoplasm (Diehl et al. 1998). Likewise, a mutant cyclin D1 protein containing an alanine-for-threonine substitution at residue 286 (D1-T286A) remains nuclear throughout the cell cycle and is not subject to GSK-3β-dependent redistribution (Diehl et al. 1998). Thus, it has been proposed that GSK-3β-dependent phosphorylation facilitates cyclin D1 nuclear export. Consistent with this notion, GSK-3β is excluded from the nucleus during G1 phase, whereas a fraction of total GSK-3β gains access to the nuclear compartment during S phase (Diehl et al. 1998). However, it is equally plausible that cytoplasmic GSK-3β phosphorylates cytosolic cyclin D1, thereby inhibiting cyclin D1 nuclear import.
Although cyclin D1 was initially cloned from a translocation break point associated with parathyroid adenoma, it is now clear that overexpression of nuclear cyclin D1 occurs at a high frequency in a variety of carcinomas, including those of breast, esophageal, and pancreatic origin (Sherr 1996). Work involving the development of mice expressing a cyclin D1 transgene indicates that cyclin D1 indeed has oncogenic capacity. Mice programmed to overexpress cyclin D1 in mammary tissue develop a protracted course of mammary neoplasia (Wang et al. 1994). In addition, overexpression of cyclin D1 in B cells augments c-Myc-induced lymphomagenesis (Bodrug et al. 1994; Lovec et al. 1994). However, immortal murine fibroblasts, which can be transformed by activated Ras, are not transformed upon overexpression of wild-type cyclin D1; cyclin D1 overexpressing cells require serum-derived mitogens for proliferation, remain contact inhibited, and do not grow in soft agar (Quelle et al. 1993). Therefore, overexpression of wild-type cyclin D1 is not sufficient to promote neoplastic transformation without cooperating mutations.
We now demonstrate that the nuclear-to-cytoplasmic relocalization of cyclin D1 during S phase is mediated by CRM1-dependent nuclear export. Our results indicate that phosphorylation of Thr-286 activates cyclin D1 nuclear export by facilitating cyclin D1-CRM1 association. Furthermore, murine fibroblasts that overexpress cyclin D1-T286A display characteristics of morphologic transformation and promote tumor growth in mice, suggesting that nuclear exclusion of cyclin D1 complexes during S phase is a critical feature in the regulation of normal cellular proliferation.
Results
Cyclin D1 is exported from the nucleus during S phase
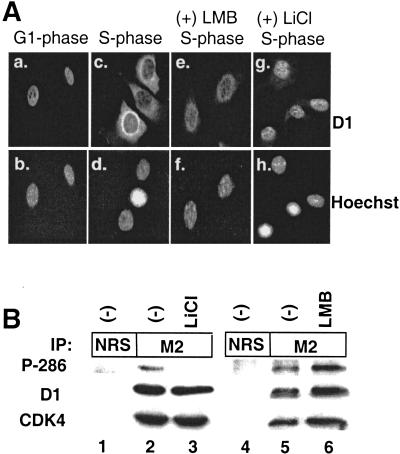
The cytoplasmic localization of cyclin D1 during S phase depends on the presence of a phosphorylatable Thr-286 (Diehl et al. 1998), implying that phosphorylation of this residue could facilitate cyclin D1 nuclear export. To test this hypothesis, we rendered NIH-3T3 cells quiescent for 36 h by removing serum; subsequently, by adding fetal calf serum (FCS), we stimulated them to synchronously re-enter the cell cycle. Localization of cyclin D1 was determined either 8 h (late G1 phase) or 16 h (S phase) after serum addition. Before fixation and immunostaining with a cyclin D1–specific monoclonal antibody, the indicated S-phase cells were treated with leptomycin B (LMB), an inhibitor of CRM1-dependent nuclear export (Nishi et al. 1994), for 2.5 h or with lithium chloride (LiCl), a specific inhibitor of GSK-3β (Stambolic et al. 1996), for 1 h. Cyclin D1 accumulated in the nucleus during G1 phase (Fig. 1A, panel a) but relocalized to the cytoplasm during S phase (panel c). The cytoplasmic relocalization of cyclin D1 was inhibited by treatment with either LMB (panel e) or LiCl (panel g), suggesting that relocalization was dependent on active GSK-3β and active CRM1.
Figure 1.
Nuclear export of cyclin D1 during S phase. (A) NIH-3T3 cells were arrested in G0 by serum deprivation for 36 h and subsequently stimulated to synchronously re-enter the cell cycle by addition of FCS. Cells were harvested and processed for immunofluorescence microscopy at 8 h (G1-phase, panels a, b) or 16 h (S-phase, panels c–h). Cyclin D1 was visualized with a cyclin D1–specific monoclonal antibody, and DNA was visualized with Hoechst dye. In certain experiments, 10 ng/mL LMB (panels e, f) or 20 mM LiCl (panels g, h) was added to culture medium before harvesting for immunofluorescence. (B) Lysates prepared from D1-3T3 cells left untreated or treated with 20 mM LiCl for 2 h or 10 ng/mL LMB for 2.5 h were precipitated (IP) with normal rabbit serum (NRS) or the M2 monoclonal antibody. Proteins were resolved on denaturing polyacrylamide gels, transferred to nitrocellulose membrane, and blotted with an antibody that specifically recognizes phosphorylated Thr-286. The membrane was subsequently stripped and reblotted with an antibody that recognizes both phosphorylated and unphosphorylated cyclin D1; then it was stripped once again and blotted with an antibody that recognizes CDK4. Sites of antibody binding were visualized by enhanced chemiluminescence.
To confirm that LiCl treatment inhibited Thr-286 phosphorylation, we precipitated cyclin D1 from cyclin D1 overexpressing NIH-3T3 cells left untreated or treated with 20 mM LiCl. Precipitates were blotted with antibodies that specifically recognize either Thr-286 phosphorylated cyclin D1 (see Fig. 5A below) or total cyclin D1. As shown in Figure 1B, cells treated with LiCl did not express detectable levels of Thr-286 phosphorylated cyclin D1 (Fig. 1B, P-286, cf. lanes 2 and 3). In contrast, LMB treatment, which inactivates CRM1, promoted the accumulation of nuclear (Fig. 1A, panel e) Thr-286 phosphorylated cyclin D1 (Fig. 1B, lane 6). Neither LMB nor LiCl affected cyclin D1/CDK4 complex formation (Fig. 1B, CDK4). These results indicate that S-phase-specific nuclear-to-cytoplasmic relocalization of cyclin D1 is positively regulated by Thr-286 phosphorylation and is the result of CRM1-dependent nuclear export.
Figure 5.
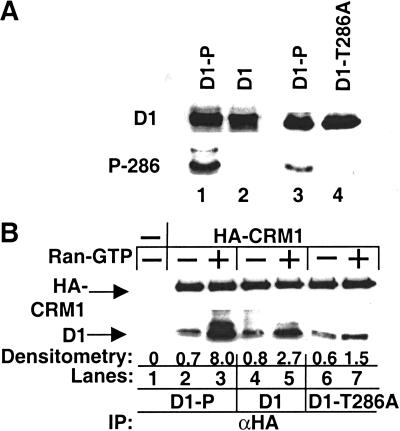
Thr-286 phosphorylation promotes cyclin D1-CRM1 binding. (A) Purified Flag-D1 phosphorylated with recombinant GSK-3β (lanes 1, 3), Flag-D1 treated with calf intestinal alkaline phosphatase (lane 2), or Flag-D1-T286A (lane 4) was separated on denaturing polyacrylamide gels and blotted with an antibody that specifically recognizes phosphorylated Thr-286 of cyclin D1. The membrane was subsequently stripped and reblotted with the cyclin D1 monoclonal antibody, which recognizes total cyclin D1. Sites of antibody binding were visualized by enhanced chemiluminescence. (B) 12CA5 Sepharose (lane 1) or recombinant HA-CRM1 bound to 12CA5 Sepharose was mixed with phosphorylated (D1-P), dephosphorylated cyclin D1 (D1), or cyclin D1-T286A in the absence or presence of Ran-GTP (lanes 2–7). After extensive washes with detergent-containing buffer, HA-CRM1-cyclin D1 complexes were denatured, resolved on polyacrylamide gels, and blotted with either the 12CA5 antibody for HA-CRM1 or the cyclin D1 monoclonal antibody. CRM1-associated D1 was quantitated by densitometry and is expressed relative to D1 binding to CRM1 in the absence of Ran-GTP. The precipitating antibody (α-HA) is indicated at bottom (IP). Sites of antibody binding were visualized by enhanced chemiluminescence.
