Neural-specific elongation of 3′ UTRs during Drosophila development (original) (raw)
Abstract
The 3′ termini of eukaryotic mRNAs influence transcript stability, translation efficiency, and subcellular localization. Here we report that a subset of developmental regulatory genes, enriched in critical RNA-processing factors, exhibits synchronous lengthening of their 3′ UTRs during embryogenesis. The resulting UTRs are up to 20-fold longer than those found on typical Drosophila mRNAs. The large mRNAs emerge shortly after the onset of zygotic transcription, with several of these genes acquiring additional, phased UTR extensions later in embryogenesis. We show that these extended 3′ UTR sequences are selectively expressed in neural tissues and contain putative recognition motifs for the translational repressor, Pumilio, which also exhibits the 3′ lengthening phenomenon documented in this study. These findings suggest a previously unknown mode of posttranscriptional regulation that may contribute to the complexity of neurogenesis or neural function.
Keywords: posttranscriptional process, maternal-to-zygotic transition, nervous system, alternative polyadenylation
The maternal-to-zygotic transition (MZT) of Drosophila embryogenesis is characterized by a number of rapid and coordinated transitions in gene expression, beginning ∼2 h after fertilization (h AF). In addition to localized patterns of zygotic transcription, the MZT deploys a series of coordinated posttranscriptional processes. For example, translation of the maternal hunchback (hb) mRNA is inhibited by Nanos (Nos), Brain tumor (Brat), and Pumilio (Pum) in posterior regions of the precellular embryo. This inhibition depends partly on the degradation of the hb polyadenylated [poly(A)] tail via Nanos-response elements located in the hb 3′ UTR (1–7). Moreover, short sequence motifs within the 3′ UTRs of several hundred maternal mRNAs are required for their clearance at the MZT. This process depends on zygotic microRNAs (miRNAs) and the RNA-binding protein Smaug (8–10).
Subcellular localization of mRNAs often depends on 3′UTRs. For example, the 3′ UTRs of several pair-rule segmentation genes mediate localization of the corresponding mRNAs to the apical surface of the embryonic syncytium (11). The 3′ UTR of oskar, a gene required for germ plasm assembly, mediates translational repression and localization of the mRNA to the posterior plasm of the oocyte (12–14). Protein null mutants for oskar induce a weaker phenotype as compared with mRNA null alleles, with both giving rise to early and late developmental defects. The early phenotype can be completely rescued by expression of the oskar 3′ UTR, suggesting that it acts as a scaffold for the formation of a ribonucleoprotein complex (15). The importance of regulated 3′ UTR expression is underscored by recent evidence showing that UTR shortening or alternative polyadenylation may enhance tumorigenicity (16).
Recently, high-density tiling microarrays assays and RNA-sequencing assays have generated single-nucleotide resolution maps of the Drosophila transcriptome (17–19). Through analysis of these datasets, we identified a set of ∼30 genes, scattered throughout the genome, that exhibit coordinate lengthening of their 3′ UTRs. These UTRs are unusually long, up to 12 kb in length, and are selectively enriched in neural tissues. Expression of the long UTRs is not associated with reduced cell proliferation or the onset of cell differentiation, as suggested by recent studies of cultured mammalian cells (16, 20). Interestingly, many of the genes encode proteins implicated in RNA binding or processing, including Argonaute1 (Ago1), Brat, Pum, insulin-like growth factor II mRNA-binding protein (Imp), and embryonic lethal abnormal vision (Elav). We propose that these unique, neural-specific 3′ extensions render the mRNAs susceptible to complex regulation, including interactions with miRNAs and RNA-binding proteins. Computational analyses identified a significant enrichment of Pum recognition sequences in the extended 3′ UTRs, suggesting coordinate posttranscriptional control in the Drosophila nervous system.
Results
A survey of whole-genome tiling microarrays and RNA-Seq datasets (17, 19) identified ∼30 genes exhibiting heavily transcribed sequences that extend several kilobases beyond the annotated 3′ termini (Table 1). These exceptionally long UTRs are first detected during early embryogenesis (e.g., see Fig. 1_A_).
Table 1.
