A Genome-Wide Meta-Analysis of Six Type 1 Diabetes Cohorts Identifies Multiple Associated Loci (original) (raw)
Abstract
Diabetes impacts approximately 200 million people worldwide, of whom approximately 10% are affected by type 1 diabetes (T1D). The application of genome-wide association studies (GWAS) has robustly revealed dozens of genetic contributors to the pathogenesis of T1D, with the most recent meta-analysis identifying in excess of 40 loci. To identify additional genetic loci for T1D susceptibility, we examined associations in the largest meta-analysis to date between the disease and ∼2.54 million SNPs in a combined cohort of 9,934 cases and 16,956 controls. Targeted follow-up of 53 SNPs in 1,120 affected trios uncovered three new loci associated with T1D that reached genome-wide significance. The most significantly associated SNP (rs539514, P = 5.66×10−11) resides in an intronic region of the LMO7 (LIM domain only 7) gene on 13q22. The second most significantly associated SNP (rs478222, P = 3.50×10−9) resides in an intronic region of the EFR3B (protein EFR3 homolog B) gene on 2p23; however, the region of linkage disequilibrium is approximately 800 kb and harbors additional multiple genes, including NCOA1, C2orf79, CENPO, ADCY3, DNAJC27, POMC, and DNMT3A. The third most significantly associated SNP (rs924043, P = 8.06×10−9) lies in an intergenic region on 6q27, where the region of association is approximately 900 kb and harbors multiple genes including WDR27, C6orf120, PHF10, TCTE3, C6orf208, LOC154449, DLL1, FAM120B, PSMB1, TBP, and PCD2. These latest associated regions add to the growing repertoire of gene networks predisposing to T1D.
Author Summary
Despite the fact that there is clearly a large genetic component to type 1 diabetes (T1D), uncovering the genes contributing to this disease has proven challenging. However, in the past three years there has been relatively major progress in this regard, with advances in genetic screening technologies allowing investigators to scan the genome for variants conferring risk for disease without prior hypotheses. Such genome-wide association studies have revealed multiple regions of the genome to be robustly and consistently associated with T1D. More recent findings have been a consequence of combining of multiple datasets from independent investigators in meta-analyses, which have more power to pick up additional variants contributing to the trait. In the current study, we describe the largest meta-analysis of T1D genome-wide genotyped datasets to date, which combines six large studies. As a consequence, we have uncovered three new signals residing at the chromosomal locations 13q22, 2p23, and 6q27, which went on to be replicated in independent sample sets. These latest associated regions add to the growing repertoire of gene networks predisposing to T1D.
Introduction
Diabetes impacts approximately 200 million people worldwide [1], with microvascular and cardiovascular disease being the primary complications. Approximately 10% of cases are type 1 diabetes (T1D) sufferers, with ∼3% increase in the incidence of T1D globally per year [2]. It is expected that the incidence is 40% higher in 2010 than in 1998 [3].
T1D is a clear example of a complex trait that results from the interplay between environmental and genetic factors. There are many lines of evidence that there is a strong genetic component to T1D, primarily due to the fact that T1D has high concordance among monozygotic twins [4] and runs strongly in families, together with a high sibling risk [5].
Prior to the era of GWAS, only five loci had been fully established to be associated with T1D. However, the majority of the other reported associations in the pre-GWAS era [6]–[8] remain highly doubtful, where an initial report of association does not hold up in subsequent replication attempts by other investigative groups. This previous hazy picture of the genetics of T1D can be put down to the use of the only methodologies that were available at the time and which were much more limited than GWAS i.e. the candidate gene approach (where genomic regions were studied based on biological reasoning) and family-based linkage methodologies. Inconsistent findings can also be attributed to small sample sizes i.e. when power is low the false discovery rate tends to be high; GWAS per se has not improved consistency, rather it has leveraged large, well powered sample sizes combined with sound statistical analyses.
It has been long established that approximately half of the genetic risk for T1D is conferred by the genomic region harboring the HLA class II genes (primarily HLA-DRB1, -DQA1 and -DQB1 genes), which encode the highly polymorphic antigen-presenting proteins. Other established loci prior to the application of GWAS are the genes encoding insulin (INS) [9]-[12], cytotoxic T-lymphocyte-associated protein 4 (CTLA4) [13]–[16], protein tyrosine phosphatase, non-receptor type 22 (PTPN22) gene [17], [18], interleukin 2 receptor alpha (IL2RA) [19]–[21] and ubiquitin-associated and SH3 domain-containing protein A (UBASH3A) [22].
The application of genome wide association studies (GWAS) has robustly revealed dozens of genetic contributors to T1D [23]–[29], the results of which have largely been independently replicated [30]–[36]. The most recently reported meta-analysis of this trait identified in excess of forty loci [29], including 18 novel regions plus confirmation of a number of loci uncovered through cross-disease comparisons [34]–[36]. As such, the risks conferred by these additional loci are relatively modest compared to the ‘low-hanging fruit’ described in the first studies and could only be ultimately uncovered when larger sample sizes were utilized.
We sought to expand further on this mode of analysis by combining our cohort with all publically released genome wide SNP datasets to identify additional loci contributing to the etiology of T1D. Unfortunately, there is a relative paucity of control genotype data in these publically available sources. To circumvent this problem, we combined individual level data from each available cohort and we then compared the cases with controls from two sources. We next separated all the individual level data into two groups, characterized by the type of genotyping platform that was used to genotype the samples, which would later be recombined using inverse-variance meta-analysis. The 6,523 cases genotyped on an Illumina BeadChip included subjects from McGill University, The Children's Hospital of Philadelphia (CHOP), The Diabetes Control and Complications Trial – Epidemiology of Diabetes Interventions and Complications (DCCT-EDIC) cohort, and the Type 1 Diabetes Genetics Consortium (T1DGC), which in turn were compared with 6,648 similarly genotyped controls recruited at CHOP. The 3,411 cases genotyped on Affymetrix arrays included subjects from the Genetics of Kidneys in Diabetes Study (GoKinD) and the Wellcome Trust Cases Control Consortium (WTCCC) that were then compared with 10,308 similarly genotyped controls, including being derived from non-autoimmune disease related cases from the WTCCC, as well as from the British 1958 Birth Cohort and the UK National Blood Service [24].
