RNA targets of wild-type and mutant FET family proteins (original) (raw)
. Author manuscript; available in PMC: 2012 Jun 1.
Published in final edited form as: Nat Struct Mol Biol. 2011 Nov 13;18(12):1428–1431. doi: 10.1038/nsmb.2163
Abstract
FUS, EWSR1 and TAF15, constituting the FET protein family, are abundant, highly conserved RNA-binding proteins with important roles in oncogenesis and neuronal disease, yet their RNA targets and recognition element are unknown. Using PAR-CLIP, we defined global RNA targets of all human FET proteins and two ALS-causing human FUS mutants. FET members displayed similar binding profiles while mutant FUS showed a drastically altered binding pattern, consistent with changes in subcellular localization.
Post-transcriptional regulatory networks controlled by microRNAs and RNA-binding proteins (RBPs) play important roles in mRNA maturation and gene regulation1–5. Dysregulation of these networks by mutation, deletion, or overexpression of ribonucleoprotein complex (RNP) components may result in disease6. We used PAR-CLIP (photoactivatable ribonucleoside-enhanced crosslinking and immunoprecipitation)7 to determine global protein-RNA interactions for all three members of the FET family of RBPs, as well as two mutant forms associated with familial amyotrophic lateral sclerosis (ALS), and provide large-scale and high-resolution target data for the entire family.
FUS, together with EWSR1 and TAF15, form a gene family (FET) of abundant, ubiquitously expressed RBPs8. FET genes are directly involved in deleterious genomic rearrangements, primarily in sarcomas and in leukemia9. Given their predominantly nuclear localization, FET family proteins have been implicated in various nuclear processes. All three associate with the transcription factor II D complex, as well as directly with RNA polymerase II9. Moreover, FUS has been attributed a role in splicing9. Recently, mutations in FUS have been described as causing familial ALS10,11, an adult-onset, rapidly progressing neurodegenerative disorder. The first reported mutations include the C-terminally located FUS-R521G and FUS-R521H, which both cause mislocalization of the physiologically mostly nuclear FUS protein to the cytoplasm10,11. Despite numerous biochemical studies addressing the function of FET proteins in various nuclear processes, the RNA recognition elements (RRE) and the molecular targets have remained unknown. The impact of FUS mutations on the RNA binding capability and effective binding spectrum has also been unclear.
We generated six stable Flp-In T-REx HEK293 cell lines with either stable or inducible expression of N-terminally FLAGHA-tagged human FUS, EWSR1 or TAF15. Additionally, we generated two cell lines stably expressing FLAGHA-tagged disease-causing mutant forms of FUS (FUS-R521G or FUS-R521H). As reported, wild-type FET proteins localized primarily to the nucleus and mutant FUS to the cytoplasm (Supplementary Fig. 1a). Cell lines were grown for 12 to 16 h in 4-thiouridine (4SU) supplemented medium to allow for its incorporation into nascent RNA transcripts, as required by the PAR-CLIP protocol7. Crosslinked RNAs were recovered from SDS-PAGE-purified FET protein immunoprecipitates (Figure 1a, see Supplementary Fig. 1 for additional PAR-CLIP controls), converted into cDNA libraries, and then Solexa-sequenced (the raw data is deposited in SRA, accession SRA025082.1). Sequence reads were preprocessed, aligned against the human genome while allowing up to one error, and annotated, essentially as described previously7 (Supplementary Table 1).
