Relationship of Axonal Voltage-gated Sodium Channel 1.8 (NaV1.8) mRNA Accumulation to Sciatic Nerve Injury-induced Painful Neuropathy in Rats (original) (raw)
Background: Painful neuropathy is an unsolved disease with increased voltage-gated sodium channel (NaV) activities.
Results: NaV1.8 shRNA treatment attenuated injury-induced pain behavior and normalized the NaV1.8 mRNA levels in the affected axons but not in somata of sensory neurons.
Conclusion: Painful neuropathy may causally involve axonal NaV1.8 mRNA.
Significance: Nerve injury may induce axonal NaV1.8 mRNA accumulation, which may offer a novel therapeutic target.
Keywords: Axon, Neurological Diseases, Rat, RNA Transport, shRNA, Sodium Channels, NaV1.8, Neuropathic Pain
Abstract
Painful peripheral neuropathy is a significant clinical problem; however, its pathological mechanism and effective treatments remain elusive. Increased peripheral expression of tetrodotoxin-resistant voltage-gated sodium channel 1.8 (NaV1.8) has been shown to associate with chronic pain symptoms in humans and experimental animals. Sciatic nerve entrapment (SNE) injury was used to develop neuropathic pain symptoms in rats, resulting in increased NaV1.8 mRNA in the injured nerve but not in dorsal root ganglia (DRG). To study the role of NaV1.8 mRNA in the pathogenesis of SNE-induced painful neuropathy, NaV1.8 shRNA vector was delivered by subcutaneous injection of cationized gelatin/plasmid DNA polyplex into the rat hindpaw and its subsequent retrograde transport via sciatic nerve to DRG. This in vivo NaV1.8 shRNA treatment reversibly and repeatedly attenuated the SNE-induced pain symptoms, an effect that became apparent following a distinct lag period of 3–4 days and lasted for 4–6 days before returning to pretreatment levels. Surprisingly, apparent knockdown of NaV1.8 mRNA occurred only in the injured nerve, not in the DRG, during the pain alleviation period. Levels of heteronuclear NaV1.8 RNA were unaffected by SNE or shRNA treatments, suggesting that transcription of the Scn10a gene encoding NaV1.8 was unchanged. Based on these data, we postulate that increased axonal mRNA transport results in accumulation of functional NaV1.8 protein in the injured nerve and the development of painful neuropathy symptoms. Thus, targeted delivery of agents that interfere with axonal NaV1.8 mRNA may represent effective neuropathic pain treatments.
Introduction
Injuries to peripheral nerves often cause chronic pain manifesting as post-herpetic neuralgia, painful diabetic neuropathy, and painful post-traumatic neuroma. Chronic peripheral neuropathy affects ∼1.5% of the general population (1) and greatly impairs quality of life, because current treatments remain unsatisfactory for most patients. Painful neuropathies commonly share clinical features such as light touch-evoked pain (allodynia) (2–5), burning sensation, exaggerated responses to noxious stimuli (hyperalgesia), and either spontaneous or evoked unpleasant abnormal sensations (dysesthesia) (6). Although several pathological mechanisms have been proposed (7–9), the precise molecular mechanisms contributing to the development and maintenance of peripheral neuropathy are still elusive.
Because the generation and propagation of action potentials in sensory neurons depend on the activity of voltage-gated sodium channels (VGSCs),2 it has been postulated that abnormal neuronal hyperexcitability caused by altered regulation of VGSCs may play a key role in the molecular pathogenesis of painful neuropathy (10, 11). Altered expression of VGSCs that are found in primary sensory neurons, e.g. NaV1.3, NaV1.7, NaV1.8, and NaV1.9, has been shown to associate at various degrees with human neuropathies (12–17) and animal neuropathy models (18–21). Among the peripheral VGSC isoforms, NaV1.8 is the predominant tetrodotoxin-resistant sodium channel expressed exclusively in primary sensory neurons with particularly high levels of expression in nociceptive neurons of small- and medium-sized soma diameters (22). NaV1.8 is involved in nociceptive signaling through up-regulation of channel expression and kinetics after tissue inflammation and its contribution to action potential propagation in nociceptive neurons (23, 24). After peripheral nerve injury, NaV1.8 appears to be redistributed preferentially to nerve axons (9, 25). Selective pharmacological inhibition of NaV1.8 function (26) or NaV1.8 expression in DRG using intrathecal administration of antisense oligodeoxynucleotides (27) has been shown to decrease mechanical allodynia and thermal hyperalgesia in rats with spinal nerve ligation (SNL). These reports underscore the role of NaV1.8 in the pathogenesis; however, the molecular mechanism of how NaV1.8 develops and maintains painful neuropathy is not yet fully established. Recently, we reported that not only NaV1.8 protein but also NaV1.8 mRNA was up-regulated in nerve axons after sciatic nerve entrapment (SNE) injury in rats (28). We have hypothesized that the NaV1.8 mRNA in the injured axon may play an important role in the pathogenesis of pain behaviors. Here we tested this hypothesis by targeted suppression of NaV1.8 mRNA using shRNA delivery to lumbar dorsal root ganglion (DRG) neurons in vivo. We report that shRNA-derived NaV1.8 knockdown attenuated SNE-induced pain behaviors in rats after a distinct lag period. A series of mechanistic experiments suggested that the axonal accumulation of NaV1.8 mRNA, possibly through an injury-activated subcellular transport system, plays a causal role in the development of painful neuropathy.
EXPERIMENTAL PROCEDURES
Animals
Adult Sprague-Dawley rats weighing 200–300 g were used throughout. The animal experiments were performed in accordance with the guidance of the National Institutes of Health on animal care and University of California at Los Angeles Animal Research Committee.
NaV1.8 shRNA Plasmid DNA and Lentiviral Vector Construction
Four siRNA target sequences to NaV1.8 mRNA (accession number NM_017247) were designed: shRNA1 (nucleotides 1203–1221); shRNA2 (nucleotides 3407–3425); shRNA3 (nucleotides 6009–6027); and shRNA4 (nucleotides 6033–6051). NaV1.8 shRNA1, 2, and 4 were previously published (29, 30), and shRNA3 was newly designed (Dharmacon, Lafayette, CO). Nucleotide BLAST verified that siRNA sequences were complimentary only to the Nav1.8 mRNA. A random shRNA served as a negative control: GCAGCAACTGGACACGTGA. To create the short hairpin constructs, sense siRNA sequences were linked to their antisense sequences by a stem loop (TTCAAGAGA) (31). The 55-nucleotide sense oligonucleotides and the 59 nucleotides of antisense oligonucleotides containing additional nucleotides at 5′ end for XhoI overhang were subcloned into pLL3.7 (Lentilox 3.7; MIT, Cambridge, MA) (see Fig. 1A). The shRNA-expressing vector contained mouse U6 promoter for shRNA expression and CMV promoter for enhanced GFP expression. The pLL3.7-NaV1.8shRNA constructs were large scale-amplified, and each lentiviral vector was produced by a triple transfection of three plasmids: pLL3.7-NaV1.8 shRNA, pΔ8.9, and pVSVG.
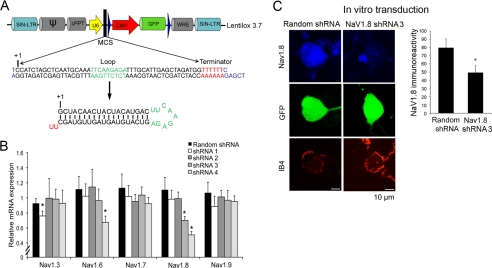
FIGURE 1.
Efficiency and specificity of RNA interference-derived knockdown of NaV1.8. A, third generation self-inactivating long terminal repeat (SIN-LTR) vector construct (Lentilox 3.7) carrying shRNA3 driven by the U6 promoter and the GFP reporter gene driven by the CMV promoter. B, RT-PCR for the steady state mRNA levels of NaV1.3, NaV1.6, NaV1.7, NaV1.8, and NaV1.9 in acutely dissociated DRG neurons in vitro after transduction of lentiviral vectors carrying NaV1.8 shRNA 1, 2, 3, 4 or control random shRNA. *, p < 0.05 against random shRNA. C, NaV1.8 immunoreactivity of DRG neurons after shRNA3 and random shRNA transduction. *, p < 0.05.