Results
We compared the power of our meta-analysis to that of the previous largest meta-analysis to date. We have more than double the power of the Barrett et al. meta-analysis to find variants with a relative risk of 1.2 and approximately three times the power to detect variants with a relative risk of 1.1 [29] (Figure S1).
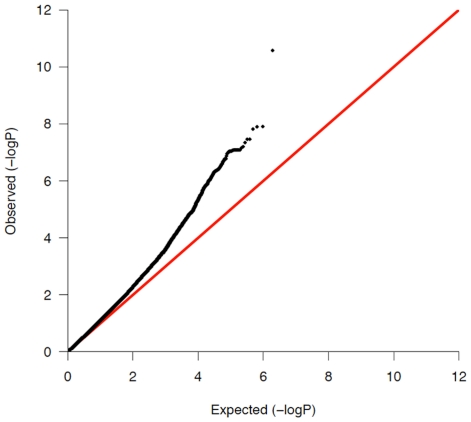
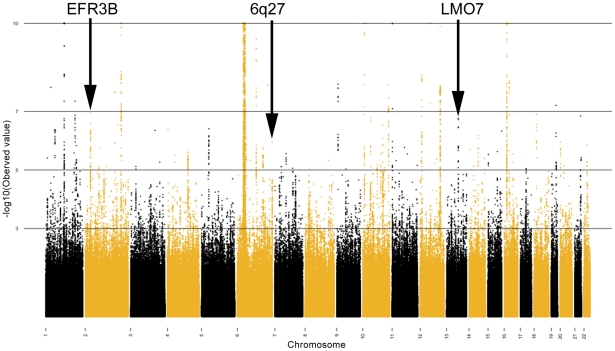
We used principal components analysis (PCA) [37] in order to minimize the potential impact of population stratification in our case/control sample sets. Eigenstrat 3.0 was employed to remove outliers and to subsequently calculate the principal components in the Illumina and Affymetrix assigned groups separately. The principal components were then used as covariates in a logistic regression, using the software PLINK [38], to compute the _P_-values, betas and standard errors which were combined in our fixed effects inverse variance meta-analysis. After controlling for population stratification, the λ in the Affymetrix and Illumina cohorts was 1.11 and 1.17, respectively (see Figure 1 for Q-Q plot). A full description of the correlation of each eigenvector with known continental ancestry appears in Text S1. Mach was used to impute ∼2.54 million SNPs, including HapMap Phase 2 SNPs in the Illumina and Affymetrix datasets in order for the statistics to be uniform and amenable to being combined [39]. Results from the meta-analysis of this resulting ‘discovery’ cohort are shown Table 1 and graphically in Figure 2.
Figure 1. QQ-plot of all previously unassociated regions in the combined meta-analysis discovery cohort.
Table 1. SNPs are shown with P<0.05 in the replication set.
| Minor Allele | Affymetrix | Illumina | Meta-Analysis | Replication | Combined | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n = 13,719 | n = 13,171 | n = 1120 | ||||||||||||
| SNP | Chr | Position | Gene/Region | Minor Allele | Frequency | OR | P | OR | P | OR | P | OR | P | P |
| Achieved GW significance overall | ||||||||||||||
| rs539514 | 13 | 75224283 | LMO7 | A | 0.499 | 0.8801 | 2.91×10−4 | 0.8769 | 1.65×10−4 | 0.8785 | 1.74×10−7 | 0.7048 | 1.16×10−5 | 5.66×10−11 |
| rs478222 | 2 | 25155259 | EFR3B | T | 0.412 | 0.8838 | 7.39×10−4 | 0.8632 | 3.79×10−5 | 0.8732 | 1.12×10−7 | 0.818 | 1.32×10−3 | 3.50×10−9 |
| rs924043 | 6 | 170220950 | 6q27 | T | 0.146 | 0.8979 | 4.44×10−2 | 0.7822 | 1.50×10−6 | 0.8353 | 1.12×10−6 | 0.7352 | 3.16×10−4 | 8.06×10−9 |
| Did not achieve GW significance overall | ||||||||||||||
| rs550448 | 7 | 28195567 | LOC100128081 | G | 0.143 | 0.8792 | 1.31×10−2 | 0.8139 | 1.12×10−4 | 0.8468 | 7.70×10−6 | 0.7621 | 3.29×10−3 | 4.68×10−7 |
| rs12679857 | 8 | 120046518 | TNFRSF11B | G | 0.309 | 0.9204 | 3.35×10−2 | 0.8465 | 1.50×10−5 | 0.8822 | 4.76×10−6 | 0.83 | 4.71×10−3 | 4.17×10−7 |
| rs6547853 | 2 | 28500305 | FOSL2 | A | 0.403 | 0.9302 | 4.76×10−2 | 0.8567 | 1.49×10−5 | 0.8919 | 7.40×10−6 | 0.8383 | 5.67×10−3 | 7.54×10−7 |
Figure 2. Fixed effects meta-analysis _P_-values shown for each SNP in the combined meta-analyzed discovery cohort.
SNPs are sorted by chromosomal location. –log10(_P_-value) are shown, where the minimum _P_-value has been capped at 1×10−10. Only the novel loci are indicated.
53 SNPs were brought forward to the replication stage based on satisfying the following criteria; however one of these SNPs consistently failed genotyping in the replication effort. The most significantly associated SNP at a given locus if the meta-analysis _P_-value was less than 1×10−5 (an independent locus was defined as a region for a given focal SNP, where we extended the region in both directions until either 250 kb had been traversed or until reaching another SNP with P<10−5), the Cochran's Q statistic _P_-value was greater than 0.05 and the locus had not been already reported from a previous GWAS of T1D. A table outlining the results for all previously described T1D associated SNPs plus our strongest associations for known regions associated with the disease are shown in Table 2 and Table S1, respectively. The replication cohort consisted of additional T1D affected trios from the T1DGC and McGill which had not been part of the original discovery cohort. The replication cohort was genotyped using the Sequenom iPLEX system and the results were analyzed using the transmission disequilibrium test in PLINK. Results for both the discovery and replication cohorts for the six SNPs that replicated with _P_≤0.05 are shown in Table 1 (the full outcomes for all SNPs tested are in Table S2).
