REGULATION OF THE MEX-5 GRADIENT BY A SPATIALLY SEGREGATED KINASE/PHOSPHATASE CYCLE (original) (raw)
. Author manuscript; available in PMC: 2012 Mar 16.
SUMMARY
Protein concentration gradients encode spatial information across cells and tissues, and often depend on spatially localized protein synthesis. Here, we report that a different mechanism underlies the MEX-5 gradient. MEX-5 is an RNA-binding protein that becomes distributed in a cytoplasmic gradient along the anterior-to-posterior axis of the 1-cell C. elegans embryo. We demonstrate that the MEX-5 gradient is a direct consequence of an underlying gradient in MEX-5 diffusion. The MEX-5 diffusion gradient arises when the PAR-1 kinase stimulates the release of MEX-5 from slow-diffusive, RNA-containing complexes in the posterior cytoplasm. PAR-1 directly phosphorylates MEX-5 and is antagonized by the spatially-uniform phosphatase PP2A. Mathematical modeling and in vivo observations demonstrate that spatially-segregated phosphorylation and dephosphorylation reactions are sufficient to generate stable protein concentration gradients in the cytoplasm. The principles demonstrated here apply to any spatially-segregated modification cycle that affects protein diffusion, and do not require protein synthesis or degradation.
INTRODUCTION
Protein gradients are an efficient way to encode spatial information within cells and across tissues. The mechanisms that generate and maintain protein gradients have been the subject of extensive theoretical and experimental analyses (Wartlick et al., 2009). Most studies have emphasized the role of a localized protein source as the foundational asymmetry underlying gradient formation. For example, in Drosophila embryos, the Bicoid protein is synthesized at one end of the egg from a localized pool of bicoid mRNA. Diffusion away from the local source and uniform protein degradation across the egg generate a concentration gradient over the course of ~2 hours (Ephrussi and St Johnston, 2004; Little et al., 2011). Extracellular gradients also depend on the localization of specialized cells that synthesize and secrete the signal (source) among cells that respond to and internalize the signal (sink) (Wartlick et al., 2009).
A spatially-segregated source/sink model can also account for the formation of phosphorylation gradients or “phosphogradients”. Phosphogradients have been implicated in the spatial organization of signal transduction pathways where phosphorylation modulates protein activity. Phosphogradients arise when a diffusing substrate is acted upon by a kinase (source) and phosphatase (sink) that are separated in space (Brown and Kholodenko, 1999). In phosphogradients, the ratio of unphosphorylated to phosphorylated substrate varies in space, but the overall concentration of the substrate is uniform (Brown and Kholodenko, 1999; Coppey et al., 2008; Fuller et al., 2008; Kalab et al., 2002; Maeder et al., 2007; Su et al., 1998). In 2008, Lipkow and Odde predicted that, if phosphorylation changes the diffusivity of the substrate, spatially segregated kinase/phosphatase cycles would also affect the overall distribution of the substrate to generate a protein concentration gradient (Lipkow and Odde, 2008). The spatial bias in the generation of the phosphorylated isoform generates a diffusion gradient that causes the substrate to concentrate in regions of low diffusivity (Lipkow and Odde, 2008). In the present study, we provide experimental evidence in support of this model in C. elegans.
The C. elegans 1-cell embryo (zygote) is a classic model for the study of intracellular asymmetries (Goldstein and Macara, 2007; St Johnston and Ahringer, 2010). After fertilization, a group of conserved polarity regulators, the PAR proteins, sort into anterior (PAR-3/PAR-6/PKC-3) and posterior (PAR-2 and PAR-1) domains in the actin-rich layer (or “cortex”) under the plasma membrane (Kemphues, 2000). In response to PAR asymmetry at the cortex, cell fate determinants become asymmetrically distributed in the cytoplasm. Among them is the RNA-binding protein MEX-5, which redistributes in 10 minutes into an anterior-high/posterior-low gradient across the length of the 50 μm zygote (Schubert et al., 2000; Tenlen et al., 2008). MEX-5, in turn, partitions other factors such as PIE-1 to the posterior cytoplasm and PLK-1 to the anterior cytoplasm (Budirahardja and Gönczy, 2008; Mello et al., 1996; Rivers et al., 2008; Schubert et al., 2000). Consequently, during the first cell division, the two daughter blastomeres inherit different determinants, which help specify their distinct fates (anterior/somatic and posterior/germline). Mutations in the PARs cause MEX-5 (and its targets) to remain symmetrically distributed (Schubert et al., 2000; Tenlen et al., 2008), but the mechanisms linking PAR asymmetry to the MEX-5 gradient are not known.
Fluorescence recovery after photobleaching (FRAP) and fluorescence correlation spectroscopy (FCS) experiments have shown that, in polarized zygotes, GFP::MEX-5 diffuses faster in the posterior cytoplasm, where MEX-5 protein concentration is lowest (Daniels et al., 2010; Tenlen et al., 2008). Fast diffusion requires par-1 activity and a C-terminal serine in MEX-5 (S458), which is phosphorylated in a par-1 and _par-4_-dependent manner in vivo (Tenlen et al., 2008). Phosphorylation of S458, however, does not correlate with gradient formation or fast diffusion, suggesting that other mechanisms regulate MEX-5 asymmetry (Tenlen et al., 2008). Two speculative models have been proposed. The first model invokes dynamic binding of MEX-5 to cytoskeletal elements asymmetrically distributed in the cytoplasm (Tenlen et al., 2008). In this model, the PARs localize MEX-5 indirectly by localizing factors, such as myosin, that retard MEX-5 diffusion in the anterior cytoplasm (Tenlen et al., 2008). A second model proposes that the PARs regulate MEX-5 distribution by forming “reactive surfaces” in the anterior and posterior cortices, which locally decrease and increase, respectively, the rate of MEX-5 diffusion (Daniels et al., 2010). How the PARs modify MEX-5 diffusion, and how differences originated at the cortex are propagated through the cytoplasm, however, is not known.
In this study, we present evidence that the MEX-5 gradient arises as a direct consequence of a complementary PAR-1 kinase activity gradient in the cytoplasm. We demonstrate that MEX-5 is a substrate of PAR-1, and identify PP2A as the opposing phosphatase in the cytoplasm. Our findings reveal an unexpected direct patterning role for PAR-1 in the cytoplasm, and provide experimental evidence for the theoretical model of Lipkow and Odde, 2008.
RESULTS
A MEX-5 diffusion gradient underlies the MEX-5 concentration gradient
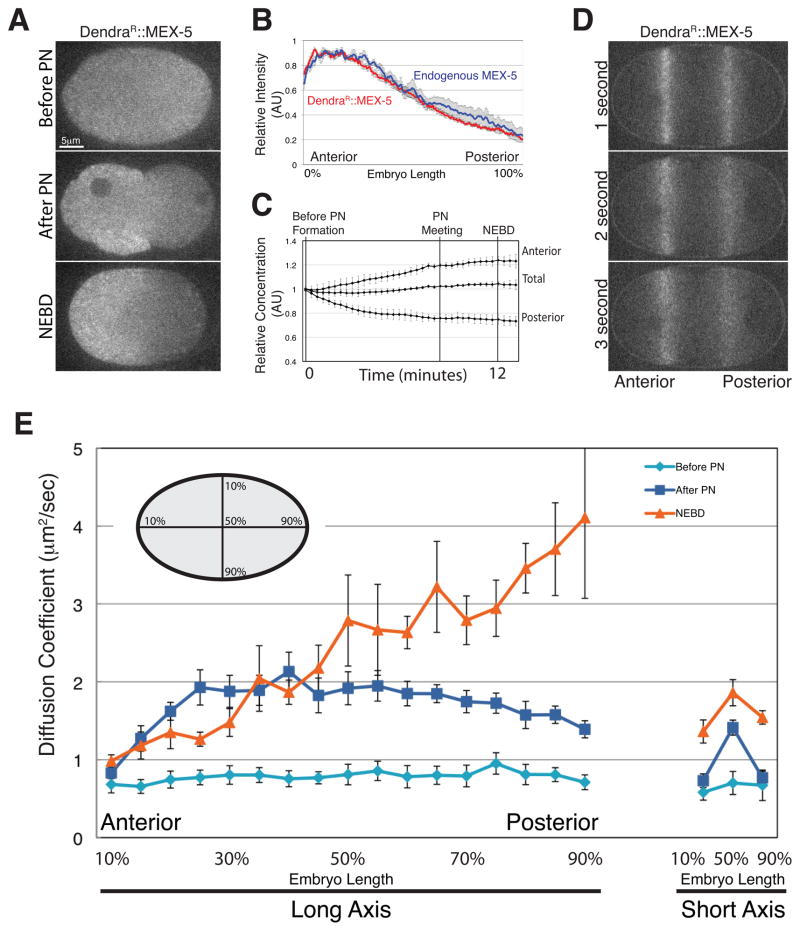
To examine MEX-5 dynamics in live zygotes, we generated a Dendra::MEX-5 fusion. Dendra is a photo-activatable fluorescent protein that is photoconverted irreversibly from green to red fluorescence by exposure to 405 nm light (Gurskaya et al., 2006). Unlike FRAP, photoconversion is a positive marking technique that can be used to measure rates of protein degradation and diffusion, without interference from new protein synthesis (Lippincott-Schwartz and Patterson, 2008). We first photoconverted Dendra::MEX-5 throughout the zygote before polarization (prior to appearance of the pronuclei). We found that photoconverted Dendra::MEX-5 (DendraR::MEX-5) formed a ~3-fold anterior-posterior gradient by nuclear envelope breakdown (NEBD, first mitotic division), as is observed for endogenous MEX-5 (Figure 1A and Figure 1B). Total levels of DendraR::MEX-5 did not change during gradient formation: levels increased in the anterior half and decreased in the posterior half by ~25% (Figure 1C). We conclude that formation of the MEX-5 gradient involves redistribution of existing MEX-5 and does not require MEX-5 synthesis or degradation.
