Stable over-expression of PPARβ/δ and PPARγ to examine receptor signaling in human HaCaT keratinocytes (original) (raw)
. Author manuscript; available in PMC: 2012 Dec 1.
Abstract
Peroxisome proliferator-activated receptor-β/δ (PPARβ/δ) function and receptor cross-talk with other nuclear receptors, including PPARγ and retinoic acid receptors (RARs), was examined using stable human HaCaT keratinocyte cell lines over-expressing PPARβ/δ or PPARγ. Enhanced ligand-induced expression of two known PPAR target genes, adipocyte differentiation-related protein (ADRP) and angiopoietin-like protein 4 (ANGPTL4), was found in HaCaT keratinocytes over-expressing PPARβ/δ or PPARγ. Over-expression of PPARβ/δ did not modulate the effect of a PPARγ agonist on up-regulation of ADRP or ANGPTL4 mRNA in HaCaT keratinocytes. All-trans retinoic acid (atRA) increased expression of a known RAR target gene, yet despite a high ratio of fatty acid binding protein 5 (FABP5) to cellular retinoic acid binding protein II, did not increase expression of ANGPTL4 or 3-phosphoinositide-dependent-protein kinase 1 (PDPK1), even in HaCaT keratinocytes expressing markedly higher levels of PPARβ/δ. While PPARβ/δ-dependent attenuation of staurosporine- or UVB-induced poly (ADP-ribose) polymerase (PARP) cleavage was not observed, PPARβ/δ- and PPARγ-dependent repression of UVB-induced expression and secretion of inflammatory cytokines was found in HaCaT keratinocytes over-expressing PPARβ/δ or PPARγ. These studies suggest that FABP5 does not transport atRA or GW0742 to PPARβ/δ and promote anti-apoptotic activity by increasing expression of PDPK1, or that PPARβ/δ interferes with PPARγ transcriptional activity. However, these studies demonstrate that stable over-expression of PPARβ/δ or PPARγ significantly increases the efficacy of ligand activation and represses UVB-induced expression of tumor necrosis factor α (TNFα), interleukin 6 (IL6), or IL8 in HaCaT keratinocytes, thereby establishing an excellent model to study the functional role of these receptors in human keratinocytes.
Keywords: human keratinocytes, PPARβ/δ, PPARγ, retinoic acid, apoptosis, inflammation
1. Introduction
It is firmly established that ligand activation of peroxisome proliferator-activated receptor-β/δ (PPARβ/δ) can increase lipid catabolism in skeletal muscle [1], reduce serum lipids levels [2], increase serum high density lipoprotein cholesterol [2, 3], improve glucose intolerance observed with dietary-induced obesity [1, 4], promote terminal differentiation (reviewed in [5, 6]), and lead to a variety of anti-inflammatory activities (reviewed in [3]). However, there are a number of examples where conflicting literature exists that prevents a more definitive understanding of PPARβ/δ function (reviewed in [4, 5]). Given the potential of targeting this nuclear receptor for the treatment and prevention of disease, there is a distinct need to delineate whether PPARβ/δ can, or cannot, be targeted by small molecules for these purposes due to possible safety issues. Towards this goal, the generation of knockout and knockdown models [9-11] and the development of highly specific agonists and antagonists [6] have proven of great value for investigating the potential for targeting PPARβ/δ for drug development.
Expression of PPARβ/δ is markedly higher in intestine and keratinocytes as compared to many other tissues in both humans and mice [7, 8]. PPARβ/δ is found predominantly in the nucleus and can be co-immunoprecipitated with its heterodimerization partner, retinoid X receptor (RXR) [7]. This suggests that PPARβ/δ has an intrinsic, constitutive function. Consistent with this idea, genetic disruption of PPARβ/δ revealed that PPARβ/δ inhibits epithelial cell proliferation [10, 18]. Three different _Pparβ/δ_-null mouse models using three different targeting approaches have been produced [9, 10, 19], and used to elucidate the developmental and physiological functions of PPARβ/δ. The phenotypes of the three different _Pparβ/δ_-null mouse models differ in some cases but are concordant for others. For example, genetic disruption of PPARβ/δ caused enhanced phorbol ester-induced epithelial hyperplasia in two different models [9]. In contrast, when Pparβ/δ_-null mice were crossed with APC_min heterozygous mice, one model exhibited no change in colon carcinogenesis [10], one model exhibited increased colon tumorigenesis [21, 22], and another model exhibited decreased colon tumorigenesis [11]. This illustrates the need for alternative approaches and/or models to study the functional role of PPARβ/δ.
In addition to null mouse models, the development of highly specific PPARβ/δ ligands has also been instrumental for evaluating the effects of PPARβ/δ activation, in particular when coupled with _Pparβ/δ_-null mouse models. For example, GW501516 selectively activates human PPARβ/δ with close to 1000-fold greater affinity as compared to human PPARα or PPARγ based on in vitro reporter assays [12]. However, this selectivity is greatly reduced for mouse PPARs where GW501516 only exhibits ≤ 62-fold greater affinity for PPARβ/δ as compared to PPARα or PPARγ [12]. This difference in ligand selectivity between species illustrates the need for controls including knockout/knockdown and/or over-expression models in order to demonstrate specificity. Collectively, despite the current availability of null mouse models and high affinity ligands with specificity toward PPARβ/δ, there remains a need for alternative approaches to study the role of PPARβ/δ, as evidenced by the conflicting literature that exists preventing a more definitive understanding of PPARβ/δ function (reviewed in [4, 5]). For these reasons, the present study characterized a new human keratinocyte model where PPARβ/δ is over-expressed to provide a new tool for elucidating the role of PPARβ/δ in cell proliferation and carcinogenesis.
2. Materials and Methods
2.1. Materials and cell culture
[4-[[[2-[3-fluoro-4-(trifluoromethyl)phenyl]-4-methyl-5-thiazolyl]methyl]thio]-2-methylphenoxy acetic acid (GW0742) was synthesized by GlaxoSmithKline (Research Triangle Park, NC) [13]. All-trans retinoic acid (atRA) was purchased (Sigma Aldrich, St. Louis, MO). GW0742 was dissolved in dimethylsulfoxide (DMSO) and atRA was dissolved in ethanol. The pMigr1 vector (Migr1) and pψ-Ampho have been previously described [14]. Briefly, the Migr1 retroviral vector encodes the murine stem cell virus promoter that drives expression of cDNA cloned into a cloning site, followed by an internal ribosome entry site (IRES) and a sequence encoding enhanced green fluorescent protein (eGFP) [14]. This vector allows for expression of a protein of interest and eGFP, which facilitates identification and sorting of cells that have stably integrated the Migr1 retroviral vector. The pcDNA3.1-hPPARβ/δ and pcDNA3.1-hPPARγ constructs were kindly provided by Dr. Curtis Omiecinski (The Pennsylvania State University, University Park, PA). Primers for quantitative real-time polymerase chain reaction (qPCR) were purchased from Integrated DNA Technologies (IDT, Coralville, IA). HaCaT cells were kindly provided by Dr. Stuart Yuspa (National Cancer Institute, Bethesda, MD), and HEK293T cells were kindly provided by Dr. Yanming Wang (The Pennsylvania State University, University Park, PA). Both cell lines were cultured in Dulbecco's minimal essential medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin at 37°C and 5% CO2.
2.2. Establishment of Migr1 stable cell lines
The Migr1-hPPARβ/δ and Migr1-hPPARγ vectors were created by subcloning the human PPARβ/δ and PPARγ cDNA sequences from pcDNA3.1-hPPARβ/δ and pcDNA3.1-hPPARγ into the Migr1 vector. The coding sequence of all constructs was confirmed by sequencing at the Penn State University Nucleic Acid Facility. Stable Migr1 (vector control), Migr1-hPPARβ/δ, and Migr1-hPPARγ cell lines were established by retrovirus spinoculation as previously described [14]. Briefly, each construct and pψ-Ampho plasmids were co-transfected into HEK293T cells to produce retrovirus using the Lipofectamine® transfection reagent and the manufacturer's recommended protocol. Forty-eight hours after transfection, the supernatant containing the retrovirus was filtered with a 0.22 μm filter and used to spinoculate HaCaT cells. eGFP-positive cells were isolated by fluorescence-activated cell sorting using an InFlux V-GS Cytometry Workbench and the Spigot software (BD Biosciences, San Jose, CA). Forward-scatter and side-scatter dot plots gave the cellular physical properties of size and granularity and allowed gating for live cells. Fluorescence was excited at 488 nm, and emission was collected using a 525 nm (eGFP) band-pass filter. Collected eGFP cells possessed a minimum of 100-fold higher eGFP expression than non-eGFP cells. Fluorescence photomicrographs were obtained with a SPOT SP100 cooled CCD camera fitted to a Nikon Eclipse TE300 upright microscope with EFD-3 episcopic fluorescence attachment. The presence of eGFP fluorescence was routinely checked using the Nikon fluorescence microscope.
2.3. Characterization of the Migr1 over-expression models
Western blot analysis was performed as described below to verify that the PPARs were over-expressed. The ability of the different cell lines to respond to ligand activation was examined by treating cells with different agonists. Ligand activation of PPARβ/δ was examined in HaCaT keratinocytes, HaCaT-Migr1 vector control cells, and HaCaT-Migr1-hPPARβ/δ cells cultured in medium with vehicle (0.02% DMSO) or the PPARβ/δ ligand GW0742 (0.01 - 10 μM) for 8 hours. Ligand activation of PPARγ was examined in HaCaT keratinocytes, HaCaT-Migr1 vector control cells, and HaCaT-Migr1-hPPARγ cells cultured in medium with vehicle (0.02% DMSO) or the PPARγ ligand rosiglitazone (0.01 - 10 μM) for 24 hours. Analysis of gene expression was performed as described below.