Genes that display long zygotic 3′ UTR extensions
| Gene symbol | PCR confirmation | Approximate length of short UTR, kb | Approximate length of extended UTR, kb |
|---|---|---|---|
| Not annotated | |||
| shep | — | 0.9 | 5.5 |
| wdb | Yes | 0.5 | 4.0 |
| g_β_13F | — | 0.1 | 3.4 |
| nej | Yes | 0.5 | 4.5 |
| (dpld) | Yes | 1.5 | 4.5 |
| brat | Yes | 1.3 | 8.5 |
| heph | — | 0.9 | 4.7 |
| hrb27C | — | 0.7 | 6.4 |
| rbp6 | — | 1.5 | 6.9 |
| step | — | 0.3 | 3.8 |
| elav | Yes | 0.9 | 7.2 |
| nmo | Yes | 0.5 | 4.2 |
| nrg | Yes | 0.1 | 3.0 |
| mei-P26 | Yes | 0.9 | 11.9 |
| bol | — | 1.1 | 5.0 |
| Annotated | |||
| ago1 | Yes | 1.4 | 4.6 |
| fas1 | Yes | 0.9 | 2.6 |
| g_α_49B | — | 1.2 | 3.6 |
| cam | Yes | 0.8 | 3.4 |
| imp | Yes | 1.1 | 8.4 |
| pum | Yes | 1.2 | 3.4 |
| adar | Yes | 0.2 | 4.3 |
| msi | — | 1.7 | 5.0 |
| pdp1 | — | 2.0 | 4.0 |
| tyf | — | 0.8 | 3.0 |
| cip4 | — | 0.5 | 1.5 |
| fne | Yes | 0.3 | 4.2 |
| mub | Yes | 1.4 | 7.1 |
| CG34360 | — | 1.1 | 7.6 |
| rbp9 | — | 1.5 | 5.0 |
Fig. 1.
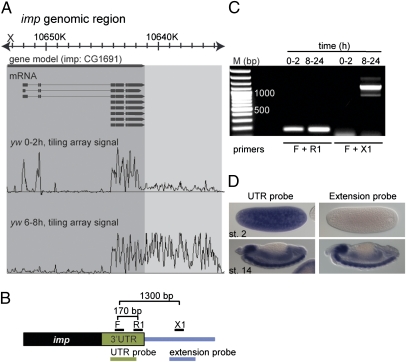
Extended 3′ UTRs. (A) Representative, whole-genome tiling array data for total Drosophila RNA from age-staged embryos produced by Manak et al. (17). Displayed is the signal from 0–2 h AF and 6–8 h AF embryos for the gene imp, aligned to the gene model. The extent of the annotated transcripts and the extent of the observed 3′ UTR extension are highlighted in dark and light gray, respectively. (B) Schematic representation of the 3′ end of imp mRNA. Indicated are the positions of PCR primers designed to amplify a fragment of the annotated imp 3′ UTR (170 bp) or a fragment consisting of 400 bp of 3′ UTR and 900 bp of the presumed extension (1,300 bp). At the bottom, probes used in D were designed to hybridize to the 3′ UTR or to the presumed extension (not to scale). (C) RT-PCR on total RNA from early (0–2 h AF) and late (8-24 h AF) embryos using the primers represented in B. The imp 3′ UTR extension can only be detected later in embryogenesis, whereas the annotated imp transcript is detected at both time points. (D) In situ hybridization for imp using probes represented in B. The 3′ UTR extension is detected only in later embryos, whereas the annotated transcript is both maternally deposited and zygotically expressed. In all figures, anterior is to the left and dorsal is at the top.
Extended 3′ UTRs Appear After the MZT.
RT-PCR assays were used to determine the time of appearance of the extended 3′ UTRs during embryogenesis (Fig. 1_B_). The short form of the imp mRNA was detected in 0- to 2-h (maternal) and 8- to 24-h embryos (zygotic), whereas the long form of the mRNA was detected only in 8- to 24-h embryos (Fig. 1_C_). Similar results were obtained for the miRNA effector gene ago1 and the tumor suppressor brat (Fig. S1 A and B). In situ hybridization assays with a probe corresponding to 1 kb of 3′ extended sequences were consistent with selective expression of the long form of the imp mRNA after the MZT (Fig. 1_D_).
