ERK7 is a negative regulator of protein secretion in response to amino-acid starvation by modulating Sec16 membrane association (original) (raw)
Abstract
RNAi screening for kinases regulating the functional organization of the early secretory pathway in Drosophila S2 cells has identified the atypical Mitotic-Associated Protein Kinase (MAPK) Extracellularly regulated kinase 7 (ERK7) as a new modulator. We found that ERK7 negatively regulates secretion in response to serum and amino-acid starvation, in both Drosophila and human cells. Under these conditions, ERK7 turnover through the proteasome is inhibited, and the resulting higher levels of this kinase lead to a modification in a site within the C-terminus of Sec16, a key ER exit site component. This post-translational modification elicits the cytoplasmic dispersion of Sec16 and the consequent disassembly of the ER exit sites, which in turn results in protein secretion inhibition. We found that ER exit site disassembly upon starvation is TOR complex 1 (TORC1) independent, showing that under nutrient stress conditions, cell growth is not only inhibited at the transcriptional and translational levels, but also independently at the level of secretion by inhibiting the membrane flow through the early secretory pathway. These results reveal the existence of new signalling circuits participating in the complex regulation of cell growth.
Keywords: ER exit sites, kinases, nutrient, RNAi screen, S2 cells
Introduction
Secretion takes place through the membrane of the secretory pathway that comprises the rough ER, ER exit sites (ERES or tER sites), where newly synthesized proteins are packaged into budding COPII vesicles, ER-Golgi intermediate compartment, Golgi apparatus and post-Golgi carriers. In Drosophila, tER sites are closely associated with individual pairs of Golgi stacks forming what we, and others have called tER-Golgi units that represent the early secretory pathway (Kondylis and Rabouille, 2009). One key protein required for tER site organization and COPII vesicle budding is the large hydrophilic protein Sec16 that localizes to the ER cup overlaying the clusters of COPII vesicles. Upon its functional disruption by depletion or mutation, tER site biogenesis is impaired and secretion is drastically inhibited (Connerly et al, 2005; Watson et al, 2005; Bhattacharyya and Glick, 2007; Ivan et al, 2008; Hughes et al, 2009).
Despite the identification of many components underlying the functional organization of the secretory pathway (Bonifacino and Glick, 2004; Spang, 2009), two recent genome-wide RNAi screens (Bard et al, 2006; Wendler et al, 2010) have led to the discovery of novel proteins required for constitutive secretion, including Tango1 (Bard et al, 2006; Saito et al, 2009), as well as Grysum and Kish (Wendler et al, 2010). However, how secretion is regulated qualitatively and quantitatively in response to changes imposed by cell growth, nutrient availability, stress and differentiation is not completely understood; In particular, the molecular mechanisms through which exogenous stimuli are sensed and relayed to the secretory machinery remains largely unknown.
The relationship between signalling and secretion has only recently started to emerge. Kinases have been recently demonstrated to reside on membrane compartments of the early secretory pathway and activate signalling cascades that modify its functional organization (for reviews, see Quatela and Philips, 2006; Omerovic and Prior, 2009; Sallese et al, 2009; Farhan and Rabouille, 2011). For instance, the budding of COPII-coated vesicles is blocked by the kinase inhibitor H89 (Aridor and Balch, 2000), ER export is inhibited by the phosphatase inhibitor okadaic acid (Pryde et al, 1998) and Akt has recently been shown to phosphorylate Sec24 (Sharpe et al, 2010). Furthermore, a siRNA screen depleting 916 human kinases and phosphatases was performed to uncover regulators of the secretory pathway. The Mitotic-Associated Protein Kinase (MAPK) Extracellularly regulated kinase (ERK) 2, which is activated by epidermal growth factor (EGF) through Ras, was shown to directly phosphorylate Sec16 on Threonine 415. This phosphorylation event led to an increase in the ERES number and secretion (Farhan et al, 2010). This reinforced the notion that the early secretory pathway is regulated by environmental conditions and that components of the secretory pathway are direct targets of signalling. Interestingly, the secretory pathway also responds to intracellular stimuli, such as increased cargo load (Guo and Linstedt, 2006; Farhan et al, 2008; Pulvirenti et al, 2008).
In order to define conserved kinases regulating the functional organization of the early secretory pathway and identify new ones, we performed a microscopy-based primary RNAi screen in Drosophila S2 cells (Kondylis et al, 2011), which show a high depletion efficiency by RNAi. Furthermore, in comparison with mammalian genomes, the Drosophila genome has less genetic redundancy, facilitating the identification of candidates that might have been missed in human cells.
Results
Primary screen and candidate validation
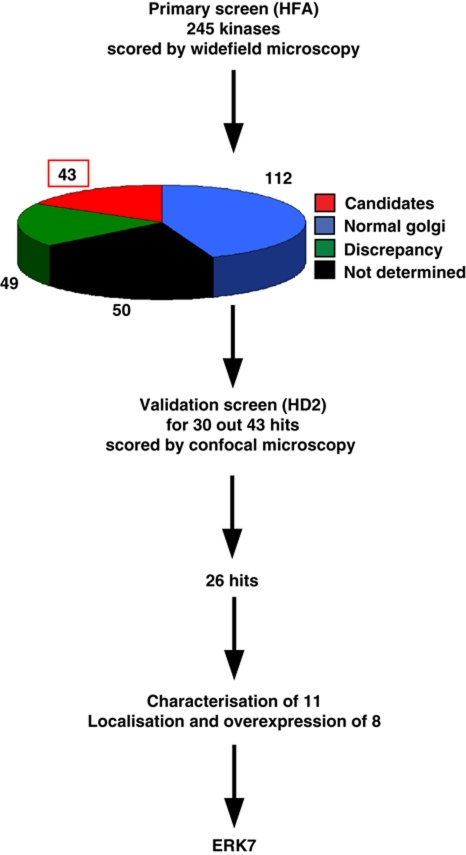
In the primary screen, we depleted 245 kinases in duplicate (Boutros et al, 2004), in addition to the positive and negative controls (depletion of apoptotic inhibitor DIAP1, Sec16, Abi and Scar) and we scored for the organization of the early secretory pathway marked by Golgi protein Fringe-GFP (as described in Kondylis et al, 2011 and Supplementary data; Figure 1). Depletion of 43 kinases significantly altered the Golgi organization in one or both plates, and the most common phenotype observed was an increase in the number of Fringe-GFP fluorescent spots (Supplementary Table S1). Depletion of 49 proteins exhibited a phenotype discrepancy between the two plates tested, while no data were obtained for another 50 proteins. These kinases together with those whose depletion did not seemingly affect the Golgi organization were not further examined (see Supplementary Materials and Methods).
Figure 1.
Overview of the microscopy-based RNAi kinase screen. The primary screen was performed in 384-well plates (in duplicate) using Fringe-GFP S2 cells and dsRNAs transcribed from the HFA library targeting 254 genes (245 kinases). The cells were immunolabelled with anti-GFP and anti α-tubulin antibodies as well as Hoechst and were viewed by widefield microscopy. Forty-three candidates (scored in the two plates or only in one) were identified, whereas 112 depletions did not lead to a Golgi phenotype. Fifty depletions led to an unclear phenotype because of a phenotypic discrepancy between the two plates examined (a Golgi phenotype was observed ‘in one plate only’). The phenotype of 49 depletions was ‘not determined’ because the data were not recorded properly (out of focus or lack of cells). The validation screen was performed using different dsRNAs transcribed from a second generation RNAi library (HD2) to target 30 out of 43 candidates. It was performed in Fringe-GFP S2 cells seeded in 24-well plates that were immunolabelled with Sec16/PDI/Dapi and viewed by confocal microscopy. In all, 26 out of 30 candidates were validated. The depletion phenotypes of 11 candidates were characterized (using a third set of independent dsRNAs) and 8 were cloned, localized and overexpressed leading to the identification of ERK7.
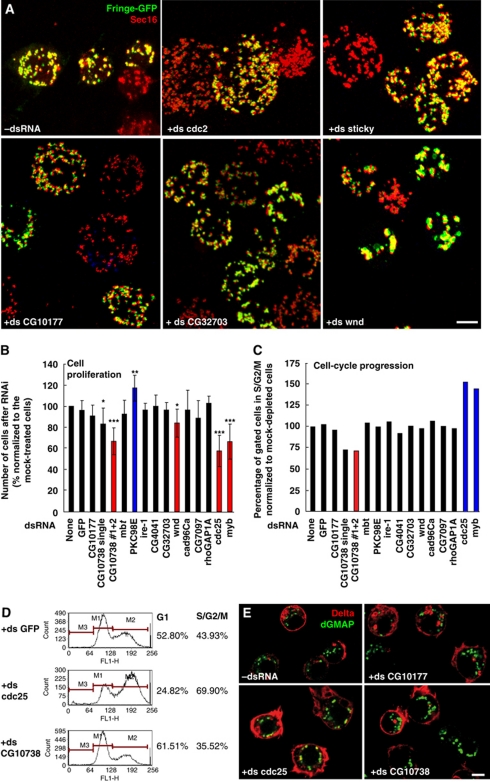
Out of these 43 candidate regulators of the early secretory pathway organization, we selected 30 that were validated using a different dsRNA after analysis by confocal microscopy (Supplementary Table S1). In all, 26 out of the 30 genes tested were confirmed, illustrating the robustness of our approach (Supplementary Tables S1 and S2). In line with the primary screen, most of the depletions increased the number of tER-Golgi units in a significant percentage of cells (Figure 2A), a phenotype thereafter referred to as MG for ‘more and smaller Golgi spots’ (Table I). Depending on the penetrance of the phenotype, the candidates were grouped as very strong (MG++++; cdc2), strong (MG+++; CG10738 and CG10177), moderate (MG++; CG32703) and weak (MG+) (Figure 2A; Supplementary Table S2). A decrease in the number of Fringe-GFP dots was more rarely observed (LS for ‘less Golgi spots’), likely due to aggregation of tER-Golgi units (Supplementary Tables S1 and S2). This was the case for Wallenda (Wnd, Figure 2A) and CG4041 (Table I). The RNAi depletion efficiency in the secondary screen was tested for four hits using cells transfected with V5-tagged versions of these analysed proteins (see below), and was found to be very efficient (Supplementary Figure S1A).
