A novel small-molecule disrupts Stat3 SH2 domain-phosphotyrosine interactions and Stat3-dependent tumor processes (original) (raw)
. Author manuscript; available in PMC: 2011 Oct 6.
Published in final edited form as: Biochem Pharmacol. 2010 Jan 11;79(10):1398–1409. doi: 10.1016/j.bcp.2010.01.001
Abstract
The molecular modeling of the phosphotyrosine (pTyr)-SH2 domain interaction in the Stat3:Stat3 dimerization, combined with in silico structural analysis of the Stat3 dimerization disruptor, S3I-201, has furnished a diverse set of analogs. We present evidence from in vitro biochemical and biophysical studies that the structural analog, S3I-201.1066 directly interacts with Stat3 or the SH2 domain, with an affinity (KD) of 2.74 nM, and disrupts the binding of Stat3 to the cognate pTyr-peptide, GpYLPQTV-NH2, with an IC50 of 23 μM. Moreover, S3I-201.1066 selectively blocks the association of Stat3 with the epidermal growth factor receptor (EGFR), and inhibits Stat3 tyrosine phosphorylation and nuclear translocation in EGF-stimulated mouse fibroblasts. In cancer cells that harbor aberrant Stat3 activity, S3I-201.1066 inhibits constitutive Stat3 DNA-binding and transcriptional activities. By contrast, S3I-201.1066 has no effect on Src activation or the EGFR-mediated activation of the Erk1/2MAPK pathway. S3I-201.1066 selectively suppresses the viability, survival, and malignant transformation of the human breast and pancreatic cancer lines and the v-Src-transformed mouse fibroblasts harboring persistently-active Stat3. Treatment with S3I-201.1066 of malignant cells harboring aberrantly-active Stat3 down-regulated the expression of c-Myc, Bcl-xL, Survivin, the matrix metalloproteinase 9, and VEGF. The in vivo administration of S3I-201.1066 induced significant antitumor response in mouse models of human breast cancer, which correlates with the inhibition of constitutively-active Stat3 and the suppression of known Stat3-regulated genes. Our studies identify a novel small-molecule that binds with a high affinity to Stat3, blocks Stat3 activation and function, and thereby induces antitumor response in human breast tumor xenografts harboring persistently-active Stat3.
Keywords: Stat3, S3I-201, S3I-201.1066, small-molecule inhibitor, antitumor effects, antitumor cell effects
1. Introduction
Signal transduction proteins have increased importance in carcinogenesis and tumor formation and represent attractive targets for the development of novel anticancer therapeutics. The Signal Transducer and Activator of Transcription (STAT) family of proteins are cytoplasmic transcription factors with important roles in the responses to cytokines and growth factors, including promoting cell growth and differentiation, and inflammation and immune responses [1, 2]. Normal STATs activation is initiated by the phosphorylation of a critical tyrosine residue upon the binding of cytokines or growth factors to cognate receptors. STATs phosphorylation is induced by growth factor receptor tyrosine kinases, or cytoplasmic tyrosine kinases, such as Janus kinases (Jaks) and Src family kinases. While pre-existing STAT dimers have been detected [3, 4], studies show that phosphorylation induces dimerization between two STAT monomers through a phosphotyrosine interaction with the SH2 domain. In the nucleus, active STAT dimers bind to specific DNA-response elements in the promoters of target genes and regulate gene expression. Normal STAT activation is transient in accordance with physiological responses. However, the persistent activation of certain STAT family members, including Stat3 is frequently observed in many human tumors. It is now well established that aberrant activation of Stat3 contributes to malignant transformation and tumorigenesis. Evidence shows that persistently-active Stat3 mediates oncogenesis and tumor formation in part by the upregulation of the expression of critical genes, the dysregulation of cell growth and survival, the promotion of angiogenesis [2, 5-11], and the induction of tumor immune-tolerance [12, 13]. Thus, the targeting of aberrant Stat3 signaling provides a novel strategy for treating the wide variety of human tumors that harbor abnormal Stat3 activity.
The critical step of dimerization [14] between two monomers within the context of STAT activation presents an attractive strategy to interfere with Stat3 signaling and functions and this approach has been exploited in prior work [15-25]. Leading agents from those earlier studies have been explored in rational design of optimized molecules, in conjunction with molecular modeling of their binding to the Stat3 SH2 domain [18, 19], per the X-ray crystal structure of the Stat3β homodimer [26]. One of those leads, S3I-201 [18] had previously been shown to exert antitumor effects against human breast cancer xenografts via mechanisms that involve the inhibition of aberrant Stat3.
In the present study, key structural information from the computational modeling of S3I-201 bound to the Stat3 SH2 domain facilitated the design of novel analogs of which S3I-201.1066 shows an improved Stat3-inhibitory activity. S3I-210.1066 inhibits Stat3 DNA-binding activity with an IC50 value of 35 μM. Current studies provide evidence that S3I-201.1066 directly interacts with the Stat3 protein in vitro, thereby disrupting Stat3 binding to cognate pTyr peptide motifs of receptors and inhibiting Stat3 phosphorylation and activation, and Stat3 nuclear localization. Furthermore, evidence is provided that S3I-201.1066 selectively induces antitumor cell effects in human breast and pancreatic cancer cells, and mouse transformed fibroblasts harboring aberrant Stat3 activity, and inhibits growth of human breast tumors in xenografts.
2. Materials and Methods
2.1 Cells and reagents
Normal mouse fibroblasts (NIH3T3) and counterparts transformed by v-Src (NIH3T3/v-Src), v-Ras (NIH3T3/v-Ras) or overexpressing the human epidermal growth factor (EGF) receptor (NIH3T3/hEGFR), and the human breast cancer (MDA-MB-231) and pancreatic cancer (Panc-1) cells have all been previously reported [15, 27-29]. The normal human pancreatic duct epithelial cells (HPDEC) was a kind gift from Dr. Tsao (OCI, UHN-PMH, Toronto) [30], the Stat3 knockout mouse embryonic fibroblasts line was generously provided by Dr. Valerie Poli (University of Turin) [31], and the ovarian cancer line, A2780S was a kind gift from Dr. Jin Q. Cheng (Moffitt Cancer Center and Research Institute). The Stat3-dependent reporter, pLucTKS3 and the Stat3-independent reporter, pLucSRE, and the v-Src transformed mouse fibroblasts that stably express pLucTKS3 (NIH3T3/v-Src/pLucTKS3) have all been previously reported [15, 32, 33]. Cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% heat-inactivated fetal bovine serum, or in the case of HPDEC, they were grown in Keratinocyte-SFM media (GIBCO, Invitrogen Corp, Carlsbad, CA) supplemented with 0.2 ng EGF and 30 μg/ml bovine pituitary extract, and containing antimycol. Antibodies against Stat3, pY705Stat3, Src, pY416Src, Jak1, pJak1, Shc, pShc, Erk1/2, pErk1/2, Survivin are from Cell Signaling Technology (Danvers, MA), and anti-EGFR and anti-VEGF from Santa Cruz Biotech (Santa Cruz, CA).
2.2. Cloning and Protein Expression
The coding regions for the murine Stat3 protein and the Stat3 SH2 domain were amplified by PCR and cloned into vectors pET-44 Ek/LIC (Novagen, EMD Chemicals, Gibbstown, NJ) and pET SUMO (Invitrogen), respectively. The primers used for amplification were: Stat3 Forward: GACGACGACAAGATGGCTCAGTGGAACCAGCTGC; Stat3 Reverse: GAGGAGAAGCCCGGTTATCACATGGGGGAGGTAGCACACT; Stat3-SH2 Forward: ATGGGTTTCATCAGCAAGGA; Stat3-SH2 Reverse: TCACCTACAGTACTTTCCAAATGC. Clones were sequenced to verify the correct sequences and orientation. His-tagged recombinant proteins were expressed in BL21(DE3) cells and purified on Ni-ion sepharose column.
2.3. Nuclear extract preparation, gel shift assays, and densitometric analysis
Nuclear extract preparations and electrophoretic mobility shift assay (EMSA) were carried out as previously described [28, 33]. The 32P-labeled oligonucleotide probes used were hSIE (high affinity sis-inducible element from the _c_-fos gene, m67 variant, 5′-AGCTTCATTTCCCGTAAATCCCTA) that binds Stat1 and Stat3 [34] and MGFe (mammary gland factor element from the bovine β-casein gene promoter, 5′-AGATTTCTAGGAATTCAA) for Stat1 and Stat5 binding [35, 36]. Except where indicated, nuclear extracts were pre-incubated with compound for 30 min at room temperature prior to incubation with the radiolabeled probe for 30 min at 30 °C before subjecting to EMSA analysis. Bands corresponding to DNA-binding activities were scanned and quantified for each concentration of compound using ImageQuant and plotted as percent of control (vehicle) against concentration of compound, from which the IC50 values were derived, as previously reported [37].
2.4. Immunoprecipitation, immunoblotting and densitometric analyses
Immunoprecipitation from whole-cell lysates, and tumor tissue lysate preparation, and immunoblotting analysis were performed as previously described [17, 18, 33, 38]. Primary antibodies used were anti-Stat3, pY705Stat3, pY416Src, Src, pErk1/2, Erk1/2, pJak1, Jak1, pShc, Shc, Grb 2, c-Myc, Bcl-xL, Survivin, MMP-9, and β-Actin (Cell Signaling), and VEGF (Santa Cruz Biotech.).