Leptomycin B inhibits GSK-3β-dependent relocalization of cyclin D1
Overexpression of GSK-3β promotes the stoichiometric redistribution of cyclin D1 from the nucleus to the cytoplasm, independent of cell cycle stage (Diehl et al. 1998). Mutation of the site of GSK-3β phosphorylation within cyclin D1 to alanine (D1-T286A) prevents both GSK-3β-dependent and S-phase-dependent nuclear-to-cytoplasmic redistribution of cyclin D1 such that the D1-T286A mutant is constitutively nuclear (Diehl et al. 1998). This redistribution of cyclin D1 could result from either phosphorylation-dependent inhibition of nuclear import or activation of nuclear export. The inhibitor experiments (Fig. 1) supported the notion that redistribution of cyclin D1 during S phase is the result of nuclear export. We therefore determined if GSK-3β-dependent relocalization of cyclin D1 is indeed regulated by CRM1-dependent nuclear export.
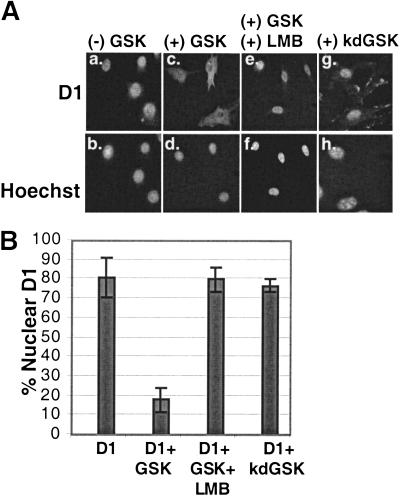
NIH-3T3 cells were transfected with expression vectors encoding Flag-tagged cyclin D1 and CDK4 alone or with vectors encoding either wild-type (wt)GSK-3β or a kinase-dead (kd)GSK-3β. Under our conditions of transfection, >60% of the cell population expressed ectopic protein (data not shown). Before immunofluorescence, the indicated cells were treated with 10 ng/mL LMB for 2.5 h to inactivate CRM1. The cells were then fixed, and the localization of cyclin D1 was visualized by indirect immunofluorescence using the M2 monoclonal antibody, which recognizes the N-terminal Flag epitope (Fig. 2A). Cells expressing cyclin D1 were counted and scored for cyclin D1 localization (Fig. 2B). As expected (Diehl and Sherr 1997), cyclin D1 localized primarily to nuclei under these conditions (Fig. 2A, panel a). In contrast, co-transfection with a vector encoding wtGSK-3β (panel c) but not kdGSK-3β (panel g) promoted the redistribution of cyclin D1 from the nucleus to the cytoplasm. Quantitation revealed that cyclin D1 was primarily nuclear in <20% of the cells upon co-transfection with GSK-3β (Fig. 2B), whereas in cells expressing only cyclin D1 and CDK4, the majority (80%) of cyclin D1 was nuclear (Fig. 2B). In contrast, treatment of the cells co-expressing D1, CDK4, and wtGSK-3β with LMB completely restored cyclin D1 nuclear localization (Fig. 2A, panel e, and B). The LMB-dependent inhibition of cyclin D1 nuclear export is likely the result of the inhibition of CRM1 activity because cyclin D1 is still phosphorylated at Thr-286 under these conditions (Fig. 1B). Therefore, GSK-3β-mediated relocalization of cyclin D1 requires CRM-1-dependent nuclear export.
Figure 2.
GSK-3β promotes cyclin D1 nuclear export. (A) Asynchronous NIH-3T3 cells transiently overexpressing Flag-D1 and CDK4 (panels a, b), Flag-D1, CDK4, and wtGSK-3β (panels c–f), or Flag-D1, CDK4, and kinase dead (kd)GSK-3β (panels g, h) were harvested and processed for immunofluorescence. Cyclin D1 was visualized with the M2 monoclonal antibody, and cellular DNA was stained with Hoechst dye. In panels e and f, LMB was added to cellular medium at a final concentration of 10 ng/mL for 2.5 h before harvest. (B) Cyclin D1 localization in cells transfected and stained as in A was scored as either exclusively nuclear (e.g., panels a, e, g) or cytoplasmic (panel c). The number of cells containing nuclear cyclin D1, from four independent transfections, were counted and are shown graphically. The bars represent standard deviation between independent transfections.
Cyclin D1 nuclear-to-cytoplasmic shuttling is dependent on CRM1 and GSK-3β
The above findings are consistent with the notion that GSK-3β-dependent phosphorylation of cyclin D1 on Thr-286 regulates its nuclear export. To further address this issue, we used an interspecies-heterokaryon shuttling assay. This assay relies on the fact that fused interphase cells maintain distinct nuclei within a contiguous cytoplasm until they enter M phase. By fusing cells from two different species, we can monitor the migration of proteins from one nucleus to another. For migration to occur, the cargo must undergo at least one round of nuclear export (from the original nucleus) and one round of nuclear import (into the heterologous nucleus). Because a protein that is defective for nuclear export will remain localized to the original nucleus, this assay can be used to assess the ability of a protein to undergo nuclear export.
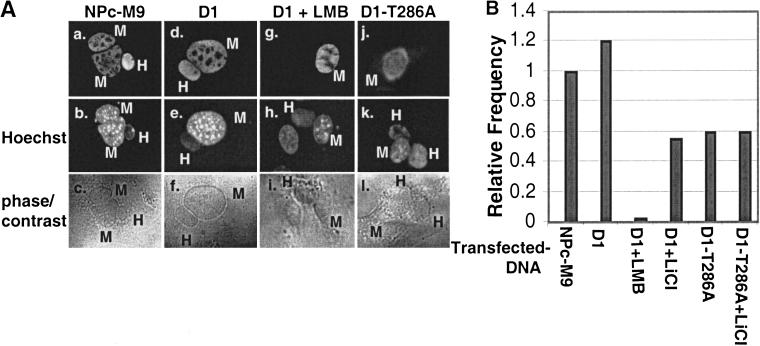
For these experiments, nucleoplasmin containing a bi-directional transport signal (M9) and a c-Myc epitope tag (NPc-M9) was used as a positive control (Michael et al. 1995; Sachdev et al. 1998). NIH-3T3 cells seeded on glass coverslips were transiently transfected with expression vectors encoding nucleoplasmin, wild-type cyclin D1, or D1-T286A. Thirty-six hours after transfection and 3 h before cellular fusion, HeLa cells were plated on top of the transfected NIH-3T3 cells and allowed to attach. Shuttling between nuclei was determined 30 min after cell fusion by indirect immunofluorescence using the 9E10 monoclonal antibody (NPc-M9) or either a monoclonal antibody that specifically recognizes murine cyclin D1 or the Flag-specific M2 monoclonal antibody. Murine and human nuclei were distinguished by staining with Hoechst dye 33258. The total number of heterokaryons displaying protein shuttling was quantitated (Fig. 3B), and representative fusions are shown (Fig. 3A). NPc-M9 was visualized in both the transfected NIH-3T3 nuclei and in nuclei of fused HeLa cells at a high frequency (Fig. 3A, panel a; the frequency of NPc-M9 shuttling was arbitrarily set at 1 and used as a point of reference). Wild-type cyclin D1 was also visualized in both the transfected NIH-3T3 nuclei and the fused HeLa nuclei (Fig. 3A, panel d) at a frequency comparable to that of NPc-M9 (Fig. 3B, cf. bars NPc-M9 and D1). Addition of LMB inhibited cyclin D1 movement between mouse and human nuclei (Fig. 3A, panel g, and B, bar D1 + LMB), consistent with the notion that cyclin D1 shuttling depends on functional CRM1. To assess the role of GSK-3β-mediated phosphorylation of Thr-286 for cyclin D1 nuclear export, we determined the ability of LiCl, which inhibits GSK-3β, to block cyclin D1 shuttling. Treatment of cells with 20 mM LiCl significantly reduced cyclin D1 shuttling (Fig. 3B, bar D1 + LiCl), as did mutation of Thr-286 to a nonphosphorylatable alanine residue (Fig. 3A, panel j, and B, bar D1-T286A). Treatment of cells expressing D1-T286A with LiCl did not further reduce its shuttling (Fig. 3B, bar D1-T286A + LiCl). Thus, GSK-3β-dependent phosphorylation of Thr-286 facilitates cyclin D1 nuclear export.
Figure 3.