2.4. Examination of putative receptor cross-talk
It was previously suggested by others that the ratio of fatty acid binding protein 5 (FABP5) to cellular retinoic acid binding protein II (CRABP-II) determines whether atRA or PPARβ/δ ligands activate either PPARβ/δ or RAR and modulate cell survival by increased expression of 3-phosphoinositide-dependent-protein kinase 1 (PDPK1) [15]. In this hypothetical model, PPARβ/δ is activated by atRA or PPARβ/δ ligands in cells where expression of FABP5 is high as compared to expression of CRABP-II due to preferential delivery of ligands via FABP5 rather than CRABP-II. This hypothesis was based in part on data obtained from HaCaT keratinocytes that exhibit a relatively high FABP5/CRABP-II ratio. This suggests that if PPARβ/δ expression was increased, then the ability of atRA to activate PPARβ/δ and modulate expression of PPARβ/δ target genes or PDPK1 should increase. This hypothesis was examined in greater detail in HaCaT keratinocytes, HaCaT-Migr1 vector control cells, and HaCaT-Migr1-hPPARβ/δ cells. Western blot analysis was performed to quantify relative expression of FABP5 and CRABP-II as described below. Cells were cultured in medium with vehicle (0.1% ethanol) or atRA (0.1 and 1.0 μM) for 8 to 16 hours and expression of PDPK1, the RAR target gene cytochrome P450 26A1 (CYP26A1), and the PPAR target genes adipocyte differentiation-related protein (ADRP) and angiopoietin-like protein 4 (ANGPTL4) was determined by qPCR and/or western blot analysis as described below.
As it was suggested that PPARβ/δ can interfere with PPARγ-dependent transcription [27, 28], this idea was examined in HaCaT keratinocytes, HaCaT-Migr1 vector control cells, HaCaT-Migr1-hPPARβ/δ cells, and HaCaT-Migr1-hPPARγ cells cultured in medium with vehicle (DMSO), GW0742 (0.1 or 1.0 μM) or rosiglitazone (1 or 10 μM) for 8 or 24 hours, respectively. ADRP and ANGPTL4 mRNA was measured because expression can be increased by activating either PPARβ/δ or PPARγ due to a PPAR response elements located near these gene [16-18].
2.5. Western blot analysis
Soluble protein lysates were isolated from 90-95% confluent 100 mm culture dishes using a modified MENG buffer (25 mM MOPS, 2 mM EDTA, 0.02% NaN3, and 10% glycerol, pH 7.5) containing 500 mM NaCl, 1% Nonidet P-40, and protease inhibitors. Fifty micrograms of protein per sample was resolved using SDS-polyacrylamide gels and transferred to a nitrocellulose membrane using an electroblotting method. The membranes were blocked with 5% dried milk in Tris buffered saline/Tween-20 and incubated overnight with primary antibodies. After incubation with biotinylated secondary antibody (Jackson ImmunoResearch Laboratories, West Grove, PA), immunoreactive proteins were detected after incubation with 125I-streptavidin. Membranes were exposed to plates and the level of radioactivity quantified with filmless autoradiographic analysis. Hybridization signals for specific proteins were normalized to the hybridization signal for lactate dehydrogenase (LDH) or ACTIN. The following antibodies were used: anti-LDH or anti-ACTIN (Rockland, Gilbertsville, PA), anti-human PPARβ/δ (ab21209, Abcam, Cambridge, MA), anti-human PPARγ (2430, Cell Signaling Technology, Danvers, MA), anti-PDPK1 (611070, BD Biosciences, San Diego, CA), anti-CYP26A1 (AB64888, Abcam, Cambridge, MA), anti-human CRABP-II (ab74365-100, Abcam, Cambridge, MA), anti-human FABP5 (RD181060100, BioVendor, Chandler, NC), anti-RXRα (SC553, Santa Cruz Biotechnology, Santa Cruz, CA) or anti-PARP (9542, Cell Signaling Technology, Danvers, MA).
2.6. Quantitative real-time polymerase chain reaction (qPCR)
Total RNA was isolated from cells using RiboZol RNA Extraction Reagent (AMRESCO, Solon, OH) and the manufacturer's recommended protocol. The mRNA encoding ANGPTL4, CYP26A1, PDPK1 and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was measured by qPCR analysis using the previously described primers [19]. The mRNA encoding ADRP (NM_001122) was measured by qPCR analysis using the following primers: forward 5’-CTGCTCTTCGCCTTTCGCT-3’ and reverse 5’-ACCACCCGAGTCACCACACT-3’. cDNA was generated from 1.25 μg of total RNA using MultiScribe Reverse Transcriptase kit (Applied Biosystems, Foster City, CA). The quantitative real-time PCR analysis was carried out using SYBR® Green PCR Supermix for IQ (Quanta Biosciences, Gaithersburg, MD) in the iCycler and detected using the MyiQ Realtime PCR Detection System (Bio-Rad Laboratories, Hercules, CA). The following PCR reaction was used for all mRNAs: 95°C for 10 s, 60°C for 30 s, and 72°C for 30 s, repeated for 45 cycles. Each PCR reaction included a no-template control reaction to control for contamination, and all real-time PCR reactions had greater than 85% efficiency. The relative mRNA value for each gene was normalized to the relative mRNA value for the housekeeping gene GAPDH.
2.7. Flow cytometry of cell cycle
HaCaT keratinocytes were seeded onto 6-well tissue culture dishes at a concentration of 250,000 cells per well and cultured in DMEM (with 10% FBS and 1% penicillin/streptomycin). Twenty-four hours after plating, cells were treated with GW0742 (0.01, 0.1, 1.0 or 10 μM) or rosiglitazone (0.01, 0.1, 1.0, 10.0 or 25.0 μM) for 24 hours. After these treatments, culture medium was removed and the cells were trypsinized. Trypsinized cells were pelleted and fixed in ice cold 70% ethanol. Prior to analysis, cells were stained with propidium iodide (PI) solution containing 1 μg PI/μL and 0.125% RNase A (Sigma Aldrich, St. Louis, MO). Approximately 10,000 cells/sample were analyzed using an EPICS-XL-MCL flow cytometer (Beckman Coulter, Miami Lakes, FL) fitted with a single 15-mW argon ion laser providing excitation at 488 nm. The percentage of cells at each phase of the cell cycle was determined with MultiCycle® analysis software. Values were calculated from a minimum of three independent samples per treatment.
2.8. Effect of PPARβ/δ and PPARγ on modulation of apoptotic signaling
While it is generally accepted that PPARγ can in some instances promote apoptosis, the role of PPARβ/δ in apoptotic signaling is confusing because there are studies showing that PPARβ/δ either, promotes, attenuates, or has no effect on apoptotic signaling (reviewed in [5]). Thus, the effect of PPARβ/δ and PPARγ on modulation of staurosporine-induced or ultraviolet B (UVB)-induced PARP cleavage, as a measure of apoptotic signaling, was examined. To determine the temporal changes of PARP cleavage, HaCaT keratinocytes, HaCaT-Migr1 vector control cells, HaCaT-Migr1-hPPARβ/δ cells or HaCaT-Migr1-hPPARγ cells were cultured until 90-95% confluent, and then treated with either 0.5 μM staurosporine or irradiated with UVB (280-315 nm, 50 mJ/cm2) in phosphate buffered saline (PBS) using a CL-1000 Ultraviolet Crosslinker (Ultra-Violet Products, Inc., Upland, CA). Protein was isolated from cells at 0, 0.5, 1, 2 or 4 hours post-staurosporine treatment or 0, 1, 2, 4 or 8 hours post-UVB irradiation. To determine the effect of ligand activation of PPARβ/δ or PPARγ on staurosporine-induced or UVB-induced PARP cleavage, cells were cultured in medium with or without GW0742 (0, 0.1 or 1.0 μM) or rosiglitazone (0, 1 or 10 μM) for one hour prior to irradiation or staurosporine treatment, and then cultured in medium with or without GW0742 or rosiglitazone. Protein was isolated from cells at 0, 1 or 2 hours post-staurosporine treatment or 0, 4 or 8 hours post-UVB treatment. Western blot analysis was performed as described above to quantify PARP cleavage.
2.9. Effect of PPARβ/δ and PPARγ on modulation of UVB-induced inflammation
The effect of PPARβ/δ and PPARγ on modulation of UVB-induced expression of inflammatory cytokines was examined in HaCaT keratinocytes, HaCaT-Migr1 vector control cells, HaCaT-Migr1-hPPARβ/δ cells or HaCaT-Migr1-hPPARγ cells. To determine the temporal changes of pro-inflammatory cytokine mRNA expression, 90-95% confluent cells were irradiated as described above. RNA was isolated from cells at 0, 1, 2, 4 or 8 hours post-irradiation. To determine the effect of ligand activation of PPARβ/δ or PPARγ on pro-inflammatory cytokine mRNA expression, cells were cultured in medium with or without GW0742 (0, 0.1 or 1.0 μM) or rosiglitazone (0, 1 or 10 μM) for one hour prior to irradiation, and then cultured in medium with or without GW0742 or rosiglitazone for four hours. Total RNA was isolated and qPCR analysis was performed as described above to quantify expression of mRNA encoding IL6, interleukin 8 (IL8) and TNFα. The following primers were used: human IL6 (NM_000600) forward: 5’-AAATTCGGTACATCCTCGACGGCA-3’, reverse: 5’-AGTGCCTCTTTGCTGCTTTCACAC-3’; human IL8 (NM_000582) forward: 5’-AGCCTTCCTGATTTCTGCAGCTCT-3’, reverse: 5’-AATTTCTGTGTTGGCGCAGTGTGG-3’; human TNFα (NM_000594) forward: 5’-ACCCACGGCTCCACCCTCTC-3’, reverse: 5’-AGGTCCCTGGGGAACTCTTCCCT-3’.
