IL28B rs12979860 Genotype and Spontaneous Clearance of Hepatitis C Virus in a Multi-Ethnic Cohort of Injection Drug Users: Evidence for a Supra-Additive Association (original) (raw)
Abstract
Among 1369 Urban Health Study participants, we evaluated genetic models for the association of IL28B genotype (rs12979860 and rs8099917) with hepatitis C virus (HCV) clearance. For rs12979860, adjusted odds ratios for spontaneous HCV clearance were as follows: _IL28B_-CC, 3.88 (P < .001); _IL28B_-CT, 1.48 (P = .08). On the basis of Akaike information criteria values and χ2 tests, a supra-additive (quadratic) model fit these data best. Models based on rs8099917 provided poorer fit. Evidence that a supra-additive rs12979860-based model best fits the association of _IL28B_-genotype with HCV clearance may improve clinical prediction models and foster a better understanding of functional mechanisms underlying this association.
Most individuals who become infected with hepatitis C virus (HCV) develop a chronic infection, which puts them at risk for cirrhosis and hepatocellular carcinoma [1]. Single-nucleotide polymorphisms (SNPs) rs12979860 and rs8099917, which are located near IL28B and in linkage disequilibrium, have been associated with spontaneous HCV clearance and response to treatment of chronic hepatitis C [2]. The identity of the functional variant underlying these associations and the specific functional mechanism are unknown.
The best genetic model to represent the association between IL28B genotype and HCV clearance is unclear. Some authors have used a recessive genetic model that compares those with beneficial genotype IL28B rs12979860-CC with those with either rs12979860-CT or rs12979860-TT [3–5]. Other reports have suggested an additive model [6, 7] or assumed no genetic model [8]. The association between IL28B genotype and HCV clearance is robust; therefore, the best genetic model was not critical for discovery studies. However, investigations of the functional mechanism underlying the IL28B genotype association may benefit from knowledge of the optimal genetic model, and, if IL28B genotype is used to guide treatment for patients with chronic hepatitis C, the best genetic model should be the basis for such decisions. We have examined alternative genetic models for the association of IL28B genotype and spontaneous HCV clearance among HCV-infected injection drug users (IDUs) enrolled in the Urban Health Study (UHS).
Methods
Participants.
As previously described, UHS recruited IDUs from San Francisco Bay, California, area neighborhoods from 1986–2002, drawing serial cross-sectional samples every 6 months [9]. Individuals ≥18 years of age were eligible for enrollment if they had injected drugs during the past 30 days or previously enrolled in UHS. After written informed consent was obtained, information was gathered through face-to-face interviews [10] and blood samples were collected. The present study includes unduplicated IDUs recruited during the period 1998 through 2000 [10]. Participants self-identified as White, Black, Asian, Pacific Islander, American Indian, Alaskan Native, other, or unknown, and as Latino/Hispanic or not. For genetic analyses, we classify ancestry as “European American” (self-reported White, not of Latino/Hispanic ethnicity), “Hispanic American” (self-reported White, Latino/Hispanic ethnicity), “African American” (self-reported Black), or “Asian American” (self-reported Asian, Pacific Islander, American Indian, or Alaskan Native). Participants who were positive for HCV antibody were divided into 2 groups on the basis of their HCV RNA result: “chronic” (positive) or “cleared” (negative). All subjects with cleared infection were included in the study and frequency matched to those with chronic infection (maximum 4:1) on the basis of self-reported ethnicity and age. Participants were not asked about treatment for HCV infection during 1998–2000, but in a survey conducted during 2002 only 3% of UHS participants reported interferon-based treatment for HCV infection [11]; thus, it is likely that the vast majority of HCV seropositive, HCV RNA–negative participants in this study had recovered spontaneously.
Study procedures were approved by the Institutional Review Board of the National Cancer Institute and the Committee on Human Subjects Research at the University of California, San Francisco.
Laboratory.
Testing for viral antibodies and HCV RNA in these participants has been described elsewhere [10]. HCV RNA was measured with branched-chain DNA assay (HCV bDNA version 3.0, Bayer [lower limit of detection, 2.5 × 103 copies/mL]). HCV antibodies were detected using HCV enzyme immunoassay version 3 (Ortho Diagnostics).