Figure 2.
Examples of different phenotypic groups from the confirmation/validation screen. (A) Visualization of tER-Golgi units (Sec16 and Fringe-GFP, respectively) upon different RNAi depletions by confocal microscopy. Typical pattern of tER-Golgi units in mock-treated cells (−dsRNA). The very strong (+ds cdc2), strong (+ds sticky; +ds CG10177) and moderate (+ ds CG32703) MG phenotype (more and smaller Golgi spots) are presented as well as the LS phenotype (less spots, +ds wallenda/wnd). The pictures represent 2D projections of confocal sections. Scale bar: 5 μm. (B) The number of S2 cells after a 5-day incubation with the indicated dsRNAs expressed as percentage relative to the number of mock-treated cells. Red and blue columns indicate genes whose depletion led to a significant decrease or increase in cell proliferation, respectively. Error bars represent s.d. from at least three independent experiments. Conditions with P<0.01, 0.01<P<0.05 and 0.05<P<0.10 are indicated with triple, double and single asterisks, respectively. (C, D) Cell-cycle distribution of live S2 cells after 5 days incubation with the indicated dsRNAs determined by staining their DNA content. The population of G1 (M1), S/G2/M (M2) or sub-G1 (M3) cells in each condition was quantified by FACS analysis. Percentage of gated cells in S/G2/M phase (4N) (normalized to the respective value of mock-treated cells, which was considered as 100%) of one representative experiment (C). Red and blue columns indicate genes whose depletion leads to a significant decrease or increase in the percentage of cells in S/G2/M phase, respectively. cdc25 and myb depletions (_n_=3) lead to an average of 152.61%±7.85 (_P_-value of 0.010) and 144.62%±12.41 (_P_-value of 0.036), respectively. For CG10738 (#1 and #2) depletion (_n_=3), the average is 71.60%±7.12 with a _P_-value of 0.014. Representative examples are shown in (D). Note the increase in G1 population upon depletion of CG10738 kinase. (E) Efficiency of anterograde transport of Delta S2 cells incubated for 5 days with the indicated dsRNAs, followed by 1-h induction of Delta with CuSO4 and 75 min chase to allow its transport to plasma membrane. Fixed cells were labelled for Delta and dGMAP (cis-Golgi marker). Scale bars: 5 μm.
Table 1. Phenotype characterisation.
| Quantification of number of Golgi spots versus cell volume | |||
|---|---|---|---|
| Average number of Fringe-GFP spotsa | Cell volume (10 μm3) | Number of Fringe-GFP spotsa/10 μm3 | |
| −ds RNA | 20.2±5.9 | 492±174 | 0.43±0.15 |
| +ds CG10117 (MG+++) | 37.3±14.6 | 1159±643 | 0.36±0.08 |
| +ds CG32703 (MG++) | 30.0±10.6 | 593±282 | 0.58±0.26 |
| +ds wnd (LS) | 17±8.8 | 490±210 | 0.40±0.19 |
In addition to the MG phenotype, the depletion of several proteins resulted in increased cell size (larger cell diameter, Table I; as described in Kondylis et al, 2011). Cdc2 depletion gave the strongest phenotype (Figure 2A), as expected considering its role in G2/M transition (de Vries et al, 2005). This phenotype was similar to that of Cdc25/string phosphatase depletion that blocks S2 cells in G2 phase and leads to a doubling in tER-Golgi number (Kondylis et al, 2007). The depletion of three other transcripts encoding the actin and cytokinesis regulators Sticky/Citron (Figure 2A), Rok/Rho kinase and Strn-Mlck that contains a myosin-light chain kinase domain (Supplementary Figure S1B; Echard et al, 2004; Eggert et al, 2004) also led to a MG+++ phenotype with a larger cell diameter and/or abnormal DNA staining indicative of cytokinesis defects (Supplementary Table S1). This is to be anticipated for cells that fail to divide their cytoplasm after having duplicated the Golgi stacks in G2, fragmented them in prophase/metaphase and reassembled them in telophase (Kondylis et al, 2007; Rabouille and Kondylis, 2007).
Characterization of candidates: cell cycle and anterograde transport
As mentioned above, the number of tER-Golgi units increases in G2 phase in Drosophila S2 cells (Kondylis et al, 2007). To assess whether the MG phenotype was due to a G2 arrest or a block in cytokinesis, we examined cell proliferation (Figure 2B) and cell-cycle distribution (Figure 2C and D) of 11 hits using a third dsRNA targeting the respective transcripts (Supplementary Tables S2 and S3). Cell proliferation was not significantly affected for most hits, with the exception of PKC98E that led to a small but significant increase in cell proliferation, and CG10738 kinase, the homologue of human single membrane-spanning atrial natriuretic peptide receptor B involved in cardiovascular and kidney homeostasis, lipid metabolism, cell proliferation and apoptosis (Martel et al, 2010). CG10738 depletion reduces cell proliferation to a similar extent as Cdc25, and Myb RNAi (Table I; Figure 2B).
Cell-cycle distribution was also assessed in depleted living cells. As previously described (Edgar and O’Farrell, 1990; Katzen et al, 1998), Cdc25- and Myb-depleted cells reduce cell proliferation by arresting the cells in G2 (Figure 2C and D). However, cell-cycle profiles for most protein depletions were similar to non- or mock-depleted cells, in line with the cell proliferation results. The cell growth inhibition observed upon CG10738 depletion appears to be due to a G1 arrest/delay, as suggested by the increase in G1 cell population (Figure 2C and D).
Last, to test whether inhibition of anterograde transport causes the disorganization of the early secretory pathway (Kondylis et al, 2011), we monitored the deposition of the transmembrane protein reporter Delta to the plasma membrane (Kondylis and Rabouille, 2003; Kondylis et al, 2005, 2007). In most cases, however, anterograde transport was unaffected (Table I; Figure 2E). Interestingly, Cdc25-depleted cells sometimes exhibited a higher amount of Delta at the plasma membrane (Figure 2E), suggesting that increased number of tER-Golgi units may perhaps support a cellular need for higher rate of transport in G2 phase.
Overall, the disorganization of the early secretory pathway observed upon kinase depletions did not correlate with changes in secretory capacity, as we have shown for the depletion of ER proteins (Kondylis et al, 2011). Furthermore, as mentioned above, in some cases, the depletions were accompanied by an increase in cell size that did not correspond to an increase in lipid droplet or activation of the TOR complex 1 (TORC1) pathway (Table I and not shown). Of note, the ratio of Golgi number per volume of cytoplasm was relatively constant (Table I), suggesting that each tER-Golgi unit is probably associated with a defined volume of cytoplasm (Kondylis et al, 2011).
ERK7 regulates Sec16 association with tER sites
The identified kinases affecting tER-Golgi unit organization could either be directly localized to these compartments, or modify substrates that reside in this pathway independently of their own localization. To test this, V5-tagged versions of 8 kinases were expressed (those highlighted in bold in Table I, Supplementary Table S4) to assess their localization and the effect of their overexpression on the secretory pathway (see detailed description for seven of them, including Wallenda, in Supplementary Table S5 and Supplementary Figures S2–S4).
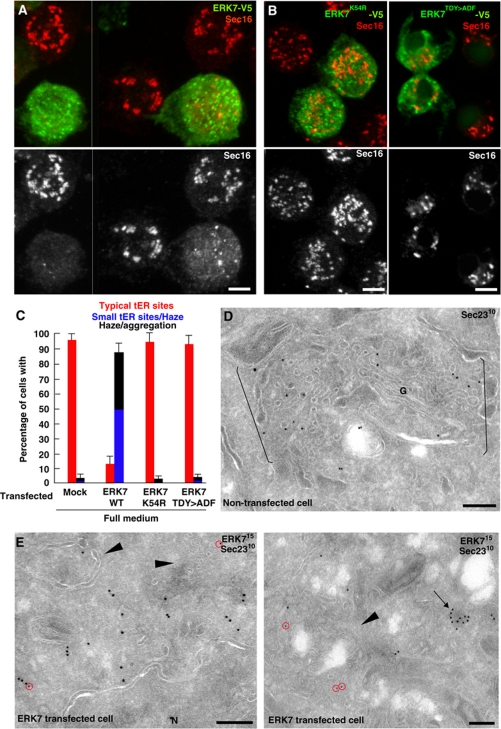
We focused on CG32703, a moderate MG hit that has significant homology to mammalian ERK7, one of the atypical MAP kinases, also called MAPK15 or ERK8 (Abe et al, 1999, 2001, 2002; Coulombe and Meloche, 2007). CG32703 has two isoforms, one of 916 amino-acid long (isoform A) and one of 451 (isoform B) (Supplementary Figure S5A). Both isoforms share a common N-terminus (aa 1–355) that contains the kinase domain and is conserved (46% identity and 68% similarity by EMBOSS Pairwise analysis; http://www.ebi.ac.uk/Tools/emboss/align/) when compared with the equivalent region of rat ERK7 and human ERK8/MAPK15 (aa 1–340). Lysine 43 at the catalytic site of rat MAPK15 corresponds to lysine 54 in Drosophila CG32703 and the kinase activation loop resides at the position 190–192 (TDY motif) whereas in the rat homologue it is located at position 175–177 (TEY motif). The C-terminal part of Drosophila ERK7 shows significantly lower similarity as compared with rat homologue, although the latter was reported to be required for full kinase activation in rat ERK7 (Abe et al, 1999). In the isoform A, this C-terminal part is also much longer (561 versus 224 amino acids in rat ERK7) and does not seem to contain the nuclear localization signal described for its mammalian homologue (Abe et al, 1999). The dsRNA used for depletion targeted both isoforms and the long form of ERK7 (referred to thereafter as ERK7 in agreement with Flybase) was used for localization and expression in S2 cells.