Figure 1. A diffusion gradient underlies the MEX-5 gradient.
A. Time-lapse photomicrographs of a zygote expressing Dendra::MEX-5 photoconverted before pronuclear formation (before polarization). All embryos in this and subsequent figures are oriented with anterior to the left and posterior to the right. PN stands for pronuclear formation, which marks the onset of polarity. NEBD stands for Nuclear Envelope BreakDown (mitosis) and occurs 10 minutes after pronuclear formation, by which time MEX-5 is maximally polarized.
B. Graph plotting the relative signal intensity of DendraR::MEX-5 (red line; n=7 embryos) and endogenous MEX-5 (blue line; n=5 embryos) along the long axis of the zygote after NEBD. Fluorescence intensity was averaged along a 20 pixel-wide box spanning the length of each zygote (0% anterior most pole, 100% posterior most pole). Maximum values for each zygote were normalized to 1. Error bars represent the standard error of the mean (SEM).
C. The relative concentration of DendraR::MEX-5 was measured in the anterior half, posterior half and throughout the zygote (total) from before pronuclear formation to 1 minute following NEBD (just prior to cytokinesis). Mid-plane images were collected every 20 seconds. Embryos were normalized to each other by setting the initial total value to 1 and averaged together (n=5 embryos). Error bars represent SEM.
D. Time-lapse photomicrographs of a zygote during polarity establishment (pronuclear migration) expressing Dendra::MEX-5 photoactivated in two stripes. Time since photoactivation is indicated. Note that the signal from the posterior stripe diffuses more rapidly.
E. Plot showing the apparent diffusion coefficient (Dc) of DendraR::MEX-5 at different positions along the long and short axis of the zygote and at different stages. Embryo schematic shows the position of the photoconversion stripes along the long and short axis. “Peripheral cytoplasm” as mentioned in the text refers to 10% and 90% embryo-length.
Next, to compare mobility of MEX-5 between the anterior and posterior, we photoconverted Dendra::MEX-5 in two stripes at ~30% and ~70% embryo-length during polarization (Figure 1D). DendraR::MEX-5 diffused symmetrically away from both stripes with no directional bias (Figure 1D). The apparent diffusivity of DendraR:MEX-5, however, appeared to differ between the two stripes, with faster diffusion in the posterior stripe (Figure 1D). These observations are consistent with earlier FRAP experiments, which showed that GFP::MEX-5 diffuses faster in the posterior cytoplasm after polarization (Daniels et al., 2010; Tenlen et al., 2008).
To examine MEX-5 mobility systematically during polarization, we measured the apparent diffusion coefficient (Dc) of DendraR::MEX-5 at 17 positions along the long (anterior-posterior) axis and 3 positions along the short axis, before polarization (before pronuclear formation), at the onset of polarization (pronuclear formation), and after polarization (NEBD). The apparent Dc of DendraR::MEX-5 was uniformly slow before pronuclear formation (average Dc between 10% and 90% embryo-length was 0.78 μm2/sec) (Figure 1E). After pronuclear formation, the apparent Dc of DendraR::MEX-5 increased to an average of 1.70 μm2/sec. This increase was observed throughout the central cytoplasm, but not in the cytoplasm nearest the cortex (peripheral cytoplasm) where DendraR::MEX-5 diffusion remained slow (Figure 1E). By NEBD, the apparent Dc of DendraR::MEX-5 was graded linearly throughout the cytoplasm, with the lowest value at the anterior-most position and the highest value at the posterior-most position, mirroring the MEX-5 protein concentration gradient (compare Figure 1B and Figure 1E). We conclude that redistribution of MEX-5 correlates temporally and spatially with changes in MEX-5 diffusion.
par-1 is necessary and sufficient to increase MEX-5 diffusion in zygotes
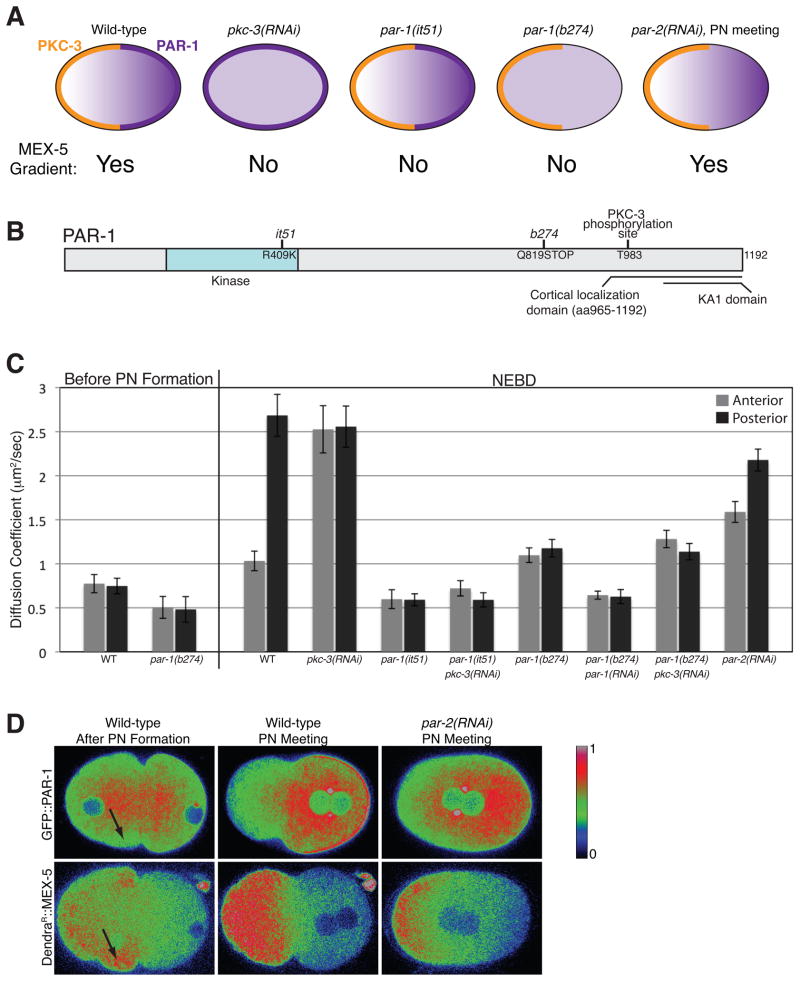
To determine whether the anterior or posterior PARs regulate MEX-5 dynamics, we monitored MEX-5 distribution and diffusion at NEBD in zygotes defective for the anterior kinase aPKC/PKC-3 or the posterior kinase PAR-1 (Figure 2A). pkc-3(RNAi) embryos lack PKC-3 and have uniform PAR-1 (Figures 2A and S1A). The par-1 allele it51 inactivates PAR-1 kinase activity but does not affect PAR-1 or PKC-3 localizations (Figures 2A, 2B and S1B) (Cheeks et al., 2004; Guo and Kemphues, 1995). Previous work has shown that in par-1(RNAi) embryos, MEX-5 mobility does not increase in the posterior cytoplasm and MEX-5 does not segregate (Tenlen et al., 2008). We found that DendraR::MEX-5 remained symmetrically distributed in both pkc-3(RNAi) and par-1(it51) zygotes (Figures S1A and S1B). Strikingly, DendraR::MEX-5 diffusion was uniformly high in pkc-3(RNAi) zygotes, and uniformly low in par-1(it51) and par-1(it51);pkc-3(RNAi) zygotes (Figure 2C). We conclude that PAR-1 functions downstream of PKC-3 and is required to stimulate MEX-5 diffusion.
Figure 2. PAR-1 is necessary and sufficient to increase MEX-5 mobility.
A. Diagrams showing the distributions of PKC-3 (orange) and PAR-1 (purple on cortex and in cytoplasm) in zygotes of the indicated genotypes. MEX-5 localizes in a gradient in wild-type and par-2(RNAi) embryos and remains symmetrically distributed in all other genotypes. par-1(it51) and par-1(b274) are mutations that, respectively, inactivate PAR-1 kinase activity and truncate the PAR-1 protein (Figure S1E) (Guo and Kemphues, 1995). Also see Figure S1A, S1B and S1E.
B. PAR-1 schematic: T983 is a conserved aPKC/PKC-3 phosphorylation site. The C-terminal domain (965-1192) contains the lipid-binding domain KA1 (Moravcevic et al., 2010) and localizes in a cytoplasmic gradient and to the posterior cortex (Motegi et al., submitted).
C. Apparent Dc of DendraR::MEX-5 measured in the anterior (25% embryo-length) and posterior (75% embryo-length) cytoplasm in zygotes of the indicated genotypes.
D. Comparison of the distribution of GFP::PAR-1 and DendraR::MEX-5 in wild-type and par-2(RNAi) zygotes. Fluorescence intensity is represented by a rainbow scale ranging from blue (low signal intensity) to red (high signal intensity). Arrows point to the subcortical region where GFP::PAR-1 is depleted and DendraR::MEX-5 accumulates after pronuclear formation. Note that in par-2(RNAi) zygotes, GFP::PAR-1 does not accumulate on the posterior cortex but still forms a cytoplasmic gradient. Also see Figures S1C, S1D and S1F.