Phosphorylation of Thr-286 facilitates cyclin D1 nuclear shuttling. (A) NIH-3T3 cells proliferating on glass coverslips and transiently expressing nucleoplasmin, NPc-M9 (panels a–c), cyclin D1 and CDK4 without (panels d–f) or with LMB (panels g–i), or D1-T286A and CDK4 (panels j–l) were treated with cycloheximide and fused with HeLa cells with polyethylene glycol. Thirty minutes after fusion, cells were harvested and processed for immunofluorescence microscopy. NPc-M9 was visualized using the 9E10 monoclonal antibody, and cyclin D1 was visualized with either a monoclonal antibody specific for murine cyclin D1 or the M2 monoclonal antibody, and DNA was visualized with Hoechst dye. The differential staining of human (homogenous) versus murine nuclei (punctate) by Hoechst dye 33258 facilitated the identification of fusions composed of murine and human nuclei. (B) The number of fusions wherein NPc-M9 and cyclin D1 were observed in HeLa nuclei was quantitated. The capacity of wild-type cyclin D1 and D1-T286A to shuttle in this assay in the presence of either LMB or LiCl was also determined. Frequencies were determined from multiple experiments and are expressed as the shuttling frequency relative to the positive shuttling control, NPc-M9.
CRM1 regulates cyclin D1 nucleocytoplasmic movement
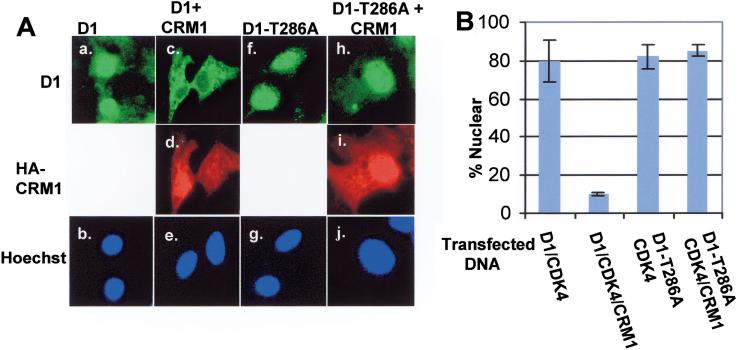
If CRM1 mediates nuclear export of Thr-286 phosphorylated cyclin D1, then overexpression of CRM1 should promote nuclear-to-cytoplasmic redistribution of cyclin D1. To test this hypothesis, we transfected NIH-3T3 cells with plasmids encoding cyclin D1 and CDK4 or cyclin D1, CDK4, and hemagglutinin antigen (HA)–tagged CRM1. In the absence of exogenous CRM1, cyclin D1 was predominantly nuclear (Fig. 4A, panel a). However, co-expression of HA-CRM1 resulted in the relocalization of cyclin D1 from the nucleus to the cytoplasm (panel c). In contrast, HA-CRM1 failed to relocalize D1-T286A to the cytoplasm (panel h), even though HA-CRM1 could be detected in both the nuclear and cytoplasmic compartments, consistent with the notion that Thr-286 phosphorylation is critical for cyclin D1 nuclear export.
Figure 4.
CRM1 promotes the cytoplasmic localization of cyclin D1. (A) NIH-3T3 cells were transfected with plasmids encoding either wild-type cyclin D1 and CDK4 (panels a, b), cyclin D1, CDK4, and HA-CRM1 (panels c_–e_), D1-T286A and CDK4 (panels f, g), or D1-T286A, CDK4, and HA-CRM1 (panels h–j). Cyclin D1 was visualized with the anti-Flag octa-probe antibody (top), HA-tagged CRM1 was visualized with the 12CA5 mouse monoclonal antibody (center), and DNA was visualized with Hoechst dye (bottom). (B) Cyclin D1 localization in cells transfected and stained as in A, was scored as either exclusively nuclear (e.g., panels a, f, h) or cytoplasmic (panel c). The number of cells containing nuclear cyclin D1, from four independent transfections, were counted and are shown graphically. The bars represent standard deviation between independent transfections.
We next considered whether phosphorylation of Thr-286 might promote cyclin D1 nuclear export via its propensity to facilitate cyclin D1-CRM1 association. To examine this possibility, we performed in vitro binding assays using purified recombinant proteins. Flag-D1 purified from baculovirus-infected Sf9 cells was either phosphorylated with GSK-3β in vitro or dephosphorylated with calf intestinal alkaline phosphatase. The presence of phosphorylated Thr-286 was confirmed by immunoblot analysis with an antibody specific for this phosphorylated residue (Fig. 5A). Purified cyclin D1 proteins were then incubated with purified HA-CRM1, and complexes were isolated by precipitation with the HA-specific 12CA5 monoclonal antibody and blotted with either the cyclin D1 monoclonal antibody or the anti-HA antibody. Both phosphorylated and dephosphorylated cyclin D1 were capable of binding to CRM1 (Fig. 5B, lanes 2, 4). However, within the nucleus, the binding of CRM1 to export substrates is facilitated by the small GTP-binding protein Ran, in its GTP-bound state, Ran-GTP (Askjaer et al. 1998). We thus tested whether the addition of Ran-GTP might differentially stimulate binding of phosphorylated versus dephosphorylated cyclin D1 to CRM1. Consistent with this notion, three- to fourfold more Thr-286 phosphorylated D1 than dephosphorylated D1 was detected in the HA-CRM1 precipitate in the presence of Ran-GTP (Fig. 5B, cf. lanes 3 and 5). The binding of the D1-T286A mutant to HA-CRM1, like dephosphorylated D1, was not significantly enhanced by Ran-GTP (Fig. 5B, lanes 6, 7). Therefore, Thr-286 phosphorylation of cyclin D1 facilitates cyclin D1-CRM1 binding under conditions of high Ran-GTP concentrations, as would be expected in the nucleus (Nigg 1997).
Cyclin D1-T286A promotes oncogenic transformation
Although cells engineered to overexpress cyclin D1 display a contracted G1 interval (Quelle et al. 1993; Resnitzky et al. 1994), these cells do not form foci, grow in soft agar, or promote tumor formation in immune-compromised mice (Quelle et al. 1993). However, even when expressed at high levels, ectopic cyclin D1 displays wild-type cell cycle–dependent subcellular localization (Diehl et al. 1998). Thus, the essentially normal characteristics of these cells may reflect their propensity to metabolize ectopically expressed cyclin D1. It was therefore reasonable to consider the possibility that failure to remove cyclin D1 from the nucleus during S phase might perturb cell homeostasis and promote cell transformation.
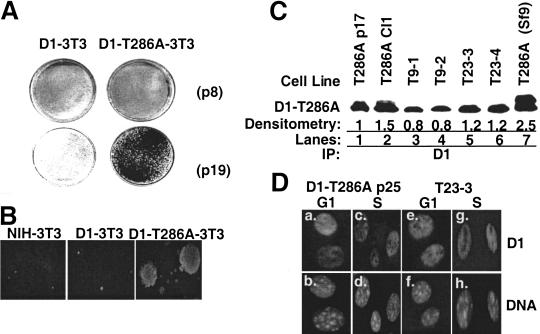
To test this hypothesis, we established NIH-3T3 cell lines that constitutively overexpress Flag epitope-tagged cyclin D1-T286A (D1-T286A-3T3) or wild-type Flag-D1 (D1-3T3) (Diehl et al. 1998). Pools of stable transfectants were subjected to an established passage protocol (Todaro and Green 1963), and various passages were analyzed for characteristics of cell transformation. Early passage ([passage 4–9] p4-p9) D1-T286A-3T3 and D1-3T3 cells were subject to contact inhibition and to growth arrest after growth factor removal (Diehl et al. 1998). To determine if long-term overexpression of cyclin D1-T286A promoted cell transformation, we examined the capacity of early versus late passage D1-3T3 and D1-T286A-3T3 cells to form foci; we found that neither early nor late passage D1-3T3 cells formed foci (Fig. 6A). In contrast, late passage (p19) D1-T286A cells readily formed large numbers of foci (Fig. 6A). Over the course of four independent experiments, cells overexpressing D1-T286A routinely formed foci after 18 passages, whereas D1-3T3 cells failed to form foci even after >40 passages (data not shown). Consistent with data from the focus formation assays is our finding that passage 18 or more D1-T286A-3T3 cells readily grew in soft agar, whereas wild-type NIH-3T3 and D1-3T3 cells were not capable of anchorage-independent growth (Fig. 6B).
Figure 6.
