Initial evaluation of a Direct Detection Device detector for single particle cryo-electron microscopy (original) (raw)
. Author manuscript; available in PMC: 2012 Dec 1.
Published in final edited form as: J Struct Biol. 2011 Sep 10;176(3):404–408. doi: 10.1016/j.jsb.2011.09.002
Abstract
We report on initial results of using a new Direct Detection Device (DDD) for single particle reconstruction of vitreous ice embedded specimens. Images were acquired on a Tecnai F20 at 200KeV and a nominal magnification of 29,000x. This camera has a significantly improved signal to noise ratio and modulation transfer function (MTF) at 200 KeV compared to a standard CCD camera installed on the same microscope. Control of the DDD has been integrated into Leginon, an automated data collection system. Using GroEL as a test specimen, we obtained images of ~30K particles with the CCD and the DDD from the same specimen sample using essentially identical imaging conditions. Comparison of the maps reconstructed from the CCD images and the DDD images demonstrates the improved performance of the DDD. We also obtained a 3D reconstruction from ~70K GroEL particles acquired using the DDD; the quality of the density map demonstrates the potential of this new recording device for cryoEM data acquisition.
Keywords: TEM, cryo-electron microscopy, direct detection device, active pixel sensor, CMOS devices
It is clear that there is an urgent need for new digital detection devices that avoid the need for light conversion as required by the current generation of CCD detectors (Faruqi and Andrews, 1997). Efforts to develop so-called direct detection devices have been the focus of intense interest from the cryoEM community over the past several years (Jin et al., 2008, Faruqi, 2009, Grigorieff and Harrison, 2011, Battaglia et al., 2009, Contarato et al., 2011, Deptuch et al., 2007, McMullan et al., 2009a, Milazzo et al., 2005). Several of these devices have been quite carefully characterized and demonstrated to have superior performance to CCD cameras (and in some cases film) (McMullan et al., 2009a, Milazzo et al., 2010). Several other promising advantages make these devices a very attractive possibility for cryoEM, including the possibility of single electron counting (McMullan et al., 2009b) and the ability to use sequential frames to correct for specimen movement or explore dose fractionation (Glaeser et al., 2011).
The Direct Detection Device (DDD) is a camera developed for transmission electron microscopy based on a radiation hardened monolithic active pixel sensor design (Jin et al., 2008, Milazzo et al., 2005, Milazzo et al., 2010). Characterization of an earlier 1K×1K pixel prototype DDD showed improved high frequency performance compared to conventional CCD cameras (Milazzo et al., 2010). However, the DQE and MTF measurements were performed using a standard edge method and did not prove the ability of the detector to image a biological specimen under low dose conditions. To our knowledge, no results have yet been published that unequivocally illustrate that the new detector is suitable for imaging samples embedded in vitreous ice. We report here the results of initial testing of the DDD, using GroEL as a test specimen, that indicate the improved performance of the DDD based on a side by side comparison to a CCD camera, and the ability of the DDD to provide images suitable for reconstructing 3D electron density maps containing high resolution features.
The GroEL specimen was generously provided by Eli Chapman, Scripps Research Institute. Grid preparation and freezing conditions were similar to those reported previously (Stagg et al., 2006). All experiments were carried out on a Tecnai F20 Twin transmission electron microscope operated at 200 KeV, with a 50μm C2 aperture and a 100μm objective aperture. Prior to data acquisition, the instrument was adjusted for coma-free alignment of less than 0.1 mrad. A Gatan side-entry 626 cryo stage was used to maintain the specimen at a temperature below −170 °C.
The two cameras compared in this experiment were a Tietz F415 4K × 4K pixel (15μm pixel pitch) CCD camera and a retractable Direct Electron DE12 3K × 4K pixel (6μm pixel pitch) DDD camera. Nominal TEM magnification used for the acquisition was 62,000X for the CCD and 29,000X for the DDD, resulting in pixel sizes at the specimen of 1.37Å (CCD) and 1.38Å (DDD). For practical reasons the DDD images were stored as integrated 3K × 3K frames, with each image composed of a sum of 10 individual frames acquired at a rate of 40 frames/sec.