ERK7 was found to be predominantly cytoplasmic, sometimes in small aggregates (arrow in Figure 3E), whether tagged at the C- (Figure 3A) or N-terminus (not shown). It was also found associated in a small extent with the nuclear envelope and ER membrane (not shown). Strikingly, when strongly overexpressed, tER sites were of smaller size and Sec16 was found significantly dispersed (Figure 3A and C). When analysed by immunoelectron microscopy, Sec23 was also dispersed and the tER-Golgi units appeared disorganized (compare Figure 3D with E). Profiles similar to those observed in Sec16, Sar1 and Sec23 depletions were observed in ERK7 transfected cells (arrowheads in Figure 3E), strengthening the notion of a tER site disorganization. This effect is specific because out of all the overexpressed kinases tested ERK7 is the only one that dispersed Sec16. Furthermore, Sec16 dispersion is due to ERK7 kinase activity, because overexpression of two kinase-dead variants (K54R and T190A/Y192F; Abe et al, 1999) did not lead to the disassembly of tER sites (Figure 3B and C).
Figure 3.
ERK7 overexpression induces Sec16 dispersion and disassembly of tER sites. (A) IF localization of ERK7-V5 (green) and effect of its overexpression on Sec16 (red). Note that ERK7 is largely cytosolic but that its expression leads to Sec16 dispersion. (B) Overexpression of kinase-dead ERK7-V5K54R and ERK7-V5TDY>ADF (green) does not lead to Sec16 dispersion (red). 2D projections of confocal sections are presented in (A) and the first panel in (B). (C) Quantification of Sec16 dispersion upon expression of WT and K54R ERK7-V5. (D, E) Localization of Sec23 by Immunoelectron microscopy (IEM) in untransfected cells (D) and ERK7-V5 expressing cells (E). Note that the tER-Golgi units in (D) (between brackets) are largely absent in (E) (arrowheads), Sec23 (red circles) is dispersed and largely absent from the remnants of tER-Golgi units, and ERK7 is sometimes present in small aggregates (arrows). Scale bars: 5 μm (A, B); 200 nm (D, E).
Serum and amino-acid starvation induce tER site disassembly
In contrast to well-studied MAPK members, such as ERK1/2, p38s and JNKs that are activated by growth factors and MAPKKs, the regulation of ERK7 activity and its physiological functions is much less explored. In particular, rat ERK7 seems to be autoactivated (Abe et al, 1999, 2001; Klevernic et al, 2006). Furthermore, rat ERK7 has relatively high, constitutive kinase activity, which is not further stimulated by the addition of serum or EGF and not inhibited by classical MEK inhibitors, such as U0126, PD98059 and YOPJ (Abe et al, 2001). A similar behaviour to growth factor signalling has also been reported for the human homologue of rat ERK7 (hERK8) in HEK-293 cells (Klevernic et al, 2006; Erster et al, 2010), although this does not seem to be the case in COS cells (Abe et al, 2002).
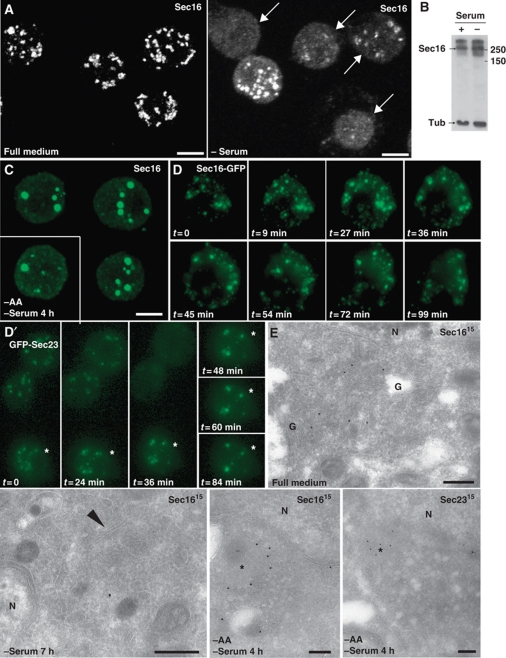
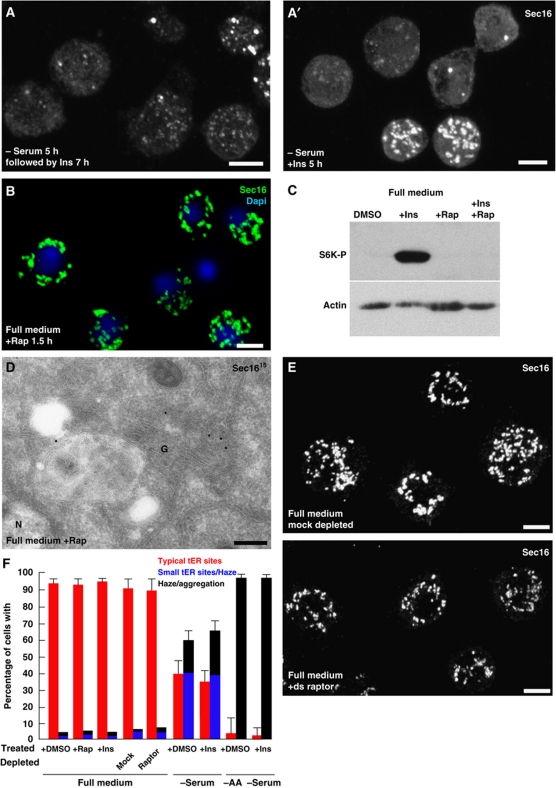
To investigate whether serum components influence the effect of ERK7 on tER sites in S2 cells, ERK7 was overexpressed in cells that were serum starved, resulting in a Sec16 dispersion as strong as in fed cells (Figure 6C). However, we noticed that serum starvation alone affected the tER site organization and Sec16 distribution. Strikingly, removing the serum from the culture medium for 5–7 h resulted in the large displacement of Sec16 from tER sites (Figure 4A), but not its degradation (Figure 4B), resulting in the disorganization of the tER-Golgi unit (Figure 4E, arrowhead), phenocopying ERK7 overexpression. In agreement with tER site disassembly and loss of Sec16 (Ivan et al, 2008), serum deprivation was accompanied by an inhibition in secretion as the delivery of the plasma membrane protein Delta was strongly impaired (Supplementary Figure S6A). The tER site disassembly (as well as the Golgi, not shown) was reversible, albeit quite slowly after re-addition of serum for 5–9 h (Supplementary Figure S6B).
Figure 4.
Serum and amino-acid starvation induces tER site disassembly. (A) IF localization of Sec16 in WT S2 cells grown in full medium and in serum-free medium for 7 h. Arrows indicate cells in which Sec16 pattern is significantly affected. Effects are quantified in Figure 6C using similar criteria as in Figure 3C. (B) Western blot of homogenates of S2 cells cultured in full medium (+ serum) and in the absence of serum (−) using anti-Sec16 and anti α-tubulin (Tub) antibodies. Note that the level of Sec16 remains similar after starvation. (C) IF localization of Sec16 in WT S2 cells starved of amino acids and serum. Note the haze and the large aggregates that are found in almost all of the cells. Effects are quantified in Figure 6C. (D, D′) Time lapse of tER sites disassembly using GFP-Sec23 and Sec16-GFP transfected S2 cells incubated in the absence of amino acids at _t_=0. Projections of the cells were made every 9–12 min. Note that in cells where expression is low (two upper cells in D′), the GFP punctuates disappear in 30–40 min of starvation and are replaced by a haze. In cells expressing higher level (D, D′, asterisks), the tER sites also disappear but are replaced by aggregates, a situation that recapitulates what is observed with endogenous Sec16. (E) Localization of endogenous Sec16 and Sec23 by IEM in S2 cells incubated in full medium, in the absence of serum and in the absence of amino acids and serum. The arrowhead indicates the remnant of Golgi that can be found in few serum starved cells whereas most of them exhibit no identifiable tER-Golgi units. In amino-acid starved cells, the asterisks indicate the Sec16 and Sec23 membrane-free aggregates that often associated with a cloud of small electron luscent vesicles. Note that this is very different from the tER-Golgi unit morphology observed in S2 cells grown in full medium. Scale bars: 5 μm (A–C) and 200 nm (E).
To test whether the tER site disassembly was also observed during amino-acid starvation, we incubated the cells in amino acid-free medium for 2–6 h. Amino-acid starvation resulted in the rapid and efficient dispersion of Sec16 (Figure 4C and D) and Sec23 (Figure 4D′), accompanied by their aggregation (Figure 4E, asterisks), leading to the disassembly of tER sites (Figure 4E). This pattern was similar to, but stronger than the effects observed upon serum starvation and ERK7 overexpression (Figures 3A and 4A). The Sec16 dispersion/aggregation triggered by amino-acid starvation was also quickly reversible (2 h, Supplementary Figure S6C). Because the presence of serum did not change the cell's response to amino-acid starvation with respect to tER site disorganization, this suggests that serum starvation might inactivate amino-acid transporters leading to a mild amino-acid starvation.
tER site disassembly by serum and amino-acid starvation is independent of TORC1
Amino-acid starvation is typically known to inhibit the activity of TORC1, which constitutes one of the major pathways involved in growth factor and nutrient regulated cell growth (see review Schmelzle and Hall, 2000; Sengupta et al, 2010). TORC1 activation stimulates protein synthesis, glucose uptake and glycolysis, lipid and sterol biosynthesis (Duvel et al, 2010). Consequently, TORC1 inhibition leads to cessation of cell growth as well as the stimulation of degradative processes, such as autophagy (Noda and Ohsumi, 1998; Cutler et al, 1999).
Secretion is a major factor also contributing to cell growth and we tested whether cessation of secretion through tER sites disassembly under serum and amino-acid starvation conditions is also mediated by TORC1. To do so, S2 cells were incubated in serum-free medium supplemented with insulin, a strong activator of TORC1 (as shown by the phosphorylation of S6K, a substrate of TORC1, Figure 5C), but this did not prevent the Sec16 dispersion observed upon starvation (Figure 5A and F). Insulin was also unable to revert the serum (Figure 5A′) and amino-acid starvation phenotype (Figure 5F).
Figure 5.
