Development and characterization of transgenic mouse models for conditional gene knockout in the blood–brain and blood-CSF barriers (original) (raw)
. Author manuscript; available in PMC: 2012 Feb 1.
Published in final edited form as: Transgenic Res. 2011 May 3;21(1):113–130. doi: 10.1007/s11248-011-9512-z
Abstract
For many CNS acting drugs, penetration into the central nervous system (CNS) is limited by the blood-CNS-barriers. In an effort to quantitate the role of the protein components that make up the blood-CNS-barriers, we created transgenic mice that allow conditional gene knockout using Cre/loxP technology. We targeted the expression of Cre-recombinase to the choroid plexus (the blood-cerebral spinal fluid barrier) using the lymphotropic papovavirus control region (LPVcr) and to brain endothelium (the blood–brain-barrier) using the proximal promoter region of the human von Willebrand Factor gene (hVWF-f). We verified that LPVcr restricts expression to the choroid plexus in adult mice by using the LPVcr to drive n-LacZ expression in transgenic mice. The LPV-Cre and hVWF-Cre plasmids were then constructed and tested for Cre-recombinase function in vitro, and subsequently used to create transgenic mice. The resulting transgenic mice were characterized for cell-type specific Cre-mediated endonuclease activity by crossing them with transgenic mice containing a loxP-flanked-LacZ/ EGFP dual reporter gene Z/EG. The dual Cre-Z/EG transgenic offspring were evaluated for the location of EGFP mRNA expression by reverse transcriptase PCR and for protein expression by immunohistochemistry. Immunohistochemistry for EGFP verified expression in the target cells, and no ectopic expression outside of the expected cell types. The LPV-Cre.0607 transgenic line expressed functional Cre only in the choroid plexus and hVWF-Cre.1304 line in brain endothelium.
Keywords: Conditional expression, Cre-recombinase, Choroid plexus, Endothelium, Brain, Von Willebrand factor, Lymphotropic papovavirus, Blood–brain barrier, Blood-CSF barrier
Introduction
For many drugs that require central nervous system (CNS) penetration for efficacy, the blood–brain barrier (BBB) and blood-cerebral spinal fluid barrier (B-CSF-B) pose significant hurdles in the path of pharmaceutical development. These structures limit drug penetration from the blood to the CNS compartment due to tight junctions (TJ) between endothelial cells, low drug uptake of capillary cells, and active cellular efflux transporters. The BBB is made up of vascular endothelium along with astrocyte cells whose foot processes make contact with the endothelial cells. The B-CSF-B is formed by the choroid plexus (CP). The CP is located in the ventricles of the brain and consists of modified ependymal cells fed by capillaries.
Conventional gene knockout mouse models have been developed to eliminate one or more of the proteins that are expressed in the endothelial cells lining the BBB and ependymal cells within the CP. Genetic knockout of drug efflux transporters (often expressed in a wide range of tissues) that regulate drug penetration into the brain have been developed in mice. These include Abcb1a [P-glycoprotein (P-gp)] (Schinkel et al. 1994), abcg2 (breast cancer resistance protein) (Jonker et al. 2002), and Mrp1 (multidrug resistance-associated protein 1) (Lorico et al. 1997). However, these genes are also expressed in many tissues involved in drug disposition such as the liver, kidney, and intestine. Thus, total genetic knockout of these transporters also abrogate the transport function across all tissues limiting the power to discern the contribution of these transporters specifically in drug penetration into the CNS. While global knockout of these genes in all tissues have showed large increases in blood to brain ratio of drug (Geyer et al. 2009; Salama et al. 2005; Jonker et al. 2002), other causes (besides the elimination of the transporters from the BBB and CP) could influence this change in brain penetration. These include altered oral absorption for oral administration, changes in excretion from the intestine and liver, distribution into other cells/organs that normally express these efflux transporters, upregulation of other transporters, as well as yet to be identified compensatory responses in these genetically modified models.
Tight junction (TJ) proteins have also been evaluated in gene knockout mice. Knockout of TJ proteins, such as zonula occluden proteins ZO-1 and ZO-2, result in death during the embryonic stage (Katsuno et al. 2008; Xu et al. 2008), while others, such as Claudin-5, survive through birth, but die soon after (Nitta et al. 2003). Claudin-5 is a major component of TJ in brain endothelial cells, and expressed in subsets of lung and kidney blood vessels (Morita et al. 1999). Administration of aqueous tracer molecules of varying size to these transgenic mice showed a size selective penetration of tracers less than 800 Daltons into the brain. However, electron microscopy of the Claudin-5 knockout mouse showed that brain endothelial cell TJ appeared normal (Nitta et al. 2003).
The global gene knockout that abrogates expression in all tissues and cell types can be useful to determine the overall contribution and function of a protein through removal of that protein. However, estimating the role of a protein specifically in the brain, and eliminating confounding factors in the body, is essential to the development of therapeutic agents targeted to the brain and CNS. Elimination of a gene in a single tissue versus multiple tissues can produce different outcomes. In addition, gene deletion across all tissues may result in lethality, such as those mentioned for the TJ protein knockouts.
Targeted gene knockouts can be used to minimize these confounding effects. The technology to ablate gene function in a temporal and/or spatial-specific manner has been extensively used in the field of developmental biology, where global gene disruption is often fatal. With regard to genes more commonly associated with drug disposition, significantly less research has been published. The methodology of choice for selective gene ablation uses the Cre-recombinase/loxP system, employing cell-type specific promoters for conditional expression of Cre-recombinase (Cre). Cre is a 38 kD protein originating from the bacteriophage P1, and catalyzes a site specific recombination reaction between two 34-base-pair loxP (locus of crossing over in P1) sites, which are not present in the mammalian genome (Sternberg and Hamilton 1981; Thyagarajan et al. 2000; Semprini et al. 2007). When two direct orientation loxP sites are present within a modified endogenous target gene or transgene, Cre will excise the intervening DNA between the sites, leaving one loxP site behind. A gene containing two loxP sites flanking essential exons can be deleted (or knocked out) in whichever cell types express Cre (Nagy 2000). Conditional activation of a transgene can also be accomplished by placing a flanked loxP (commonly referred to as a floxed) stop cassette between the promoter and gene. The stop cassette is removed upon Cre-mediated excision, and results in transgene expression (Lakso et al. 1992).
While over 500 Cre transgenic mouse lines expressing Cre in various tissues have been created (Nagy and Mar 2001; Nagy et al. 2009), there is no transgenic mouse that expresses Cre exclusively in the brain endothelium or CP. Therefore, the focus of this project is to develop a mouse model that will allow for gene knockout specifically at the BBB or B-CSF-B. To accomplish this, our aim was to create transgenic mice that express Cre in the brain endothelium or CP in a cell-type specific manner. To target the BBB, we chose the −487 to +247 fragment of the human von Willebrand Factor gene (promoter and first exon) (referred to as hVWF-f) that has been used to target gene expression specifically to brain endothelium (Jahroudi and Lynch 1994; Aird et al. 1995; Jahroudi et al. 2003). For CP selection, we chose the lymphotropic papovavirus control region (referred to as LPVcr) that was shown to produce tumors in the CP when driving expression of the SV40 Large T antigen (Chen et al. 1989; Chen and Van Dyke 1991). However, because the LPVcr-SV40 Large T antigen mice died between 1 and 3 months due to tumor formation, we evaluated the LPVcr expression profile in adult mice, using it to control the n-LacZ gene to ensure safety, prior to constructing the Cre constructs. Evaluation of tissue selective Cre expression and function under the control of hVWF-f and LPVcr was achieved using the dual-reporter Z/EG (loxP-nLacZ-neo-PolyA-loxP/EGFP) construct and transgenic mouse line (Novak et al. 2000). Our results suggest that the hVWF-f and LPVcr provide brain endothelium and CP selective Cre expression, respectively.
Materials and methods
Cells and animals
The bEnd.3 and K562 were purchased from American Type Culture Collection (ATCC, Manassa, VA). The cells were maintained in DMEM (Gibco-BRL, Rockville, MD) containing 10% FCS (Gibco-BRL) and 1% antibiotic/antimycotic (Gibco-BRL). The Z/EG (B6.Cg-Tg(CAG-Bgeo/GFP)21Lbe/J) and EIIa-Cre (FVB/N-Tg(EIIa-cre)C5379Lmgd/J) transgenic mice were purchased from The Jackson Laboratory (Bar Harbor, Maine). C57/B16 wild type mice were purchased from Charles River Laboratories (Wilmington, MA). All animal work was conducted under IACUC approved protocols. All reagents used were analytical grade or higher.
Plasmid construction
LPV-LacZ and LPV-Cre
The LPV control region (LPVcr) was removed from the pLST plasmid (Chen and Van Dyke 1991) by _Hin_dIII digestion. The LPVcr was then ligated into the multiple cloning site (MCS) of the pBluescript II KS (−) plasmid (Stratagene Products, La Jolla, CA) at the _Hin_dIII site, resulting in plasmid pEK2-2. The n-LacZ gene was removed from the pLacF plasmid and subcloned into pEK2-2 to create the LPV-LacZ construct.