2.10. Enzyme-linked immunosorbent assay (ELISA)
ELISAs were performed to quantify the concentration of tumor necrosis factor α (TNFα) and interleukin 6 (IL6), in culture medium using commercially available kits (TNFα kit was purchased from R&D Systems, Minneapolis, MN; IL6 kit was purchased from Biolegend, San Diego, CA).
2.11. Data analysis
Data were analyzed for statistical significance using one-way analysis of variance (ANOVA) and the Bonferroni's multiple comparison tests, or Student's T-test as described in the figure legends. All data are presented as the mean ± standard error of the mean (SEM) using Prism 5.0 (GraphPad Software Inc., La Jolla, CA).
3. Results
3.1. Enhanced receptor activity in HaCaT keratinocyte over-expressing PPARβ/δ or PPARγ
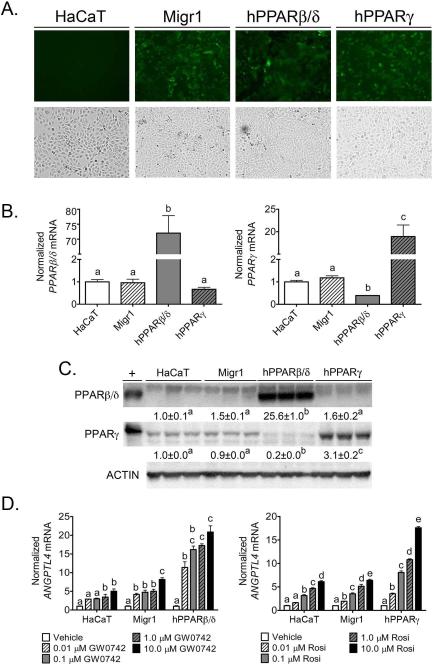
The Migr1 retroviral system [14] was used to generate HaCaT cells over-expressing PPARβ/δ and PPARγ for gain-of-function models to study PPAR signaling in human keratinocytes. After eGFP sorting and propagation of heterogeneous cell populations, stable cell lines were characterized for eGFP fluorescence, PPAR protein expression, and ligand-dependent transcriptional regulation. HaCaT-Migr1 vector control, HaCaT-Migr1-hPPARβ/δ, and HaCaT-Migr1-hPPARγ cells exhibited strong eGFP fluorescence that was not observed in control, uninfected HaCaT keratinocytes (Fig. 1A). No macroscopic changes in cell morphology were observed in any of these cell lines as compared to the parent HaCaT keratinocytes (Fig. 1A). Stable integration of the expression constructs for PPAR-specific proteins led to increased PPARβ/δ or PPARγ mRNA in respective cell lines as compared to controls (Fig. 1B). Interestingly, expression of PPARγ mRNA was lower in HaCaT-Migr1-hPPARβ/δ cells as compared to controls (Fig. 1B). A marked increase of PPARβ/δ protein was found in HaCaT keratinocytes infected with Migr1-hPPARβ/δ as compared to HaCaT-Migr1 vector control cells and the control HaCaT keratinocytes (Figure 1C). Increased expression of PPARγ protein was found in HaCaT keratinocytes infected with Migr1-hPPARγ as compared to HaCaT-Migr1 vector control cells and the parent HaCaT keratinocytes (Fig. 1C). Consistent with the observed decrease in mRNA encoding PPARγ mRNA, expression of PPARγ protein was also relatively lower in HaCaT-Migr1-hPPARβ/δ cells as compared to controls (Fig. 1C). To determine whether the increase in expression of PPARs led to functional changes in their ability to modulate ligand-dependent transcriptional regulation, the effect of ligand activation was examined using the high affinity PPARβ/δ ligand, GW0742, or the PPARγ ligand, rosiglitazone. The target gene ANGPTL4 was used for this analysis as expression of this gene can be increased by ligand activation of both PPARβ/δ or PPARγ, depending on expression of receptor and the presence of specific ligands [18]. A dose dependent increase in expression of ANGPTL4 mRNA was observed in parent HaCaT keratinocytes and HaCaT-Migr1 vector control cells in response to 0.01 μM to 10 μM GW0742 (Fig. 1D). Markedly higher increases in ligand induced expression of ANGPTL4 mRNA was observed in HaCaT-Migr1-hPPARβ/δ cells in response to 0.01 μM to 10 μM GW0742 as compared to both parent HaCaT keratinocytes and HaCaT-Migr1 vector control cells (Fig. 1D). Similarly, a dose dependent increase in expression of ANGPTL4 mRNA was observed in parent HaCaT keratinocytes and HaCaT-Migr1 vector control cells in response to 0.01 μM to 10 μM rosiglitazone (Fig. 1D). Additionally, higher increases in ligand induced expression of ANGPTL4 mRNA were observed in HaCaT-Migr1-hPPARγ cells in response to 0.01 μM to 10 μM rosiglitazone as compared to both control HaCaT keratinocytes and HaCaT-Migr1 vector control cells (Fig. 1D). It is also worth noting that a significant difference in ligand-induced expression of ANGPTL4 mRNA was not observed between the parent HaCaT keratinocyte cell line and the Migr1 vector control cell lines (Fig. 1D). Combined, these data establish that over-expression of PPARβ/δ or PPARγ in HaCaT keratinocytes can cause enhanced ligand-induced receptor activity and provides a useful model for examining the functional roles of these receptors.
Fig. 1.
Characterization of Migr1 over-expression of PPARβ/δ and PPARγ. HaCaT cells were infected with empty vector (Migr1), hPPARβ/δ, or hPPARγ, and stable GFP positive cells were sorted by flow cytometry and propagated as described in Materials and Methods. (A) GFP fluorescence of sorted HaCaT keratinocytes, HaCaT-Migr1 vector control cells (Migr1), HaCaT-Migr1-hPPARβ/δ cells (hPPARβ/δ) and HaCaTMigr1-hPPARγ cells (hPPARγ). (B) mRNA expression of PPARβ/δ and PPARγ in the Migr1 cell lines as compared to HaCaT keratinocytes. qPCR was performed to examine the expression of mRNA encoding PPARβ/δ and PPARγ. (C) Protein expression of PPARβ/δ and PPARγ was assessed by Western blotting in the Migr1 cell lines as compared to HaCaT keratinocytes. (D) Ligand response of HaCaT cell lines to the PPARβ/δ ligand GW0742 for 8 hours and the PPARγ ligand rosiglitazone for 24 hours. qPCR was performed to examine the expression of the mRNA encoding ANGPTL4 normalized to the mRNA encoding GAPDH. Fold induction of ANGPTL4 mRNA was calculated by normalization to vehicle control for each cell line. Data represents triplicate independent sample means ± SEM. Values with different letters are significantly different (P ≤ 0.05) using Bonferroni's multiple comparison.
3.2. Over-expression of PPARβ/δ does not interfere with PPARγ activity in HaCaT keratinocytes
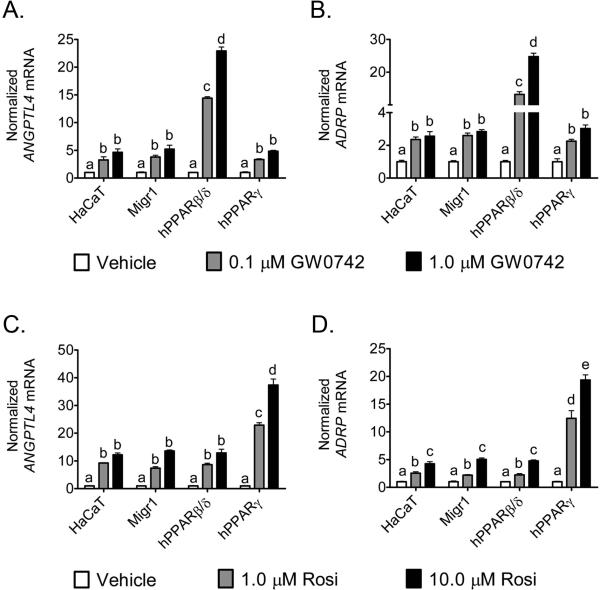
Previous in vitro studies suggest that PPARβ/δ can interfere or inhibit PPARγ-dependent gene expression [27, 28]. However, these studies are limited since this idea was based on examination of reporter assays and limited analysis (n=1) of an endogenous target gene [27, 28]. Thus, the effect of ligand activation of PPARβ/δ or PPARγ on expression of two well-characterized PPAR target genes (ADRP and ANGPTL4) was examined in HaCaT keratinocytes over-expressing PPARβ/δ or PPARγ by direct comparison in the same experiment. A dose-dependent increase in expression of ANGPTL4 and ADRP mRNA was found in parent HaCaT keratinocytes, HaCaTMigr1 vector control cells, and HaCaT-Migr1-hPPARγ cells in response to 0.1 and 1.0 μM GW0742, and this increase in expression was similar between these three cell lines (Fig. 2A,B). In HaCaT-Migr1-hPPARβ/δ cells, ligand activation of PPARβ/δ with GW0742 resulted in a dose-dependent increase in ANGPTL4 and ADRP mRNA that was markedly greater as compared to the effect of ligand in parent HaCaT keratinocytes, HaCaT-Migr1 vector control cells, and HaCaT-Migr1-hPPARγ cells (Fig. 2A,B). Similarly, increased expression of ANGPTL4 and ADRP mRNA was found in parent HaCaT keratinocytes, HaCaT-Migr1 vector control cells, and HaCaT-Migr1-hPPARβ/δ cells in response to 1.0 and 10.0 μM rosiglitazone, and this increase in expression was similar between these three cell lines (Fig. 2C,D). In HaCaT-Migr1-hPPARγ cells, ligand activation of PPARγ with rosiglitazone resulted in a marked increase in ANGPTL4 and ADRP mRNA as compared to the parent HaCaT keratinocytes, HaCaT-Migr1 vector control cells and HaCaT-Migr1-hPPARβ/δ cells (Fig. 2C,D). While the relative expression of PPARγ was lower in HaCaT-Migr1-hPPARβ/δ cells as compared to controls (Fig. 1B,C), this change was not reflected in the ability of rosiglitazone to activate PPARγ target gene expression as the efficacy of ANGPTL4 and ADRP mRNA induction by rosiglitazone was comparable between the parent HaCaT keratinocytes, HaCaT-Migr1 vector control cells and HaCaT-Migr1-hPPARβ/δ cells (Fig. 2C,D).