Amino-acid starvation is mediated by a TORC1-independent pathway. (A, A′) IF visualization of Sec16 in cells incubated in the absence of serum followed by insulin or simultaneous serum withdrawal and insulin addition (A′). Note that Insulin neither rescues nor protects from serum starvation. (B) IF visualization of Sec16 (green) in S2 cells grown full medium supplemented by rapamycin (Rap) for 1.5 h. Note that Sec16 localization is unaffected by drug treatment. (C) Western blot of Phospho-S6K on lysates of S2 cells incubated in DMSO, rapamycin, insulin, insulin+rapamycin showing the effectiveness of the drugs (see also Kondylis et al, 2011). (D) IEM localization of Sec16 in S2 cells treated with rapamycin for 2 h. Note that the structure of the early secretory pathway is intact and Sec16 localizes as in non-treated cells (compared with Figure 4E). (E) IF visualization of Sec16 in mock or raptor depleted S2 cells for 5 days. Note that raptor depletion does not affect Sec16 localization. (F) Quantitation of the different treatments on S2 cells with respect to Sec16 phenotypes. Error bars represent s.d. from five independent experiments (+DMSO, +Rap, + Ins in full serum and in − serum) and three independent experiments for the rest of the conditions. Scale bars: 5 μm.
To test TORC1 involvement further, cells in full medium were incubated in the presence of rapamycin, a specific TORC1 inhibitor (Loewith et al, 2002). In S2 cells, rapamycin completely inhibits the insulin-induced S6K phosphorylation (Figure 5C). This, however, did not induce the dispersion of Sec16 whose pattern was undistinguishable from non-treated fed cells (Figure 5B, D and F), strengthening the notion that TORC1 inhibition is not what mediates tER site disassembly upon starvation. However, since rapamycin can, in some cases, inhibit TORC1 activity only towards a subset of substrates (Thoreen et al, 2009), we also depleted fed cells of Raptor by RNAi. Raptor is a subunit of the TORC1 complex critical for its activity (Hara et al, 2002; Kim et al, 2002). Although the cell number after 5 days of depletion was markedly decreased (about 40%), the tER site organization was unaffected (Figure 5E and F). Starvation effect on Sec16 was also identical in the Raptor depleted as in non-depleted cells (not shown). Altogether, these data strongly indicates that TORC1 is not involved in the tER site disassembly observed upon starvation.
ERK7 mediates tER site disassembly upon starvation
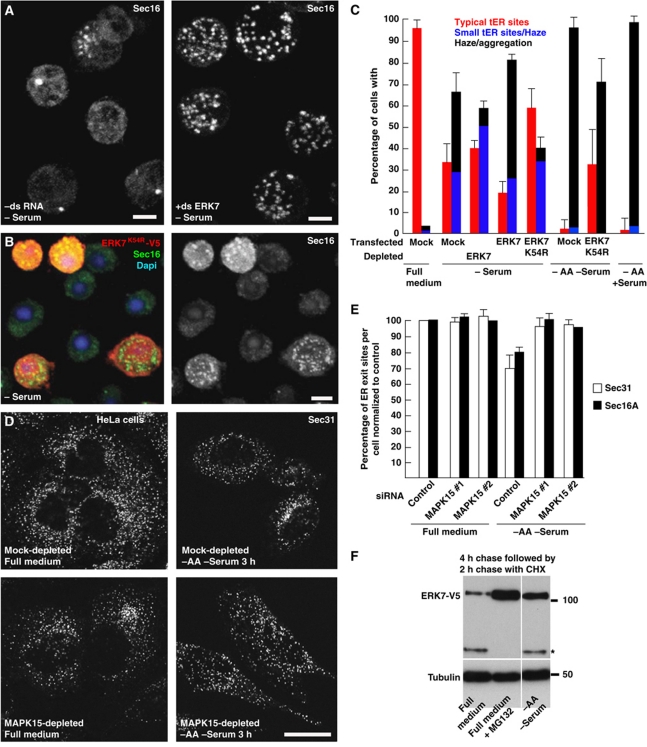
The phenotypic similarity between ERK7 overexpression, serum deprivation and amino-acid starvation with respect to tER site disassembly prompted us to test whether ERK7 was required for the cellular response to serum and amino-acid starvation. To this end, S2 cells were serum deprived after either ERK7 depletion by RNAi (Figure 6A), or overexpression of a kinase-dead version of ERK7 (Figure 6B). In both cases, the serum deprivation phenotype was partially prevented (Figure 6C). It should be noted that ERK7 depletion prevented Sec16 dispersion but the cells exhibited the MG phenotype reported above (see Figure 2 and Table I). Conversely, WT ERK7 expression in serum-starved cells accentuated Sec16 dispersion (Figure 6C). Amino-acid starvation was also partially rescued by ERK7 kinase-dead overexpression (Figure 6C). These results show that ERK7 is involved in the signalling pathway sensing/relaying the amino-acid starvation, which leads to tER disassembly and inhibition of secretion.
Figure 6.
The disassembly of ER exit sites is rescued by loss of ERK7. (A) IF localization of Sec16 in mock-treated (−dsRNA) or ERK7-depleted S2 cells (+ds ERK7) in the absence of serum. (B) IF localization of Sec16 (green) in S2 cells overexpressing ERK7K54R-V5 in the absence of serum. Transfected cells are marked by the V5 labelling (red) and the nuclei are stained with DAPI. (C) Quantitation of Sec16 dispersion upon serum starvation in non-transfected, WT ERK7-V5 and ERK7K54R-V5 transfected cells and in cells depleted of ERK7, and upon amino-acid starvation in WT cells and cells expressing ERK7K54R-V5. Note that removing ERK7 activity partially rescues the loss of ER exit sites upon starvation. (D) IF visualization of the ER exit site marker Sec31 upon serum and amino-acid starvation in mock- and MAKP15-depleted HeLa cells. (E) Quantitation of the number of ER exit sites in the conditions indicated in (D) using Sec31 and Sec16 as ER exit sites markers. Note that removing MAPK15 totally rescues the loss of ER exit sites upon starvation. (F) Western blot of lysates of S2 cells overexpressing ERK7-V5 in full medium for 4 h followed by a 2-h chase in the presence of cycloheximide in full medium, in full medium supplemented with the proteasome inhibitor MG132, and in Ringer/glucose (−AA −serum). Note that ERK7-V5 (upper band) is severely reduced in cells incubated in full medium when compared with starved cells. The band marked by an asterisk is recognized by the V5 antibody and corresponds to a ERK7-V5 truncation occurring normally in cells. Error bars in (C) and (E) represent SD from three and five independent experiments, respectively. Scale bars: 5 μm (A, B) and 25 μm (D).
We then tested whether amino-acid starvation has the same effect on ER exit sites of mammalian (HeLa) cells and whether this was also mediated by ERK7. In the absence of amino acid and serum for 3 h, the number of ER exit sites per cell was consistently reduced by 20–30% and this was almost completely rescued by depleting the ERK7 human homologue MAPK15 (Figure 6D and E), underlying the conservation of the cellular response mechanism to serum and amino-acid starvation via this atypical MAP kinase.
These results show that cessation of secretion upon amino-acid starvation is an active process, requiring, at least in part, ERK7. In this respect, a recent study has identified human ERK2 as a regulator of tER sites biogenesis in response to EGF via Ras signalling (Farhan et al, 2010). ERK2 directly phosphorylates human Sec16 on Threonine 415 that is situated in the NC1 domain (Supplementary Figure S5B), that in turn increases its recruitment to ER exit sites. Conversely, removing growth factors (or depleting ERK2) decreases Sec16 recruitment to ER exit sites and reduces anterograde transport (Farhan et al, 2010; Supplementary Figure S8). Therefore, in mammalian cells, serum starvation decreases secretory capacity via ERK2 inhibition, whereas amino-acid starvation inhibits secretion via ERK7 activation both leading to ER exit sites functional disorganization.
We, therefore, tested whether tER site organization in Drosophila is also regulated by the classical MAPK/ERK. As mentioned above, the ERK2 site is situated in NC1 of the human Sec16 (Supplementary Figure S5B) and NetPhos predicted putative ERK sites in the equivalent domain of Drosophila Sec16. To test whether NC1 has a role in the cell's response to starvation, we truncated it in the Drosophila Sec16, and showed that this truncation behaved as endogenous Sec16 upon serum starvation, suggesting that this domain is not involved in the serum starvation response (Supplementary Figure S7A). To test further whether ERK1/2 signalling is involved, S2 cells were incubated in full medium supplemented by the MEK inhibitors PD98059 and UO126 (at concentrations known to inhibit ERK1/2 phosphorylation, see Materials and methods; Supplementary Figure S7B), but this did not lead to Sec16 dispersion, suggesting that the classical ERK pathway is not involved, at least in a measurable level, using our approach. In S2 cells, therefore, tER site organization does not seem regulated by classical ERKs whereas it does involve ERK7.
ERK7 is stabilized by amino-acid starvation
We next investigated what are the effects of serum and amino-acid starvation on ERK7. In mammalian cells, ERK7 has been shown to be unstable and degraded extremely quickly through the proteasome after its ubiquitination by E3 ligase Cullin (Kuo et al, 2004). We, therefore, hypothesized that amino-acid starvation stabilizes ERK7. To test this, we monitored the stability of ERK7-V5 in cells incubated in the presence and absence of amino acids. We found that ERK7-V5 was rapidly degraded in cells grown in full medium in a proteasome-dependent manner, as addition of the proteasome inhibitor MG132 to the medium, stabilized ERK7-V5. Incubation of cells in the absence of amino acid stabilized ERK7-V5 to a similar level as incubation with MG132 (Figure 6F). This suggests that endogenous ERK7 stabilization by amino-acid starvation could be one of the mechanisms leading to Sec16 dispersion and tER site disassembly.