Genotyping.
Genotyping for rs12979860 and rs8099917 was performed on the ABI 7900HT platform (ABI) using custom-designed allelic discrimination TaqMan assays. Details for these assays are available at http://variantgps.nci.nih.gov/cgfseq/pages/snp500.do.
Statistical Analyses.
For each ancestry group, the adjusted odds ratio (aOR) and corresponding 95% confidence interval (CI) was determined in logistic regression models that included age, sex, hepatitis B virus (HBV) status, and human immunodeficiency virus (HIV) status. We also combined data from all 4 groups to calculate overall aORs from logistic regression models that added ancestry as a variable. Homozygosity for rs12979860-T or rs8099917-T served as the reference genotype for analyses. To investigate whether ancestry modifies the association between IL28B genotype and HCV clearance, we created models with an IL28B genotype–ancestry (cross product) interaction term and assessed potential (multiplicative) interaction with the Wald test.
We examined 5 potential genetic models: general, dominant, recessive, additive, and supra-additive (quadratic) [12]. The general model (2 degrees of freedom) estimates separate aORs for heterozygous and nonreference homozygous genotypes and thus makes no assumptions about allele dosage. Other models are based on these assumptions: dominant, 1 allele suffices for the effect; recessive model, effect requires 2 alleles; additive, 2:1 allele dosage effect; and quadratic, 4:1 allele dosage effect.
We used the Akaike information criterion (AIC) [13] to assess which SNP and genetic model fit the data best. The AIC value is proportional to the number of parameters in a model minus the maximized value of the log-likelihood function for the estimated model. AIC allows comparison of the fit of nonnested models on the basis of the lowest value. No statistical test is available to compare AIC values; therefore, we also used Wald χ2 tests to assess the appropriateness of dominant, recessive, additive, and quadratic models for coefficients obtained from the general model. For example, to assess the recessive model, we tested whether the log-OR estimate for the heterozygous genotype equaled zero. For these tests, a significant P value suggests lack of fit. Analyses were performed using SAS version 9.1 (SAS Institute).
Results
Descriptive Data.
Table 1 presents the study population characteristics. Among African American participants, age, sex, and duration of injection drug use were similar in those with chronic or cleared HCV infection. Compared with those with chronic HCV infection, European American participants who cleared HCV infection were somewhat younger (despite matching); median age for participants with chronic and cleared infection status was very similar in the other groups. In all 4 groups, individuals with chronic HCV infection were more likely to be male and to have HIV infection. Except among Asian American participants, those with chronic HCV infection were less likely to have chronic HBV infection.
Table 1.
Characteristics and Genotype Distribution Among Injection Drug Users With Chronic or Cleared Hepatitis C Virus (HCV) Infection, by Ancestry, Urban Health Study—1998–2000
| African American | European American | Hispanic | Asian | |||||
|---|---|---|---|---|---|---|---|---|
| Characteristic | Chronic (n = 348) | Cleared (n = 111) | Chronic (n = 392) | Cleared (n = 157) | Chronic (n = 88) | Cleared (n = 33) | Chronic (n = 55) | Cleared (n = 25) |
| Age, median years (IQR) | 46 (42–51) | 46 (42–49) | 42 (36–48) | 39.5 (33–47) | 46 (39–50.5) | 47 (42–51) | 43 (38–52) | 43 (39–48) |
| Duration of injection drug use, median years (IQR) | 27 (21–31) | 25 (19–31) | 23 (16–29) | 20 (11.5–29) | 27 (20–33) | 27 (21–33) | 23 (14.5–33) | 25 (21–31) |
| HCV log10 viral load, median (IQR) | 6.48 (6.05–6.92) | … | 6.42 (5.89–6.84) | … | 6.53 (5.86–6.88) | … | 6.25 (5.43–6.74) | … |
| Male (%) | 62.64 | 63.06 | 73.21 | 66.88 | 79.55 | 66.67 | 69.09 | 60.00 |
| HIV-1 infection (%) | 12.64 | 9.91 | 12.24 | 5.10 | 9.09 | 0 | 12.73 | 8.00 |
| Chronic HBV infection (%) | 2.92 | 10.91 | 3.59 | 3.85 | 1.14 | 9.09 | 5.45 | 0 |
| rs12979860 genotype (%) | ||||||||
| TT | 35.34 | 22.52 | 10.46 | 3.82 | 11.36 | 6.06 | 12.73 | 4.00 |
| CT | 51.15 | 48.65 | 46.94 | 24.84 | 54.55 | 27.27 | 34.55 | 32.00 |
| CC | 13.51 | 28.83 | 42.60 | 71.34 | 34.09 | 66.67 | 52.73 | 64.00 |
| rs8099917 genotype (%) | ||||||||
| TT | 88.79 | 93.69 | 61.48 | 82.17 | 48.86 | 72.73 | 67.27 | 76.00 |
| GT | 10.92 | 6.31 | 32.91 | 17.20 | 47.73 | 24.24 | 25.45 | 24.00 |
| GG | 0.29 | 0 | 5.61 | 0.64 | 3.41 | 3.03 | 7.27 | 0 |
HCV Clearance by rs12979860 Genotype.