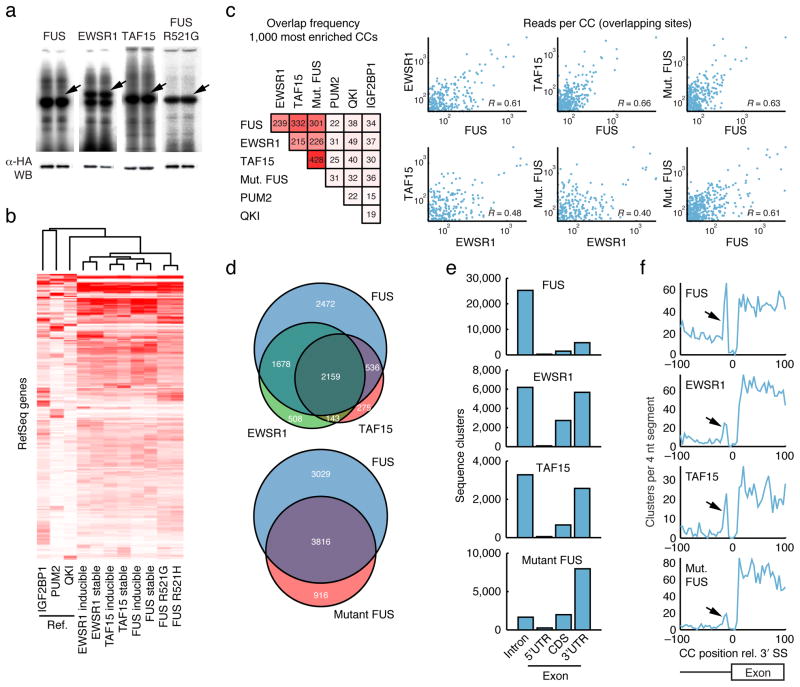
Figure 1.
Protein-RNA interaction maps of FET (FUS, EWSR1, TAF15) proteins. (a) Phosphorimages of SDS-PAGE gels that resolved 32P-labeled RNA–FLAGHA–FUS or EWSR1 or TAF15 PAR-CLIP immunoprecipitates. Excised regions are indicated by arrows. Protein identities of these bands were confirmed by mass spectrometry (not shown). Western blots (WB) were probed with an anti-HA antibody. (b) Hierarchical clustering diagram of binding patterns based on the number of reads per gene and Spearman correlation. Three unrelated reference datasets were included for comparison7. Binding profiles were mean intensity normalized. Similar results were obtained when datasets were size-normalized (data not shown). Stable, constitutive expression of the indicated protein; inducible, inducible expression of the indicated protein. (c) Overlap frequencies based on the top 1000 crosslinked clusters (CCs) of each protein, based on the number of sequence reads. CCs were considered overlapping when center positions were within 10 nucleotides (nt). Scatter plots show the reproducibility in number of reads per overlapping site. Correlations (Pearson’s R) were calculated based on log-transformed values. (d) Venn diagrams that illustrate overlaps between genes targeted by the three FET proteins, as well as between FUS and mutant FUS. (e) Distribution of CCs across intronic and exonic regions of RefSeq mRNAs. (f) Positional distribution of CCs near intron-exon junctions show enriched binding upstream of the 3′ splice site (3′ SS, arrow). The Y-axis indicates the number of observed CCs per 4 nt segment. The _P_-value for observing a peak of similar magnitude or higher anywhere in a 10.000 nt region upstream of the SS was in all cases < 0.025 (based on randomization of CC positions within introns).
As an initial quality control we computed quantitative binding profiles for each dataset based on the total number of uniquely mapped sequence reads per RefSeq gene, without yet introducing thresholds to identify individual binding sites. Clustering revealed a high degree of similarity between replicates (Rs = 0.84 to 0.87, Spearman’s rank correlation), as well as gross similarities in the binding spectra of the FET proteins relative to three unrelated reference RBPs7 (Figure 1b). The binding patterns of the two mutant FUS proteins closely resembled each other (Rs = 0.91), while also being similar to, but yet distinguishable from, the wild-type FET proteins (Rs = 0.63 to 0.71).