Fig. 2.
Effect of modulated PPARβ/δ and PPARγ expression on receptor-dependent transcriptional regulation. Quantitative real-time PCR (qPCR) for ANGPTL4 and ADRP mRNA in HaCaT keratinocytes, HaCaT-Migr1 vector control cells (Migr1), HaCaTMigr1-hPPARβ/δ cells (hPPARβ/δ) and HaCaT-Migr1-hPPARγ cells (hPPARγ) treated with the PPARβ/δ ligand GW0742 for 8 hours (A,B) or the PPARγ rosiglitazone (Rosi) for 24 hours (C,D). mRNA was isolated from the cells following indicated treatments and qPCR was performed to examine the expression of the mRNA encoding ANGPTL4 and ADRP, as normalized to the mRNA encoding GAPDH. Fold induction of ANGPTL4 and ADRP mRNA was calculated by normalization to vehicle control for each cell line. Data represents triplicate independent sample means ± SEM. Values with different letters are significantly different (P ≤ 0.05) using Bonferroni's multiple comparison.
3.3. Over-expression of PPARβ/δ does not increase FABP5 shuttling of atRA or PPARβ/δ ligands to PPARβ/δ
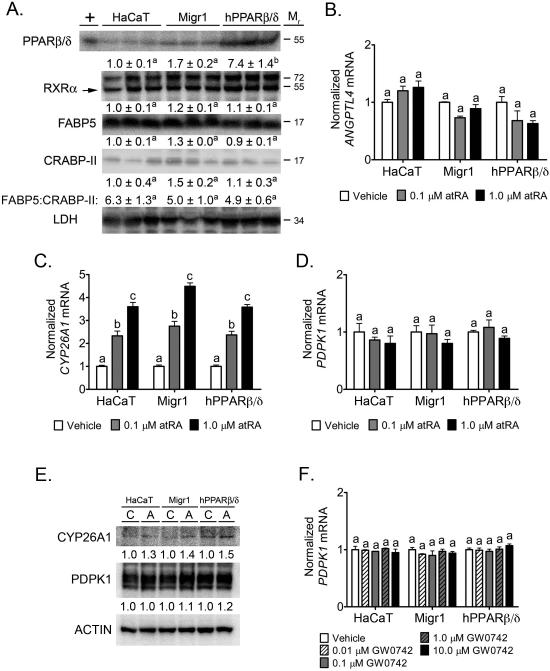
It was suggested that the biological effects of atRA or PPARβ/δ ligands could be modulated by differential shuttling of atRA or PPARβ/δ ligands between RAR and PPARβ/δ [15]. In this hypothetical model, shuttling of atRA towards RAR occurs in cells expressing more cellular retinoic acid binding protein II (CRABP-II) as compared to fatty acid binding protein 5 (FABP5), whereas atRA (or PPARβ/δ ligands) is shuttled toward PPARβ/δ in cells expressing more FABP5 as compared to CRABP-II leading to activation of PPARβ/δ, increased expression of target genes and PDPK1, and increased cell survival [15]. Expression of FABP5 was approximately 5-6X higher as compared to CRABP-II in parent HaCaT keratinocytes, HaCaT-Migr1 vector control cells, and HaCaT-Migr1-hPPARβ/δ cells (Fig. 3A). Expression of PPARβ/δ was more than 5-6X higher in HaCaT-Migr1-hPPARβ/δ as compared to parent HaCaT keratinocytes and HaCaT-Migr1 vector control cells (Fig. 3A). Expression of RXRα was similar in HaCaT keratinocytes, HaCaT-Migr1 vector control cells, and HaCaT-Migr1-hPPARβ/δ cells (Fig. 3A). HaCaT keratinocytes, HaCaT-Migr1 vector control cells, and HaCaT-Migr1-hPPARβ/δ cells cultured in the presence of atRA exhibited no change in expression of the PPAR target gene ANGPTL4 (Fig. 3B), while comparable dose dependent increases in mRNA expression of the RAR target gene CYP26A1 was observed in all three cell lines (Fig. 3C). The change in CYP26A1 mRNA was confirmed at the protein level by western blot analysis (Fig. 3E). Expression of PDPK1 mRNA or protein was unchanged in response to atRA (Fig. 3D,E) in HaCaT keratinocytes, HaCaT-Migr1 vector control cells, and HaCaT-Migr1-hPPARβ/δ cells. Further, expression of PDPK1 mRNA was unchanged in response GW0742 (Fig. 3F) in HaCaT keratinocytes, HaCaT-Migr1 vector control cells, and HaCaT-Migr1-hPPARβ/δ cells. Thus, despite over-expression of functional PPARβ/δ in cells expressing a high FABP5 to CRABP-II ratio, increased shuttling of atRA or the PPARβ/δ ligand GW0742 towards PPARβ/δ was not observed.
Fig. 3.
Effect of modulated PPARβ/δ expression on retinoic acid signaling. (A) Western blot analysis of PPARβ/δ, RXRα, FABP5 and CRABP-II. Lysates from HaCaT keratinocytes, HaCaT-Migr1 vector control cells (Migr1) or HaCaT-Migr1-hPPARβ/δ cells (hPPARβ/δ) were prepared and probed for PPARβ/δ, RXRα, FABP5, CRABP-II, and LDH expression. Cells were treated with retinoic acid for 8 hours and qPCR was performed to quantify mRNAs for the (B) PPARβ/δ-dependent target gene ANGPTL4, (C) the RAR-dependent target gene CYP26A1, and (D) the putative PPARβ/δ-dependent target gene PDPK1. Fold induction of mRNA was calculated from data normalized to GAPDH mRNA relative to vehicle control for each cell line. (E) Cells were treated with retinoic acid (1.0 μM) for 16 hours and western blot analysis was performed to quantify expression of CYP26A1 or PDPK1. (F) PDPK1 mRNA expression in cells following ligand activation of PPARβ/δ with GW0742 for 8 hours. Data represents triplicate independent sample means ± SEM (A-D,F). Values with different letters are significantly different (P ≤ 0.05) using Bonferroni's multiple comparison.
3.4 Effect of over-expression of PPARβ/δ and PPARγ on cell cycle progression in HaCaT keratinocytes
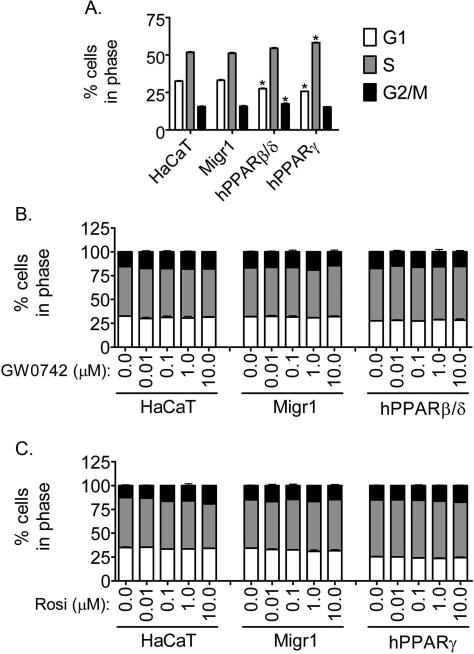
In response to ligand activation of PPARβ/δ or PPARγ, HaCaT keratinocytes or human keratinocytes exhibit modest inhibition of cell proliferation mediated in part by a small increase in apoptosis [19, 20], but these effects are not as strong as compared to more potent inhibitors of cell proliferation such as ultraviolet radiation [21]. However, it was also suggested that activating PPARβ/δ in HaCaT keratinocytes promotes cell survival [15]. To determine whether over-expression of PPARβ/δ or PPARγ could modulate cell cycle progression, flow cytometric analysis was performed. Over-expression of PPARβ/δ in HaCaT keratinocytes caused a decrease in the percentage of cells at the G1 phase and an increase in the percentage of cells at the G2/M phase of the cell cycle as compared to control HaCaT keratinocytes (Fig. 4A). Over-expression of PPARγ in HaCaT keratinocytes caused a decrease in the percentage of cells in the G1 phase and the percentage of cells in S phase was increased as compared to control HaCaT keratinocytes (Fig. 4A). Ligand activation of PPARβ/δ with GW0742 (0.01 – 10.0 μM) had no further effect on the distribution of cells in the different phases of the cell cycle in HaCaT keratinocytes, HaCaT-Migr1 vector control cells, and HaCaT-Migr1-hPPARβ/δ cells (Fig. 4B). Similarly, ligand activation of PPARγ with rosiglitazone (0.01 – 25.0 μM) had no further effect on the distribution of cells in the different phases of the cell cycle in HaCaT keratinocytes, HaCaT-Migr1 vector control cells, and HaCaT-Migr1-hPPARγ cells (Fig. 4C, data not shown for 25 μM).