Table 2. Discovery set _P_-values and odd ratios are shown for known T1D associated autosomal SNPs.
| SNP | CHR | Position | Gene/Region | Effect Allele | _P_-Value | OR | References |
|---|---|---|---|---|---|---|---|
| rs2476601 | 1 | 114179091 | PTPN22 | A | 5.93E-80 | 1.96 | [23], [25], [29] |
| rs2816316 | 1 | 190803436 | RGS1 | C | 8.52E-04 | 0.89 | [34] |
| rs3024505 | 1 | 205006527 | IL10 | A | 2.09E-08 | 0.82 | [29] |
| rs9653442 | 2 | 100191799 | AFF3 | C | 5.89E-04 | 1.09 | [25] |
| rs1990760 | 2 | 162832297 | IFIH1 | C | 2.21E-08 | 0.87 | [25], [29] |
| rs7574865 | 2 | 191672878 | STAT4 | T | 0.0544 | 1.06 | [35] |
| rs3087243 | 2 | 204447164 | CTLA4 | A | 1.42E-13 | 0.83 | [27], [29] |
| rs11711054 | 3 | 46320615 | CCR5 | G | 0.0399 | 1.06 | [34] |
| rs10517086 | 4 | 25694609 | 4p15.2 | A | 2.13E-04 | 1.10 | [29] |
| rs4505848 | 4 | 123351942 | IL2 | G | 2.26E-05 | 1.12 | [27], [29] |
| rs9268645 | 6 | 32516505 | HLA | G | 3.94E-136 | 1.91 | [24] |
| rs3757247 | 6 | 91014184 | BACH2 | T | 1.62E-08 | 1.15 | [27]-[29] |
| rs9388489 | 6 | 126740412 | 6q22.32 | G | 4.10E-06 | 1.12 | [29] |
| rs10499194 | 6 | 138044330 | TNFAIP3 | T | 7.92E-04 | 0.91 | [35] |
| rs1738074 | 6 | 159385965 | TAGAP | T | 9.48E-05 | 0.91 | [34] |
| rs7804356 | 7 | 26858190 | SKAP2 | C | 0.0101 | 0.93 | [29] |
| rs4948088 | 7 | 50994688 | 7p12.1 | NA | NA | NA | [29] |
| rs10758593 | 9 | 4282083 | GLIS3 | A | 1.18E-08 | 1.15 | [28], [29] |
| rs12251307 | 10 | 6163501 | IL2RA | T | 1.22E-08 | 0.79 | [27], [29] |
| rs11258747 | 10 | 6512897 | PRKCQ | T | 2.24E-05 | 1.13 | [27], [29] |
| rs10509540 | 10 | 90013013 | 10q23.31 | C | 2.83E-06 | 0.88 | [29] |
| rs3741208 | 11 | 2126350 | INS | A | 6.33E-08 | 1.16 | [23], [25], [29] |
| rs4763879 | 12 | 9801431 | 12p13.31 | A | 6.45E-07 | 1.14 | [24], [29] |
| rs1701704 | 12 | 54698754 | 12q13.2 | G | 1.08E-30 | 1.35 | [24]-[27], [29] |
| rs10877012 | 12 | 56448352 | CYP27B1 | NA | NA | NA | [54] |
| rs3184504 | 12 | 110368991 | SH2B3 | C | 1.77E-21 | 0.79 | [29] |
| rs9585056 | 13 | 98879767 | GPR183 | C | 1.27E-03 | 1.09 | [55] |
| rs1465788 | 14 | 68333352 | 14q24.1 | T | 1.79E-06 | 0.87 | [29] |
| rs4900384 | 14 | 97568704 | 14q32.2 | G | 0.0972 | 1.05 | [29] |
| rs941576 | 14 | 100375798 | DLK1 | G | 9.33E-05 | 0.91 | [56] |
| rs17574546 | 15 | 36689768 | RASGRP1 | C | 3.19E-03 | 1.09 | [57] |
| rs3825932 | 15 | 77022501 | 15q25.1 | T | 5.15E-05 | 0.90 | [27], [29] |
| rs2903692 | 16 | 11146284 | 16p13.13 | A | 4.21E-15 | 0.81 | [23]-[25], [27], [29] |
| rs4788084 | 16 | 28447349 | IL27 | T | 7.55E-04 | 0.92 | [29] |
| rs7202877 | 16 | 73804746 | 16q23.1 | G | 1.84E-05 | 1.19 | [29] |
| rs2290400 | 17 | 35319766 | ORMDL3 | T | 3.55E-03 | 0.93 | [29] |
| rs7221109 | 17 | 36023812 | 17q21.2 | T | 6.46E-04 | 0.92 | [29] |
| rs478582 | 18 | 12825976 | PTPN2 | C | 7.72E-04 | 0.92 | [24], [25], [27], [29] |
| rs763361 | 18 | 65682622 | CD226 | T | 1.17E-04 | 1.10 | [25] |
| rs2304256 | 19 | 10336652 | TYK2 | NA | NA | NA | [56] |
| rs425105 | 19 | 51900321 | 19q13.32 | C | 5.51E-06 | 0.85 | [29] |
| rs2281808 | 20 | 1558551 | 20p13 | T | 2.06E-06 | 0.88 | [29] |
| rs9976767 | 21 | 42709459 | UBASH3A | G | 1.69E-05 | 1.11 | [28], [29] |
| rs5753037 | 22 | 28911722 | 22q12.2 | T | 0.0164 | 1.06 | [29] |
| rs229541 | 22 | 35921264 | IL2RB | A | 3.67E-06 | 1.12 | [27], [29] |
We combined the ‘discovery’ and ‘replication’ meta-analysis _P_-values using Fisher's combined _P_-value method implemented in Haploview, comparable to what has been previously employed by others [40]. Three of the SNPs, namely rs539514, rs478222 and rs924043, had combined _P_-values <5×10−8, the statistical threshold for genome wide significance, while the remaining three, namely rs550448, rs12679857 and rs6547853, failed to reach this bar but were suggestive of association as the alleles yielded both a consistent direction of effect and _P_-values <0.05 in the replication cohort. These two categories of outcome are summarized in Table 1; in addition, these six SNPs were further investigated with respect to adjustments of the discovery and met-analysis _P_-values based on the lambdas of each respective cohort (Table S3).
Discussion
We have carried out the largest meta-analysis of genome wide genotyped datasets for T1D to date. The replication of three loci using the stratification-free TDT with minimal Mendelian error clearly indicates that they are not false positives due to artifacts such as uncorrected systematic error from stratification or genotyping bias.