The Cre expression plasmid was made by digesting the expression plasmid pTurboCre (Washington University ES Cell Core) with _Eco_RI to excise the NLS-Cre fragment. The NLS-Cre fragment was separated from the vector backbone using agarose gel electrophoresis and purified from the gel via electro-elution. The mammalian expression plasmid pCI (Promega, Madison, WI) was linerized with _Eco_RI and ligated with the NLS-Cre fragment and used to transform electrocompetant DH-5 E-coli (Strategene, La Jolla, CA). The transformed E-coli were seeded onto LB agar plates with ampicillin (100 ug/ml). Colony lifts using nitrocellulose filters were hybridized with a Cre DNA probe labeled with 50 μCi [_α_-32P]dCTP, 3,000 Ci/mmol (New England Nuclear, Boston, MA) using the Random Primers DNA Labeling System (Invitrogen, Carlsbad, CA) to detect positive clones. Positive colonies were picked, grown in LB-ampicillin growth medium and the pCI-Cre plasmid was extracted using the Qiagen Plasmid Mini Prep kit (Qiagen, Valencia, CA). Directionality was determined with restriction digestion using _Eco_RI, _Bam_HI, and _Cla_I. PCR amplification of the Intron-NLS-Cre-PolyA sequence was done using the following primers:
- pCI-Intron-For: GTA AGT ATC AAG GTT ACA AGA CAG G
- pCI-PolyA-Rev: CCA CTA ATG TAG AGG TTT TAC ATG C
The Intron-NLS-Cre-PolyA PCR product was subcloned into pEK2-2 by blunted SalI site ligation within the MCS to generate LPV-Cre. Colony lifts, clone selection, growth, and plasmid extraction were done as above. Directionality was verified using _Not_I and _Xho_I digestion. Sequence verification was completed using the following primers:
- pCI-Cre-1310-For: CGC AGA ACC TGA AGA TGT TCG
- pCI-Cre-1873-For: CCA GCC AGC TAT CAA CTC GCG
- LPV-3′: TAA GGC CAC CTA GGT AAT TAA
- M13(-20): GTA AAA CGA CGG CCA GT
- M13 Reverse: AAC AGC TAT GAC CAT G
hVWF-Cre
The −487 to +247 fragment of the hVWF gene was removed from the pHGH-K plasmid (Jahroudi and Lynch 1994) by SalI digestion, and inserted into the SalI site of pBluescript II KS (−) plasmid. The directionality was determined by restriction digestion using _Hin_dIII. The resulting plasmid was named pEK8-3. The PCR amplified Intron-NLS-Cre-PolyA sequence from above was subcloned into pEK 8-3 by blunted _Eco_RV site ligation within the MCS and named hVWF-Cre. Colony lifts, clone selection, growth, and plasmid extraction were done as above. Directionality was verified using _Bam_HI digestion. The plasmid integrity was then verified by sequencing.
Microscopy
Microscopy was performed using an Zeiss AxioVert 200 inverted microscope. Images were captured using Zeiss Axiovision software and a Zeiss MRc color camera.
In vitro evaluation of Cre expression plasmids
K562 and bEnd.3 cells were transfected with 10 μg of Z/EG plasmid (gift from Corrinne Lobe) or mock plasmid by electroporation (Gene Pulser II, BioRad, Hercules, CA) with the voltage and capacitor parameters set at 250 V/975 μF and 275 V/960 μF, respectively, using 0.4 cm cuvette. Starting 2 days post transfection, cells were subjected to G418 selection starting at 200 μg/ml and increasing to 800 μg/ml over 2 weeks. To verify Z/EG expression, the K562 and bEnd.3 cells were seeded into six-well plates and grown overnight. Cells were fixed in 4% paraformaldehyde for 15 min, and washed three times with PBS, then stained in LacZ stain solution (0.5 mg/ml X-gal, 5 mM potassium ferrocyanide, and 5 mM potassium ferricyanide in PBS) for 6 h at 37°C. Cells were washed three times in PBS and photographed using a light microscope with a 20× objective.
To assess the cell-type specificity of the LPV-Cre and hVWF-Cre plasmids, K562-Z/EG and bEnd.3-Z/ EG cells were transfected with 10 μg of pCI (mock), pTurboCre (under the control of the chicken beta-actin promoter), LPV-Cre, or hVWF-Cre plasmid with Lipofectamine 2,000 (Invitrogen, Carlsbad, CA) as per the manufacturer's instructions. The cells were then seeded into six-well plates and grown. Six days post transfection, cells were visualized on a fluorescent microscope for enhanced green fluorescent protein (EGFP) expression with a 20× objective.
Transgenic mouse production
The LPV-LacZ injection construct was excised from the LPV-LacZ plasmid with _Pvu_II restriction enzyme to remove the pBluescript-II KS (−) backbone. The fragments were separated using agarose gel electrophoresis and the LPV-LacZ fragment was removed from the gel via electro-elution, phenol–chloroform extracted, ethanol precipitated and resuspended in sterile double-distilled water. The LPVcr-Intron-NLS-Cre-PolyA fragment was excised from the LPV-Cre plasmid with _Pvu_II restriction enzymes to remove the pBluescript-II KS (−) backbone. The fragments were separated using agarose gel electrophoresis and the 2.3 kb fragment containing the Cre fragment was removed from the gel via electro-elution. The hVWF-f-Intron-NLS-Cre-PolyA fragment was amplified by PCR with the M13-20fwd and T3rev primers, phenol–chloroform extracted, ethanol precipitated, and resuspended in sterile double-distilled water.
T3Rev: ATT AAC CCT CAC TAA AGG GA
Mouse lines were generated by the Transgenic Animal Core of the Center for Ecogenetiics and Environmental Health (P30 ES0733). These three DNA fragments were each given to the University of Washington Department of Comparative Medicine Transgenic Resource Program for pronuclear microinjection into embryos produced by mating B6C3 females to C57BL/6 males to generate founder animals which are 75% C57BL/6 and 25% C3H. Offspring were weaned and DNA was isolated from tail snips or ear punches using the DNeasy Tissue Kit (Qiagen, Valencia, CA) for genotyping. Four carriers of each transgene from the F0 generation were mated with C57Bl6 mates. LPV-Cre, hVWF-Cre, or EIIa-Cre (Lakso et al. 1996) (used as a Cre positive control) transgenic mice were then mated with Z/EG stock mice [The Jackson Laboratory, Bar Harbor, Maine. (B6.Cg-Tg(ACTB-Bgeo/GFP)21Lbe/J; JAX #3920)] to characterize the location of Cre-mediated activity.
Genotyping
Genomic DNA was isolated from tail snips using the DNeasy Tissue Kit (Qiagen, Valencia, CA). Genotyping was performed by PCR amplification [2 min at 95°C, 38 cycles of (30 s at 95°C, 30 s at 59°C, 2 min at 72°C), 10 min at 72°C] and visualized by agarose gel electrophoresis. To verify that the LPV-Cre founder mice had the full-length construct, PCR amplification of LPVcr-Intron-NLS-Cre-PolyA was done using the LPV3FWD and pCI-PolyA-Rev primers.
- LPV3FWD: TTC AGC AGC AAT TTC AGA AAT ATT
- pCI-PolyA-Rev: CCA CTA ATG TAG AGG TTT TAC ATG C
LPV-LacZ mice were genotyped using the LacZ-111FWD and LacZ-611REV primers to PCR amplify a 500 base pair fragment of the n-LacZ gene.
- LacZ-111FWD: TAA TAG CGA AGA GGC CCG C
- LacZ-611REV: CGC CAC ATA TCC TGA TCT TCC
To genotype the LPV-Cre and hVWF-Cre transgenic offspring for the Cre transgene, the Cre-408-FOR and Cre-408-Rev primers were used to PCR amplify a 408 base pair fragment of the Cre gene.
- Cre-408-FOR: GCA TTA CCG GTC GAT GCA ACG AGT GAT GAG
- Cre-408-REV: GAG TGA ACG AAC CTG GTC GAA ATC AGT GCG
The Z/EG and Cre × Z/EG dual transgenic mice were genotyped using the LacZ-111FWD and LacZ-611REV for the n-LacZ gene and EGFP-294F and EGFP-594R to amplify a 300 base pair fragment of the EGFP gene.
- EGFP-294F: CTA CGG CGT GCA GTG CTT C
- EGFP-594R: GAA GTT CAC CTT GAT GCC GTT C
Fatty acid binding protein I (FABPI) was used as the positive control for DNA extractions and PCR reactions using the FABPI-500-fwd and FABPI-500-rev primers (Stratman et al. 2003).
- FABPI-500-fwd: CCT CCG GAG CGC AGC GAT TAA AAG TGT CAG
- FABPI-500-rev: TAG AGC TTT GCC ACA TCA CAG GTC ATT CAG
Transgenic mouse tissue collection and preparation
Dual transgenic mice were anesthetized by CO2 asphyxiation followed by cardiac exsanguination. The brain, liver, kidney, intestine and spleen were removed and either frozen on dry ice for mRNA and protein extraction, or embedded in OCT and frozen on dry ice for histology.
Southern blot of Cre transgenic mice
One F1 and one F2 mouse from each mouse line was sacrificed and the liver was taken for DNA. Genomic DNA was obtained by phenol/chloroform extraction and rehydrated in TE buffer. The genomic DNA was then cut with either _Eco_RI, _Nco_I, and _Hpa_I restriction endonucleases. Fifteen micrograms of digested DNA was then separated by agarose gel electorphoresis. The gel was depurinated in 0.2 N HCl, denatured in 0.5 M NaOH, 1.5 M NaCl and neutralized in 1 M Tris pH 7.4, 1.5 M NaCl. The DNA was then transferred to a nylon membrane and cross-linked to the membrane with UV light. The 1.1 kb _Eco_RI Cre containing fragment from the pTurboCre plasmid was freshly labeled with 50 μCi [_α_-32P]dCTP, 3000 Ci/ mmol (New England Nuclear, Boston, MA) using the Random Primers DNA Labeling System (Invitrogen, Carlsbad, CA) at a specific activity of > 108 CPM/ug and denatured at 95°C for 5 min. The blot was pre-hybridized with Quickhyb (Stratagene Products, La Jolla, CA) at 68°C for 30 min. Ten microliters of 7 × 105 CPM/ml probe was then added and hybridized for 3 h at 68°C. This was followed by two 15 min washes in 2× SCC, 0.1% SDS at room temperature, and one 15 min wash in 0.2× SCC, 0.1% SDS at room temperature. The blot was exposed to X-ray film for 5 days at −70°C with an intensifying screen before development.