2.5. Cell viability and proliferation assay
Cells in culture in 6-well or 96-well plates were treated with or without S3I-201.1066 for 24-144 h and subjected to CyQuant cell proliferation assay (Invitrogen Corp/Life Technologies Corp), or harvested, and the viable cells counted by trypan blue exclusion with phase contrast microscopy.
2.6. Immunofluorescence imaging/confocal microscopy
NIH3T3/hEGFR cells were grown in multi-cell plates, serum-starved for 8 h and treated with or without S3I-201.1066 for 30 min prior to stimulation by rhEGF (1 μg/ml) for 10 min. Cells were fixed with ice-cold methanol for 15 min, washed 3 times in phosphate buffered saline (PBS), permeabilized with 0.2% Triton X-100 for 10 min, and further washed 3-4 times with PBS. Specimens were then blocked in 1% bovine serum albumin (BSA) for 30 min and incubated with anti-EGFR (Santa Cruz) or anti-Stat3 (Cell Signaling) antibody at 1:50 dilution at 4 °C overnight. Subsequently, cells were rinsed 4-5 times in PBS, incubated with Alexa fluor 546 rat antibody for EGFR detection and Alexa fluor 488 rabbit antibody for Stat3 detection (Invitrogen) for 1 h at room temperature in the dark. Specimens were then washed 5 times with PBS, covered with cover slides with VECTASHIELD mounting medium containing DAPI (Vector Lab, Inc, Burlingame, CA), and examined immediately under a Leica TCS SP5 confocal microscope (Germany) at appropriate wavelengths. Images were captured and processed using the Leica TCS SP 5 software.
2.7. Soft-agar colony formation assay
Colony formation assays were carried out in 6-well dishes, as described previously [16, 37]. Briefly, each well contained 1.5 ml of 1% agarose in Dulbeco’s modified Eagle’s medium as the bottom layer and 1.5 ml of 0.5% agarose in Dulbeco’s modified Eagle’s medium containing 4-6 × 103 NIH3T3/v-Src, NIH3T3/v-Ras, A2780S, MDA-MB-231 or Panc-1 cells, as the top layer. Treatment with S3I-201.1066 was initiated 1 day after seeding cells by adding 80 μl of medium with or without S3I-201.1066, and repeating every 2 or 3 days, until large colonies were evident. Colonies were quantified by staining with 20 μl of 1 mg/ml crystal violet (ThermoFisher, Waltham, MA), incubating at 37 °C overnight, and counting the next day under phase contrast microscope.
2.8. Fluorescence Polarization Assay
Fluorescence Polarization (FP) Assay was conducted as previously reported [23], with some modification using the phospho-peptide, 5-carboxyfluorescein-GpYLPQTV-NH2 (where pY represents phospho-Tyr) as probe and Stat3. A fixed concentration of the fluorescently-labeled peptide probe (10 nM) was incubated with increasing concentration of the Stat3 protein for 30 min at room temperature in the buffer, 50 mM NaCl, 10 mM HEPES, 1 mM EDTA, 0.1% Nonidet P-40, and the fluorescent polarization measurements were determined using the POLARstar Omega (BMG LABTECH, Durham, NC), with the set gain adjustment at 35 mP. The Z’ value was derived per the equation Z’=1−(3SDbound+3SDfree)/(mPbound−mPfree), where SD is the standard deviation and mP is the average of fluorescence polarization. In the “bound” state, 10 nM 5-carboxyfluorescein-GpYLPQTV-NH2 was incubated with 150 nM purified Stat3 protein, while the “free” (unbound) state represents the same mixture, but incubated with an additional 10 μM unlabeled Ac-GpYLPQTV-NH2. For evaluating agents, Stat3 protein (150 nM) was incubated with serial concentrations of S3I-201.1066 at 30 °C for 60 min in the indicated assay buffer conditions. Prior to the addition of the fluorescent probe, the protein:S3I-201.1066 mixtures were allowed to equilibrate at room temperature for 15 min. Probe was added at a final concentration of 10 nM and incubated for 30 min at room temperature following which the FP measurements were taken using the POLARstar Omega, with the set gain adjustment at 35 mP.
2.9. Surface Plasmon Resonance Analysis
SensiQ and its analysis software Qdat (ICX Technologies, Oklahoma City, OK) were used to analyze the interaction between agents and the Stat3 protein and to determine the binding affinity. Purified Stat3 was immobilized on a HisCap Sensor Chip by injecting 50 μg/ml of Stat3 onto the chip. Various concentrations of S3I-201.1066 in running buffer (1X PBS, 0.5% DMSO) were passed over the sensor chip to produce response signals. The association and dissociation rate constants were calculated using the Qdat software. The ratio of the association and dissociation rate constants was determined as the affinity (KD).
2.10. Colony Survival Assay
This was performed as previously reported [39]. Briefly, cells were seeded as single-cell in 6-cm dishes (500 cells per well), treated once the next day with S3I-201.1066 for 48 h, and allowed to culture until large colonies were visible. Colonies were stained with crystal violet (ThermoFisher) for 4 h and counted under phase-contrast microscope.
2.11. Wound healing assay for migration
Wounds were made using pipette tips in monolayer cultures of cells in six-well plates. Cells were treated with or without increasing concentrations of S3I-201.1066 and allowed to migrate into the denuded area for 12-24 hours. The migration of cells was visualized at a 10X magnification using an Axiovert 200 Inverted Fluorescence Microscope (Zeiss, Göttingen, Germany), with pictures taken using a mounted Canon Powershot A640 digital camera (Canon USA, Lake Success, NY). Cells that migrated into the denuded area were quantified.
2.12. Mice and in vivo tumor studies
Six-week-old female athymic nude mice were purchased from Harlan and maintained in the institutional animal facilities approved by the American Association for Accreditation of Laboratory Animal Care. Athymic nude mice were injected subcutaneously in the left flank area with 5 × 106 human breast cancer MDA-MB-231 cells in 100 μL of PBS. After 5 to 10 days, tumors of a diameter of 3 mm were established. Animals were grouped so that the mean tumor sizes in all groups were nearly identical, then given S3I-201.1066, i.v., at 3 mg/kg every 2 or every 3 days for 17 days and monitored every 2 or 3 days, and tumor sizes were measured with calipers. Tumor volume was calculated according to the formula V=0.52 × a2 × b, where a, smallest superficial diameter, b, largest superficial diameter. For each treatment group, the tumor volumes for each set of measurements were statistically analyzed in comparison to the control (non-treated) group. Upon completion of the study, tumors were extracted and tumor tissue lysates were prepared for immunoblotting and gel shit analyses.
2.13. Statistical analysis
Statistical analysis was performed on mean values using Prism GraphPad Software, Inc. (La Jolla, CA). The significance of differences between groups was determined by the paired _t-_test at p <0.05*, <0.01**, and < 0.001***.
3. Results
3.1. Computer-aided design of S3I-201 analogs as Stat3 inhibitors
Close structural analysis of the lowest Genetic Optimization for Ligand Docking (GOLD) [40] conformation of the lead Stat3 inhibitor, S3I-201 (green) (IC50 = 86 μM for inhibition of Stat3:Stat3 [18]) (Fig. 1A & C) bound within the Stat3 SH2 domain (Fig. 1C), per the X-ray crystal structure of DNA-bound Stat3β homodimer [26] showed significant complementary interactions between the protein surface and the compound and identified key structural requirements for tight binding. Docking studies permitted in silico structural design of analogs of differing Stat3 SH2 domain-binding characteristics in order to derive Stat3 inhibitors of improved potency and selectivity. GOLD docking studies showed limited structural occupation of the Stat3-SH2 domain, identifying a potential means for improving inhibitor potency. The SH2 domain is broadly composed of three sub-pockets, only two of which are accessed by S3I-201 (Fig. 1C). Lead agent, S3I-201 (Fig. 1A) has a glycolic acid scaffold with its carboxylic acid condensed with hetero-trisubstituted aromatic species to furnish the amide bond, and a hydroxyl moiety that has been tosylated. The _ortho_-hydroxybenzoic acid component is a known pTyr mimetic, and low energy GOLD docking studies consistently placed this unit in the pTyr-binding site, making hydrogen bonds and electrostatic interactions with Lys591, Ser611, Ser613 and Arg609. Due to the strength of such interactions between oppositely charged ions, it is likely that a considerable portion of the binding between the SH2 domain and S3I-201 arises from the pTyr mimetic. The O-tosyl group binds in the mostly-hydrophobic pocket that is derived from the tetramethylene portion of the side chain of Lys592 and the trimethylene portion of the side chain of Arg595, along with Ile597 and Ile634. Given the potency of S3I-201 towards Stat3 inhibition, a rational synthetic program was undertaken to modify and optimize the core scaffold to furnish more potent analogs. We additionally exploited key hydrophobic interactions with Phe716, Ile659, Val637 and Trp623 (Fig. 1C, see arrow) in generating compounds made of N-substituted (_para_-cyclohexyl)benzyl analogs [41], including S3I-201.1066 (Fig. 1B). Present study of the analog S3I-201.1066 (Fig. 1B & D) was undertaken to derive biochemical and biophysical evidence of binding to Stat3 and to define the mechanisms of inhibition of Stat3 and its functions in the context of Stat3-dependent malignant transformation and tumorigenesis.