The ‘serum/amino-acid starvation responsive domain’ of Sec16 is in its C-terminus
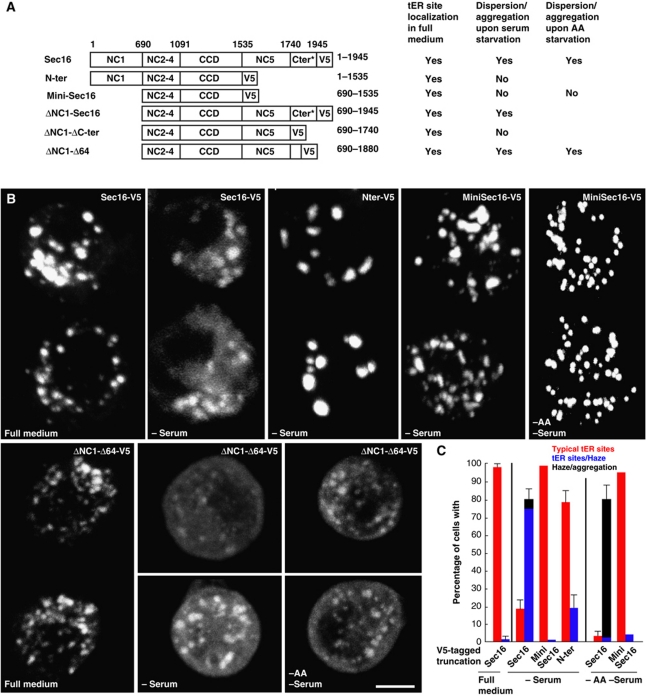
The starvation-induced Sec16 release from tER sites is likely due to post-translational modifications, for instance phosphorylation, either on Sec16 or on its ER receptor (yet to be identified). To test this, we expressed the region of Sec16 (V5-tagged) that is necessary and sufficient to localize to tER sites (mini-Sec16, aa 690–1535 that we have called NC2.3.4-CCD in Ivan et al, 2008) and examined its behaviour upon serum and amino-acid starvation. As we have previously shown mini-Sec16 localized to tER sites but this localization was not affected by starvation (Figure 7A–C), eliminating the hypothesis of the putative receptor modification. In contrast, Sec16-V5 did disperse (Figure 7A–C) under serum starvation, recapitulating the behaviour of endogenous Sec16. To narrow down the potential domains involved in the starvation-induced Sec16 dispersion/aggregation, we showed that N-ter (Sec16 lacking the C-terminus) behaved in a similar manner to mini-Sec16 (Figure 7A–C) upon serum starvation, suggesting that NC1 (aa 1–690) is not involved. Accordingly, Sec16 lacking NC1 (ΔNC1-Sec16) behaved like endogenous Sec16 (Supplementary Figure S7A). Altogether, these results reveal that the 410 C-terminal amino acids (1535 and 1945) comprising the NC5 and C-ter domains are required for the starvation-induced Sec16 dispersion.
Figure 7.
Response of Sec16 truncations to starvation. (A) Schematic representation of the V5-tagged Sec16 truncations (Sec16 (1–1945); N-ter; MiniSec16, minimum region of Sec16 necessary for its localization to tER sites (aa 690–1535) as described in Ivan et al, (2008); ΔNC1-Sec16; ΔNC1-ΔCter; and ΔNC1-Δ64) with regards to their ability to localize to tER sites and to disperse/aggregate upon serum and amino-acid starvation. The asterisks indicate that the Sec16 construct used in 75 amino acids shorter than endogenous Sec16 (2021). (B) IF visualization of V5-tagged Sec16 truncations (using an anti-V5 antibody) in transfected cells in full medium, serum-free medium (−serum) for 7 h, and serum and amino acid-free medium (−AA –Serum) for 4 h. (C) Quantitation of dispersion/aggregation of V5-tagged Sec16 truncations upon serum and AA starvation. Error bars represent SD from three independent experiments. Scale bars: 5 μm.
To further pin-point the starvation responsive domain, a Sec16 truncated mutant lacking NC1 and C-ter (ΔNC1-ΔC-ter) was expressed but did not disperse in the absence of serum (Figure 7A and B), suggesting that the C-ter is critical for the response to starvation. This domain is predicted to contain numerous PKC phosphorylation sites (mostly clustered between amino acids 1881 and 1945, using NetPhos). To test whether these sites were targets of PKC phosphorylation (that could act downstream of ERK7 effector), we checked whether the PKC inhibitor Go6983 inhibits the serum starvation-induced Sec16 dispersion. But, it was not the case (not shown), suggesting that PKC activation is not involved in Sec16 dispersion. We also expressed a mutant version of Sec16 lacking these last 64 amino acids (ΔNC1-Δ64, thus removing the clusters of potential PKC phosphorylation sites), and monitor its dispersion/aggregation upon serum starvation. This behaved as the endogenous Sec16 (Figure 7A and B).
As a result of these observations, we analysed the 141 amino acids situated between 1741 and 1880 forming the ‘serum and amino-acid starvation Sec16 responsive domain’ that contains 14 putative phosphorylation sites (Supplementary Figure S5B). Analysis by NetPhos identified two potential casein kinase (CK) sites (CK1, S1782 and CK2, S1859, the latest conserved in most Drosophila and insect species, in red in Supplementary Figure S5B). We, therefore, tested whether Sec16 dispersion/aggregation observed upon serum and amino-acid starvation, as well as ERK7 overexpression, was inhibited by the CK2 inhibitor, TBB (Sarno et al, 2001), but this was not the case (not shown). This suggests that Sec16 is a not a CK2 substrate, at least in this pathway. CK1 inhibitor also did not have any effect. Given that Sec16 dispersion cannot be inhibited by any of the drugs used in this study, we suggest that the ‘starvation responsive domain’ could be either directly modified by ERK7 and/or act as a landing platform for ERK7 that would phosphorylate other sites in the Sec16 molecule, for instance those in the localization domain (see Discussion).
Taken together, we have identified ERK7 as a key kinase transducing signals elicited by serum and amino-acid starvation, independently of TORC1, which leads to the downregulation of secretion by targeting a major regulator of the organization of the tER sites, Sec16, in both Drosophila and mammalian cells.
Discussion
Over the past three decades genetic, biochemical and morphological analyses have advanced our understanding of the molecular machineries mediating the functional organization of the early secretory pathway. Key factors have been identified to have a role in the ER to Golgi trafficking and the architecture of this pathway. In particular, Sec16 has been identified as a large hydrophilic protein that is tightly associated with the cup-shaped ER structures that characterize the ER exit sites, where it sustains its function in the recruitment of COPII components to increase vesicular budding and transport (Connerly et al, 2005; Bhattacharyya and Glick, 2007; Ivan et al, 2008; Hughes et al, 2009). To gain insight on how signalling molecules regulate the organization of the early secretory pathway, 245 Drosophila kinases were depleted by RNAi and their effect on tER-Golgi units was assessed by a microscopy-based screen (Kondylis et al, 2011).
About 10% of the depleted kinases (26 out of 245) were confirmed to alter the organization of tER-Golgi units. Most of the depletions led to an increase in their number (that we refer to as MG phenotype). We sometimes observed a clustering of the tER-Golgi units but not their complete disassembly, similar to the results obtained in the screen for ER proteins (Kondylis et al, 2011). In addition, for a number of kinases, we also observed an increase in the cell size (Kondylis et al, 2011) that remains unexplained by the parameters we tested (cell-cycle inhibition, anterograde transport, TORC1 activation and lipid droplet biogenesis).
Several kinases were identified both in our screen and a recently published similar screen performed in human cells (Farhan et al, 2010). The overlap between the two screens is small (see Supplementary Table S6). Although technical differences in the screening methods, cell-type specificity and genetic redundancy could explain this paucity, this might also suggest that despite the functional organization of the early secretory pathway mediated by a relatively conserved set of factors (see Discussion in Wendler et al, 2010), its regulation by signalling molecules may vary from organism to organism.
ERK7: a novel kinase mediating the amino-acid starvation-induced disorganization of the early secretory pathway
Our screen identified ERK7 that encodes a protein homologous to mammalian ERK7/8 also known as MAPK15 (Abe et al, 1999, 2001, 2002; Klevernic et al, 2006; Saelzler et al, 2006). Interestingly, the screen in HeLa cells did not pick MAPK15 because the phenotype is only obvious upon starvation, not in normal growth conditions (Farhan et al, 2010). When depleted from S2 cells, ERK7 led to an MG phenotype and when overexpressed, it downregulated secretion. As secretion is a key factor in cell growth, it suggests that ERK7 has an inhibitory role on cell growth. In support of this, ERK7 expression is very low (and even downregulated) during larval stages that are characterized by massive growth (http://www.flybase.org). Growth inhibition by ERK7 has also been observed for mammalian ERK7/8. For instance, overexpression of MAPK15 has been found to negatively regulate hippocampal neurite growth (Supplementary Table 1 in Buchser et al, 2010). As neurite growth is linked to ER to Golgi trafficking in Drosophila (Ye et al, 2007) and in mammals (Aridor and Fish, 2009), the neurite growth phenotype could be linked to disassembly of ER exit sites. In addition, a decreased expression of ERK8 has been reported in breast cancer cell lines, indicating that this kinase could function as tumour suppressor by reducing cell growth (Henrich et al, 2003). Overexpressing ERK7 and ERK8 also inhibits DNA synthesis in COS cells (Abe et al, 1999) and HEK cell proliferation (Erster et al, 2010), although the mechanism underlying growth inhibition is clearly different. Drosophila ERK7 has not been genetically characterized during development, but it has been identified in screens searching for regulators of cell cycle (Bettencourt-Dias et al, 2004; Bjorklund et al, 2006) and calcium signalling (Vig et al, 2006).
Strikingly, ERK7 overexpression has a very similar effect on Sec16 (and Sec23) to that of serum and amino-acid starvation. Furthermore, lowering ERK7 activity partially rescues the tER site disassembly phenotype induced by serum and amino-acid starvation, in both S2 and HeLa cells, underlying the central role of ERK7 in this event. Moreover, we have shown that ERK7 is stabilized upon amino-acid deprivation, which protects it from degradation through the proteasome. Interestingly, ERK7 harbours two βTRCP phospho-degrons. βTRCP is known to interact with SCF and the E3 ligase Cullin, to promote the phosphorylation-dependent ubiquitination and proteasomal degradation of its targets (Winston et al, 1999). As mammalian ERK7 is one of the targets (Kuo et al, 2004), we hypothesize that amino-acid starvation could stabilize ERK7 by preventing its targeting by the SCFβTRCP. This will need to be further investigated.
However, inhibiting endogenous ERK7 degradation by treating S2 cells cultured in full medium with MG132 did not lead to Sec16 dispersion (not shown). This could be due to a number of reasons. One is that MG132 inhibits the proteasome without preventing the ubiquitination of the targets, and ERK7 ubiquitination could possibly compromise its kinase activity. The second is that starvation could also result in the upregulation of a factor needed for ERK7 localization to ER exit sites so that it can modify Sec16. Third, amino-acid starvation might not only lead to ERK7 stabilization, but also to its activation. All this, of course, could be overridden by ERK7 overexpression, as it leads to the dispersion of Sec16 even in the presence of nutrient and growth factors. Last, amino-acid starvation is likely to activate parallel pathways and this would explain why lowering ERK7 only partially rescues the induced Sec16 dispersion/aggregation phenotype. In this regard, it should be noted that serum and amino-acid starvation of HeLa cells results in a milder ER exit site disassembly but is completely rescued by MAPK15 depletion.