Dot blot analysis of copy number in Cre transgenic mice
One F1 and one F2 mouse from each mouse line was sacrificed and the liver was taken for DNA. The EIIa-Cre mouse as a positive control, and a wild-type C57/ Bl6/J as the negative control. Genomic DNA was obtained by phenol/chloroform extraction and rehydrated in TE buffer. Three micrograms of DNA was denatured and spotted onto a nylon membrane along with serial dilutions of the 1.1 kb _Eco_RI Cre containing fragment from the pTurboCre plasmid and were cross-linked to the membrane with UV light. Hybridization and washing were preformed using the same probe and method as the Southern blot described above.
RNA purification, reverse transcription and amplification
Tissue samples (∼ 50 mg) were homogenized and lysed in 500 μl TRI-Reagent and incubated for 5 min at RT. The lysate was centrifuged at 12K × gravity for 10 min at 4°C. The supernatant was transferred to a clean tube and 50 μl BCP (bromo chloropropane) Phase Separation Reagent was added, shaken for 15 s, incubated for 10 min at room temperature, and centrifuged at 12K×g for 15 min at 4°C. The top, clear, aqueous layer was transferred to a clean tube, 250 μl isopropanol was added, vortexed for 15 s, and incubated for 10 min at room temperature. This was then centrifuged at 12K×g for 8 min at 4°C to pellet the RNA. The supernatant was removed and 500 μl 75% cold ethanol was added to wash the pellet. The tube was centrifuged at 7,500×g for 5 min at 4°C, and the pellet was rehydrated in 50 μl TE buffer. Residual genomic DNA was removed using the Turbo DNA-Free assay kit (Applied Biosystems, Foster City, CA). The RNA concentration was determined by UV absorption at 260 nm and 280 nm.
One microgram of RNA was reverse transcribed to cDNA with the Applied Biosystems Reverse Transcription Kit (Applied Biosystems, Foster City, CA, catalog # N808-0234) using random hexamers. The cDNA was analyzed by either Real-Time PCR (thermocycler conditions: [2 min at 95°C, 40 cycles of (15 s at 95°C, 60 s at 60°C]) for nLacZ, and mouse Gusb, or PCR (thermocycler conditions: [2 min at 95°C, 38 cycles of (30 s at 95°C, 30 s at 59°C, 2 min at 72°C), 10 min at 72°C]) using the genotyping primers for EGFP, LacZ, or Cre.
- mGusB: ABI TaqMan® Gene Expression Assay Mm00446953_m1
- LacZ.tmp: 6FAM TAC CCC GTA CGT CTT CCC GAG CG TAMRA
- LacZ-taq.rev: CTG TTG ACT GTA GCG GCT GAT G
- LacZ-taq.for: GGA TCT GCC ATT GTC AGA CAT G
Fractionation of mouse tissue
Frozen tissue was homogenized with a tissue grinder in three volumes of KPi homogenization buffer (10 mM KH2PO4, 250 mM sucrose, pH 7.4 with 1 M KOH). The homogenate was centrifuged at 10,000×g for 30 min at 4°C. The supernatant was transferred to a clean tube and protein concentration was measured with the BioRad Dc Protein determination assay.
Western immunoblot
SDS–PAGE electrophoresis was performed with 40 μg of tissue homogenate per lane in a 4–20% gradient SDS–PAGE gel. The protein was transferred to a PDFV membrane and blocked in 5% NFDM/ tris-buffered saline-tween20 (TBST) for 1 h at RT or overnight at 4°C. The blot was incubated with primary antibody diluted in 5% non-fat dry milk (NFDM)/TBST for 90 min. The blot was washed 3 times for 5 min each in TBST, followed by incubation with the horseradish peroxidase (HRP) labeled secondary antibody in 5% NFDM/TBST for 60 min. It was then washed three times for 5 min in TBST. The blot was incubated in ECL-Plus for 5 min, exposed to X-ray film, and developed.
Immunohistochemistry (IHC)
The brain, liver, kidney, intestine and spleen of the transgenic mice were dissected and embedded directly into OCT (Tissue Tek, Sakura Finetec, Torrance, CA), frozen on dry ice and cryosectioned at 25 μm and placed onto Superfrost Plus slides and dried overnight. The slides were fixed in ice-cold acetone for 5 min, dried for 30 min, and rehydrated in TBS buffer for 15 min. The tissue was blocked for 30 min with 5% serum from the secondary antibody host. The tissue was incubated with the primary antibody at 25°C for 2 h. The slides were washed 3 times for 5 min each in TBS and incubated with the secondary antibody for 60 min. Slides were washed three 3 for 5 min each in TBS. The HRP labeled secondary antibody was developed with 3,3′-Diaminobenzidine (DAB) reagent, resulting in brown precipitant, followed by washing in tap water for 2 min. Slides were dehydrated in 90, 95 and 100% ethanol and cleared in three changes of Xylene for 5 min each. The slides were coverslipped with Permount and viewed on a light microscope with a 20× objective.
Results
Evaluation of the LPV control region for brain specific expression
Due to the lack of information for the expression profile of the LPVcr in adult mice, we evaluated the LPVcr as a candidate for B-CSF-B expression using the _β_-galactosidase (_β_-gal) reporter gene (n-LacZ) under the control of the LPVcr. Following pronuclear microinjection of the LPV-LacZ construct into fertilized mouse oocytes, three independent transgenic lines of LPV-LacZ mice were generated. The LPV-LacZ.3753 line did not produce any DNA positive offspring. The LPV-LacZ.3750 line did not have detectable n-LacZ mRNA transcripts in any tissue tested by Real-Time RT–PCR, while the LPV-LacZ.3746 line expressed n-LacZ mRNA in the brain and spleen but not the liver, kidney, or intestine (Fig. 1a). Histochemical staining with the LacZ specific fluorescent compound Ima-gene Red showed CP specific expression of the _β_-gal enzyme within the brain when compared to LacZ positive and negative control mice (data not shown).
Fig. 1.
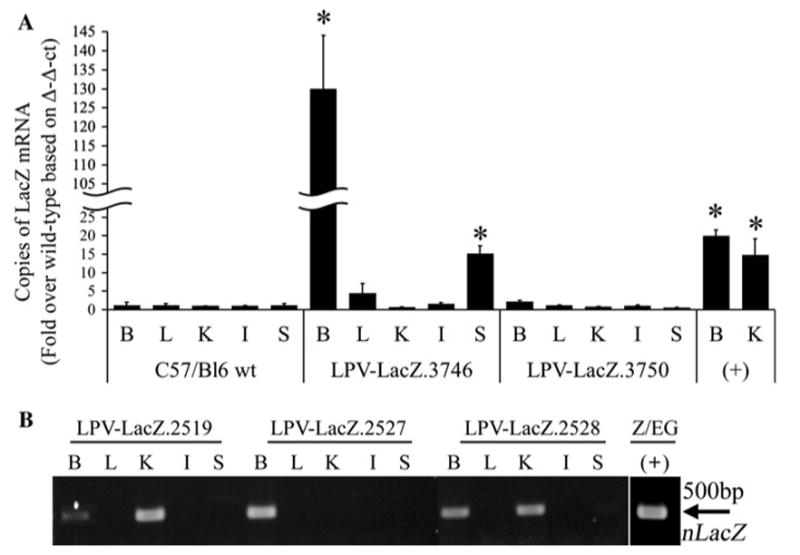
Analysis of LacZ mRNA transcripts in LPV-LacZ transgenic mice. Brain (B), liver (L), kidney (K), intestine (I), and spleen (S) tissue samples were homogenized and RNA was extracted using TRI-Reagent and isopropanol precipitation. The mRNA was reverse transcribed to cDNA and analyzed by Real-Time PCR for LacZ (a) or by PCR using the LacZ-111FWD and LacZ-611REV primers (b). * P < 0.01 compared to C57/Bl6 wild type
A second microinjection of LPV-LacZ was completed and resulted in five DNA positive founders that could be evaluated. RT–PCR was performed on the RNA from the brain, liver, kidney, intestine and spleen. Three of the five founders (LPV-LacZ.2519, LPV-LacZ.2527, and LPV-LacZ.2528) were positive for n-LacZ RNA in the brain (Fig. 1b). While there was no ectopic expression in the LPV-LacZ.2527 mouse liver, kidney, intestine or spleen, there was ectopic kidney expression in the LPV-LacZ.2519 and LPV-LacZ.2528 lines. Immunohistochemical analysis of the three LPV-LacZ RNA positive mice was performed to determine the location of _β_-gal expression within the brain. Strong positive staining was shown in nuclei of the epithelial cells lining the ventricles that form the choroid plexus, but not in any other cell types of the brain (Fig. 2).
Fig. 2.
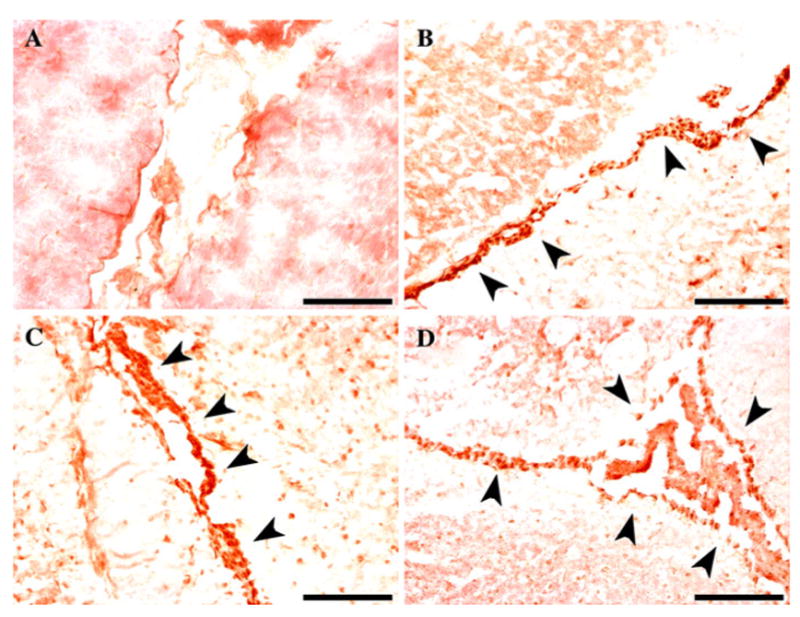
Immunohistochemistry detection of _β_-gal in the choroid plexus of LPV-LacZ transgenic mice. Filled arrow indicates positive LacZ immunostaining in the brain of C57/Bl6 wild type (a), LPV-LacZ.2519 (b), LPV-LacZ.2527 (c), and LPV-LacZ.2528 (d) mice Black bar = 100 μm
The data from the LPV-LacZ.3746 and LPV-LacZ.2527 mice supported the published expression profile for the LPVcr, showing CP and spleen expression (Chen et al. 1989; Chen and Van Dyke 1991′). Therefore, the LPVcr was deemed a viable candidate to drive B-CSF-B targeted expression of future constructs.