Figure 1.
(A and B), Structures of (A) S3I-201, (B) S3I-201.1066; (C and D), GOLD docking of (C) S3I-201 (green), and (D) S3I-201 (green) and S3I-201.1066 (yellow) to the SH2 domain of Stat3; arrow denotes potential binding sub-pocket accessed by S3I-201.1066, but not S3I-201.
3.2. Inhibition of Stat3 DNA-binding activity
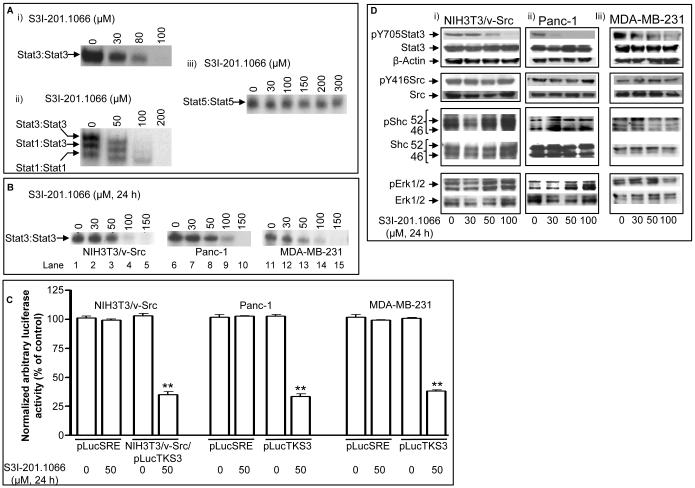
S3I-201 analogs derived per in silico structural optimization and molecular modeling of the binding to the Stat3 SH2 domain were synthesized and evaluated in Stat3 DNA-binding assay in vitro, as previously done [18]. Nuclear extracts containing activated Stat3 prepared from v-Src-transformed mouse fibroblasts (NIH3T3/v-Src) that harbor aberrantly-active Stat3 were incubated for 30 min at room temperature with or without increasing concentrations of the analog, S3I-201.1066, prior to incubation with the radiolabeled hSIE probe that binds to Stat3 and Stat1 and subjecting to electrophoretic mobility shift assay (EMSA) analysis [18]. Stat3 DNA-binding activity was inhibited in a dose-dependent manner by S3I-201.1066 (Figure 2A(i)), with average IC50 value of 35 ± 09 μM. This value represents 2-3 fold improvement over the activity of the lead agent, S3I-201 (IC50 of 86 μM) [18]. For selectivity, nuclear extracts containing activated Stat1, Stat3 and Stat5 prepared from EGF-stimulated NIH3T3/hEGFR (mouse fibroblasts over-expressing the human epidermal growth factor receptor) were pre-incubated at room temperature with or without increasing concentrations of S3I-201.1066 for 30 min, prior to incubation with the radiolabeled oligonucleotide probes and subjecting to EMSA analyses, as previously done [18]. EMSA results of the binding studies using the hSIE probe shows the strongest complex of Stat3:Stat3 with the probe (Fig. 2A(ii) upper band, lanes 1 and 2), which is significantly disrupted at 50 μM S3I-201.1066 and completely disrupted at 100 μM S3I-201.1066 (Fig. 2A(ii), upper band, lanes 2 and 3). EMSA analysis further shows a less intense Stat1:Stat3 complex (intermediate band), which is similarly repressed at 50 μM and completely disrupted at 100 μM S3I-201.1066 (Fig. 2A(ii), lanes 2 and 3). By contrast, we observe no significant inhibition of the Stat1:Stat1 complex that is of the lowest intensity (lower band) at 50 μM S3I-201.1066, a moderate inhibition at 100 μM S3I-201.1066, but a complete inhibition at 200 μM S3I-201.1066 (Fig. 2A(ii), lower band). Of importance, at the 100 μM S3I-201.1066 concentration at which only a moderate inhibition of Stat1:Stat1 complex occurred, the larger Stat3:Stat3 complex is completely dissociated (Fig. 2A(ii), lane 3). Moreover, EMSA analysis showed no effect on Stat5:Stat5 complex with the MGFe probe, up to 300 μM S3I-201.1066 (Fig. 2A(iii)). Thus, S3I-201.1066 preferentially inhibits DNA-binding activity of Stat3 over that of Stat1 and Stat5.
Figure 2. Effects of S3I-201.1066 on the activities of STATs, Src, Shc, and Erks.
(A) Nuclear extracts of equal total protein containing activated Stat1, Stat3, and/or Stat5 were pre-incubated with or without S3I-201.1066 for 30 min at room temperature prior to the incubation with the radiolabeled (i) and (ii) hSIE probe that binds Stat1 and Stat3 or the (iii) MGFe probe that binds Stat5 and subjecting to EMSA analysis; (B) Nuclear extracts of equal total protein prepared from the designated malignant cells following 24-h treatment with or without S3I-201.1066 were subjected to in vitro DNA-binding assay using the radiolabeled hSIE probe and analyzed by EMSA; (C) Cytosolic extracts of equal total protein were prepared from 24 h, S3I-201.1066-treated or untreated NIH3T3/v-Src fibroblasts that stably express the Stat3-dependent luciferase reporter (pLucTKS3) or from treated or untreated NIH3T3/v-Src fibroblasts, the human pancreatic (Panc-1) and breast (MDA-MB-231) carcinoma lines that are transiently-transfected with pLucSRE or pLucTKS3 and analyzed for luciferase activity using a luminometer; and (D) SDS-PAGE and Western blotting analysis of whole-cell lysates of equal total protein prepared from S3I-201.1066-treated or untreated NIH3T3/v-Src, Panc-1 and MDA-MB-231 cells probing for pY705Stat3, Stat3, pY416Src, Src, pShc, Shc, pErk1/2 and Erk1/2. Positions of STATs:DNA complexes or proteins in gel are labeled; control lanes (0) represent nuclear extracts treated with 0.05% DMSO, or nuclear extracts or whole-cell lysates prepared from 0.05% DMSO-treated cells; luciferase activities were normalized to β-galactosidase activity. Data are representative of 3-4 independent determinations. **p - <0.05.
3.3. Inhibition of intracellular Stat3 activation
Stat3 is constitutively-activated in a variety of malignant cells, including human breast and pancreatic cancer cells [9, 10, 20]. Given the effect against Stat3 DNA-binding activity in vitro, we evaluated S3I-201.1066 in v-Src transformed mouse fibroblasts (NIH3T3/v-Src), human breast cancer (MDA-MB-231) and human pancreatic cancer (Panc-1) lines that harbor aberrant Stat3 activity. Twenty-four hours after treatment, nuclear extracts were prepared from cells and subjected to Stat3 DNA-binding assay in vitro using the radiolabeled hSIE probe and analyzed by EMSA. Compared to the control (0.05% DMSO-treated cells, lane 1), nuclear extracts from S3I-201.1066-treated NIH3T3/v-Src, Panc-1 and MDA-MB-231 cells showed dose-dependent decreases of constitutive Stat3 activation, with significant inhibition at 50 μM S3I-201.1066 (Fig. 2B, compare lanes 2-5, 8-10, and 12-15 to their respective controls (0)). Luciferase reporter studies were performed to further determine the effect of S3I-201.1066 on Stat3 transcriptional activity. Results show that treatment with S3I-201.1066 of the v-Src transformed mouse fibroblasts (NIH3T3/v-Src) that stably express the Stat3-dependent luciferase reporter (NIH3T3/v-Src/pLucTKS3) [15, 32, 33] significantly (** p<0.01) repressed the induction of the Stat3-dependent reporter (Fig. 2C, left panel, NIH3T3/v-Src/pLucTKS3). Similar results were obtained when the human pancreatic cancer, Panc-1 and breast cancer, MDA-MB-231 cells harboring aberrant Stat3 activity were transiently transfected with the Stat3-dependent reporter, pLucTKS3 and treated with S3I-201.1066 (Fig. 2C, middle and right panels, pLucTKS3). By contrast, a similar treatment of malignant cells that are transiently transfected with the Stat3-independent luciferase reporter, pLucSRE, which is driven by the serum response element (SRE) of the c-fos promoter, had no observable effect on the reporter induction (Fig. 2C, pLucSRE). Moreover, immunoblotting analysis showed a concentration-dependent reduction of pTyr705Stat3 levels in NIH3T3/v-Src (Fig. 2D(i), top panel), Panc-1 cells (Fig. 2D(ii), top panel), and MDA-MB-231 (Fig. 2D(iii), top panel) cells upon treatment with S3I-201.1066 for 24 h, presumably through the blockade of Stat3 binding to pTyr motifs of receptors and the prevention of de novo phosphorylation by tyrosine kinases. By contrast, immunoblotting analysis showed no significant effects of S3I-201.1066 on the phosphorylation of Src (pY416Src), Shc (pShc), and Erk1/2 (pErk1/2) under the same treatment conditions (Fig. 2D (i)-(iii), panels 2-4 from the top). In spite of the inhibition of aberrant Stat3 activity, no observable change in total Stat3 protein was made, consistent with previous report [18, 19]. Also, total Src, Shc and Erk1/2 protein levels remained unchanged. We infer that at the concentrations that inhibit Stat3 activity, S3I-201.1066 has minimal effect on Src, Shc and Erk1/2 activation.