Culture of DRG Neurons
DRG neuron cultures (32) were used to determine the efficiency and specificity of shRNA constructs. Harvested rat lumbar DRG were transferred to ice-cold Hanks' balanced salt solution (HBSS) containing 20% FBS and cut into small pieces. Tissues were washed with cold 20% FBS-HBSS and then HBSS before incubating in collagenase solution (1.25 mg of collagenase P/5 ml of HBSS and 0.2 mg of DNase I/ml of HBSS) for 75 min. Next, 1.25 mg/ml trypsin and 0.5 mg of DNase I/ml in HBSS were added and incubated for 5 min at 37 °C followed by wash with 20% FBS-HBSS followed by HBSS. After washing, the pellet was resuspended in dissociation solution (12 mm MgSO4 and 0.2 mg of DNase I/ml of HBSS) and triturated with a silicone-coated Pasteur pipette until the solution appeared uniformly cloudy. After centrifugation, the cell pellet was collected, resuspended with 1000 ml of DRG culture medium/well (10% FBS, 1% antibiotic-antimycotics, 0.5% of 1.5 mg/ml uridine and 0.5% of 3.5 mg/ml floxuridine in minimum essential medium), and plated on a 6-well plate precoated with Matrigel (Becton Dickinson, Franklin Lakes, NJ). The cells were subjected to experiments 24 h after seeding. The medium was changed every 2 days.
NaV1.8 shRNA lentiviral vectors (1 mg of p24 = 5 × 107 infectious units in 293T cells/ml of minimum essential medium without FBS), polybrene (8 mg/ml), 1.5 mg/ml uridine (final v/v 0.5%), and 3.5 mg/ml floxuridine (final v/v 0.5%) were added to DRG cells and incubated for 24 h. DRG cells were washed and supplemented with DRG culture medium. The cells were collected 48 h after transduction for FACS, RT-PCR, and immunocytochemistry analyses (see below).
FACS Analysis
DRG cells were dissociated from the plate with 2 ml of trypsin solution. The cells were collected and resuspended with cold PBS. The cells were stained with 10 μg/ml of isolectin B4 (IB4) conjugated with Alexa Fluor 647 (Invitrogen). Enhanced GFP and Alexa Fluor 647 were measured by FACS (EPICS Elite ESP; Beckman Coulter, Fullerton, CA).
Quantitative Real Time PCR
Total RNA from DRG cultures was isolated and treated with DNase I (Ambion, Austin, TX). The steady state mRNA levels of NaV1.3, NaV1.6, NaV1.7, NaV1.8, and NaV1.9 were determined by TaqMan-based real time PCR (RT-PCR): Nav1.3-Rn00565438_m1; Nav1.6-Rn00570506_m1, Nav1.7-Rn00581647_m1; NaV1.8-Rn_00568393_m1; and Nav1.9-Rn00570487_m1. The mRNA expression levels were normalized using the comparative _C_T method.
DRG Culture Immunocytochemistry
DRG culture medium was decanted, and cultures were rinsed with PBS at 37 °C for 1 min and fixed with 3.7% formaldehyde for 10 min. Next, the cells were treated with 0.2% Triton X-100 for 5 min and background masking solution (ImageIT FX; Invitrogen) for 30 min, followed by incubation with primary antibody against rat C-terminal NaV1.8 peptide at 4 °C overnight (1:200 dilution; Sigma) and followed by secondary antibody-conjugated with Alexa Fluor 647 (Invitrogen) at room temperature for 1 h. After staining with 10 μg/ml Alexa Fluor 594 conjugated with IB4 (Invitrogen) at room temperature for 30 min, the slides were coverslipped with mounting medium (ProLong Gold; Invitrogen). A confocal laser-scanning microscope (Carl Zeiss LSM 310) was used to scan 1-μm focal layers of each specimen at 63× magnification. Individual sections were then digitally reconstructed and analyzed with digital software (ImageJ 1.43; National Institutes of Health, Bethesda, MD).
In Vivo Transfection with Cationized Gelatin (CG)/NaV1.8 shRNA Plasmid DNA Polyplexes
CG/shRNA plasmid DNA polyplex was prepared as previously described (32). Briefly, 85–100 μl of CG/DNA polyplex injectates were prepared at 7.5:1 CG-to-DNA mass ratio containing 17–25 μg/injection. The CG/DNA polyplex injectates were incubated at 37 °C for 30 min prior to injection.
The rats were anesthetized with isoflurane (2%), and the vector/DNA polyplex was slowly (∼1 min) injected subcutaneously into the center of the plantar surface of the left hindpaw with 27-gauge needles. The needle was removed, and the injection site was immediately sealed with liquid Band-Aid (Johnson & Johnson, New Brunswick, NJ).
Detection of Synthesized NaV1.8 siRNA Molecules
DRG and nerve tissues were harvested 2.5 and 7 days after in vivo injection of CG/NaV1.8 shRNA polyplexes. Small-size RNA specimens were prepared from DRG and nerves (mirVaNaTM small RNA isolation kit; Ambion). Synthesized siRNA was detected by TaqMan based stem-loop RT-PCR (33). RNA (100 ng) was used for reverse transcription with custom stem-loop primer for siRNA3 with 100 mm dNTPs, 50 units/μl multiscribe reverse transcriptase, 10× reverse transcriptase buffer, and 20 units/μl RNase inhibitor, followed by RT-PCR. 4.5 S RNA served as an endogenous control. Spiked samples of siRNA3 molecules were obtained by adding known amounts of synthetic siRNA3 (500, 200, 100, 50, 10, and 1 pmol) to total RNA of rat DRG and nerves. The amount of mature siRNA3 from experimental tissues was based on a comparison of _C_T values between experimental and spiked samples using the standard curve.
In Vivo Efficiency of shRNA-derived NaV1.8 Knockdown
DRG tissues were harvested 2.5 days after in vivo injection of CG/shRNA3 polyplex and acutely dissociated as described above. DRG culture was subjected to immunostaining with NaV1.8 antibody and GFP antibody, as well as IB4 staining. The NaV1.8 immunofluorescence intensity was determined in GFP+/IB4+ and GFP−/IB4+ neurons.
Sciatic Nerve Entrapment
Surgical procedure for the SNE model was described previously (34, 35). Briefly, in anesthetized rats, the left sciatic nerve was surgically exposed, and three polyethylene cuffs (1 mm long, 2.28-mm outer diameter, and 0.76-mm inner diameter) were loosely fitted to the sciatic nerve proximal to the trifurcation of common peroneal, tibial, and sural nerves (see Fig. 3A). Muscle and skin were separately closed.
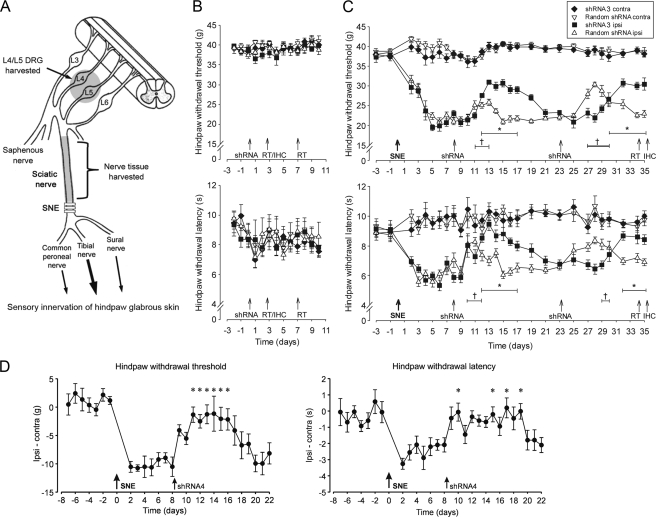
FIGURE 3.
Reversal of SNE-induced neuropathy symptoms after the injection of NaV1. 8 shRNA or random shRNA. A, diagram of SNE-induced neuropathic pain model representing location of polyethylene cuffs. In some experiments, L4/L5 DRG and sciatic nerve tissues proximal to the SNE site were harvested. B, hindpaw withdrawal thresholds (mean ± S.E.) to mechanical stimuli and hindpaw withdrawal latencies (mean ± S.E.) to thermal stimuli for the ipsilateral and contralateral sides of two groups of naïve rats (n = 8 each) injected (day 0) with shRNA3 or random shRNA. C, changes in hindpaw withdrawal thresholds to mechanical stimuli (left) and withdrawal latencies to thermal stimuli (right) after unilateral SNE surgery (day 0) and after shRNA3 or random shRNA injections (days 8 and 23). The data are presented as the means ± S.E. (n = 9 shRNA3, n = 10 random shRNA). D, changes in hindpaw withdrawal thresholds to mechanical stimuli (left) and withdrawal latencies to thermal stimuli (right) after unilateral SNE surgery (day 0) and after shRNA4 injection (day 8). The data are presented as the means ± S.E. (n = 8) of differences between ipsilateral and contralateral thresholds and latencies. *, p < 0.05 (repeated measures ANOVA).