Development of Cre-recombinase transgenic mice under the control of LPVcr and hVWF-f
In vitro expression and function of Cre-recombinase constructs
Following the evaluation of the LPVcr for B-CSF-B expression, and based on previous data supporting the BBB targeted expression by the hVWF-f, we created two constructs using these promoters to drive Cre expression. Assessment of the LPV-Cre and hVWF-Cre plasmids for Cre expression and function was completed in vitro. The bEnd.3 and K562 cells were stably transfected with the Z/EG plasmid and selected with G418 for stable expression. The bEnd.3-Z/EG and K562-Z/EG cells were then transiently transfected with pTurboCre, LPV-Cre or hVWF-Cre, and the cells were examined for expression of EGFP, indicating Cre-mediated endonuclease activity. These studies showed that pTurboCre was able to invoke gene switching from LacZ to EGFP in both cell types (Fig. 3). The hVWF-Cre plasmid produced LacZ to EGFP gene switching only in the bEnd.3-Z/EG cells. The LPV-Cre mediated gene switching from LacZ to EGFP was shown in both K562-Z/EG and bEnd.3-Z/ EG cells (Fig. 3). These data suggest that both the LPV-Cre and hVWF-Cre plasmids were capable of expressing functional Cre enzyme.
Fig. 3.
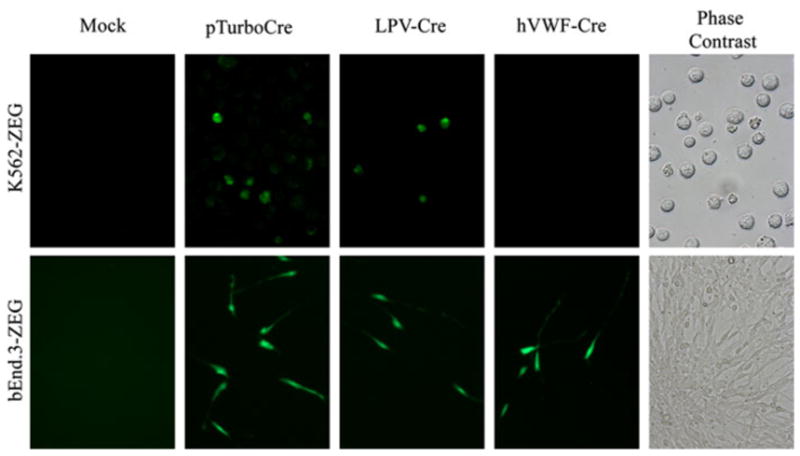
Transient transfection of bEnd.3-Z/EG and K562-Z/EG cells with Cre expression plasmids
Cre-recombinase transgenic mouse production and progeny
The LPVcr-Intron-Cre-PolyA fragment was microinjected into fertilized oocytes to create LPV-Cre transgenic mice, generating two DNA positive founder lines (F0). However, the F0 mice were infertile, producing no offspring (F1 generation). The LPV-Cre construct was submitted for a second microinjection, which resulted in twelve live births; four DNA pups were positive for the Cre transgene DNA. The four founders were mated with C57/Bl6 mice to generate hemizygotic F1 generations. Two LPV-Cre founders mated but did not produce DNA positive offspring (LPV-Cre.0600 and LPV-Cre.0610). Two LPV-Cre lines successfully mated and produced DNA positive F1 offspring (LPV-Cre.0605 and LPV-Cre.0607) according to Mendelian genetics. The transgene carrying F1 mice were back-crossed with C57Bl6 mice to produce the F2 generation.
The hVWF-f-Intron-Cre-PolyA fragment was amplified by PCR from the hVWF-Cre plasmid and microinjected into fertilized oocytes. This injection yielded thirty-two pups, with four pups DNA positive for the transgene. These four mice were mated with C57/Bl6 mice to generate the hemizygotic F1 generation mice. One hVWF-Cre founder was infertile (hVWF-Cre.1332), while another hVWF-Cre founder did not produce any DNA positive pups (hVWF-Cre.1301). The remaining two hVWF-Cre lines (hVWF-Cre.1304 and hVWF-Cre.1329) successfully mated and produced DNA positive F1 offspring according to Mendelian genetics. The transgene carrying F1 mice were backcrossed with C57/Bl6 mice to produce the F2 generation.
Determination of stable integration of the LPV-Cre and hVWF-Cre transgenes into the mouse genome
To evaluate the number of transgene copies that are present in the genomes of the Cre transgenic lines, a dot blot analysis was performed with genomic DNA from the F1 and F2 generations of each line. These results showed that when bred as hemizygotes, the LPV-Cre.605 and hVWF-Cre.1304 each carried three copies of their respective transgene, and LPV-Cre.607 carried four copies.
To evaluate whether the LPVcr-Intron-Cre-PolyA and hVWF-f-Intron-Cre-PolyA transgenes were stably integrated into the genome, Southern blot was performed with genomic DNA from the F1 and F2 generations of each line. The endonucleases _Hpa_I and _Nco_I are single cutters within both transgenes, and were chosen to allow visualization of the 5′ and 3′ tails of the Cre gene, respectively (Fig. 4). While _Hpa_I cuts within the PolyA sequence, the _Nco_I enzyme cuts 27 bp into the 1.1 Kb Cre sequence. To verify that the _Hpa_I and _Nco_I endonucleases cleaved the DNA in the correct location, a double digestion of the LPV-Cre and hVWF-Cre plasmids was performed. The correct size bands of approximately 1,250 base pairs (bp) and 3,900 bp for LPV-Cre, and, 1,250 and 4,025 bp for hVWF-Cre were visualized after agarose gel electrophoresis (data not shown).
Fig. 4.
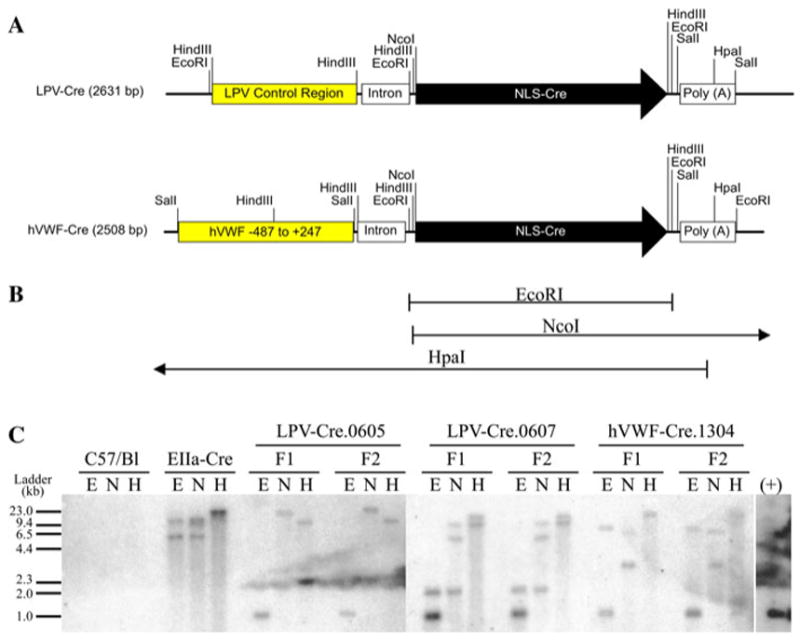
LPV-Cre and hVWF-Cre Transgene maps and Southern blot for Cre. a DNA maps of LPV-Cre and hVWF-Cre injected transgenes. b Expected bands for Southern blot restriction enzyme digestion. c Southern blot of F1 and F2 mice using genomic DNA cut with _Eco_RI (E), _Nco_I (N), and _Hpa_I (H) restriction endonucleases
The Southern blot restriction digestion pattern for each line was consistent between F1 and F2 generations (Fig. 4). There was no signal for Cre in the wild type mouse. The size of the DNA fragments for EIIa-Cre were approximately 6 and 15 Kb for _Eco_RI, 6 and 15 Kb for _Nco_I, and 23 Kb for _Hpa_I. The sizes of the DNA fragments for LPV-Cre.0605 were approximately 1 Kb for _Eco_RI, 2 and 23 Kb for _Nco_I, and 15 Kb for _Hpa_I. The sizes of the DNA fragments for LPV-Cre.0607 were approximately 1 and 2 Kb for _Eco_RI, 2, 5, and 10 Kb for _Nco_I, and 10 and 15 Kb for _Hpa_I. The sizes of the DNA fragments for hVWF-Cre.1304 were approximately 1 and 9.5 Kb for _Eco_RI, 3 and 9.5 Kb for _Nco_I, and 23 Kb for _Hpa_I. These data indicate that the injected Cre transgenes were incorporated into the mouse genome, and exhibited stable germ line transmission.