The most significantly associated SNP (rs539514, P = 5.66×10−11) resides in an intronic region of the LMO7 (LIM domain only 7) gene on 13q22. We investigated the associated region using LocusZoom [41] and determined that it is the only gene residing within the block of linkage disequilibrium harboring the signal (Figure S3). Regional plots showing _P_-values, linkage disequilibrium, and recombination rate for all SNPs in Table 1 are outlined in the Figures S2, S3, S4, S5, S6, S7. LMO7 encodes a protein that contains multiple domains, including a calponin homology domain, a PDZ domain and a LIM domain. There are multiple LMO7 isoforms already known but their full nature and the actual extent of different isoforms remains unclear [42]. Mice with homozygous deletions of LMO7 display retinal, muscular, and growth retardation [43]. Although the function of LMO7 doesn't clearly relate to the etiology of T1D, LMO7 is expressed in pancreatic islets and thus is a possible biological candidate at this locus [44]; however it should be noted that the retinal, muscular development and islet patterns are a key element in Emery-Dreifuss Muscular Dystrophy, caused by mutations in LMO7 [45], but bears very little similarity to T1D.
The second most significantly associated SNP (rs478222, P = 3.50×10−9) resides in an intronic region of the EFR3B (protein EFR3 homolog B) gene on 2p23; however the region of linkage disequilibrium is approximately 800 kb and harbors additional multiple genes, including 3_NCOA1, C2orf79, CENPO, ADCY3, DNAJC27, POMC_, and DNMT3A. (Figure S2). A previous meta-analysis of a subset of the data used in this current study found suggestive association with T1D in the same LD block with the independent SNP, rs2165738(r2 = 0.115), but did not achieve genome wide significance at that time (P = 3.65×10−6) [27]; however, we only found modest evidence of association with rs2165738 (P = 4.78×10−3) in our discovery cohort. There has also been association to inflammatory bowel disease [46] height [47], [48] and BMI [49] reported at this locus, where in both cases the risk allele for increased height or BMI was protective for T1D risk.
The third most significantly associated SNP (rs924043, P = 8.06×10−9) lies in an intergenic region on 6q27, where the region of association is approximately 900 kb and harbors multiple genes including WDR27, C6orf120, PHF10, TCTE3, C6orf208, LOC154449, DLL1, FAM120B, PSMB1, TBP and PCD2 (Figure S5). In addition, despite not reaching the bar for genome wide significance, we did observe evidence for association at three additional loci (Table 1) containing the candidate genes LOC100128081, TNFRSF11B and FOSL2. Of these, it is notable that TNFRSF11B is a strongly associated locus with bone mineral density, also as a consequence of GWAS [50], [51]. In addition, the locus harboring LOC100128081 has also been reported in the context of a GWAS of SLE [52]. Further work will be required to fully validate the role of these particular loci in the pathogenesis of T1D.
The Barrett et al. meta-analysis was able to use British controls with British cases and American controls with American cases [29]. We did not have the same control data to be able to make the same comparisons. In the case of the Affymetrix analysis, some American cases were analyzed with purely British controls and, in the case of the Illumina analysis, some British cases with purely American controls. As such, we were forced to make our corrections using eigenvectors as covariates in our analysis; this will have the effect of modestly weakening the level of significance for associations that vary in allele frequency between the cases and controls, as now the case and controls will both vary with the eigenvectors to some degree. This in effect will make our analysis overly conservative with estimating the true effect of a SNP, and in fact every SNP that had a _P_-value less than 0.05 in the replication set did indeed have a greater effect than that which was estimated from the discovery set.
In summary, we provide convincing evidence for the existence of three additional loci associated with the T1D, adding to the repertoire of over 50 loci already demonstrated to be associated with the disease.
Materials and Methods
Ethical statement
The study was approved by the institutional review board and the ethics committee of each institution. Written informed consent was obtained from each participant in accordance with institutional requirements and the Declaration of Helsinki Principles.
Samples
Cases in the discovery set were obtained from four publically available resources and combined with those from a previous publication for the meta-analysis. Samples descriptions are available on dbgap (http://www.ncbi.nlm.nih.gov/sites/entrez?db=gap) for the T1DGC (http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000180.v1.p1), GoKinD (http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000088.v1.p1), and DCCT-EDIC (http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000086.v2.p1) patients. The WTCCC sample information is available from [24]. Samples from the T1D segment of the WTCCC were used as cases, while controls were derived from the 1958 Birth Cohort, UK Blood Service, Bipolar disorder, Coronary heart disease, Hypertension, and Type 2 Diabetes segments. The remaining cases used in the meta-analysis were previously described [23].
The total number of individuals used in the meta-analysis discovery set was 26,890 (9,934 cases/16,956 controls). The replication set consisted of 1120 case-parent trios from the T1DGC and those identified through pediatric diabetes clinics in Canada. The replication set was identical to that used in Hakonarson et al. with an extension of patients identified through pediatric diabetes clinics in Montreal, Toronto, Ottawa, and Winnipeg. All individuals were of Caucasian ancestry. A breakdown of the number of samples in each cohort in the discovery phase and a comparison with the numbers used in the Barrett et al. meta-analysis are shown in Table 3 [29]. The minor variation in the number of cases reflects that, despite using slight differences in both quality control and methods for dealing with population stratification, we have comparable numbers of cases from this cohort remaining in our analysis. Primarily, this small difference is due to the fact that we strictly accounted for relatedness and duplicates within and across cohorts in this current setting.
Table 3. A comparison of the number of samples used in each discovery cohort from the current meta-analysis and those used in the previously reported meta-analysis [29] .
| WTCCC | GoKinD/NIMH | DCCT-EDIC | T1DGC | CHOP-McGill | Totals | |
|---|---|---|---|---|---|---|
| Cases in Barrett et al meta-analysis | 1,930 | 1,601 | 0 | 3,983 | 0 | 7,514 |
| Controls in Barrett et al meta-analysis | 3,342 | 1,704 | 0 | 3,999 | 0 | 9,045 |
| Cases in current meta-analysis | 1,920 | 1,491 | 1,363 | 4,029 | 1,131 | 9,934 |
| Controls in current meta-analysis | 10,308 | 0 | 0 | 0 | 6,648 | 16,956 |
Power analysis
Power analysis was performed with the genetic analysis calculator which can be found at (http://pngu.mgh.harvard.edu/~purcell/gpc/) [53]. Various assumption were made included perfect LD between the causative variant and the markers that were genotyped, an additive genetic model, a disease prevalence of 0.0033 and an alpha of 1×10−5.