Cyclin D1-T286A promotes cellular transformation. (A) Early (p4) or late (p19) passage cell lines were plated in complete medium containing 5% FCS at 4 × 105 cells per 60-mm dish. Cells were grown for an additional 21 d, and foci were counted after staining with Giemsa. (B) NIH-3T3 (p22), D1-3T3 (p22), and D1-T286A-3T3 (p23) were plated in semisolid medium and cultured in a humidified environment for 21 d. Colonies were visualized by phase-contrast microscopy. Colonies >50 cells were scored. (C) Whole-cell lysates were prepared from cell lines established from D1-T286A-derived soft agar colonies or prepared from tumors that arose in SCID mice injected with either p9 or p23 D1-T286A-3T3 cells were precipitated with the cyclin D1 monoclonal antibody (lanes 1–6) and subjected to direct Western blotting with the cyclin D1 monoclonal antibody. Flag-D1 expressed in Sf9 cells served as a positive Western blotting control (lane 7). Sites of antibody binding were visualized by enhanced chemiluminescence. Relative levels of expression were determined by densitometry and are noted below the gel. (D) D1-T286A-3T3 cells or cells isolated from a D1-T286A-derived tumor (T23-3) were synchronized by serum deprivation and contact inhibition for 36 h and subsequently stimulated to synchronously re-enter the cell cycle by addition of FCS. Cells were harvested and processed for immunofluorescence microscopy at 8 h (G1 phase) or 16 h (S phase). Cyclin D1 was visualized with the cyclin D1–specific monoclonal antibody, and DNA was visualized with Hoechst dye.
To address whether fibroblasts that overexpressed D1-T286A are tumorigenic, D1-T286A-3T3 cells were tested for their ability to form subcutaneous tumors in SCID mice. As shown in Table 1, 7 out of 7 mice injected with passage 23 D1-T286A-3T3 cells formed tumors within 22 d, whereas 7 of 7 mice injected with RasVal12-3T3 cells developed tumors within 10 d. In addition, even early passage D1-T286A-expressing cells formed tumors, albeit at a protracted rate (Table 1). Similar to the in vitro transformation assays, neither early nor late passage NIH-3T3 nor D1-3T3 cells were able to form tumors (Table 1). To ensure that colonies that formed in soft agar and tumors that arose in SCID mice expressed D1-T286A, soft agar colonies were picked and expanded in vitro (Fig. 6C, lane 2), and whole-cell protein lysates were prepared. In parallel, protein lysates were prepared from tumors that arose in SCID mice injected with early (T9-1 and -2; Fig. 6C, lanes 3, 4) or late passage (T23-3 and -4; Fig. 6C, lanes 5, 6) D1-T286A-3T3 cells. Cyclin D1 was precipitated from lysates, resolved on a denaturing polyacrylamide gel, and transferred to membrane. Immunoblot analysis with the cyclin D1 monoclonal antibody confirmed that D1-T286A was expressed at comparable levels in both cloned cell lines and in the tumor samples (Fig. 6C, lanes 2–7).
Table 1.
In vivo tumor growth of D1-T286A expressing cells
| Cell line | Passage no. | No. of mice with tumors | Time until tumor development |
|---|---|---|---|
| RasVa112-3T3 | N/D | 7 of 7 | 10 d |
| NIH-3T3 | p10 | 0 of 7 | N/A |
| NIH-3T3 | p20 | 0 of 7 | N/A |
| D1-3T3 | p8 | 0 of 7 | N/A |
| D1-3T3 | p22 | 0 of 7 | N/A |
| D1-T286A-3T3 | p9 | 6 of 7 | 40 d |
| D1-T286A-3T3 | p23 | 7 of 7 | 22 d |
We have previously shown that D1-T286A remains exclusively nuclear throughout the cell cycle (Diehl et al. 1998), unlike wild-type cyclin D1, which is cytoplasmic in S phase (see Fig. 1). To confirm that D1-T286A was still a nuclear protein in tumor-derived cells, we established cell lines from the D1-T286A-induced tumors. The localization of D1-T286A in G1- or S-phase cells for one such tumor-derived cell line is shown in Figure 6D. As previously shown (Diehl et al. 1998), the D1-T286A protein was nuclear during G1 and S phase in the D1-T286A-3T3 cells (Fig. 6D, panels a, c). The D1-T286A protein was also nuclear in tumor-derived cells during both G1 and S phase (Fig. 6D, panels e, g).
Because the D1-T286A mutant has a half-life approximately sevenfold longer than wild-type cyclin D1 (Diehl et al. 1997), the differential growth properties of the D1-3T3 versus D1-T286A-3T3 could reflect different levels of the respective cyclin D1 proteins. Ectopic D1 and D1-T286A can be distinguished from endogenous cyclin D1 as a result of the presence of the N-terminal Flag epitope, which results in the reduced mobility of ectopic cyclin D1 (Diehl et al. 1997). Whole-cell lysates were prepared from either parental NIH-3T3, D1-3T3, or D1-T286A-3T3 cells and blotted with the cyclin D1 monoclonal antibody. Both early and late passage D1-3T3 and D1-T286A-3T3 cells expressed equivalent levels of the respective Flag-tagged D1 proteins (Fig. 7A). In agreement, cyclin D1 complexes precipitated from the respective cell lines phosphorylated recombinant Rb to the same extent in an in vitro kinase assay (Fig. 7B).
Figure 7.
Cellular transformation correlates with constitutive nuclear accumulation of cyclin D1-T286A. (A) Whole-cell lysates prepared from early (p11) or late (p18) NIH-3T3 or derivatives overexpressing either wild-type cyclin D1 or D1-T286A were subjected to direct Western blotting with the cyclin D1 monoclonal antibody. Sites of antibody binding were visualized by enhanced chemiluminescence. (B) Lysates were prepared as above and precipitated with either a control antibody (NRS) or the cyclin D1 monoclonal antibody. Precipitates were then assayed for their ability to phosphorylate recombinant GST-Rb. After SDS-PAGE, phosphorylated GST-Rb was visualized by autoradiography. (C) D1-3T3 (lanes 1–4) or D1-T286A-3T3 (lanes 5–7) cells were pulse-labeled with [35S]methionine for the indicated intervals. Cyclin D1 was immunoprecipitated from lysates, resolved on a denaturing gel, and visualized by autoradiography. Accumulation of newly synthesized cyclin D1 protein was quantitated by densitometry (bottom). (D) Asynchronously proliferating early and late passages of the indicated cell lines were harvested and stained with propidium iodide; DNA content was determined by flow cytometry.
Given the extended half-life of the D1-T286A protein relative to wild-type cyclin D1, we presumed that the steady-state levels of each protein in the respective cell lines must reflect biosynthetic rate (which is determined by plasmid uptake during the transfection to establish the cell lines). To confirm this, D1-3T3 and D1-T286A-3T3 cells were pulse-labeled with [35S]methionine, and cyclin D1 was precipitated from cellular lysates and resolved on a polyacrylamide gel. Significantly more radiolabeled cyclin D1 was synthesized during the course of this experiment than was D1-T286A (Fig. 7C, cf. lanes 2–4 with 5–7), which is consistent with the idea that the high level of accumulation of wild-type cyclin D1 results from its increased synthesis. The fact that there appears to be an upper limit for total protein accumulation, be it wild-type or mutant D1, likely reflects the fact that overexpression of cyclin D1 can be associated with apoptosis (Sofer-Levi and Resnitzky 1996).
These results suggest that the differential growth of the D1-T286A-3T3 cells was not simply the result of increased levels of the D1-T286A protein. Oncogenic transformation, particularly that which targets the p53 pathway, is frequently associated with genomic instability (Levine 1997). However, analysis of the cellular DNA content by flow cytometry revealed that the transformed D1-T286A cell lines remained pseudo-diploid (Fig. 7D). Therefore, D1-T286A-dependent transformation does not appear to be associated with cellular aneuploidy.
Discussion
Regulation of cyclin D1 nuclear export by phosphorylation
GSK-3β-dependent phosphorylation of Thr-286 promotes the stoichiometric redistribution of wild-type cyclin D1 from the nucleus to the cytoplasm during S phase (Diehl et al. 1998). Consistent with this finding, a cyclin D1 mutant containing an alanine for threonine substitution at residue 286 (D1-T286A) remains nuclear throughout the cell cycle and is not subject to GSK-3β-dependent regulation (Diehl et al. 1998). We considered two possible models that address the mechanism whereby phosphorylation of Thr-286 modulates cyclin D1 localization (Diehl et al. 1998). In the first, phosphorylation of nuclear cyclin D1 increases its rate of nuclear export relative to nuclear import. In the second model, GSK-3β phosphorylates cytoplasmic cyclin D1, thereby inhibiting cyclin D1 nuclear import.