The _Hpa_I digestions are valuable in showing consistency of integration between generations. However, the _Hpa_I banding pattern can be difficult to discern as this enzyme is influenced by methylation of the DNA. Therefore, using this enzyme to predict transgene orientation and conformation was not done. Nevertheless, the _Eco_RI and _Nco_I digestions, together with the transgene copy number afford us the ability to discern the likely transgene orientations. The LPV-Cre.605 banding pattern can be explained by having the three copies of the transgene inserted into the genome in a head-to-tail conformation (−>/ −>/ −>/). Taking into account the transgene copy numbers, based on the banding pattern in the LPV-Cre.607 and hVWF-Cre.1304, we can see multiple potential configurations. One scenario for the four copies in the LPV-Cre.607 line is that there is a 5′ head to head configuration, followed by two additional head to tail arrangements. This would explain the 5 and 10 Kb _Nco_I bands. Additionally, the 2 kB _Eco_RI fragment may be due to the loss of the 3′ end of the final transgene, removing the _Eco_RI restriction site, during the integration event (5′ \<− −>/ − >/ −>× 3′). We believe the three hVWF-Cre.1304 transgene copies are in a tail-to-tail, head to head configuration. In the case of _Nco_I, the 3 Kb band represents the tail to tail fragment, and the 9.5 Kb fragment represents the distal 3′ end. For _Eco_RI, we feel the 9.5 Kb band may represent the loss of 3′ _Eco_RI restriction side due loss of the 3′ end during the integration event (5′ −>/ \<− −>× 3′).
Characterization of tissue and cell-type specific Cre excision in transgenic mice
The expression and function of the LPV-Cre and hVWF-Cre transgenes were characterized for cell-type specific Cre excision by crossing the transgenic mice with dual reporter gene Z/EG mice. In non-Cre expressing cells, β-geo (a fusion of LacZ and neomycin phosphoribosyl transferase) is expressed and transcription is stopped via the three SV40 polyadenylation sequences immediately following β-geo. In cells types that express the Cre enzyme, excision of the loxP-β-geo-PolyA-loxP sequence within the Z/EG transgene will occur. The removal of the PolyA stop sequence will activate EGFP expression. Mice were genotyped, and those carrying both the Z/EG and Cre transgenes (LPV-Cre.605-Z/EG, LPV-Cre.607-Z/ EG, hVWF-Cre.1304-Z/EG, and hVWF-Cre.1329-Z/ EG) were necropsied and the tissues were evaluated for tissue and cell-type expression of EGFP using mRNA, protein, and histology. As controls, one EIIa-Cre-Z/EG dual transgenic mouse, one Z/EG, and one wild type littermate control were also necropsied for evaluation.
The presence of EGFP mRNA was determined by reverse transcription of RNA followed by PCR analysis of EGFP (Fig. 5a, b). EIIa-Cre-Z/EG mice express EGFP mRNA in all tested tissues. Transcripts of EGFP were also found in the brain and spleen of the LPV-Cre.605-Z/EG line, brain of the LPV-Cre.607-Z/EG line, brain of the hVWF-Cre.1304-Z/EG line, and liver of the hVWF-Cre.1329-Z/EG line.
Fig. 5.
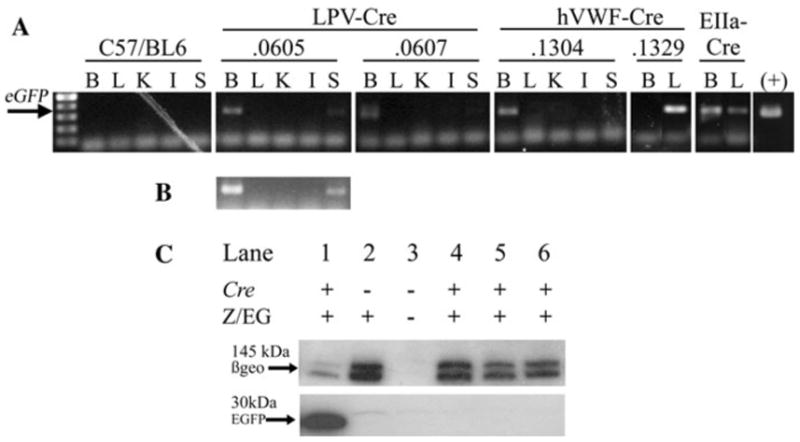
Analysis of EGFP and n-LacZ expression in Cre-Z/EG dual transgenic mice. a RT–PCR and amplification of EGFP mRNA from brain (B), liver (L), kidney (K), intestine (I), and spleen (S) tissue samples from Cre × Z/EG dual transgenic mice. b Overexposure of LPV-Cre.605 to show spleen expression. c Western immunoblot analysis by SDS–PAGE electrophoresis of tissue homogenate from the brain of EIIa-Cre-Z/EG (1), Z/EG (2), C57/BL6 wt (3), LPV-Cre.0605-Z/EG (4), LPV-Cre.0607-Z/EG (5), and hVWF-Cre.1304-Z/EG (6)
The homogenized tissues were analyzed for protein expression of EGFP and _β_-gal by Western immunoblot analysis. Western immunoblot analysis of _β_-gal expression was positive for all mice except the wild type negative control, confirming that the mice were expressing the Z/EG gene (Fig. 5). The EGFP protein was expressed in all tissues of the EIIa-Cre-Z/EG line and in the liver of the hVWF-Cre.1329-Z/EG line. EGFP protein in the tissues from the hVWF-Cre.1304-Z/EG or LPV driven Cre mice was undetectable (Fig. 5). Due to the ectopic expression of the Cre transgene in the liver of the hVWF-Cre.1329-Z/EG line, it was excluded from further evaluation.
Immunohistochemistry was performed to detect EGFP protein in tissue sections using a rabbit polyclonal anti-EGFP ab6556 antibody (Table 1). Adjacent sections were stained for either vWF (endothelium marker) or glial fibrillary acidic protein (GFAP) (astrocyte marker). EGFP specific staining was not detected in the liver, kidney, or intestine of the LPV-Cre.605-Z/EG, or LPV-Cre.607-Z/EG, and hVWF-Cre.1304-Z/EG mice. Positive staining for EGFP was shown in the spleen of the LPV-Cre.605-Z/EG mouse line (in agreement with the mRNA data) but not in the LPV-Cre.0607-Z/EG or hVWF-Cre.1304-Z/EG lines (Fig. 6). If ectopic expression due to the hVWF-f promoter expressing in all endothelial cells was present, this would have shown an expression pattern similar to that shown by the vWF specific antibody (Fig. 6b). Antibody specific staining for EGFP was seen in the brains of the LPV-Cre.605-Z/EG, LPV-Cre.607-Z/EG mice, and hVWF-Cre.1304-Z/EG mice. LPV-Cre.605-Z/EG and LPV-Cre.607-Z/EG brains both showed positive EGFP immunostaining that was restricted to the epithelium of the CP (Fig. 7). The LPV-Cre.607-Z/EG line showed stronger staining of the CP than the LPV-Cre.605-Z/EG line. The brain of the hVWF-Cre.1304-Z/EG line showed EGFP specific staining in the general brain endothelium, including some endothelial cells located in the CP. The immunostaining of vWF protein in adjacent sections showed overlapping locations to that of the EGFP for both the CP (Fig. 7) and endothelium (Fig. 8) in the hVWF-Cre.1304-Z/EG mouse. No EGFP expression overlapped with the GFAP expression in any of the lines, verifying that Cre, and consequently EGFP, is not expressed in the astrocytes.
Table 1. Primary antibodies used for Western immunoblot and immunohistochemistry.
| Antibody | Antigen | Dilution | Source | |
|---|---|---|---|---|
| WB | IHC | |||
| Ab6556 | Green fluorescent protein | 1:5000 | 1:1000 | Abcam |
| Ab616 | _β_-Galactosidase | 1:1000 | 1:500 | Abcam |
| VWF (C-20) | von Willebrand factor | n.a. | 1:50 | Santa Cruz Biotech |
| GFAP (C-19) | Glial fibrillary acidic protein | n.a. | 1:50 | Santa Cruz Biotech |
Fig. 6.
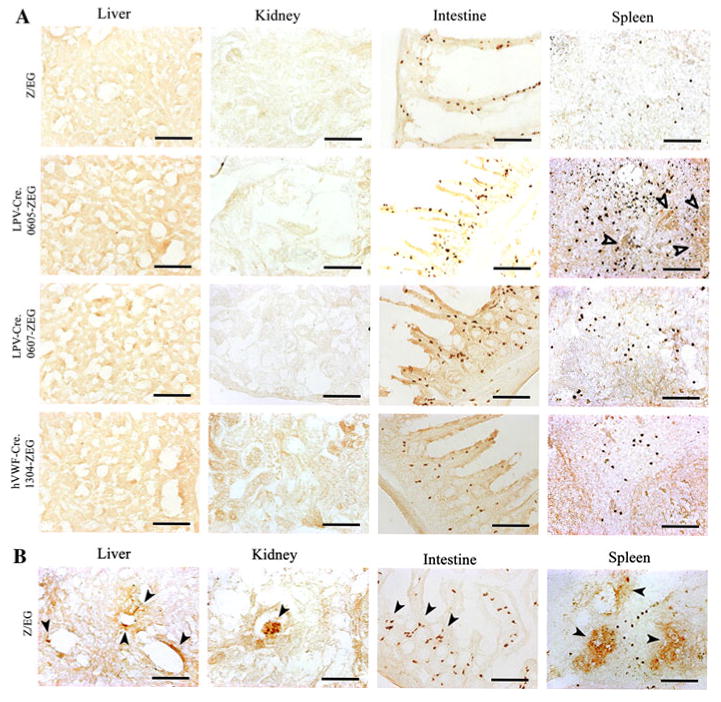
Immunohistochemistry of the liver, kidney, intestine and spleen Cre-Z/EG dual transgenic mice. Immunohistochemistry was performed with EGFP (a) or vWF (b) specific antibodies Hollow arrow indicates positive EGFP immunostaining. Filled arrow indicates positive vWF immunostaining. Black bar = 100 μm
Fig. 7.