Data were collected using Leginon (Suloway et al., 2005) in two separate experiments. The first experiment acquired data only to the DDD. In the second experiment, Leginon was used to randomly select either the DDD or the CCD cameras for acquiring images of targeted holes. Thus, the same grid was used under essentially identical conditions to acquire images on the two devices. Image exposure time was 400 ms, total dose per image was ~20e−/Å2, defocus was randomly varied in a range from 0.8μm to 2.5μm.
A total of 1293 and 2966 DDD images were acquired in the first and second experiments, respectively. In the comparison experiment, 879 CCD images were acquired; fewer CCD images were required to achieve an equivalent number of particles from the CCD and DD images due to the larger field of view of the CCD camera.
After rejecting unsuitable micrographs, a total of 583 images from the CCD and 2313 images from the DDD were available for the comparison measurement, and 1981 images for the combined DDD reconstruction. Appion (Lander et al., 2009) was used in all processing steps prior to projection-matching refinement. CTF estimation was performed using CTFFind (Mindell and Grigorieff, 2003); particles were selected with FindEM (Roseman, 2004) using templates of side, top and tilted views. Some additional filtering steps were used to reject classes of selected objects that were obviously not GroEL particles. Stacks were created by boxing out the selected particles from the micrographs and flipping the phases using the CTFFIND estimations. The initial model used for the reconstructions was a GroEL map low pass filtered to 30Å. XMIPP was used for the projection refinement analysis (Sorzano et al., 2004) with D7 symmetry imposed; no masking was applied during the projection refinement. 14 iterations were performed with an inner radius for rotational correlation of 0 pixels and an outer radius for rotational correlation of 80 pixels. The angular distance between neighboring projection points for the first 4 iterations was 10 degrees. The next 10 iterations had the angular distance decremented to 5, 3, 2, 1, and 0.5 degrees every 2 iterations. The maximum change in rotation and tilt was unlimited for the first 4 iterations and constrained to 20 degrees for next 2 iterations, 9 degrees for the next 2 iterations, and 6 degrees for the final 6 iterations. The XMIPP weighted back projection method was used for the 3D reconstruction. FSC0.5 was calculated from even-odd volumes. Amplitude correction was applied to the final volumes using X-ray scattering data from GroEL as implemented in SPIDER where an enhancement curve from the X-ray data is applied as a filter to the EM volume (Frank et al., 1996). After amplitude correction, a Gaussian low pass filter was applied. Data were visualized using Chimera (Pettersen et al., 2004).
The cryo-EM density raw and post processed maps from the CCD camera micrographs are available from the EMDB as EMDB ID 5336 and EMDB ID 5340, respectively. The cryo-EM density raw and post processed maps from the DDD camera to compare to the CCD camera are available from the EMDB as EMDB ID 5337 and EMDB ID 5339, respectively. The cryo-EM density map from the combined DDD reconstruction is available from the EMDB as EMDB ID 5338.
A comparison between a DDD camera and a CCD camera under the same conditions shows improved performance using the DDD
Bright field imaging of carbon film is a standard technique to compare different imaging devices for TEM. The Fourier transform of such images can be used to calculate a power spectrum where the amplitude as a function of frequency represents the contrast transfer of the TEM and the transfer function of the camera (Zhang et al., 2003).