To define individual top target sites, we combined replicate reads for each of the three FET proteins as well as of the mutant forms. PAR-CLIP identifies sites of crosslinking between protein and RNA by scoring for T-to-C mutations in clusters of cDNA sequence reads7. Top target sites were defined as clusters of 10 or more overlapping reads of which at least 25% contained T-to-C mutations (referred to below as “crosslinked clusters”, CCs). We obtained 39984, 19020, 8678 and 14953 such CCs for FUS, EWSR1, TAF15 and mutant FUS, respectively; 82% of these were within RefSeq transcripts (CCs are provided in BED format for display in e.g. the UCSC Genome Browser12, Supplementary Data 1). Power analysis showed that we did not reach saturation of sites at our depth of Solexa sequencing (Supplementary Fig. 2). Intersection of the 1000 most highly ranked CCs in each dataset revealed large site-level overlaps between FET proteins (215–332 CCs) as well as between FET proteins and mutant FUS (226–428 CCs), while overlaps with unrelated reference RBPs were small (22–49 CCs, Figure 1c). The number of reads per overlapping site was positively correlated between different FET proteins (Figure 1c).
To complement the site-level analysis, CCs were summarized on a per-gene basis (Supplementary Data 2). FUS, EWSR1 and TAF15 each targeted 6845, 4488 and 3113 different genes, and these sets were largely overlapping (Figure 1d). Mutant FUS, which targeted 4732 genes, had an elevated fraction of unique targets compared to EWSR1 and TAF15, pointing toward an altered, rather than disrupted, binding profile (Figure 1d). We also resampled library sequence reads to compare data sets with similar target gene numbers and obtained similar results (Supplementary Table 2). Overall, transcripts bound by wild-type FET proteins were often bound at multiple positions, on average one CC every 13379 nucleotides. A large fraction of CCs for wild-type FET proteins fell within intronic regions, consistent with their nuclear localization (FUS: 78%, EWS: 39%, TAF15: 47% of mRNA clusters, Figure 1e). In contrast, mutant FUS had few intronic sites (13%) and bound predominantly to 3′ UTRs (61%). The distribution of CCs across mRNA regions (UTRs and CDS) was markedly different from those of reference RBPs7 and also deviated from the relative sizes of these regions in RefSeq (Supplementary Fig. 3). Taken together, gene- and site-level analyses showed similarities in the binding patterns of FET proteins, and indicated that the RNA-binding properties of mutant FUS were not impaired or altered but that a different spectrum of target RNAs was accessed as a result of altered subcellular localization.
By investigating the positional distribution of FET CCs in relation to splice sites, we observed an increased frequency of intronic binding immediately upstream of 3′ splice sites (Figure 1f) but not downstream of 5′ splice sites (not shown). This pattern was not observed in the three reference datasets (Supplementary Fig. 4). The G-rich intron-exon junction was essentially void of crosslinked clusters, which may be due to specific cleavage 3′ to G residues by RNase T1 in the PAR-CLIP protocol.
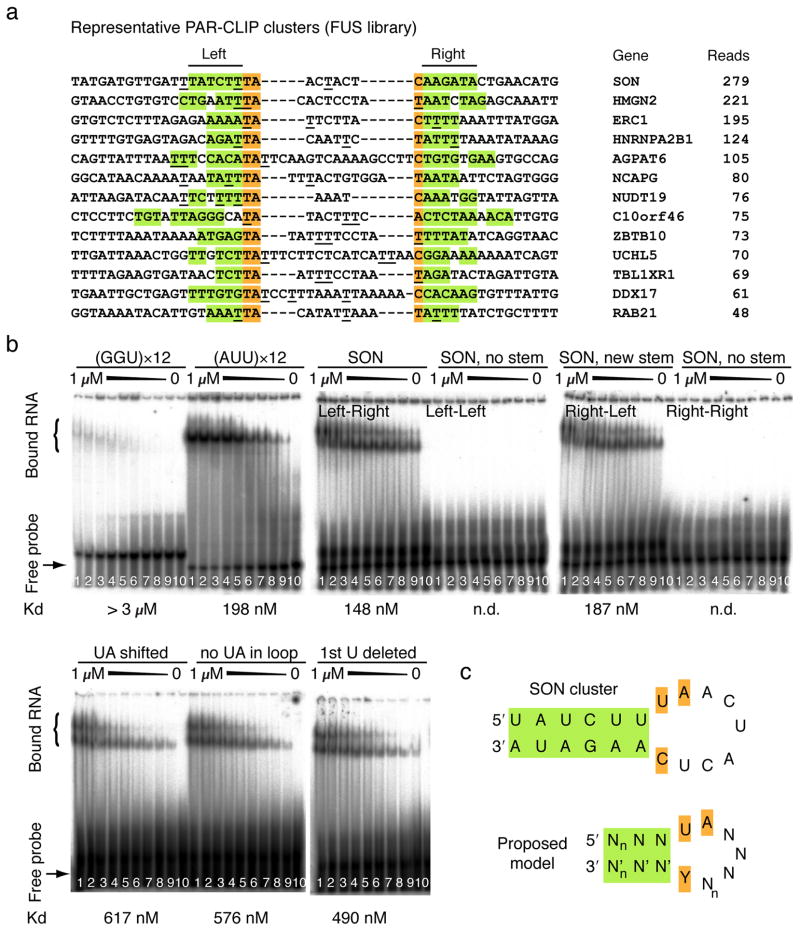