Fig. 4.
Effect of over-expression of PPARβ/δ and PPARγ on cell cycle progression in HaCaT keratinocytes. Cell cycle progression was examined in HaCaT keratinocytes, HaCaT-Migr1 vector control cells (Migr1), HaCaT-Migr1-hPPARβ/δ cells (hPPARβ/δ) and HaCaT-Migr1-hPPARγ cells (hPPARγ) by flow cytometry. (A) Effect of over-expression of hPPARβ/δ or hPPARγ on cell cycle progression. (B) Effect of ligand activation of PPARβ/δ on cell cycle progression. Cells were treated for 24 hours with 0.01 – 10 μM GW0742. (C) Effect of ligand activation of PPARγ on cell cycle progression. Cells were treated for 24 hours with 0.01 – 10 μM rosiglitazone (Rosi). Values represent the mean ± SEM. from three independent samples. *Significantly different than control, P ≤ 0.05.
3.5. Effect of PPARβ/δ and PPARγ on induced apoptosis in HaCaT keratinocytes
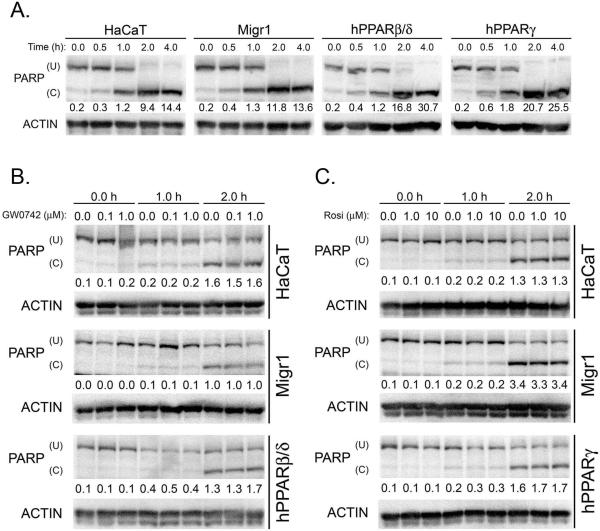
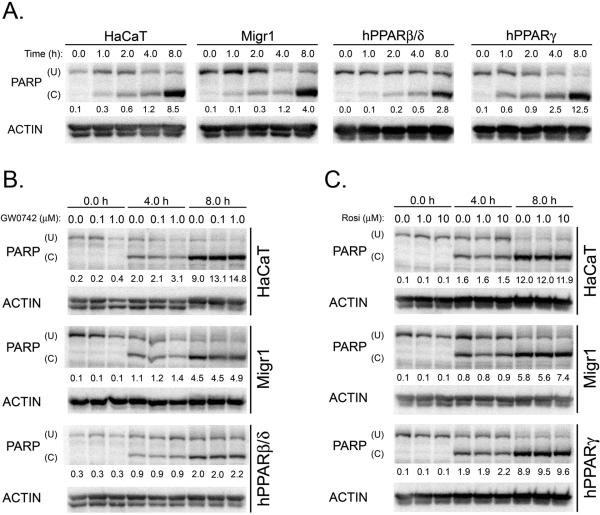
While there is evidence that ligand activation of PPARβ/δ in HaCaT keratinocytes increases apoptosis [19], it was also suggested that activating PPARβ/δ in HaCaT keratinocytes promotes cell survival [15]. Further, docosahexaenoic acid, which can activate PPARβ/δ [22], can enhance UV-induced apoptosis in HaCaT keratinocytes [23]. Thus, the effect of staurosporine-induced or UVB-induced apoptosis was examined in HaCaT keratinocytes, HaCaT-Migr1 vector control cells, HaCaT-Migr1-hPPARβ/δ cells and HaCaT-Migr1-hPPARγ cells. Maximal PARP cleavage occurred by four hours post-staurosporine treatment in all four cells lines (Fig. 5A). Over-expression of PPARβ/δ or PPARγ in HaCaT keratinocytes had no effect on staurosporine-induced PARP cleavage (Fig. 5A). Ligand activation of PPARβ/δ with GW0742 (0.1 or 1.0 μM) had no effect on staurosporine-induced PARP cleavage in either HaCaT keratinocytes, HaCaT-Migr1 vector control cells or HaCaT-Migr1-hPPARβ/δ cells (Fig. 5B). Moreover, ligand activation of PPARγ with rosiglitazone (1.0 or 10.0 μM) did not influence staurosporine-induced PARP cleavage in either HaCaT keratinocytes, HaCaT-Migr1 vector control cells or HaCaT-Migr1-hPPARγ cells (Fig. 5C). Maximal PARP cleavage occurred by 8 hours post-UVB treatment in all four cells lines (Fig. 6A). UVB-induced PARP cleavage was not different between HaCaT keratinocytes, HaCaT-Migr1 vector control cells, HaCaT-Migr1-hPPARβ/δ cells or HaCaT-Migr1-hPPARγ cells (Fig. 6A). Ligand activation of PPARβ/δ with GW0742 (0.1 or 1.0 μM) had no effect on UVB-induced PARP cleavage in either HaCaT keratinocytes, HaCaT-Migr1 vector control cells or HaCaT-Migr1-hPPARβ/δ cells (Fig. 6B). Similarly, ligand activation of PPARγ with rosiglitazone (1.0 or 10.0 μM) did not influence UVB-induced PARP cleavage in either HaCaT keratinocytes, HaCaT-Migr1 vector control cells or HaCaT-Migr1-hPPARγ cells (Fig. 6C).
Fig. 5.
Effect of over-expression and ligand activation of PPARβ/δ and PPARγ in HaCaT keratinocytes on staurosporine-induced PARP cleavage. (A) Quantitative Western blotting for PARP was performed using cell lysates from HaCaT keratinocytes, HaCaT-Migr1 vector control cells (Migr1), HaCaT-Migr1-hPPARβ/δ cells (hPPARβ/δ) and HaCaT-Migr1-hPPARγ cells (hPPARγ) treated with staurosporine (0.5 μM) for the indicated times. (B) Effect of ligand activation of PPARβ/δ on staurosporine-induced PARP cleavage. Cells were treated with GW0742 (0.1 or 1.0 μM) for the indicated times and quantitative Western blotting was performed. (C) Effect of ligand activation of PPARγ on staurosporine-induced PARP cleavage. Cells were treated with rosiglitazone (1 or 10 μM) for the indicated times and quantitative Western blotting was performed. (U) indicates uncleaved PARP and (C) indicates cleaved PARP. The ratio of C to U PARP was calculated and is presented below each sample.
Fig. 6.
Effect of over-expression and ligand activation of PPARβ/δ and PPARγ in HaCaT keratinocytes on UVB-induced PARP cleavage. (A) Quantitative Western blotting for PARP was performed using cell lysates from HaCaT keratinocytes, HaCaTMigr1 vector control cells (Migr1), HaCaT-Migr1-hPPARβ/δ cells (hPPARβ/δ) and HaCaT-Migr1-hPPARγ cells (hPPARγ) irradiated with UVB for the indicated times. (B) Effect of ligand activation of PPARβ/δ on UVB-induced PARP cleavage. Cells were treated with GW0742 (0.1 or 1.0 μM) for the indicated times and quantitative Western blotting was performed. (C) Effect of ligand activation of PPARγ on UVB-induced PARP cleavage. Cells were treated with rosiglitazone (1 or 10 μM) for the indicated times and quantitative Western blotting was performed. (U) indicates uncleaved PARP and (C) indicates cleaved PARP. The ratio of C to U PARP was calculated and is presented below each sample.