Behavioral Testing
For a set of control and SNE rats, daily measurements of hindpaw withdrawal thresholds to mechanical stimuli and withdrawal latencies to thermal stimuli were obtained in both naïve controls and SNE rats as described in detail previously (35).
In Vivo Transfection with CG/NaV1.8 shRNA Plasmid DNA Polyplexes
After the stable pain behavior was established; CG/shRNA plasmid DNA polyplex was injected to the ipsilateral hindpaw as described above.
DRG, Nerve, and Hindpaw Skin Harvested from SNE Rats
After completion of behavioral testing, a set of SNE and shRNA-treated rats were anesthetized with pentobarbital (80 mg/kg) and perfusion-fixed for histological processing of tissues. A different set of rats were anesthetized with isoflurane (3%) and tissues (DRG, sciatic nerve, and hindpaw skin) collected for molecular studies.
Immunohistochemistry of Nerve and DRG Tissue
The rats were anesthetized with pentobarbital (80 mg/kg) and perfused through the ascending aorta with 300 ml of 0.9% NaCl, followed by 300 ml of ice-cold freshly prepared 4% (w/v) paraformaldehyde in 0.1 m phosphate buffer, pH 7.4. The L4/L5 DRG and sciatic nerves proximal to the injury were harvested and post-fixed at 4 °C for 2–4 h. All DRG and sciatic nerves were cryoprotected through sucrose gradient concentrations in 0.1 m phosphate buffer at 4 °C overnight and then embedded in the frozen mold using tissue freezing medium (TFMTM; Triangle Biomedical Sciences, Durham, NC). Specimens were cryosectioned at 25 μm (DRG) or 20 μm (sciatic nerve) and mounted on gelatin-coated precleaned microscopic slides. To minimize variability between specimens, ipsilateral and contralateral tissues were processed simultaneously. Four specimens from ipsilateral and contralateral tissues were mounted on the same slide. For immunostaining, tissue sections were fixed in cold acetone (−20 °C) for 10 min, rinsed with Tris-buffered saline (TBS) four times (3, 5, 7, and 7 min, respectively), and air-dried. The sections were incubated (30 min) in blocking solution (1% BSA + 2% normal donkey serum + 0.2% Tween) and then incubated in mouse monoclonal antiserum targeting the C-terminal residues (positions 1724–1956) of NaV1.8 (1:500 dilution; Neuromab, Davis, CA) at 4 °C overnight. Then sections were washed and incubated in secondary antibody (Alexa Fluor 488-conjugated donkey anti-mouse antiserum; Invitrogen) (1:500 dilution) for 1 h at room temperature and then in IB4-AlexaFluor 594, 1:250 dilution) for 30 min at room temperature. All of the sections were coverslipped with mounting medium (Vectashield; Vector Laboratories, Burlingame, CA).
The images were acquired using a Leica confocal SP2 1P-FCS microscope (Leica Microsystems, Bannockburn, IL) and captured using the same parameter settings (i.e. gain, pinhole, thickness of scanned images). For each DRG and nerve, four sections were used for confocal scanning. Captured images were 512 × 512 pixels. ImageJ software was used for quantitative analysis. DRG neurons positive to IB4 and with clearly visible nuclei were included for analysis. Only cytoplasmic immunoreactivity of NaV1.8 in IB4+ DRG neurons was outlined for quantification, making each region of interest correspond to the cytoplasmic profile of a single DRG neuron. Pixel intensities of image NaV1.8 immunoreactivity ranged from 0 (darkest) to 255 (lightest). The mean immunoreactivity for each cytoplasmic profile of DRG cells and nerves was converted to relative immunoreactivity using the formula (36); [(mean immunoreactivity value − MIN)/(MAX − MIN)]×100, where MAX and MIN are the maximum and minimum mean immunoreactivity values, respectively. Relative immunoreactivity values were used to generate scatter plots. In addition, the mean immunoreactivity from DRG neuron and nerve sections was used for plotting bar graphs. DRG neurons were sorted based on size <700 μm2 and 700–1200 μm2. DRG neurons >1200 μm2 were not included in the analyses because of the generally low levels of NaV1.8 expression.
Scn10a Gene Transcription
Total RNA was extracted from collected tissues (DRG, sciatic nerve, and skin). To evaluate the transcriptional activity of Scn10A gene (encoding NaV1.8), the NaV1.8 heteronuclear RNA (hnRNA, also called pre-mRNA) assay was performed as a surrogate measurement (37). TaqMan-based primers were designed targeting the intron between exons 27 and 28 (5′-TCATGGCTTGAGACACTGATTAGAC-3′) and exon 28 (5′-CAGTGACTTAAGGATTGCAGAAAACA-3′). Total RNA (1 μg) from DRG of SNE-injured and shRNA-treated rats were reverse transcribed using a random hexamer primer and subjected to RT-PCR analysis using the hnRNA primer/probe set. The initial validation of hnRNA RT-PCR assay was performed in vitro using acutely dissociated DRG neuron cultures treated with different doses of NGF (0, 10, 50, or 100 ng/ml) for 48 h.
Quantitative Real Time PCR
The steady state NaV1.8 mRNA levels of DRG, sciatic nerve, and skin were determined using TaqMan-based RT-PCR. GAPDH served as internal control. The data were expressed as a ratio of ipsilateral mRNA level over contralateral mRNA level within the same animals. In a separate experiment, the steady state mRNA levels of NaV1.6, 1.7, 1.8, and 1.9 were determined in DRG and nerve contralateral and ipsilateral to SNE.
Statistical Analyses
Student's t test for two-group comparison and one- or two-way analysis of variance (ANOVA) with Tukey's multiple comparisons for multi-group comparison were used to analyze RT-PCR and immunohistochemistry data with p < 0.05 accepted as statistically significant. Repeated measures ANOVA analysis was used to compare hindpaw sensitivity to mechanical and thermal stimuli in naïve and SNE-injured rats. Ipsilateral and contralateral hindpaw data were standardized by subtracting the ipsilateral and contralateral data points to get absolute difference. For each animal, the baseline value (i.e. average standardized score, post-SNE, or preinjection) was subtracted from the corresponding value (absolute difference value) at each subsequent time point (post-injection) to adjust for any potential baseline difference between groups. The change from baseline means was compared parametrically using repeated measures ANOVA. For each outcome, estimated mean, S.E., and p value for between group comparisons across time were reported. 5% least significant difference (LSD) for within group comparison was calculated. The mean difference between any two-time points must be larger than LSD.
RESULTS
shRNA-mediated NaV1.8 Knockdown in Vitro
We designed four RNA interference knockdown vectors containing U6 promoter-driven shRNA and CMV promoter-driven enhanced GFP reporter (Fig. 1A). The efficacy and selectivity of NaV1.8 mRNA knockdown was determined in cultured DRG neurons after lentivirus-assisted transduction of the shRNA vectors. Transduction efficiency assessed by detection of enhanced GFP in IB4-positive neurons using FACS was determined to be 69.5% (data not shown). Significant NaV1.8 mRNA knockdown was achieved by shRNA3 and shRNA4 (Fig. 1B). The effect of all shRNAs on other NaV isoforms (NaV1.3, NaV1.5, NaV1.6, NaV1.7, and NaV1.9) that share some sequence similarities with NaV1.8 was also examined. Off target knockdown of NaV1.6 was observed with shRNA4, whereas shRNA1 decreased NaV1.3 mRNA (Fig. 1B). Thus, effective and specific NaV1.8 knockdown was achieved only by shRNA3, which contained the 19-bp target sequence for siRNA within the coding region of NaV1.8 mRNA corresponding to nucleotide positions 6009–6027 (CCATCTAGCTCAATGCAAA; accession number NM_017247). The reduction of NaV1.8 protein expression by shRNA3 was measured and confirmed (p < 0.05 from random shRNA) by immunohistochemistry of transfected IB4+ DRG neurons in culture (Fig. 1C).
In Vivo Administration of shRNA3 in Naïve Rats
We have previously developed an in vivo nonviral gene transfer protocol whereby subcutaneous injection of CG/plasmid DNA polyplex in the hindpaw resulted in its uptake, retrograde transport, and subsequent expression in L4/L5 DRG neurons (Fig. 2A) (32). This protocol was used to test the effects of the selected shRNA3 construct in vivo. The shRNA3 plasmid DNA was polyplexed with CG and subcutaneously injected into the rat hindpaw.