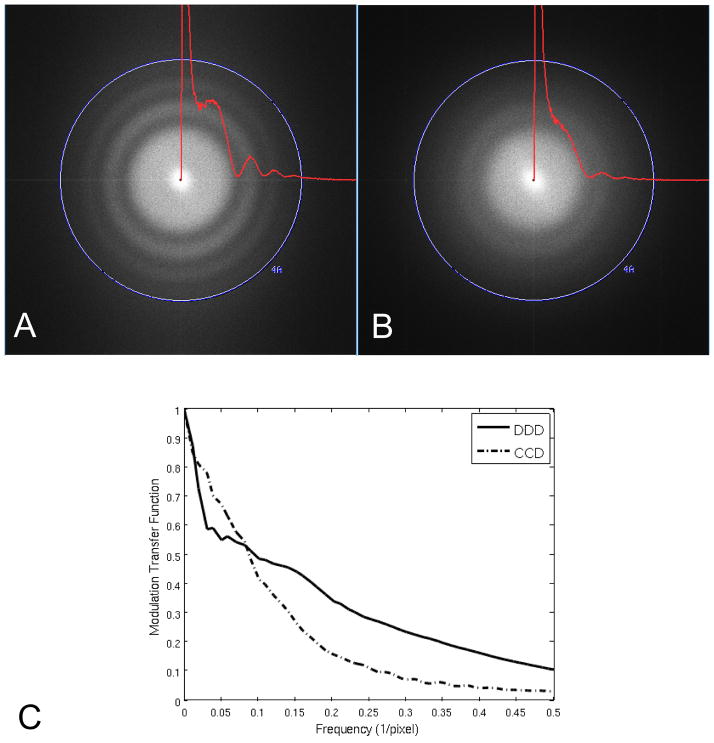
Figure 1 shows the Fourier transforms of 3K × 3K pixel images of carbon film obtained with the DDD in (A) and the CCD in (B). Imaging conditions were as similar as possible except those related to magnification so as to match the pixel size at the specimen. Overlaid on the Fourier transforms are the 2D power spectra of the carbon image radially averaged without smoothing. A similar radially averaged power spectra from an image of an empty area was used for background subtraction after being scaled to match the carbon scattering value at the Nyquist limit. The Thon rings on the DDD extend out to ~4Å while the Thon rings on the CCD camera extend to ~5Å. This comparison using carbon film clearly demonstrates the improved performance of the DDD over the CCD in the high frequency range. Figure 1(C) shows the MTF calculated using the edge method (Meyer and Kirkland, 2000) for the DDD and CCD under the same conditions, which indicates an improved MTF for the DDD compared to the CCD camera above a frequency of ~0.085 1/pixel.
Figure 1.
Comparison of detection limit between cameras installed on the same microscope using identical instrument conditions and almost matching pixel size. Fourier transforms and radially averaged power spectra of 3K × 3K cropped images from: (A) DDD (1.38Å/pixel) and (B) CCD (1.37Å/pixel). High tension 200 KeV, electron dose ~20e−/Å2, exposure time 400ms. The modulation transfer function is shown in (C) for the CCD and DDD cameras calculated using the edge method.
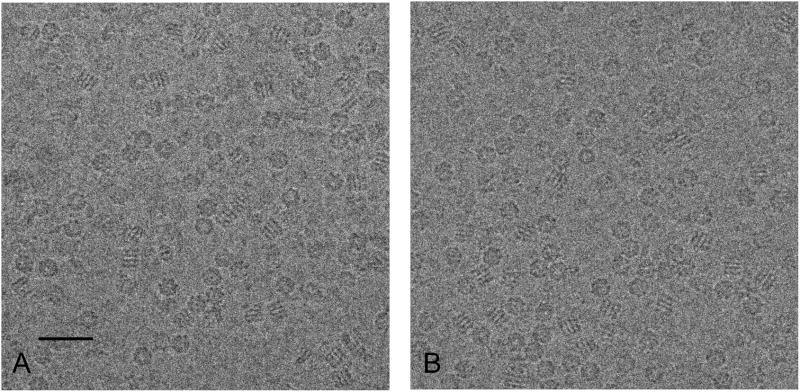
A careful side-by-side comparison of low dose imaging of a biological specimen preserved in vitreous ice using the DDD and a CCD camera establishes a benchmark for the performance of the DDD for cryoEM. GroEL is a dual ringed tetradecamer molecule that has been extensively used as a test-bed for cryoEM reconstructions (Danev and Nagayama, 2008, Ludtke et al., 2008, Stagg et al., 2006, Stagg et al., 2007). Figure 2 shows representative (cropped 2000 × 2000 pixel regions) images of GroEL obtained using the DDD in (A) and the CCD camera in (B) during the comparison experiment. The main morphological features of different views of the GroEL protein are visible in both micrographs. We note, however, that this sample preparation was not optimal for achieving a high resolution 3D map in that many of the fields contained particles that were somewhat crowded or embedded in vitreous ice somewhat thicker than desirable. We also noted in class averages of end on views of the GroEL that it apparently contained a client protein, indicating that it was actively chaperoning and thus potentially conformationally heterogeneous (Falke et al., 2005).