TORC1 independence
Serum starvation-induced tER site disassembly is not inhibited by MEK inhibitors, nor is it by PKC and p38 inhibitors; neither does it involve pre-translational events, as blocking translation with cycloheximide addition while serum starving the cells exacerbates the starvation effect. More surprisingly, although amino-acid starvation is well known to inhibit TORC1 in many organisms, resulting in growth inhibition through decreased protein synthesis and increased protein degradation (through stimulation of autophagy; Sengupta et al, 2010; Yang and Xu, 2011). TORC1 does not seem to be involved in the cessation of secretion through the tER disassembly, as shown by the lack of effect of rapamycin, and Raptor depletion or the addition of insulin. We, therefore, argue that although secretion is clearly a mechanism controlling cell growth in response to nutrients, the cell uses a signalling pathway involving ERK7, but distinct from TORC1, to downregulate protein transport upon starvation. Taken together, it means that nutrient starvation controls cell growth not only through translation inhibition and downregulation of biosynthetic pathway as documented by others, but also through secretion inhibition by using a novel signalling pathway that we have started to identify in this study. Interestingly, secretion rate has also been shown to be inhibited by 50% in yeast upon amino-acid starvation (Geng et al, 2010) and yeast could prove a useful model organism to dissect the signalling cascade involved in transducing nutrient restriction.
Sec16 is an MAP kinase signalling integrating platform for the ER exit site organization and the control of secretion
ERK7 exerts its inhibitory role on secretion through the Sec16 release from tER sites, in a kinase-dependent manner, leading to their disassembly. Since the possibility that ERK7 modifies the putative Sec16 receptor at the tER sites is ruled out, Sec16 itself is likely the target of the ERK7 signalling pathway. It could be indirect with other kinases downstream of ERK7, such as CK2 but this has been ruled out. Furthermore, despite the large battery of inhibitors we used (see Materials and methods), none have inhibited serum starvation triggered Sec16 dispersion downstream of ERK7. This, therefore, opens the possibility that Sec16 is a direct ERK7 substrate. One serine/threonine residue situated in the ‘starvation responsive domain’ (aa 1740–1880) could be phosphorylated by ERK7. Alternatively, this domain could act as a landing platform for ERK7 that could in turn phosphorylate residues situated in other domains of Sec16. A more extensive proteomics and mutation analysis will be required to fully elucidate this issue.
In the same vein, the Sec16 responsive domain may act as a landing platform for a phosphatase. Drosophila Sec16 harbours many phosphorylation sites (Bodenmiller et al, 2007; Zhai et al, 2008) that are mostly located in the minimal domain for tER site localization (our analysis). This putative phosphatase could specifically dephosphorylate specific residues of the localization domain, resulting in the Sec16 release. However, incubation with the broad-spectrum protein phosphatase inhibitors (sodium orthovanadate, inhibitor of protein phosphor-tyrosine and alkaline phosphatases and okadaic acid, inhibitor of phospho-serine/threonine protein phosphatases 1 and 2) did not protect against the tER disassembly induced by serum starvation (not shown).
Although this work points to Sec16 as an ERK7 target (whether direct or not) and to tER site maintenance as a way to control secretion, other key proteins of the secretory pathway might also be targets, leading to an inhibition of secretion at multiple levels within the secretory pathway. This will need further research.
ERK2 versus ERK7 in the regulation of secretion
The results presented here also strengthen the relationship between the secretory pathway and signalling (Farhan and Rabouille, 2011), and in particular the role of ERKs on ER exit sites. In HeLa cells, ERK2 was identified to directly phosphorylate human Sec16 on Threonine 415 (T415) both in vivo and in vitro followed by its recruitment to ERESs, increased ERES number and anterograde ER to Golgi trafficking (Farhan et al, 2010). Although ERK2 is clearly important in human cells, our results show that it does not seem to have a role in S2 cells. This suggests that in S2 cells, inhibiting secretion when serum and/or amino acids are missing is not only a passive mechanism of not stimulating ERK2, but an active mechanism involving ERK7. Furthermore, HeLa cells also exhibit this active mechanism (our results).
In short, ERK2 has an opposite effect on Sec16 from our proposed function for ERK7/MAPK15 (Supplementary Figure S8): (1) Growth factors stimulate Ras and ERK2 that directly phosphorylates Sec16 on T415. (2) This results in an increased mobility of Sec16 (Sec16 recruitment to ERES is increased, either from the cytosol or from the general ER membrane). (3) The number of ERES as well as the secretory capacity increase (Farhan et al, 2010). Conversely, (1′) Amino-acid starvation stabilizes ERK7 in a TORC1-independent pathway. This induces Sec16 phosphorylation in a ‘starvation responsive domain’-dependent manner. (2′) This results in Sec16 release from the tER sites leading to (3′) tER site disassembly and ER–plasma membrane transport inhibition, thus negatively regulating cell growth.
Taken together, our results point towards ERK7 as a novel mediator of nutrient availability in the control of secretion and provide a framework for a better mechanistic understanding of the regulation of protein secretion and cell growth as a response to environmental stimuli.
Materials and methods
Cell lines and culture conditions, primary and validation screen, antibodies, imaging data analysis and quantitation of cell proliferation, cell-cycle distribution by flow cytometry and statistical analysis
The reagents, cell lines and RNAi screen design, data acquisition and analysis are described in detail in Supplementary data and Kondylis et al, 2011.
dsRNA designing for the characterization of selected hits
The dsRNAs used for the characterization of selected screen hits were independently designed and each probe was evaluated for its efficiency and potential off-target effects on the website http://e-rnai.dkfz.de. Only probes with 100% specificity for the targeted gene were used. The primers and dsRNA sizes of each targeted gene are mentioned in Supplementary Table S3.
Cloning/Sec16 truncations
To test the subcellular localization of selected hits, the full-length coding sequences were amplified by PCR and cloned into pMT/V5-HisA, B, C vectors (Invitrogen). The expressed proteins were C-terminally tagged. The primers and restriction sites used to clone each gene are mentioned in Supplementary Table S4.
Wallenda K188A, CG32703/ERK7 K54R and T195A/Y197F mutants were created using the QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA, USA) and confirmed by sequencing (Supplementary Table S4). For expression in HeLa cells, Drosophila Wallenda-V5 was subcloned into pcDNA3.1 vector (Invitrogen).
Sec16 truncations were cloned in pMT/V5-HisB or pRmeGFP (Farkas et al, 2004) using the primers and restriction sites mentioned in Supplementary Table S4.
Transient transfections
S2 cells were transiently transfected for 2 days in 35 mm dish as previously described (Kondylis et al, 2005). The expression of each tagged protein was induced for 2 h with CuSO4 followed by a 2-h chase, sometimes in the presence of cycloheximide. HeLa cells were transfected with Effectene (Qiagen) for 24 h as described by the manufacture then fixed in 4% PFA for immunofluorescence.
Immunoelectron microscopy
S2 cells were fixed and processed for immunoelectron microscopy as described in Kondylis and Rabouille (2003).
Serum and amino-acid starvation of S2 cells and incubations
Two million cells were plated on coverslips in 3 cm plastic dishes in full Schneider medium (including 10% fetal calf serum) for 16–24 h. The medium was replaced by serum-free medium for 4–7 h at 26°C (−serum) or 2–4 h in Ringer buffer supplemented by 2 g/l glucose (−amino acid −serum). In one experiment, 10% serum has been added to Ringer/glucose.
When starvation follows transfections, protein expression was first induced with CuSO4 in full medium for 2 h. The medium was replaced with serum-free or ringer/glucose medium. In this set of experiments, cycloheximide was not added.
To test recovery after starvation, the cells were serum or amino acid starved as described above followed by incubation with serum for 1–9 h or amino acid for 2–4 h, respectively, before being fixed and processed for IF.
To test whether insulin would revert serum starvation effect, cells were plated with full medium overnight followed by incubation in serum-free medium supplemented or not with 3 μg/ml insulin for 5 h (Miron et al, 2003). To test whether insulin could protect against serum starvation, cells were incubated with a medium in which serum was replaced by 3 μg/ml insulin for 5–7 h before being fixed and processed for IF.
Rapamycin was used at 10 μg/ml for 1.5 and 16 h in serum-rich medium. The potency of insulin and rapamycin as TORC1 activator and inhibitor were tested using the detection of S6k-p as described in Kondylis et al (2011).
Quantification of ERK7 and starvation effects on Sec16
Coverslips of immunolabelled cells were examined under a confocal microscope (Zeiss 5.10 and Leica SPE). Stacks of confocal sections of about 50 cells per conditions were collected, and the Sec16 pattern was assessed and ranked according to three categories:
- A typical wild-type pattern of tER sites as described in Kondylis and Rabouille (2003) and Ivan et al (2008) (typical tER sites, red bars).
- An intermediate pattern where Sec16 was clearly dispersed but some tER sites, often smaller, were also visible (tER sites/Haze, blue bars).
- A dispersed pattern where no or almost no tER sites are visible with sometimes (especially in the amino-acid starvation) degrees of aggregation characterized by very round and bright dots (haze/aggregation, black bars). Each category is expressed as a percentage of the total number of cells examined.
Anterograde protein transport assay
Anterograde transport of Delta was assessed as previously described (Kondylis and Rabouille, 2003). To test it upon serum deprivation conditions, Delta S2 cells were serum starved for 4 h as described above. In all, 1 mM CuSO4 was added to the serum-free medium to induce the synthesis of Delta for 1 h after which the medium was replaced with fresh serum-free medium for 1.5 h without addition of cycloheximide. Cells were then processed for IF and labelled for Delta as described before (Kondylis and Rabouille, 2003).
Time lapse of ER exit site dispersion/aggregation upon amino-acid starvation
The expression of GFP-Sec23 or ΔNC1-Sec16-GFP was induced by addition of 1 mM CuSO4 in full medium for 1 and 2 h, respectively, to transfected S2 cells. The medium was then replaced by ringer/glucose and projections of the cells were collected immediately (_t_=0) and every 9/12 min under a Leica AF7000 microscope to capture the GFP signal.
