Identification of DHX33 as a Mediator of rRNA Synthesis and Cell Growth (original) (raw)
Abstract
In this report, we employed a lentiviral RNA interference screen to discover nucleolar DEAD/DEAH-box helicases involved in RNA polymerase I (Pol I)-mediated transcriptional activity. Our screen identified DHX33 as an important modulator of 47S rRNA transcription. We show that DHX33 is a cell cycle-regulated nucleolar protein that associates with ribosomal DNA (rDNA) loci, where it interacts with the RNA Pol I transcription factor upstream binding factor (UBF). DHX33 knockdown decreased the association of Pol I with rDNA and caused a dramatic decrease in levels of rRNA synthesis. Wild-type DHX33 overexpression, but not a DNA binding-defective mutant, enhanced 47S rRNA synthesis by promoting the association of RNA polymerase I with rDNA loci. In addition, an NTPase-defective DHX33 mutant (K94R) acted as a dominant negative mutant, inhibiting endogenous rRNA synthesis. Moreover, DHX33 deficiency in primary human fibroblasts triggered a nucleolar p53 stress response, resulting in an attenuation of proliferation. Thus, we show the mechanistic importance of DHX33 in rRNA transcription and proliferation.
INTRODUCTION
RNA is a highly structured macromolecule whose secondary and tertiary conformations facilitate an array of specific interactions with proteins. The DEAD/DEAH-box family of RNA helicases (here referred to as DDX/DHX) (3) is one such classification of RNA binding proteins that are capable of modifying the higher-ordered structures of RNA through the hydrolysis of ATP/nucleoside triphosphate (NTP) (41). DDX/DHX proteins often form large multiprotein complexes that participate in fundamental biological activities such as RNA transcription, RNA editing, pre-mRNA splicing, ribosome biogenesis, and RNA decay (3).
DDX/DHX helicases are named and characterized by the conserved DEAD/DEAH motif common among all family members. Through site-directed mutagenesis analysis, DEAD/DEAH along with seven conserved peptide motifs have been found to participate in ATP/NTP binding, hydrolysis, and substrate binding (28). Despite the conservation of these peptide motifs, the remaining sequences within each RNA helicase family member vary widely. Specifically, differences exist between the two categories of DDX and DHX proteins. DDX proteins contain a unique Q motif at their N termini that distinguishes them from DHX proteins. It was proposed previously that the Q motif might sense the state of ATP in vivo (40), given that DHX-box proteins are promiscuous in their ability to utilize NTP (16).
Ribosome biogenesis is a complex multistep process, the majority of which occurs in the nucleolus of the cell (24, 43). The transcription of ribosomal DNA (rDNA) is the initial and rate-limiting step in ribosome biogenesis, and as such, it is influenced by multiple levels of regulation (25). One of the key regulators of rDNA transcription is the upstream binding factor (UBF), a transcriptional transactivator that binds to the upstream core element of rDNA and subsequently bends rDNA (37). This change in the rDNA structure favors the binding of SL.1 as well as other associating factors to the rDNA locus (2). This allows for the recruitment of RNA polymerase I (Pol I) to initiate rDNA transcription (14, 23, 34). Recently, multiple functions have been found to be associated with UBF in rRNA transcription, including promoter clearance, the displacement of histone H1, and an enhancement of elongation (15, 27, 35). UBF also binds to rDNA regions outside transcribed regions, and its overexpression causes a global decondensation of rDNA chromatin structures (6). The activities of various transcription factors in rRNA transcription can be altered by posttranslational modifications involving the phosphorylation and acetylation of UBF, SL.1, and TIF-IA (25). In mammalian cells, a single precursor rRNA transcript, 47S rRNA (14.3 kb), is transcribed from rDNA by the RNA polymerase I complex. This large polycistronic transcript encompasses 18S, 5.8S, and 28S rRNAs and includes several spacer regions.
In the present report, we screened a group of nucleolar DDX/DHX-box proteins for their influence on pre-rRNA transcription by utilizing lentiviral RNA interference (RNAi) knockdown analysis. Various degrees of perturbation in rRNA transcription were observed by reducing the expression levels of numerous nucleolar DDX/DHX proteins. One of these nucleolar family members, DHX33, had a dramatic impact on pre-rRNA transcription and on nucleolar structure upon its knockdown, which could be rescued only by a helicase-competent and DNA binding-competent DHX33 protein. We found that DHX33 localized to nucleoli, where it associated with rDNA and UBF. The DHX33 knockdown caused a dramatic reduction of RNA polymerase I-mediated transcriptional activity. The overexpression of DHX33 stimulated rDNA transcription by promoting the rDNA occupancy of RNA polymerase I, all of which required NTPase activity and an rDNA binding capacity. Thus, DHX33 appears to be an intricate player in rRNA transcription and nucleolar organization.
MATERIALS AND METHODS
Cell culture.
Primary human BJ fibroblasts were cultured in Dulbecco's modified essential medium (DMEM) and M199 medium (4:1; Sigma) supplemented with 15% fetal bovine serum (FBS) and penicillin-streptomycin. HeLa cells, _p53_-null mouse embryonic fibroblast (MEF) cells, and BT549 breast cancer cells were cultured in complete DMEM supplemented with 10% FBS and penicillin-streptomycin. All cells were maintained at 37°C in 5% CO2.
Antibodies.
Anti-DHX33 antibody (rabbit polyclonal, catalog number NB100-2581) was obtained from Novus Biologicals. Anti-UBF mouse monoclonal (F-9, catalog number sc-13125), rabbit polyclonal (H-300, catalog number sc-9131), anti-phospho-UBF (pUBF) (Ser484) (catalog number sc-21638), anti-gamma-tubulin (catalog number sc-7396), anti-p21 (F-5, catalog number sc-6246), anti-p53 (DO-1, catalog number sc-126), anti-fibrillarin (H-140, catalog number sc-25397), anti-cyclin D1 (catalog number sc-450), anti-RPA194 (H-300, catalog number sc-28714), anti-RRN-3 (Y-23, catalog number sc-133978), anti-transcription termination factor (TTF) (24) (catalog number sc-136371), anti-TATA binding protein (TBP) (catalog number sc-204), and anti-epidermal growth factor receptor (EGFR) (catalog number sc-03) antibodies were obtained from Santa Cruz Biotechnology. Anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody (A300-641A) was obtained from Bethyl Laboratories. Anti-FLAG (M2) antibody was obtained from Sigma.
Immunoprecipitation and Western blots.
Cells were lysed in buffer containing 1% NP-40, 50 mM Tris (pH 7.5), and 150 mM NaCl and supplemented with protease and phosphatase inhibitor cocktails (Sigma). After incubation on ice for 10 min, cell lysates were further sonicated to ensure complete disruption. Lysates were then centrifuged for 10 min at 13,000 rpm, and supernatants were subjected to a protein quantification assay. For Western blots, 50 μg of cell lysate was loaded onto a precast minigel (Bio-Rad), followed by transfer onto a polyvinylidene difluoride (PVDF) membrane. For immunoprecipitation, cell lysates were diluted to approximately 1 mg/ml with lysis buffer; 500 μg of the total cell lysate was incubated with 2 μg of the indicated antibody for 2 h at 4°C, followed by the addition of protein A/G-Sepharose beads, and further incubated for 1 h at 4°C. After centrifugation at 1,500 rpm for 2 min, the beads were washed 3 times with cell lysis buffer before analysis.
Plasmids.
The full-length coding sequence for DHX33 (Mammalian Gene Collection [MGC]) was purchased from Open Biosystems. Primers were designed to clone the open reading frame (ORF) of DHX33 into p3XFLAG-CMV10.0 at HindIII/BamHI sites. To subclone DHX33 into pLVX-IRES-hygromycin, p3XFLAG-CMV10.0-DHX33 was further digested with SacI/BamHI to release the 3× FLAG-DHX33 fragment, treated with Klenow fragment, and then inserted into pLVX-IRES-hygromycin at the blunted XbaI site. A QuikChange site-directed mutagenesis kit (Stratagene) was used to carry out the mutation of K94N and K94R as well as the DNA binding box deletion mutants of DHX33 in plasmid pLVX-DHX33. The human UBF1 ORF with a Myc tag and a Flag tag (pCMV-6-UBF1) was purchased from Origene. To subclone UBF1-Myc into the pLVX vector, UBF1-Myc was cut by BamHI/EcoRV from pCMV6-UBF1, blunt ended by Klenow fragment, and then ligated into the pLVX vector at the blunt-ended XbaI site. The human UBF2 ORF was purchased from Origene and was subcloned into the pLVX vector at the XbaI/BamHI site. A QuikChange site-directed mutagenesis kit was used to carry out the mutation of S484D and S484A in UBF1/2. Five different pLKO.1 plasmids containing short hairpin RNA (shRNA) to target each of the indicated DEAD/DEAH-box proteins (human) were obtained from the Genome Sequencing Center at Washington University. A pLKO.1 scrambled shRNA control plasmid was purchased from Addgene.
Immunofluorescence microscopy.
For 5-bromo-2-deoxyuridine (BrdU) analysis, cells were incubated with BrdU for 18 h, followed by 10% formalin-10% methanol fixation for 15 min at room temperature. Cells were blocked with 5% FBS and incubated with mouse anti-BrdU antibody (Clontech). Rhodamine-conjugated goat anti-mouse antibody was then applied and incubated for an additional 30 min at room temperature. Cell nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI). To analyze DHX33 localization, cells were transfected with p3XFLAG-CMV-DHX33. Transfected cells were fixed with 10% formalin-10% methanol. Cells were then incubated with mouse anti-FLAG (M2; Sigma) antibody at a 1:200 dilution. Goat anti-mouse antibody-fluorescein isothiocyanate (FITC) was applied to facilitate the visualization of the DHX33 protein. To mark cell nucleoli, rabbit antifibrillarin antibody was used at a dilution of 1:100, followed by incubation with goat anti-rabbit antibody-rhodamine.
47S rRNA synthesis analysis.
Cells were pulsed with [3H]uridine (Amersham) at a concentration of 2.5 μCi/ml for 30 min and chased with unlabeled uridine-containing medium at a concentration of 5 mM for the indicated time points. Cells were normalized based on equal cell numbers. Total RNA was isolated by using RNAsolv (Omega Biotek) according to the manufacturer's instructions. RNA was loaded onto 1% agarose-formaldehyde gels and was transferred onto a nylon Hybond membrane (Millipore). After UV cross-linking and spraying with Enhancer (Amersham), the membrane was exposed to film and subjected to autofluorography. For in situ run-on assays, the protocol was performed as previously described (18). Briefly, cells were incubated in complete medium containing 2 mM 5′-fluorouridine (FUrd) for 10 min at 37°C in 5% CO2 and fixed in 10% formalin-10% methanol. The incorporated FUrd was visualized by incubating cells with monoclonal anti-BrdU antibody (Sigma) for 1 h and with goat anti-mouse antibody coupled to rhodamine for 30 min.