Figure 2.
Representative cropped 2000 × 2000 pixel micrographs of GroEL. (A) DDD image (1.38Å/pixel), (B) CCD image (1.37Å/pixel). Defocus value is approximately 2μm for both images. Scale bar is 40nm.
Figure 3 shows the results of comparing the 3D reconstructions of GroEL from 30,711 particles from the DDD in (A) and the CCD camera in (B). The 3D volumes in (a) and (e) have not been low pass filtered or amplitude corrected and show that the unprocessed maps from each camera are visually very similar. In (b) and (f) the volumes were amplitude corrected, using a GroEL X-ray scattering curve, out to 8Å and low pass filtered to 7.5Å; the two maps once again provide similar overall features. However, once the maps are amplitude corrected with a scattering curve out to 5Å and low pass filtered to 6Å, secondary structures such as alpha helices are clearly visible in the DDD map (c), whereas the CCD map (g) indicates excessive sharpening at this resolution. The resolution (FSC0.5) calculated from even-odd maps using XMIPP, was 6.7Å and 7.3Å for the DDD (d) and CCD (h) data sets, respectively. The obvious qualitative difference between the volumes in the high frequency domain and the quantitative improvement provided by the FSC0.5 values is encouraging preliminary evidence that the DDD camera provides improved performance over a standard CCD camera for cyoEM imaging at 200 KeV.
Figure 3.
GroEL reconstructions from ~30K particle data sets comparing DDD in (A) and the CCD camera in (B). The final volumes with no post processing are shown in (a) and (e). Maps shown in (b) and (f) were amplitude corrected with an X-ray scattering curve out to 8Å and low pass filtered to 7.5Å. Volumes shown in (c) and (g) were amplitude corrected with an X-ray scattering curve out to 5Å and low pass filtered to 6Å. The Fourier Shell Correlation plots for (a) and (e) are shown in (d) and (h).
A “high resolution” map is obtained from a 72K GroEL particle data set using the DDD
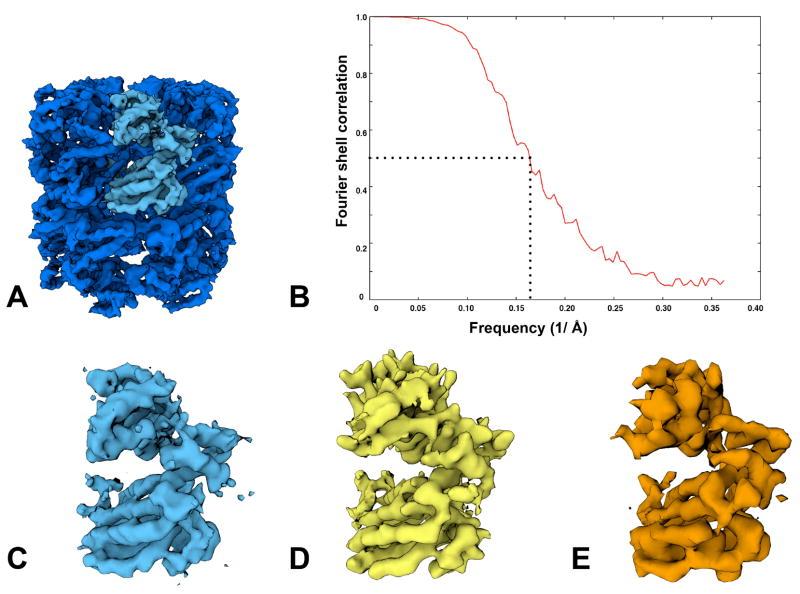
A total of 72,316 particles obtained from the DDD images combined from both experiments were used to calculate a 3D map of GroEL (figure 4). Figure 4(A) shows a map of GroEL amplitude corrected using a GroEL X-ray scattering curve out to 5Å and low pass filtered to 4.5Å resolution. Figure 4(B) shows the FSC curve of even-odd reconstructions with an FSC0.5 of 6.1Å. In Figure 4(C), we show an asymmetric subunit from the top DDD density and previous published structures of GroEL at a stated resolution of 4Å (Ludtke et al., 2008) in (D) and 5.4Å (Stagg et al., 2008) in (E). All maps were set to a threshold to represent equivalent molecular weights. Visual inspection indicates that the DDD map provides many of the high resolution secondary features visible in the previously published structures, including distinct alpha helices with discernable pitch.