In polarized zygotes, PAR-1 kinase is present both in the cytoplasm and on the posterior cortex (Guo and Kemphues, 1995). To examine PAR-1 dynamics during polarization, we imaged zygotes expressing a full-length GFP::PAR-1 fusion. Before pronuclear formation, GFP::PAR-1 was uniformly distributed in the cytoplasm and weakly on the cortex (data not shown). At pronuclear formation, GFP::PAR-1 levels increased in the central cytoplasm and decreased in the peripheral cytoplasm (Figure 2D). This relocalization coincided temporally and spatially with an increase in DendraR::MEX-5 diffusion in the central cytoplasm (Figure 1E), and an increase in DendraR::MEX-5 levels in the peripheral cytoplasm (Figure 2D). During pronuclear migration, GFP::PAR-1 levels remained low in the anterior peripheral cytoplasm but increased in the posterior cytoplasm and on the posterior cortex. By NEBD, GFP::PAR-1 was enriched on the posterior cortex and formed a 3-fold anterior low/posterior high gradient in the cytoplasm, paralleling the MEX-5 diffusion gradient (Figure 2D and Figure S1C). Immunostaining of wild-type embryos with an anti-PAR-1 antibody confirmed the presence of a PAR-1 gradient in the cytoplasm of zygotes at NEBD (Figure S1D). We conclude that PAR-1 dynamics in the cytoplasm correlate with MEX-5 diffusion dynamics and that MEX-5 responds quickly to changes in PAR-1 distribution.
To explore whether cytoplasmic PAR-1 is sufficient to stimulate MEX-5 diffusion, we analyzed the par-1 allele b274. par-1(b274) zygotes do not localize PAR-1 to the cortex and do not segregate MEX-5, but are positive for pS458, suggesting that this allele retains some par-1 kinase activity (Figure S1B) (Guo and Kemphues, 1995; Tenlen et al., 2008). We sequenced par-1(b274) and found a premature stop codon at residue Q819 between the kinase domain and the domain that localizes PAR-1 to the cortex (Figure 2B). Western blotting and immunofluorescence analyses confirmed the presence of a truncated PAR-1 protein, expressed at 14% of wild-type levels and uniformly cytoplasmic (Figure S1B and S1E) (Hurd and Kemphues, 2003). Before pronuclear formation, DendraR::MEX-5 mobility was uniformly low in par-1(b274) zygotes, as in wild-type and par-1(it51) zygotes. By NEBD, however, DendraR::MEX-5 diffusion had increased throughout the cytoplasm to a value intermediate between that of par-1(it51) and pkc-3(RNAi) zygotes (Figure 2C). In par-1(b274) zygotes, PKC-3 became enriched on the anterior cortex as in wild-type, whereas DendraR::MEX-5 remained symmetrically distributed (Figure S1B) (Tenlen et al., 2008). The intermediate DendraR::MEX-5 diffusion in par-1(b274) zygotes was dependent on PAR-1 but not on PKC-3 (Figure 2C). We conclude that PAR-1 kinase activity in the cytoplasm is sufficient to increase MEX-5 diffusion after pronuclear formation.
We also examined the distribution of PAR-1 and MEX-5 in par-2 zygotes, which localize anterior PARs to the anterior cortex before, but not after, NEBD and which never enrich PAR-1 on the posterior cortex (Boyd et al., 1996; Cuenca et al., 2003). We found that GFP::PAR-1 still formed a cytoplasmic gradient by pronuclear meeting in par-2(RNAi) zygotes (Figure 2D and Figure S1C). The GFP::PAR-1 gradient was transient and became less pronounced following NEBD (Figure S1C). Remarkably, DendraR::MEX-5 also formed a gradient by pronuclear meeting, which weakened following NEBD (Figure 2D and S1F). DendraR::MEX-5 diffusion was also asymmetric in par-2(RNAi) zygotes (Figure 2C). We conclude that formation of a cytoplasmic PAR-1 gradient is sufficient to change MEX-5 diffusion and drive the formation of a complementary MEX-5 gradient.
PAR-1 phosphorylates MEX-5 on two residues: S458 and S404
Phosphorylation of S458 depends on par-1 activity in vivo, raising the possibility that MEX-5 is a PAR-1 substrate (Tenlen et al., 2008). To test this possibility directly, we expressed the PAR-1 kinase domain (aa 1-492) fused to maltose binding protein (MBP) in E. coli. We also included the activating mutation T325E in the kinase activation loop (Lizcano et al., 2004). MBP:PAR-1(aa1-492, T325E) phosphorylated MBP:MEX-5, but not MBP or MBP:PIE-1 (Figure 3A). Replacement of S458 with alanine reduced, but did not abolish, phosphorylation of MBP:MEX-5 (Figure 3A). Using a combination of deletion and alanine mutagenesis, we identified S404 as a second PAR-1 phosphorylation site in MEX-5 (Figure 3A). MEX-5 mutated at both S404 and S458 was no longer a substrate for MBP:PAR-1(aa1-492, T325E) (Figure 3A). To determine whether S404 is phosphorylated by PAR-1 in vivo, we generated an antibody specific for pS404 (Figure S2A). Anti-pS404 immunoprecipitated ~5% of total MEX-5 from extracts prepared from wild-type hermaphrodites and only ~1.7% from extracts prepared from par-1(RNAi) hermaphrodites (Figure 3B). We conclude that PAR-1 phosphorylates MEX-5 on S458 and S404 in vitro and in vivo.
Figure 3. PAR-1 phosphorylates MEX-5 on S404 in vitro and in vivo.
A. SDS-PAGE gel of kinase reactions using MBP:PAR-1(aa1-492; T325E) and the indicated MBP substrates. Reactions were performed in the presence of 32P-ATP for 30 minutes. Top panel shows 32P incorporation and bottom panel is coomassie blue staining of the same gel.
B. Western blot analysis of immunoprecipitates from whole worm extracts obtained with anti-MEX-5 pS404 antibodies. 5% of extract used for immunoprecipitation was loaded in input lanes. In the bottom panel, extract was probed with anti-PAR-1 antibodies to demonstrate depletion by par-1(RNAi).
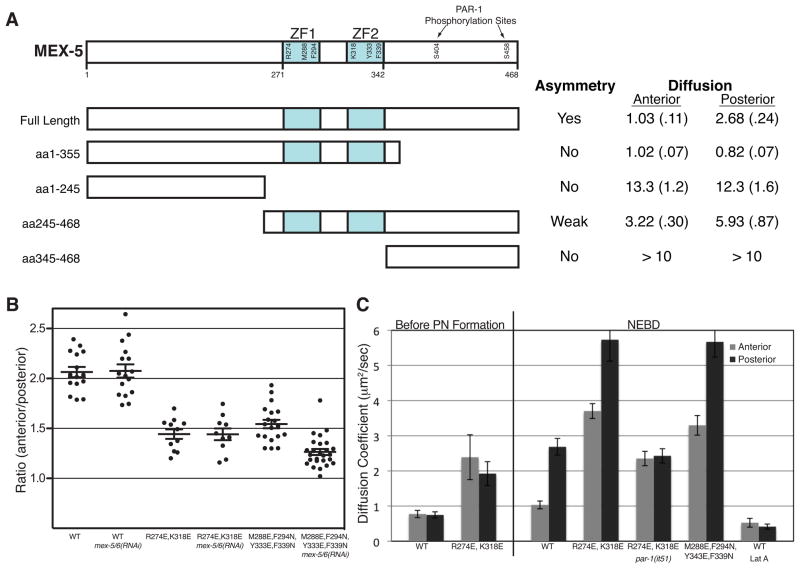
C. Ratio of mean anterior and posterior fluorescence intensities for embryos expressing the indicated DendraR::MEX-5 fusions at NEBD. Each dot represents an individual embryo. Long bars represents the mean ratio and short bars represent the SEM.
D. Apparent Dc of DendraR::MEX-5 mutants at NEBD. Dendra::MEX-5 was photoconverted along the anterior-posterior axis and apparent Dc was calculated at 25% (anterior) and 75% (posterior) embryo-length. Error bars represent SEM. The results for wild-type embryos are also displayed in Figure 2C and 4D. Also see Figures S2B and S2C.
E. Dynamics of pS404 and pS458 in vitro. Left Panel: MBP:MEX-5 was incubated with MBP:PAR-1 (aa1-492; T325E) and analyzed by western blot with phosphospecific pS458 and pS404 antibodies at the indicated times. Right Panel: Phosphorylated MBP::MEX-5 was incubated with embryonic extract in the presence or absence of 200 nM okadaic acid (+ OA) and analyzed by western blot with pS404 and pS458 phospho-specific antibodies. Note the rapid dephosphorylation at S404. Also see Figure S2A.
Reversible phosphorylation of S404 is required to form the MEX-5 gradient
To investigate the role of S404 and S458 phosphorylation in vivo, we examined the distribution of MEX-5(S404A) and MEX-5(S458A) fusions. As reported in (Tenlen et al., 2008), the distribution of MEX-5(S458A) was variable from embryo to embryo, with a minority of embryos forming a shallow MEX-5 gradient. In contrast, DendraR::MEX-5(S404A) was symmetrically distributed in all embryos examined (Figure 3C). The double mutant S404A/S458A behaved like S404A (data not shown). DendraR::MEX-5(S404A) diffusion was slow, comparable to that of wild-type DendraR::MEX-5 in par-1(it51) (Figure 3D and Figure 2C). DendraR::MEX-5(S404A) remained slow diffusing in pkc-3(RNAi) and in par-1(b274) zygotes, indicating that this fusion is no longer sensitive to changes in PAR-1 activity or localization (Figure 3D). We conclude that the MEX-5 protein and diffusion gradients depend primarily on phosphorylation of S404 by PAR-1.
