microRNA Therapeutics (original) (raw)
. Author manuscript; available in PMC: 2012 Jun 1.
Published in final edited form as: Gene Ther. 2011 Apr 28;18(12):1104–1110. doi: 10.1038/gt.2011.50
Abstract
MicroRNAs (miRNAs) provide new therapeutic targets for many diseases, while their myriad roles in development and cellular processes make them fascinating to study. We still do not fully understand the molecular mechanisms by which miRNAs regulate gene expression nor do we know the complete repertoire of mRNAs each miRNA regulates. However, recent progress in the development of effective strategies to block miRNAs suggests that anti-miRNA drugs may soon be used in the clinic.
Keywords: miRNA, antagomir, anti-miR, antisense, ASO, Argonaute, RNA silencing, miRNA inhibition, miRNA replacement
MicroRNAs (miRNAs) are 21-23 nt long RNAs that direct Argonaute proteins to bind to and repress complementary mRNA targets. The human genome contains more than 500 miRNAs, and each miRNA can repress hundreds of genes, regulating almost every cellular process1,2. Individual miRNAs are often produced only in specific cell types or developmental stages.
Inappropriate miRNA expression has been linked to a variety of diseases3,4. For example, the _let_-7 miRNA prevents proliferation of cancer stem cells. miRNAs have roles in metabolic diseases such as obesity and diabetes; differentiation of adipocytes is promoted by miR-143 and insulin secretion is regulated by miR-375 in pancreatic-islet cells. Mutation of just a single nucleotide in the sequence of a miRNA or its mRNA target can eliminate target regulation. Mutation of the fifth nucleotide of miR-96 is associated with autosomal dominant, progressive, high-frequency hearing loss in humans; the mutation decreases the levels of miR-96 and impairs target mRNA repression5. A different mutation in miR-96 was discovered in a mouse mutant with hair cell defects and progressive hearing loss6. In contrast to mutation of miRNAs, normal miR-122 participates in the development of liver disease: hepatitis C virus (HCV) hijacks this miRNA, making miR-122 required for HCV to replicate in the liver7. Some viruses express their own miRNAs, presumably to repress cellular mRNAs that would otherwise interfere with viral infection8. Tissue-specific miRNAs may also be involved in the pathogenesis of cardiovascular, muscular and neurodegenerative diseases. Thus, molecules that alter the function or abundance of specific miRNAs represent a new strategy for treating human disease.
miRNAs are transcribed by RNA polymerase II and matured by RNase III enzymes in two steps
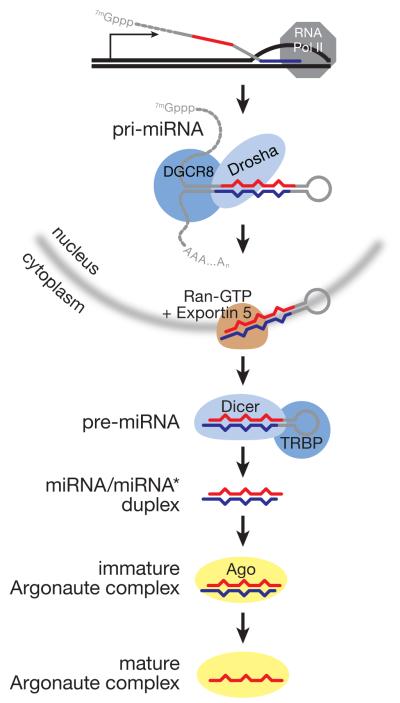
miRNAs are transcribed from their own genes by RNA polymerase II. Consequently, miRNA primary transcripts (pri-miRNAs) begin with 5′ 7-methylguanosine caps and end with 3′ poly(A) tails. The pre-miRNA, a ~65 nt stem-loop structure that contains the miRNA and its corresponding miRNA* within its stem, resides within the pri-miRNA (Figure 1). Cleavage of the pri-miRNA by the ribonuclease III (RNase III) enzyme, Drosha, releases the pre-miRNA stem-loop, which bears the 2 nt 3′ overhanging ends characteristic of RNase III enzymes. The pre-miRNA is then exported to the cytoplasm, where its loop is removed by a second, RNase III enzyme, Dicer, that specifically recognizes the pre-miRNA structure, including its 2 nt 3′ overhanging end. The resulting miRNA/miRNA* duplex is then loaded into a member of the Argonaute family of proteins. Subsequently, the miRNA* strand departs from the Argonaute protein, producing a mature, active miRNA:protein complex.