3.6. Inhibition of UVB-induced expression of cytokines in HaCaT keratinocytes by PPARβ/δ and PPARγ
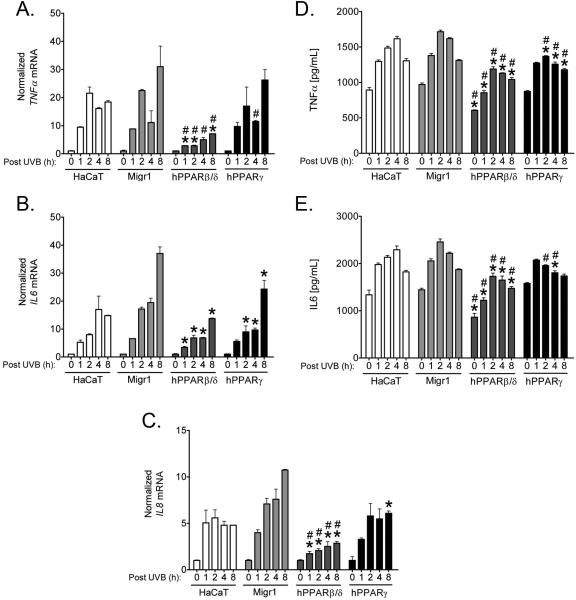
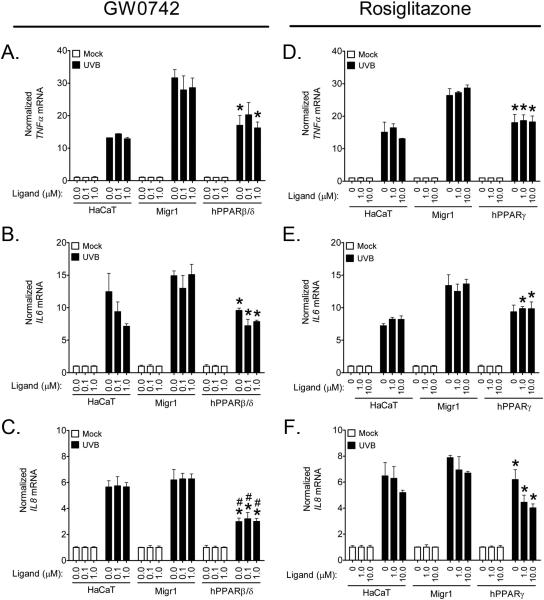
Since PPARβ/δ and PPARγ can inhibit inflammatory signaling through both receptor-dependent and/or ligand-dependent mechanisms (reviewed in [3]), expression of mRNAs encoding inflammatory cytokines was examined in HaCaT keratinocytes, HaCaT-Migr1 vector control cells, HaCaT-Migr1-hPPARβ/δ cells and HaCaT-Migr1-hPPARγ cells. UVB-induced expression of TNFα, IL6 and IL8 mRNA was markedly repressed in HaCaT-Migr1-hPPARβ/δ cells as compared to either HaCaT keratinocytes and/or HaCaT-Migr1 vector control cells (Fig. 7). Similar, but less striking repression of UVB-induced expression of these mRNAs encoding inflammatory cytokines in HaCaTMigr1-hPPARγ cells as compared to HaCaT-Migr1 vector control cells was also observed (Fig. 7). These changes in mRNA expression were reflected by the concentrations of TNFα and IL6 in the culture medium (Fig. 7D,E). Ligand activation of PPARβ/δ with GW0742 did not further repress the UVB-induced expression of TNFα, IL6 or IL8 mRNA further than that found with PPARβ/δ over-expression alone (Fig. 8A-C). As compared to HaCaT-Migr1 vector control cells, UVB-induced expression of TNFα, IL6 or IL8 mRNA was lower in HaCaT-Migr1-hPPARγ cells, but treatment with rosiglitazone did not further reduce expression (Fig. 8D-F).
Fig. 7.
Effect of over-expression of PPARβ/δ and PPARγ in HaCaT keratinocytes on UVB-induced expression and secretion of inflammatory cytokines. HaCaT keratinocytes, HaCaT-Migr1 vector control cells (Migr1), HaCaT-Migr1-hPPARβ/δ cells (hPPARβ/δ) and HaCaT-Migr1-hPPARγ cells (hPPARγ) were irradiated with UVB for the indicated times and mRNA expression of the inflammatory cytokines (A) TNFα, (B) IL6, and (C) IL8, or the concentration of culture medium (D) TNFα and (E) IL6, was examined. Fold induction of cytokine mRNA was calculated by normalization to the non-UVB control for each cell line. Data represents triplicate independent sample means ± SEM. *statistically different as compared to the HaCaT-Migr1 vector control cell line by Student's T-test at each time point (P < 0.05). #statistically different compared to the HaCaT keratinocytes by Student's T-test at each time point (P < 0.05).
Fig. 8.
Effect of ligand activation of PPARβ/δ and PPARγ on UVB-induced expression of inflammatory cytokines in HaCaT keratinocytes over-expressing of PPARβ/δ and PPARγ. HaCaT keratinocytes, HaCaT-Migr1 vector control cells (Migr1), HaCaT-Migr1-hPPARβ/δ cells (hPPARβ/δ) and HaCaT-Migr1-hPPARγ cells (hPPARγ) were treated with either 0, 0.1 or 1.0 μM GW0742, or 0, 1 or 10 μM rosiglitazone and irradiated with UVB, and mRNA expression of the inflammatory cytokines (A,D) TNFα, (B,E) IL6, and (C,F) IL8 was examined 4 hours post-UVB treatment. Fold induction of cytokine mRNA was calculated by normalization to the non-UVB control for each cell line. Data represents triplicate independent sample means ± SEM. *statistically different as compared to the HaCaT-Migr1 vector control cell line by Student's T-test at each time point (P < 0.05). #statistically different compared to the HaCaT keratinocytes by Student's T-test at each time point (P < 0.05).
4. Discussion
Results from the present study establish the first stable, gain-of-function model for PPARβ/δ and PPARγ in HaCaT keratinocytes, a cell type where PPARβ/δ and PPARγ are known to play an important role in differentiation, cell proliferation and apoptosis [24] as reviewed in [5, 6, 8, 40]. The Migr1 retroviral system [14] is an ideal approach to over-express PPARs and isolate cell populations with high expression due to the ability to sort based on eGFP expression. Stable over-expression of PPARβ/δ and PPARγ resulted in HaCaT keratinocytes that exhibited markedly enhanced expression and function of these nuclear receptors. Thus, these cells will be useful for future studies to delineate the functional role of PPARβ/δ and PPARγ in human keratinocytes, and both complement and improve alternative approaches including the use of knockdown/knockout models and highly specific PPAR ligands.
Earlier studies suggested that PPARβ/δ might cross-talk with PPARγ [27, 28]. For example, over-expression of PPARβ/δ in the monkey CV1 or COS1 cell lines represses PPARγ ligand-induced reporter activity [27, 28]. Further, over-expression of PPARβ/δ in human NIH 3T3 cells represses PPARγ ligand-induced expression of PPARγ-dependent target gene expression [25]. Thus, the present study examined this hypothetical cross-talk by comparing the effect of ligand activation of PPARγ in HaCaT keratinocytes over-expressing PPARβ/δ. In contrast to previous reports [27, 28], PPARγ target gene expression was not repressed when expression of PPARβ/δ was markedly increased in HaCaT-Migr1-hPPARβ/δ cells. This suggests that over-expression of PPARβ/δ does not interfere with PPARγ-dependent transcription. This is consistent with the observation that PPARγ ligand-induced expression of PPARγ target genes required for adipocyte differentiation is not enhanced in _Pparβ/δ_-null adipocytes [26]. In fact, in this model, expression of PPARβ/δ potentiated PPARγ-dependent adipocyte differentiation [26], suggesting that PPARβ/δ alters ligand-dependent function of PPARγ through an additive mechanism. The finding from the present work that PPARβ/δ does not interfere with PPARγ-dependent transcription is also consistent with previous in vivo analysis of _Pparβ/δ_-null mice where ligand activation of PPARγ in _Pparβ/δ_-null mouse colon (a tissue known to express PPARβ/δ and PPARγ at very high levels [42, 43]) does not lead to enhanced expression of PPARγ target genes [27]. These observations suggest that physiologically, it is unlikely that PPARβ/δ interferes with PPARγ transcription as suggested by in vitro studies. Since previous studies suggesting that PPARβ/δ interferes with PPARγ transcription based this interpretation in part on data from reporter constructs [27, 28], it remains a possibility that the observed differences were due to receptor-dependent modulation of transcriptional events observed with plasmid constructs that lack chromatin structure associated with endogenous target genes. Another possible explanation for the differences between the present study showing no change in PPARγ target gene expression when PPARβ/δ is over-expressed, and others showing inhibition of PPARγ target gene expression when PPARβ/δ is over-expressed, is that over-expression of PPARβ/δ could deplete co-effectors required by PPARγ for transcription (e.g. co-activators) in COS1 or CV1 cells while this does not occur in HaCaT keratinocytes. It is thus important to note that the level of over-expression of PPARβ/δ in these models may not reflect physiological achievable concentrations. If this is true, then the observed inhibition of PPARγ-dependent transcription may be an artifact of the in vitro model that does model normal physiology. This idea is supported by the in vivo analysis showing that disruption of expression of PPARβ/δ in tissues that express very high levels of both PPARβ/δ and PPARγ does not alter PPARγ ligand inducibility of PPARγ target gene expression [26].
The hypothetical signaling proposed by others suggesting that the biological effects of atRA can be modulated by differential shuttling between RAR and PPARβ/δ [15] based on the expression of FABP5 and CRABP-II led to the examination of this signaling in HaCaT keratinocytes in the present study. HaCaT keratinocytes are ideal for this purpose because they express high levels of FABP5 as compared to CRABP-II, and HaCaT is the same cell line used to suggest this hypothetical signaling [15]. As noted above, the potency of ligand activation and efficacy of PPARβ/δ target expression was enhanced in HaCaT keratinocytes over-expressing PPARβ/δ in response to a highly specific PPARβ/δ ligand. However, despite the fact that atRA was capable of activating RAR in control HaCaT keratinocytes, HaCaT-Migr1 vector control cells, and HaCaT-Migr1-hPPARβ/δ cells (as shown by increased expression of CYP26A1), no evidence of enhanced shuttling of atRA towards activating PPARβ/δ and increasing PPARβ/δ target gene expression was observed. These results are consistent with previous studies showing that: 1) atRA does not increase Angptl4 mRNA expression in HaCaT keratinocytes or mouse keratinocytes [19], 2) atRA does not cause PPARβ/δ-dependent anti-apoptotic activities in mouse keratinocytes [19], 3) atRA does not cause an increase in PPARβ/δ-dependent reporter assays [28], and 4) atRA does not cause association of PPARβ/δ with co-activators based on a time-resolved fluorescence resonance energy transfer assay [28]. The present studies advance the former findings because even when expression of PPARβ/δ is markedly increased in HaCaT keratinocytes where the FABP5:CRABP-II is high, atRA does not activate PPARβ/δ; moreover, atRA or GW0742 do not increase expression of PDPK1, and GW0742 does not attenuate UVB-induced PARP cleavage. Collectively, these observations and results from the present study demonstrate that no shuttling of atRA towards activating PPARβ/δ occurs in cells expressing high levels of FABP5 as compared to CRABP-II, even when PPARβ/δ expression is markedly increased. This reinforces the notion that the hypothesis that the biological effects of atRA can be modulated by differential shuttling between RAR and PPARβ/δ based on the expression of FABP5 and CRABP-II [15], should be rigorously re-examined.