Pulse-chase with [_methyl_-3H]methionine for rRNA processing.
Cells were first incubated with methionine-cysteine-free medium for 30 min, and [_methyl_-3H]methionine was then added at a concentration of 50 μCi/ml and incubated for 30 min at 37°C. Cells were chased with unlabeled medium containing 10 μM methionine and incubated at 37°C for various times. Approximately 1.0 × 106 cells were pelleted and dissolved in RNAsolv for RNA extraction. RNA was loaded onto a denaturing gel, transferred onto a nylon membrane, and subjected to autofluorography as described above.
Nucleolus fractionation.
Approximately 108 cells were collected for cell fractionation. Cells were washed with phosphate-buffered saline (PBS) twice and resuspended in 1 ml buffer (10 mM HEPES-KOH [pH 7.9], 1.5 M MgCl2, 10mM KCl, and 0.5 mM dithiothreitol [DTT]) for 30 min on ice. Phenylmethylsulfonyl fluoride (PMSF) was added to a final concentration of 0.2 mM, and the mixture was then Dounce homogenized until all cytoplasmic membranes were disrupted. For cytosolic isolation, cells were centrifuged at 1,190 rpm for 5 min at 4°C to obtain the supernatant. The resulting pellet was resuspended in 0.45 ml of 0.25 M sucrose-10 mM MgCl2, layered onto 0.45 ml of 0.35 M sucrose-0.5 mM MgCl2, and centrifuged at 2,790 rpm for 5 min at 4°C. Pelleted nuclei were then resuspended in 0.75 ml of 0.35 M sucrose-0.5 mM MgCl2 with protease and phosphatase inhibitors (Sigma). Nuclei were sonicated to disrupt the nuclear membrane. The nuclear isolate was layered on top of 0.75 ml of 0.88 M sucrose-0.5 mM MgCl2 and centrifuged at 2,800 × g for 10 min at 4°C. The pellet was resuspended in 0.5 ml of 0.35 M sucrose-0.5 mM MgCl2, and sucrose layering was repeated as described above. Nucleoli were fractionated as the subsequent pellet.
FACS analysis.
Cells were trypsinized and washed with PBS. Cells were then resuspended in PBS, and 100% ethanol was added dropwise to obtain a final ethanol concentration of 75%. Cells were centrifuged at 2,000 rpm at 4°C for 2 min. Cells were then washed with PBS and resuspended in propidium iodide (PI) working solution (PBS containing 1% FBS, 250 μg/ml of RNase A, and 30 μg/ml of propidium iodide). Cells were filtered through a 35-μm strainer cap (Becton Dickinson) before being subjected to fluorescence-activated cell sorter (FACS) analysis.
qRT-PCR.
The primers were all designed by use of Primer Express 2.0 software and purchased from Integrated DNA Technologies. Total RNA was extracted by use of a NucleoSpin II (Clontech) RNA isolation kit and was reverse transcribed into cDNA by use of a SuperScript III first-strand synthesis kit (Invitrogen). PCRs were performed with a Bio-Rad C1000 thermal cycler and managed with Bio-Rad CFX96 software. For analysis of 47S rRNA transcript levels, SYBR green FastMix (Quanta Biosciences) was used, and transcript quantification was performed by comparison with standard curves generated from dilution series of cDNA of human 47S rRNA (cloned into pCR2.1Topo). SYBR green mix from Bio-Rad was used for all other quantitative real-time PCR (qRT-PCR) analyses. Transcript quantification was calculated based on the ΔΔ_CT_ value after normalization to GAPDH values. Melt curve analysis confirmed that single products were amplified.
Chromatin immunoprecipitation.
Trypsinized cells were washed with PBS and fixed with 1% formaldehyde at room temperature for 10 min. To stop cross-linking, 1.25 M l-glycine was added to a final concentration of 0.125 M. After washing with 1× PBS, cells were resuspended in lysis buffer containing 1% SDS, 10 mM EDTA, and 50 mM Tris (pH 8.1) with protease and phosphatase inhibitors. To shear chromatin, cell lysates were sonicated extensively, centrifuged to pellet debris, and then diluted in buffer containing 0.5% NP-40, 50 mM Tris (pH 7.5), and 150 mM NaCl at a 1:5 ratio. Cell lysates were precleared by incubation with 2.5 μg of sheared salmon sperm DNA and 50 μl protein A/G beads for 30 min at 4°C. Following incubation with 5 μg of antibody overnight at 4°C, 2.5 μg sheared salmon sperm DNA and 50 μl of protein A/G beads were added and further incubated for 1 h. The beads were then washed twice in radioimmunoprecipitation assay (RIPA) buffer, twice in RIPA buffer containing 500 mM NaCl, and once with buffer containing 0.5% NP-40, 50 mM Tris (pH 7.5), and 150 mM NaCl. The beads were then extracted three times with a solution containing 1% SDS and 0.1 M NaHCO3. To reverse the cross-linking, 6 M NaCl was added into the pooled extraction samples to a final concentration of 0.3 M, and samples were heated at 65°C for 5 h. DNA fragments were extracted by use of a Qiagen QuickSpin column and eluted. Quantitative PCR (qPCR) was performed with these purified DNA samples.
Cell size and volume.
Primary human BJ fibroblasts (2 × 106 cells) were harvested by trypsinization, washed with 1× PBS, and resuspended in 10 ml DMEM. One milliliter of the cell suspension was mixed with 20 ml Isoton diluent (Beckman) and analyzed on a Multisizer III instrument according to the manufacturer's protocol (Beckman). Over 75,000 particles were analyzed in triplicate from three independent isolations using a 100-μm aperture.
Ag staining of nuclealar organizing region (AgNOR staining).
Cells were seeded onto glass coverslips overnight and were fixed and stained the following day. Cells were fixed in 2% glutaraldehyde, followed by postfixation in a 3:1 ethanol-acetic acid solution. Cells were stained with a 0.33% formic acid-33.3% silver nitrate solution in 0.66% gelatin and mounted onto slides with Vectashield (Vector Labs).
Subcellular protein fractionation.
Subcellular proteins were fractionated by use of a subcellular protein fractionation kit from Pierce. BJ cells (2 × 106 cells) were fractionated into cytosolic, membrane-bound, soluble nuclear, as well as chromatin-bound parts according to standard protocols.
RESULTS
Screening of nucleolar RNA helicases involved in rRNA synthesis.
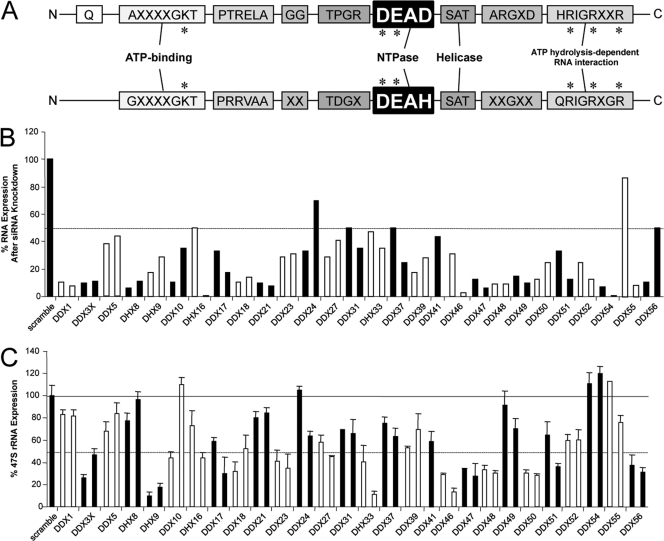
Ribosome biogenesis is an evolutionarily conserved cellular activity that is vital for normal cell growth and proliferation. Several nuclear/nucleolar DEAD/DEAH-box proteins (represented in Table 1, together with their orthologs in Saccharomyces cerevisiae) were chosen based on the human nucleolar proteome (http://www.lamondlab.com) (1). They have been shown to participate in various aspects of ribosome biogenesis, including rRNA processing and ribosome assembly (41). However, none of the currently characterized family members have been shown to participate in pre-rRNA transcription. We performed a lentiviral RNA interference screen in order to identify a set of nucleolar DDX/DHX RNA helicases that were necessary for RNA polymerase I-mediated pre-rRNA synthesis. First, five unique shRNAs for each DDX/DHX protein were screened to validate their knockdown efficiencies. Two validated shRNAs were then delivered into primary human fibroblast cells by lentiviral infection. The knockdown efficiency for each DDX/DHX mRNA was detected by quantitative real-time PCR (qRT-PCR) reactions, as shown in Fig. 1 B. Pre-rRNA transcription was assayed by monitoring the production of the short-lived 47S rRNA precursor by qRT-PCR (7). Changes in 47S rRNA transcript levels correlated with the efficiency of each DDX/DHX helicase knockdown by qRT-PCRs. As shown in Fig. 1C, the knockdown of several DDX/DHX proteins correlated with decreased pre-rRNA synthesis although with various degrees of perturbation. A significant decrease in the rRNA transcription level (up to 10-fold) compared to that of the control was observed for DHX33, DHX9, and DDX46 following the shRNA delivery (Fig. 1C), while up to a 3-fold to 4-fold decrease was seen for DDX23, DDX48 (eukaryotic initiation factor 4A [eIF4A]), DDX18, DDX47, DDX56, DDX50, DDX51, DDX3X, and DDX48 (Fig. 1C). Table 2 shows the ratios between the change of 47S rRNA expression and the change of mRNA expression for each protein after small interfering RNA (siRNA) knockdown. Notably, although the DHX33 mRNA expression level was decreased by only 60%, we detected a tremendous 10-fold reduction of rRNA synthesis. Given the extreme sensitivity of rRNA synthesis to lower DHX33 expression levels, we chose to further explore the potential role of DHX33 in rRNA transcription.
Table 1.