Figure 4.
A high resolution DDD reconstruction from a 72,316 particle dataset is shown in (A), low pass filtered to 4.5Å. In (B), the Fourier Shell Correlation, derived from reconstructed even-odd particle data sets, gives the FSC0.5 as 6.1Å. For comparison, we show an asymmetric subunit from the DDD map in (C) and previously published asymmetric subunits from a 4Å map acquired using film in (D) (Ludtke et al., 2008) and a 5.4Å map from a CCD camera in (E) (Stagg et al., 2008).
Further exploration of high resolution single particle cryoEM with a DDD camera is warranted based on these initial results
The first cryoEM reconstruction of GroEL using a DDD camera shows that high resolution 3D reconstruction using low dose images is possible. It was our intention with this study to demonstrate that the DDD camera could be used under practical or real-world circumstances to produce a useful 3D map.
Given that the specimen used for this analysis was not optimal and that the direct detector can also be further optimized, we are optimistic about the potential of these new detection devices for high throughput and high resolution molecular microscopy. We also note that resolution estimates in cryoEM are not straightforward, and depend to some extent on the details of the processing algorithms used to reconstruct the map (e.g. the use of masks, high pass filters, etc.), and the method used to compute the FSC curves. Thus, our optimism in the potential of this new camera is based on the appearance of the details in the 3D maps (figure 4) rather than on the absolute resolution based on the FSC0.5 measures. We also note that integration of the DDD within the Leginon infrastructure was important for conducting a comparison of the CCD to the DDD under closely controlled conditions.
We conclude that, despite past concern about increased noise in the low frequency range due to backscattered electrons (Milazzo et al., 2010), there is strong evidence that the improved performance of the DDD results will translate into higher quality 3D density maps of biological specimens derived from low dose imaging and single particle reconstruction methods. The new detectors provide advantages over currently used CCD cameras in both the spatial resolution and the signal to noise ratio of the acquired images. Furthermore, as the data is acquired as a set of multiple image frames, each corresponding to a fraction of the total dose, the DDD offers the potential of correcting for specimen drift and for calculating maps from very limited dose exposures (Glaeser et al., 2011). Exploiting this potential will require the development of supporting infrastructure, algorithms and procedures.
Acknowledgments
We thank Dr. Reza Khayat for helpful advice and discussion. A.M. received support from the NIH Roadmap Award P50 GM073197. The work presented here was conducted at the National Resource for Automated Molecular Microscopy, which is supported by the National Institutes of Health through the National Center for Research Resources’ P41 program (RR17573). This work is also partially supported by the National Institutes of Health Research Resource grants RR018841 and RR004050.