Inhibition of HaCaT cell proliferation is found in response to ligand activation of PPARβ/δ, but these changes are modest and only occur after 72 hours of treatment [19]. Over-expression of PPARβ/δ in HaCaT keratinocytes did not markedly alter cell cycle progression as compared to controls, but a decrease in the percentage of cells in the G1 phase and an increase in the percentage of cells at the G2/M phase was noted; an effect that was not further influenced by ligand activation of PPARβ/δ. This is in contrast to results observed in N/TERT-1 keratinocytes where ligand activation of PPARβ/δ inhibits cell cycle progression by increasing the percentage of cells in the G1 phase and decreasing the percentage of cells in S phase [29]. The reason why an increase in the percentage of cells in the G2/M phase and a decrease in the percentage of cells in the G1 phase of the cell cycle was found in HaCaT-Migr1-hPPARβ/δ keratinocytes while an increase in the percentage of cell in the G1 phase of the cell cycle was found in N/TERT-1 keratinocytes cannot be determined from these studies. However, this could be due in part to differences in the genetic alterations causing immortalization of HaCaT (p53) and N/TERT-1 keratinocytes (hTERT), or the fact that HaCaT keratinocytes are resistant to induced growth inhibition [21, 30, 31].
Over-expression of PPARγ also caused a decrease in the percentage of cells in the G1 phase of the cell cycle, but this change was associated with an increase in the percentage of cells in S phase of the cells cycle. Ligand activation of PPARγ did not further influence these kinetics. Previous studies have shown that activating PPARγ in mouse and human keratinocytes can inhibit cell proliferation [36, 37, 49]. Results from the present studies are inconsistent with these studies, but could also be influenced by the relative resistance to growth inhibition observed in HaCaT keratinocytes [21]. However, it is of interest to note that expression of PPARγ is increased during the resolution phase of wound healing [32], during which a transition between active cell proliferation and apoptosis is observed. Thus, the finding that over-expression of PPARγ in HaCaT keratinocytes caused a modest increase in the percentage of cells in the S phase of the cell cycle may reflect this model. Further studies with the HaCaTMigr1-hPPARγ keratinocytes may be suitable to examine this in greater detail. Alternatively, the increase in the percentage of cells in the S phase resulting from over-expression of PPARγ could be influenced by the mutant p53 gene in HaCaT keratinocytes [31].
Ligand activation of PPARβ/δ in HaCaT keratinocytes can modestly increase apoptosis [19], but it was also suggested that activating PPARβ/δ in HaCaT keratinocytes promotes cell survival [15]. Results from the present studies demonstrate that over-expression of PPARβ/δ or PPARγ did not markedly alter PARP cleavage induced by either staurosporine or UVB. Further, ligand activation of either PPARβ/δ or PPARγ, in the presence or absence of over-expressed receptor, did not influence either staurosporine- or UVB-induced PARP cleavage. These results are in contrast to studies suggesting that activating PPARβ/δ causes decreased expression of PTEN, increased expression of ILK and PDPK1 causing phosphorylation of AKT and enhanced cell survival in HaCaT keratinocytes [33]. If activating PPARβ/δ caused anti-apoptotic activity, then reduced PARP cleavage would be expected, and this was not found in the present study. These results are however consistent with a recent study showing that ligand activation of PPARβ/δ did not prevent non-steroidal anti-inflammatory drug (NSAID)-induced apoptosis or increase the number of viable cells in human colon cancer cells [34]. Moreover, the lack of change in PDPK1 expression following ligand activation of PPARβ/δ in the presence or absence of over-expressed PPARβ/δ in HaCaT keratinocytes is also consistent with other studies where no change in expression of PTEN, PDPK1, ILK and/or phosphorylated AKT were found in mouse keratinocytes, N/TERT-1 keratinocytes or HaCaT keratinocytes [30, 54]. Collectively, these results demonstrate that activating PPARβ/δ, with or without over-expression of PPARβ/δ in HaCaT keratinocytes does not prevent staurosporine- or UVB-induced apoptosis.
Expression of inflammatory cytokine expression was also examined in HaCaT keratinocytes because PPARβ/δ and PPARγ can inhibit inflammatory signaling through both receptor-dependent and/or ligand-dependent mechanisms (reviewed in [3]). Over-expression of PPARβ/δ or PPARγ both reduced UVB-induced mRNA expression of TNFα, IL6 and IL8, but this effect was more striking in cells over-expressing PPARβ/δ. Consistent with these observations, UVB-induced secretion of TNFα and IL6 was also repressed in cells over-expressing PPARβ/δ or PPARγ. Ligand activation of either PPARβ/δ or PPARγ did not modulate the receptor-dependent repression of UVB-induced expression of TNFα, IL6 or IL8 mRNA. This observation is similar to PPARβ/δ-dependent repression of dextran sodium sulfate (DSS)-induced colitis where DSS-induced colitis is exacerbated in _Pparβ/δ_-null mice as compared to wild-type mice, but ligand activation of PPARβ/δ did not further influence DSS-induced colitis [35]. Since receptor-dependent repression of UVB-induced expression of inflammatory cytokines in HaCaT keratinocytes is not influenced by exogenous ligands, it remains possible that high affinity endogenous ligands prevent any further modulation due to differences in relative receptor affinity. It is also possible that PPAR-dependent repression of UVB-induced expression of inflammatory cytokines does not require ligand activation and is mediated through mechanisms facilitated by PPARs interacting with other transcription factors such as the p65 subunit of NF-κB thereby attenuating NF-κB-dependent signaling. Further studies are needed to examine this hypothesis and the HaCaT-Migr1-hPPARβ/δ and the HaCaT-Migr1-hPPARγ keratinocytes are excellent models for this purpose.
In conclusion, results from the present study demonstrate the feasibility of the Migr1 system to over-express PPARs in HaCaT keratinocytes in order to generate a model to delineate the functional roles of PPARs in these cells. This approach is likely suitable for other cell lines as well. These models will complement knockout and knockdown approaches and provide an alternative approach to determine receptor-dependent and ligand-dependent function of receptors with great promise for therapeutic targets.
Research Highlights.
- The Migr1-PPAR retroviral system allows for efficient sorting of cell over-expressing PPARs.
- HaCaT cells over-expressing PPARs exhibit enhanced ligand-induced gene expression.
- No change in PPARβ/δ target gene expression by retinoic acid occurs in HaCaT cells over-expressing PPARβ/δ.
- Over-expression of PPARβ/δ does not interfere with PPARγ ligand induced target gene expression.
- Over-expression of PPARβ/δ inhibits pro-inflammatory cytokine expression and secretion.
Acknowledgements
We gratefully acknowledge Drs. Andrew N. Billin and Timothy M. Willson for providing GW0742, Dr. Gary H. Perdew for the use of a fluorescent microscope, and Susan Magargee and Nicole Bem from the Center for Quantitative Cell Analysis at the Huck Institutes of Life Sciences of The Pennsylvania State University for their technical support in sorting the fluorescent cells. This work supported in part by the National Institutes of Health Grants CA124533, CA126826, CA141029, CA140369 (JMP) and the National Cancer Institute Intramural Research Program ZIABC005561, ZIABC005562, ZIABC005708 (FJG).
Abbreviations
ADRP
adipocyte differentiation-related protein
atRA
all-trans retinoic acid
ANGPTL4
angiopoietin-like protein 4
CRABP-II
cellular retinoic acid binding protein II
CYP26A1
cytochrome P450 26A1
DMEM
Dulbecco's minimal essential medium
DMSO
dimethylsulfoxide
eGFP
enhanced green fluorescent protein
ELISA
enzyme-linked immunosorbent assay
FABP5
fatty acid binding protein 5
FBS
fetal bovine serum
GAPDH
glyceraldehyde 3-phosphate dehydrogenase
IL6
interleukin 6
IL8
interleukin 8
IRES
internal ribosome entry site
NSAID
non-steroidal anti-inflammatory drug
PDPK1
3-phosphoinositide-dependent-protein kinase 1
PPAR
peroxisome proliferator-activated receptor
PARP
poly (ADP-ribose) polymerase
qPCR
quantitative real-time polymerase chain reaction
RAR
retinoic acid receptor
RXR
retinoid X receptor
TNFα
tumor necrosis factor α
UVB
ultraviolet B
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions: MB designed and performed the experiments, interpreted the data and wrote the manuscript. CK performed experiments and reviewed the manuscript. PPA helped make the Migr1-hPPARβ/δ construct, performed experiments and reviewed the manuscript. BZ helped make the Migr1-hPPARβ/δ and Migr1-hPPARγ construct and reviewed the manuscript. TSL helped make the Migr1-hPPARγ construct and reviewed the manuscript. CL performed experiments and reviewed the manuscript. FJG and JMP designed experiments, interpreted the data and wrote the manuscript.