DEAD/DEAH RNA helicase family
| Human/mouse helicase | Yeast ortholog | Ribosome biogenesis |
|---|---|---|
| DDX1 | ||
| DDX3X | Dbp1p | + |
| DDX5 | Dbp2p | |
| DHX8 | Prp22p | |
| DHX9 | ||
| DDX10 | Dbp4p | + |
| DHX16 | Prp2p | |
| DDX17 | Dbp2p | |
| DDX18 | Has1p | + |
| DDX2 | + | |
| DDX23 | Prp28p | |
| DDX24 | Mak5p | + |
| DDX27 | Drs1p | + |
| DDX31 | Dbp7p | + |
| DHX33 | ||
| DDX37 | Dhr1p | + |
| DDX39 | Sub2p | |
| DDX41 | ||
| DDX46 | Prp5p | |
| DDX47 | Rrp3p | + |
| DDX48 | Fal1p | + |
| DDX49 | Dbp8p | + |
| DDX50 | + | |
| DDX51 | Dbp6p | + |
| DDX52 | Rok1p | + |
| DDX54 | Dbp10p | + |
| DDX55 | Spb4p | + |
| DDX56 | Dbp9p | + |
Fig. 1.
Lentivirus-mediated RNAi screen of DDX/DHX-box proteins reveals a requirement for several helicases in RNA polymerase I-mediated transcription. (A) Schematic diagram for conserved peptide motifs of DDX and DHX proteins. Critical amino acids for helicase function are marked with an asterisk. (B) Total RNA extracted from human fibroblast BJ cells 4 days after lentiviral infection with each indicated shRNA. The knockdown efficiency of each shRNA for a specific DDX/DHX protein was analyzed by qRT-PCR using scrambled shRNA as a negative control. The dashed line marks 50% expression. (C) Total RNA from BJ cells analyzed by qRT-PCR for the 47S rRNA transcript copy number. The graph shows representative results of two independent screenings performed in triplicate. All values were normalized based on the scrambled control, which was set as 100%. Bars represent standard deviations for three separate qRT-PCR analyses. The solid line marks 100% expression, and the dashed line marks 50% expression.
Table 2.
DDX/DHX knockdown efficiencies and 47S rRNA transcription ratios
| Gene | Mean % mRNA | Mean % 47S rRNA | Ratio of 47S rRNA/mRNA |
|---|---|---|---|
| DDX1 | 7.69 | 81.4 | 10.58518 |
| DDX3X | 10 | 26.6 | 2.66 |
| DDX5 | 38 | 68.3 | 1.797368 |
| DHX8 | 6.2 | 77.2 | 12.45161 |
| DHX9 | 18 | 9.6 | 0.533333 |
| DDX10 | 10.5 | 44.4 | 4.228571 |
| DHX16 | 1 | 43.8 | 43.8 |
| DDX17 | 18 | 29.5 | 1.638889 |
| DDX18 | 10.8 | 32.2 | 2.981481 |
| DDX2 | 10 | 80.2 | 8.02 |
| DDX23 | 30.7 | 34.8 | 1.13355 |
| DDX24 | 33 | 105 | 3.181818 |
| DDX27 | 35.3 | 66.4 | 1.88102 |
| DDX31 | 29 | 58 | 2 |
| DHX33 | 35 | 11.2 | 0.32 |
| DDX37 | 25 | 63.6 | 2.544 |
| DDX39 | 18 | 53.2 | 2.955556 |
| DDX41 | 43.5 | 59.1 | 1.358621 |
| DDX46 | 29 | 31 | 1.068966 |
| DDX47 | 6.25 | 28 | 4.48 |
| DDX48 | 9 | 30.8 | 3.422222 |
| DDX49 | 10 | 70.5 | 7.05 |
| DDX50 | 12.5 | 30.9 | 2.472 |
| DDX51 | 13 | 36.4 | 2.8 |
| DDX52 | 12.5 | 60.1 | 4.808 |
| DDX54 | 7 | 111 | 15.85714 |
| DDX55 | 8.24 | 76 | 9.223301 |
| DDX56 | 10.8 | 37.7 | 3.490741 |
DHX33 is essential for pre-rRNA synthesis and is a cell cycle-regulated protein.
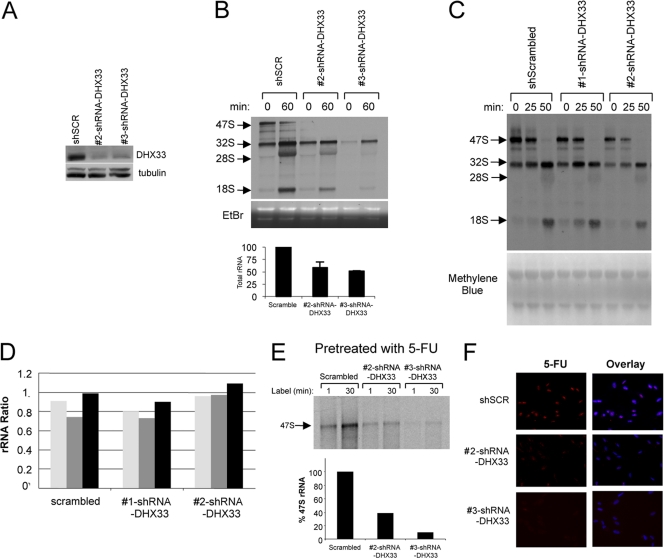
Given that the nucleolus is the site of rRNA synthesis, processing, and assembly (24), we monitored newly synthesized rRNA species in both control and DHX33 knockdown cells using a pulse-chase of [3H]uridine (39). The DHX33 knockdown efficiency was verified by Western blots, as shown in Fig. 2 A. Steady-state levels of rRNA harvested from equal numbers of control and DHX33 knockdown cells showed significant reductions of steady-state levels of mature 28S rRNA and 18S rRNA in DHX33 knockdown cells (Fig. 2A, middle and bottom). While there was a significant reduction in levels of all rRNA species, including 47S, 32S, 28S, and 18S rRNAs (Fig. 2B, top), we observed no accumulation of rRNA precursors (Fig. 2B, top), nor did we find any changes in the ratios of each rRNA species when we performed additional [_methyl_-3H]methionine pulse-chase experiments (39) to label newly synthesized pre-rRNA (Fig. 2C and D), indicating that DHX33 is not involved in pre-rRNA processing. To dissect the mechanism of DHX33 in pre-rRNA synthesis, and to further exclude the possibility of rRNA processing defects due to the DHX33 knockdown, BJ fibroblasts were pretreated with 5-fluorouracil to halt rRNA processing for 15 min prior to a [3H]uridine pulse to label newly synthesized 47S rRNA (9, 39). Using equal numbers of cells, we found that the synthesis of 47S rRNA was greatly diminished in DHX33 knockdown cells following a 30-min pulse with [3H]uridine (Fig. 2E).
Fig. 2.
DHX33 is essential for pre-rRNA synthesis but not processing. (A) BJ cells were infected with scrambled or DHX33 shRNAs. Total cell lysates from each were prepared at 4 days postinfection and subjected to Western blot analysis with the indicated antibodies. (B) From the above-mentioned infected cells, equal numbers of BJ cells (2 × 106 cells) were pulsed with [3H]uridine and then chased with unlabeled uridine and harvested at the indicated times. Total RNA was extracted, separated, transferred onto membranes, and subjected to autofluorography. Newly synthesized 47S, 32S, 28S, and 18S rRNAs are shown (top). Ethidium bromide (EtBr)-stained agarose gels show overall 28S and 18S rRNAs in these samples (middle). Quantification of mature rRNAs is shown in the bottom panel. Bars represents standard deviations from 3 independent experiments. (C) BJ cells infected with the 2 different shRNA-DHX33-encoding lentiviruses were labeled with [_methyl_-3H]methionine and chased with cold methionine for the indicated times. Total RNA was extracted from equal numbers of cells, separated, transferred onto membranes, and subjected to autoradiography. Newly synthesized and labeled rRNA species are indicated by arrows. The bottom panel shows the methylene blue-stained membrane to show the loading of each sample. (D) Densitometry quantitation of ratios between different rRNA species in each BJ cell sample depicted in a bar graph. (E) The BJ cells described above (2 × 106 cells) were pretreated with 5-fluorouracil (5-FU) to halt rRNA processing and then pulse-chased with [3H]uridine to monitor the total levels of 47S rRNA over a 30-min period. Total RNA was extracted, separated, transferred onto membranes, and subjected to autoradiography. Newly synthesized 47S rRNA is shown. (F) The BJ cells described above were pulsed with 5-fluorouridine (5-FUrd), fixed, and stained with an anti-BrdU antibody recognizing incorporated 5-FUrd (red). Nuclei were demarcated with DAPI, and an overlay of the signals is shown. Representative images from three independent experiments are shown.
In addition, RNA polymerase I transcription was more directly assessed by using a 5-fluorouridine (FUrd) incorporation in situ run-on assay. In this assay, active RNA polymerase I transcription is correlated directly with the amount of incorporated FUrd in nucleoli (18). In control BJ cells, we observed high levels of FUrd incorporation at nucleolar sites using an anti-BrdU antibody recognizing incorporated FUrd (Fig. 2F). In contrast, the knockdown of DHX33 with two separate shRNAs elicited a pronounced decrease, based on equal exposure times of immunofluorescent signals, in levels of nucleolar FUrd incorporation (Fig. 2F).
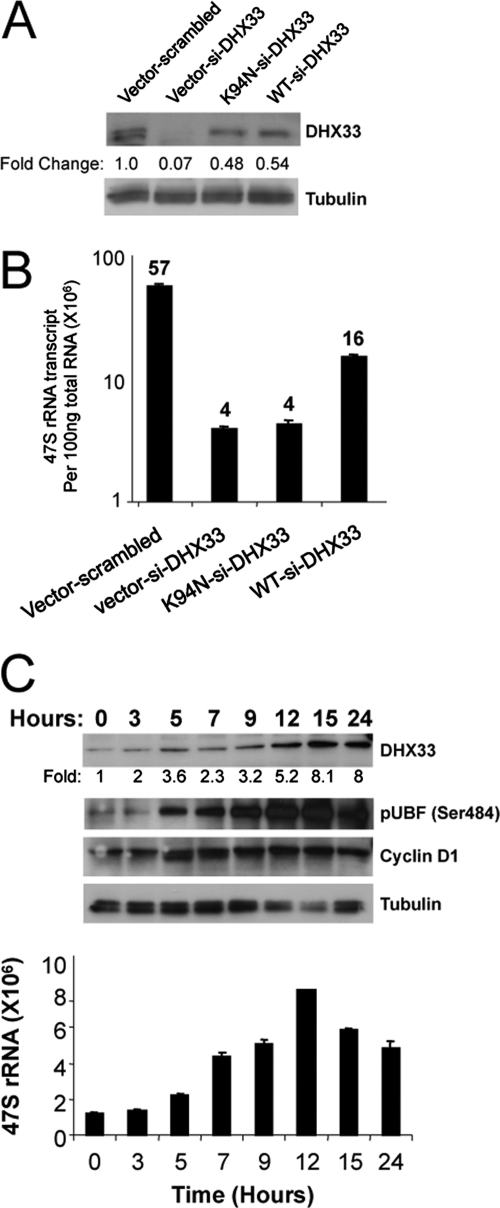
DHX33, along with its conserved family members, is an NTP-dependent RNA helicase (32). To determine whether NTPase activity was required for DHX33's rRNA transcriptional regulation, we performed site-directed mutagenesis on a critical and conserved amino acid, K94, in the NTPase domain of DHX33; the resulting single point mutation is K94N. The mutation of this critical lysine residue was shown previously to severely cripple RNA NTPase activity among DHX family members (28). BJ fibroblasts were infected with shRNAs to reduce levels of DHX33 and were rescued with the lentiviral delivery of either wild-type or K94N mouse DHX33. Three base pair mismatches exist between the murine DHX33 sequence and the human-targeted shRNA sequence, making the murine overexpression construct moderately resistant to shRNA-directed knockdown. As shown in Fig. 3 A, the overexpression of wild-type and K94N mutant DHX33 rescued DHX33 protein levels to approximately 50% of scrambled shRNA control levels. Importantly, the wild-type but not the NTPase-dead mutant DHX33 expression construct could rescue pre-rRNA synthesis to an appreciable degree (4-fold increase over DHX33 knockdown) (Fig. 3B).