Footnotes
Conflicts of Interest
Prior to the research for this publication, A.-C.M. received consulting fees from Direct Electron for unrelated professional services.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Battaglia M, Contarato D, Denes P, Doering D, Giubilato P, et al. A rad-hard CMOS active pixel sensor for electron microscopy. Nucl Instrum Methods A. 2009;598:642–649. [Google Scholar]
- Chen JZ, Settembre EC, Aoki ST, Zhang X, Bellamy AR, et al. Molecular interactions in rotavirus assembly and uncoating seen by high-resolution cryo-EM. Proc Natl Acad Sci U S A. 2009;106:10644–10648. doi: 10.1073/pnas.0904024106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng A, Fellmann D, Pulokas J, Potter CS, Carragher B. Does contamination buildup limit throughput for automated cryoEM? J Struct Biol. 2006;154:303–311. doi: 10.1016/j.jsb.2006.03.005. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Walz T. The advent of near-atomic resolution in single-particle electron microscopy. Annu Rev Biochem. 2009;78:723–742. doi: 10.1146/annurev.biochem.78.070507.140543. [DOI] [PubMed] [Google Scholar]
- Contarato Devis, Denes Peter, Doering Dionisio, Joseph John, Krieger Brad. Direct detection in Transmission Electron Microscopy with a 5 micron pitch CMOS pixel sensor. Nucl Instrum Methods Phys Res A. 2011;635:69–73. [Google Scholar]
- Danev R, Nagayama K. Single particle analysis based on Zernike phase contrast transmission electron microscopy. J Struct Biol. 2008;161:211–218. doi: 10.1016/j.jsb.2007.10.015. [DOI] [PubMed] [Google Scholar]
- Deptuch G, Besson A, Rehak P, Szelezniak M, Wall J, et al. Direct electron imaging in electron microscopy with monolithic active pixel sensors. Ultramicroscopy. 2007;8:674–684. doi: 10.1016/j.ultramic.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Falke S, Tama F, Brooks CL, 3rd, Gogol EP, Fisher MT. The 13 angstroms structure of a chaperonin GroEL-protein substrate complex by cryo-electron microscopy. J Mol Biol. 2005;348:219–230. doi: 10.1016/j.jmb.2005.02.027. [DOI] [PubMed] [Google Scholar]
- Faruqi AR, Andrews HN. Cooled CCD camera with tapered fibre optics for electron microscopy. Nucl Instrum Methods A. 1997;392:233–236. [Google Scholar]
- Faruqi AR. Principles and prospects of direct high resolution electron image acquisition with CMOS detectors at low energies. J Phys Condens Matter. 2009;21:314004–314013. doi: 10.1088/0953-8984/21/31/314004. [DOI] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Glaeser RM, McMullan G, Faruqi AR, Henderson R. Images of paraffin monolayer crystals with perfect contrast: Minimization of beam-induced specimen motion. Ultramicroscopy. 2011;111:90–100. doi: 10.1016/j.ultramic.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigorieff N, Harrison S. Near-atomic resolution reconstructions of icosahedral viruses from electron cryo-microscopy. Curr Opin Struct Biol. 2011;21:265–273. doi: 10.1016/j.sbi.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludtke SJ, Baker ML, Chen DH, Song JL, Chuang DT, et al. De novo backbone trace of GroEL from single particle electron cryomicroscopy. Structure. 2008;16:441–448. doi: 10.1016/j.str.2008.02.007. [DOI] [PubMed] [Google Scholar]
- Jiang W, Baker ML, Jakana J, Weigele PR, King J, et al. Backbone structure of the infectious epsilon15 virus capsid revealed by electron cryomicroscopy. Nature. 2008;451:1130–1134. doi: 10.1038/nature06665. [DOI] [PubMed] [Google Scholar]
- Jin L, Milazzo A, Kleinfelder S, Li S, Leblanc P, et al. Applications of direct detection device in transmission electron microscopy. J Struct Biol. 2008;161:352–358. doi: 10.1016/j.jsb.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander GC, Stagg SM, Voss NR, Cheng A, Fellmann D, et al. Appion: an integrated, database-driven pipeline to facilitate EM image processing. J Struct Biol. 2009;166:95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Jin L, Koh SB, Atanasov I, Schein S, et al. Atomic structure of human adenovirus by cryo-EM reveals interactions among protein networks. Science. 2010;329:1038–1043. doi: 10.1126/science.1187433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Zhang Q, Murata K, Baker ML, Sullivan MB, et al. Structural changes in a marine podovirus associated with release of its genome into Prochlorococcus. Nat Struct Mol Biol. 2010;17:830–836. doi: 10.1038/nsmb.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludtke SJ, Baker ML, Chen DH, Song JL, Chuang DT, et al. De novo backbone trace of GroEL from single particle electron cryomicroscopy. Structure. 2008;16:441–448. doi: 10.1016/j.str.2008.02.007. [DOI] [PubMed] [Google Scholar]