Genotyping, quality control, and imputation
Discovery samples from Philadelphia, Canada, T1DGC, and DCCT-EDIC were genotyped on a mixture of the Illumina HumanHap 550v1, 2, and 3, whereas samples from GoKinD and WTCCC were genotyped on the Affymetrix 500 K Chip. Sequenom iPlex was used to replicate the findings of the meta-analysis in 1,120 affected offspring trios from the T1DGC and from Canada.
All individuals needed an individual genotyping call rate greater than 0.98 to be included in the analysis pre-imputation and individuals were removed that showed evidence of cryptic relatedness and duplication within and across cohorts using identity-by-state. SNP quality control was performed on all samples pre-imputation. SNPs were excluded from the analysis if the minor allele was below 1%, the genotyping call rate was less than 95%, or the Hardy Weinberg equilibrium _P_-value was less than 0.00001.
To control for population stratification, Eigenstrat 3.0 was used to compute the top 10 principal components of the individuals genotyped on the Illumina SNP chips and the Affymetrix SNP chips separately [37]. Individuals were removed from the analysis if they were 6 standard deviations away from the mean of one of the top 10 principal components. After controlling for population stratification, the estimated lambda in the Affymetrix data was 1.11 and 1.17 in the Illumina data.
Mach 1.0 was used to impute ∼2.54 millions SNPs from the HapMap CEU panel for all individuals [39]. SNPs were excluded after imputation if they had a minor allele frequency less than 0.01 and an r2 value less than 0.3.
Genome-wide association and meta-analysis
PLINK [38] was used to perform a logistic regression using the 10 principal components as covariates, T1D status as the outcome, and in the case of the Affymetrix cohort, an extra dummy covariate specifying WTCCC or GoKinD cohort membership. Results from the logistic regression of 2,436,110 SNPs from the Affymetrix samples and 2,062,307 SNPs from the Illumina samples separately were combined using inverse-variance meta-analysis in PLINK. A fixed effects meta-analysis was performed and 53 SNPs were chosen for replication who had a fixed effects _P_-value <0.00001, a Cochran's Q statistic _P_-value greater than 0.05 and were not previously known to be associated with type 1 diabetes. However one of the SNPs consistently failed during the replication effort.
Supporting Information
Figure S1
Comparison of plot of power for previous and current meta-analyses. a: Plot of power (y-axis) for variants from the previously reported meta-analysis [29] with various allele frequencies (x-axis) and relative risks. Plots assume disease prevalence of 0.0033, an additive genetic model, perfect LD between causative variant and marker, and are shown for an alpha of 1×10−5. b: Plot of power (y-axis) in the current meta-analysis for variants with various allele frequencies (x-axis) and relative risks. Plots assume disease prevalence of 0.0033, an additive genetic model, perfect LD between causative variant and marker, and are shown for an alpha of 1×10−5.
(DOC)
Figure S2
Regional plot of the EFRB associated region. –log10(_P_-values) are shown for all SNPs in the region and color of circles indicates degree of LD with the most associated SNP in the region. Recombination rate is overlaid on the figure and the position with respect to genes is shown at the bottom.
(DOC)
Figure S3
Regional plot of the LMO7 associated region. –log10(_P_-values) are shown for all SNPs in the region and color of circles indicates degree of LD with the most associated SNP in the region. Recombination rate is overlaid on the figure and the position with respect to genes is shown at the bottom.
(DOC)
Figure S4
Regional plot of the LOC100128081 associated region. –log10(_P_-values) are shown for all SNPs in the region and color of circles indicates degree of LD with the most associated SNP in the region. Recombination rate is overlaid on the figure and the position with respect to genes is shown at the bottom.
(DOC)
Figure S5
Regional plot of the Chromosome 6 associated region. –log10(_P_-values) are shown for all SNPs in the region and color of circles indicates degree of LD with the most associated SNP in the region. Recombination rate is overlaid on the figure and the position with respect to genes is shown at the bottom.
(DOC)
Figure S6
Regional plot of the Chromosome 8 associated region. –log10(_P_-values) are shown for all SNPs in the region and color of circles indicates degree of LD with the most associated SNP in the region. Recombination rate is overlaid on the figure and the position with respect to genes is shown at the bottom.
(DOC)
Figure S7
Regional plot of the Chromosome 2 FOSL2 associated region. –log10(_P_-values) are shown for all SNPs in the region and color of circles indicates degree of LD with the most associated SNP in the region. Recombination rate is overlaid on the figure and the position with respect to genes is shown at the bottom.
(DOC)
Table S1
Discovery set _P_-values and odd ratios are shown for strongest associated SNP in known T1D associated regions. The list of known associated regions was collected from references cited in the references column and shown below.
(DOC)
Table S2
_P_-values and odds ratios of discovery and replication cohort are shown for all SNPs taken forward to replication stage. Combined _P_-values are shown for all SNPs that had the same direction of effect. _P_-values were combined using the Fishers combined _P_-value method implemented in Haploview. NA refers to a different direction of effect and the _P_-value was never computed. One SNP, rs722988, which failed in the genotyping assay is not shown.
(DOC)
Table S3
_P_-values for the six SNPs highlighted in Table 1 following adjustment for lambdas.
(DOC)
Text S1
Correlation outcomes of Eigenvectors with known continental ancestry.
(DOC)
Acknowledgments
We would like to thank all participating subjects and families. We also thank Smari Kristinsson, Larus Arni Hermannsson, and Asbjörn Krisbjörnsson of Raförninn ehf for their extensive software design and contribution.
Footnotes
The authors have declared that no competing interests exist.