Fig. 3.
DHX33 helicase activity is required for pre-rRNA synthesis. (A and B) BJ cells were first infected with wild-type (WT) or helicase-dead K94N DHX33 mutant constructs, followed by a secondary infection with lentiviruses containing the indicated siRNA knockdown constructs (scrambled or DHX33 specific). Cells were then selected in both puromycin- and hygromycin-containing media and analyzed by Western blotting for DHX33 and tubulin (A) and by qRT-PCR for 47S rRNA transcript levels (B) of BJ cells (n = 3). Numbers of 47S rRNA transcripts appear above each bar. (C) BJ cells were grown arrested in serum-free medium for 48 h and stimulated with 15% serum for the indicated times. Whole-cell lysates were subjected to Western blot analysis using antibodies recognizing DHX33, pUBF (Ser484), cyclin D1, and tubulin. Total RNA from the same samples was subjected qRT-PCR analysis for 47S pre-rRNA levels and is reported per 100 ng RNA.
Previous studies have shown that rRNA transcription is a growth factor-stimulated process (17). We observed a steady increase in 47S transcription levels as BJ cells reentered the cell cycle following serum stimulation (Fig. 3C). Additionally, expression levels of DHX33 increased upon serum stimulation and reached maximal levels concomitant with maximum phosphorylated UBF Ser484 and 47S rRNA transcription levels (Fig. 3C), indicating that DHX33 responds to growth stimuli in a manner similar to that of the canonical rRNA transcriptional apparatus.
DHX33 localizes to the cell nucleolus and associates with NORs during mitosis.
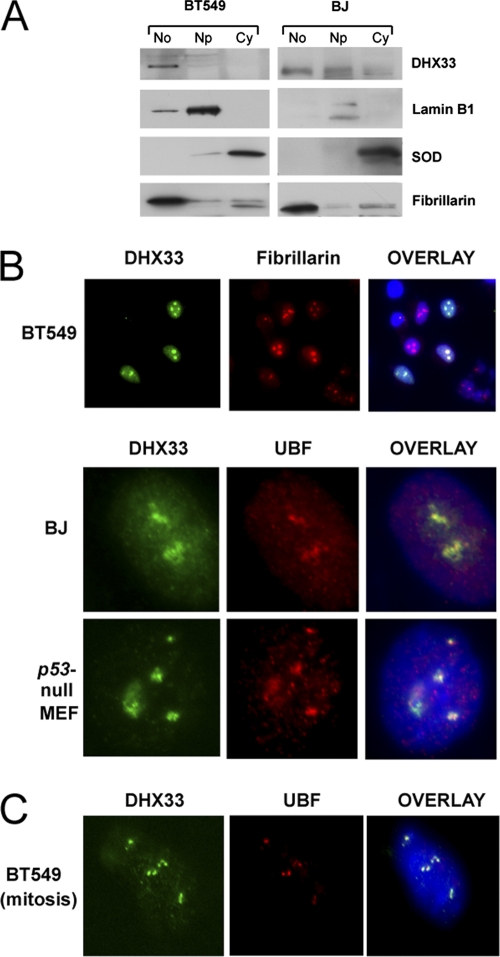
Although DHX33 is an uncharacterized member of the DEAH-box family, a previous proteomic analysis of HeLa cells identified DHX33 as a nucleolar protein (1). To verify the nucleolar localization of endogenous human DHX33, cellular fractionation was performed on two different human cell lines: BT549 breast cancer cells and diploid BJ fibroblasts. We found that DHX33 localized to cell nucleoli in both BT549 and BJ cells, although it was not entirely excluded from the nucleoplasm (Fig. 4 A). The nucleolar localization of DHX33 was further confirmed by the costaining of BT549 cells that transiently overexpress FLAG-DHX33 with fibrillarin (Fig. 4B). Colocalization between UBF and DHX33 was detected in both murine _p53_-null MEFs and human BJ fibroblasts (Fig. 4B). As cells progress through the G2/M border, nucleoli disassemble, with only a few proteins that are critical for RNA polymerase I transcription remaining associated with nucleolar organizing regions (NORs). UBF1 is one of these proteins (51). To check the localization of DHX33 in mitosis, cells were treated with nocodazole for 18 h to arrest the cell cycle at prometaphase. Under this cell cycle setting, we found that DHX33 and UBF1 again colocalized at the NOR (Fig. 4C).
Fig. 4.
DHX33 localizes to cell nucleoli. (A) Equal numbers of BT549 cells and BJ human primary fibroblasts (1 × 108 cells) were separated into nucleolar (No), nucleoplasmic (Np), and cytoplasmic (Cy) fractions. Proteins isolated from each fraction were analyzed by Western blotting using antibodies recognizing DHX33, cytosolic superoxide dismutase (SOD), nucleolar fibrillarin, and nuclear lamin B1. (B) BT549 cells, BJ cells, or _p53_-null MEFs were infected with a lentivirus encoding FLAG-tagged DHX33. At 4 days postinfection, transduced cells were fixed and stained with antibodies recognizing FLAG (DHX33) (green), fibrillarin (red), or UBF (red). Nuclei were demarcated with DAPI. Transfected BT549 cells showed an overexpression of FLAG-DHX33 in the nucleoli, as marked by fibrillarin at a ×20 magnification. An overlay of signals (yellow) is also shown for DHX33-UBF colocalization at a ×100 magnification for BJ cells and _p53_-null MEFs. (C) The BT549 cells described above were treated with nocodazole at 1 μM for 18 h, and cells were then fixed and costained with anti-UBF (red) and anti-FLAG (green) antibodies as described above. DHX33 and UBF colocalized at the NOR, as shown by the overlaid yellow signal at a ×100 magnification.
DHX33 associates with rDNA and UBF.
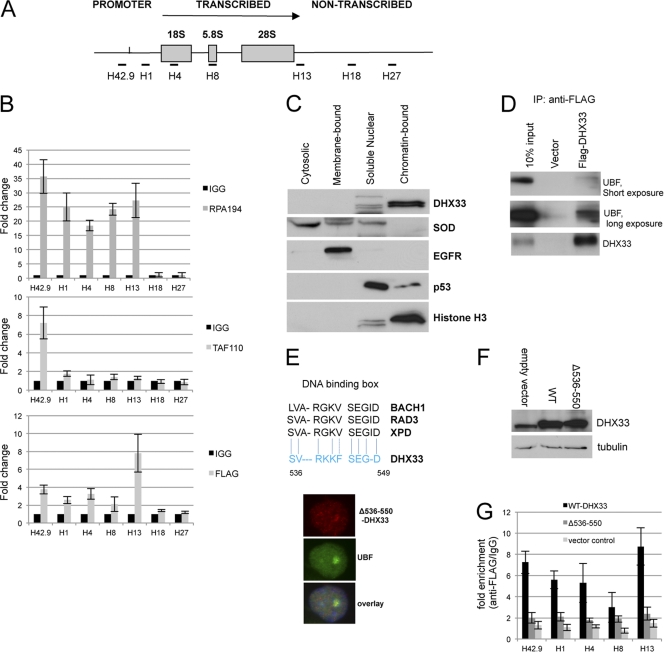
To further dissect the mechanism of DHX33's role in RNA polymerase I transcription, chromatin immunoprecipitation (ChIP) was performed with BT549 human breast cancer cells that overexpressed the FLAG epitope-tagged DHX33 protein. Figure 5 A depicts the position of the primer sets used for ChIP analysis along the rDNA locus. We observed a 2- to 7-fold increase in the association between DHX33 and the transcribed/promoter rDNA regions, demonstrating an association between DHX33 and the rDNA locus (Fig. 5B). As two positive controls, we also compared DHX33 with SL.1 and the RNA Pol I large subunit RPA194. SL.1 displayed an interaction only with the promoter region, with enrichment up to 8-fold, while RPA194 showed enrichment up to 40-fold at both promoter and transcribed regions (Fig. 5B). The fact that DHX33 associated with rDNA across the entire promoter and transcribed regions implies that its function may not be confined to transcriptional initiation. To further confirm the interaction between DHX33 and rDNA, we performed a cell fractionation analysis of BJ fibroblast cells. DHX33 was released only by micrococcal nuclease, and this occurred predominantly in chromatin-bound fractionation (Fig. 5C), indicating that DHX33 could be an architecture protein that associates with rDNA chromatin and modulates its function. In addition, we found that overexpressed FLAG-tagged DHX33 interacted with endogenous UBF, as demonstrated by the coimmunoprecipitation of UBF with FLAG antibodies (Fig. 5D). To examine this property more closely, we generated a deletion mutant of DHX33 that lacks 15 amino acids (Δ536-550). The fragment (positions 536 to 550) was found to be fairly similar to a conserved DNA binding box shared by several DEAH DNA helicases (Fig. 5E). The subcellular localization of this deletion mutant still remained in the nucleolus but with diffuse staining apparent in the nucleoplasm (Fig. 5E). Protein expression levels of mutant and wild-type DHX33 are shown in Fig. 5F. We found that the deletion mutant severely crippled its DNA binding activity, as detected by ChIP analysis (Fig. 5F). Taken together, these findings demonstrate the association of DHX33 with rDNA loci, through a critical 15-amino-acid motif, and UBF, a known rDNA binding protein.