The experimental data provided here support the first model. Treatment of cells with LMB, an inhibitor of the nuclear export receptor CRM1, inhibited GSK-3β-dependent nucleo-cytoplasmic redistribution of cyclin D1. Using heterokaryon shuttling assays, we found that cyclin D1 shuttled between heterologous nuclei. Cyclin D1 shuttling was inhibited by LMB, demonstrating that movement represented at least one round of nuclear export. In addition, cyclin D1 shuttling was reduced by inhibiting GSK-3β or by mutation of Thr-286 to alanine. We also found that S-phase-specific nuclear-to-cytoplasmic movement of cyclin D1 was prevented by inhibition of GSK-3β (LiCl treatment) or CRM1 (LMB treatment), thereby implicating phosphorylation-dependent nuclear export of cyclin D1 during S phase. Finally, overexpression of CRM1 promotes the cytoplasmic localization of cyclin D1 under conditions in which cyclin D1 is normally nuclear. Once again, CRM1-dependent relocalization of cyclin D1 was dependent on the presence of Thr-286, because D1-T286A remained nuclear even in the presence of overexpressed wild-type GSK-3β and CRM1. These data demonstrate that localization of cyclin D1 is at least in part determined by the efficiency of nuclear export. Furthermore, phosphorylation of Thr-286 appears to act as a required switch that facilitates cyclin D1 nuclear export such that cyclin D1 is predominantly cytoplasmic during S phase of the cell cycle.
CRM1 regulates cyclin D1 nuclear export
Transit of proteins between nuclear and cytoplasmic compartments occurs via nuclear pores (Nigg 1997; Gorlich 1998). For cytoplasmic to nuclear translocation through the nuclear pore, proteins must first interact with soluble, cytoplasmic import factors (importins), followed by the docking of this complex to the cytoplasmic face of the nuclear pore. The mechanisms governing nuclear export are analogous, with nuclear export substrates binding to soluble, nuclear export factors (exportins). CRM1 is one such exportin that binds to leucine-rich nuclear export signals (NES) (Fornerod et al. 1997; Fukada et al. 1997; Stade et al. 1997). CRM1 likely mediates nuclear export of several cell cycle regulatory proteins, including cyclin B1 (Yang et al. 1998), CDC25 (Lopez-Girona et al. 1999), and p27Kip1 (Tomoda et al. 1999), and the data presented here demonstrate that cyclin D1 should be added to this list.
Site-specific phosphorylation can either positively or negatively regulate nuclear export. For example, phosphorylation of Pho4 is necessary for association with its nuclear exportin, Msn5 (Kaffman et al. 1998). Analogous with Pho4, two observations herein suggest that phosphorylation of Thr-286 regulates cyclin D1-CRM1 association and thereby determines the rate of cyclin D1 nuclear export. First, CRM1-dependent nuclear-to-cytoplasmic transport of cyclin D1 is dependent on active GSK-3β and on the presence of phosphorylatable Thr-286. Second, the physical association of cyclin D1 and CRM1 was greatly enhanced by Thr-286 phosphorylation. Overall, these data provide direct evidence that phosphorylation of Thr-286 facilitates the binding of cyclin D1 with CRM1 and thereby promotes cyclin D1 nuclear export.
The binding of CRM1 to a leucine-rich NES within the cargo is an early step in the nuclear export process (Gorlich 1998). Because the exact spacing of the leucine/hydrophobic residues within each substrate is subject to variation, it can be difficult to define an NES signal on this basis alone (Nigg 1997). Examination of cyclin D1 revealed two sequences that closely resemble characterized NES signals. The first is located within the “cyclin box” from residues 87–94 (RFLSLEPL), and the second is located at the C terminus beginning with Thr-286 (TPTDVRDVDI). Deletion of either of these sequences alone was not sufficient to abolish CRM1-dependent nuclear-to-cytoplasmic relocalization of cyclin D1 (negative data not shown). The possibility remains that cyclin D1 may have more than one functional NES, as has been demonstrated for the adenomatous polyposis coli (APC) tumor suppressor (Henderson 2000; Neufeld et al. 2000; Rosen-Arbesfeld et al. 2000). Accordingly, removal of any one NES would not be sufficient to abolish CRM1-mediated nuclear export. Alternately, D1-CRM1 binding may be mediated by a novel type of interaction. Further investigation of cyclin D1-CRM1 binding is clearly warranted.
Constitutively nuclear cyclin D1-T286A promotes cell transformation
The capacity of the cell to direct nuclear import of cyclin D1 during G1 phase and nuclear export during S phase suggests that movement of cyclin D1 must be considered bi-directional. Consequently, cyclin D1 subcellular distribution is in fact an equilibrium established between nuclear import and nuclear export. The observed overexpression of nuclear cyclin D1 in a variety of cancers (Hall and Peters 1996; Sherr 1996) suggests that its overexpression provides cells with a distinct growth advantage. The development of mammary hyperplasia and cancer in mice bearing a cyclin D1 transgene is consistent with this hypothesis (Wang et al. 1994). Yet, cell lines engineered to overexpress wild-type cyclin D1 remain growth factor dependent and fail to exhibit overt signs of cell transformation (Quelle et al. 1993; Resnitzky et al. 1994). Thus, cyclin D1 overexpression by itself is not sufficient to promote uncontrolled cell growth. In contrast, we have shown that the constitutively nuclear cyclin D1-T286A protein promotes cell transformation. This single-point mutation alters two known properties of cyclin D1. First, D1-T286A is resistant to polyubiquitination, resulting in an extended half-life relative to wild-type cyclin D1 (Diehl et al. 1997). Second, this mutation inhibits the nuclear export of cyclin D1 during S phase (data herein). The steady-state levels of wild-type versus mutant cyclin D1 are comparable in the respective cell lines, resulting in roughly equivalent levels of D-type cyclin kinase activity (Fig. 7). Therefore, increased protein stability is likely not the primary contributor to cell transformation by D1-T286A. Rather, the oncogenicity of D1-T286A correlates with its constitutive nuclear localization.
These observations highlight the possibility that the tolerance of cells to wild-type cyclin D1 may reflect their ability to “restrain” cyclin D1 through regulation of its subcellular localization. In fact, the subcellular localization of overexpressed wild-type cyclin D1 mirrors that of endogenous cyclin D1 (Diehl et al. 1998), being nuclear in G1 and cytoplasmic during S phase. This is significant given that the critical functions of cyclin D1 are nuclear (Sherr 1996).
What then is the mechanism whereby D1-T286A promotes cell transformation? The cyclin D1/CDK4 complex is thought to regulate cell cycle progression via its propensity to promote Rb phosphorylation. However, the presence of D1-T286A/CDK4 complexes in the nucleus during S phase suggests that it could function downstream of Rb. One possibility is that the inability of a cell to eliminate nuclear cyclin D1 during S phase perturbs the fidelity of DNA synthesis, perhaps the result of phosphorylation of proteins that are normally substrates of the CDK2 kinase. High-fidelity DNA replication depends on appropriate timing and activation of the DNA replication machinery (Krude et al. 1997). The cyclin E/CDK2 and cyclin A/CDK2 kinases play an active role in regulating and activating DNA synthesis. Phosphorylation of these substrates by the D1-T286A/CDK4 kinase could result in the premature or inappropriate activation of these CDK2 substrates. Implicit to this model is the possibility that D1-T286A-dependent transformation occurs downstream of Rb phosphorylation and thus may be Rb independent. The capacity of cyclin D1–dependent kinase to promote S-phase entry independent of Rb phosphorylation is consistent with this notion (Leng et al. 1997). Furthermore, it has been reported that wild-type cyclin D1 can promote focal gene amplification in cells containing wild-type p53 (Zhou et al. 1996). Although our analysis reveals that D1-T286A cells remain diploid and do not show overt signs of genetic instability, it is possible that expression of D1-T286A leads to specific gene amplification events undetectable by fluorescence-activated cell sorting.
Alternately, the constitutively nuclear D1-T286A/CDK4 complex may perturb the normal temporal and spatial course of the Rb phosphorylation/dephosphorylation cycle. The growth-suppressive activity of Rb is intimately associated with its ability to bind to and inactivate a large number of growth-promoting proteins, including members of the E2F family of transcription factors (Lees et al. 1993), Myc family proteins (Rustgi et al. 1991), and even proteins that directly participate in DNA replication such as MCM7 (Sterner et al. 1998). Phosphorylation of Rb triggers the release of these bound proteins, thereby permitting activation of their growth-promoting functions. Dephosphorylation of Rb during mitosis is an important step in the re-activation of Rb and likely serves to reset the cell cycle clock for the ensuing G1 interval. Perturbation of either phosphorylation or dephosphorylation of Rb through overexpression of D1-T286A would likely perturb the critical temporal and spatial timing of Rb-dependent growth control, resulting in neoplastic growth.