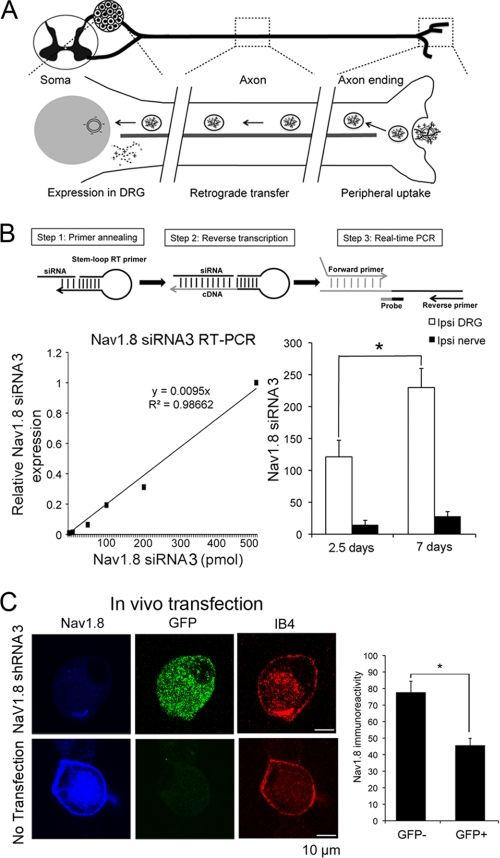
FIGURE 2.
RNA interference-derived knockdown of NaV1. 8 in DRG neurons in vivo. A, diagram of the retrograde gene transfer protocol using CG and plasmid DNA polyplex via subcutaneous peripheral injection of the rat hindpaw. This protocol was used to deliver shRNAs in vivo. B, quantitative measurement of the mature siRNA synthesized from shRNA plasmid DNA using stem-loop primer. The standard curve of relative siRNA3 expression corresponded to the known amount of siRNA3 added in DRG total RNA (left). Shown are the amounts of synthesized mature siRNA3 in DRG and nerve 2.5 and 7 days after shRNA3 injection in naïve rats (n = 4, right). *, p < 0.05. C, NaV1.8 immunocytology of IB4-labeled DRG neurons acutely dissociated 2.5 days after injection of the CG/NaV1.8 shRNA3 polyplex. Significantly reduced NaV1.8 immunoreactivity was observed in GFP+/IB4+ neurons, compared with that in GFP−/IB4+ neurons. *, p < 0.05.
Based on previous evidence of reporter gene expression in DRG at 2.5 and 6 days after hindpaw injection (32), we wanted to determine whether synthesis of the mature form of siRNA3 molecules (21 nucleotides with 3′overhangs) would follow a similar pattern. To determine the levels of synthesized siRNA molecules, we designed a set of stem-loop RT primer, forward/reverse primers, and probe for TaqMan-based RT-PCR. Standards were obtained by supplementing the total RNA samples of naïve DRG with 10, 50, 100, 200, or 500 pmol of synthetic siRNA3 and subjecting them to stem-loop RT-PCR. There was a linear correlation between the concentration of synthetic siRNA3 and RT-PCR readings (_R_2 = 0.99), resulting in the formulation of a simple conversion equation (Fig. 2B). Using this method, ∼120 pmol of mature siRNA3 was detected in DRG 2.5 days after the shRNA3 polyplex injection in naïve rats. The amount of mature siRNA3 increased 2-fold in DRG collected 7 days after injection (Fig. 2B). The amount of mature siRNA3 in the sciatic nerve was relatively low and did not significantly change between 2.5 and 7 days after injections.
We further examined the effect of shRNA3 injection on NaV1.8 immunoreactivity. L4/L5 DRG were harvested 2.5 days after injection of CG/NaV1.8 shRNA polyplex and acutely dissociated. DRG were cultured on Matrigel-coated glass chamber slides and immunocytology with NaV1.8 antibody, GFP antibody and IB4 staining was performed. Analysis indicated that NaV1.8 immunoreactivity of GFP+/IB4+ DRG neurons was significantly reduced by ∼40% as compared with GFP−/IB4+ DRG neurons (Fig. 2C).
In Vivo Administration of shRNA3 Does Not Affect Mechanical or Thermal Sensitivity in Naïve Rats
In separate sets of naïve rats, daily behavioral testing of mechanical withdrawal thresholds and thermal withdrawal latency showed that neither mechanical nor thermal sensitivity was affected by unilateral injections of NaV1.8 shRNA or injections of the control random sequence shRNA (Fig. 3B).
In Vivo Administration of shRNA3 or shRNA4 Reverses SNE-induced Neuropathy Symptoms
Rats destined for SNE surgery were monitored daily for mechanical withdrawal thresholds and thermal withdrawal latencies. Once stable neuropathic pain symptoms were established, CG/shRNA3 polyplex was subcutaneously injected in the hindpaw of one randomly selected group (n = 9) ipsilateral to SNE, and the other group received the CG/random shRNA injection (n = 10). After a lag period of 3–4 days, the mechanical withdrawal thresholds and thermal withdrawal latencies in the shRNA3-treated group rapidly improved and reached their maximum levels 4–5 days after the shRNA3 injection (Fig. 3C). The uninjured contralateral side was used to standardize the threshold and latency, and the LSDs of the mean changes of threshold and latency from the baseline (prior to the first injection) were determined using the multiple comparisons one-way ANOVA test. The LSD of the mechanical withdrawal threshold was achieved during the 4-day period from 5 to 8 days after the injection, whereas the LSD of the thermal withdrawal latency was achieved earlier and lasted 6 days. After the return of pain behaviors to preinjection levels (23 days post-SNE), a second injection achieved a similar magnitude of pain alleviation (Fig. 3C). Importantly, no signs of anaphylactic reaction or inflammation at the injection site were detected in any of the rats.
The random shRNA-treated group, to our surprise, produced a short period of pain alleviation after the first and the second injections (Fig. 3C). However, repeated measures ANOVA for the period between the first and second shRNA injections revealed that the effect of shRNA3 was significantly different from the random shRNA for the mechanical withdrawal thresholds (group p = 0.0001; time p < 0.0001; group − time p < 0.0001) and the thermal withdrawal latencies (group p = 0.0370; time p = 0.0002; group × time p = 0.0006).
In a separate experiment, SNE-treated rats (n = 8) were injected with shRNA4 polyplexed with CG. Similar to shRNA3, there was a significant attenuation of neuropathic pain symptoms after shRNA4 (Fig. 3D). However, the onset of analgesic effects occurred faster with shRNA4. Maximum analgesic effects were observed after 2–3 days and lasted for 6 days (mechanical withdrawal thresholds) or 10 days (thermal withdrawal latencies). In addition, peak relief of mechanical allodynia symptoms was greater with shRNA4 (∼75%) compared with shRNA3 (∼50%).
Effect of NaV1.8 shRNA3 Injection on NaV1.8 Protein Expression
DRG and sciatic nerves were harvested during maximal suppression of neuropathy symptoms 12 days after the second NaV1.8 shRNA3 injection. Analysis of NaV1.8 immunoreactivity (NaV1.8-ir) in DRG sections co-stained with IB4 (38) revealed a reduction of NaV1.8-ir in clusters of IB4+ neurons (Fig. 4A). Scatter plot of NaV1.8-ir intensity measured from a total of 857 IB4+ DRG neurons demonstrated that there are essentially two clusters: a high intensity cluster between 45 and 60% and a low intensity cluster less than 40% (Fig. 4B). The high intensity cluster was composed of all groups, whereas the low intensity cluster contained predominantly the SNE/shRNA3 group. The low intensity cluster accounted for 9.8–18.2% of neurons in the SNE/NaV1.8 shRNA group compared with only 1.5% of neurons from the SNE/random shRNA group. The average NaV1.8 immunoreactivity was significantly decreased in the SNE/shRNA3 group compared with SNE/random shRNA group of small (<700 μm2) and medium (700–1200 μm2) size IB4+ DRG neurons (Fig. 4_D_). The expression of NaV1.8 in larger diameter neurons (>1200 μm2) was not included in this analysis because of their generally low level NaV1.8 expression that could confound the measurements of decreases in NaV1.8 protein from shRNA3 injection.