The ∼8.5-kb extended 3′ UTR seen for the imp mRNA is one of the longest 3′ UTRs in the Drosophila genome and is equivalent to the longest 3′ UTRs in the human genome (21). The length of the imp extension is not unique among the genes identified in this study. For example, the extended forms of brat and mei-P26 mRNAs are also exceptionally long (∼8.5 kb and 12 kb, respectively; Fig. S1_C_). The precise 3′ termini of several of the extended UTRs were determined by 3′ RACE assays (SI Text). The most recent update of the Drosophila genome assembly (FlyBase release 5.39, July 2011) is consistent with several of the extended transcript isoforms.
Coordinate Appearance of Extended 3′ UTRs.
The preceding assays suggest that the long forms of imp and brat arise after the MZT. To pinpoint the timing of their appearance, quantitative RT-PCR (qRT-PCR) assays were performed with tightly staged embryos (Fig. 2). Short isoforms for most of the mRNAs were detected at the earliest surveyed embryonic stages (0–2 h, stages 1–4), indicating maternal expression of the corresponding genes [e.g., ago1, nejire (nej), and _brat_]. However, some of the genes (e.g., elav) exhibit exclusive zygotic expression, beginning at 2–4 h AF. The levels of the short 3′ UTRs remain constant relative to the levels of the associated coding sequences throughout development (Fig. 2 B–E). In contrast, there is a sudden appearance of the extended 3′ UTRs at 4–6 h AF (stages 9–11) (Fig. 2 B and C), which coincides with enhanced expression of intronic sequences. Thus, the long forms of the mRNAs containing the extended 3′ UTRs appear to be derived exclusively from zygotic transcripts. Similar results were obtained for all of the genes tested in this survey but not for genes used as negative controls, including staufen and roundabout (Figs. S2 and S3).
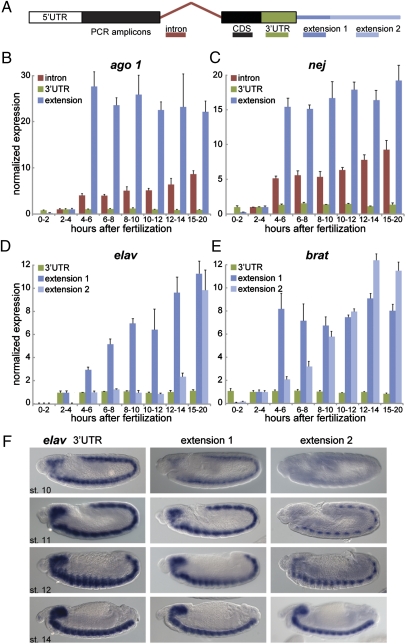
Fig. 2.
Timing of 3′ UTR isoform expression during embryogenesis. (A) Schematic of qRT-PCR experiments. Primers were designed to amplify target sequences in the intron (red), coding region (CDS; black), 3′ UTR (green), and proximal (extension 1; dark blue) or distal (extension 2; light blue) portions of the extension. Note that the primer pairs “intron,” “CDS,” and “UTR” detect both the short and the long forms, whereas “extension” primer pairs are specific to the long form. (B–E) Quantification of indicated transcripts by qRT-PCR using primer combinations shown in A. RNA was extracted from embryos at different times in embryonic development (indicated in h AF). At indicated time points, expression levels were calculated for individual primer sets by normalization to rp49 mRNA (constitutively expressed). Baseline expression was established by setting the value at 2–4 h AF to 1. Represented is the normalized expression relative to levels obtained with the CDS primer pair. The increase in extension levels compared with CDS levels indicates an increase in the ratio of the long form to the short form at later time points, whereas UTR sequences remain unchanged compared with the coding region. Error bars represent mean ± SD of three to four biological replicates for each time point. (F) In situ hybridization for elav using probes against the 3′ UTR (Left) or an extension (Center and Right) at indicated embryonic stages (st.).
Several of the genes that were investigated display sequential, phased lengthening of their 3′ UTRs during embryogenesis. For example, elav shows an initial extension at 4–6 h, with the first appearance of a 4.7-kb 3′ UTR. An even longer 3′ UTR (7.2 kb) appears several hours later, between 12 and 14 h AF (Fig. 2_D_). Previous studies suggest that these UTRs are important for autoregulation of elav gene activity during neurogenesis (22, 23). Several other genes involved in RNA processing or metabolism also exhibit phased, extended UTR isoforms, including the elav paralog found in neurons (fne), brat, mei-P26, and ago1 (Fig. 2_E_ and Figs. S1 and S2). brat and mei-P26 belong to the Drosophila tripartite motif and Ncl-1, HT2A, and Lin-4 domain (TRIM-NHL) family of proteins, which includes dappled (dpld)/wech. The dpld/wech 3′ UTR is ∼3 kb longer than expected based on the latest fly genome release. However, unlike brat and mei-P26, the dpld/wech gene expresses the long 3′ UTR isoform maternally and does not appear to be developmentally regulated (Fig. S2). That brat, mei-P26, and ago1 exhibit similar regulation is particularly intriguing because the encoded proteins form a common complex that facilitates miRNA function (24).