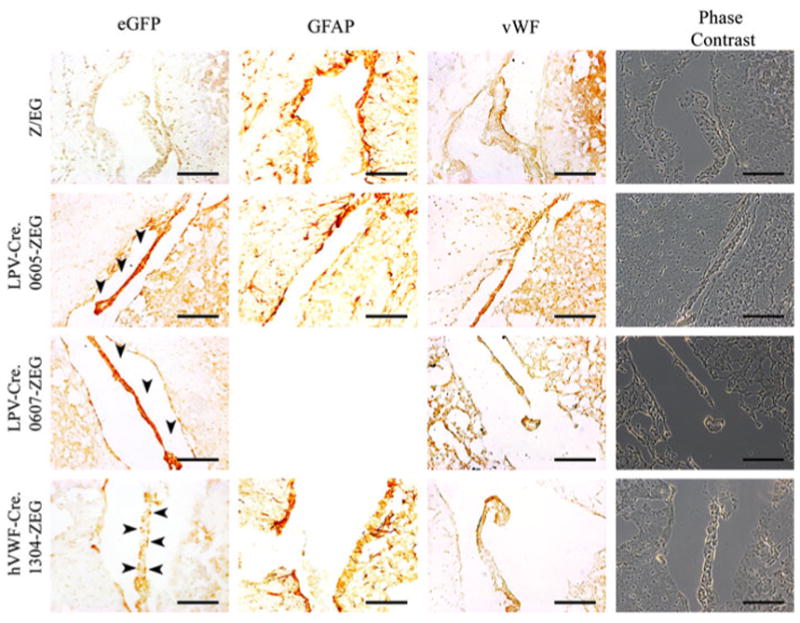
Immunohistochemistry of the choroid plexus of Cre-Z/ EG dual transgenic mice. Filled arrow indicates positive EGFP immunostaining in the brain from Z/EG, hVWF-Cre.1304-Z/EG, LPV-Cre.0605-Z/EG, and LPV-Cre.0607-Z/EG mice. Black bar = 100 μm
Fig. 8.
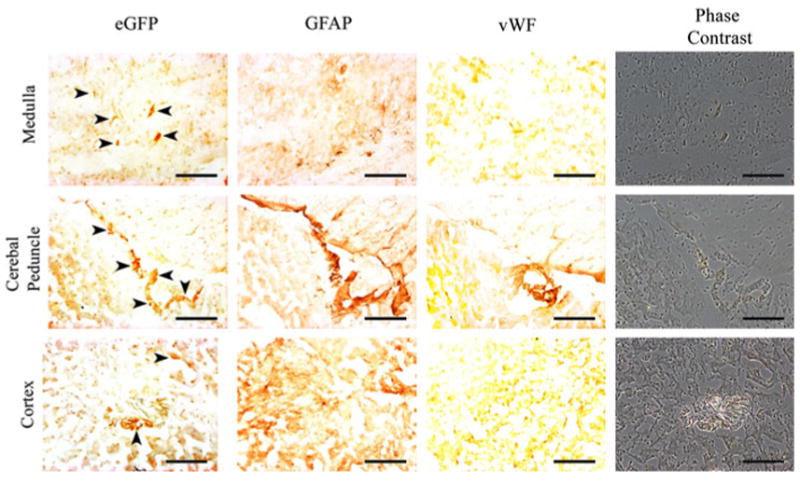
Immunohistochemistry of the hVWF-Cre.1304-Z/EG dual transgenic brain. Filled arrow indicates positive EGFP immunostaining in the brain from Z/EG, hVWF-Cre.1304-Z/EG, LPV-Cre.0605-Z/EG, and LPV-Cre.0607-Z/EG mice. Black bar = 100 μm
Collectively, the data show that the LPVcr and hVWF-f do not exhibit expression of the Cre gene in the liver, kidney or intestine. The LPVcr restricts expression to the CP in the 0607 line and the brain and spleen in the 0605 line, while the hVWF-f restricts expression to the brain endothelium.
Discussion
The focus of this research was to develop transgenic mouse models that will allow for gene knockout specifically at the BBB or B-CSF-B. We created transgenic mice that express Cre-recombinase in the brain endothelium or choroid plexus in a cell-type specific manner using the −487 to +247 fragment of the hvWF gene and the LPVcr respectively. The resulting LPV-Cre and hVWF-Cre mice were then evaluated for cell-type targeted Cre expression and function by crossing them with the dual reporter mouse Z/EG. Our results from mRNA and IHC suggest that the hVWF-f and LPVcr provide brain endothelium and CP selective Cre expression, respectively.
Compared to other endothelium specific promoters such as the endothelium receptor tyrosine kinase Tek promoter/enhancer (Koni et al. 2001) or VE-Cadherin (Cdh5) mouse promoter (Alva et al. 2006), which limit expression to endothelium throughout the vasculature, the hVWF-f targets only brain endothelium in adult mice (Jahroudi and Lynch 1994; Aird et al. 1995; Jahroudi et al. 2003). Choroid plexus expression can be seen using the transthyretin _cis_-regulatory elements that direct visceral endoderm expression in the mouse embryo, but they also show hepatocyte expression in addition to the CP in adults (Kwon and Hadjantonakis 2009). The FOXJ1 promoter was shown to have CP expression, but also drives expression in ependyma, oviduct, testis, and ciliated epithelial cells present in pulmonary airways (Zhang et al. 2007).
Our focus is specifically on the BBB and B-CSF-B, as limited penetration into the brain for many CNS active drugs remains an obstacle for drug delivery. Research has focused on delineating the extent to which differing protein components of the BBB and B-CSF-B function to reduce penetration of compounds into the brain, and ways to overcome them. Gene knockout mouse models have attempted to quantitate this. However, the elimination of a gene from the entire genome can be lethal (i.e. TJ proteins), or have confounding effects due to elimination of the gene from other tissue types (i.e. efflux transporters). To this extent, we created two mouse models for conditional gene knockout specifically at the BBB or the B-CSF-B.
Although previous studies with LPVcr driving the SV40 Large T antigen demonstrated that these sequences resulted in tumor formation in the CP (Chen and Van Dyke 1991), further confirmation of the restriction of this promoter's activity to the CP was warranted. There is also documented atypical tumor formation in the ependymal tissue seen with SV40 T Antigen transgenes (Brinster et al. 1984; Messing et al. 1988; Chen and Van Dyke 1991; Perraud et al. 1992; Feigenbaum et al. 1992). Evaluation of tissue expression in adulthood in the LPV-SV40 T antigen mice could not be completed as these mice died within 3 month of birth due to the tumor formations. In addition, some transgenic mice that used the LPVcr to drive the LPV Large T antigen developed tumors in the spleen in addition to the CP. Additional evaluation of hVWF-f was not necessary as this sequence has been well characterized driving multiple reporter genes (Aird et al. 1995; Jahroudi et al. 2003).
We generated a construct with n-LacZ under the control of the LPVcr to create LPV-LacZ transgenic mice and analyzed the tissue specific expression of _β_-gal in adult mice. Four of the resulting LPV-LacZ transgenic lines (LPV-LacZ.3746, LPV-LacZ.2519, LPV-LacZ.2527, and LPV-LacZ.2528) showed evidence of n-LacZ mRNA in the brain. IHC showed that within the brain, expression was limited to the epithelium of the CP (Fig. 2). However, the mRNA also confirmed that n-LacZ was expressed ectopically in the spleen of LPV-LacZ.3746 and kidney of LPV-LacZ.2519 and LPV-LacZ.2528 (Fig. 1). Expressing a transgene ectopically in founder mice occurs often as integration into the host genome occurs at random locations, often at locations of double-strand chromosomal breaks (Brinster et al. 1985). At the site of integration, regulatory elements of neighboring genes may function as an enhancer of the transgene promoter, giving rise to unexpected expression (Auerbach 2004). Therefore, as demonstrated here, it is important to generate several independent lines for a single transgene construct to ensure the correct expression profile for the promoter. Collectively, we found that the tissue-specific pattern of the LPVcr in adult LPV-LacZ transgenic mice was consistent with previous reports.
Construction of the LPV-Cre and hVWF-Cre transgenes were achieved by subcloning the 1.1 Kb NLS-Cre fragment from the pTurboCre plasmid into the pCI MCS, and subsequently subcloning the Intron-Cre-polyA fragment into pBluescript containing either the LPVcr or hVWF-f. For the in vitro functional evaluation of the LPV-Cre and hVWF-Cre plasmids, bEnd.3 cells were chosen due to their brain endothelium origin to test the hVWF-Cre vector specificity, while K562 were chosen due to their lymphotropic origin to test the LPV-Cre vector specificity (Fig. 3). As predicted, the pTurboCre plasmid driving Cre with the chicken beta-actin promoter induced gene switching to EGFP in both the K562-Z/EG and the bEnd.3-Z/EG cells. The hVWF-Cre plasmid was only able to induce gene switching in the bEnd.3-Z/EG cells, as expected. The LPV-Cre showed activity in both the K562-Z/EG and bEnd.3-Z/EG cells. The expression in the bEnd.3-Z/ EG cells was not expected based on previous data for the LPVcr expression profile. However, the bEnd.3 cells are polyoma middle T antigen transformed brain endothelial cell line. These transformed and immortal cells may not act identically to brain endothelial cells in vivo. Based on the LPV-LacZ transgenic data, where the LPVcr did not express in vivo in brain endothelium, the expression of the LPV-Cre plasmid in the bEnd.3 cells did not result in removal of the LPV-Cre plasmid from consideration.