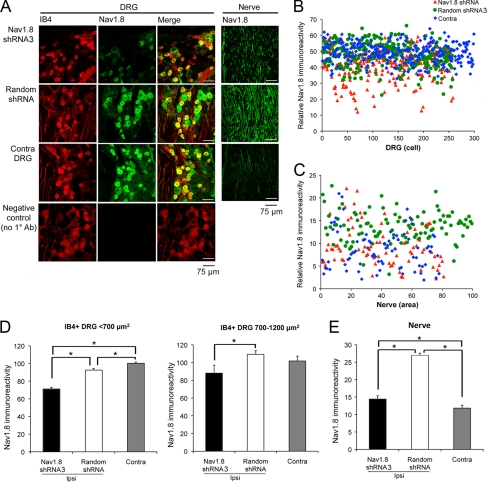
FIGURE 4.
NaV1.8 immunostaining in DRG and nerve tissues harvested from SNE rats injected with either shRNA3 or random shRNA. A, NaV1.8 immunoreactivity and IB4 staining in representative ipsilateral/contralateral sections from L4/L5 DRG and sciatic nerves proximal to the SNE site. Note the decreased NaV1.8 immunoreactivity in DRG and nerve after shRNA3 injections compared with random shRNA injections. B, scatter plot of relative NaV1.8 immunofluorescence intensity of IB4+ L4/L5 DRG neurons ipsilateral (Ipsi) to shRNA3 injection (red triangles, n = 297 cells, four rats), random shRNA injection (green circles, n = 258 cells, four rats), and contralateral (Contra) DRG (blue diamonds, n = 302 cells, eight rats). C, scatter plot of relative NaV1.8 immunofluorescence intensity of nerve sections ipsilateral to shRNA3 injection (red triangles, n = 81 areas, 27 sections), ipsilateral to random shRNA injection (green circles, n = 102 areas, 34 sections), and in contralateral nerve (blue diamonds, n = 78 areas, 26 sections). D, average NaV1.8 immunofluorescence intensity of IB4+ DRG neurons sorted by cell area < 700 and 700–1200 μm2 (mean ± S.E.). *, p < 0.05. E, average NaV1.8 immunofluorescence intensity of nerve sections. *, p < 0.05.
Sciatic nerve sections revealed a robust increase of NaV1.8-ir in the SNE/random shRNA group, confirming previous findings of SNE-induced increases in NaV1.8-ir (28). By contrast, NaV1.8-ir in SNE-injured nerve was markedly reduced by the shRNA3 injection (Fig. 4A). Scatter plot of a total of 261 randomly selected microscopic fields of sciatic nerve illustrates the uniform increases in ipsilateral NaV1.8-ir and its reduction by the shRNA3 injection to the range comparable with that of the contralateral (uninjured) sciatic nerve (Fig. 4C). The average NaV1.8-ir was significantly higher in the SNE-injured/random shRNA-injected group than in the uninjured contralateral group. Injection of shRNA3 effectively attenuated the increased NaV1.8-ir, although falling short of completely normalizing the average NaV1.8-ir to the level of uninjured contralateral sciatic nerves (Fig. 4E).
Effect of shRNA3 Injection on NaV1.8 Gene Expression
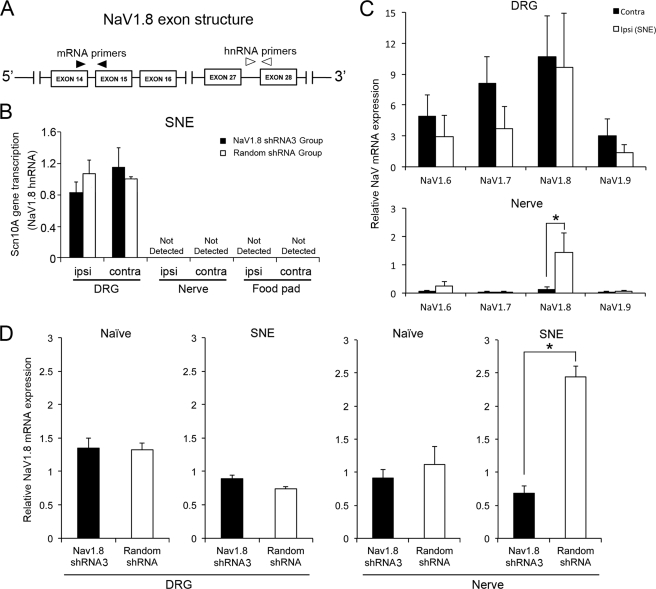
We first determined whether SNE- and shRNA-induced changes in NaV1.8 were due to altered transcription of the Scn10A gene, which encodes NaV1.8. Because of difficulties experienced with a conventional nuclear run-off assay, we used an alternative method (37) in which heterogeneous nuclear RNA (hnRNA) or pre-mRNA of NaV1.8 was measured using primers designed across the intron-exon boundaries of the Scn10A gene (Fig. 5A). Measurements in the DRG of SNE rats (n = 4) revealed similar levels of NaV1.8 hnRNA among all groups, suggesting that neither SNE nor shRNA injections affected the Scn10A gene transcription (Fig. 5B). Because NaV1.8 hnRNA was not detected in sciatic nerve (Fig. 5B), local non-neuronal cells were unlikely to be the source of NaV1.8 mRNA in sciatic nerves. In a separate group (n = 4) of SNE-treated rats without shRNA injections, the steady state NaV1.8 mRNA in DRG was not affected by SNE, whereas NaV1.8 mRNA, but not NaV1.6, NaV1.7, or NaV1.9 mRNA, was significantly increased in the affected nerve (Fig. 5C), confirming previous findings (28).
FIGURE 5.
Effects of SNE and shRNA3 treatment on NaV1. 8 gene transcription and steady state mRNA levels in DRG and nerve. A, diagram of relevant exon structures Scn10A encoding NaV1.8 and the locations of PCR primers. targeting NaV1.8 mRNA (forward primer in exon 14 and reverse primer in exon 15) and NaV1.8 hnRNA (forward primer-intron, reverse primer-exon 28). B, Scn10A gene transcription was measured by NaV1.8 hnRNA in SNE-treated rats with NaV1.8 shRNA (n = 4) or random shRNA (n = 4) injection. The data are presented with the contralateral tissue to SNE of the random shRNA group as the standard. The transcriptional activity of Scn10a in DRG was not affected by SNE injury and the injection of NaV1.8 shRNA. There was no NaV1.8 hnRNA detected in the nerve tissues as well as in the food pad tissue where cationized gelatin/plasmid DNA polyplex was injected. C, mRNA levels of NaV1.6, 1.7, 1.8, and 1.9 in the DRG and sciatic nerve of the SNE rat model. The relative mRNA level was normalized by the housekeeping gene expression in DRG. *, p < 0.05. D, steady state NaV1.8 mRNA levels in DRG and sciatic nerve harvested from naïve or SNE-injured rats treated with shRNA3 (black bars) or random shRNA (white bars). RT-PCR data were compared using the untreated contralateral tissue as the standard in each group (n = 4). *, p < 0.05. ipsi, ipsilateral; contra, contralateral.
Next, we addressed the effect of the shRNA3 treatment on the steady state NaV1.8 mRNA. L4/L5 DRG and sciatic nerve tissues were harvested from naïve (n = 4) or SNE-injured (n = 4) rats after shRNA3 or random shRNA injections. Naïve rat tissues were obtained 11 days after shRNA injections. Tissues from SNE rats were harvested 11 days after the second shRNA3 injection, when alleviation of SNE-induced pain symptoms was still evident (Fig. 5D). Compared with naïve rats, SNE decreased the steady state NaV1.8 mRNA levels in the DRG. Surprisingly, NaV1.8 mRNA levels in the DRG were not affected by shRNA3 injection as compared with the random shRNA injection (Fig. 5D). However, the increased NaV1.8 mRNA level by SNE injury in the nerve was significantly attenuated by the shRNA3 treatment to the levels observed in the naïve nerve (Fig. 5D).
DISCUSSION
The present study demonstrated that NaV1.8 knockdown using peripheral administration of shRNA in the SNE-injured rats resulted in significant reduction of neuropathic pain behaviors. This confirms and extends previous studies in which pharmacological blockade of NaV1.8 function or expression was shown to decrease mechanical allodynia and thermal hyperalgesia in experimental neuropathies. The peripheral neuropathy induced by SNE is highly comparable with that of chronic constriction injury (CCI), which uses chromic gut suture materials instead of polyethylene cuffs (25, 39, 40). SNE was demonstrated to produce consistent pain behaviors (34, 41), a bona fide transient loss of varicosities in nociceptive fibers (42), and increases in evoked excitability of sciatic nerve compound action potentials (28). The increased excitability of the injured sciatic nerve likely contributes to the ectopic activity in the injured nerve and the exaggerated afterdischarge of wide dynamic range neurons in the spinal cord evoked by mechanical cutaneous stimulation in this model (43). The hyperexcitability and ectopic burst discharge of primary sensory neurons are widely considered to be the major contributors to pain symptomatology of peripheral neuropathy models.