Cyclin D1 and human neoplasia
There have been no reported mutations at Thr-286 of cyclin D1 in human neoplasia. However, overexpression of cyclin D1 in certain tumors cannot be explained by gene amplification or chromosomal rearrangements. In these tumors, it is reasonable to propose that the regulatory circuitry controlling cyclin D1 nuclear accumulation is deregulated. For example, in patients having colorectal adenocarcinoma and in mice bearing intestinal adenomas, high levels of nuclear cyclin D1 are observed, with no apparent genetic abnormalities in the cyclin D1 gene (Hall and Peters 1996; Sherr 1996). Inactivating mutations in the APC tumor suppressor, a target of Wnt signaling pathways (Kinzler and Vogelstein 1996) occurs with a high frequency in such carcinomas (Powell et al. 1992). The demonstration that deregulation of Wnt-dependent signaling promotes overexpression of nuclear cyclin D1 and cooperates with activated MEK1 in cell transformation (Rimerman et al. 2000) supports the concept that cellular defects that impinge on cyclin D1 subcellular localization contribute to oncogenesis. These observations, coupled with the ability of constitutively nuclear D1-T286A to promote cellular transformation, raise the intriguing possibility that deregulation of cyclin D1 localization may be a previously unappreciated contributor to human neoplasia.
Materials and methods
Cell culture conditions and transfections
NIH-3T3 cells and HeLa cells were maintained in Dulbecco's modified Eagle's medium (DMEM) containing glutamine supplemented with antibiotics (Cellgro) and 10% FCS (BioWhitaker). Insect Sf9 cells were grown in Grace's medium supplemented with 10% heat-inactivated FCS. For expression in insect Sf9 cells, HA-tagged CRM1 was inserted into pVL1393 as an _Sma_I-_Not_I fragment. Procedures for manipulation of baculoviruses were previously described (Summers and Smith 1987). Derivatives of NIH-3T3 cells engineered to overexpress flag-tagged cyclin D1 and flag-tagged cyclin D1-T286A were previously described (Diehl et al. 1998). Cell lines were maintained on a passage protocol wherein 5 × 109 cells were passaged per 60-mm dish every third day (Todaro and Green 1963). Transient expression of Myc-tagged GSK-3β and Flag-tagged cyclin D1 in NIH-3T3 cells or Flag-tagged cyclin D1 along with HA-tagged CRM1 was performed as previously described (Diehl et al. 1998). For determination of cellular DNA content, cells were trypsinized, stained with propidium iodide, and analyzed by flow cytometry.
Immunoblotting and kinase assays
For Western analysis, cells were lysed in EBC buffer (50 mM Tris-HCl at pH 7.5, 120 mM NaCl, 0.5 % NP-40, 1 mM phenylmethylsulfonyl fluoride [PMSF], 20 U/mL aprotinin, 5 μg/mL leupeptin, 0.4 mM NaVO4, 0.4 mM NaF). Total cellular proteins (200 μg) were resolved on denaturing polyacrylamide gels (tumor lysates were subjected to precipitation with the cyclin D1 antibody before electrophoresis), electrophoretically transferred to nitrocellulose membranes (MSI, Westborough, MA), and blotted with the cyclin D1 monoclonal antibody (D1-17-13G). For detection of cyclin D1–dependent kinase activity, cells were harvested in Tween 20 IP buffer (50 mM HEPES at pH 7.5, 150 mM NaCl, 10 mM MgCl2, 1 mM EDTA, 0.1% Tween 20, 1 mM PMSF, 20 U/mL aprotinin, 5 μg/mL leupeptin, 0.4 mM NaVO4, 0.4 mM NaF). After precipitation with the cyclin D1 monoclonal antibody, protein kinase assays using 1 μg of recombinant GST-Rb were performed as previously described (Diehl and Sherr 1997; Rimerman et al. 2000).
Biosynthetic labeling
Subconfluent cells were incubated for 30 min in methionine-free DMEM (BioWhittaker) and shifted to medium containing 200 μCi/mL Tran-35S-label (ICN) for the indicated periods of time. Proteins were precipitated from cell lysates normalized for total protein concentration with the cyclin D1 monoclonal antibody, and radiolabeled proteins were resolved on denaturing polyacrylamide gels and visualized by autoradiography.
Expression and purification of proteins
HA-CRM1 and Flag-D1 were cloned into the pVDL-1393 baculoviral expression vector (Pharmingen), and virus was isolated according to established procedures (Summers and Smith 1987). After infection of insect Sf9 cells at high multiplicity, cells were lysed in EBC buffer and clarified by sedimentation in a microcentrifuge for 10 min. Recombinant proteins were purified by affinity chromatography with antibodies that recognize either the HA or Flag epitope tags. HA-CRM1 was left attached to Sepharose beads for use in the in vitro interaction assays. Before the elution of Flag-D1 from beads with excess Flag peptide, Flag-D1 was either dephosphorylated by treatment with calf intestinal alkaline phosphatase for 15 min at 30°C or mixed with recombinant GSK-3β and ATP as previously described (Diehl et al. 1998) to ensure stoichiometric phosphorylation of Thr-286.
A pQE32-derived expression vector encoding a hexahistidine-tagged canine Ran cDNA was introduced into BLR[pRep4] cells. The cells were grown to an O.D. of 0.5, and expression of Ran was induced by the addition of 1mM isopropyl-β-D-thiogalactopyranoside (IPTG). The cell pellet was resuspended in lysis buffer (50 mM Tris at pH 8.0, 150 mM NaCl, 10 mM imidazole, 10% glycerol, 1% Triton X-100, 2 mM β-mercaptoethanol, 0.2 mM PMSF, 5 μg/mL each of antipain, leupeptin, pepstatin, aprotinin, soybean trypsin inhibitor). The cells were lysed by gentle sonication and clarified by centrifugation at 10,000_g_. The supernatant was passed over a metal-chelate affinity column (Invitrogen) and washed with lysis buffer containing 60 mM imidazole. The bound proteins were eluted with 450 mM imidazole and dialyzed against 10 mM HEPES (pH 7.3), 160 mM potassium acetate, 5 mM magnesium acetate containing 1 mM dithiothreitol, 1 mM PMSF.
In vitro interaction assays
HA-CRM1 coupled to 12CA5-protein A Sepharose was mixed with purified cyclin D1 in the absence or presence of RanGTP and incubated at 4°C for 2 h. Complexes were washed four times with Tween 20 IP buffer and denatured in sample buffer. Proteins were resolved on denaturing polyacrylamide gels, electrophoretically transferred to nitrocellulose membranes (Millipore), and blotted with the cyclin D1 monoclonal antibody (D1-17-13G). Sites of antibody binding were visualized with either protein-conjugated horseradish peroxidase (HRP; EY Laboratories), anti-mouse-conjugated HRP, or anti-rabbit-conjugated HRP (Amersham Pharmacia Biotech).
Immunofluorescence
NIH-3T3 cells seeded on glass coverslips were transfected with expression vectors encoding the indicated DNAs. Cells were fixed at 48 h after transfection, using either 3% paraformaldehyde or methanol-acetone (1:1). For visualization of cyclin D1 in the interspecies heterokaryons, coverslips were stained with either a mouse-specific cyclin D1 monoclonal antibody (D1-17-13G) or the Flag-specific M2 monoclonal (Sigma) in PBS containing 1% dry milk. Secondary antibody staining was performed for 30 min using fluorescein isothiocyanate (FITC)–conjugated anti-mouse (Amersham Pharmacia Biotech). For visualization of Flag-tagged cyclin D1 and HA-tagged CRM1 in the same cell, cells were stained with a goat antibody specific for the Flag epitope (10 μg/mL) (Santa Cruz Biotechnology), followed by rabbit anti-goat (10 μg/mL) and FITC-conjugated anti-rabbit (1:100). HA-CRM1 was detected by staining with the 12CA5 mouse monoclonal (10 μg/mL) followed by biotinylated anti-mouse (1:500) and subsequently streptavidin Texas Red (1:500). In all cases, DNA was visualized using Hoescht 33258 dye at a 1:500 dilution. Coverslips were mounted on glass slides with Vectashield mounting medium (Vector Laboratories).