Two LPV-Cre (LPV-Cre.0605 and LPV-Cre.0607) and two hVWF-Cre (hVWF-Cre.1304, and hVWF-Cre.1329) founder lines were able to pass a stably integrated transgene on to their offspring (Fig. 4). By mating these mice with Z/EG transgenic mice to create dual Cre-Z/EG mice, we were able to characterize the location of Cre-mediated excision at the organ and tissue level using mRNA and protein expression of EGFP. Both the mRNA and protein expression data for the hVWF-Cre.1329-Z/EG mouse line showed the presence of EGFP only in the liver (Fig. 5). Due to ectopic expression, the hVWF-Cre.1329 line was removed from further evaluation, leaving the single hVWF-Cre.1304 line.
While EGFP mRNA transcripts were present in the brains of the transgenic lines, the level of EGFP protein was below the limit of detection for Western immunoblot for all but the EIIa-Cre mice (Fig. 5). This was most likely due to the Cre transgenes expressing only within a small subset of cells within the brain. Immunohistochemistry for EGFP protein expression demonstrated that expression in the brain was limited to the CP in the LPV-Cre.607-Z/EG line, the CP and spleen in the LPV-Cre.605-Z/EG line, and brain endothelium in hVWF-Cre.1304-Z/EG line (Figs. 6, 7, 8). These data are also in agreement with previously published data for their respective expression profiles.
The utility of these cell-type specific Cre transgenic mice should not be underestimated. The future goal of our project is to delineate the contributions of proteins expressed at the BBB and B-CSF-B that control drug penetration into the brain. This can include, but is not limited to, efflux transporters, TJ proteins, uptake transporters, receptors, and drug metabolizing or conjugating enzymes.
Transmembrane spanning efflux transporters at the blood-CNS barriers have become a major focus due to their function in limiting drug delivery to the brain. The endothelium that forms the BBB expresses both MRP and P-gp efflux transporters on the luminal (blood) side, limiting penetration of many drugs into the brain. However, in the CP, while the choroidal epithelial cells express MRP on the blood side, P-gp is expressed on the opposite side, pumping drug into the CSF (Rao et al. 1999; de Lange 2004). The rationale for P-gp expression on the CSF side has not been fully elucidated. The expected impact would be P-gp transporting drug out of the CP epithelia and into the CSF. In addition, drug concentration in the CSF does not always correlate with drug concentration in the brain tissue. In studies dosing the MRP1 substrate etoposide in M_rp1_ deficient mice, the data showed a 10-fold increase in CSF drug concentration, but no significant difference in drug concentration in the brain tissue 1 h after injection (Wijnholds et al. 2000). The opposite is also true; where chemical inhibition of P-gp using the highly specific drug zosuquidar in non-human primates showed a 146-fold increase in brain concentration of the HIV protease inhibitor Nelfinavir, but no significant change in CSF concentration (Kaddoumi et al. 2007). Whether the data can hold true for mice with brain tissue and cell-selective knockout of P-gp remains to be evaluated.
P-gp and other transporters located in the BBB and CP are expressed throughout the body in tissues that are involved in drug disposition (i.e. liver, kidney, intestine). As such, while current P-gp knockout models show large increases in brain penetration (Schinkel et al. 1995), the effect of P-gp specifically in the CP on CSF drug concentrations, without any other variables, cannot be quantified due to systemic changes in disposition (i.e. increased plasma concentration, reduced elimination, increased brain penetration through brain endothelium). However, creating CP knockouts by crossing a transgenic mouse carrying a floxed Abcb1a gene (which encodes mouse P-gp) with the LPV-Cre transgenic, it would be possible to enumerate the impact of P-gp efflux on drug distribution into the CSF and brain.
Similarly, drug metabolizing and conjugating enzymes such as the cytochrome P450-dependent monooxygenases, NADPH–cytochrome P450 reductase, epoxide hydrolase, UDP-glucuronoxyltransferase, gluthathione S-transferase, UDP-glucuronosyl-transferase, and epoxide hydrolase have been found in the BBB and B-CSF-B (de Lange 2004). Site-specific knockout of these enzymes in the brain would show if they limit entry of parent drug into the brain without effecting the systemic metabolism and disposition.
It has been well established that tissue selective Cre-mediated gene excision can circumvent embryonic lethal phenotypes caused by global gene knockout (reviewed by (Kos 2004)). Using the hVWF-Cre and LPV-Cre mice, BBB and B-CSF-B specific knockouts of TJ proteins such as Claudin-5, ZO-1 and ZO-2 could be created. These mice could then be used to study the extent to which these proteins contribute to limiting brain penetration, and determine if they are possible targets for future studies for drug delivery technologies.
Cell-type specific activation of a dormant transgene is another important use of the Cre/loxP system as a research tool. A transgene containing a floxed stop cassette [loxP-stop-loxP (LSL)] between a promoter and gene can be activated upon Cre excision. While it is possible to create a transgenic mouse expressing a gene of interest in a cell-type dependent manner, a new mouse line would need to be created for each new cell type that is to be studied; each with a different promoter driving the gene. However, with a single LSL-gene mouse line, one could mate it with any number of the over 500 currently available Cre lines to activate that gene in specific cell types, dependent on the Cre line chosen. This LSL approach was first demonstrated by Lakso et al. (1992) to form malignant lens tumors. Similarly, the LPV-Cre transgenic mouse may also be useful in assessing the tumorigenicity of oncogenes implicated in the formation of CP tumors by selectively activating them in the CP. The LSL technique has also been used to look at gene expression and induction of P-gp in the mouse. Gu et al. engineered an LSL transgene controlling luciferase with the endogenous Abcb1a enhancer and promoter. Through tissue selective Cre excision of the LSL, they are able to monitor regulated Abcb1a expression and induction in vivo (Gu et al. 2009).
Conclusions
The aim of this research was to create two transgenic mouse models that would allow for conditional gene knockout at the BBB and B-CSF-B. We targeted the expression of Cre to the CP and brain endothelium by using the LPVcr and −487 to +247 fragment of the hvWF gene, respectively. The results showed that the LPV-Cre.0605 line expressed Cre in the CP and spleen, while the LPV-Cre.0607 line expressed Cre only in the CP. The hVWF-Cre line restricted expression of Cre to the brain endothelium. To date, no other Cre expressing transgenic mouse line has these specific patterns of expression.
These mice will be important for future knockout studies in multiple research areas. From a drug disposition perspective, many proteins are expressed at the blood-CNS barriers that regulate drug penetration into the brain including drug metabolizing and conjugating enzymes, and drug transporters. These mice would also be extremely useful to discern the impact of tight junction proteins at the BBB and B-CSF-B, as systemic lack of functional tight junction proteins is lethal. Additionally, the LPV-Cre transgenic mouse may also be useful in assessing the tumorigenicity of oncogenes implicated in the formation of CP tumors using LSL transgenes.
Acknowledgments
We thank Terry Kavanagh for the use of the Cryocut cryomicrotome, Richard Palmiter for the pLacF plasmid, Nancy Jahroudi for the pHGH-K plasmid, Terry Van Dyke for the pLST plasmid, Corrinne Lobe for the Z/EG plasmid, Washington University ES Cell Core for the pTurboCre plasmid, and Kerstin Verdina for assistance in editing. This work was supported by the National Institute of Health grants NS39178, AI077390, and AI52663. Rodney J. Y. Ho is also supported by the Milo Gibaldi Endowment.