Selective Expression of Long 3′ UTRs in Neural Tissues.
Many of the genes examined in this study are specifically expressed in neural tissues (e.g., elav). However, a number of the genes are broadly expressed throughout the embryo and not restricted to the nervous system. In situ hybridization assays were performed to determine whether the long mRNA isoforms encoded by such genes nonetheless exhibit restricted expression in neural tissues.
ago1 exhibits near-ubiquitous expression at all embryonic stages. However, a hybridization probe directed to the extended 3′ UTR detects transcripts primarily in neural tissues, both the CNS and peripheral sensory cells (Fig. 3_A_). A similar trend is observed for brat expression, except that the extended 3′ UTR probe detects transcripts more specifically in the nervous system (Fig. 3_B_). The broadly distributed hybridization signals identified with brat coding sequences correspond to authentic expression because an intronic probe detects nascent transcripts throughout the embryo (Fig. S4). Neural-specific expression of the extended mRNA isoforms, but not the short isoforms, was observed for most other genes tested (Fig. S5).
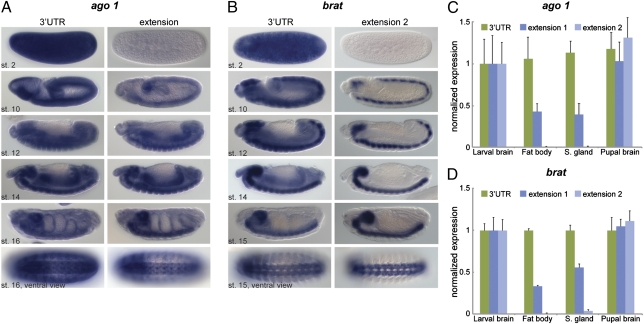
Fig. 3.
Neural enrichment of long 3′ UTR isoforms. (A and B) In situ hybridization for ago1 (A) and brat (B) using probes that hybridize to the coding region and 3′ UTR (Left) or an extension (Right). (C and D) Quantification of ago1 (C) and brat (D) transcripts by qRT-PCR using primer combinations shown in Fig. 2A. RNA was extracted from brains, fat body, and salivary glands of third-instar larvae as well as from pupal brains (48 h after puparium formation). Levels were normalized to rp49 mRNA, and expression in larval brains was set to the value 1 for each primer pair. Represented is the fold change relative to levels obtained with the CDS primer pair. Error bars represent mean ± SD of four to six biological replicates for each tissue.
3′ Extensions Correlate with Cell Type, Not Proliferation.
Recent studies in mammalian cells implicate long 3′ UTR isoforms in the inhibition of cell proliferation during cell differentiation (20). Indeed, imp mRNA exhibits a shortened 3′ UTR in proliferative human cancer cell lines (16). To determine whether the appearance of the 3′ UTR extensions correlates with the loss of cell proliferation, we surveyed RNAs extracted from proliferative and nonproliferative tissues. RNA was prepared from the brains of larvae (proliferative) and late pupae (nonproliferative) as well as from additional nonproliferative larval tissues, including salivary glands and fat bodies. qRT-PCR assays were used to measure the occurrence of the short and long forms of the brat and ago1 mRNAs in these tissues. Expression levels of 3′ UTR extensions relative to coding regions were comparable in differentiated and proliferating neural tissues. Proximal extensions exhibited reduced levels in nonneural tissues, whereas the distal-most 3′ UTR extensions were expressed almost exclusively in brains (Fig. 3 C and D). Thus, the extended UTR isoforms are enriched in neural tissues in both embryos and larvae. Similar results were obtained for all other genes tested (Figs. S5 and S6). We conclude that the key feature of the extended 3′ UTRs is neural-specific expression, not the loss of cell proliferation, in contrast to results observed in mammalian cells (16, 20).