3.4. In vitro evidence that S3I-201.1066 interacts with Stat3 (or SH2 domain) and selectively disrupts Stat3 binding to cognate pTyr peptide motif of receptor
Given the computational modeling prediction that S3I-201.1066 interacts with the Stat3 SH2 domain, we deduce that S3I-201.1066 blocks Stat3 DNA-binding activity by binding to the Stat3 SH2 domain, thereby disrupting Stat3:Stat3 dimerization. To determine therefore if the Stat3 SH2 domain could interact with S3I-201.1066, we tested whether the addition of purified recombinant Stat3 SH2 domain into the DNA-binding assay mixture could intercept the inhibitory effect of the agent on Stat3 activity, as observed in Fig. 2A(i). The purified histidine-tagged Stat3 SH2 domain was added at increasing concentrations (1-500 ng) to the nuclear extracts containing activated Stat3 and the mixed extracts were pre-incubated with 100 μM S3I-201.1066 for 30 min at room temperature and subjected to DNA-binding assay in vitro for the study of the effect of S3I-201.1066, as was done in Fig. 2A(i). EMSA analysis shows a strong inhibition by S3I-201.1066 of Stat3 DNA-binding activity, as shown in Fig. 2A(i), when no purified Stat3 SH2 domain was added to the nuclear extracts (Fig. 3A, lanes 2, 7, and 9, compared to lane 1). By contrast, the observed S3I-201.0166-mediated inhibition of Stat3 DNA-binding activity was progressively eliminated by the presence of an increasing concentration of the purified Stat3 SH2 domain (Stat3 SH2), leading to the full recovery of Stat3 activity when the recombinant SH2 domain protein was present at 125-500 ng (Fig. 3A, lanes 3-6, 8 and 10).
Figure 3. Studies of the interaction of S3I-201.1066 with Stat3 or the Stat3 SH2 domain.
(A) EMSA analysis of in vitro binding activity of Stat3 to the radiolabeled hSIE probe using nuclear extracts containing activated Stat3 pre-incubated with 0 or 100 μM S3I-201.1066 in the presence or absence of 0-500 ng of purified His-tagged Stat3 SH2 domain; (B) Surface Plasmon Resonance analysis of the binding of (i) 0-5 μM GYLPQTV-NH2 (unphosphorylated, gp-130 peptide), (ii) 0-5 μM GpYLTQTV-NH2 (phosphorylated, high affinity gp-130 peptide), or (iii) 0-50 μM S3I-201.1066 (or 50 μM S3I-201.1066, insert) as the analyte to the purified His-tagged Stat3 protein immobilized on HisCap Sensor Chip; and (C) Fluorescence Polarization assay of the binding to the 5-carboxyfluorescein-GpYLPQTV-NH2 probe of (i) an increasing concentration of purified His-Stat3, or (ii)-(iv) a fixed amount of purified His-Stat3 (150 nM) in the presence of increasing concentrations of (ii) GpYLPQTV-NH2, (iii) GYLPQTV-NH2 or (iv) S3I-201.1066. Stat3:DNA complexes in gel are shown, control (-) lane or zero (0) represent 0.05% DMSO. Data are representative of 2-4 independent determinations.
The preceding studies suggest that S3I-201.1066 interacts with the Stat3 SH2 domain. However, the studies do not demonstrate a direct binding to the Stat3 SH2 domain. To provide definitive evidence of direct binding to Stat3, biophysical studies were performed. His-tagged Stat3 protein (or SH2 domain; 50 ng) was immobilized on a Ni-NTA sensor chip surface for Surface Plasmon Resonance (SPR) analysis of the binding of S3I-201.1066 as the analyte. Association and dissociation measurements were taken and the binding affinity of S3I-201.1066 for Stat3 was determined using Qdat software. Results showed gradual increase and decrease with time in the signals (response unit, RU) for the association and dissociation, respectively, of the agent upon its addition to the immobilized His-Stat3 (Fig. 3B(iii)), indicative of the binding of S3I-201.1066 to and dissociation from the Stat3 protein, with a binding affinity, _K_D of 2.74 μM. These data provide the first definitive evidence of the direct binding of Stat3 to derivatives of S3I-201. This SPR analysis of the conformational changes in His-Stat3 was validated by using the high affinity Stat3-binding phosphoTyr (pY) peptide, GpYLPQTV-NH2, derived from the interleukin-6 receptor (IL-6R) subunit, gp-130 [22, 23] (with a KD of 24 nM) (Fig. 3B(ii)), and its non-phosphorylated counterpart, GYLPQTV-NH2, which showed no significant binding to Stat3 (Fig. 3B(i)). Interestingly, the dissociation curve for S3I-201.1066 showed a large residual binding of S3I-201.1066 to Stat3 at 500-1000 s (Fig. 3B(iii), 10-50 μM, 500-1000 s), which gradually dissipated over a period longer than 6000 s (Fig. 3B(iii), insert). The natural dissociation time of S3I-201.1066 from Stat3 was determined to be 103 min. This contrasts the rapid dissociation of the high affinity phosphopeptide, GpYLPQTV-NH2 from Stat3 (Fig. 3B(ii)). The slower “off” rate for S3I-201.1066 could impact its overall functional effects, with implications for its in vivo therapeutic application. Differences in the physicochemical properties would account for the different behaviors of the interactions with the Stat3 protein.
The studies so far demonstrate that S3I-201.1066 interacts with Stat3 or the Stat3 SH2 domain (data not shown). The interaction with the Stat3 SH2 domain could block the binding of Stat3 to cognate pTyr peptide motifs of receptors. To verify that S3I-201.1066 disrupts pTyr-Stat3 SH2 domain interactions, hence Stat3:Stat3 dimerization, we set up a fluorescence polarization (FP) study based on the binding of Stat3 to the high affinity phospho-peptide, GpYLPQTV-NH2 [22, 23]. It has previously been reported that Stat3 binds to GpYLPQTV-NH2 with a higher affinity than to the Stat3-derived pTyr peptide, PpYLKTK. It is also reported that this high affinity peptide disrupted Stat3 DNA-binding activity in vitro with an IC50 value of 0.15 μM [22]. The FP assay utilizing the 5-carboxyfluorescein-GpYLPQTV-NH2 as a probe showed increasing fluorescence polarization signal (mP) with increasing concentration (in μM) of purified His-Stat3 for a robust Z’ value of 0.84 (Fig. 3C(i)), which closely matches the previously reported value of 0.87 [23]. The test of the non-phosphorylated, unlabeled GYLPQTV-NH2 in the FP assay showed no evidence of inhibition (Fig. 3C(iii)), while as expected, the phosphorylated, unlabeled counterpart, GpYLPQTV-NH2 induced a complete inhibition with an IC50 value of 0.3 μM (Fig. 3C(ii)), consistent with the previously reported value of 0.25 ± 0.03 μM [23]. The FP assay was used to further test the ability of S3I-201.1066 to disrupt the Stat3 interaction with cognate pTyr peptide (GpYLPQTV-NH2), which showed a concentration-dependent inhibition of the fluorescent polarization signal (Fig. 3C(iv)). Inhibitory constant (IC50 value) was derived to be 20 ± 7.3 μM, which is within the range for the IC50 value (35 ± 9 μM) determined for the inhibition of Stat3 DNA-binding activity (Fig. 2A(i)). These findings together demonstrate that S3I-201.1066 binds to Stat3 or the Stat3 SH2 domain and disrupts the interaction of Stat3 with cognate pTyr peptide motifs. This mode of action underlies the blocking Stat3 DNA-binding activity by S3I-201.1066.