Immunofluorescence experiments using a phosphospecific antibody have shown that S458 is phosphorylated during oogenesis, and MEX-5 phosphorylated on S458 becomes enriched in the anterior in zygotes as does total MEX-5 (Tenlen et al., 2008). These observations suggest that pS458 is relatively stable and does not respond to changes in PAR-1 localization during polarization. In contrast, we were not able to visualize pS404 by immunofluorescence, even though our phosphospecific antibody could immunoprecipitate MEX-5 from extracts (Figure 3B). We detected pS404 in extracts from fem-3(e2006) females, which contain oocytes but no embryos, suggesting that like S458, S404 is already phosphorylated during oogenesis (data not shown). To examine phosphorylation and dephosphorylation dynamics at S458 and S404, we phosphorylated MEX-5 in vitro using MBP::PAR-1(aa1-492; T325E), and incubated phosphorylated MEX-5 with embryo extract. While both sites were phosphorylated at similar rates in vitro, S404 was dephosphorylated significantly faster than S458 in embryo extracts (Figure 3E). Dephosphorylation was inhibited by 200 nM okadaic acid, consistent with the presence of phosphatases in the extract (Figure 3E). We conclude that embryos contain a phosphatase activity that efficiently reverses S404 phosphorylation.
The okadaic acid-sensitive phosphatase PP2A has been implicated as a PAR-1 antagonist in Drosophila and C. elegans (Kao et al., 2004; Krahn et al., 2009; Nam et al., 2007; Yoder et al., 2004). PP2A is a heterotrimeric phosphatase consisting of structural, catalytic and regulatory subunits. In C. elegans, the catalytic subunit, LET-92, is distributed throughout the cytoplasm, on centrosomes, and on P granules (Schlaitz et al., 2007). To test whether PP2A influences MEX-5 dynamics, we analyzed let-92(RNAi) embryos. let-92(RNAi) increased the mobility and decreased the asymmetry of wild-type DendraR::MEX-5 (Figure 3C and Figure 3D). Consistent with PP2A acting primarily via S404, let-92(RNAi) only slightly increased the mobility of DendraR::MEX-5 (S404A) (Figure 3D). let-92(RNAi) did not affect the posterior localization of PAR-1 (Figure S2B and Figure S2C). We conclude that PP2A, and possibly other phosphatases, antagonize PAR-1-dependent phosphorylation of MEX-5 to return MEX-5 to a slow diffusing state.
RNA-binding limits MEX-5 diffusion
The apparent Dc of DendraR::MEX-5 before and after polarization is 10–20 fold lower than that of DendraR alone (data not shown). To determine which domains of MEX-5 retard its mobility, we compared the localization and diffusion behavior of a Dendra::MEX-5 deletion series (Figure 4A). A C-terminal truncation lacking S404 and S458 [Dendra::MEX-5(1-355)] was symmetrically distributed and uniformly slow diffusing even after polarization of the zygote (Figure 4A). An N-terminal truncation [DendraR::MEX-5(245-468)] showed a moderate increase in mobility in the anterior and posterior cytoplasm, and a shallower but still detectable gradient (Figure 4A). In contrast, fusions lacking the CCCH fingers [DendraR::MEX-5(aa1-244) and DendraR::MEX-5(aa345-468)] diffused >10 times faster and lacked all asymmetry (Figure 4A). Consistent with these findings, a GFP::MEX-5 fusion lacking only the CCCH fingers was uniformly distributed and fast diffusing (Tenlen et al., 2008). We conclude that MEX-5 localization and slow mobility depends primarily on the CCCH fingers, with an additional contribution from the N-terminal domain.
Figure 4. Regulation of MEX-5 mobility by RNA-binding.
A. Schematic showing the MEX-5 truncations. Each construct was expressed as a Dendra fusion and its localization and apparent Dc (μm2/sec) were determined at NEBD (SEM in parentheses). The diffusion of DendraR::MEX-5(aa345-468) could not be determined accurately because of its rapid diffusion and relatively low expression, but exceeded 10 μm2/sec.
B. Ratio of anterior and posterior fluorescence intensities for embryos expressing the indicated Dendra::MEX-5 fusions. Each dot represents an individual embryo. Long bars represent the mean ratio and short bars represent SEM.
C. Apparent Dc of DendraR::MEX-5 mutants measured before pronuclear (PN) formation (before polarization) and at NEBD (after polarization). The results for wild-type embryos are also presented in Figure 2C and 3D. Error bars represent SEM.
The CCCH fingers of MEX-5 mediate RNA binding in vitro (Pagano et al., 2007). To test whether RNA binding retards MEX-5 mobility, we examined missense mutations in the CCCH fingers. Studies on the TIS11 family of CCCH finger proteins identified key amino acids that contact RNA, mutations in which disrupt RNA binding (Hudson et al., 2004; Lai et al., 2002). The corresponding mutations in MEX-5 are M288E, M294N, Y333E, F339N. In vitro, MEX-5 binds preferentially to poly-U tracks, a sequence common in C. elegans 3′ UTRs (Pagano et al., 2007). R274E and K318E decrease MEX-5 affinity for poly-U by 35-fold, but only modestly reduces MEX-5 ability to bind to a related sequence (UUAUUUAUU) (Pagano et al., 2007). We found that both DendraR::MEX-5(M288E, M294N, Y333E, F339N) and DendraR::MEX-5 (R274E, K318E) formed a shallower gradient than wild-type and exhibited increased diffusion in both the anterior and posterior (Figure 4B and Figure 4C). DendraR::MEX-5 (R274E, K318E) diffusion was reduced in par-1(RNAi) but remained higher than wild-type MEX-5, indicating that DendraR::MEX-5 (R274E, K318E) is still regulated by PAR-1 but is intrinsically more mobile than wild-type DendraR::MEX-5 (Figure 4C). We conclude that RNA binding retards MEX-5 mobility.
Tenlen et al., 2008 reported that cysteine-to-serine substitutions predicted to disrupt folding of the CCCH fingers do not affect the MEX-5 gradient (Tenlen et al., 2008). We also found that DendraR::MEX-5(C286S,C292S,C331S,C337S) forms a gradient similar to wild-type DendraR::MEX-5. DendraR::MEX-5(C286S,C292S,C331S,C337S), however, diffused faster than DendraR::MEX-5 and was dependent on endogenous wild-type MEX-5 and MEX-6 to form a gradient (Figure S3A and S3B) (Tenlen et al., 2008). In contrast, the diffusive behaviors of DendraR::MEX-5(WT) and DendraR::MEX-5(R274E, K318E) were not dependent on endogenous MEX-5 or MEX-6 (Figure 4B). These observations suggest that, in addition to RNA binding, interactions among MEX-5 and MEX-6 molecules can also contribute to MEX-5’s diffusive behavior.
The actin cytoskeleton, which becomes enriched in the anterior cytoplasm during polarization, has been proposed as another candidate for retarding MEX-5 mobility (Tenlen et al., 2008). To test this possibility, we treated zygotes with Latrunculin A, which depolymerizes F-actin and blocks polarization of the PARs (Severson and Bowerman, 2003). Latrunculin A treatment resulted in uniformly slow MEX-5 diffusion and blocked DendraR::MEX-5 gradient formation (Figure 4C and data not shown), indicating that F-actin is not essential to retard MEX-5 mobility.
MEX-5 associates with large complexes in a RNA-dependent manner
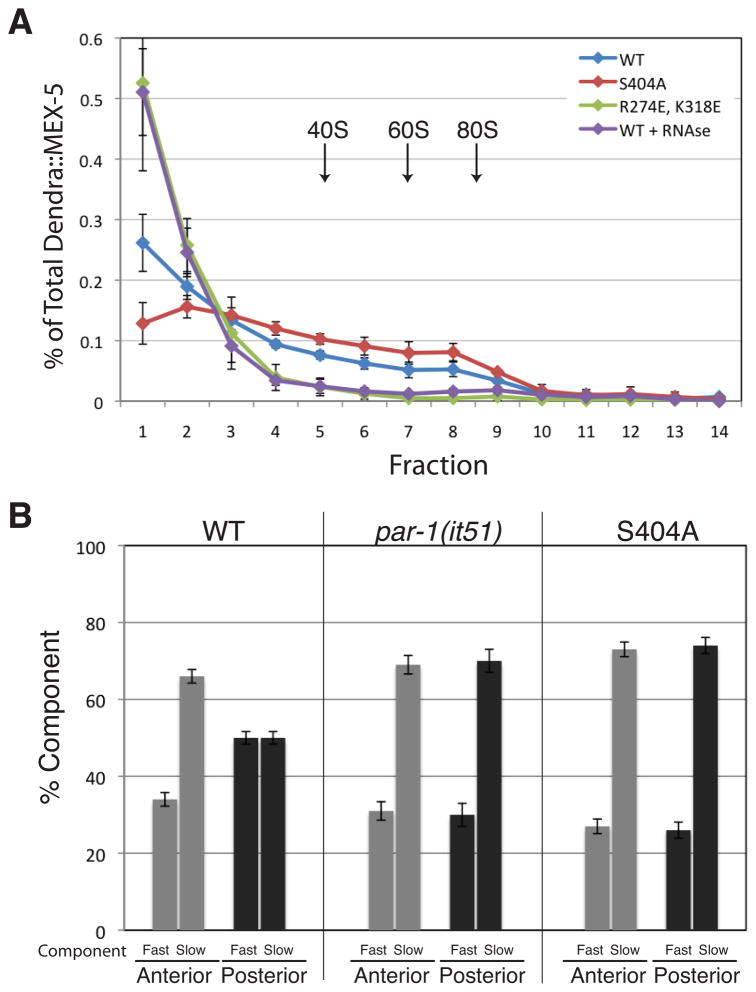
To directly investigate whether MEX-5 associates with RNA in vivo, we examined the distribution of Dendra::MEX-5 in worm extracts fractionated on a 10–45% sucrose gradient. Dendra::MEX-5 was detected in both light and heavy fractions, including fractions containing 80S ribosomes (Fraction 8 and 9, Figure 5A and Figure S4A). In contrast, Dendra alone was found primarily in the lightest fractions (Figure S4A). RNAse treatment that eliminated the polysome RNA peaks, but not the 80S peaks, caused the Dendra::MEX-5 to shift towards the lighter fractions, indicating that the association of MEX-5 with large complexes is RNA-dependent (Figure 5A and Figure S4A).