Fig. 5.
DHX33 associates with rDNA and UBF. (A) Diagram depicting rDNA transcribed, nontranscribed, and promoter regions and the location for primer sets used for chromatin immunoprecipitation (ChIP). (B) BT549 cells transduced with FLAG-tagged DHX33 were lysed and incubated with nonimmune mouse IgG or antibodies recognizing the FLAG epitope, RPA194, as well as TAF110. PCR analysis using the above-mentioned rDNA primers was used to detect DNA sequences associated with immunoprecipitation. (C) BT549 cells were fractionated, and equal amounts of protein lysates were analyzed by Western blotting with the indicated antibodies. (D) BT549 cells stably expressing either FLAG-tagged DHX33 or an empty vector were immunoprecipitated (IP) with anti-FLAG antibody and immunoblotted with the indicated antibodies. (E) Alignment of DHX33 amino acids 536 to 550 with the DNA binding boxes of several known DEAH DNA helicases. BT549 cells were infected by a lentivirus encoding a DHX33 deletion mutant (Δ536-550). Cells were fixed and stained with anti-FLAG antibody for the detection of the localization of the DHX33 deletion mutant (red) and anti-UBF antibody (green) as a nucleolar marker by immunofluorescence. (F) BT549 cells were infected by a lentivirus encoding either wild-type (WT) DHX33 or its deletion mutant (Δ536-550). Whole-cell lysates were prepared at 4 days postinfection and analyzed by Western blotting with the indicated antibodies. (G) The above-mentioned cells were fixed and subjected to ChIP analysis. Equal amounts of cell lysate were immunoprecipitated with either anti-FLAG antibody or mouse IgG. (Error bars are taken from data from 2 independent experiments.)
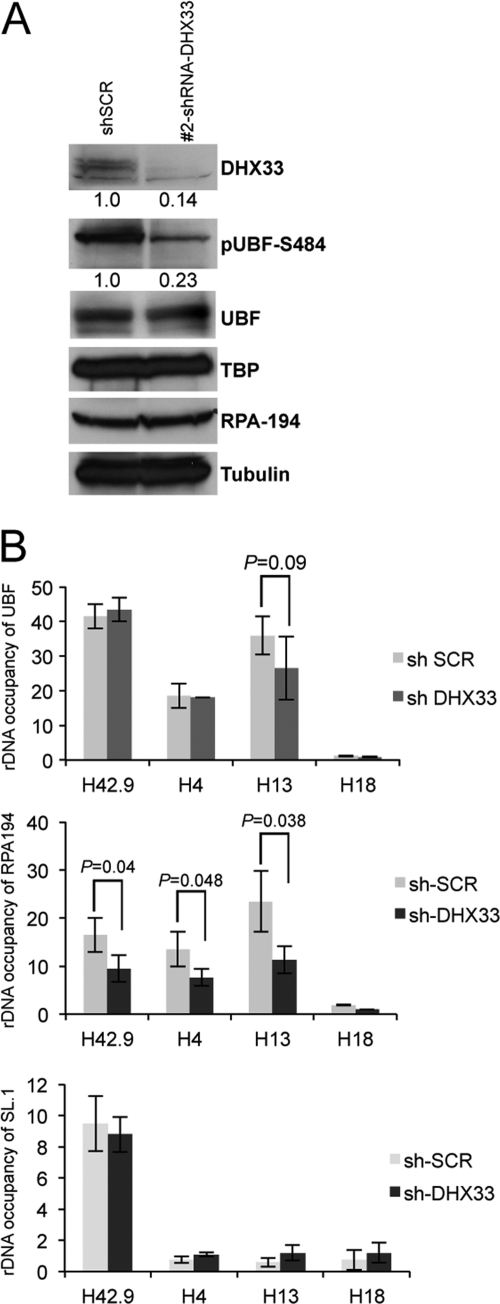
DHX33 deficiency reduces RPA194 recruitment to the rDNA locus.
UBF is a critical player in the transcription of pre-rRNA. Its phosphorylation has been shown to be required for its activation and participation in RNA polymerase I transcription (38, 42, 45). We found that the DHX33 knockdown significantly reduced the phosphorylation of UBF on Ser484 without affecting the total cellular expression levels of UBF (Fig. 6 A). Moreover, the phosphorylation of UBF on Ser484 was shown previously to be required for the recruitment of RNA polymerase I proteins to the rDNA locus (44). In DHX33 knockdown cells, RPA194, a core component of RNA polymerase I (11), displayed no reduction in expression levels (Fig. 6A). However, RPA194 exhibited more than a 50% reduction in its rDNA binding activity, as measured by chromatin immunoprecipitation (Fig. 6B), implying that in the absence of DHX33, the UBF phosphorylation level was reduced, with a concomitant failure to recruit RPA194 to rDNA loci. As parallel controls in the analysis, we also analyzed rDNA occupancy for UBF and SL.1 across the promoter/transcribed region. We found no significant change for UBF and SL.1 rDNA occupancy between scrambled control and DHX33 knockdown cells, indicating that the DHX33 knockdown specifically reduced RNA Pol I loading at rDNA (Fig. 6B).
Fig. 6.
DHX33 deficiency reduces RPA194 recruitment to the rDNA locus. (A) BJ cells were infected with the indicated lentiviruses, and whole-cell lysates were subjected to Western blot analysis using antibodies recognizing the indicated proteins. (B) Equal amounts of the cell lysates described above were subjected to ChIP analysis using anti-RPA194 or anti-SL.1 and anti-UBF antibodies, IgG, and the indicated rDNA primer sets (H42.9, H4, H13, and H18). The fold change for each antibody or IgG is shown across the rDNA locus. Bars represent standard deviations from 3 independent experiments.
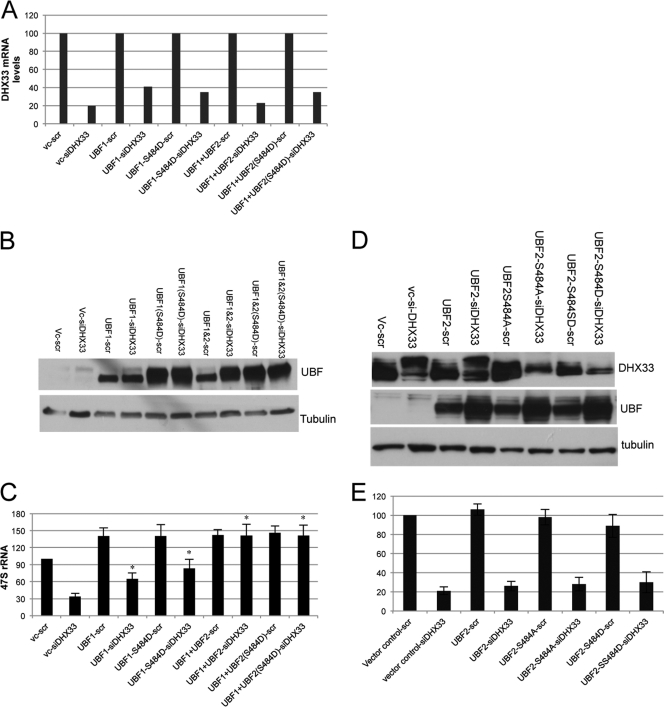
To further explore the mechanism for the reduced pre-rRNA synthesis in DHX33 knockdown BJ cells, we hypothesized that the phosphorylation of UBF resided downstream of DHX33 and upstream of efficient rDNA transcription. Thus, we rescued levels of phosphorylated UBF at Ser484 with phosphomimetic mutants of both UBF1 and UBF2 in DHX33 knockdown cells. UBF2 is a splice variant of UBF1 but has a truncated second high-mobility-group (HMG) box and has a decidedly weaker activity than UBF1 in bending DNA (19, 36). Knockdown levels for DHX33 were precisely evaluated by qPCR analysis after normalization with GAPDH mRNA (Fig. 7 A). Overexpression levels of UBF were analyzed by Western blotting (Fig. 7B and D). We found that in DHX33 knockdown cells, the weaker UBF2 (S484D) mutant did not restore rRNA transcription (Fig. 7E). However, the overexpression of UBF1 (wild type and S484D) was able to restore 47S rRNA levels approximately 70 to 80% in DHX33 knockdown cells (Fig. 7C). When both UBF1 and UBF2 (wild type and S484D) were transduced into DHX33 knockdown cells, Pol I transcription was restored (Fig. 7C) to a level comparable to that of the scrambled shRNA control, indicating that saturating the system with large amounts of UBF overcomes the deleterious effects of the DHX33 knockdown.
Fig. 7.
Overexpression of UBF restores 47S rRNA levels in the absence of DHX33. (A) BJ cells were first infected with the indicated lentiviruses encoding the empty vector or UBF1/UBF2 (wild type or S484 mutant) and then infected with a lentivirus encoding either scrambled or shRNA-DHX33 lentivirus. Total RNA was prepared 4 days after the last infection, and the total mRNA level of DHX33 is shown after normalization to GAPDH mRNA levels. vc, vector control. (B) Whole-cell lysates were prepared from the above-mentioned cells and were subjected to Western blot analysis with anti-UBF and anti-tubulin antibodies. (C). RNA polymerase I transcriptional activity was detected by qRT-PCR analysis of human 47S rRNA transcripts from equal amounts of total RNA samples (*, P ≤ 0.01 for n = 3). (D) BJ cells were first infected with the indicated lentiviruses encoding the empty vector or UBF2 (wild type or S484 mutants) and then infected with a lentivirus encoding either scrambled or shRNA-DHX33 lentivirus. Whole-cell lysates were prepared 4 days after the second infections and were subjected to Western blotting with the indicated antibodies. (E) RNA polymerase I transcriptional activity was detected by qRT-PCR analysis of human 47S rRNA transcripts from equal amounts of total RNA samples.
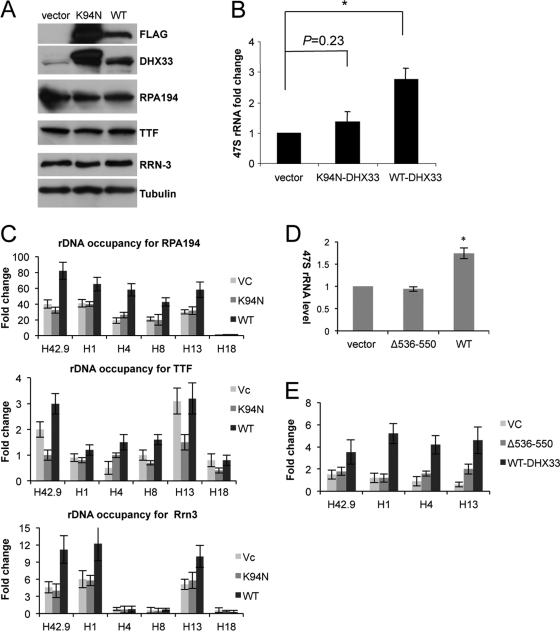
DHX33 overexpression increases RNA polymerase I-mediated transcription, a process that requires DHX33 rDNA binding activity.