All genotyping and other aspects of the study were funded by an Institutional Development to the Center for Applied Genomics from CHOP. SFAG, CP, and HH are funded in part by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) award (DP3 DK085708) and SFAG and HH by a Development award from the Cotswold Foundation. This research was financially supported by the Children′s Hospital of Philadelphia, Genome Canada through the Ontario Genomics Institute, and the Juvenile Diabetes Research Foundation. This research utilizes resources provided by the Type 1 Diabetes Genetics Consortium, a collaborative clinical study sponsored by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), National Institute of Allergy and Infectious Diseases (NIAID), National Human Genome Research Institute (NHGRI), National Institute of Child Health and Human Development (NICHD), and Juvenile Diabetes Research Foundation International (JDRF) and supported by U01 DK062418. The data from the T1DGC were supplied by the NIDDK Central Repositories. This manuscript was not prepared in collaboration with investigators of the T1DGC study, except for those listed as authors on the current manuscript, and does not necessarily reflect the opinions or views of the T1DGC study, the NIDDK Central Repositories, or the NIDDK. The Diabetes Control and Complications Trial (DCCT) and its follow-up the Epidemiology of Diabetes Interventions and Complications (EDIC) study were conducted by the DCCT/EDIC Research Group and supported by National Institute of Health grants and contracts and by the General Clinical Research Center Program, NCRR. The data from the DCCT/EDIC study were supplied by the NIDDK Central Repositories. This manuscript was not prepared under the auspices of the DCCT/EDIC study and does not represent analyses or conclusions of the DCCT/EDIC study group, the NIDDK Central Repositories, or the NIH. The Genetics of Kidneys in Diabetes (GoKinD) Study was conducted by the GoKinD Investigators and supported by the Juvenile Diabetes Research Foundation, the CDC, and the Special Statutory Funding Program for Type 1 Diabetes Research administered by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK). The data from the GoKinD study were supplied by the NIDDK Central Repositories. This manuscript was not prepared in collaboration with investigators of the GoKinD study and does not necessarily reflect the opinions or views of the GoKinD study, the NIDDK Central Repositories, or the NIDDK. This study makes use of data generated by the Wellcome Trust Case Control Consortium. A full list of the investigators who contributed to the generation of the data is available from www.wtccc.org.uk. Funding for the project was provided by the Wellcome Trust under award 076113. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Steyn NP, Lambert EV, Tabana H. Conference on "Multidisciplinary approaches to nutritional problems". Symposium on "Diabetes and health". Nutrition interventions for the prevention of type 2 diabetes. Proc Nutr Soc. 2009;68:55–70. doi: 10.1017/S0029665108008823. [DOI] [PubMed] [Google Scholar]
- 2.EURODIAB ACE Study Group. Variation and trends in incidence of childhood diabetes in Europe. EURODIAB ACE Study Group. Lancet. 2000;355:873–876. [PubMed] [Google Scholar]
- 3.Onkamo P, Vaananen S, Karvonen M, Tuomilehto J. Worldwide increase in incidence of Type I diabetes-the analysis of the data on published incidence trends. Diabetologia. 1999;42:1395–1403. doi: 10.1007/s001250051309. [DOI] [PubMed] [Google Scholar]
- 4.Redondo MJ, Yu L, Hawa M, Mackenzie T, Pyke DA, et al. Heterogeneity of type I diabetes: analysis of monozygotic twins in Great Britain and the United States. Diabetologia. 2001;44:354–362. doi: 10.1007/s001250051626. [DOI] [PubMed] [Google Scholar]
- 5.Clayton DG. Prediction and interaction in complex disease genetics: experience in type 1 diabetes. PLoS Genet. 2009;5:e1000540. doi: 10.1371/journal.pgen.1000540. doi: 10.1371/journal.pgen.1000540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo D, Li M, Zhang Y, Yang P, Eckenrode S, et al. A functional variant of SUMO4, a new I kappa B alpha modifier, is associated with type 1 diabetes. Nat Genet. 2004;36:837–841. doi: 10.1038/ng1391. [DOI] [PubMed] [Google Scholar]
- 7.Mirel DB, Valdes AM, Lazzeroni LC, Reynolds RL, Erlich HA, et al. Association of IL4R haplotypes with type 1 diabetes. Diabetes. 2002;51:3336–3341. doi: 10.2337/diabetes.51.11.3336. [DOI] [PubMed] [Google Scholar]
- 8.Biason-Lauber A, Boehm B, Lang-Muritano M, Gauthier BR, Brun T, et al. Association of childhood type 1 diabetes mellitus with a variant of PAX4: possible link to beta cell regenerative capacity. Diabetologia. 2005;48:900–905. doi: 10.1007/s00125-005-1723-5. [DOI] [PubMed] [Google Scholar]
- 9.Bell GI, Horita S, Karam JH. A polymorphic locus near the human insulin gene is associated with insulin-dependent diabetes mellitus. Diabetes. 1984;33:176–183. doi: 10.2337/diab.33.2.176. [DOI] [PubMed] [Google Scholar]
- 10.Bennett ST, Lucassen AM, Gough SC, Powell EE, Undlien DE, et al. Susceptibility to human type 1 diabetes at IDDM2 is determined by tandem repeat variation at the insulin gene minisatellite locus. Nat Genet. 1995;9:284–292. doi: 10.1038/ng0395-284. [DOI] [PubMed] [Google Scholar]
- 11.Vafiadis P, Bennett ST, Todd JA, Nadeau J, Grabs R, et al. Insulin expression in human thymus is modulated by INS VNTR alleles at the IDDM2 locus. Nat Genet. 1997;15:289–292. doi: 10.1038/ng0397-289. [DOI] [PubMed] [Google Scholar]
- 12.Barratt BJ, Payne F, Lowe CE, Hermann R, Healy BC, et al. Remapping the insulin gene/IDDM2 locus in type 1 diabetes. Diabetes. 2004;53:1884–1889. doi: 10.2337/diabetes.53.7.1884. [DOI] [PubMed] [Google Scholar]
- 13.Kristiansen OP, Larsen ZM, Pociot F. CTLA-4 in autoimmune diseases-a general susceptibility gene to autoimmunity? Genes Immun. 2000;1:170–184. doi: 10.1038/sj.gene.6363655. [DOI] [PubMed] [Google Scholar]
- 14.Ueda H, Howson JM, Esposito L, Heward J, Snook H, et al. Association of the T-cell regulatory gene CTLA4 with susceptibility to autoimmune disease. Nature. 2003;423:506–511. doi: 10.1038/nature01621. [DOI] [PubMed] [Google Scholar]