To extend the studies to verify that S3I-201.1066 could disrupt the binding of Stat3 to receptors, mouse fibroblasts over-expressing the EGF receptor (NIH3T3/hEGFR) were treated with or without the compound prior to stimulation with EGF for 10 min. Cells were then subjected to immunofluorescence staining for EGFR (red) and Stat3 (green) and confocal microscopy for the EGF-induced colocalization of Stat3 and EGFR and the Stat3 nuclear translocation. In the resting NIH3T3/hEGFR fibroblasts, EGFR (red) is widely localized at the plasma membrane, Stat3 (green) is localized at both the plasma membrane and in the cytoplasm, with no visible presence in the nucleus (stained blue for DAPI), while the colocalization of Stat3 with EGFR is minimal at the plasma membrane (Fig. 4A, upper panel). The stimulation by EGF of untreated cells induced a strong nuclear presence of Stat3 (cyan for merged Stat3 (green) and DAPI (blue)), as well as the colocalizations of EGFR and Stat3 (yellow for merged EGFR (red) and Stat3 (green)) at the plasma membrane, cytoplasm, and peri-nuclear space, and in the nucleus (Fig. 4A, bottom left). Both of the EGF-stimulated colocalization between EGFR and Stat3 and the Stat3 nuclear localization events were strongly blocked when cells were pre-treated with S3I-201.1066 before stimulating with EGF (Fig. 4A, bottom right compared to non-treated, bottom left), indicating that the compound disrupts Stat3 binding to EGFR. We infer that by blocking Stat3 binding to the receptor, S3I-201.1066 attenuates Stat3 phosphorylation/activation and thereby prevents Stat3 nuclear translocation. To investigate further the Stat3 interaction with the EGFR receptor and the effect of S3I-201.1066, co-immunoprecipitation with immunoblotting studies were performed in which EGFR immunecomplex prepared from whole-cell lysates of treated and untreated cancer cells were blotted for Stat3, and for Shc and Grb 2 as negative control. Results showed that the EGFR immunecomplex from the untreated Panc-1 and MDA-MB-231 cells contained Stat3, Shc and Grb 2 (Fig. 4B(i), lanes 1 and 3, i.p. EGFR, blot Stat3, Shc, and Grb 2). By contrast, treatment of both cell lines with S3I-201.1066 significantly diminished the level of Stat3 that associated with EGFR immunecomplex of equal total protein, without affecting the levels of Shc or Grb 2 (Fig. 4B(i), lanes 2 and 4, i.p. EGFR, blot Stat3, Shc and Grb 2). Western blotting of whole-cell lysates of equal total protein shows that the activated and total Erk1/2 levels are unaffected by the treatment of cells with S3I-201.1066 (Fig. 4B(i), input, blot pErk and Erk), and that the levels of Stat3 protein were the same (Fig. 4B(i), input, blot Stat3). To further analyze the effect of S3I-201.1066 on Stat3 binding to EGFR, a sequential immunecomplex precipitation study was performed in which EGFR and Stat3 immunecomplexes were independently prepared from whole-cell lysates of untreated Panc-1 cells. Immunecomplexes of equal total protein were directly treated with 0, 30, 50, and 100 μM S3I-201.1066 for 3 h, and then subjected to a second EGFR or Stat3 immunecomplex precipitation and immunoblotting analysis. Compared to untreated samples (Fig. 4B(ii), lane 1), results show that the direct treatment with S3I-201.1066 of the EGFR immunecomplex dramatically diminished the level of Stat3 protein that remained associated with EGFR in the complex (Fig. 4B(ii), i.p. EGFR, blot Stat3, lanes 2-4), but had no visible effect on the levels of Shc or Grb 2 (Fig. 4B(ii), i.p., EGFR, blot Shc or Grb 2). The EGFR levels in the immunecomplexes are the same (Fig. 4B(ii), upper band). Similarly, the Stat3 immunecomplex that is directly treated with S3I-201.1066 and blotted for EGFR showed strongly reduced EGFR levels, compared to the untreated Stat3 immunecomplex of equal total protein (Fig. 4B(ii), i.p. Stat3, blot EGFR, compare lanes 2-4 to lanes 1). The Stat3 levels in the immunecomplexes are the same (Fig. 4B(ii), i.p. Stat3, blot Stat3). Altogether, these findings strongly demonstrate that S3I-201.1066 selectively disrupts the binding of Stat3 to cognate receptor motifs. By this mode of activity, S3I-201.1066 could block Stat3 phosphorylation and hence, nuclear translocation.
Figure 4. Effect of S3I-201.1066 on the colocalization or association of Stat3 with EGF receptor and on Stat3 nuclear translocation.
(A) Immunofluorescence imaging/confocal microscopy of Stat3 colocalization with EGFR and Stat3 nuclear localization in EGF-stimulated (1 μg/ml; 10 min) NIH3T3/hEGFR pre-treated with or without 50 μM S3I-201.1066 for 30 min; or (B) Immunoblotting analysis of (i) EGFR immunecomplex (upper panel) or whole-cell lysates (lower panel) from S3I-201.1066-treated Panc-1 and MDA-MB-231 cells, or (ii) immunecomplexes of EGFR (upper panel) or Stat3 (lower panel) were treated with the indicated concentrations of S3I-201.1066, and subsequent immunecomplexes of EGFR or Stat3 were probed for EGFR, Stat3, Shc, Grb 2, or Erk1/2MAPK. Data are representative of 3 independent studies.
3.5. S3I-201.1066 blocks growth, viability, malignant transformation, and the migration of malignant cells harboring constitutively-active Stat3
Constitutively-active Stat3 promotes malignant cell proliferation, survival and malignant transformation [10, 20, 42]. We asked the question whether S3I-201.1066 is able to selectively decrease the viability and growth of malignant cells that harbor aberrant Stat3 activity. The human breast (MDA-MB-231) and pancreatic (Panc-1) cancer lines and the v-Src-transformed mouse fibroblasts (NIH3T3/v-Src) that harbor constitutively-active Stat3, and cells that do not harbor aberrant Stat3 activity (Stat3 knockout mouse embryonic fibroblasts (MEFs) (Stat3-/-) [31], normal human pancreatic duct epithelial cells (HPDEC) [30], and the ovarian cancer line, A2780S in culture were treated with or without an increasing concentration of S3I-201.1066 for up to 6 days and analyzed for viable cell numbers by CyQuant cell proliferation/viability kit or trypan blue exclusion/phase-contrast microscopy. Compared to the control (DMSO-treated) cells, the mouse fibroblasts transformed by v-Src (NIH3T3/v-Src), and the human breast cancer, MDA-MB-231 and pancreatic cancer, Panc-1 lines showed significantly reduced viable cell numbers (Fig. 5A) and were growth inhibited (data not shown) following treatment with increasing concentrations of S3I-201.1066 for 24-48 h. By contrast, the viability and growth of the Stat3-null MEFs (Stat3-/-), the ovarian cancer line, A2780S and the normal human pancreatic duct epithelial cells (HPDEC) that do not harbor aberrant Stat3 activity were not significantly altered by S3I-201.1066 treatment (Fig. 5A, and data not shown), with derived IC50 values that are >100 μM, compared to values of 35, 48, and 37 μM for the inhibition of NIH3T3/v-Src, Panc-1, and MDA-MB-231, respectively (Fig. 5A, lower panel). These findings indicate that S3I-201.1066 exerts preferential biological effects on malignant cells that harbor constitutively-active Stat3, with little effects on non-target cells at concentrations that inhibit Stat3 activity.
Figure 5. S3I-201.1066 suppresses viability, survival, malignant transformation and migration of malignant cells that harbor persistently-active Stat3.
(A and B) Human breast (MDA-MB-231), pancreatic (Panc-1), and ovarian (A2780S) cancer cells, the v-Src transformed mouse fibroblasts (NIH3T3/v-Src) and their v-Ras transformed counterparts (NIH3T3/v-Ras), the Stat3 null mouse embryonic fibroblasts (Stat3-/-), and the normal human pancreatic duct epithelial cells (HPDEC) were treated once or untreated with 30-100 μM S3I-201.1066 for 24-48 h. Cells were (A) assayed for viability using CyQuant cell proliferation kit; IC50 values (bottom panel) were derived from graphical representation, or (B) allowed to culture until large colonies were visible, which were stained with crystal violet and enumerated; (C) Cells (NIH3T3/v-Src, NIH3T3/v-Ras, A2780S, MDA-MB-231, and Panc-1) growing in soft-agar suspension were treated with or without 30-100 μM S3I-201.1066 every 2-3 days until large colonies were visible, which were stained with crystal violet and enumerated; and (D) Cells (MDA-MB-231, Panc-1, NIH3T3/v-Src and NIH3T3/v-Ras in culture were wounded and treated with or without 50 μM S3I-201.1066 for 12 or 24 h and allowed to migrate into the denuded area in a wound healing assay. Cultures were visualized at 10X magnification by light microscopy and (i) cells that migrated into the denuded area counted and plotted against the concentration of S3I-201.1066 or (ii) cultures were photographed. Values are the mean and S.D. of 3-4 independent determinations, data are representative of 4 independent studies. p values, * - <0.05 , and ** - <0.01.
We extended these studies to examine the effect of S3I-201.1066 in colony survival assay performed as previously reported [39]. Cultured single-cells were untreated or treated once with S3I-201.1066 and allowed to grow until large colonies were visible, which were stained and enumerated. Results showed a dose-dependent suppression of the number of colonies for the v-Src transformed mouse fibroblasts (NIH3T3/v-Src), and the human pancreatic cancer, Panc-1 and breast cancer, MDA-MB-231 cells (Fig. 5B(iii)-(v)) (paired _t_-test was used to compare treated samples to their respective untreated controls). By contrast, minimal effect was observed on the colony numbers for mouse fibroblasts transformed by v-Ras (NIH3T3/v-Ras) or the ovarian cancer line, A2780S that do not harbor constitutively-active Stat3 (Fig. 5B(i) and (ii)). Furthermore, growth in soft-agar suspension of NIH3T3/v-Src, MDA-MB-231 and Panc-1 cells treated with S3I-201.1066 was significantly inhibited (Fig. 5C(iii)-(v)), compared to the minimal effect on the soft-agar growth of NIH3T3/v-Ras and the ovarian cancer line, A2780S at concentrations that inhibit Stat3 activity (Fig. 5C(i) and (ii)). Thus, S3I-201.1066 selectively blocks Stat3-dependent malignant transformation.
Studies also demonstrate that Stat3 is important for tumor progression [43, 44]. To further investigate the biological effects of S3I-201.1066 and to assess the ability to block Stat3-dependent tumor progression processes, a wound healing study was performed as a measure of the migration of malignant cells. Significantly reduced numbers of MDA-MB-231, Panc-1 and NIH3T3/v-Src cells migrating into the denuded area were observed following 12-24 h treatment with S3I-201.1066 (Fig. 5D and data not shown), with statistically significant lower numbers observed at 30 μM S3I-201.1066 treatment (data not shown). By contrast, the migration of NIH3T3/v-Ras fibroblasts was minimally affected by the same treatment conditions (Fig. 5D). In the 12-24 h treatment duration, there was no evidence of apoptosis of the treated cells (data not shown). These findings demonstrate that S3I-201.1066 selectively suppresses the migration of malignant cells that harbor aberrant Stat3.