Figure 5. MEX-5 associates with multiple complexes in vivo.
A. Distribution of Dendra::MEX-5 fusions following sucrose gradient fractionation and detection by anti-Dendra western blot. Light fractions are on the left and heavy fractions are on the right. Approximate positions of the 40S, 60S and 80S ribosomal subunits are indicated. See Figure S4A for UV trace.
B. Percentage of fast and slow MEX-5 complexes detected by FCS. Note that fast and slow components were detected in all measurements. Error bars represent SEM (wild-type n=24, par-1(it51) n=5; S404A, n=8 embryos). See Figures S4B and S4C.
To test whether MEX-5’s behavior on sucrose gradients correlates with MEX-5’s diffusive behavior in vivo, we examined Dendra::MEX-5(R274E, K318E) and Dendra::MEX-5(S404A). We found that the profile of the fast-diffusing Dendra::MEX-5(R274E, K318E) was shifted towards the lighter fraction, whereas the profile of slow diffusing MEX-5(S404A) was shifted towards the heavy fractions (Figure 5A and Figure S4A). We conclude that MEX-5 exists in both light and heavy complexes, and that association with the latter depends on RNA and correlates with slower diffusion.
MEX-5 exists in multiple diffusive complexes in vivo
The broad distribution of MEX-5 in sucrose gradients suggests that MEX-5 exists in multiple complexes in vivo. To test this hypothesis directly, we used fluorescence correlation spectroscopy (FCS) to measure the diffusive behavior of individual GFP::MEX-5 molecules in live zygotes. We monitored GFP::MEX-5 at 30% and 70% embryo-length in 24 zygotes at NEBD. Autocorrelation curves were fit to 3-dimensional models containing 1, 2 or 3 diffusive components. One-component models yielded Dc values that were significantly slower than observed with DendraR::MEX-5 in both the anterior and posterior (anterior = 0.26 +/− 0.05 μm2/sec; posterior = 0.37 +/− 0.1 μm2/sec) (Figure S4B). Two component models yielded fast and slow components with ~100-fold difference in Dc values, whose weighted averages (an estimate of the population Dc) were in good agreement with those observed with DendraR::MEX-5 (anterior = 1.40 +/− 0.29 μm2/sec; posterior = 3.13 +/− 0.37 μm2/sec) (Figure S4B and S4C). The concentration ratio of slow:fast components was significantly higher in the anterior cytoplasm (66:34) compared to the posterior cytoplasm (50:50) (Figure 5B). We conclude that, as suggested by the sucrose gradients, MEX-5 exists in multiple complexes in the cytoplasm, with a bias towards slower complexes in the anterior.
To examine the effect of phosphorylation by PAR-1, we repeated the FCS measurements in par-1(it51) zygotes and in wild-type zygotes expressing MEX-5(S404A). We obtained similar FCS profiles for both genotypes. As described above for wild-type GFP::MEX-5, one-component model yielded Dc values that were significantly lower than those observed with DendraR:MEX-5 in par-1(it51) embryos or DendraR::MEX-5(S404A) in wild-type embryos (Figure S4B). 2-component models, in contrast, yielded Dc values consistent with the DendraR values (Figure 5B and Figure S4B). The Dc and concentration ratios of fast and slow complexes were similar to those observed in the anterior of wild-type embryos (Figure 5B and Figure S4C). These results suggest that MEX-5 distributes between slow and fast complexes even in the absence of PAR-1, and that phosphorylation by PAR-1 shifts the distribution of MEX-5 in favor of faster complexes.
Our FCS results indicate that MEX-5 fast and slow complexes exhibit dramatically different rates of diffusion: 5.15 μm2/sec (10–90th percentile range of 1.73 to 10.7 μm2/s) for the fast class, and 0.086 μm2/s (10–90th percentile range of 0.025 to 0.16 μm2/s) for the slow class. Daniels et al. 2010 reported a similar range of mobilities for wild-type MEX-5, but did not report the relative concentration of the slowest species and only considered species within the fast range in their modeling of the MEX-5 gradient (Daniels et al., 2010). Our analysis, however, indicate that the slow species contributes significantly to the overall diffusive behavior of MEX-5 (~70% of MEX-5 complexes in the anterior). Omission of the slow-diffusing species when calculating population Dc yields values that do not match those observed experimentally using photoactivation (this work) or FRAP (Daniels et al., 2010). We conclude that the slow-diffusing species cannot be excluded from a description of MEX-5’s diffusive behavior.
Modeling of the MEX-5 gradient
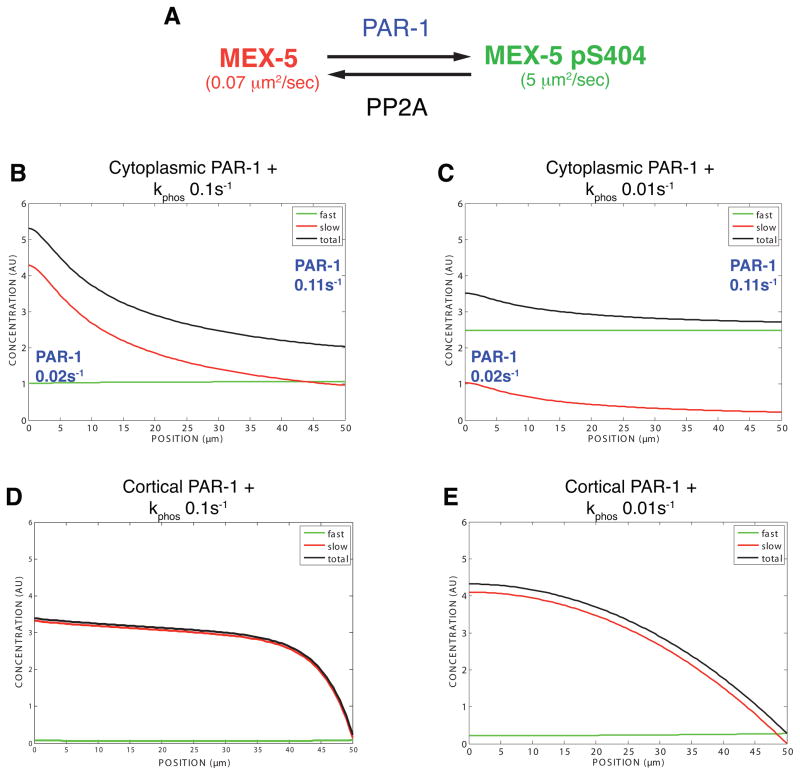
To determine whether our experimental results can be integrated into a self-consistent theoretical framework, we developed a mathematical model for the reaction-diffusion dynamics of MEX-5 (Figure 6A). The model is based on the principle that steady-state protein gradients form if 1) different phosphostates exhibit different diffusion coefficients and 2) interconversion between phosphostates is mediated by spatially-segregated kinase and phosphatase reactions (Lipkow and Odde, 2008). We approximated MEX-5 diffusion dynamics by allowing for a fast species (Dfast= 5 μm2/sec) and a slow species (Dslow =0.07 μm2/sec) whose interconversion is regulated by a phosphorylation cycle mediated by PAR-1 and PP2A (and possibly other phosphatases) (Figure 6A). Because the relative activity of cortical and cytoplasmic PAR-1 are not known, we independently considered how cytoplasmic and cortical PAR-1 affect MEX-5 segregation. Phosphatase activity was assumed to be uniform in the cytoplasm such that, in both scenarios, the kinase and phosphatase activities are spatially distinct from each other. Unsteady-state analysis and the sensitivity of the cytoplasmic and cortical PAR-1 models to changes in individual parameters are presented in Figures S5 and S6 and described in Extended Experimental Procedures.
Figure 6. Mathematical modeling of the MEX-5 gradient.
A. Model reactions. PAR-1 and PP2A are assumed to regulate interconversion between fast and slow MEX-5 species through a phosphorylation cycle. See Table 1 for assumptions used in the model.
B–E: Graphs showing the model-generated distribution of MEX-5 at steady-state along the anterior-posterior axis (anterior end, 0 micron; posterior end, 50 micron). See Extended Experimental Procedures and Figures S5 and S6 for unsteady-state analysis.
B. Cytoplasmic PAR-1 model. PAR-1 activity is assumed to be linearly distributed in the cytoplasm (low in anterior; high in posterior). This imposes an oppositely-oriented MEX-5 gradient with the fast and slow species approximately equal in concentration in the posterior and the slow species enriched in the anterior. The total MEX-5 gradient primarily reflects the gradient in slow-diffusing MEX-5. The rapid diffusion of the fast-diffusing species effectively counteracts its asymmetric formation in the posterior. See Figure S5B for unsteady-state analysis.