Use of standard bioinformatic tools (see Supplementary Methods) did not return a significant RRE motif for any of the FET proteins, indicating that RNA structure may play a role in recognition. Many FUS CCs contained a conventional stem-loop structure (Figure 2a) that frequently opens with a U-U or U-C non-Watson-Crick base pair, where the U at the 5′ end of the loop is followed by an A in the loop (67% in a 60 nucleotide window around cluster centers). This pattern was less frequent in randomly selected intronic and 3′ UTR regions (14% and 39%) but similar in shuffled sequences. Furthermore, the FUS CCs have low G and high AU content, which could reflect the binding preferences of the protein, but also, at least in part, a methodological preference7,13. We therefore experimentally tested the ability of FUS protein to bind to AU-rich stem-loop structures, and evaluated 37-nucleotide oligoribonucleotides corresponding to a predicted stem-loop sequence within a CC of the SON transcript (Supplementary Fig. 5) by electrophoretic mobility shift assays. We additionally evaluated a GGU repeat, since FUS had previously been shown to bind to GGUG-containing RNAs in a G-rich context14. The dissociation constant of the SON stem-loop (148 nM) was at least 15-fold higher than for the GGU repeat RNA (Figure 2b). Disruption of the SON stem-loop abolished FUS binding, while the compensatory sequence change restoring the disrupted stem-loop also restored binding. Altering the UA residues opening the loop also decreased binding (Figure 2b). An AUU trinucleotide repeat sequence, which approximates the nucleotide distribution of our CCs, and is predicted to accommodate a stem-loop structure comprising non-Watson-Crick U-U base pairs, had a binding constant similar to the SON stem-loop (198 nM). Together, our results suggest that FUS protein binds AU-rich stem-loops and that especially the stem but also specific loop residues contribute to binding (Figure 2c). Structural studies using natural and non-natural high-affinity target RNAs can now be initiated to test our interaction model.
Figure 2.
RNA-binding preferences of wild-type FET and mutant FUS proteins. (a) Genomic sequences of representative FET CCs. All clusters are present in all wild-type FET and mutant FUS datasets. Green shading indicates stems (“left” and “right”), orange shading the nucleotides closing the loop. The most frequent crosslinking positions are underscored. Gene name and number of reads are indicated. (b) Phosphorimage of native PAGE resolving complexes of recombinant FUS protein with different RNA oligoribonucleotides (all at 1 nM): GGU×12, AUU×12, SON (stem in natural left-right configuration as indicated in panel A), altered SON (non-complementary left-left stem), altered SON (non-complementary right-right stem), and altered SON (reconstituted right-left stem). Additionally, the effects of changing the “UA” opening the loop are shown (UA shifted, no UA in loop, 1st U deleted). Concentrations of FUS protein ranged from 1000 nM to 0 nM (lanes 1 to 10; fractions of bound vs. unbound protein can be found in Supplementary Data 3). Dissociation constants (Kd) are indicated; n.d., non-determinable. (c) Proposed model of the FET protein RRE. n is an integer ≥ 1 indicating variable stem and loop length.