3.6. S3I-201.1066 represses the expression of c-Myc, Bcl-xL, VEGF, Survivin, and MMP-9
Known Stat3 downstream target genes are critical to the dysregulated biological processes promoted by aberrantly-active Stat3 [9, 20, 42]. We sought to validate the inhibitory effect of S3I-201.1066 on aberrant Stat3 signaling and to define the underlying molecular mechanisms for the antitumor cell effects of the agent by investigating the changes in the induction of known Stat3-regulated genes. In the human breast carcinoma, MDA-MB-231 and pancreatic cancer, Panc-1 lines, and the mouse fibroblasts transformed by v-Src, which harbor constitutively-active Stat3, immunoblotting analysis of whole-cell lysates shows that treatment with 50 μM S3I-201.1066 for 24 h down-regulated the expression of c-Myc, Bcl-xL, VEGF, Survivin, and MMP-9 proteins (Fig. 6A). Bands were quantified, normalized to β-Actin, and the values corresponding to the band intensities for the samples from treated cells relative to the respective control (set at 1) are reported in parenthesis. These data indicate that S3I-201.1066 sufficiently represses the constitutive induction of Stat3-regulated genes. We infer that in doing so, S3I-201.1066 is able to thwart the ability of aberrant Stat3 to promote the dysregulation of growth and survival of malignant cells. These findings are in agreement with the results in Fig. 2C and together support the ability of S3I-201.1066 to block Stat3 transcriptional activity.
Figure 6. S3I-201.1066 suppresses c-Myc, Bcl-xL, Survivin, MMP-9 and VEGF expression in vitro and in vivo and inhibits growth of human breast tumor xenografts.
(A) SDS-PAGE and Western blotting analysis of whole-cell lysates prepared from the human breast cancer, MDA-MB-231 and pancreatic cancer, Panc-1 cells, and the v-Src-transformed mouse fibroblasts (NIH3T3/v-Src) untreated (DMSO, control) or treated with 50 μM S3I-201.1066 for 24 h and probing with anti-Myc, Bcl-xL, MMP-9, Survivin, VEGF or β-Actin antibodies; (B) Human breast (MDA-MB-231) tumor-bearing mice were given S3I-201.1066 (3 mg kg−1) or vehicle (0.1% DMSO in PBS) i.v. every 2 or 3 days. Tumor sizes, measured every 2 or 3 days, were converted to tumor volumes and plotted against treatment days; (C) Tumor tissue lysates prepared from extracted tumor tissues from one control (C) and three treated (T1-T3) mice were subjected to (i) Stat3 DNA-binding activity and EMSA analysis or (ii) immunoblotting analysis for pY705Stat3, Stat3, c-Myc, Bcl-xL, VEGF, and β-Actin. Positions of proteins in gel are shown. Data are representative of 2-3 independent determinations, values in parenthesis represent the band intensities for the samples from treated cells relative to the respective control (set at 1), data are the mean and S.D. from replicates of 12 tumor-bearing mice in each group. p values, *<0.05.
3.7. S3I-201.1066 inhibits growth of human breast tumor xenografts
Given Stat3’s importance in tumor growth and tumor progression, we evaluated S3I-201.1066 in xenograft models of the human breast cancer (MDA-MB-231) cells that harbor aberrant Stat3 activity. Compared to control (vehicle-treated) tumor-bearing mice, treatment (i.v. injection) with S3I-201.1066 at 3 mg/kg every 2 or 3 days for 17 days induced significant decrease in tumor growth (Fig. 6B). At the dosing schedule used, the drug was well tolerated and the animals showed no obvious signs of toxicity. The underlying premise of the antitumor effects is the ability of S3I-201.1066 to inhibit aberrant Stat3 activity. To determine whether the treatment with S3I-201.1066 modulated the in vivo activity and function of aberrant Stat3 in the human breast tumor xenografts, we evaluated the status of Stat3 activity and the expression of known Stat3-regulated genes in vivo. Upon the completion of the study, control tumors and residual tumors from treated mice were harvested and tissue lysates were prepared and analyzed by electrophoretic mobility shift assay using the radiolabeled hSIE probe that binds Stat3 (Fig. 6C(i)) or immunoblotting (Fig. 6C(ii)). Representative data for one control, untreated tumor and three treated tumor tissues showed both decreased phosphorylation (pY705Stat3) (Fig. 6C(ii), upper band) and DNA-binding activity (Fig. 6C(i)) of Stat3 in tumors from treated mice (T1-T3, versus C). Furthermore, immunoblotting analysis showed diminished expression of c-Myc, Bcl-xL, VEGF, and Survivin in the tumor tissues from treated mice compared to control (Fig. 6C(ii)). These data indicate that the i.v. administration of S3I-201.1066 at the dosing schedule used achieved sufficient intra-tumoral levels of S3I-201.1066, which led to the suppression of Stat3 tyrosine phosphorylation, DNA-binding and transcriptional activities. These findings together demonstrate that S3I-201.1066 inhibits constitutive Stat3 activation, leading to decreased expression of known Stat3-regulated genes, and hence inducing antitumor cell effects and tumor regression.
4. Discussion
Computational modeling of the interactions of the Stat3 SH2 domain with the previously reported Stat3 inhibitor lead, S3I-201 [18], derived key structural information for lead optimization and a rational synthetic program that furnished exciting new analogs. Analog, S3I-201.1066 shows improved Stat3-inhibitory potency and selectivity in vitro, with intracellular Stat3-inhibitory activity that is enhanced 2-3-fold. Moreover, S3I-201.1066 exhibited improved target selectivity, with minimal inhibitory effect on the phosphorylation of Src, Erk1/2MAPK and Shc proteins at concentrations (30-50 μM) that inhibit intracellular Stat3 activation, despite there being SH2 domains involved in the mechanisms leading to the activation of these other proteins. Per molecular modeling, the improved activity could in part be due to the enhanced interactions with the Stat3 protein, possibly by the (_para_-cyclohexyl)benzyl moiety that extends from the scaffold amide nitrogen and makes important contacts with the hydrophobic residues Trp623, Ile659, Val637 and Phe716 within the unexplored pocket.
The native Stat3 peptide inhibitor, PpYLKTK and its peptidomimetic analogs [15, 16] and several other Stat3 SH2 domain-binding and dimerization disrupting peptides and derivatives have been reported [21, 22, 25]. Previous studies have utilized the fluorescence polarization analysis to characterize the binding of the native, high affinity phosphopeptide, GpYLPQTV-NH2 (as 5-carboxyfluorescein-GpYLPQTV-NH2) to the Stat3 protein [22, 23]. Using this assay platform and SPR analysis, we provide definitive evidence for the physical interaction of S3I-201.1066 with Stat3 or the Stat3 SH2 domain, with an affinity (_K_D) of 2.74 μM. The analysis of the interaction reveals a slower kinetics of the association and dissociation events, which contrasts the more rapid binding and dissociation of the native, high affinity peptide, GpYLPQTV-NH2 to and from Stat3, with a corresponding affinity (KD) of 24 nM. The second supporting evidence for the interaction of S3I-201.1066 with Stat3 comes by way of the disruption by S3I-201.1066 of the Stat3 binding to the pTyr peptide in a fluorescent polarization assay, with a derived IC50 of 20 μM. By comparison, the unlabeled, native phosphopeptide disrupts the Stat3 biding to the pTyr peptide probe, with an IC50 value of 0.3 μM, which is in line with the reported affinity of 0.15 ± 0.01 μM [23] or the IC50 value of 0.290 ± 0.063 μM [21]. The higher affinity of the native peptide for the protein should be expected, given the more favorable physicochemical properties that will facilitate a stronger binding to the Stat3 protein. Nonetheless, data suggesting a slower dissociation of S3I-201.1066 from Stat3 suggests this drug is likely to show a more prolonged effect on the target and its function per a given dose.
Current study provides support for the binding of S3I-201.1066 to Stat3 and for the disruption of the interaction between Stat3 and pTyr peptide. Given the disruption of the Stat3 binding to the cognate peptide, GpYLPQTV-NH2, we infer that inside cells, S3I-201.1066 could interfere with the ability of Stat3 (via SH2 domain) to bind to cognate pTyr motifs on receptors and thereby block de novo phosphorylation by tyrosine kinases, as well as disrupt pre-existing Stat3:Stat3 dimers, particularly in malignant cells that harbor aberrantly-active Stat3. Accordingly, we present evidence that both of the association of Stat3 with EGFR and the Stat3 nuclear localization in ligand-stimulated cells are strongly blocked by the treatment of cells with S3I-201.1066. Although other Stat3 dimerization disruptors have been previously identified through molecular modeling [19, 45], the present study is the first to provide biophysical evidence for a direct interaction of a small-molecule, dimerization disruptor with the Stat3 protein.