To more directly determine the involvement of DHX33 in RNA polymerase I-mediated transcription, we utilized lentiviral infection to overexpress DHX33 in human BT549 cells. Western blot analysis showed the overexpression of both wild-type DHX33 and its NTPase-dead mutant (K94N) (Fig. 8 A). However, only wild-type DHX33 stimulated 47S rRNA synthesis (3-fold) (Fig. 8B), further demonstrating the positive role of DHX33 and its NTPase activity in rRNA transcription. To study the mechanism of the DHX33-mediated enhancement of rRNA synthesis, we performed ChIP analyses for cells overexpressing wild-type DHX33, an empty vector, or K94N mutant DHX33. Wild-type but not K94N mutant DHX33 increased the rDNA occupancy of RNA Pol I 2-fold (Fig. 8C). Additionally, we measured the rDNA occupancy of Rrn-3, a transcription-competent subunit of the RNA polymerase I complex, and transcription termination factor (TTF) and found that the rDNA occupancy of RRN-3 was also increased by 2-fold in the presence of wild-type DHX33 but not the K94N mutant. We found no significant change of rDNA occupancy for TTF-1 in the presence of either wild-type or K94N mutant DHX33 (Fig. 8C).
Fig. 8.
DHX33 overexpression increases rRNA transcription. (A) BT549 cells were infected with lentiviruses encoding FLAG-tagged wild-type (WT) DHX33, K94N mutant DHX33, or an empty vector. Cell lysates were analyzed for DHX33 expression by Western blot analysis using antibodies recognizing the FLAG epitope, DHX33, RPA194, TTF, Rrn-3, and tubulin. (B) BT549 cells were selected in hygromycin for 3 days and harvested 48 h after selection for total RNA extraction. RNA polymerase I transcriptional activity was detected by qRT-PCR analysis of human 47S rRNA transcripts (*, P ≤ 0.01 for n = 3). (C) The BT549 cells described above were fixed and immunoprecipitated for ChIP analysis with anti-RPA194, anti-Rrn, and anti-TTF antibodies and IgG. The indicated primer sets (H42.9, H1, H4, H8, H13, and H18) were used for PCR amplifications. Error bars are taken from 2 independent experiments. (D) BT549 cells were infected with lentiviruses encoding FLAG-tagged wild-type (WT) DHX33 or Δ536-550 mutant DHX33. Total RNA was extracted. RNA polymerase I transcriptional activity was detected by qRT-PCR analysis of human 47S rRNA transcripts from equal amounts of total RNA samples (*, P ≤ 0.01 for n = 3). (E) The above-mentioned cells were fixed and subjected to ChIP analysis with anti-RPA194 antibody. Equal amounts of cell lysates were used for immunoprecipitation, and data presented are fold enrichments of anti-RPA194/IgG across the indicated rDNA loci. Error bars are taken from data from 3 independent experiments.
The overexpression of the DHX33 Δ536-550 DNA binding mutant caused a significant reduction in rRNA synthesis compared to that of the wild-type DHX33 control (Fig. 8D), which correlated with a severe inhibition of RNA polymerase I recruitment to rDNA loci (Fig. 8E). This further supports the idea that DHX33 binds to the rDNA locus through its DNA binding domain to recruit RNA Pol I and promote rRNA transcription.
An NTPase-defective mutant of DHX33 (K94R) acts in a dominant negative manner to inhibit rRNA synthesis.
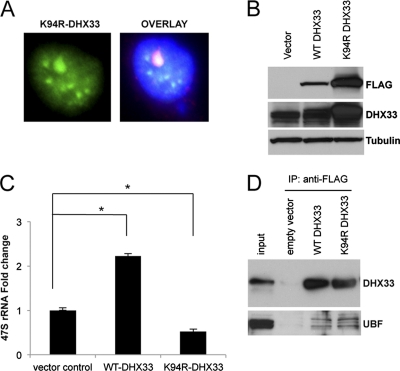
To provide more evidence for the participation of DHX33 and the importance of its NTPase activity in rRNA transcription, we generated an additional NTPase-defective mutation of DHX33, K94R. The DHX33 K94R mutant localized properly to nucleoli (Fig. 9 A). BT549 cells were transduced with a lentivirus expressing either the wild type or the DHX33 K94R mutant (Fig. 9B) and were assayed for rRNA transcription. We found that wild-type DHX33 was able to enhance rRNA synthesis by more than 2-fold, while the K94R mutant inhibited rRNA synthesis by 2-fold compared to cells transduced with an empty vector (Fig. 9C), indicating that endogenous DHX33 was competitively inhibited by the overexpressed DHX33 K94R mutant. We hypothesized that mutant DHX33 might therefore bind equally well to UBF. Indeed, the coimmunoprecipitation of UBF with FLAG-tagged wild-type and mutant DHX33 showed that the K94R mutant of DHX33 bound to UBF as well as wild-type DHX33 (Fig. 9D), providing further mechanistic evidence that the NTPase activity of DHX33 is critical for its function in rRNA synthesis.
Fig. 9.
A helicase-defective mutant of DHX33 inhibits endogenous rRNA synthesis in a dominant negative manner. (A) BT549 cells were infected with a lentivirus encoding either wild-type DHX33, the K94R DHX33 mutant, or an empty vector. Cells that were infected with the K94R DHX33 mutant were fixed and stained with anti-FLAG antibody to check for the localization of mutant DHX33 (green) and UBF (red). Nuclei were demarcated with DAPI, and the overlaid image is shown. The DHX33 K94R mutant colocalized with UBF, as shown in yellow in the nucleolus. (B) The BT549 cells described above were harvested for whole-cell extraction. Equal amounts of protein lysate were analyzed by Western blotting to detect the expression of the wild type or the DHX33 K94R mutant with the indicated antibodies. (C) The BT549 cells described above were harvested as described above for total RNA extraction. Endogenous rRNA transcription was monitored by qPCR analysis of human 47S rRNA transcript levels (P < 0.001 [n = 3]). (D) The BT549 cells described above were immunoprecipitated with anti-FLAG tag antibody and immunoblotted with the indicated antibodies.
DHX33 is required for maintenance of nucleolar integrity.
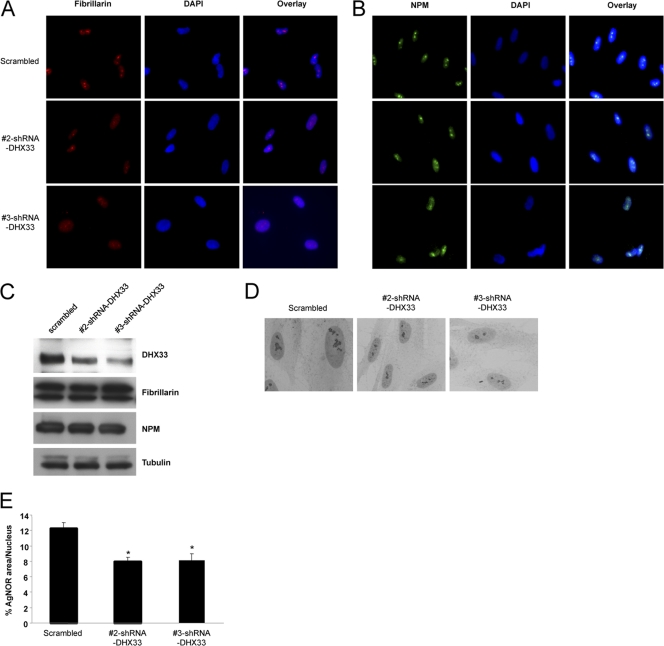
Previous studies have shown that the transcriptional repression of rRNA synthesis alters the normal nucleolar structure (33). To establish the consequences of the DHX33 knockdown on nucleolar morphology, two different nucleolar markers, nucleophosmin (NPM) and fibrillarin, were used to mark the nucleolus after DHX33 knockdown in BJ fibroblasts. NPM- and fibrillarin-marked nucleoli became less distinct and much smaller following the DHX33 knockdown (Fig. 10 A). We verified that the contraction of nucleoli was not due to decreased protein expression levels for either NPM or fibrillarin (Fig. 10B). Furthermore, silver staining for nucleolar organizing regions (30) was also performed on both control and DHX33 knockdown cells to further document changes in nucleolar morphology. Following the DHX33 knockdown (Fig. 10C), the nuclear area occupied by AgNOR staining was significantly reduced (Fig. 10D), consistent with previous studies linking changes in pre-rRNA transcription with nucleolar morphology (5).
Fig. 10.
Altered nucleolar morphology in the absence of DHX33. (A and B) BJ cells infected with the indicated shRNA-encoding lentiviruses were subjected to immunofluorescence microscopy with either antifibrillarin (A) or anti-NPM (B) antibody. A typical image is shown for each sample. (C) The above-mentioned cells were harvested, and whole-cell lysates were analyzed by Western blotting using antibodies recognizing DHX33, fibrillarin, NPM, and tubulin. (D) Infected BJ cells were fixed and stained with silver. Silver-stained nucleoli were visualized by phase-contrast microscopy. (E) Quantitation of silver-stained nucleolar area versus total nuclear area for >100 cells, given as percentages by use of MetaMorph software (*, P ≤ 0.001 for n = 3).
DHX33 is essential for cell growth, with a DHX33 deficiency inducing p53-dependent cell cycle arrest.