Regulatory Motifs That Are Enriched in Extended 3′ UTRs.
We undertook a computational search for regulatory motifs within the 3′ UTR extensions, including 219 motifs involved in posttranscriptional regulation, such as miRNA 5′ seed sequences (i.e., the reverse complement of nucleotides 2–8) (25) and recognition sequences for RNA-binding proteins (26). Such motifs are expected to occur at higher frequencies in extended 3′ UTRs compared with short UTRs. Therefore, we compared the long UTRs to control sequences, including all Drosophila 3′ UTRs and their reverse-complementary sequences. Enrichment was normalized for sequence length (Materials and Methods and Table S1). Several motifs are significantly enriched in 3′ UTR extensions, and some of these exhibit significant conservation in divergent drosophilids (Tables S1 and S2). For example, the Pum motif, the poly(A) signal and recognition sequences for miRNAs including the neural-specific miR-315 (27), as well as miR-137 and miR-190 are found 5- to 10-fold more often in extensions than in the short 3′ UTRs. The Pum motif is found 9–16 times in the extensions of pum, elav, brat, and imp, whereas it is absent or represented only once in the respective short mRNA forms (Table S3 and Fig. S7).
It is possible that the extended 3′ UTRs regulate the associated mRNAs by adding recognition sequences for miRNAs or RNA-binding proteins. However, it is unclear why expression of these UTRs is restricted to neural tissues as well as being enriched for genes implicated in RNA binding and processing. It is conceivable that genes with large 3′ UTR isoforms are subject to a unique mode of regulation in neural tissues. Although the degree of UTR elongation is less dramatic, the Drosophila Hox genes may be regulated by a similar mechanism. Ubx, abd-A, Abd-B, and Antp mRNAs contain short 3′ UTRs during early development but acquire longer 3′ UTRs at later stages. These extensions are thought to be the primary targets of one or more miRNAs (28).
Discussion
We identified ∼30 genes that exhibit developmental regulation of their 3′ UTRs. As a class, the expressed transcripts contain some of the longest 3′ UTRs in the Drosophila genome and are comparable to the largest 3′ UTRs known in mammals. All of the genes undergo this posttranscriptional transition shortly after the onset of zygotic transcription, with the first detection of the long isoforms at 2–4 h AF. Perhaps the loss or gain of specialized RNA-processing factors during the MZT leads to the extension of the 3′ UTRs. Alternatively, depletion of one or more components of the general mRNA poly(A) machinery at the MZT or in neural tissues could lead to weakened poly(A) and mRNA cleavage efficiency, therefore promoting the synthesis of longer transcripts. Such a mechanism, diminished levels of the essential poly(A) factor Cstf-64, promotes the formation of longer isoforms of IgM in B lymphocytes (29).
Previous studies suggest that Drosophila 3′ UTRs are longest during early development (9). The genes identified in this study do not conform to this general trend but are consistent with recent whole-genome studies in vertebrates that suggest a statistical enrichment for longer 3′ UTRs at later stages in development (30). In mammals, the expression of long 3′ UTR isoforms has been correlated with the loss of cell proliferation and the onset of differentiation (16, 20). The genes we describe do not fit this model (e.g., see Fig. 3) and may instead be responding to a specific developmental cue during neurogenesis. The key correlation for the large 3′ extensions identified in this study is neural expression, irrespective of the state of proliferation. However, we cannot exclude the occurrence of 3′ elongation events at additional genes in other tissues because the datasets used for this analysis made use of whole-embryo RNA samples at various developmental stages.
A significant fraction of the genes with extended 3′ UTRs encode proteins implicated in RNA binding or processing, including ago1, adar, pumilio, brat, mei-P26, shep, imp, fne, and elav. Some of these genes, like ago1, are broadly expressed in a variety of tissues. Nonetheless, the extended isoforms of ago1 mRNAs are specifically enriched in neural tissues, a known hotbed of posttranscriptional regulation, including regulation by miRNAs (31) and differential splicing. For example, Dscam is thought to produce tens of thousands of spliced isoforms in the Drosophila CNS (32). Furthermore, in Drosophila, directed transport of mRNAs, like bicoid, requires functional elements within the 3′ UTR (33). Whether RNA binding factors such as Pum participate in a network of cross-regulation by repression, activation, or transport awaits further study.