References
- 1.Tanaka T, Yamamoto J, Iwasaki S, Asaba H, Hamura H, Ikeda Y, Watanabe M, Magoori K, Ioka RX, Tachibana K, Watanabe Y, Uchiyama Y, Sumi K, Iguchi H, Ito S, Doi T, Hamakubo T, Naito M, Auwerx J, Yanagisawa M, Kodama T, Sakai J. Proc Natl Acad Sci U S A. 2003;100(26):15924–15929. doi: 10.1073/pnas.0306981100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oliver WR, Jr., Shenk JL, Snaith MR, Russell CS, Plunket KD, Bodkin NL, Lewis MC, Winegar DA, Sznaidman ML, Lambert MH, Xu HE, Sternbach DD, Kliewer SA, Hansen BC, Willson TM. A selective peroxisome proliferator-activated receptor δ agonist promotes reverse cholesterol transport. PNAS. 2001:5306–5311. doi: 10.1073/pnas.091021198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kilgore KS, Billin AN. Curr Opin Investig Drugs. 2008;9(5):463–469. [PubMed] [Google Scholar]
- 4.Peraza MA, Burdick AD, Marin HE, Gonzalez FJ, Peters JM. Toxicol. Sci. 2006;90(2):269–295. doi: 10.1093/toxsci/kfj062. [DOI] [PubMed] [Google Scholar]
- 5.Peters JM, Gonzalez FJ. Biochim Biophys Acta. 2009;1796(2):230–241. doi: 10.1016/j.bbcan.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Palkar PS, Borland MG, Naruhn S, Ferry CH, Lee C, Sk UH, Sharma AK, Amin S, Murray IA, Anderson CR, Perdew GH, Gonzalez FJ, Muller R, Peters JM. Mol Pharmacol. 2010 doi: 10.1124/mol.110.065508. doi:10.1124/mol.110.065508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Girroir EE, Hollingshead HE, He P, Zhu B, Perdew GH, Peters JM. Biochem Biophys Res Commun. 2008;371(3):456–461. doi: 10.1016/j.bbrc.2008.04.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berglund L, Bjorling E, Oksvold P, Fagerberg L, Asplund A, Szigyarto CA, Persson A, Ottosson J, Wernerus H, Nilsson P, Lundberg E, Sivertsson A, Navani S, Wester K, Kampf C, Hober S, Ponten F, Uhlen M. Mol Cell Proteomics. 2008;7(10):2019–2027. doi: 10.1074/mcp.R800013-MCP200. [DOI] [PubMed] [Google Scholar]
- 9.Kim DJ, Akiyama TE, Harman FS, Burns AM, Shan W, Ward JM, Kennett MJ, Gonzalez FJ, Peters JM. J. Biol. Chem. 2004;279(22):23719–23727. doi: 10.1074/jbc.M312063200. [DOI] [PubMed] [Google Scholar]
- 10.Barak Y, Liao D, He W, Ong ES, Nelson MC, Olefsky JM, Boland R, Evans RM. PNAS. 2002;99(1):303–308. doi: 10.1073/pnas.012610299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang D, Wang H, Guo Y, Ning W, Katkuri S, Wahli W, Desvergne B, Dey SK, Dubois RN. Proc Natl Acad Sci U S A. 2006;103(50):19069–19074. doi: 10.1073/pnas.0607948103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shearer BG, Hoekstra WJ. Current Medicinal Chemistry. 2003;10(4):267–280. doi: 10.2174/0929867033368295. [DOI] [PubMed] [Google Scholar]
- 13.Sznaidman ML, Haffner CD, Maloney PR, Fivush A, Chao E, Goreham D, Sierra ML, LeGrumelec C, Xu HE, Montana VG, Lambert MH, Willson TM, Oliver WR, Sternbach DD. Bioorganic & Medicinal Chemistry Letters. 2003;13(9):1517–1521. doi: 10.1016/s0960-894x(03)00207-5. [DOI] [PubMed] [Google Scholar]
- 14.Pear WS, Miller JP, Xu L, Pui JC, Soffer B, Quackenbush RC, Pendergast AM, Bronson R, Aster JC, Scott ML, Baltimore D. Blood. 1998;92(10):3780–3792. [PubMed] [Google Scholar]
- 15.Schug TT, Berry DC, Shaw NS, Travis SN, Noy N. Cell. 2007;129(4):723–733. doi: 10.1016/j.cell.2007.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chawla A, Lee CH, Barak Y, He W, Rosenfeld J, Liao D, Han J, Kang H, Evans RM. PNAS. 2003;100(3):1268–1273. doi: 10.1073/pnas.0337331100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gupta RA, Brockman JA, Sarraf P, Willson TM, DuBois RN. J. Biol. Chem. 2001;276(32):29681–29687. doi: 10.1074/jbc.M103779200. [DOI] [PubMed] [Google Scholar]
- 18.Mandard S, Zandbergen F, Tan NS, Escher P, Patsouris D, Koenig W, Kleemann R, Bakker A, Veenman F, Wahli W, Muller M, Kersten S. J Biol Chem. 2004;279(33):34411–34420. doi: 10.1074/jbc.M403058200. [DOI] [PubMed] [Google Scholar]
- 19.Borland MG, Foreman JE, Girroir EE, Zolfaghari R, Sharma AK, Amin S, Gonzalez FJ, Ross AC, Peters JM. Mol Pharmacol. 2008;74(5):1429–1442. doi: 10.1124/mol.108.050609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Westergaard M, Henningsen J, Svendsen ML, Johansen C, Jensen UB, Schroder HD, Kratchmarova I, Berge RK, Iversen L, Bolund L, Kragballe K, Kristiansen K. J. Invest. Dermatol. 2001;116(5):702–712. doi: 10.1046/j.1523-1747.2001.01329.x. [DOI] [PubMed] [Google Scholar]
- 21.Henseleit U, Rosenbach T, Kolde G. Arch Dermatol Res. 1996;288(11):676–683. doi: 10.1007/BF02505277. [DOI] [PubMed] [Google Scholar]
- 22.Forman BM, Chen J, Evans RM. PNAS. 1997;94(9):4312–4317. doi: 10.1073/pnas.94.9.4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Serini S, Donato V, Piccioni E, Trombino S, Monego G, Toesca A, Innocenti I, Missori M, De Spirito M, Celleno L, Fasano E, Ranelletti FO, Calviello G. J Nutr Biochem. 2011 doi: 10.1016/j.jnutbio.2010.08.004. 10.1016/j.nutbio.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 24.Ellis CN, Varani J, Fisher GJ, Zeigler ME, Pershadsingh HA, Benson SC, Chi Y, Kurtz TW. Arch. Derm. 2000;136(5):609–616. doi: 10.1001/archderm.136.5.609. [DOI] [PubMed] [Google Scholar]
- 25.Shi Y, Hon M, Evans RM. PNAS. 2002;99(5):2613–2618. doi: 10.1073/pnas.052707099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsusue K, Peters JM, Gonzalez FJ. PPARβ/δ; potentiates PPARγ-stimulated adipocyte differentiation. FASEB J. 2004:04–1944fje. doi: 10.1096/fj.04-1944fje. [DOI] [PubMed] [Google Scholar]
- 27.Marin HE, Peraza MA, Billin AN, Willson TM, Ward JM, Kennett MJ, Gonzalez FJ, Peters JM. Cancer Research. 2006;66(8):4394–4401. doi: 10.1158/0008-5472.CAN-05-4277. [DOI] [PubMed] [Google Scholar]
- 28.Rieck M, Meissner W, Ries S, Muller-Brusselbach S, Muller R. Mol Pharmacol. 2008;74(5):1269–1277. doi: 10.1124/mol.108.050625. [DOI] [PubMed] [Google Scholar]
- 29.Burdick AD, Bility MT, Girroir EE, Billin AN, Willson TM, Gonzalez FJ, Peters JM. Cell Signal. 2007;19(6):1163–1171. doi: 10.1016/j.cellsig.2006.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dickson MA, Hahn WC, Ino Y, Ronfard V, Wu JY, Weinberg RA, Louis DN, Li FP, Rheinwald JG. Mol Cell Biol. 2000;20(4):1436–1447. doi: 10.1128/mcb.20.4.1436-1447.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lehman TA, Modali R, Boukamp P, Stanek J, Bennett WP, Welsh JA, Metcalf RA, Stampfer MR, Fusenig N, Rogan EM, et al. Carcinogenesis. 1993;14(5):833–839. doi: 10.1093/carcin/14.5.833. [DOI] [PubMed] [Google Scholar]
- 32.Kapoor M, Kojima F, Yang L, Crofford LJ. Prostaglandins Leukot. Essent. Fatty Acids. 2007;76(2):103–112. doi: 10.1016/j.plefa.2006.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Di-Poi N, Tan NS, Michalik L, Wahli W, Desvergne B. Molecular Cell. 2002;10(4):721–733. doi: 10.1016/s1097-2765(02)00646-9. [DOI] [PubMed] [Google Scholar]
- 34.Foreman JE, Chang W-C, Palkar PS, Zhu B, Borland MG, Williams JL, Kramer LR, Clapper ML, Gonzalez FJ, Peters JM. Mol Carcinog. 2011 doi: 10.1002/mc.20757. DOI: 10.1002/mc.20757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hollingshead HE, Morimura K, Adachi M, Kennett MJ, Billin AN, Willson TM, Gonzalez FJ, Peters JM. Dig Dis Sci. 2007;52(11):2912–2919. doi: 10.1007/s10620-006-9644-9. [DOI] [PubMed] [Google Scholar]