Figure 1.
miRNA biogenesis in mammals.
miRNAs provide the specificity determinants for Argonaute proteins
Binding of a miRNA:Argonaute protein complex to the 3′ untranslated region (UTR) of an mRNA silences its expression1. The human genome encodes four closely related Argonaute proteins, Ago1, Ago2, Ago3, and Ago4, and most tissues and cultured mammalian cell lines express all four, albeit in different proportions. Argonaute proteins are structural homologs of the DNA-guided ribonuclease, RNase H. Ago2 can cleave its target RNAs (after the nucleotide paired to the tenth base of the small RNA guide), but the other three human Argonaute proteins have lost the capacity for such site-specific, small RNA-directed, endonucleolytic target cleavage.
Argonaute-catalyzed target cleavage requires extensive, but not complete, complementarity between the miRNA guide and an mRNA. However, human miRNAs generally base pair only partially with their target mRNAs. In fact, as few as six base pairs between a special region of the miRNA, the “seed sequence” (miRNA nucleotides 2 through 7 or 8), and an mRNA, may suffice to recruit Argonaute to repress the mRNA (Figure 2). Consequently, most miRNAs do not direct Argonaute to slice the target mRNA, but instead the miRNA:Argonaute complex triggers general degradation of the target mRNA9. In some cases, the miRNA may block the translation of the mRNA into protein10. Because a miRNA need only pair only partially with its mRNA target, a single miRNA can repress hundreds of genes10-16. Most mRNAs contain multiple potential miRNA-binding sites in their 32 UTRs1,12. Current computational estimates suggest that more than half of all human genes are regulated by miRNAs at some time or place in human development.
Figure 2.
miRNAs bind target mRNAs via their seed sequence. Typical miRNA-binding sites also feature an adenosine (underlined) across from the first nucleotide of the miRNA, even though the structure of a miRNA bound to an Argonaute protein precludes base pairing at this position.
Viral miRNAs target cellular and viral mRNAs
Mammalian viruses produce at least 66 distinct miRNAs. Most miRNA-producing viruses are double-stranded DNA (dsDNA) viruses from the herpes virus family; no miRNAs have been detected from RNA viruses such as retroviruses or flaviviruses, or the papillomavirus, a dsDNA virus17,18. One known function of viral miRNAs is to thwart the immune response during infection by regulating the expression of the major histocompatibility complex class I chain-related molecule B (MICB), a natural killer cell ligand19. Epstein-Barr virus (EBV), Kaposi’s sarcoma-associated herpesvirus (KSHV) and human cytomegalovirus (HCMV) express miRNAs that target separate sites in the MICB mRNA to prevent its expression. Blocking these miRNAs might permit a more robust immune response to herpesvirus infection.
HCMV expresses a miRNA, miR-UL112, that targets the 32 UTR of MICB at a site overlapping the binding site for the cellular miRNA, miR-373. Interestingly, the MICB mRNA also contains nearby sites for two other cellular miRNAs, miR-376a and miR-433. After 72 hours of HCMV infection, the viral miR-UL112 and the cellular miR-376a collaborate to silence expression of MICB20. KSHV express three miRNAs, each with the same seed sequence as a cellular miRNA. One of the viral miRNAs, miR-K12-11, represses the same set of mRNAs as its cellular counterpart, miR-155.