It is currently unclear whether the long forms of mRNAs produce less protein than the short forms in Drosophila, as seen in mammalian cells (20). However, enrichment of Pum recognition motifs in the extended 3′ UTRs of elav, brat, and pumilio suggests regulation by repression because Pum and Brat are known to form localized translation repression complexes essential for anterior–posterior body patterning in early embryogenesis (1, 5). Such regulation may have particular relevance in the Drosophila nervous system because Pum is required for dendrite morphogenesis (34). We propose that neural-specific isoforms of the genes identified in this study comprise elements of an interactive RNA-processing network that mediates some of the distinctive posttranscriptional processes seen in the nervous system.
Materials and Methods
Tissue and Embryo Collection for RNA Extraction.
Flies carrying a histone-GFP construct were raised by standard procedures. Embryos were collected for 2 h, aged, dechorionated in 50% bleach, and observed under a Zeiss Axio Imager.A2 microscope. Embryos of appropriate ages were manually selected and immediately transferred to liquid nitrogen. Each 2-h embryo collection consisted of ∼25 embryos. Three to four embryo collections (biological replicates) were carried out for each time point. Pupal brains (excluding eye discs) were dissected at 48 h after puparium formation in cold PBS. Brain tissue, salivary glands, and fat body tissue were simultaneously dissected from wandering third-instar larvae in cold PBS. After dissection, tissues were immediately transferred to RNAlater RNA Stabilization Reagent (Qiagen). Four to six independent dissections were carried out for each tissue.
RNA Extraction, Reverse Transcription, and qRT-PCR.
Total RNA was extracted from staged embryos or from dissected tissues with TRIzol (Invitrogen) or an RNeasy Micro Kit (Qiagen). For both reverse transcription and qRT-PCR, 100–200 ng of total RNA was treated with RNase-free DNase (Promega). First-strand synthesis used random hexamer primers and SuperScript III Reverse Transcriptase (Invitrogen). For reverse transcription, cDNA was used in a 1:10 dilution, and amplification was performed with standard Taq polymerase (Roche) and the primers described in Table S4. For qRT-PCR, samples were treated with RNase H after the reverse transcription reaction and used in a 1:100 dilution. qRT-PCR was performed and monitored in a 7300 Real-Time PCR System using SYBR Green Master Mix (Applied Biosystems). Primer pairs for qRT-PCR are listed in Table S5.
In Situ Hybridization.
Embryos (0–20 h) were collected and fixed following standard procedures (34). All templates for synthesis of RNA probes were obtained from PCR-amplified genomic fragments cloned into pGEM-T Easy Vector (Promega) and confirmed by sequencing. PCR primer sequences and amplicon lengths are listed in Table S6. For each template, antisense RNA probes were in vitro-transcribed with T7 or SP6 RNA polymerase and digoxigenin-UTP or biotin-UTP (Roche). Fixed embryos were hybridized with the riboprobes according to standard protocols (34). Colorimetric detection of RNA probes was carried out with anti–digoxigenin-AP and nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate (NBT/BCIP) substrate. Fluorescent detection of RNA probes was carried out with sheep anti-digoxigenin and mouse anti-biotin primary antibodies and fluorescent secondary antibodies (Invitrogen). Imaging of embryos after in situ hybridization was performed on a Zeiss Axio Imager.A2 microscope (colorimetric) or a Zeiss LSM 700 confocal microscope (fluorescent).
3′ RACE.
We performed 3′ end sequencing as described in ref. 35. Forward primers are listed in Table S7.
Sequence Motif Analysis.
We scanned all annotated Drosophila melanogaster 3′ UTR sequences (FlyBase release 5.35) and the 3′ UTR sequences described here on both strands for miRNA 5′ seed motifs and predicted 3′ UTR regulatory motifs as described previously (26). miRNA 5′ seed motifs were defined as the reverse-complementary sequence to positions 2–8 of the mature miRNA sequence (25) from miRBase (36) (release 16). We also assessed the sequence conservation of each motif occurrence in a strand-specific manner by branch-length score (BLS) as described previously (26, 37). For each gene for which we describe an extended 3′ UTR, we counted the number of motif occurrences on the plus strand in the short and long 3′ UTR form at BLS cutoffs of 0–100% (in steps of 10%) of the entire Drosophila phylogeny. We report the raw numbers for each gene and as a fold increase across all genes. To assess the statistical significance of the motif occurrences in the added sequence (i.e., the extension itself), we calculated a P value for the enrichment of motifs on the plus strand of the 3′ UTR extensions compared with all 3′ UTR sequences in D. melanogaster by the cumulative hypergeometric distribution, normalizing for length differences. We similarly assessed the enrichment of motif matches over matches to the 3′ UTR minus strands. Furthermore, we assessed for BLS cutoffs of 10–100% (in steps of 10%) of the entire Drosophila phylogeny the fraction of conserved motif occurrences in the extensions compared with the fraction expected given the motifs’ conservation in all 3′ UTRs and 3′ UTR minus strands (37).