Table 1 also presents IL28B genotype distributions, by ancestry and HCV status. Among 459 African American participants, individuals with the rs12979860-CC genotype cleared HCV infection more often than did those with genotype rs12979860-TT (aOR, 3.12; P < .001); the aOR for rs12979860-CT was 1.41 (95% CI, 0.82–2.41; _P_ = .22). Similarly, among 549 European American participants, individuals with rs12979860-CC were much more likely to have cleared HCV infection (aOR = 4.67; _P_ < .001), and for rs12979860-CT genotype the aOR was 1.46 (_P_ = .43). Among Hispanic participants (n = 121), the aOR for the rs12979860-CC genotype was 6.27 (_P_ = .06); for genotype rs12979860-CT, the aOR was 1.53 (_P_ = .66). Among the 80 IDUs of Asian or Amerindian ancestries, aORs were 4.13 and 2.76 for genotypes rs12979860-CC and rs12979860-CT, respectively (_P_ > .20, each comparison).
In analyses that combined racial/ethnic groups, there was no interaction between IL28B genotype and ancestry overall, nor was there any interaction in an analysis restricted to African American and European American subjects (general and quadratic models, _P_interaction > .30 for all comparisons), which suggests a similar pattern for genotype associations in all groups. In the overall analysis, rs12979860-CC homozygotes had cleared HCV about 4-fold more often than TT homozygotes (aOR, 3.88 [95% CI, 2.49–6.07]; P < .001); for participants with the rs12979860-CT genotype, the aOR was 1.48 (95% CI, .96–2.28; P = .08). An analysis that excluded individuals with either chronic hepatitis B or HIV infection yielded similar aORs (rs12979860-CC, 4.24; rs12979860-CT, 1.37).
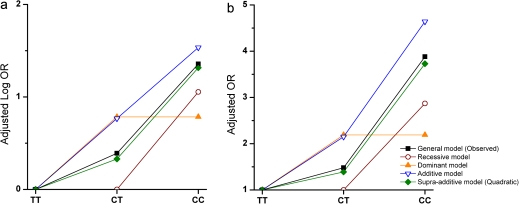
Figure 1 shows aORs observed for IL28B rs12979860 genotypes and those expected under alternative genetic models; aORs predicted by the quadratic model (rs12979860-CC, 3.73; rs12979860-CT, 1.39) fit the observed data more closely than those expected under alternative models. Under the recessive model, which presumes that the aOR for rs12979860-CT equals 1, the expected aOR for rs12979860-CC was 2.87. For the additive model, expected aORs were 4.64 for rs12979860-CC and 2.15 for rs12979860-CT. Comparing AIC values, the quadratic model yielded the lowest value (1338.1), indicating the best fit. AIC values were slightly greater for recessive (1340.0) and additive (1341.6) models, and considerably greater for the dominant model (1379.9). The results of χ2 tests were consistent with AIC rankings: the dominant model was rejected (<0.001); recessive (P = .08) and additive models (P = .06) were borderline; the quadratic model showed no lack of fit (P = .78). Results for each separate ancestry group were consistent (data not shown).