- 15.Anjos SM, Tessier MC, Polychronakos C. Association of the cytotoxic T lymphocyte-associated antigen 4 gene with type 1 diabetes: evidence for independent effects of two polymorphisms on the same haplotype block. J Clin Endocrinol Metab. 2004;89:6257–6265. doi: 10.1210/jc.2004-0881. [DOI] [PubMed] [Google Scholar]
- 16.Nistico L, Buzzetti R, Pritchard LE, Van der Auwera B, Giovannini C, et al. The CTLA-4 gene region of chromosome 2q33 is linked to, and associated with, type 1 diabetes. Belgian Diabetes Registry. Hum Mol Genet. 1996;5:1075–1080. doi: 10.1093/hmg/5.7.1075. [DOI] [PubMed] [Google Scholar]
- 17.Bottini N, Musumeci L, Alonso A, Rahmouni S, Nika K, et al. A functional variant of lymphoid tyrosine phosphatase is associated with type I diabetes. Nat Genet. 2004;36:337–338. doi: 10.1038/ng1323. [DOI] [PubMed] [Google Scholar]
- 18.Smyth D, Cooper JD, Collins JE, Heward JM, Franklyn JA, et al. Replication of an association between the lymphoid tyrosine phosphatase locus (LYP/PTPN22) with type 1 diabetes, and evidence for its role as a general autoimmunity locus. Diabetes. 2004;53:3020–3023. doi: 10.2337/diabetes.53.11.3020. [DOI] [PubMed] [Google Scholar]
- 19.Vella A, Cooper JD, Lowe CE, Walker N, Nutland S, et al. Localization of a type 1 diabetes locus in the IL2RA/CD25 region by use of tag single-nucleotide polymorphisms. Am J Hum Genet. 2005;76:773–779. doi: 10.1086/429843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qu HQ, Montpetit A, Ge B, Hudson TJ, Polychronakos C. Toward further mapping of the association between the IL2RA locus and type 1 diabetes. Diabetes. 2007;56:1174–1176. doi: 10.2337/db06-1555. [DOI] [PubMed] [Google Scholar]
- 21.Lowe CE, Cooper JD, Brusko T, Walker NM, Smyth DJ, et al. Large-scale genetic fine mapping and genotype-phenotype associations implicate polymorphism in the IL2RA region in type 1 diabetes. Nat Genet. 2007;39:1074–1082. doi: 10.1038/ng2102. [DOI] [PubMed] [Google Scholar]
- 22.Concannon P, Onengut-Gumuscu S, Todd JA, Smyth DJ, Pociot F, et al. A human type 1 diabetes susceptibility locus maps to chromosome 21q22.3. Diabetes. 2008;57:2858–2861. doi: 10.2337/db08-0753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hakonarson H, Grant SFA, Bradfield JP, Marchand L, Kim CE, et al. A genome-wide association study identifies KIAA0350 as a type 1 diabetes gene. Nature. 2007;448:591–594. doi: 10.1038/nature06010. [DOI] [PubMed] [Google Scholar]
- 24.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Todd JA, Walker NM, Cooper JD, Smyth DJ, Downes K, et al. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet. 2007;39:857–864. doi: 10.1038/ng2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hakonarson H, Qu HQ, Bradfield JP, Marchand L, Kim CE, et al. A novel susceptibility locus for type 1 diabetes on Chr12q13 identified by a genome-wide association study. Diabetes. 2008;57:1143–1146. doi: 10.2337/db07-1305. [DOI] [PubMed] [Google Scholar]
- 27.Cooper JD, Smyth DJ, Smiles AM, Plagnol V, Walker NM, et al. Meta-analysis of genome-wide association study data identifies additional type 1 diabetes risk loci. Nat Genet. 2008;40:1399–1401. doi: 10.1038/ng.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grant SF, Qu HQ, Bradfield JP, Marchand L, Kim CE, et al. Follow-up analysis of genome-wide association data identifies novel loci for type 1 diabetes. Diabetes. 2009;58:290–295. doi: 10.2337/db08-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barrett JC, Clayton DG, Concannon P, Akolkar B, Cooper JD, et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet. 2009;41:703–707. doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qu HQ, Bradfield JP, Li Q, Kim C, Frackelton E, et al. In silico replication of the genome-wide association results of the Type 1 Diabetes Genetics Consortium. Hum Mol Genet. 2010;19:2534–2538. doi: 10.1093/hmg/ddq133. [DOI] [PubMed] [Google Scholar]
- 31.Wang K, Baldassano R, Zhang H, Qu HQ, Imielinski M, et al. Comparative genetic analysis of inflammatory bowel disease and type 1 diabetes implicates multiple loci with opposite effects. Hum Mol Genet. 2010;19:2059–2067. doi: 10.1093/hmg/ddq078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cooper JD, Walker NM, Smyth DJ, Downes K, Healy BC, et al. Follow-up of 1715 SNPs from the Wellcome Trust Case Control Consortium genome-wide association study in type I diabetes families. Genes Immun. 2009;10(Suppl 1):S85–94. doi: 10.1038/gene.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barrett JC, Clayton DG, Concannon P, Akolkar B, Cooper JD, et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet. 2009 doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smyth DJ, Plagnol V, Walker NM, Cooper JD, Downes K, et al. Shared and distinct genetic variants in type 1 diabetes and celiac disease. N Engl J Med. 2008;359:2767–2777. doi: 10.1056/NEJMoa0807917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fung EY, Smyth DJ, Howson JM, Cooper JD, Walker NM, et al. Analysis of 17 autoimmune disease-associated variants in type 1 diabetes identifies 6q23/TNFAIP3 as a susceptibility locus. Genes Immun. 2009;10:188–191. doi: 10.1038/gene.2008.99. [DOI] [PubMed] [Google Scholar]
- 36.Cooper JD, Walker NM, Healy BC, Smyth DJ, Downes K, et al. Analysis of 55 autoimmune disease and type II diabetes loci: further confirmation of chromosomes 4q27, 12q13.2 and 12q24.13 as type I diabetes loci, and support for a new locus, 12q13.3-q14.1. Genes Immun. 2009;10(Suppl 1):S95–120. doi: 10.1038/gene.2009.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 38.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Y, Willer C, Sanna S, Abecasis G. Genotype imputation. Annu Rev Genomics Hum Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 41.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pruitt KD, Tatusova T, Maglott DR. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33:D501–504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Semenova E, Wang X, Jablonski MM, Levorse J, Tilghman SM. An engineered 800 kilobase deletion of Uchl3 and Lmo7 on mouse chromosome 14 causes defects in viability, postnatal growth and degeneration of muscle and retina. Hum Mol Genet. 2003;12:1301–1312. doi: 10.1093/hmg/ddg140. [DOI] [PubMed] [Google Scholar]