Although the behavioral symptoms of the SNE/CCI models are superficially similar to the model of segmental deafferentation induced by L5/L6 SNL (44), these models differ in many important respects. In contrast to the SNE/CCI models, where injured and uninjured axons commingle in the sciatic nerve, the tight ligation (deafferentation) of L5/L6 spinal nerve segments in the SNL model results in complete segregation of the deafferented neuronal somata in L5/L6 DRG and the “uninjured” somata and axons of L4 DRG neurons. The phenotypic alterations in SNL include large decreases in NaV1.8 mRNA, protein, and function in the deafferented L5 DRG and large increases in NaV1.8 mRNA, protein, and function in the uninjured L4 DRG (45), concomitant with large increases in NaV1.8 protein, function and resistance to tetrodotoxin blockade of action potential conduction in the uninjured sciatic nerve axons (9, 27). By contrast, the SNE/CCI produce modest decreases in NaV1.8 mRNA and protein within L4/L5 DRG (25, 28, 29) and increases in immunohistochemical detection of axonal NaV1.8 protein (27, 28). Functionally, only modest increases in resistance to tetrodotoxin conduction block are observed after SNE (28). Moreover, the demonstrated increases in sciatic nerve NaV1.8 mRNA after SNE (28) and the lack of nerve NaV1.8 mRNA increases after the L5 spinal nerve ligation3 suggest differences in the molecular mechanisms of axonal NaV1.8 accumulation in the two models.
Targeted Gene Delivery to the Sciatic Nerve via Subcutaneous Injection of CG/DNA Polyplex
We used a simple gene transfer protocol of subcutaneous CG/DNA polyplex injection in the hindpaw (32) to deliver shRNA plasmid DNA targeting NaV1.8 mRNA to sciatic nerve, which resulted in retrograde transfection and synthesis of mature siRNA molecules in L4/L5 DRG (Fig. 2B). Unlike other studies using intrathecal injections of antisense oligodeoxynucleotides (9, 27) or epidural injections of siRNA targeting NaV1.8 (29), the expression of plasmid DNA by injection of CG/DNA polyplex is limited to the DRG and sciatic nerve innervating the subcutaneous injection site (32). In this study, ∼10% of small and medium DRG cells were found positive for the reporter GFP. Puigdellívol-Sánchez et al. (46) estimated only 20–30% of DRG neurons to innervate the hindpaw. Therefore, the relevant transfection efficiency of the present method may exceed 40%.
Significant alleviation of mechanical allodynia and thermal hyperalgesia in SNE-injured rats was achieved with a distinct latent period of 3–4 days after hindpaw injection of shRNA3 specific to NaV1.8 (Fig. 3C). Following neuronal uptake, the injected CG/DNA polyplex may undergo retrograde axonal transport along the sciatic nerve (10–15 cm) before reaching the soma in the DRG. The rate of microtubule-mediated retrograde axonal transport along the sciatic and spinal nerves has been estimated to be 10–25 cm/day (47, 48). Thus, CG/DNA polyplex should reach DRG neuronal nuclei within a day, and the mature siRNA should be synthesized. Indeed, the synthesis of mature siRNA as well as measurable NaV1.8 knockdown in DRG neurons was evident 2.5 days after the NaV1.8 shRNA injection but doubled by 7 days (Fig. 2B). Therefore, the latent period of 3–4 days is likely due to the continued accumulation of the siRNA and resultant NaV1.8 knockdown.
In rats injected with shRNA4, the late onset pain alleviation appeared to be consistent with the pain alleviation pattern of shRNA3, but the effect was more significant (Fig. 3D). Because both shRNA3 and shRNA4 were shown to decrease NaV1.8 mRNA levels, the late onset pain alleviation may likely be due to NaV1.8 knockdown, and the robust pain alleviation by shRNA4 may directly relate to its larger knockdown efficiency (Fig. 1B). It was noted, however, that significant pain relief was faster with shRNA4. Because shRNA4 also affected NaV1.6 (Fig. 1B), it is tempting to speculate that the knockdown of NaV1.6 could contribute to the earlier onset of pain relief. Further studies are needed.
The U6 promoter in our shRNA constructs is widely used in mammalian expression systems; however, when applied in vivo, its transcriptional activity was shown to be silenced after 1∼2 weeks (49). Therefore, the gradual return of pain symptoms after shRNA injection may primarily be due to the silencing of promoters in the expression plasmid.
In the random shRNA-injected group, we unexpectedly observed small decreases in neuropathy symptoms of relatively short duration (Fig. 3C). Suspecting an off target effect, we repeated a BLAST search using the 19 nucleotides of the random sequence siRNA; however, none of the mRNA species in the database were detected. It is possible that the seed sequence (nucleotides 2–7) of random siRNA could behave as a miRNA and promote translational inhibition of the off target gene(s) (50). Within the scope of this study, possible off target molecules were not determined.
NaV1.8 Expression in DRG and Sciatic Nerve after SNE Injury
Decreases in expression of NaV1.8 in the DRG were demonstrated after SNL (27), CCI (25, 29), as well as SNE (28), whereas expression of NaV1.8 in peripheral nerve increases (9, 25, 28). The present study confirmed these previous reports (Fig. 4) and also demonstrated the accumulation of NaV1.8 mRNA in sciatic nerve ipsilateral to SNE injury (Fig. 5C).
A recent study suggested that chronic nerve compression resulted in the up-regulation of NaV1.8 immunoreactivity in Schwann cells (51). Notably, the chronic nerve compression model does not induce mechanical allodynia or thermal hyperalgesia, instead resulting in progressive decreases in mechanical sensitivity (52). In our study, the lack of detectable NaV1.8 hnRNA in the SNE-injured nerve (Fig. 5B) strongly suggests that the source of NaV1.8 mRNA is not the non-neuronal cells, such as Schwann cells, but the axons themselves.
The present study further addressed whether increases in NaV1.8 transcription from the Scn10A gene in DRG contributed to the axonal NaV1.8 mRNA accumulation. The NaV1.8 hnRNA level was not influenced by SNE injury (Fig. 5B), and thus de novo Scn10A gene transcription should not have a direct mechanistic role in the accumulation of axonal NaV1.8 mRNA. Also, we previously determined that the accumulation of axonal NaV1.8 was not due to the increase of the mRNA half-life evaluated by poly(A) tail elongation (28).
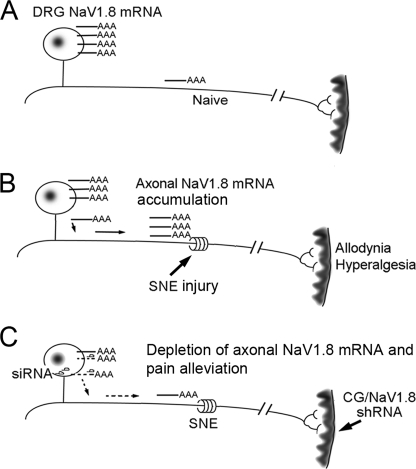
It has been established that mRNAs are actively transported to subcellular sites (53). For example, subcellular localization of β-actin mRNA occurs during neuronal regeneration facilitated by the binding of zip code protein (ZBP1) to its 3′-UTR (54). Therefore, it is reasonable to postulate that NaV1.8 mRNA may be post-transcriptionally transported to the SNE-injured axons from their DRG somata (Fig. 6). Increased transport of NaV1.8 mRNA to axons in the absence of increased somatic mRNA production (Fig. 5B) might be expected to result in decreased steady state levels of somatic NaV1.8 mRNA ipsilateral to the SNE treatment. We have previously observed a trend of decreased somatic NaV1.8 mRNA ipsilateral to SNE and significant decreases in somatic NaV1.8 immunoreactivity (28). A similar trend of decreased somatic NaV1.8 mRNA may be seen in DRG ipsilateral to SNE compared with contralateral or naïve rat DRG in Fig. 5D. It must also be noted that the amount of NaV1.8 mRNA in the axons is ∼1/10 to 1/50 of the DRG NaV1.8 mRNA (Fig. 5C). Therefore, relatively large increases in axonal NaV1.8 mRNA may result from relatively small decreases in the amount of somatic NaV1.8 mRNA.