C. PP2A depletion. Reducing the phosphatase rate by 10-fold weakens the MEX-5 gradient and increases the proportion of phosphorylated MEX-5. See Figure S5D for unsteady-state analysis.
D. Posterior cortical PAR-1 plus uniform phosphatase (kphos=0.1 s_−_1). See Figure S6B for unsteady-state analysis.
E. Posterior cortical PAR-1 plus uniform phosphatase (kphos=0.01 s_−_1). See Figure S6C for unsteady-state analysis.
We first considered a model in which PAR-1 activity exists in a linear 5.5-fold gradient in the cytoplasm (low anterior, high posterior). The PAR-1 and phosphatase rates were matched in the posterior to yield slow:fast ratios of 1:1 in the posterior and 2:1 in the anterior, as observed in our FCS measurements (Figure 6B). This model gives rise to a temporally stable ~2.9-fold MEX-5 concentration gradient as is observed in vivo. Given a phosphatase rate of 0.1 s−1 (within the range reported in the literature of 0.1–100 s−1 (Brown and Kholodenko, 1999), the time scale of gradient formation is ~160 seconds (Figure S5B), consistent with the kinetics observed in vivo (Figure 1). Coordinately changing the absolute kinase and phosphatase rates over two orders of magnitude has little effect on the strength of the gradient. For example, increasing or decreasing both the kinase and phosphatase rates by a factor of 10 generates 3.0 and 2.8-fold MEX-5 gradients, respectively (Figures S5G and S5H). If only the kinase or phosphatase rate is changed, the MEX-5 gradient is lost (Figure S5C – S5F). For example, reducing phosphatase activity while maintaining PAR-1 activity increases the proportion of fast-diffusing species and flattens the MEX-5 gradient (Figure 6C and Figure S5D), as observed in let-92(RNAi) embryos (Figure 3C and Figure 3D). Interestingly, the MEX-5 concentration gradient is always weaker than the PAR-1 activity gradient (see Discussion).
We next considered a model where PAR-1 is entirely cortical and instantaneously phosphorylates MEX-5. In this cortical-only PAR-1 model, the extent of PAR-1’s influence on MEX-5 is determined by the phosphatase rate. For example, at a kphos= 0.1 s−1, the effect on MEX-5 drops off sharply within 10 micron of the cortex (Figure 6D and S6B). A phosphatase rate of kphos=0.01 s−1 would generate a ~3-fold MEX-5 gradient (Figure 6E and S6C). However, nearly all MEX-5 would be in the slow-diffusing state, in contrast to our FCS observations. A gradient with the observed proportions of fast and slow diffusing MEX-5 species is only obtained at a phosphatase rate of kphos=0.0001 s−1. However, the approach to steady-state would be ~ 17 minutes, far slower than what is observed in vivo (Figures 6K and S6D). Thus, the cortical-only PAR-1 model is not able to simultaneously explain the relative proportions of fast and slow species while also maintaining a rapid approach to steady-state. Taken together, the modeling analyses support a critical role for cytoplasmic PAR-1, and demonstrate that the MEX-5 diffusion gradient is sufficient to account for the MEX-5 protein gradient.
DISCUSSION
In this study, we present evidence that the antagonistic activities of PAR-1 and PP2A regulate MEX-5 diffusion to establish the MEX-5 protein gradient. We propose the following model. MEX-5 is in dynamic, local equilibrium between different diffusive RNA complexes in the cytoplasm. Phosphorylation of S404 by PAR-1 biases MEX-5 towards faster-diffusing complexes, and dephosphorylation by PP2A returns MEX-5 into slower-diffusing complexes. Before polarization, PP2A activity dominates over PAR-1, pS404 levels are low, and the majority of MEX-5 molecules are in slow diffusing complexes. At polarity onset, an unknown mechanism favors PAR-1 activity over PP2A, causing pS404 levels to rise and MEX-5 to enter faster complexes. During polarization, the PP2A/PAR-1 balance is changed along the anterior-posterior axis as PAR-1 becomes enriched in a posterior-to-anterior gradient in the cytoplasm and on the posterior cortex, causing MEX-5 to switch from phosphorylated (on average faster-diffusing) to unphosphorylated (on average slower-diffusing), as it diffuses down the PAR-1 gradient. As a result, MEX-5 redistributes in a gradient opposite PAR-1. We consider each aspect of this model in turn.
MEX-5 diffusion is retarded by binding to RNA throughout the cytoplasm
Our FCS analysis indicates that MEX-5 distributes between two classes of diffusive complexes: a “fast” class averaging 5.15 μm2/sec and a “slow” class averaging 0.086 μm2/s. Both classes are present throughout the cytoplasm, but the slow class is distributed in an anterior/high to posterior/low gradient. Because FCS analysis only constrains the minimum number of diffusive species, we cannot distinguish whether MEX-5 participates in two or more complexes. The broad range of diffusion coefficients for the “fast” and “slow” components and the broad distribution of Dendra::MEX-5 in sucrose gradients suggest, in fact, that MEX-5 may interact with a large range of complexes.
Several lines of evidence suggest that MEX-5’s association with slow diffusive complexes depends on binding to RNA. First, mutations in the CCCH fingers that reduce MEX-5 affinity for RNA increase MEX-5 diffusion and reduce the steepness of the MEX-5 gradient. Second, sucrose gradient fractionation demonstrates that MEX-5 associates with high-density complexes (comparable to 80S ribosomes) in a manner dependent on RNA and the MEX-5 RNA-binding domain. Third, the Dc for the slow species is consistent with mRNP diffusion rates (0.01–0.09 μm2/sec) in the cytoplasm of E. coli and in the nucleus of mammalian cells (Golding and Cox, 2004; Shav-Tal et al., 2004). Because mutations that block MEX-5 phosphorylation (S404A) cause the slow MEX-5 species to be symmetrically distributed even in wild-type zygotes, we do not favor a model where MEX-5 diffusion is retarded by binding to a subclass of asymmetrically localized mRNAs. Rather, we suggest that MEX-5 interacts dynamically with many mRNAs throughout the cytoplasm. Consistent with MEX-5 functioning as a broad-spectrum RNA-binding protein, MEX-5 binds to poly-U tracks which are common in C. elegans 3′ UTRs (Pagano et al., 2007), and activates maternal mRNA turnover in somatic blastomeres after the 2-cell stage (Gallo et al., 2008).
Our mutational analysis also indicates that the amino terminus of MEX-5 contributes to, but is not sufficient for, slow MEX-5 diffusion. This region is rich in poly-glutamine stretches, which could mediate MEX-5 self-association. One possibility is that, as proposed for Bruno in Drosophila, MEX-5 uses self-interactions and RNA-binding to assemble into large ribonucleoprotein particles with retarded diffusion (Chekulaeva et al., 2006).
Phosphorylation of S404 by PAR-1 biases MEX-5 towards fast complexes, and dephosphorylation by PP2A returns MEX-5 into slow complexes
In the absence of PAR-1, fast and slow MEX-5 complexes are distributed in a 30:70 constant ratio throughout the cytoplasm, indicating that phosphorylation enhances, but is not essential, for the formation of fast MEX-5 complexes. Because conditions predicted to reduce [_par-1(it51)_] or increase [_let-92(RNAi)_] phosphorylation have opposite effects on MEX-5 diffusion, we suggest that phosphorylation promotes the shifting of MEX-5 from slow to fast-diffusing complexes. Consistent with this view, MEX-5(S404A) was enriched in the heavier sucrose gradient fractions compared to wild-type MEX-5. In our simulation of the MEX-5 gradient, MEX-5 must switch multiple times between phosphostates as it diffuses across the embryo (see discussion in Extended Experimental Procedures). Consistent with this possibility, we find pS404 is highly labile in embryo extracts. We suggest that the rapid turnover of pS404 renders MEX-5 exquisitely sensitive to changes in PAR-1 distribution.
Cytoplasmic PAR-1 is essential for the formation of the MEX-5 gradient
Our simulations also demonstrate the importance of cytoplasmic PAR-1 in specifying the MEX-5 gradient. In the cortical-only PAR-1 model, high kphos values generate MEX-5 gradients that drop off sharply from the posterior cortex while low kphos values yield MEX-5 gradients that form too slowly. In contrast, in the presence of a cytoplasmic PAR-1 activity gradient, a broad range of kinase and phosphatase activities could generate the MEX-5 gradient. Cytoplasmic PAR-1, therefore, eliminates the tradeoff between gradient scale and response time. Our in vivo observations confirm that cytoplasmic PAR-1 is sufficient to regulate MEX-5 distribution: most notably, MEX-5 forms a gradient in par-2(RNAi) zygotes, which enrich PAR-1 in the posterior cytoplasm but not on the cortex. That PAR-1 can function off the cortex has also been suggested by Boyd et al. 1996, who noted that par-2 mutant zygotes localize P granules, a function requiring par-1 activity (Boyd et al., 1996; Cheeks et al., 2004).