Supplementary Material
Supporting Information
Acknowledgments
We thank Kevin Tsui for technical assistance. V.H. is supported by a Long-Term Fellowship from the European Molecular Biology Organization (EMBO). This work was funded by National Institutes of Health Grant GM34431.
Footnotes
The authors declare no conflict of interest.
References
- 1.Sonoda J, Wharton RP. Drosophila Brain Tumor is a translational repressor. Genes Dev. 2001;15:762–773. doi: 10.1101/gad.870801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murata Y, Wharton RP. Binding of Pumilio to maternal hunchback mRNA is required for posterior patterning in Drosophila embryos. Cell. 1995;80:747–756. doi: 10.1016/0092-8674(95)90353-4. [DOI] [PubMed] [Google Scholar]
- 3.Irish V, Lehmann R, Akam M. The Drosophila posterior-group gene nanos functions by repressing hunchback activity. Nature. 1989;338:646–648. doi: 10.1038/338646a0. [DOI] [PubMed] [Google Scholar]
- 4.Wharton RP, Struhl G. RNA regulatory elements mediate control of Drosophila body pattern by the posterior morphogen nanos. Cell. 1991;67:955–967. doi: 10.1016/0092-8674(91)90368-9. [DOI] [PubMed] [Google Scholar]
- 5.Wreden C, Verrotti AC, Schisa JA, Lieberfarb ME, Strickland S. Nanos and pumilio establish embryonic polarity in Drosophila by promoting posterior deadenylation of hunchback mRNA. Development. 1997;124:3015–3023. doi: 10.1242/dev.124.15.3015. [DOI] [PubMed] [Google Scholar]
- 6.Sonoda J, Wharton RP. Recruitment of Nanos to hunchback mRNA by Pumilio. Genes Dev. 1999;13:2704–2712. doi: 10.1101/gad.13.20.2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chagnovich D, Lehmann R. Poly(A)-independent regulation of maternal hunchback translation in the Drosophila embryo. Proc Natl Acad Sci USA. 2001;98:11359–11364. doi: 10.1073/pnas.201284398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bushati N, Stark A, Brennecke J, Cohen SM. Temporal reciprocity of miRNAs and their targets during the maternal-to-zygotic transition in Drosophila. Curr Biol. 2008;18:501–506. doi: 10.1016/j.cub.2008.02.081. [DOI] [PubMed] [Google Scholar]
- 9.Tadros W, et al. SMAUG is a major regulator of maternal mRNA destabilization in Drosophila and its translation is activated by the PAN GU kinase. Dev Cell. 2007;12:143–155. doi: 10.1016/j.devcel.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 10.Liang HL, et al. The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature. 2008;456:400–403. doi: 10.1038/nature07388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis I, Ish-Horowicz D. Apical localization of pair-rule transcripts requires 3′ sequences and limits protein diffusion in the Drosophila blastoderm embryo. Cell. 1991;67:927–940. doi: 10.1016/0092-8674(91)90366-7. [DOI] [PubMed] [Google Scholar]
- 12.Kim-Ha J, Webster PJ, Smith JL, Macdonald PM. Multiple RNA regulatory elements mediate distinct steps in localization of oskar mRNA. Development. 1993;119:169–178. doi: 10.1242/dev.119.1.169. [DOI] [PubMed] [Google Scholar]
- 13.Kim-Ha J, Kerr K, Macdonald PM. Translational regulation of oskar mRNA by Bruno, an ovarian RNA-binding protein, is essential. Cell. 1995;81:403–412. doi: 10.1016/0092-8674(95)90393-3. [DOI] [PubMed] [Google Scholar]
- 14.Chekulaeva M, Hentze MW, Ephrussi A. Bruno acts as a dual repressor of oskar translation, promoting mRNA oligomerization and formation of silencing particles. Cell. 2006;124:521–533. doi: 10.1016/j.cell.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 15.Jenny A, et al. A translation-independent role of oskar RNA in early Drosophila oogenesis. Development. 2006;133:2827–2833. doi: 10.1242/dev.02456. [DOI] [PubMed] [Google Scholar]
- 16.Mayr C, Bartel DP. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manak JR, et al. Biological function of unannotated transcription during the early development of Drosophila melanogaster. Nat Genet. 2006;38:1151–1158. doi: 10.1038/ng1875. [DOI] [PubMed] [Google Scholar]