- 44.Kutlu B, Burdick D, Baxter D, Rasschaert J, Flamez D, et al. Detailed transcriptome atlas of the pancreatic beta cell. BMC Med Genomics. 2009;2:3. doi: 10.1186/1755-8794-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Holaska JM, Rais-Bahrami S, Wilson KL. Lmo7 is an emerin-binding protein that regulates the transcription of emerin and many other muscle-relevant genes. Hum Mol Genet. 2006;15:3459–3472. doi: 10.1093/hmg/ddl423. [DOI] [PubMed] [Google Scholar]
- 46.Franke A, McGovern DP, Barrett JC, Wang K, Radford-Smith GL, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lango Allen H, Estrada K, Lettre G, Berndt SI, Weedon MN, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–838. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gudbjartsson DF, Walters GB, Thorleifsson G, Stefansson H, Halldorsson BV, et al. Many sequence variants affecting diversity of adult human height. Nat Genet. 2008;40:609–615. doi: 10.1038/ng.122. [DOI] [PubMed] [Google Scholar]
- 49.Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Richards JB, Rivadeneira F, Inouye M, Pastinen TM, Soranzo N, et al. Bone mineral density, osteoporosis, and osteoporotic fractures: a genome-wide association study. Lancet. 2008;371:1505–1512. doi: 10.1016/S0140-6736(08)60599-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rivadeneira F, Styrkarsdottir U, Estrada K, Halldorsson BV, Hsu YH, et al. Twenty bone-mineral-density loci identified by large-scale meta-analysis of genome-wide association studies. Nat Genet. 2009;41:1199–1206. doi: 10.1038/ng.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gateva V, Sandling JK, Hom G, Taylor KE, Chung SA, et al. A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus. Nat Genet. 2009;41:1228–1233. doi: 10.1038/ng.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Purcell S, Cherny SS, Sham PC. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19:149–150. doi: 10.1093/bioinformatics/19.1.149. [DOI] [PubMed] [Google Scholar]
- 54.Bailey R, Cooper JD, Zeitels L, Smyth DJ, Yang JH, et al. Association of the vitamin D metabolism gene CYP27B1 with type 1 diabetes. Diabetes. 2007;56:2616–2621. doi: 10.2337/db07-0652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Heinig M, Petretto E, Wallace C, Bottolo L, Rotival M, et al. A trans-acting locus regulates an anti-viral expression network and type 1 diabetes risk. Nature. 467:460–464. doi: 10.1038/nature09386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wallace C, Smyth DJ, Maisuria-Armer M, Walker NM, Todd JA, et al. The imprinted DLK1-MEG3 gene region on chromosome 14q32.2 alters susceptibility to type 1 diabetes. Nat Genet. 42:68–71. doi: 10.1038/ng.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Qu HQ, Grant SF, Bradfield JP, Kim C, Frackelton E, et al. Association of RASGRP1 with type 1 diabetes is revealed by combined follow-up of two genome-wide studies. J Med Genet. 2009;46:553–554. doi: 10.1136/jmg.2009.067140. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1
Comparison of plot of power for previous and current meta-analyses. a: Plot of power (y-axis) for variants from the previously reported meta-analysis [29] with various allele frequencies (x-axis) and relative risks. Plots assume disease prevalence of 0.0033, an additive genetic model, perfect LD between causative variant and marker, and are shown for an alpha of 1×10−5. b: Plot of power (y-axis) in the current meta-analysis for variants with various allele frequencies (x-axis) and relative risks. Plots assume disease prevalence of 0.0033, an additive genetic model, perfect LD between causative variant and marker, and are shown for an alpha of 1×10−5.
(DOC)
Figure S2
Regional plot of the EFRB associated region. –log10(_P_-values) are shown for all SNPs in the region and color of circles indicates degree of LD with the most associated SNP in the region. Recombination rate is overlaid on the figure and the position with respect to genes is shown at the bottom.
(DOC)
Figure S3
Regional plot of the LMO7 associated region. –log10(_P_-values) are shown for all SNPs in the region and color of circles indicates degree of LD with the most associated SNP in the region. Recombination rate is overlaid on the figure and the position with respect to genes is shown at the bottom.
(DOC)
Figure S4
Regional plot of the LOC100128081 associated region. –log10(_P_-values) are shown for all SNPs in the region and color of circles indicates degree of LD with the most associated SNP in the region. Recombination rate is overlaid on the figure and the position with respect to genes is shown at the bottom.
(DOC)
Figure S5
Regional plot of the Chromosome 6 associated region. –log10(_P_-values) are shown for all SNPs in the region and color of circles indicates degree of LD with the most associated SNP in the region. Recombination rate is overlaid on the figure and the position with respect to genes is shown at the bottom.
(DOC)
Figure S6
Regional plot of the Chromosome 8 associated region. –log10(_P_-values) are shown for all SNPs in the region and color of circles indicates degree of LD with the most associated SNP in the region. Recombination rate is overlaid on the figure and the position with respect to genes is shown at the bottom.
(DOC)
Figure S7
Regional plot of the Chromosome 2 FOSL2 associated region. –log10(_P_-values) are shown for all SNPs in the region and color of circles indicates degree of LD with the most associated SNP in the region. Recombination rate is overlaid on the figure and the position with respect to genes is shown at the bottom.
(DOC)
Table S1
Discovery set _P_-values and odd ratios are shown for strongest associated SNP in known T1D associated regions. The list of known associated regions was collected from references cited in the references column and shown below.
(DOC)
Table S2
_P_-values and odds ratios of discovery and replication cohort are shown for all SNPs taken forward to replication stage. Combined _P_-values are shown for all SNPs that had the same direction of effect. _P_-values were combined using the Fishers combined _P_-value method implemented in Haploview. NA refers to a different direction of effect and the _P_-value was never computed. One SNP, rs722988, which failed in the genotyping assay is not shown.
(DOC)
Table S3
_P_-values for the six SNPs highlighted in Table 1 following adjustment for lambdas.
(DOC)
Text S1
Correlation outcomes of Eigenvectors with known continental ancestry.
(DOC)