PAR-CLIP results show that FET proteins bind RNA at high frequency, including the majority of cell-expressed mRNAs. In support of their proposed role in pre-mRNA splicing9, we detected preferential binding near splice acceptors, but these events represent a minor fraction of all CCs. In addition, global changes in transcript abundance in response to FUS silencing in HEK293 cells were weak and not correlated with FUS binding (Supplementary Fig. 6). Mutant FUS showed drastically altered distribution of CCs across transcript regions, consistent with translocation to the cytoplasm, while still maintaining its RNA-binding capability and specificity. This supports a model in which the deleterious effect of dominant ALS-causing FUS mutations, such as R521G and R521H, may in part be caused by a gain-of-function effect due to either increased interaction with cytoplasmic RNA targets or independent cellular stress caused by mislocalization of an abundant nuclear protein. Interestingly, we found endoplasmatic reticulum and ubiquitin-proteasome-related target gene categories to be overrepresented among transcripts uniquely targeted by mutant FUS (Figure 1d, Supplementary Table 3), which further supports the idea that protein synthesis and degradation represents major pathways perturbed in ALS15. Comprehensive mapping of the molecular targets of FET proteins, as well as disease-causing mutant FUS forms, (Supplementary Data 1 and 2) might facilitate future studies related to ALS and the evaluation of models of this disease.
Supplementary Material
1
2
3
4
Acknowledgments
J.I.H. is supported by the Deutsche Forschungsgemeinschaft, E.L. by the Swedish Research Council and M.H. by the Charles Revson Jr. Foundation. T.T. is an HHMI investigator, work in his laboratory was supported by NIH grant MH08442 and the Starr Foundation.
Footnotes
Author Contributions
J.I.H., stable cell line generation (FUS, EWSR1, FUS mutants), PAR-CLIP experiments (FUS, EWSR1, FUS mutants), siRNA knockdowns, gel shift experiments; E.L., all computational analysis; S.R., generation of TAF15 stable cell lines and TAF15 PAR-CLIP; J.N., recombinant protein purification and gel shift experiments; S.D., Western blot experiments; T.A.F. and M.H., siRNA knockdowns. A.B., C.S. and T.T., supervision of the project; E.L., J.I.H., and T.T. wrote the paper.
Competing financial interests
T.T. is cofounder and scientific advisor of Alnylam Pharmaceuticals and advisor to Regulus Therapeutics.
References and notes
- 1.Keene JD. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet. 2007;8:533–43. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- 2.Licatalosi DD, Darnell RB. RNA processing and its regulation: global insights into biological networks. Nat Rev Genet. 2010;11:75–87. doi: 10.1038/nrg2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martin KC, Ephrussi A. mRNA localization: gene expression in the spatial dimension. Cell. 2009;136:719–30. doi: 10.1016/j.cell.2009.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 5.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–45. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell. 2009;136:777–93. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hafner M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–41. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu C, et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10:R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tan AY, Manley JL. The TET family of proteins: functions and roles in disease. J Mol Cell Biol. 2009;1:82–92. doi: 10.1093/jmcb/mjp025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kwiatkowski TJ, Jr, et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–8. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- 11.Vance C, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–11. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kishore S, et al. A quantitative analysis of CLIP methods for identifying binding sites of RNA-binding proteins. Nat Methods. 2011 doi: 10.1038/nmeth.1608. [DOI] [PubMed] [Google Scholar]
- 14.Lerga A, et al. Identification of an RNA binding specificity for the potential splicing factor TLS. J Biol Chem. 2001;276:6807–16. doi: 10.1074/jbc.M008304200. [DOI] [PubMed] [Google Scholar]
- 15.Okamoto K, Fujita Y, Mizuno Y. Pathology of protein synthesis and degradation systems in ALS. Neuropathology. 2010;30:189–93. doi: 10.1111/j.1440-1789.2009.01088.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2
3
4