Figure 1.
Adjusted log odds ratios (a) and adjusted odds ratios (b) observed for IL28B rs12979860 genotypes (”general model”) and expected under alternative genetic models, all ancestries combined, Urban Health Study—1998–2000.
HCV Clearance by rs8099917 Genotype.
Combining ancestral groups, the aOR for rs8099917-GG was 0.13 (95% CI, .03–.56; P = .006), and that for rs8099917-GT genotype was 0.44 (95% CI, .31–.63; P < .001). In each ancestral group, persons with the rs8099917-GT genotype cleared HCV less often than did those with genotype rs8099917-TT (Supplementary Table 1), although this finding was not statistically significant among African American (aOR, 0.57; P = .19) or Asian American (aOR,0.73; P = .61) participants. Comparison of the overall AIC values revealed that rs8099917-based genetic models were less informative than were those for rs12979860; the lowest AIC value for a genotype model based on rs8099917 was 1363.4 (additive).
Discussion
In this multi-ethnic cohort of HCV-infected IDUs, the association between variation in IL28B genotype and spontaneous clearance of HCV was best described by a quadratic genetic model based on rs12979860. This nonlinear allele dosage effect suggests that the beneficial rs12979860-C allele and the unfavorable rs12979860-T allele contribute unequally to HCV clearance, such that the unfavorable allele contributes more than the favorable allele in people with the heterozygous genotype.
Our results are generally consistent with the pattern seen in previous studies of the association of IL28B genotype with spontaneous or treatment-induced HCV clearance. For spontaneous HCV clearance, Thomas et al reported unadjusted ORs for rs12979860-CC of 4.68 and for rs12979860-CT of 1.88 among African Americans and for rs12979860-CC of 2.35 and for rs12979860-CT of 0.84 among European Americans [3]. Among German women who received HCV-contaminated anti-D immunoglobulin in the late 1970s, frequencies of spontaneous clearance were as follows: rs12979860-CC, 43 (64.2%) of 67; CT, 22 (24.4%) of 90; and TT, 2 (6.1%) of 33 (rs12979860-CT vs rs12979860-TT: P = .02, Fisher exact test) [5]. Several larger studies of treatment response among individuals infected with HCV genotype 1 reported data separately for all 3 rs12979860 genotypes. For the IDEAL study, ORs (rs12979860-CC/rs12979860-CT) based on reported data were as follows: European American, 5.93/1.30; African American, 6.18/1.18; Hispanic American, 3.43/1.63 [4]. Among European American participants enrolled in the HALT-C Trial, who were retreated with pegylated interferon-α/ribavirin after previous treatment failure, aORs were 5.29 for rs12979860-CC and 1.49 for rs12979860-CT [8]. Similar ORs were seen in a group of Asian subjects on the basis of reported data: 6.60 for rs12979860-CC and 1.93 for rs12979860-CT [14]. To summarize, as in our study, prior studies found a strong association with rs12979860-CC genotype and a weaker association with 12979860-CT genotype that was not statistically significant at the P < .05 level.
Strengths and limitations of our study should be noted. UHS participants had almost no access to therapy during the study period; hence, our results are not distorted by selection for unfavorable IL28B genotypes by treatment-induced clearance. UHS is a multi-ethnic cohort in which the predominant risk factor for HCV infection is drug injection; therefore, differences between ethnic groups are less likely to be confounded by factors associated with socioeconomic strata. Regarding limitations, the numbers of Hispanic American and Asian American participants were too few to make precise estimates within these groups. In this study, viral clearance was defined by means of an HCV bDNA assay with a higher detection limit than that of assays in current clinical use. Most persons with chronic hepatitis C have high viral levels, but it is possible that a small number of participants whom we classified as cleared, in fact, had low levels of HCV RNA in the blood. The consistency of our results with those of other studies of IL28B genotype and HCV clearance suggests that, if such misclassification exists, any resulting distortion is likely minor. Finally, rs12979860 is likely only a marker for 1 or more as yet unidentified functional variants. The genetic model for a functional variant may differ from that for rs12979860.