FIGURE 6.
A hypothetical molecular mechanism of painful neuropathy involving axonal NaV1. 8 mRNA. A, NaV1.8 mRNA is largely localized in DRG in the uninjured neurons. B, SNE injury induces axonal accumulation of NaV1.8 mRNA, likely through active mRNA transport, not by the increase of gene transcription or axonal mRNA half-life. C, retrograde transport of NaV1.8 shRNA facilitates RNAi-derived NaV1.8 mRNA degradation in DRG, which eventually normalizes axonal NaV1.8 mRNA, leading to the attenuation of neuropathic pain symptoms after a distinct lag period.
Because all of the necessary molecular components for protein translation exist in peripheral axons (55, 56), the accumulated axonal NaV1.8 mRNA may, in part, contribute to the increased functional expression of NaV1.8 in the injured nerve. The present study further demonstrated that injection of shRNA3 achieved a normalization of SNE-induced NaV1.8 mRNA in the nerve during the time (Fig. 5D) when suppression of pain symptoms was maximal (Fig. 3C). From these observations, NaV1.8 mRNA in the nerve, not in the DRG, appears to play a more relevant role in the pathogenesis of this painful neuropathy.
Mechanism of NaV1.8 Knockdown and Neuropathic Pain Alleviation
Most of the mature siRNA3 was detected in the DRG (Fig. 2B), suggesting that the somata of sensory neurons in the DRG may be the main site of siRNA-derived NaV1.8 mRNA degradation. If so, the attenuation of pain symptoms would occur after the depletion of axonally transported NaV1.8 mRNA within the somata of DRG neurons and subsequent decreases in axonal NaV1.8 synthesis (Fig. 6). However, it has also been well documented that RNAi machinery exists in the peripheral nerve axons (57, 58). Therefore, siRNA-mediated degradation of NaV1.8 mRNA could also occur in peripheral axons.
Maximum relief of mechanical allodynia with shRNA3 in the present study was ∼50% and was less effective than the relief of thermal hyperalgesia, which reached nearly 100% (Fig. 3C). Mechanical stimulation activates low threshold mechanoreceptors (e.g. Meissner's corpuscle and Merkel disk receptors) on the encapsulated terminals of Aα or Aβ fibers (59). These fibers terminate subcutaneously at the epidermal-dermal junction and are thickly myelinated. By contrast, thinly myelinated Aδ fibers and unmyelinated C fibers have bare nerve endings, and both highly innervate the glabrous skin of hindpaw. Although Aδ fibers are high threshold mechanosensors and constitute the afferent portion of the reflex arc that results in withdrawal from noxious and mechanical stimuli, C fibers are polymodal and respond to thermal (both heat and cold), mechanical, and chemical stimuli (59). Subcutaneously injected CG/DNA polyplex may be better endocytosed into bare nerve endings of Aδ fiber and C fiber than into the encapsulated terminals of Aα and Aβ fibers. Because NaV1.8 is primarily synthesized in Aδ and C nociceptors (60), the nearly complete NaV1.8 knockdown observed in the sciatic nerve tissue may reflect the preferential CG/shRNA uptake by these neurons, reflected in the nearly complete alleviation of thermal hyperalgesia.
Conclusions
Taken together, this study demonstrated that NaV1.8 in the affected sciatic nerve, not in DRG, plays a significant role in the development and maintenance of painful neuropathy symptoms. We propose that the molecular mechanism of SNE-induced neuropathy includes injury-induced post-transcriptional NaV1.8 mRNA transport, axonal accumulation of NaV1.8 mRNA, and its local protein translation leading to the increased NaV1.8 functional expression in injured nerve. This study further suggests that axonal NaV1.8 mRNA may be an attractive therapeutic target for painful neuropathy, and the subcutaneous injection of CG/DNA polyplex at the pain site may present a novel therapeutic modality.
Acknowledgments
We thank Prof. Yasuhiko Tabata (Institute for Frontier Medical Science, Kyoto University, Japan) for providing cationized gelatin nanoparticles, Dr. Emmanuelle Faure-Kuma (Vector Core, UCLA) for lentivirus production, Dr. Mathew Schibler (California NanoSystems Institute and Brain Research Institute microscopy core facility, UCLA) for confocal laser scanning micrography, Mariane Cilluffo (Brain Research Institute microscopy core facility) for immunohistochemistry consultation, and (Daniela Markovic of Department of Biomathematics), and David Geffen (School of Medicine at UCLA) for statistical consultation. We also thank Prof. David Wong (Associate Dean of Research, UCLA School of Dentistry) for generous funding support for statistical consultations.
*
This work was supported, in whole or in part, by National Institutes of Health Grants NS049137 and DA023163. This work was also supported by a Royal Thai Government scholarship from Thailand, a UCLA School of Dentistry seed grant, and Research Facilities Improvement Program Grant C06 RR014529 from NCRR, National Institutes of Health.
3
D. K. Thakor, S. Mitrirattanakul, I. Spigelman, and I. Nishimura, unpublished observations.
2
The abbreviations used are:
VGSC
voltage-gated sodium channel
NaV
voltage-gated sodium channel
SNE
sciatic nerve entrapment
DRG
dorsal root ganglia
SNL
spinal nerve ligation
HBSS
Hanks' balanced salt solution
IB4
isolectin B4
CG
cationized gelatin
ANOVA
analysis of variance
LSD
least significant difference
hnRNA
heterogeneous nuclear RNA
CCI
chronic constriction injury.
REFERENCES
- 1.Taylor R. S. (2006) Pain Pract. 6, 22–26 [DOI] [PubMed] [Google Scholar]
- 2.Baron R., Tölle T. R., Gockel U., Brosz M., Freynhagen R. (2009) Pain 146, 34–40 [DOI] [PubMed] [Google Scholar]
- 3.England J. D., Happel L. T., Kline D. G., Gamboni F., Thouron C. L., Liu Z. P., Levinson S. R. (1996) Neurology 47, 272–276 [DOI] [PubMed] [Google Scholar]
- 4.Kretschmer T., Nguyen D. H., Beuerman R. W., Happel L. T., England J. D., Tiel R. L., Kline D. G. (2002) J. Neurosurg. 97, 1424–1431 [DOI] [PubMed] [Google Scholar]
- 5.Delaney A., Colvin L. A., Fallon M. T., Dalziel R. G., Mitchell R., Fleetwood-Walker S. M. (2009) Neurotherapeutics 6, 630–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gilron I., Watson C. P., Cahill C. M., Moulin D. E. (2006) CMAJ 175, 265–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Julius D., Basbaum A. I. (2001) Nature 413, 203–210 [DOI] [PubMed] [Google Scholar]
- 8.Milligan E. D., Watkins L. R. (2009) Nat. Rev. Neurosci. 10, 23–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gold M. S., Weinreich D., Kim C. S., Wang R., Treanor J., Porreca F., Lai J. (2003) J. Neurosci. 23, 158–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lai J., Porreca F., Hunter J. C., Gold M. S. (2004) Annu. Rev. Pharmacol. Toxicol. 44, 371–397 [DOI] [PubMed] [Google Scholar]
- 11.Ji R. R., Strichartz G. (2004) Sci. STKE 2004, reE14. [DOI] [PubMed] [Google Scholar]
- 12.Coward K., Plumpton C., Facer P., Birch R., Carlstedt T., Tate S., Bountra C., Anand P. (2000) Pain 85, 41–50 [DOI] [PubMed] [Google Scholar]