Interspecies heterokaryon assay
NIH-3T3 cells plated at 5 × 105 cells per 60-mm dish on glass coverslips at 8 h before transfection with expression vectors encoding CDK4 along with either cyclin D1 or the D1-T286A mutant. After 48 h, HeLa cells were seeded on top of the transfected NIH-3T3 cells at 3 × 106 cells per dish and allowed to attach for 3 h. Cycloheximide was added at 50 μg/mL at 5 min before fusion. Cellular fusions were induced with 50% polyethylene glycol 8000 for 1 min 53 sec. Coverslips were placed back in media containing cycloheximide for 30 min and fixed in 3% paraformaldehyde. Cyclin D1 localization was visualized by immunofluorescence, as indicated above. In certain experiments, LMB was added to media at 50 μg/mL or 20 mM LiCl after co-culture with HeLa cells.
Cell transformation and tumorgenicity assays
Early and late passage NIH-3T3 cell lines were plated at 3 × 105 cells per 60-mm dish. After attachment, cells were cultured in media containing 5% FCS. Foci were visualized after 21 d with Wright Giemsa stain (Sigma). Anchorage-dependent growth of early and late passage NIH-3T3, D1-3T3, and D1-T286A-3T3 was determined by analyzing cellular growth in semisolid medium. Five thousand cells were placed in Iscove's media containing 0.65% noble agar containing 10% FCS. Cells were allowed to grow for 3 wk in 8% CO2 , and we analyzed formation of colonies containing >50 cells. Individual colonies were picked, expanded, and analyzed for expression of Flag-D1-T286A by Western analysis. For experiments examining tumor formation in SCID mice, cells were harvested by trypsinization and washed extensively in PBS. Ten million cells were then resuspended in 0.2 mL of PBS and injected subcutaneously into the right flank of 6-wk-old male SCID mice (Charles River).
Acknowledgments
We thank Charles J. Sherr (St. Jude Children's Research Hospital) for providing NIH-3T3 cells and cyclin D1 specific antibodies (total and phospho-specific); Martine F. Roussel for providing the Ras-transformed NIH-3T3 cells; and Charles Kuszynski for his expertise with flow cytometry. The authors gratefully acknowledge the excellent technical assistance of Chunying Yang, Ronald Rimerman, and Edward Arvisais. This work was supported in part by NCI Cancer Center support grant P30 CA36727 (Eppley Institute), American Cancer Society grant RPG-00-303-01 (to J.A.D.); National Institutes of Health (NIH) training grant T32 CA09476 (to J.R.A.); NIH grants DK44158 and CA76379 (to J.L.C.); NIH grant GM59213 (to M.H.); and the American Lebanese Syrian Associated Charities. J.A.D. is a 2000 V Scholar.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL adiehl@unmc.edu; FAX (402) 559-4651.
References
- Aktas H, Cai H, Cooper GM. Ras links growth factor signaling to the cell cycle machinery via regulation of cyclin D1 and the cdk inhibitor p27. Mol Cell Biol. 1997;17:3850–3857. doi: 10.1128/mcb.17.7.3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albanese C, Johnson J, Watanabe G, Eklund N, Vu D, Arnold A, Pestell RG. Transforming p21ras mutants and c-Ets-2 activate the cyclin D1 promoter through distinguishable regions. J Biol Chem. 1995;270:23589–23597. doi: 10.1074/jbc.270.40.23589. [DOI] [PubMed] [Google Scholar]
- Askjaer P, Jensen TH, Nilsson J, Englmeier L, Kjems J. The specificity of the CRM1-Rev nuclear export signal interaction is mediated by RanGTP. J Biol Chem. 1998;273:33414–33422. doi: 10.1074/jbc.273.50.33414. [DOI] [PubMed] [Google Scholar]
- Baldin V, Lukas J, Marcote MJ, Pagano M, Draetta G. Cyclin D1 is a nuclear protein required for cell cycle progressin in G1. Genes & Dev. 1993;7:812–821. doi: 10.1101/gad.7.5.812. [DOI] [PubMed] [Google Scholar]
- Bodrug SE, Warner BJ, Bath ML, Lindeman GJ, Harris AW, Adams JM. Cyclin D1 transgene impedes lymphocyte maturation and collaborates in lymphomagenesis with the myc gene. EMBO J. 1994;13:2124–2130. doi: 10.1002/j.1460-2075.1994.tb06488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C, Olivier P, Diehl JA, Fero M, Roussel MF, Roberts JM, Sherr CJ. The p21Cip1 and p27Kip1 CDK 'inhibitors' are essential activators of cyclin D–dependent kinases in murine fibroblasts. EMBO J. 1999;18:1571–1583. doi: 10.1093/emboj/18.6.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng M, Sexl V, Sherr CJ, Roussel MF. Assembly of cyclin D–dependent kinase and titration of p27Kip1 regulated by mitogen-activated protein kinase kinase (MEK1) Proc Natl Acad Sci. 1998;95:1091–1096. doi: 10.1073/pnas.95.3.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl JA, Sherr CJ. A dominant-negative cyclin D1 mutant prevents nuclear import of cyclin-dependent kinase 4 (CDK4) and its phosphorylation by CDK-activating kinase. Mol Cell Biol. 1997;17:7362–7374. doi: 10.1128/mcb.17.12.7362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl JA, Zindy F, Sherr CJ. Inhibition of cyclin D phosphorylation on threonine-286 prevents its rapid degradation via the ubiquitin-proteasome pathway. Genes & Dev. 1997;11:957–972. doi: 10.1101/gad.11.8.957. [DOI] [PubMed] [Google Scholar]
- Diehl JA, Cheng M, Roussel MF, Sherr CJ. Glycogen synthase kinase-3β regulates cyclin D1 proteolysis and subcellular localization. Genes & Dev. 1998;12:3499–3511. doi: 10.1101/gad.12.22.3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- Franke TF, Yang TO, Chan K, Datta A, Kazlauskas DK, Morrison DK, Kaplan DR, Tsichlis PN. The protein kinase encoded by the Akt proto-oncogene is a target of the PKGF-activated phosphatidylinositol 3-kinase. Cell. 1995;81:727–736. doi: 10.1016/0092-8674(95)90534-0. [DOI] [PubMed] [Google Scholar]
- Franke TF, Kaplan DR, Cantley LC, Toker A. Direct regulation of the Akt proto-oncogene product by phosphatidylinositol-3-4-bisphosphate. Science. 1997;275:665–668. doi: 10.1126/science.275.5300.665. [DOI] [PubMed] [Google Scholar]
- Fukada M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature. 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- Gorlich D. Transport into and out of the cell nucleus. EMBO J. 1998;17:2721–2727. doi: 10.1093/emboj/17.10.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall M, Peters G. Genetic alterations of cyclins, cyclin-dependent kinases, and cdk inhibitors in human cancer. Adv Cancer Res. 1996;68:67–108. doi: 10.1016/s0065-230x(08)60352-8. [DOI] [PubMed] [Google Scholar]
- Henderson BR. Nuclear-cytoplasmic shuttling of APC regulates β-catenin subcellular localization and turnover. Nat Cell Biol. 2000;2:653–660. doi: 10.1038/35023605. [DOI] [PubMed] [Google Scholar]
- Kaffman A, Rank NM, O'Neill EM, Huang LS, O'Shea EK. The receptro Msn5 exports the phosphorylated transcription factor Pho4 out of the nucleus. Nature. 1998;396:482–486. doi: 10.1038/24898. [DOI] [PubMed] [Google Scholar]
- Kauffmann-Zeh A, Rodrigues-Viciana P, Ulrich E, Gilbert C, Coffer P, Downward J, Evan G. Suppression of c-Myc-induced apoptosis by Ras signalling through PI(3)K and PKB. Nature. 1997;385:544–548. doi: 10.1038/385544a0. [DOI] [PubMed] [Google Scholar]
- Kerkhoff E, Rapp UR. Induction of cell proliferation in quiescent NIH 3T3 cells by oncogenic c-Raf-1. Mol Cell Biol. 1997;17:2576–2586. doi: 10.1128/mcb.17.5.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- Klinghoffer RA, Duckworth B, Valius M, Cantley L, Kazlauskas A. Platelet-derived growth factor-dependent activation of phosphatidylinositol 3-kinase is regulated by receptor binding of SH2-domain-containing proteins which influence ras activity. Mol Cell Biol. 1996;16:5909–5914. doi: 10.1128/mcb.16.10.5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krude T, Jackman MPJ, Laskey RA. Cyclin/cdk-dependent initiation of DNA replication in a human cell-free system. Cell. 1997;88:109–119. doi: 10.1016/s0092-8674(00)81863-2. [DOI] [PubMed] [Google Scholar]