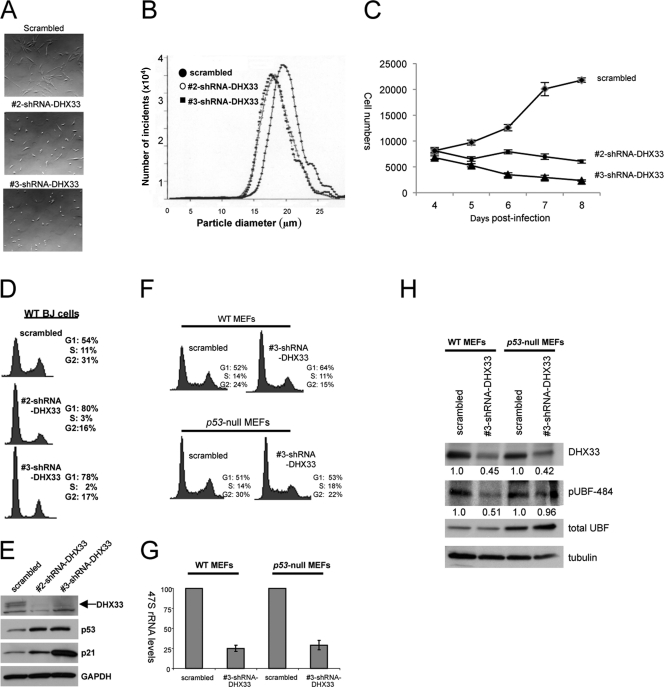
During our initial RNA interference screening, we observed changes in cellular morphology in BJ cells transduced with shRNAs targeting DHX33. Moreover, previous studies have shown clear links between nucleolar integrity and p53 activation, with altered nucleoli resulting in a p53-dependent stress response (31). Compared to control cells, DHX33 knockdown cells were significantly smaller (Fig. 11 A). This diminished size was further verified by measuring the diameter of individual cells. BJ cells transduced with scrambled control shRNAs measured 19.6 μm in diameter, while transduction with shRNA 2 and shRNA 3 targeting DHX33 resulted in cell diameters of 18.1 and 17.9 μm, respectively (P ≤ 0.05) (Fig. 11B). These findings correlate with our initial screen, which identified DHX33 expression as a requirement for maintaining 47S rRNA transcript levels (Fig. 11C); reduced levels of rRNA transcripts would predict a commensurate reduction in the overall cell size.
Fig. 11.
DHX33 is essential for cell growth, and DHX33 deficiency induces cell cycle arrest. (A) BJ cells were infected with lentiviruses encoding scrambled or two DHX33 short hairpins. Live cells were visualized 5 days after lentiviral infection. (B) The infected cells described above were trypsinized and subjected to size analysis using the Coulter method. (C) BJ cells were infected with two different shRNA-DHX33 lentiviruses, selected for 3 days, replated (7.5 × 103 cells), and manually counted daily for 5 days. (D) The BJ cells described above were trypsinized and fixed with 75% ethanol. Cell cycle analysis were performed with a FACSCalibur instrument after propidium iodide staining. (E) Whole-cell lysates from the above-mentioned BJ cells were subjected to Western blot analysis using antibodies recognizing DHX33, p53, p21, and tubulin. (F) Wild-type MEFs (WT MEFs) or _p53_-null MEFs were infected with lentiviruses encoding scrambled or DHX33 short hairpin RNA. Cells were trypsinized and subjected to cell cycle analysis at 5 days postinfection. (G) Total RNAs were extracted from the above-described infected cells and analyzed for 47S pre-rRNA by qRT-PCR based on 100 ng of total RNA and normalized to the scrambled control of wild-type MEFs. Bars represent standard deviations from 3 separate experiments. (H) Total cell lysates were prepared from the above-mentioned BJ cells and subjected to Western blot analysis using antibodies recognizing DHX33, phospho-UBF (Ser484), UBF, and tubulin.
Recent studies have emphasized a new role for the nucleolus as a sensor of cellular stress (21, 22, 31, 50). In this context, a disruption of rRNA synthesis results in a prototypical p53 response, often triggering cell cycle arrest (29, 31). We sought next to determine whether the loss of DHX33 would trigger a similar response. Proliferation curves of human primary fibroblast cells were generated following infection with lentiviruses encoding shRNAs targeting DHX33 (Fig. 11C). Cell proliferation was reduced upon the DHX33 knockdown (Fig. 11C). The DHX33 reduction also induced a significant G1 cell cycle arrest following FACS analysis of transduced BJ cells (Fig. 11D).
The reduction in DHX33 expression levels in BJ fibroblasts resulted in a significant induction of both p53 and p21 protein expression (Fig. 11E). To determine whether the cell cycle arrest following the DHX33 loss was solely p53 dependent, we infected _p53_-null mouse embryonic fibroblasts (MEFs), compared to wild-type MEFs, with lentiviruses encoding shRNAs targeting DHX33. For _p53_-null MEFs, we did not observe any significant differences in the cell cycle profiles between scrambled and DHX33 shRNA-infected cells (Fig. 11F) despite our ability to knock down DHX33 by 50% (Fig. 11H). Additionally, we did not observe a decrease in UBF Ser484 phosphorylation in _p5_3-null MEFs exhibiting a reduction in DHX33 protein expression levels (Fig. 11H), suggesting that the inhibition of UBF phosphorylation upon the loss of DHX33 is p53 dependent and is an indirect result of cell cycle arrest following the depletion of DHX33. This further supports the notion that DHX33 is essential for pre-rRNA synthesis regardless of the UBF phosphorylation status. Previous reports have shown that p53 induction can repress RNA polymerase I-mediated transcription under certain conditions (4, 52). However, in _p53_-null MEFs, the synthesis of 47S rRNA was still decreased by up to 75% upon the DHX33 knockdown (Fig. 11G), indicating that the attenuation of 47S rRNA synthesis following the loss of DHX33 is p53 independent.
DISCUSSION
The DEAD/DEAH-box protein family consists of 57 members; family members are encoded as relatively large proteins of more than 400 amino acids in length. In addition to a core helicase domain, there are also extending N-terminal and C-terminal domains that are thought to be involved in protein-protein/RNA interactions (20). Although generally believed to contain probable RNA helicase/unwindase activity, some members of the DDX/DHX family, such as DHX9 (also called RNA helicase A), have also been shown to bind DNA and function as a DNA helicase (53). In fact, DEAH RNA helicases contain some degree of structural homology to DNA helicases (13). Other than their functions in general RNA metabolism, including RNA splicing, translation, rRNA processing, and mRNA decay, some DDX/DHX proteins, including DHX9 (26), DDX5/DDX17 (46, 48), DHX20 (49), and DDX21 (47), have been found to play important roles in transcriptional regulation (8). Specifically, DHX9 has been shown to bind directly to RNA polymerase II through the concerted efforts of its helicase core domain and N-terminal domain (26). Other DDX proteins, such as DDX5, DDX20, and DDX21, can function as transcriptional coactivators or repressors through direct protein-protein interactions (8). Our large-scale screen in this study focused on important players in RNA polymerase I transcription in the nucleolus, a cellular activity which none of the DDX/DHX family of proteins had been associated with to date. Within the entire DEAD/DEAH-box protein family, over half of all DDX/DHX proteins localize to the nucleolus, making them prime candidates for RNA polymerase I-mediated transcriptional regulation.
The regulation of RNA polymerase I transcription succumbs to multiple forms of intracellular stress or perturbations in signaling events involved in cell growth (10). Additionally, the nucleolus can act as a sensor of DNA damage, nutrient availability, and stress signals to shut down rRNA synthesis (10, 21, 31). In this report, we observed decreases in levels of rRNA synthesis after knocking down many members of the nucleolar DEAD/DEAH-box protein family. Whether all of these proteins are directly required for RNA polymerase I-mediated transcription remains a critical question. Given the feedback loop between cellular stress and reduced nucleolar function, it is possible that a reduced expression level of DEAD/DEAH proteins elicits a stress signal that ultimately reduces rRNA synthesis and cell growth.
Our findings provide a direct link between critical roles of DHX33 in cell growth and rRNA synthesis. The DHX33 knockdown exhibited a great degree of rRNA synthesis inhibition (10-fold) in relation to the extent of the knockdown achieved for the entire nucleolar helicase family screened. This drastic degree of reduction of rRNA synthesis upon a DHX33 deficiency implicates its pivotal role in RNA polymerase I-mediated transcription. We have shown that DHX33 participates in rRNA transcription through its interaction with one of the key players in RNA polymerase I-mediated transcription, UBF, an rDNA architectural protein.
Ser484 of UBF is phosphorylated by two G1-specific protein kinase holoenzymes, cyclin D1-CDK4 and cyclin E-CDK2 (44). We originally postulated that the loss of DHX33, which induced a significant G1 cell cycle arrest in wild-type cells, might result in a loss of UBF phosphorylation through an indirect means. Specifically, G1-arrested DHX33 knockdown cells should exhibit decreased CDK4 and CDK2 activities, thus reducing UBF Ser484 phosphorylation. Moreover, we showed that the loss of DHX33 triggered a potent p53 response with a rapid induction of p21, a well-known inhibitor of CDK2 activity (12). However, we demonstrated that the DHX33 loss in _p53_-null MEFs did not cause a reduction of UBF Ser484 phosphorylation but still diminished rRNA transcription levels without cell cycle arrest. Thus, it appears more likely that DHX33 exerts a more direct effect on rRNA transcription through influencing the rDNA occupancy of RNA polymerase I, an effect that we have also shown requires DHX33's NTPase activity.
Our data have identified a novel protein, DHX33, as a critical player in ribosome RNA transcription. We have shown that DHX33 is a nucleolar chromatin binding protein, where it associates with UBF and rDNA loci. UBF, as an rDNA chromatin-modulating protein, binds across the entire rDNA promoter and the transcribed as well as the nontranscribed regions to facilitate changes in the rDNA conformation in order to fully activate rRNA transcription. Our findings are consistent with the idea that DHX33, through both its NTPase activity and DNA binding, might facilitate the conformational change of rDNA through the hydrolysis of ATP. Thus, DHX33 would act as a crucial interaction partner for UBF during this essential process in order to promote rDNA transcription. Further elucidation of the extent of the regulation imposed on the cell by this family of RNA helicases could deepen our understanding of several essential biological processes.
ACKNOWLEDGMENTS
We thank the members of the Weber laboratory for their advice and technical assistance. The Genome Institute and Children's Discovery Institute at Washington University provided lentiviral RNAi library constructs.
J.T.F. was supported by NIH grant 5T32 GM007067. A.P.M. was supported by Department of Defense Breast Cancer Research Program award X81XWH-08-BCRP-PREDOC. This work was supported by NIH grant CA120436 and an Era of Hope scholar award in breast cancer research (BC007304) to J.D.W.
Footnotes
▿
Published ahead of print on 19 September 2011.