MAPK15 depletion and starvation of HeLa cells
HeLa cells were transfected with 10 nM of MAPK15 siRNA as described in Farhan et al (2010). After 72 h, cells were treated as indicated (cultured in full medium and in PBS supplemented with 4 g/l glucose for 3 h), fixed with 4% paraformaldehyde for 20 min at room temperature and processed for immunofluorescence using the mouse monoclonal anti-Sec31antibody (BD, 1:1000) and the rabbit polyclonal anti-Sec16 antibody from (Bethyl, 1:400). Images were acquired using a Leica-SP5 confocal microscope and the number of ERES was evaluated using ImageJ.
Incubation with inhibitors
S2 cells were incubated with full medium or serum starved for 7 h in the presence and absence of MEK inhibitors UO126 (10 μM; Kim et al, 2004) and PD98059 (50 μM; Bond and Foley, 2009); the p38 inhibitor SD202190 (20 μM, Chen et al, 2006); the PKC inhibitor Go1983 (100 nM, Baek et al, 2010); the CKI inhibitor D4476 (100 μM, Bryja et al, 2007) and CK2 inhibitor TBB (10 μM, Stark et al, 2011). The inhibitor of protein phosphotyrosine and alkaline phosphatases, sodium orthovanadate was used at 1 μM, and the inhibitor of phospho-serine/threonine protein phosphatases 1 and 2, okadaic acid was used at 100 nM.
ERK7-V5 stabilization assay
In all, 1.5 × 106 S2 cells were transiently transfected with ERK7-V5 and induced for 4 h with CuSO4, followed by a chase of 2 h in the presence of cycloheximide in full medium, in full medium supplemented with 10 μM MG132 and in absence of amino acid and serum.
Cell lysate and western blotting
Cells were lysed in 50 mM Tris–HCl pH 7.5, 150 mM NaCl, 1% Triton X-100, 50 mM NaF, 1 mM Na3VO4, 25 mM Na2-β-glycerophosphate supplemented with a protease inhibitors (Roche) and proteins were separated on SDS–PAGE followed by western blot using anti-V5 and anti-tubulin monoclonal antibodies and detection by ECL.
Supplementary Material
Supplementary Information
Supplementary Table 1
Review Process File
Acknowledgments
We thank our colleagues of the Dept of Cell Biology for discussion and Fulvio Reggiori and Adam Grieve for critically reading the manuscript. We would also like to thank Ody Sibon (University of Groningen) for providing the WT S2 cells used for cell-cycle progression analysis and Jan van Linden (UMC Utrecht) for his help with the FACS analysis. We acknowledge the use of Flybase (http://www.flybase.org) NetPHOS (www.cbs.dtu.dk/services/NetPhos/). VK was supported by a short-term EMBO fellowship and supported by a Horizon Programme from National Regie-Orgaan Genomics (050-71-029) to CR. YT was supported by a ‘Aard en Levenswetenschappen’ grant from the Nederlandse Organisatie voor Wetenschappelijke Onderzoek (NWO, 816.02.004). HF is supported by Swiss National Science Foundation, by the German Science Foundation and by an AFF Grant from the University of Konstanz. Research in the laboratory of MB was supported by an Emmy-Nother Grant from the German Research Council and the Helmholtz Alliance for Systems Biology.
Author contributions: MZ performed the experiments related to serum starvation, ERK7 overexpression and the different constructs of Sec16. VK performed the primary screen in the laboratory of Michael Boutros with the help of FF as well as the validation screen and the characterization of the candidates. YT performed the cloning and the localization, assisted by VK and identified ERK7. HF performed the MAPK15 depletion in HeLa cells and subsequent analyses. DX performed the immunoelectron microscopy. The work was done in the laboratory of CR who designed, initiated and supervised the project. VK and CR wrote the manuscript with critical reading from the other authors.
Footnotes
The authors declare that they have no conflict of interest.
References
- Abe MK, Kahle KT, Saelzler MP, Orth K, Dixon JE, Rosner MR (2001) ERK7 is an autoactivated member of the MAPK family. J Biol Chem 276: 21272–21279 [DOI] [PubMed] [Google Scholar]
- Abe MK, Kuo WL, Hershenson MB, Rosner MR (1999) Extracellular signal-regulated kinase 7 (ERK7), a novel ERK with a C-terminal domain that regulates its activity, its cellular localization, and cell growth. Mol Cell Biol 19: 1301–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe MK, Saelzler MP, Espinosa R III, Kahle KT, Hershenson MB, Le Beau MM, Rosner MR (2002) ERK8, a new member of the mitogen-activated protein kinase family. J Biol Chem 277: 16733–16743 [DOI] [PubMed] [Google Scholar]
- Aridor M, Balch WE (2000) Kinase signaling initiates coat complex II (COPII) recruitment and export from the mammalian endoplasmic reticulum. J Biol Chem 275: 35673–35676 [DOI] [PubMed] [Google Scholar]
- Aridor M, Fish KN (2009) Selective targeting of ER exit sites supports axon development. Traffic 10: 1669–1684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek EB, Jin C, Park SJ, Park KS, Yoo HY, Jeon JH, Earm YE, Kim SJ (2010) Differential recruitment of mechanisms for myogenic responses according to luminal pressure and arterial types. Pflugers Arch 460: 19–29 [DOI] [PubMed] [Google Scholar]
- Bard F, Casano L, Mallabiabarrena A, Wallace E, Saito K, Kitayama H, Guizzunti G, Hu Y, Wendler F, Dasgupta R, Perrimon N, Malhotra V (2006) Functional genomics reveals genes involved in protein secretion and Golgi organization. Nature 439: 604–607 [DOI] [PubMed] [Google Scholar]
- Bettencourt-Dias M, Giet R, Sinka R, Mazumdar A, Lock WG, Balloux F, Zafiropoulos PJ, Yamaguchi S, Winter S, Carthew RW, Cooper M, Jones D, Frenz L, Glover DM (2004) Genome-wide survey of protein kinases required for cell cycle progression. Nature 432: 980–987 [DOI] [PubMed] [Google Scholar]
- Bhattacharyya D, Glick BS (2007) Two mammalian Sec16 homologues have nonredundant functions in endoplasmic reticulum (ER) export and transitional ER organization. Mol Biol Cell 18: 839–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorklund M, Taipale M, Varjosalo M, Saharinen J, Lahdenpera J, Taipale J (2006) Identification of pathways regulating cell size and cell-cycle progression by RNAi. Nature 439: 1009–1013 [DOI] [PubMed] [Google Scholar]
- Bodenmiller B, Malmstrom J, Gerrits B, Campbell D, Lam H, Schmidt A, Rinner O, Mueller LN, Shannon PT, Pedrioli PG, Panse C, Lee HK, Schlapbach R, Aebersold R (2007) PhosphoPep—a phosphoproteome resource for systems biology research in Drosophila Kc167 cells. Mol Syst Biol 3: 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond D, Foley E (2009) A quantitative RNAi screen for JNK modifiers identifies Pvr as a novel regulator of Drosophila immune signaling. PLoS Pathog 5: e1000655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino JS, Glick BS (2004) The mechanisms of vesicle budding and fusion. Cell 116: 153–166 [DOI] [PubMed] [Google Scholar]
- Boutros M, Kiger AA, Armknecht S, Kerr K, Hild M, Koch B, Haas SA, Paro R, Perrimon N, Heidelberg Fly Array Consortium (2004) Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science 303: 832–835 [DOI] [PubMed] [Google Scholar]
- Bryja V, Schulte G, Rawal N, Grahn A, Arenas E (2007) Wnt-5a induces dishevelled phosphorylation and dopaminergic differentiation via a CK1-dependent mechanism. J Cell Sci 120: 586–595 [DOI] [PubMed] [Google Scholar]
- Buchser WJ, Slepak TI, Gutierrez-Arenas O, Bixby JL, Lemmon VP (2010) Kinase/phosphatase overexpression reveals pathways regulating hippocampal neuron morphology. Mol Syst Biol 6: 391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HB, Shen J, Ip YT, Xu L (2006) Identification of phosphatases for Smad in the BMP/DPP pathway. Genes Dev 20: 648–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connerly PL, Esaki M, Montegna EA, Strongin DE, Levi S, Soderholm J, Glick BS (2005) Sec16 is a determinant of transitional ER organization. Curr Biol 15: 1439–1447 [DOI] [PubMed] [Google Scholar]
- Coulombe P, Meloche S (2007) Atypical mitogen-activated protein kinases: structure, regulation and functions. Biochim Biophys Acta 1773: 1376–1387 [DOI] [PubMed] [Google Scholar]
- Cutler NS, Heitman J, Cardenas ME (1999) TOR kinase homologs function in a signal transduction pathway that is conserved from yeast to mammals. Mol Cell Endocrinol 155: 135–142 [DOI] [PubMed] [Google Scholar]
- de Vries HI, Uyetake L, Lemstra W, Brunsting JF, Su TT, Kampinga HH, Sibon OC (2005) Grp/DChk1 is required for G2-M checkpoint activation in Drosophila S2 cells, whereas Dmnk/DChk2 is dispensable. J Cell Sci 118: 1833–1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvel K, Yecies JL, Menon S, Raman P, Lipovsky AI, Souza AL, Triantafellow E, Ma Q, Gorski R, Cleaver S, Vander Heiden MG, MacKeigan JP, Finan PM, Clish CB, Murphy LO, Manning BD (2010) Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell 39: 171–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echard A, Hickson GR, Foley E, O’Farrell PH (2004) Terminal cytokinesis events uncovered after an RNAi screen. Curr Biol 14: 1685–1693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar BA, O’Farrell PH (1990) The three postblastoderm cell cycles of Drosophila embryogenesis are regulated in G2 by string. Cell 62: 469–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggert US, Kiger AA, Richter C, Perlman ZE, Perrimon N, Mitchison TJ, Field CM (2004) Parallel chemical genetic and genome-wide RNAi screens identify cytokinesis inhibitors and targets. PLoS Biol 2: e379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erster O, Seger R, Liscovitch M (2010) Ligand interaction scan (LIScan) in the study of ERK8. Biochem Biophys Res Commun 399: 37–41 [DOI] [PubMed] [Google Scholar]