In conclusion, our results suggest that the association between rs12979860 genotype and HCV clearance is better described by a supra-additive genetic model rather than by the recessive model, which has been the basis for some prior reports. If confirmed, these findings may inform IL28B genotype–based clinical prediction models for treatment of chronic hepatitis C and the search for functional IL28B variants.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://www.oxfordjournals.org/our_journals/jid/).
Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Supplementary Data
Notes
Financial support.
This work was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Division of Cancer Epidemiology and Genetics. This research was also supported by federal funds from the National Institute on Drug Abuse (R01-DA09532, R01-DA12109, R01-DA13245); the National Cancer Institute, National Institutes of Health (N01-CO-12400); the Substance Abuse and Mental Health Services Administration (H79-TI12103); and the San Francisco Department of Public Health. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government.
Potential conflicts of interest.
All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.NIH Consensus Statement on Management of Hepatitis C: 2002. NIH Consens State Sci Statements. 2002;19:1–46. [PubMed] [Google Scholar]
- 2.Balagopal A, Thomas DL, Thio CL. IL28B and the control of hepatitis C virus infection. Gastroenterology. 2010;139:1865–76. doi: 10.1053/j.gastro.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thomas DL, Thio CL, Martin MP, et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461:798–801. doi: 10.1038/nature08463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thompson AJ, Muir AJ, Sulkowski MS, et al. Interleukin-28B polymorphism improves viral kinetics and is the strongest pretreatment predictor of sustained virologic response in genotype 1 hepatitis C virus. Gastroenterology. 2010;139:120–9. doi: 10.1053/j.gastro.2010.04.013. [DOI] [PubMed] [Google Scholar]
- 5.Tillmann HL, Thompson AJ, Patel K, et al. A polymorphism near IL28B is associated with spontaneous clearance of acute hepatitis C virus and jaundice. Gastroenterology. 2010;139:1586–92. doi: 10.1053/j.gastro.2010.07.005. [DOI] [PubMed] [Google Scholar]
- 6.Rauch A, Kutalik Z, Descombes P, et al. Genetic variation in IL28B is associated with chronic hepatitis C and treatment failure: a genome-wide association study. Gastroenterology. 2010;138:1338–45. doi: 10.1053/j.gastro.2009.12.056. [DOI] [PubMed] [Google Scholar]
- 7.Mangia A, Thompson AJ, Santoro R, et al. An IL28B polymorphism determines treatment response of hepatitis C virus genotype 2 or 3 patients who do not achieve a rapid virologic response. Gastroenterology. 2010;139:821–7827. doi: 10.1053/j.gastro.2010.05.079. [DOI] [PubMed] [Google Scholar]
- 8.O'Brien TR, Everhart JE, Morgan TR, et al. An IL28B genotype-based clinical prediction model for treatment of chronic hepatitis C. PLoS One. 2011;6 doi: 10.1371/journal.pone.0020904. e20904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Watters JK, Bluthenthal RN, Kral AH. HIV seroprevalence in injection drug users. JAMA. 1995;273:1178. [PubMed] [Google Scholar]
- 10.Brown EE, Zhang M, Zarin-Pass R, et al. MBL2 and hepatitis C virus infection among injection drug users. BMC Infect Dis. 2008;8:57. doi: 10.1186/1471-2334-8-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seal KH, Kral AH, Lorvick J, Gee L, Tsui JI, Edlin BR. Among injection drug users, interest is high, but access low to HCV antiviral therapy. J Gen Intern Med. 2005;20:171. [Google Scholar]
- 12.Lewis CM. Genetic association studies: design, analysis and interpretation. Brief Bioinform. 2002;3:146–53. doi: 10.1093/bib/3.2.146. [DOI] [PubMed] [Google Scholar]
- 13.Lindsey JK, Jones B. Choosing among generalized linear models applied to medical data. Stat Med. 1998;17:59–68. doi: 10.1002/(sici)1097-0258(19980115)17:1<59::aid-sim733>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 14.Hayes CN, Kobayashi M, Akuta N, et al. HCV substitutions and IL28B polymorphisms on outcome of peg-interferon plus ribavirin combination therapy. Gut. 2011;60:261–7. doi: 10.1136/gut.2010.223495. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data