Substantive evidence demonstrates that aberrant Stat3 activity promotes cancer cell growth and survival [15, 16, 29, 46, 47], and induces tumor angiogenesis [48, 49] and metastasis [43, 49]. Accordingly, inhibitors of Stat3 activation and signaling have been shown to induce antitumor cell effects consistent with the abrogation of Stat3 function [15-19, 37, 50-52]. The present study parallels those published reports in showing that a newly-derived agent, S3I-201.1066 induces the growth inhibition and the loss of viability and survival of the human pancreatic cancer, Panc-1 and breast cancer, MDA-MB-231 cells, and transformed mouse fibroblasts (NIH3T3/v-Src) that harbor aberrant Stat3 activity, while having minimal effects on normal human pancreatic duct epithelial cells, the Stat3-null mouse embryonic fibroblasts [31], the ovarian cancer line, A2780S, and the viral Ras-transformed mouse fibroblasts that do not harbor aberrant Stat3 activity. Moreover, the S3I-201.1066-induced antitumor cell effects on malignant cells harboring aberrant Stat3 activity occurred at significantly lower concentrations, 30-50 μM than the 100 μM cellular activity previously reported for the lead agent [18]. Mechanistic insight into the biological effects of S3I-201.1066 reveal the suppression of the constitutive expression of known Stat3-regulated genes, including c-Myc, Bcl-xL, VEGF, Survivin, and MMP-9, which control cell growth and apoptosis, promote tumor angiogenesis, or modulate tumor cell invasion [19, 43, 46, 49, 53, 54]. Furthermore, the effect of S3I-201.1066 on Stat3 oncogenic function is shown by the significant antitumor response induced in human breast tumor xenografts following the in vivo administration of this agent. Data also suggest that at the dosing schedule used, the i.v. administration of S3I-201.1066 achieved intra-tumoral levels sufficient to modulate activated Stat3 and its function.
We report the application of computational modeling in conjunction with rational, structure-based virtual design approach for the optimization of S3I-201. The new agent, S3I-201.1066 binds to Stat3, disrupts Stat3 SH2 domain:pTyr interactions, and hence Stat3:Stat3 dimerization and Stat3 binding to receptor, thereby inhibiting Stat3 phosphorylation, nuclear translocation and oncogenic functions, and inducing antitumor cell effects in vitro and antitumor effects in vivo.
Acknowledgements
We thank all colleagues and members of our laboratory for the stimulating discussions. We also thank Vijay Shahani at the University of Toronto for the GOLD molecular docking studies. This work was supported by the National Cancer Institute Grants CA106439 (JT) and CA128865 (JT), and by the Leukemia and Lymphoma Society of Canada (PTG) and the University of Toronto (PTG).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Bromberg J. Signal transducers and activators of transcription as regulators of growth, apoptosis and breast development. Breast Cancer Res. 2000;2:86–90. doi: 10.1186/bcr38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Darnell JE., Jr. Transcription factors as targets for cancer therapy. Nat Rev Cancer. 2002;2:740–9. doi: 10.1038/nrc906. [DOI] [PubMed] [Google Scholar]
- [3].Schröder M, Kroeger K, Volk HD, Eidne KA, Grütz G. Preassociation of nonactivated STAT3 molecules demonstrated in living cells using bioluminescence resonance energy transfer: a new model of STAT activation. J Leukoc Biol. 2004;75:792–7. doi: 10.1189/jlb.1003496. [DOI] [PubMed] [Google Scholar]
- [4].Sehgal PB. Paradigm shifts in the cell biology of STAT signaling. Semin Cell. Dev Biol. 2008;19:329–40. doi: 10.1016/j.semcdb.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Bromberg J, Darnell JE., Jr. The role of STATs in transcriptional control and their impact on cellular function. Oncogene. 2000;19:2468–73. doi: 10.1038/sj.onc.1203476. [DOI] [PubMed] [Google Scholar]
- [6].Bowman T, Garcia R, Turkson J, Jove R. STATs in oncogenesis. Oncogene. 2000;19:2474–88. doi: 10.1038/sj.onc.1203527. [DOI] [PubMed] [Google Scholar]
- [7].Turkson J, Jove R. STAT proteins: novel molecular targets for cancer drug discovery. Oncogene. 2000;19:6613–26. doi: 10.1038/sj.onc.1204086. [DOI] [PubMed] [Google Scholar]
- [8].Buettner R, Mora LB, Jove R. Activated STAT signaling in human tumors provides novel molecular targets for therapeutic intervention. Clin Cancer Res. 2002;8:945–54. [PubMed] [Google Scholar]
- [9].Yu H, Jove R. The STATS of Cancer-New molecular targets come of age. Nat Rev Cancer. 2004;4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- [10].Turkson J. STAT proteins as novel targets for cancer drug discovery. Expert Opin Ther Targets. 2004;8:409–22. doi: 10.1517/14728222.8.5.409. [DOI] [PubMed] [Google Scholar]
- [11].Darnell JE. Validating Stat3 in cancer therapy. Nat Med. 2005;11:595–6. doi: 10.1038/nm0605-595. [DOI] [PubMed] [Google Scholar]
- [12].Kortylewski M, Yu H. Stat3 as a potential target for cancer immunotherapy. J Immunother. 2007;30:131–9. doi: 10.1097/01.cji.0000211327.76266.65. [DOI] [PubMed] [Google Scholar]
- [13].Kortylewski M, Yu H. Role of Stat3 in suppressing anti-tumor immunity. Curr Opin Immunol. 2008;20:228–33. doi: 10.1016/j.coi.2008.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Shuai K, Horvath CM, Huang LH, Qureshi SA, Cowburn D, Darnell JE., Jr. Interferon activation of the transcription factor Stat91 involves dimerization through SH2-phosphotyrosyl peptide interactions. Cell. 1994;76:821–8. doi: 10.1016/0092-8674(94)90357-3. [DOI] [PubMed] [Google Scholar]
- [15].Turkson J, Ryan D, Kim JS, Zhang Y, Chen Z, Haura E, et al. Phosphotyrosyl peptides block Stat3-mediated DNA-binding activity, gene regulation and cell transformation. J Biol Chem. 2001;276:45443–55. doi: 10.1074/jbc.M107527200. [DOI] [PubMed] [Google Scholar]
- [16].Turkson J, Kim JS, Zhang S, Yuan J, Huang M, Glenn M, et al. Novel peptidomimetic inhibitors of signal transducer and activator of transcription 3 dimerization and biological activity. Mol Cancer Ther. 2004;3:261–9. [PubMed] [Google Scholar]
- [17].Siddiquee K, Glenn M, Gunning P, Katt WP, Zhang S, Schroeck C, et al. An oxazole-based small-molecule Stat3 inhibitor modulates Stat3 stability and processing and induces antitumor cell effects. ACS Chem Biol. 2007;2:787–98. doi: 10.1021/cb7001973. [DOI] [PubMed] [Google Scholar]
- [18].Siddiquee K, Zhang S, Guida WC, Blaskovich MA, Greedy B, Lawrence H, et al. Selective chemical probe inhibitor of Stat3, identified through structure-based virtual screening, induces antitumor activity. Proc Natl Acad Sci U S A. 2007;104:7391–6. doi: 10.1073/pnas.0609757104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Song H, Wang R, Wang S, Lin J. A low-molecular-weight compound discovered through virtual database screening inhibits Stat3 function in breast cancer cells. Proc Natl Acad Sci U S A. 2005;102:4700–5. doi: 10.1073/pnas.0409894102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Yue P, Turkson J. Targeting STAT3 in cancer: how successful are we? Expert Opin Investig Drugs. 2009;18:45–56. doi: 10.1517/13543780802565791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Coleman DRI, Ren Z, Mandal PK, Cameron AG, Dyer GA, Muranjan S, et al. Investigation of the Binding Determinants of Phosphopeptides Targeted to the Src Homology 2 Domain of the Signal Transducer and Activator of Transcription 3. Development of a High-Affinity Peptide Inhibitor. J Med Chem. 2005;48:6661–70. doi: 10.1021/jm050513m. [DOI] [PubMed] [Google Scholar]
- [22].Ren Z, Cabell LA, Schaefer TS, McMurray JS. Identification of a high-affinity phosphopeptide inhibitor of stat3. Bioorg Med Chem Lett. 2003;13:633–6. doi: 10.1016/s0960-894x(02)01050-8. [DOI] [PubMed] [Google Scholar]
- [23].Schust J, Berg T. A high-throughput fluorescence polarization assay for signal transducer and activator of transcription 3. Anal Biochem. 2004;330:114–8. doi: 10.1016/j.ab.2004.03.024. [DOI] [PubMed] [Google Scholar]