- 13.Dib-Hajj S. D., Estacion M., Jarecki B. W., Tyrrell L., Fischer T. Z., Lawden M., Cummins T. R., Waxman S. G. (2008) Mol. Pain 4, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kretschmer T., Happel L. T., England J. D., Nguyen D. H., Tiel R. L., Beuerman R. W., Kline D. G. (2002) Acta Neurochir. (Wien) 144, 803–810 [DOI] [PubMed] [Google Scholar]
- 15.Shembalkar P. K., Till S., Boettger M. K., Terenghi G., Tate S., Bountra C., Anand P. (2001) Eur. J. Pain 5, 319–323 [DOI] [PubMed] [Google Scholar]
- 16.Siqueira S. R., Alves B., Malpartida H. M., Teixeira M. J., Siqueira J. T. (2009) Neuroscience 164, 573–577 [DOI] [PubMed] [Google Scholar]
- 17.Yiangou Y., Birch R., Sangameswaran L., Eglen R., Anand P. (2000) FEBS Lett. 467, 249–252 [DOI] [PubMed] [Google Scholar]
- 18.Matzner O., Devor M. (1994) J. Neurophysiol. 72, 349–359 [DOI] [PubMed] [Google Scholar]
- 19.Waxman S. G. (1999) Pain 6, S133–S140 [DOI] [PubMed] [Google Scholar]
- 20.Akopian A. N., Sivilotti L., Wood J. N. (1996) Nature 379, 257–262 [DOI] [PubMed] [Google Scholar]
- 21.Black J. A., Nikolajsen L., Kroner K., Jensen T. S., Waxman S. G. (2008) Ann. Neurol. 64, 644–653 [DOI] [PubMed] [Google Scholar]
- 22.Fukuoka T., Kobayashi K., Yamanaka H., Obata K., Dai Y., Noguchi K. (2008) J. Comp. Neurol. 510, 188–206 [DOI] [PubMed] [Google Scholar]
- 23.Akopian A. N., Souslova V., England S., Okuse K., Ogata N., Ure J., Smith A., Kerr B. J., McMahon S. B., Boyce S., Hill R., Stanfa L. C., Dickenson A. H., Wood J. N. (1999) Nat. Neurosci. 2, 541–548 [DOI] [PubMed] [Google Scholar]
- 24.Renganathan M., Cummins T. R., Waxman S. G. (2001) J. Neurophysiol. 86, 629–640 [DOI] [PubMed] [Google Scholar]
- 25.Novakovic S. D., Tzoumaka E., McGivern J. G., Haraguchi M., Sangameswaran L., Gogas K. R., Eglen R. M., Hunter J. C. (1998) J. Neurosci. 18, 2174–2187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jarvis M. F., Honore P., Shieh C. C., Chapman M., Joshi S., Zhang X. F., Kort M., Carroll W., Marron B., Atkinson R., Thomas J., Liu D., Krambis M., Liu Y., McGaraughty S., Chu K., Roeloffs R., Zhong C., Mikusa J. P., Hernandez G., Gauvin D., Wade C., Zhu C., Pai M., Scanio M., Shi L., Drizin I., Gregg R., Matulenko M., Hakeem A., Gross M., Johnson M., Marsh K., Wagoner P. K., Sullivan J. P., Faltynek C. R., Krafte D. S. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 8520–8525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lai J., Gold M. S., Kim C. S., Bian D., Ossipov M. H., Hunter J. C., Porreca F. (2002) Pain 95, 143–152 [DOI] [PubMed] [Google Scholar]
- 28.Thakor D. K., Lin A., Matsuka Y., Meyer E. M., Ruangsri S., Nishimura I., Spigelman I. (2009) Mol. Pain 5, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dong X. W., Goregoaker S., Engler H., Zhou X., Mark L., Crona J., Terry R., Hunter J., Priestley T. (2007) Neuroscience 146, 812–821 [DOI] [PubMed] [Google Scholar]
- 30.Mikami M., Yang J. (2005) Anesthesiology 103, 828–836 [DOI] [PubMed] [Google Scholar]
- 31.Rubinson D. A., Dillon C. P., Kwiatkowski A. V., Sievers C., Yang L., Kopinja J., Rooney D. L., Zhang M., Ihrig M. M., McManus M. T., Gertler F. B., Scott M. L., Van Parijs L. (2003) Nat. Genet. 33, 401–406 [DOI] [PubMed] [Google Scholar]
- 32.Thakor D., Spigelman I., Tabata Y., Nishimura I. (2007) Mol. Ther. 15, 2124–2131 [DOI] [PubMed] [Google Scholar]
- 33.Chen C., Ridzon D. A., Broomer A. J., Zhou Z., Lee D. H., Nguyen J. T., Barbisin M., Xu N. L., Mahuvakar V. R., Andersen M. R., Lao K. Q., Livak K. J., Guegler K. J. (2005) Nucleic Acids Res. 33, e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mosconi T., Kruger L. (1996) Pain 64, 37–57 [DOI] [PubMed] [Google Scholar]
- 35.Matsuka Y., Ono T., Iwase H., Mitrirattanakul S., Omoto K. S., Cho T., Lam Y. Y., Snyder B., Spigelman I. (2008) Mol. Pain 4, 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoffman E. M., Schechter R., Miller K. E. (2010) J. Histochem. Cytochem. 58, 329–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao J., Lee M. C., Momin A., Cendan C. M., Shepherd S. T., Baker M. D., Asante C., Bee L., Bethry A., Perkins J. R., Nassar M. A., Abrahamsen B., Dickenson A., Cobb B. S., Merkenschlager M., Wood J. N. (2010) J. Neurosci. 30, 10860–10871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu J., Hu J. Y., Wu F., Schwartz J. H., Schacher S. (2006) J. Neurosci. 26, 5204–5214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kajander K. C., Bennett G. J. (1992) J. Neurophysiol. 68, 734–744 [DOI] [PubMed] [Google Scholar]
- 40.Bennett G. J., Xie Y. K. (1988) Pain 33, 87–107 [DOI] [PubMed] [Google Scholar]
- 41.Pitcher G. M., Ritchie J., Henry J. L. (1999) Pain 83, 37–46 [DOI] [PubMed] [Google Scholar]
- 42.Bailey A. L., Ribeiro-da-Silva A. (2006) Neuroscience 138, 675–690 [DOI] [PubMed] [Google Scholar]
- 43.Pitcher G. M., Henry J. L. (2008) Exp. Neurol. 214, 219–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim S. H., Chung J. M. (1992) Pain 50, 355–3631333581 [Google Scholar]
- 45.Zhang X. F., Zhu C. Z., Thimmapaya R., Choi W. S., Honore P., Scott V. E., Kroeger P. E., Sullivan J. P., Faltynek C. R., Gopalakrishnan M., Shieh C. C. (2004) Brain Res. 1009, 147–158 [DOI] [PubMed] [Google Scholar]
- 46.Puigdellívol-Sánchez A., Valero-Cabré A., Prats-Galino A., Navarro X., Molander C. (2002) J. Neurosci. Methods 115, 115–127 [DOI] [PubMed] [Google Scholar]
- 47.Brown A. (2003) J. Cell Biol. 160, 817–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tomishima M. J., Smith G. A., Enquist L. W. (2001) Traffic 2, 429–436 [DOI] [PubMed] [Google Scholar]
- 49.Löser P., Jennings G. S., Strauss M., Sandig V. (1998) J. Virol. 72, 180–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jackson A. L., Burchard J., Schelter J., Chau B. N., Cleary M., Lim L., Linsley P. S. (2006) RNA 12, 1179–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Frieboes L. R., Palispis W. A., Gupta R. (2010) J. Orthop. Res. 28, 753–761 [DOI] [PubMed] [Google Scholar]
- 52.Gupta R., Rummler L. S., Palispis W., Truong L., Chao T., Rowshan K., Mozaffar T., Steward O. (2006) Exp. Neurol. 200, 418–429 [DOI] [PubMed] [Google Scholar]
- 53.Kiebler M. A., Bassell G. J. (2006) Neuron 51, 685–690 [DOI] [PubMed] [Google Scholar]
- 54.Chao J. A., Patskovsky Y., Patel V., Levy M., Almo S. C., Singer R. H. (2010) Genes Dev. 24, 148–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang D. O., Kim S. M., Zhao Y., Hwang H., Miura S. K., Sossin W. S., Martin K. C. (2009) Science 324, 1536–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang W., van Niekerk E., Willis D. E., Twiss J. L. (2007) Dev. Neurobiol. 67, 1166–1182 [DOI] [PubMed] [Google Scholar]
- 57.Hengst U., Cox L. J., Macosko E. Z., Jaffrey S. R. (2006) J. Neurosci. 26, 5727–5732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Murashov A. K., Chintalgattu V., Islamov R. R., Lever T. E., Pak E. S., Sierpinski P. L., Katwa L. C., Van Scott M. R. (2007) FASEB J. 21, 656–670 [DOI] [PubMed] [Google Scholar]
- 59.Garder E. S., Martin J. H., Jessell J. M. (2000) Principles of Neural Science (Kandel E. R., Schwartz J. H., Jessell T. M. eds) 4th Ed., pp. 430–450, McGraw-Hill Book Co., New York [Google Scholar]
- 60.Djouhri L., Fang X., Okuse K., Wood J. N., Berry C. M., Lawson S. N. (2003) J. Physiol. 550, 739–752 [DOI] [PMC free article] [PubMed] [Google Scholar]