One striking aspect of our model is that the amplitude of the MEX-5 gradient will always be smaller than the PAR-1 activity gradient (a ~2.9-fold MEX-5 gradient requires a 5.5-fold PAR-1 activity gradient). GFP::PAR-1 forms a ~3 to 4-fold cytoplasmic concentration gradient and regulation of PAR-1 kinase activity along the anterior-posterior axis could also contribute to an overall PAR-1 activity gradient. PAR-1 kinase activity has been suggested to be regulated by several mechanisms (Marx et al., 2010), including inhibition by aPKC (Hurov et al., 2004). aPKC phosphorylates PAR-1 in vitro on a conserved threonine (T983) required for PAR-1 asymmetry in vivo (Motegi et al., submitted). One possibility is that phosphorylation by anteriorly-enriched PKC-3 regulates both PAR-1 activity and levels along the anterior-posterior axis. In polarized T cells, PAR1b/EMK/MARK2 forms a cytoplasmic gradient near the immunological synapse that depends on PKC phosphorylation sites (Lin et al., 2009). These observations raise the possibility that the polarizing effects of the PAR network depend on the formation of cytoplasmic PAR-1 gradients in several cell types.
The “reactive surface” model of Daniels et al., 2010 proposes that MEX-5 diffusion is regulated at the cortex by interactions with both anterior and posterior PARs. Anterior PARs convert phosphorylated MEX-5 into a slower-diffusive form (0.4–1 μm2/s), which must be dephosphorylated before conversion back into a faster (~15 μm2/s) form by the posterior PARs (Daniels et al., 2010). The “reactive surface” model does not consider the behavior of the slowest MEX-5 species, which in our FCS analysis account for >50% of total MEX-5 (average 0.086 μm2/s, range 0.025 to 0.16 μm2/s). Furthermore, this model predicts that loss of phosphatase activity should lower MEX-5 diffusion, whereas we find that loss of the PP2A phosphatase increases MEX-5 diffusion. This model also predicts that, under conditions where MEX-5 is phosphorylated (PAR-1 active), loss of anterior PARs should increase MEX-5 diffusion. In contrast, we find that pkc-3(RNAi) has no effect on MEX-5 diffusion in par-1(b274) zygotes (Figure 2C). Our genetic analyses demonstrate that par-1 is fully epistatic to pkc-3 with respect to MEX-5 diffusion, making a direct contribution by anterior PARs unlikely. Rather, our data indicate that anterior PARs regulate MEX-5 diffusion indirectly, by controlling the distribution (and possibly the activity) of PAR-1 along the anterior-posterior axis.
Formation of concentration gradients by spatially-segregated modification enzymes
The model of Lipkow and Odde, 2008 can be used to form gradients at any cellular scale by varying diffusion and phosphatase rates. The MEX-5 gradient is established in a ~50 μm zygote, but the same principles could account for the apparent CheY gradient that emerges in the cytoplasm of E. coli (~5-micron) upon uncoupling of the phosphatase/kinase pair CheZ/CheA (Vaknin and Berg, 2004). Spatial segregation of kinase and phosphatase activities have been shown to lead to phosphogradients in many cell types from bacteria to eukaryotic cells (Brown and Kholodenko, 1999; Coppey et al., 2008; Fuller et al., 2008; Kalab et al., 2002; Maeder et al., 2007; Su et al., 1998). Our modeling analyses demonstrate that a spatially biased kinase and phosphatase cycle can give rise to protein concentration gradients even under conditions where the phosphogradient is weak. Despite higher PAR-1 activity in the posterior, phosphorylated MEX-5 is predicted to distribute almost evenly across the zygote due to its faster diffusion (Figure 6). In principle, any post-translational modification cycle could generate a protein concentration gradient, as long as the opposing enzymes are spatially segregated and the modification affects protein diffusion rates. We suggest that the mechanism we uncover here for MEX-5 can be applied broadly to understanding rapid changes in the distribution of cytoplasmic proteins in a variety of cell types.
EXPERIMENTAL PROCEDURES
Detailed experimental procedures are described in the Extended Experimental Procedures.
C. elegans Strains
Transgenic worms used in this study are listed in Table S1.
Determination of Dendrared::MEX-5 Diffusion Coefficients
Dendra::MEX-5 was photoconverted in a stripe with UV light and imaged on a spinning disk confocal microscope. Intensity values were fit to Gaussian distributions for each time point (GraphPad Prism) and the change in variance over time was used to calculate Dc (Berg, 1993).
Recombinant Protein Purification, Kinase Assays and Dephosphorylation Assays
MBP:MEX-5 and MBP:PAR-1 (1-492, T325E) were partially purified from E. coli and incubated at 30°C in the presence of 32P ATP or cold ATP. For non-isotopic phosphorylation and dephosphorylation assays, kinase reactions were terminated with 20 nM staurosporine before embryonic extract was added.
Immunoprecipitations
MEX-5 pS404 phospho-specific antibodies coupled to ProteinG dynabeads were used to immunoprecipitate from whole worm extracts.
Sucrose Gradient Fractionation
Cycloheximide-treated whole worm extracts were fractionated over 10-45% linear sucrose gradients at 39,000 rpm for 3 hours. Fractions were collected after passing the gradient through a UV detector, and the distribution of Dendra::MEX-5 was determined by western blot with anti-Dendra antibodies (Axxora).
Fluorescence Correlation Spectroscopy
GFP::MEX-5 levels were reduced by partial GFP RNAi depletion prior to imaging. Embryos were imaged on a Zeiss LSM 510 Confocal microscope equipped with a Confocor 3 FCS. Autocorrelation curves were analyzed within the Zeiss Confocor 3 software package.
Modeling of the MEX-5 gradient
Parameters used in the models are listed in Table 1. A detailed description of model and the contribution of individual parameters to the steady-state and unsteady-state models are provided in the Extended Experimental Procedures.
Table 1.
Parameters and Variables for MEX-5 Models
| Parameter/Variable | Symbol | Value | Units | Notes |
|---|---|---|---|---|
| Slow Diffusion Coefficient | Dslow | 0.07 | μm2/s | |
| Fast Diffusion Coefficient | Dfast | 5 | μm2/s | |
| Kinase (PAR-1) rate constant | kkin(x) | 0.02–0.11 | s−1 | Linear rise along A/P axis |
| Phosphatase rate constant | kphos | 0.1 | s−1 | Uniform along A/P axis |
| Embryo Length | L | 50 | μm |
Supplementary Material
01
02
Acknowledgments
We thank F. Motegi for providing the Dendra2 construct, S. He for help with sucrose gradients and N. Perkins and E. Pryce at the JHU Integrated Imaging Center for assistance with FCS. We thank K. Kemphues for providing strains and antibodies. We thank S. Kuo for helpful conversations and members of the Seydoux lab for helpful comments on the manuscript. This work was supported by the National Institutes of Health (Grants R01HD37047 [G.S.] and R01 GM71522 [D.O.]) and the American Cancer Society (Grant PF-08-158-01-DDC [EG]). G.S. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Berg HC. Random Walks in Biology, Expanded Edition edn. Princeton, New Jersey: Princeton University Press; 1993. [Google Scholar]
- Boyd L, Guo S, Levitan D, Stinchcomb DT, Kemphues KJ. PAR-2 is asymmetrically distributed and promotes association of P granules and PAR-1 with the cortex in C. elegans embryos. Development. 1996;122:3075–3084. doi: 10.1242/dev.122.10.3075. [DOI] [PubMed] [Google Scholar]
- Brown GC, Kholodenko BN. Spatial gradients of cellular phospho-proteins. FEBS Lett. 1999;457:452–454. doi: 10.1016/s0014-5793(99)01058-3. [DOI] [PubMed] [Google Scholar]
- Budirahardja Y, Gönczy P. PLK-1 asymmetry contributes to asynchronous cell division of C. elegans embryos. Development. 2008;135:1303–1313. doi: 10.1242/dev.019075. [DOI] [PubMed] [Google Scholar]
- Cheeks RJ, Canman JC, Gabriel WN, Meyer N, Strome S, Goldstein B. C. elegans PAR proteins function by mobilizing and stabilizing asymmetrically localized protein complexes. Curr Biol. 2004;14:851–862. doi: 10.1016/j.cub.2004.05.022. [DOI] [PubMed] [Google Scholar]
- Chekulaeva M, Hentze MW, Ephrussi A. Bruno acts as a dual repressor of oskar translation, promoting mRNA oligomerization and formation of silencing particles. Cell. 2006;124:521–533. doi: 10.1016/j.cell.2006.01.031. [DOI] [PubMed] [Google Scholar]
- Coppey M, Boettiger AN, Berezhkovskii AM, Shvartsman SY. Nuclear trapping shapes the terminal gradient in the Drosophila embryo. Curr Biol. 2008;18:915–919. doi: 10.1016/j.cub.2008.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuenca AA, Schetter A, Aceto D, Kemphues K, Seydoux G. Polarization of the C. elegans zygote proceeds via distinct establishment and maintenance phases. Development. 2003;130:1255–1265. doi: 10.1242/dev.00284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels BR, Dobrowsky TM, Perkins EM, Sun SX, Wirtz D. MEX-5 enrichment in the C. elegans early embryo mediated by differential diffusion. Development. 2010;137:2579–2585. doi: 10.1242/dev.051326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ephrussi A, St Johnston D. Seeing is believing: the bicoid morphogen gradient matures. Cell. 2004;116:143–152. doi: 10.1016/s0092-8674(04)00037-6. [DOI] [PubMed] [Google Scholar]