- [24].Gunning PT, Glenn MP, Siddiquee KA, Katt WP, Masson E, Sebti SM, et al. Targeting protein-protein interactions: suppression of Stat3 dimerization with rationally designed small-molecule, nonpeptidic SH2 domain binders. Chembiochem. 2008;9:2800–3. doi: 10.1002/cbic.200800291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Fletcher S, Turkson J, Gunning PT. Molecular approaches towards the inhibition of the signal transducer and activator of transcription 3 (Stat3) protein. ChemMedChem. 2008;3:1159–68. doi: 10.1002/cmdc.200800123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Becker S, Groner B, Muller CW. Three-dimensional structure of the Stat3beta homodimer bound to DNA. Nature. 1998;394:145–51. doi: 10.1038/28101. [DOI] [PubMed] [Google Scholar]
- [27].Johnson PJ, Coussens PM, Danko AV, Shalloway D. Overexpressed pp60c-src can induce focus formation without complete transformation of NIH 3T3 cells. Mol Cell Biol. 1985;5:1073–83. doi: 10.1128/mcb.5.5.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Yu CL, Meyer DJ, Campbell GS, Larner AC, Carter-Su C, Schwartz J, et al. Enhanced DNA-binding activity of a Stat3-related protein in cells transformed by the Src oncoprotein. Science. 1995;269:81–3. doi: 10.1126/science.7541555. [DOI] [PubMed] [Google Scholar]
- [29].Garcia R, Bowman TL, Niu G, Yu H, Minton S, Muro-Cacho CA, et al. Constitutive activation of Stat3 by the Src and JAK tyrosine kinases participates in growth regulation of human breast carcinoma cells. Oncogene. 2001;20:2499–513. doi: 10.1038/sj.onc.1204349. [DOI] [PubMed] [Google Scholar]
- [30].Ouyang H, Mou LJ, Luk C, Liu N, Karaskova J, Squire J, et al. Immortal human pancreatic duct epithelial cell lines with near normal genotype and phenotype. Am J Pathol. 2000;157:1623–31. doi: 10.1016/S0002-9440(10)64800-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Maritano D, Sugrue ML, Tininini S, Dewilde S, Strobl B, Fu X, et al. The STAT3 isoforms alpha and beta have unique and specific functions. Nat Immunol. 2004;5:401–9. doi: 10.1038/ni1052. [DOI] [PubMed] [Google Scholar]
- [32].Turkson J, Bowman T, Adnane J, Zhang Y, Djeu JY, et al. Requirement for Ras/Rac1-mediated p38 and c-Jun N-terminal kinase signaling in Stat3 transcriptional activity induced by the Src oncoprotein. Mol Cell Biol. 1999;19:7519–7528. doi: 10.1128/mcb.19.11.7519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Turkson J, Bowman T, Garcia R, Caldenhoven E, De Groot RP, Jove R. Stat3 activation by Src induces specific gene regulation and is required for cell transformation. Mol Cell Biol. 1998;18:2545–52. doi: 10.1128/mcb.18.5.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Wagner M, Kleeff J, Friess H, Buchler MW, Korc M. Enhanced expression of the type II transforming growth factor-beta receptor is associated with decreased survival in human pancreatic cancer. Pancreas. 1999;19:370–6. doi: 10.1097/00006676-199911000-00008. [DOI] [PubMed] [Google Scholar]
- [35].Gouilleux F, Moritz D, Humar M, Moriggl R, Berchtold S, Groner B. Prolactin and interleukin-2 receptors in T lymphocytes signal through a MGF-STAT5-like transcription factor. Endocrinology. 1995;136:5700–8. doi: 10.1210/endo.136.12.7588326. [DOI] [PubMed] [Google Scholar]
- [36].Seidel HM, Milocco LH, Lamb P, Darnell JE, Jr., Stein RB, Rosen J. Spacing of palindromic half sites as a determinant of selective STAT (signal transducers and activators of transcription) DNA binding and transcriptional activity. Proc Natl Acad Sci U S A. 1995;92:3041–5. doi: 10.1073/pnas.92.7.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Turkson J, Zhang S, Mora LB, Burns A, Sebti S, Jove R. A novel platinum compound inhibits constitutive Stat3 signaling and induces cell cycle arrest and apoptosis of malignant cells. J Biol Chem. 2005;280:32979–88. doi: 10.1074/jbc.M502694200. [DOI] [PubMed] [Google Scholar]
- [38].Zhang Y, Turkson J, Carter-Su C, Smithgall T, Levitzki A, Kraker A, et al. Activation of Stat3 in v-Src Transformed Fibroblasts Requires Cooperation of Jak1 Kinase Activity. J Biol Chem. 2000;275:24935–44. doi: 10.1074/jbc.M002383200. [DOI] [PubMed] [Google Scholar]
- [39].Zhao S, Venkatasubbarao K, Lazor JW, Sperry J, Jin C, Cao L, et al. Inhibition of STAT3Tyr705 Phosphorylation by Smad4 Suppresses Transforming Growth Factor β–Mediated Invasion and Metastasis in Pancreatic Cancer Cells. Cancer Res. 2008;68:4221–8. doi: 10.1158/0008-5472.CAN-07-5123. [DOI] [PubMed] [Google Scholar]
- [40].Jones G, Willett P, Glen RC, Leach AR, Taylor R. Development and validation of a genetic algorithm for flexible docking. J Mol Biol. 1997;267:727–48. doi: 10.1006/jmbi.1996.0897. [DOI] [PubMed] [Google Scholar]
- [41].Fletcher S, Jardeephi S, Zhang X, Yue P, Page BD, Sharmeen S, et al. Disruption of Transcriptionally Active Stat3 Dimers with Non-Phosphorylated, Salicylic Acid-Based Small Molecules: Potent in vitro and Tumour Cell Activities. ChemBioChem. 2009;10:1959–64. doi: 10.1002/cbic.200900172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Siddiquee KAZ. STAT3 as a target for inducing apoptosis in solid and hematological tumors. Cell Res. 2008;18:254–67. doi: 10.1038/cr.2008.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Xie TX, Wei D, Liu M, Gao AC, Ali-Osman F, Sawaya R, et al. Stat3 activation regulates the expression of matrix metalloproteinase-2 and tumor invasion and metastasis. Oncogene. 2004;23:3550–60. doi: 10.1038/sj.onc.1207383. [DOI] [PubMed] [Google Scholar]
- [44].Huang C, Cao J, Huang KJ, Zhang F, Jiang T, Zhu L, et al. Inhibition of STAT3 activity with AG490 decreases the invasion of human pancreatic cancer cells in vitro. Cancer Sci. 2006;97:1417–23. doi: 10.1111/j.1349-7006.2006.00340.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Bhasin D, Cisek K, Pandharkar T, Regan N, Li C, Pandit B, et al. Design, synthesis, and studies of small molecule STAT3 inhibitors. Bioorg Med Chem Lett. 2008;18:391–5. doi: 10.1016/j.bmcl.2007.10.031. [DOI] [PubMed] [Google Scholar]
- [46].Catlett-Falcone R, Landowski TH, Oshiro MM, Turkson J, Levitzki A, Savino R, et al. Constitutive activation of Stat3 signaling confers resistance to apoptosis in human U266 myeloma cells. Immunity. 1999;10:105–15. doi: 10.1016/s1074-7613(00)80011-4. [DOI] [PubMed] [Google Scholar]
- [47].Mora LB, Buettner R, Seigne J, Diaz J, Ahmad N, Garcia R, et al. Constitutive activation of Stat3 in human prostate tumors and cell lines: direct inhibition of Stat3 signaling induces apoptosis of prostate cancer cells. Cancer Res. 2002;62:6659–66. [PubMed] [Google Scholar]
- [48].Niu G, Wright KL, Huang M, Song L, Haura E, Turkson J, et al. Constitutive Stat3 activity upregulates VEGF expression and tumor angiogenesis. Oncogene. 2002;21:2000–8. doi: 10.1038/sj.onc.1205260. [DOI] [PubMed] [Google Scholar]
- [49].Wei D, Le X, Zheng L, Wang L, Frey JA, Gao AC, et al. Stat3 activation regulates the expression of vascular endothelial growth factor and human pancreatic cancer angiogenesis and metastasis. Oncogene. 2003;22:319–29. doi: 10.1038/sj.onc.1206122. [DOI] [PubMed] [Google Scholar]
- [50].Fuh B, Sobo M, Cen L, Josiah D, Hutzen B, Cisek K, et al. LLL-3 inhibits STAT3 activity, suppresses glioblastoma cell growth and prolongs survival in a mouse glioblastoma model. Br J Cancer. 2009;100:106–12. doi: 10.1038/sj.bjc.6604793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Blaskovich MA, Sun J, Cantor A, Turkson J, Jove R, Sebti SM. Discovery of JSI-124 (cucurbitacin I), a selective Janus kinase/signal transducer and activator of transcription 3 signaling pathway inhibitor with potent antitumor activity against human and murine cancer cells in mice. Cancer Res. 2003;63:1270–9. [PubMed] [Google Scholar]
- [52].Sun J, Blaskovich MA, Jove R, Livingston SK, Coppola D, Sebti SM. Cucurbitacin Q: a selective STAT3 activation inhibitor with potent antitumor activity. Oncogene. 2005;24:3236–45. doi: 10.1038/sj.onc.1208470. [DOI] [PubMed] [Google Scholar]
- [53].Real PJ, Sierra A, De Juan A, Segovia JC, Lopez-Vega JM, Fernandez-Luna JL. Resistance to chemotherapy via Stat3-dependent overexpression of Bcl-2 in metastatic breast cancer cells. Oncogene. 2002;21:7611–8. doi: 10.1038/sj.onc.1206004. [DOI] [PubMed] [Google Scholar]
- [54].Gritsko T, Williams A, Turkson J, Kaneko S, Bowman T, Huang M, et al. Persistent activation of stat3 signaling induces survivin gene expression and confers resistance to apoptosis in human breast cancer cells. Clin Cancer Res. 2006;12:11–9. doi: 10.1158/1078-0432.CCR-04-1752. [DOI] [PubMed] [Google Scholar]