- 18.Roy S, et al. modENCODE Consortium Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010;330:1787–1797. doi: 10.1126/science.1198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Graveley BR, et al. The developmental transcriptome of Drosophila melanogaster. Nature. 2011;471:473–479. doi: 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mignone F, Gissi C, Liuni S, Pesole G. Untranslated regions of mRNAs. Genome Biol. 2002;3:REVIEWS0004. doi: 10.1186/gb-2002-3-3-reviews0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Samson ML. Evidence for 3′ untranslated region-dependent autoregulation of the Drosophila gene encoding the neural nuclear RNA-binding protein ELAV. Genetics. 1998;150:723–733. doi: 10.1093/genetics/150.2.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Borgeson CD, Samson ML. Shared RNA-binding sites for interacting members of the Drosophila ELAV family of neural proteins. Nucleic Acids Res. 2005;33:6372–6383. doi: 10.1093/nar/gki942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neumüller RA, et al. Mei-P26 regulates microRNAs and cell growth in the Drosophila ovarian stem cell lineage. Nature. 2008;454:241–245. doi: 10.1038/nature07014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 26.Stark A, et al. Harvard FlyBase curators; Berkeley Drosophila Genome Project Discovery of functional elements in 12 Drosophila genomes using evolutionary signatures. Nature. 2007;450:219–232. doi: 10.1038/nature06340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aboobaker AA, Tomancak P, Patel N, Rubin GM, Lai EC. Drosophila microRNAs exhibit diverse spatial expression patterns during embryonic development. Proc Natl Acad Sci USA. 2005;102:18017–18022. doi: 10.1073/pnas.0508823102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomsen S, Azzam G, Kaschula R, Williams LS, Alonso CR. Developmental RNA processing of 3’UTRs in Hox mRNAs as a context-dependent mechanism modulating visibility to microRNAs. Development. 2010;137:2951–2960. doi: 10.1242/dev.047324. [DOI] [PubMed] [Google Scholar]
- 29.Takagaki Y, Seipelt RL, Peterson ML, Manley JL. The polyadenylation factor CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. Cell. 1996;87:941–952. doi: 10.1016/s0092-8674(00)82000-0. [DOI] [PubMed] [Google Scholar]
- 30.Ji Z, Lee JY, Pan Z, Jiang B, Tian B. Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc Natl Acad Sci USA. 2009;106:7028–7033. doi: 10.1073/pnas.0900028106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao FB. Context-dependent functions of specific microRNAs in neural development. Neural Dev. 2010;5:25. doi: 10.1186/1749-8104-5-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmucker D, et al. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell. 2000;101:671–684. doi: 10.1016/s0092-8674(00)80878-8. [DOI] [PubMed] [Google Scholar]
- 33.Martin KC, Ephrussi A. mRNA localization: Gene expression in the spatial dimension. Cell. 2009;136:719–730. doi: 10.1016/j.cell.2009.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ye B, et al. Nanos and Pumilio are essential for dendrite morphogenesis in Drosophila peripheral neurons. Curr Biol. 2004;14(4):314–321. doi: 10.1016/j.cub.2004.01.052. [DOI] [PubMed] [Google Scholar]
- 35.Scotto-Lavino E, Du G, Frohman MA. 3′ End cDNA amplification using classic RACE. Nat Protoc. 2006;1:2742–2745. doi: 10.1038/nprot.2006.481. [DOI] [PubMed] [Google Scholar]
- 36.Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004;32(Database issue):D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kheradpour P, Stark A, Roy S, Kellis M. Reliable prediction of regulator targets using 12 Drosophila genomes. Genome Res. 2007;17:1919–1931. doi: 10.1101/gr.7090407. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information