- Farhan H, Rabouille C (2011) Signalling to and from the secretory pathway. J Cell Sci 124: 171–180 [DOI] [PubMed] [Google Scholar]
- Farhan H, Weiss M, Tani K, Kaufman RJ, Hauri HP (2008) Adaptation of endoplasmic reticulum exit sites to acute and chronic increases in cargo load. EMBO J 27: 2043–2054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhan H, Wendeler MW, Mitrovic S, Fava E, Silberberg Y, Sharan R, Zerial M, Hauri HP (2010) MAPK signaling to the early secretory pathway revealed by kinase/phosphatase functional screening. J Cell Biol 189: 997–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas A, Tompa P, Schad E, Sinka R, Jekely G, Friedrich P (2004) Autolytic activation and localization in Schneider cells (S2) of calpain B from Drosophila. Biochem J 378: 299–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng J, Nair U, Yasumura-Yorimitsu K, Klionsky DJ (2010) Post-Golgi Sec Proteins Are Required for Autophagy in Saccharomyces cerevisiae. Mol Biol Cell 21: 2257–2269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Linstedt AD (2006) COPII-Golgi protein interactions regulate COPII coat assembly and Golgi size. J Cell Biol 174: 53–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K, Maruki Y, Long X, Yoshino K, Oshiro N, Hidayat S, Tokunaga C, Avruch J, Yonezawa K (2002) Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell 110: 177–189 [DOI] [PubMed] [Google Scholar]
- Henrich LM, Smith JA, Kitt D, Errington TM, Nguyen B, Traish AM, Lannigan DA (2003) Extracellular signal-regulated kinase 7, a regulator of hormone-dependent estrogen receptor destruction. Mol Cell Biol 23: 5979–5988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes H, Budnik A, Schmidt K, Palmer KJ, Mantell J, Noakes C, Johnson A, Carter DA, Verkade P, Watson P, Stephens DJ (2009) Organisation of human ER-exit sites: requirements for the localisation of Sec16 to transitional ER. J Cell Sci 122: 2924–2934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivan V, de Voer G, Xanthakis D, Spoorendonk KM, Kondylis V, Rabouille C (2008) Drosophila Sec16 mediates the biogenesis of tER sites upstream of Sar1 through an arginine-rich motif. Mol Biol Cell 19: 4352–4365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzen AL, Jackson J, Harmon BP, Fung SM, Ramsay G, Bishop JM (1998) Drosophila myb is required for the G2/M transition and maintenance of diploidy. Genes Dev 12: 831–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM (2002) mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110: 163–175 [DOI] [PubMed] [Google Scholar]
- Kim SE, Cho JY, Kim KS, Lee SJ, Lee KH, Choi KY (2004) Drosophila PI3 kinase and Akt involved in insulin-stimulated proliferation and ERK pathway activation in Schneider cells. Cell Signal 16: 1309–1317 [DOI] [PubMed] [Google Scholar]
- Klevernic IV, Stafford MJ, Morrice N, Peggie M, Morton S, Cohen P (2006) Characterization of the reversible phosphorylation and activation of ERK8. Biochem J 394: 365–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondylis V, Rabouille C (2009) The Golgi apparatus: lessons from Drosophila. FEBS Lett 583: 3827–3838 [DOI] [PubMed] [Google Scholar]
- Kondylis V, Rabouille C (2003) A novel role for dp115 in the organization of tER sites in Drosophila. J Cell Biol 162: 185–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondylis V, Spoorendonk KM, Rabouille C (2005) dGRASP localization and function in the early exocytic pathway in Drosophila S2 cells. Mol Biol Cell 16: 4061–4072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondylis V, Tang Y, Fuchs F, Boutros M, Rabouille C (2011) Identification of ER proteins involved in the functional organisation of the early secretory pathway in Drosophila cells by a targeted RNAi screen. PLoS One 6: e17173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondylis V, van Nispen tot Pannerden HE, Herpers B, Friggi-Grelin F, Rabouille C (2007) The golgi comprises a paired stack that is separated at G2 by modulation of the actin cytoskeleton through Abi and Scar/WAVE. Dev Cell 12: 901–915 [DOI] [PubMed] [Google Scholar]
- Kuo WL, Duke CJ, Abe MK, Kaplan EL, Gomes S, Rosner MR (2004) ERK7 expression and kinase activity is regulated by the ubiquitin-proteosome pathway. J Biol Chem 279: 23073–23081 [DOI] [PubMed] [Google Scholar]
- Loewith R, Jacinto E, Wullschleger S, Lorberg A, Crespo JL, Bonenfant D, Oppliger W, Jenoe P, Hall MN (2002) Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol Cell 10: 457–468 [DOI] [PubMed] [Google Scholar]
- Martel G, Hamet P, Tremblay J (2010) GREBP, a cGMP-response Element-binding Protein Repressing the Transcription of Natriuretic Peptide Receptor 1 (NPR1/GCA). J Biol Chem 285: 20926–20939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miron M, Lasko P, Sonenberg N (2003) Signaling from Akt to FRAP/TOR targets both 4E-BP and S6K in Drosophila melanogaster. Mol Cell Biol 23: 9117–9126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T, Ohsumi Y (1998) Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J Biol Chem 273: 3963–3966 [DOI] [PubMed] [Google Scholar]
- Omerovic J, Prior IA (2009) Compartmentalized signalling: Ras proteins and signalling nanoclusters. FEBS J 276: 1817–1825 [DOI] [PubMed] [Google Scholar]
- Pryde JG, Farmaki T, Lucocq JM (1998) Okadaic acid induces selective arrest of protein transport in the rough endoplasmic reticulum and prevents export into COPII-coated structures. Mol Cell Biol 18: 1125–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulvirenti T, Giannotta M, Capestrano M, Capitani M, Pisanu A, Polishchuk RS, San Pietro E, Beznoussenko GV, Mironov AA, Turacchio G, Hsu VW, Sallese M, Luini A (2008) A traffic-activated Golgi-based signalling circuit coordinates the secretory pathway. Nat Cell Biol 10: 912–922 [DOI] [PubMed] [Google Scholar]
- Quatela SE, Philips MR (2006) Ras signaling on the Golgi. Curr Opin Cell Biol 18: 162–167 [DOI] [PubMed] [Google Scholar]
- Rabouille C, Kondylis V (2007) Golgi ribbon unlinking: an organelle-based G2/M checkpoint. Cell Cycle 6: 2723–2729 [DOI] [PubMed] [Google Scholar]
- Saelzler MP, Spackman CC, Liu Y, Martinez LC, Harris JP, Abe MK (2006) ERK8 down-regulates transactivation of the glucocorticoid receptor through Hic-5. J Biol Chem 281: 16821–16832 [DOI] [PubMed] [Google Scholar]
- Saito K, Chen M, Bard F, Chen S, Zhou H, Woodley D, Polischuk R, Schekman R, Malhotra V (2009) TANGO1 facilitates cargo loading at endoplasmic reticulum exit sites. Cell 136: 891–902 [DOI] [PubMed] [Google Scholar]
- Sallese M, Giannotta M, Luini A (2009) Coordination of the secretory compartments via inter-organelle signalling. Semin Cell Dev Biol 20: 801–809 [DOI] [PubMed] [Google Scholar]
- Sarno S, Reddy H, Meggio F, Ruzzene M, Davies SP, Donella-Deana A, Shugar D, Pinna LA (2001) Selectivity of 4,5,6,7-tetrabromobenzotriazole, an ATP site-directed inhibitor of protein kinase CK2 (′casein kinase-2′). FEBS Lett 496: 44–48 [DOI] [PubMed] [Google Scholar]
- Schmelzle T, Hall MN (2000) TOR, a central controller of cell growth. Cell 103: 253–262 [DOI] [PubMed] [Google Scholar]
- Sengupta S, Peterson TR, Sabatini DM (2010) Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress. Mol Cell 40: 310–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe LJ, Luu W, Brown AJ (2010) Akt phosphorylates Sec24: New clues into the regulation of ER-to-Golgi trafficking. Traffic 12: 19–27 [DOI] [PubMed] [Google Scholar]
- Spang A (2009) On vesicle formation and tethering in the ER-Golgi shuttle. Curr Opin Cell Biol 21: 531–536 [DOI] [PubMed] [Google Scholar]
- Stark F, Pfannstiel J, Klaiber I, Raabe T (2011) Protein kinase CK2 links polyamine metabolism to MAPK signalling in Drosophila. Cell Signal 23: 876–882 [DOI] [PubMed] [Google Scholar]
- Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, Reichling LJ, Sim T, Sabatini DM, Gray NS (2009) An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem 284: 8023–8032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vig M, Peinelt C, Beck A, Koomoa DL, Rabah D, Koblan-Huberson M, Kraft S, Turner H, Fleig A, Penner R, Kinet JP (2006) CRACM1 is a plasma membrane protein essential for store-operated Ca2+ entry. Science 312: 1220–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson P, Forster R, Palmer KJ, Pepperkok R, Stephens DJ (2005) Coupling of ER exit to microtubules through direct interaction of COPII with dynactin. Nat Cell Biol 7: 48–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendler F, Gillingham AK, Sinka R, Rosa-Ferreira C, Gordon DE, Franch-Marro X, Peden AA, Vincent JP, Munro S (2010) A genome-wide RNA interference screen identifies two novel components of the metazoan secretory pathway. EMBO J 29: 304–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston JT, Strack P, Beer-Romero P, Chu CY, Elledge SJ, Harper JW (1999) The SCFbeta-TRCP-ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IkappaBalpha and beta-catenin and stimulates IkappaBalpha ubiquitination in vitro. Genes Dev 13: 270–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Xu T (2011) Molecular mechanism of size control in development and human diseases. Cell Res 21: 715–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye B, Zhang Y, Song W, Younger SH, Jan LY, Jan YN (2007) Growing dendrites and axons differ in their reliance on the secretory pathway. Cell 130: 717–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai B, Villen J, Beausoleil SA, Mintseris J, Gygi SP (2008) Phosphoproteome analysis of Drosophila melanogaster embryos. J Proteome Res 7: 1675–1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Supplementary Table 1
Review Process File