- McMullan G, Chen S, Henderson R, Faruqi AR. Detective quantum efficiency of electron area detectors in electron microscopy. Ultramicroscopy. 2009;109:1126–1143. doi: 10.1016/j.ultramic.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMullan G, Clark AT, Turchetta R, Faruqi AR. Enhanced imaging in low dose electron microscopy using electron counting. Ultramicroscopy. 2009;109:1411–1416. doi: 10.1016/j.ultramic.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer RR, Kirkland AI. Characterisation of the signal and noise transfer of CCD cameras for electron detection. Microsc Res Tech. 2000;49:269–280. doi: 10.1002/(SICI)1097-0029(20000501)49:3<269::AID-JEMT5>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Milazzo AC, Leblanc P, Duttweiler F, Jin L, Bouwer JC, et al. Active pixel sensor array as a detector for electron microscopy. Ultramicroscopy. 2005;104:152–159. doi: 10.1016/j.ultramic.2005.03.006. [DOI] [PubMed] [Google Scholar]
- Milazzo AC, Moldovan G, Lanman J, Jin L, Bouwer JC, et al. Characterization of a direct detection device imaging camera for transmission electron microscopy. Ultramicroscopy. 1995;110:741–744. doi: 10.1016/j.ultramic.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Roseman AM. FindEM—a fast, efficient program for automatic selection of particles from electron micrographs. J Struct Biol. 2004;145:91–99. doi: 10.1016/j.jsb.2003.11.007. [DOI] [PubMed] [Google Scholar]
- Settembre EC, Chen JZ, Dormitzer PR, Grigorieff N, Harrison SC. Atomic model of an infectious rotavirus particle. Embo J. 2011;30:408–416. doi: 10.1038/emboj.2010.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorzano COS, Marabini R, Velazquez-Muriel J, Bilbao-Castro JR, Scheres SHW, et al. XMIPP: a new generation of an open-source image processing package for electron microscopy. J Struct Biol. 2004;148:194–204. doi: 10.1016/j.jsb.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, et al. Automated molecular microscopy: the new Leginon system. J Struct Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Stagg SM, Lander GC, Quispe J, Voss NR, Cheng A, et al. A test-bed for optimizing high-resolution single particle reconstructions. J Struct Biol. 2008;163:29–39. doi: 10.1016/j.jsb.2008.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf M, Garcea RL, Grigorieff N, Harrison SC. Subunit interactions in bovine papillomavirus. Proc Natl Acad Sci U S A. 2010;107:6298–6303. doi: 10.1073/pnas.0914604107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Jin L, Zhou ZH. 3.88 A structure of cytoplasmic polyhedrosis virus by cryo-electron microscopy. Nature. 2008;453:415–419. doi: 10.1038/nature06893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Borgnia MJ, Mooney P, Shi D, Pan M, et al. Automated image acquisition and processing using a new generation of 4K × 4K CCD cameras for cryo electron microscopic studies of macromolecular assemblies. J Struct Biol. 2003;143:135–144. doi: 10.1016/s1047-8477(03)00124-2. [DOI] [PubMed] [Google Scholar]
- Zhang X, Settembre E, Xu C, Dormitzer PR, Bellamy R, et al. Near-atomic resolution using electron cryomicroscopy and single-particle reconstruction. Proc Natl Acad Sci USA. 2008;105:1867–1872. doi: 10.1073/pnas.0711623105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Jin L, Fang Q, Hui WH, Zhou ZH. 3.3 A cryo-EM structure of a nonenveloped virus reveals a priming mechanism for cell entry. Cell. 2010;141:472–482. doi: 10.1016/j.cell.2010.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]