References
- Aird WC, Jahroudi N, Weiler-Guettler H, Rayburn HB, Rosenberg RD. Human von Willebrand factor gene sequences target expression to a subpopulation of endothelial cells in transgenic mice. Proc Natl Acad Sci USA. 1995;92(10):4567–4571. doi: 10.1073/pnas.92.10.4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alva JA, Zovein AC, Monvoisin A, Murphy T, Salazar A, Harvey NL, Carmeliet P, Iruela-Arispe ML. Ve-cadherin-cre-recombinase transgenic mouse: a tool for lineage analysis and gene deletion in endothelial cells. Dev Dyn. 2006;235(3):759–767. doi: 10.1002/dvdy.20643. [DOI] [PubMed] [Google Scholar]
- Auerbach AB. Production of functional transgenic mice by DNA pronuclear microinjection. Acta Biochim Pol. 2004;51(1):9–31. [PubMed] [Google Scholar]
- Brinster RL, Chen HY, Messing A, van Dyke T, Levine AJ, Palmiter RD. Transgenic mice harboring sv40 t-antigen genes develop characteristic brain tumors. Cell. 1984;37(2):367–379. doi: 10.1016/0092-8674(84)90367-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinster RL, Chen HY, Trumbauer ME, Yagle MK, Palmiter RD. Factors affecting the efficiency of introducing foreign DNA into mice by microinjecting eggs. Proc Natl Acad Sci USA. 1985;82(13):4438–4442. doi: 10.1073/pnas.82.13.4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JD, Van Dyke T. Uniform cell-autonomous tumorigenesis of the choroid plexus by papovavirus large t antigens. Mol Cell Biol. 1991;11(12):5968–5976. doi: 10.1128/mcb.11.12.5968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JD, Neilson K, Van Dyke T. Lymphotropic papovavirus early region is specifically regulated transgenic mice and efficiently induces neoplasia. J Virol. 1989;63(5):2204–2214. doi: 10.1128/jvi.63.5.2204-2214.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange EC. Potential role of abc transporters as a detoxification system at the blood-csf barrier. Adv Drug Deliv Rev. 2004;56(12):1793–1809. doi: 10.1016/j.addr.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Feigenbaum L, Hinrichs SH, Jay G. Jc virus and simian virus 40 enhancers and transforming proteins: role in determining tissue specificity and pathogenicity in transgenic mice. J Virol. 1992;66(2):1176–1182. doi: 10.1128/jvi.66.2.1176-1182.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer J, Gavrilova O, Petzinger E. Brain penetration of ivermectin and selamectin in mdr1a, b p-glycoprotein-and bcrp- deficient knockout mice. J Vet Pharmacol Ther. 2009;32(1):87–96. doi: 10.1111/j.1365-2885.2008.01007.x. [DOI] [PubMed] [Google Scholar]
- Gu L, Tsark WM, Brown DA, Blanchard S, Synold TW, Kane SE. A new model for studying tissue-specific mdr1a gene expression in vivo by live imaging. Proc Natl Acad Sci USA. 2009;106(13):5394–5399. doi: 10.1073/pnas.0807343106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahroudi N, Lynch DC. Endothelial-cell-specific regulation of von Willebrand factor gene expression. Mol Cell Biol. 1994;14(2):999–1008. doi: 10.1128/mcb.14.2.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahroudi N, Schmaier A, Srikanth S, Mahdi F, Lutka FA, Bowser R. VonWillebrand factor promoter targets the expression of amyloid beta protein precursor to brain vascular endothelial cells of transgenic mice. J Alzheimers Dis. 2003;5(2):149–158. doi: 10.3233/jad-2003-5209. [DOI] [PubMed] [Google Scholar]
- Jonker JW, Buitelaar M, Wagenaar E, Van Der Valk MA, Scheffer GL, Scheper RJ, Plosch T, Kuipers F, Elferink RP, Rosing H, Beijnen JH, Schinkel AH. The breast cancer resistance protein protects against a major chlorophyll-derived dietary phototoxin and protoporphyria. Proc Natl Acad Sci USA. 2002;99(24):15649–15654. doi: 10.1073/pnas.202607599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaddoumi A, Choi SU, Kinman L, Whittington D, Tsai CC, Ho RJ, Anderson BD, Unadkat JD. Inhibition of p-glycoprotein activity at the primate blood-brain barrier increases the distribution of nelfinavir into the brain but not into the cerebrospinal fluid. Drug Metab Dispos. 2007;35(9):1459–1462. doi: 10.1124/dmd.107.016220. [DOI] [PubMed] [Google Scholar]
- Katsuno T, Umeda K, Matsui T, Hata M, Tamura A, Itoh M, Takeuchi K, Fujimori T, Nabeshima Y, Noda T, Tsukita S. Deficiency of zonula occludens-1 causes embryonic lethal phenotype associated with defected yolk sac angiogenesis and apoptosis of embryonic cells. Mol Biol Cell. 2008;19(6):2465–2475. doi: 10.1091/mbc.E07-12-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koni PA, Joshi SK, Temann UA, Olson D, Burkly L, Flavell RA. Conditional vascular cell adhesion molecule 1 deletion in mice: impaired lymphocyte migration to bone marrow. J Exp Med. 2001;193(6):741–754. doi: 10.1084/jem.193.6.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kos CH. Cre/loxP system for generating tissue-specific knockout mouse models. Nutr Rev. 2004;62(6 Pt 1):243–246. doi: 10.1301/nr2004.jun243-246. [DOI] [PubMed] [Google Scholar]
- Kwon GS, Hadjantonakis AK. Transthyretin mouse transgenes direct rfp expression or cre-mediated recombination throughout the visceral endoderm. Genesis. 2009;47(7):447–455. doi: 10.1002/dvg.20522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakso M, Sauer B, Mosinger B, Jr, Lee EJ, Manning RW, Yu SH, Mulder KL, Westphal H. Targeted oncogene activation by site-specific recombination in transgenic mice. Proc Natl Acad Sci USA. 1992;89(14):6232–6236. doi: 10.1073/pnas.89.14.6232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakso M, Pichel JG, Gorman JR, Sauer B, Okamoto Y, Lee E, Alt FW, Westphal H. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc Natl Acad Sci USA. 1996;93(12):5860–5865. doi: 10.1073/pnas.93.12.5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorico A, Rappa G, Finch RA, Yang D, Flavell RA, Sartorelli AC. Disruption of the murine mrp (multidrug resistance protein) gene leads to increased sensitivity to etoposide (vp-16) and increased levels of glutathione. Cancer Res. 1997;57(23):5238–5242. [PubMed] [Google Scholar]
- Messing A, Pinkert CA, Palmiter RD, Brinster RL. Developmental study of sv40 large t antigen expression in transgenic mice with choroid plexus neoplasia. Oncogene Res. 1988;3(1):87–97. [PubMed] [Google Scholar]
- Morita K, Sasaki H, Furuse M, Tsukita S. Endothelial claudin: Claudin-5/tmvcf constitutes tight junction strands in endothelial cells. J Cell Biol. 1999;147(1):185–194. doi: 10.1083/jcb.147.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A. Cre recombinase: the universal reagent for genome tailoring. Genesis. 2000;26(2):99–109. [PubMed] [Google Scholar]
- Nagy A, Mar L. Creation and use of a cre recombinase transgenic database. Methods Mol Biol. 2001;158:95–106. doi: 10.1385/1-59259-220-1:95. [DOI] [PubMed] [Google Scholar]
- Nagy A, Mar L, Watts G. Creation and use of a cre recombinase transgenic database. Methods Mol Biol. 2009;530:365–378. doi: 10.1007/978-1-59745-471-1_19. [DOI] [PubMed] [Google Scholar]
- Nitta T, Hata M, Gotoh S, Seo Y, Sasaki H, Hashimoto N, Furuse M, Tsukita S. Size-selective loosening of the blood-brain barrier in claudin-5-deficient mice. J Cell Biol. 2003;161(3):653–660. doi: 10.1083/jcb.200302070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak A, Guo C, Yang W, Nagy A, Lobe CG. Z/e.g, a double reporter mouse line that expresses enhanced green fluorescent protein upon Cre-mediated excision. Genesis. 2000;28(3–4):147–155. [PubMed] [Google Scholar]
- Perraud F, Yoshimura K, Louis B, Dalemans W, Ali-Hadji D, Schultz H, Claudepierre MC, Chartier C, Danel C, Bellocq JP, et al. The promoter of the human cystic fibrosis transmembrane conductance regulator gene directing sv40 t antigen expression induces malignant proliferation of ependymal cells in transgenic mice. Oncogene. 1992;7(5):993–997. [PubMed] [Google Scholar]
- Rao VV, Dahlheimer JL, Bardgett ME, Snyder AZ, Finch RA, Sartorelli AC, Piwnica-Worms D. Choroid plexus epithelial expression of mdr1 p glycoprotein and multidrug resistance-associated protein contribute to the blood-cerebrospinal-fluid drug-permeability barrier. Proc Natl Acad Sci USA. 1999;96(7):3900–3905. doi: 10.1073/pnas.96.7.3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salama NN, Kelly EJ, Bui T, Ho RJ. The impact of pharmacologic and genetic knockout of p-glycoprotein on nelfinavir levels in the brain and other tissues in mice. J Pharm Sci. 2005;94(6):1216–1225. doi: 10.1002/jps.20344. [DOI] [PubMed] [Google Scholar]
- Schinkel AH, Smit JJ, van Tellingen O, Beijnen JH, Wagenaar E, van Deemter L, Mol CA, van der Valk MA, Robanus-Maandag EC, te Riele HP, et al. Disruption of the mouse mdr1a p-glycoprotein gene leads to a deficiency in the blood-brain barrier and to increased sensitivity to drugs. Cell. 1994;77(4):491–502. doi: 10.1016/0092-8674(94)90212-7. [DOI] [PubMed] [Google Scholar]
- Schinkel AH, Wagenaar E, van Deemter L, Mol CA, Borst P. Absence of the mdr1a p-glycoprotein in mice affects tissue distribution and pharmacokinetics of dexamethasone, digoxin, and cyclosporin a. J Clin Invest. 1995;96(4):1698–1705. doi: 10.1172/JCI118214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semprini S, Troup TJ, Kotelevtseva N, King K, Davis JR, Mullins LJ, Chapman KE, Dunbar DR, Mullins JJ. Cryptic loxP sites in mammalian genomes: genome-wide distribution and relevance for the efficiency of bac/pac recombineering techniques. Nucleic Acids Res. 2007;35(5):1402–1410. doi: 10.1093/nar/gkl1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg N, Hamilton D. Bacteriophage p1 site-specific recombination I. Recombination between loxP sites. J Mol Biol. 1981;150(4):467–486. doi: 10.1016/0022-2836(81)90375-2. [DOI] [PubMed] [Google Scholar]
- Stratman JL, Barnes WM, Simon TC. Universal pcr genotyping assay that achieves single copy sensitivity with any primer pair. Transgenic Res. 2003;12(4):521–522. doi: 10.1023/a:1024225408961. [DOI] [PubMed] [Google Scholar]
- Thyagarajan B, Guimaraes MJ, Groth AC, Calos MP. Mammalian genomes contain active recombinase recognition sites. Gene. 2000;244(1–2):47–54. doi: 10.1016/s0378-1119(00)00008-1. [DOI] [PubMed] [Google Scholar]
- Wijnholds J, deLange EC, Scheffer GL, van den Berg DJ, Mol CA, van der Valk M, Schinkel AH, Scheper RJ, Breimer DD, Borst P. Multidrug resistance protein 1 protects the choroid plexus epithelium and contributes to the blood-cerebrospinal fluid barrier. J Clin Invest. 2000;105(3):279–285. doi: 10.1172/JCI8267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Kausalya PJ, Phua DC, Ali SM, Hossain Z, Hunziker W. Early embryonic lethality of mice lacking zo-2, but not zo-3, reveals critical and nonredundant roles for individual zonula occludens proteins in mammalian development. Mol Cell Biol. 2008;28(5):1669–1678. doi: 10.1128/MCB.00891-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Huang G, Shornick LP, Roswit WT, Shipley JM, Brody SL, Holtzman MJ. A transgenic FOXJ1-cre system for gene inactivation in ciliated epithelial cells. Am J Respir Cell Mol Biol. 2007;36(5):515–519. doi: 10.1165/rcmb.2006-0475RC. [DOI] [PMC free article] [PubMed] [Google Scholar]