REFERENCES
- 1.Andersen J. S., et al. 2002. Directed proteomic analysis of the human nucleolus. Curr. Biol. 12:1–11 [DOI] [PubMed] [Google Scholar]
- 2.Bell S. P., Learned R. M., Jantzen H. M., Tjian R. 1988. Functional cooperativity between transcription factors UBF1 and SL1 mediates human ribosomal RNA synthesis. Science 241:1192–1197 [DOI] [PubMed] [Google Scholar]
- 3.Bleichert F., Baserga S. J. 2007. The long unwinding road of RNA helicases. Mol. Cell 27:339–352 [DOI] [PubMed] [Google Scholar]
- 4.Budde A., Grummt I. 1999. p53 represses ribosomal gene transcription. Oncogene 18:1119–1124 [DOI] [PubMed] [Google Scholar]
- 5.Caperta A. D., Neves N., Viegas W., Pikaard C. S., Preuss S. 2007. Relationships between transcription, silver staining, and chromatin organization of nucleolar organizers in Secale cereale. Protoplasma 232:55–59 [DOI] [PubMed] [Google Scholar]
- 6.Chen D., Belmont A. S., Huang S. 2004. Upstream binding factor association induces large-scale chromatin decondensation. Proc. Natl. Acad. Sci. U. S. A. 101:15106–15111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cui C., Tseng H. 2004. Estimation of ribosomal RNA transcription rate in situ. Biotechniques 36:134–138 [DOI] [PubMed] [Google Scholar]
- 8.Fuller-Pace F. V. 2006. DExD/H box RNA helicases: multifunctional proteins with important roles in transcriptional regulation. Nucleic Acids Res. 34:4206–4215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghoshal K., Jacob S. T. 1994. Specific inhibition of pre-ribosomal RNA processing in extracts from the lymphosarcoma cells treated with 5-fluorouracil. Cancer Res. 54:632–636 [PubMed] [Google Scholar]
- 10.Grummt I. 2003. Life on a planet of its own: regulation of RNA polymerase I transcription in the nucleolus. Genes Dev. 17:1691–1702 [DOI] [PubMed] [Google Scholar]
- 11.Grummt I. 1999. Regulation of mammalian ribosomal gene transcription by RNA polymerase I. Prog. Nucleic Acid Res. Mol. Biol. 62:109–154 [DOI] [PubMed] [Google Scholar]
- 12.Harper J. W., Adami G. R., Wei N., Keyomarsi K., Elledge S. J. 1993. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 75:805–816 [DOI] [PubMed] [Google Scholar]
- 13.He Y., Andersen G. R., Nielsen K. H. Structural basis for the function of DEAH helicases. EMBO Rep. 11:180–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jantzen H. M., Chow A. M., King D. S., Tjian R. 1992. Multiple domains of the RNA polymerase I activator hUBF interact with the TATA-binding protein complex hSL1 to mediate transcription. Genes Dev. 6:1950–1963 [DOI] [PubMed] [Google Scholar]
- 15.Kermekchiev M., Workman J. L., Pikaard C. S. 1997. Nucleosome binding by the polymerase I transactivator upstream binding factor displaces linker histone H1. Mol. Cell. Biol. 17:5833–5842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim S. H., Smith J., Claude A., Lin R. J. 1992. The purified yeast pre-mRNA splicing factor PRP2 is an RNA-dependent NTPase. EMBO J. 11:2319–2326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klein J., Grummt I. 1999. Cell cycle-dependent regulation of RNA polymerase I transcription: the nucleolar transcription factor UBF is inactive in mitosis and early G1. Proc. Natl. Acad. Sci. U. S. A. 96:6096–6101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kruhlak M., et al. 2007. The ATM repair pathway inhibits RNA polymerase I transcription in response to chromosome breaks. Nature 447:730–734 [DOI] [PubMed] [Google Scholar]
- 19.Kuhn A., et al. 1994. Functional differences between the two splice variants of the nucleolar transcription factor UBF: the second HMG box determines specificity of DNA binding and transcriptional activity. EMBO J. 13:416–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Linder P. 2006. Dead-box proteins: a family affair—active and passive players in RNP-remodeling. Nucleic Acids Res. 34:4168–4180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maggi L. B., Jr., Weber J. D. 2005. Nucleolar adaptation in human cancer. Cancer Invest. 23:599–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mayer C., Bierhoff H., Grummt I. 2005. The nucleolus as a stress sensor: JNK2 inactivates the transcription factor TIF-IA and down-regulates rRNA synthesis. Genes Dev. 19:933–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McStay B., Frazier M. W., Reeder R. H. 1991. xUBF contains a novel dimerization domain essential for RNA polymerase I transcription. Genes Dev. 5:1957–1968 [DOI] [PubMed] [Google Scholar]
- 24.Melese T., Xue Z. 1995. The nucleolus: an organelle formed by the act of building a ribosome. Curr. Opin. Cell Biol. 7:319–324 [DOI] [PubMed] [Google Scholar]
- 25.Moss T., Stefanovsky V. Y. 2002. At the center of eukaryotic life. Cell 109:545–548 [DOI] [PubMed] [Google Scholar]
- 26.Nakajima T., et al. 1997. RNA helicase A mediates association of CBP with RNA polymerase II. Cell 90:1107–1112 [DOI] [PubMed] [Google Scholar]
- 27.Panov K. I., Friedrich J. K., Russell J., Zomerdijk J. C. 2006. UBF activates RNA polymerase I transcription by stimulating promoter escape. EMBO J. 25:3310–3322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pause A., Sonenberg N. 1992. Mutational analysis of a DEAD box RNA helicase: the mammalian translation initiation factor eIF-4A. EMBO J. 11:2643–2654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pestov D. G., Strezoska Z., Lau L. F. 2001. Evidence of p53-dependent cross-talk between ribosome biogenesis and the cell cycle: effects of nucleolar protein Bop1 on G(1)/S transition. Mol. Cell. Biol. 21:4246–4255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roussel P., Hernandez-Verdun D. 1994. Identification of Ag-NOR proteins, markers of proliferation related to ribosomal gene activity. Exp. Cell Res. 214:465–472 [DOI] [PubMed] [Google Scholar]
- 31.Rubbi C. P., Milner J. 2003. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J. 22:6068–6077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwer B. 2001. A new twist on RNA helicases: DExH/D box proteins as RNPases. Nat. Struct. Biol. 8:113–116 [DOI] [PubMed] [Google Scholar]
- 33.Shav-Tal Y., et al. 2005. Dynamic sorting of nuclear components into distinct nucleolar caps during transcriptional inhibition. Mol. Biol. Cell 16:2395–2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith S. D., et al. 1990. Interaction of RNA polymerase I transcription factors with a promoter in the nontranscribed spacer of rat ribosomal DNA. Nucleic Acids Res. 18:1677–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stefanovsky V., Langlois F., Gagnon-Kugler T., Rothblum L. I., Moss T. 2006. Growth factor signaling regulates elongation of RNA polymerase I transcription in mammals via UBF phosphorylation and r-chromatin remodeling. Mol. Cell 21:629–639 [DOI] [PubMed] [Google Scholar]
- 36.Stefanovsky V. Y., Moss T. 2008. The splice variants of UBF differentially regulate RNA polymerase I transcription elongation in response to ERK phosphorylation. Nucleic Acids Res. 36:5093–5101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stefanovsky V. Y., Pelletier G., Bazett-Jones D. P., Crane-Robinson C., Moss T. 2001. DNA looping in the RNA polymerase I enhancesome is the result of non-cooperative in-phase bending by two UBF molecules. Nucleic Acids Res. 29:3241–3247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stefanovsky V. Y., et al. 2001. An immediate response of ribosomal transcription to growth factor stimulation in mammals is mediated by ERK phosphorylation of UBF. Mol. Cell 8:1063–1073 [DOI] [PubMed] [Google Scholar]
- 39.Sugimoto M., Kuo M. L., Roussel M. F., Sherr C. J. 2003. Nucleolar Arf tumor suppressor inhibits ribosomal RNA processing. Mol. Cell 11:415–424 [DOI] [PubMed] [Google Scholar]
- 40.Tanner N. K. 2003. The newly identified Q motif of DEAD box helicases is involved in adenine recognition. Cell Cycle 2:18–19 [DOI] [PubMed] [Google Scholar]
- 41.Tanner N. K., Linder P. 2001. DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol. Cell 8:251–262 [DOI] [PubMed] [Google Scholar]
- 42.Tuan J. C., Zhai W., Comai L. 1999. Recruitment of TATA-binding protein-TAFI complex SL1 to the human ribosomal DNA promoter is mediated by the carboxy-terminal activation domain of upstream binding factor (UBF) and is regulated by UBF phosphorylation. Mol. Cell. Biol. 19:2872–2879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Venema J., Tollervey D. 1999. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 33:261–311 [DOI] [PubMed] [Google Scholar]
- 44.Voit R., Hoffmann M., Grummt I. 1999. Phosphorylation by G1-specific cdk-cyclin complexes activates the nucleolar transcription factor UBF. EMBO J. 18:1891–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Voit R., Kuhn A., Sander E. E., Grummt I. 1995. Activation of mammalian ribosomal gene transcription requires phosphorylation of the nucleolar transcription factor UBF. Nucleic Acids Res. 23:2593–2599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Watanabe M., et al. 2001. A subfamily of RNA-binding DEAD-box proteins acts as an estrogen receptor alpha coactivator through the N-terminal activation domain (AF-1) with an RNA coactivator, SRA. EMBO J. 20:1341–1352 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 47.Westermarck J., et al. 2002. The DEXD/H-box RNA helicase RHII/Gu is a co-factor for c-Jun-activated transcription. EMBO J. 21:451–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wilson B. J., et al. 2004. The p68 and p72 DEAD box RNA helicases interact with HDAC1 and repress transcription in a promoter-specific manner. BMC Mol. Biol. 5:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yan X., Mouillet J. F., Ou Q., Sadovsky Y. 2003. A novel domain within the DEAD-box protein DP103 is essential for transcriptional repression and helicase activity. Mol. Cell. Biol. 23:414–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yao Z., et al. 2010. B23 acts as a nucleolar stress sensor and promotes cell survival through its dynamic interaction with hnRNPU and hnRNPA1. Oncogene 29:1821–1834 [DOI] [PubMed] [Google Scholar]
- 51.Young D. W., et al. 2007. Mitotic occupancy and lineage-specific transcriptional control of rRNA genes by Runx2. Nature 445:442–446 [DOI] [PubMed] [Google Scholar]
- 52.Zhai W., Comai L. 2000. Repression of RNA polymerase I transcription by the tumor suppressor p53. Mol. Cell. Biol. 20:5930–5938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou K., et al. 2003. RNA helicase A interacts with dsDNA and topoisomerase IIalpha. Nucleic Acids Res. 31:2253–2260 [DOI] [PMC free article] [PubMed] [Google Scholar]