- Fuller BG, Lampson MA, Foley EA, Rosasco-Nitcher S, Le KV, Tobelmann P, Brautigan DL, Stukenberg PT, Kapoor TM. Midzone activation of aurora B in anaphase produces an intracellular phosphorylation gradient. Nature. 2008;453:1132–1136. doi: 10.1038/nature06923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo CM, Munro E, Rasoloson D, Merritt C, Seydoux G. Processing bodies and germ granules are distinct RNA granules that interact in C. elegans embryos. Dev Biol. 2008;323:76–87. doi: 10.1016/j.ydbio.2008.07.008. [DOI] [PubMed] [Google Scholar]
- Golding I, Cox EC. RNA dynamics in live Escherichia coli cells. Proc Natl Acad Sci U S A. 2004;101:11310–11315. doi: 10.1073/pnas.0404443101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein B, Macara IG. The PAR proteins: fundamental players in animal cell polarization. Dev Cell. 2007;13:609–622. doi: 10.1016/j.devcel.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Kemphues KJ. par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed. Cell. 1995;81:611–620. doi: 10.1016/0092-8674(95)90082-9. [DOI] [PubMed] [Google Scholar]
- Gurskaya NG, Verkhusha VV, Shcheglov AS, Staroverov DB, Chepurnykh TV, Fradkov AF, Lukyanov S, Lukyanov KA. Engineering of a monomeric green-to-red photoactivatable fluorescent protein induced by blue light. Nat Biotechnol. 2006;24:461–465. doi: 10.1038/nbt1191. [DOI] [PubMed] [Google Scholar]
- Hudson BP, Martinez-Yamout MA, Dyson HJ, Wright PE. Recognition of the mRNA AU-rich element by the zinc finger domain of TIS11d. Nat Struct Mol Biol. 2004;11:257–264. doi: 10.1038/nsmb738. [DOI] [PubMed] [Google Scholar]
- Hurd DD, Kemphues KJ. PAR-1 is required for morphogenesis of the Caenorhabditis elegans vulva. Dev Biol. 2003;253:54–65. doi: 10.1006/dbio.2002.0866. [DOI] [PubMed] [Google Scholar]
- Hurov JB, Watkins JL, Piwnica-Worms H. Atypical PKC phosphorylates PAR-1 kinases to regulate localization and activity. Curr Biol. 2004;14:736–741. doi: 10.1016/j.cub.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Kalab P, Weis K, Heald R. Visualization of a Ran-GTP gradient in interphase and mitotic Xenopus egg extracts. Science. 2002;295:2452–2456. doi: 10.1126/science.1068798. [DOI] [PubMed] [Google Scholar]
- Kao G, Tuck S, Baillie D, Sundaram MV. C. elegans SUR-6/PR55 cooperates with LET-92/protein phosphatase 2A and promotes Raf activity independently of inhibitory Akt phosphorylation sites. Development. 2004;131:755–765. doi: 10.1242/dev.00987. [DOI] [PubMed] [Google Scholar]
- Kemphues K. PARsing embryonic polarity. Cell. 2000;101:345–348. doi: 10.1016/s0092-8674(00)80844-2. [DOI] [PubMed] [Google Scholar]
- Krahn MP, Egger-Adam D, Wodarz A. PP2A antagonizes phosphorylation of Bazooka by PAR-1 to control apical-basal polarity in dividing embryonic neuroblasts. Dev Cell. 2009;16:901–908. doi: 10.1016/j.devcel.2009.04.011. [DOI] [PubMed] [Google Scholar]
- Lai WS, Kennington EA, Blackshear PJ. Interactions of CCCH zinc finger proteins with mRNA: non-binding tristetraprolin mutants exert an inhibitory effect on degradation of AU-rich element-containing mRNAs. J Biol Chem. 2002;277:9606–9613. doi: 10.1074/jbc.M110395200. [DOI] [PubMed] [Google Scholar]
- Lin J, Hou KK, Piwnica-Worms H, Shaw AS. The polarity protein Par1b/EMK/MARK2 regulates T cell receptor-induced microtubule-organizing center polarization. J Immunol. 2009;183:1215–1221. doi: 10.4049/jimmunol.0803887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipkow K, Odde DJ. Model for Protein Concentration Gradients in the Cytoplasm. Cell Mol Bioeng. 2008;1:84–92. doi: 10.1007/s12195-008-0008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Patterson GH. Fluorescent proteins for photoactivation experiments. Methods Cell Biol. 2008;85:45–61. doi: 10.1016/S0091-679X(08)85003-0. [DOI] [PubMed] [Google Scholar]
- Little SC, Tka ik G, Kneeland TB, Wieschaus EF, Gregor T. The Formation of the Bicoid Morphogen Gradient Requires Protein Movement from Anteriorly Localized mRNA. PLoS Biol. 2011;9:e1000596. doi: 10.1371/journal.pbio.1000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizcano JM, Göransson O, Toth R, Deak M, Morrice NA, Boudeau J, Hawley SA, Udd L, Mäkelä TP, Hardie DG, et al. LKB1 is a master kinase that activates 13 kinases of the AMPK subfamily, including MARK/PAR-1. EMBO J. 2004;23:833–843. doi: 10.1038/sj.emboj.7600110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder CI, Hink MA, Kinkhabwala A, Mayr R, Bastiaens PI, Knop M. Spatial regulation of Fus3 MAP kinase activity through a reaction-diffusion mechanism in yeast pheromone signalling. Nat Cell Biol. 2007;9:1319–1326. doi: 10.1038/ncb1652. [DOI] [PubMed] [Google Scholar]
- Marx A, Nugoor C, Panneerselvam S, Mandelkow E. Structure and function of polarity-inducing kinase family MARK/Par-1 within the branch of AMPK/Snf1-related kinases. FASEB J. 2010;24:1637–1648. doi: 10.1096/fj.09-148064. [DOI] [PubMed] [Google Scholar]
- Mello CC, Schubert C, Draper B, Zhang W, Lobel R, Priess JR. The PIE-1 protein and germline specification in C. elegans embryos. Nature. 1996;382:710–712. doi: 10.1038/382710a0. [DOI] [PubMed] [Google Scholar]
- Moravcevic K, Mendrola JM, Schmitz KR, Wang YH, Slochower D, Janmey PA, Lemmon MA. Kinase associated-1 domains drive MARK/PAR1 kinases to membrane targets by binding acidic phospholipids. Cell. 2010;143:966–977. doi: 10.1016/j.cell.2010.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam SC, Mukhopadhyay B, Choi KW. Antagonistic functions of Par-1 kinase and protein phosphatase 2A are required for localization of Bazooka and photoreceptor morphogenesis in Drosophila. Dev Biol. 2007;306:624–635. doi: 10.1016/j.ydbio.2007.03.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagano JM, Farley BM, McCoig LM, Ryder SP. Molecular basis of RNA recognition by the embryonic polarity determinant MEX-5. J Biol Chem. 2007;282:8883–8894. doi: 10.1074/jbc.M700079200. [DOI] [PubMed] [Google Scholar]
- Rivers DM, Moreno S, Abraham M, Ahringer J. PAR proteins direct asymmetry of the cell cycle regulators Polo-like kinase and Cdc25. J Cell Biol. 2008;180:877–885. doi: 10.1083/jcb.200710018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaitz AL, Srayko M, Dammermann A, Quintin S, Wielsch N, MacLeod I, de Robillard Q, Zinke A, Yates JR, Müller-Reichert T, et al. The C. elegans RSA complex localizes protein phosphatase 2A to centrosomes and regulates mitotic spindle assembly. Cell. 2007;128:115–127. doi: 10.1016/j.cell.2006.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert CM, Lin R, de Vries CJ, Plasterk RH, Priess JR. MEX-5 and MEX-6 function to establish soma/germline asymmetry in early C. elegans embryos. Mol Cell. 2000;5:671–682. doi: 10.1016/s1097-2765(00)80246-4. [DOI] [PubMed] [Google Scholar]
- Severson AF, Bowerman B. Myosin and the PAR proteins polarize microfilament-dependent forces that shape and position mitotic spindles in Caenorhabditis elegans. J Cell Biol. 2003;161:21–26. doi: 10.1083/jcb.200210171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shav-Tal Y, Darzacq X, Shenoy SM, Fusco D, Janicki SM, Spector DL, Singer RH. Dynamics of single mRNPs in nuclei of living cells. Science. 2004;304:1797–1800. doi: 10.1126/science.1099754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Johnston D, Ahringer J. Cell polarity in eggs and epithelia: parallels and diversity. Cell. 2010;141:757–774. doi: 10.1016/j.cell.2010.05.011. [DOI] [PubMed] [Google Scholar]
- Su TT, Sprenger F, DiGregorio PJ, Campbell SD, O’Farrell PH. Exit from mitosis in Drosophila syncytial embryos requires proteolysis and cyclin degradation, and is associated with localized dephosphorylation. Genes Dev. 1998;12:1495–1503. doi: 10.1101/gad.12.10.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenlen JR, Molk JN, London N, Page BD, Priess JR. MEX-5 asymmetry in one-cell C. elegans embryos requires PAR-4- and PAR-1-dependent phosphorylation. Development. 2008;135:3665–3675. doi: 10.1242/dev.027060. [DOI] [PubMed] [Google Scholar]
- Vaknin A, Berg HC. Single-cell FRET imaging of phosphatase activity in the Escherichia coli chemotaxis system. Proc Natl Acad Sci U S A. 2004;101:17072–17077. doi: 10.1073/pnas.0407812101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wartlick O, Kicheva A, González-Gaitán M. Morphogen gradient formation. Cold Spring Harb Perspect Biol. 2009;1:a001255. doi: 10.1101/cshperspect.a001255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder JH, Chong H, Guan KL, Han M. Modulation of KSR activity in Caenorhabditis elegans by Zn ions, PAR-1 kinase and PP2A phosphatase. EMBO J. 2004;23:111–119. doi: 10.1038/sj.emboj.7600025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01
02