- LaBaer J, Garrett MD, Stevenson LF, Slingerland JM, Sandhu C, Chou HS, Fattaey A, Harlow E. New functional activities for the p21 family of inhibitors. Genes & Dev. 1997;11:847–862. doi: 10.1101/gad.11.7.847. [DOI] [PubMed] [Google Scholar]
- Lavoie JN, L'Allemain G, Brunet A, Muller R, Pouyssegur J. Cyclin D1 expression is regulated positively by the p42/p44MAPK and negatively by the p38/HOGMAPK pathway. J Biol Chem. 1996;271:20608–20616. doi: 10.1074/jbc.271.34.20608. [DOI] [PubMed] [Google Scholar]
- Lees JA, Saito M, Vidal MVM, Look T, Harlow E, Dyson N, Helin K. The retinoblastoma protein binds to a family of E2F transcription factors. Mol Cell Biol. 1993;13:7813–7825. doi: 10.1128/mcb.13.12.7813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng X, Connell-Crowley L, Goodrich D, Harper JW. S-phase entry upon ectopic expression of G1 cyclin-dependent kinases in the absence of retinoblastoma protein phosphorylation. Curr Biol. 1997;9:709–712. doi: 10.1016/s0960-9822(06)00301-0. [DOI] [PubMed] [Google Scholar]
- Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- Lopez-Girona A, Furnari B, Modesert O, Russell P. Nuclear localization of Cdc25 is regulated by DNA damage and a 14-3-3 protein. Nature. 1999;397:172–175. doi: 10.1038/16488. [DOI] [PubMed] [Google Scholar]
- Lovec H, Grzeschiczek A, Kowalski M-B, Moroy T. Cyclin D1/bcl-1 cooperates with myc genes in the generation of B-cell lymphoma in transgenic mice. EMBO J. 1994;13:3487–3495. doi: 10.1002/j.1460-2075.1994.tb06655.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael WM, Choi M, Dreyfuss G. A nuclear export signal in hnRNP A1: A signal-mediated, temperature-dependent nuclear protein export pathway. Cell. 1995;83:415–422. doi: 10.1016/0092-8674(95)90119-1. [DOI] [PubMed] [Google Scholar]
- Neufeld KL, Nix DA, Bogerd H, Kang Y, Beckerle MC, Cullen BR, White RL. Adenomatous polyposis coli protein contains two nuclear export signals and shuttles between the nucleus and cytoplasm. Proc Natl Acad Sci. 2000;97:12085–12090. doi: 10.1073/pnas.220401797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg EA. Nucleocytoplasmic transport: Signals, mechanisms and regulation. Nature. 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- Nishi K, Yoshida M, Fujiwara D, Nishikawa M, Horinouchi S, Beppu T. Leptomycin B targets a regulatory cascade for crm1, a fission yeast nuclear protein, involved in control of higher order chromosome structure and gene expression. J Biol Chem. 1994;267:6320–6324. [PubMed] [Google Scholar]
- Powell SM, Zilz N, Beazer-Barclay Y, Bryan TM, Hamilton SR, Thibodeau SN, Vogelstein B, Kinzler KW. APC mutations occur early during colorectal tumorigenesis. Nature. 1992;359:235–237. doi: 10.1038/359235a0. [DOI] [PubMed] [Google Scholar]
- Quelle DE, Ashmun RA, Shurtleff SE, Kato JY, Bar-Sagi D, Roussel MF, Sherr CJ. Overexpression of mouse D-type cyclins accelerates G1 phase in rodent fibroblasts. Genes & Dev. 1993;7:1559–1571. doi: 10.1101/gad.7.8.1559. [DOI] [PubMed] [Google Scholar]
- Resnitzky D, Gossen M, Bujard H, Reed SI. Acceleration of the G1/S hase transition by expression of cyclins D1 and E with an inducible system. Mol Cell Biol. 1994;14:1669–1679. doi: 10.1128/mcb.14.3.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimerman RA, Gellert-Randleman A, Diehl JA. Wnt1 and MEK1 cooperate to promote cyclin D1 accumulation and cellular transformation. J Biol Chem. 2000;275:14736–14742. doi: 10.1074/jbc.m910241199. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Viciana P, Warne PH, Dhand R, Gout I, Fry MJ, Waterfield M, Downward J. Phosphatidylinositol-OH-kinase as a direct target of Ras. Nature. 1994;370:527–532. doi: 10.1038/370527a0. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Viciana P, Warne PH, Khwaja A, Marte BM, Pappin D, Das P, Waterfield MD, Ridley A, Downward J. Role of phosphoinisitide-3-OH kinase in cell transformation and control of the actin cytoskeleton by Ras. Cell. 1997;89:457–467. doi: 10.1016/s0092-8674(00)80226-3. [DOI] [PubMed] [Google Scholar]
- Rosen-Arbesfeld R, Townsley F, Bienz M. The APC tumour suppressor has a nuclear export function. Nature. 2000;406:1009–1012. doi: 10.1038/35023016. [DOI] [PubMed] [Google Scholar]
- Rustgi AK, Dyson N, Bernards R. Amino-terminal domains of c-myc and N-myc proteins mediate binding to the retinoblastoma gene product. Nature. 1991;352:541–544. doi: 10.1038/352541a0. [DOI] [PubMed] [Google Scholar]
- Sachdev S, Hoffmann A, Hannink M. Nuclear localization of IκBα is mediated by the second ankyrin repeat: The IκBα ankyrin repeats define a novel class of cis-acting nuclear import sequences. Mol Cell Biol. 1998;18:2524–2534. doi: 10.1128/mcb.18.5.2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr CJ. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- Sherr CJ, Roberts JM. CDK inhibitors: Positive and negative regulators of G1-phase progression. Genes & Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- Sofer-Levi Y, Resnitzky D. Apoptosis induced by ectopic expression of cyclin D1 but not cyclin E. Oncogene. 1996;13:2431–2437. [PubMed] [Google Scholar]
- Stade K, Ford CS, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- Stambolic V, Ruel L, Woodgett JR. Lithium inhibits glycogen synthase kinase-3 activity and mimics Wingless signalling in intact cells. Curr Biol. 1996;6:1664–1668. doi: 10.1016/s0960-9822(02)70790-2. [DOI] [PubMed] [Google Scholar]
- Sterner JM, Dew-Knight S, Musahl C, Kornbluth S, Horowitz JM. Negative regulation of DNA replication by the retinoblastoma protein is mediated by its association with MCM7. Mol Cell Biol. 1998;18:2747–2757. doi: 10.1128/mcb.18.5.2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers MD, Smith GE. Tex. Agric. Exp. St. Bull. Vol. 1555 1987. A manual of methods for baculovirus vectors and insect culture procedures. [Google Scholar]
- Todaro GJ, Green H. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J Cell Biol. 1963;17:299–313. doi: 10.1083/jcb.17.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomoda K, Kubota Y, Kato J. Degradation of the cyclin-dependent-kinase inhibitor p27Kip1 is instigated by Jab1. Nature. 1999;398:160–165. doi: 10.1038/18230. [DOI] [PubMed] [Google Scholar]
- Vanhaesebroeck B, Leevers SJ, Panayotou B, Waterfield M. Phosphoinositide 3-kinases: A conserved family of signal transducers. Trends Biochem Sci. 1997;22:267–272. doi: 10.1016/s0968-0004(97)01061-x. [DOI] [PubMed] [Google Scholar]
- Wang TC, Cardiff RD, Zukerberg L, Lees E, Arnold A, Schmidt EV. Mammary hyperplasia and carcinoma in MMTV-cyclin D1 transgenic mice. Nature. 1994;369:669–671. doi: 10.1038/369669a0. [DOI] [PubMed] [Google Scholar]
- Weber JD, Raben DM, Phillips PJ, Baldassare JJ. Sustained activation of extracellular-signal-regulated kinase 1 (ERK1) is required for the continued expression of cyclin D in G1 phase. Biochem J. 1997;326:61–68. doi: 10.1042/bj3260061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg RA. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- Winston JT, Coats SR, Wang Y-Z, Pledger WJ. Regulation of the cell cycle machinery by oncogenic ras. Oncogene. 1996;12:127–134. [PubMed] [Google Scholar]
- Yang J, Bardes ESG, Moore JD, Brennan J, Powers MA, Kornbluth S. Control of cyclin B1 localization through regulated binding of the nuclear export factor CRM1. Genes & Dev. 1998;12:2131–2143. doi: 10.1101/gad.12.14.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Weghorst CM, Weinstein IB. Overexpression of cyclin D1 enhances gene amplification. Cancer Res. 1996;56:36–39. [PubMed] [Google Scholar]