A second function for viral miRNAs may be to repress host mRNAs so as to maintain viral latency21,22. KSHV miR-9* binds the 32 UTR of the major lytic-switch mRNA, preventing expression of the protein that controls viral reactivation from latency23. Expression of two cellular miRNAs, miR-200b and miR-429, correlates with EBV lytic gene expression. These two microRNAs cause EBV to enter the lytic phase by repressing ZEB1 and ZEB2 protein expression24. miRNA inhibitors may some day be used to coax latent viruses into the more therapeutically tractable replicating state, allowing the elimination of reservoirs that enable viral reemergence after anti-viral therapy is completed.
Artificially introducing or inhibiting miRNAs provides clues to their function
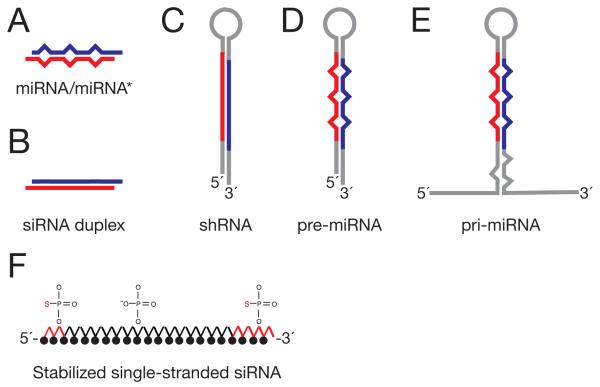
To understand the role of miRNAs in normal cellular processes and in human disease, we need tools to increase and decrease miRNA function or abundance. Expression of exogenous small RNAs in cells is possible through transient or stable transfection or viral transduction of a pri-miRNA transgene, pre-miRNA, mature miRNA/miRNA*, small interfering RNA (siRNA) or short hairpin RNA (Figure 3). Using this strategy, intratumoral injection of exogenous _let_-7 was found to block tumor development in a mouse model of non-small cell lung cancer25-28. Similarly, reintroduction of miR-26a in a mouse model of liver cancer caused regression of tumors29. The opposite strategy—targeting miRNAs for inhibition—has yielded interesting results in vivo. For example, inhibition of miRNA-132 prevented angiogenesis in an orthotopic mouse model of ovarian and breast carcinoma and inhibition of miRNA-21, a miRNA that promotes oncogenesis, led to regression of malignant pre-B lymphoid tumors30-32.
Figure 3.
miRNA replacement strategies: (A) mature miRNA/miRNA* duplex; (B) small interfering RNA duplex; (C) small hairpin RNA; (D) pre-miRNA; (E) pri-miRNA; (F) modified single stranded RNA.
Antisense oligonucleotides (ASOs) are short, single-stranded RNA or DNA molecules that bind other nucleic acids by Watson-Crick base pairing. Traditional ASOs target a specific mRNA in order to block its translation into protein (e.g., morpholino ASOs) or to trigger its destruction by recruiting RNase H to hydrolyze the RNA strand of an RNA:DNA duplex (“gapmer” ASOs). ASOs are used in vitro and in vivo to discover gene function, and some ASOs are being tested in clinical trials. ASOs were first shown to inhibit specific miRNAs in cultured cells and in invertebrates in 2004. Subsequent studies have examined various chemical modifications of ASOs to improve their in vitro and in vivo stability and to improve their in vivo delivery. Moreover, some ASO chemistries can trigger the destruction of a miRNA through a mechanism recently discovered by using miRNA inhibitors. The stability of a miRNA is determined by the Argonaute protein with which it associates and the degree of sequence complementarity between the miRNA and its target mRNA33. When a miRNA encounters a target to which it can pair extensively—including miRNA inhibitor oligonucleotides, it is tailed with adenosines or uridines and subsequently degraded. This type of regulatory mechanism, dependent on the presence of a target and the extent of pairing to a complementary RNA, is critical to strategies that aim to inhibit or replace miRNAs to discover their roles in cellular processes or pathogenic mechanisms.
Modified antisense oligonucleotides can help define the molecular function of an individual miRNA
The molecular function of an individual miRNA can be discovered by inhibiting it and measuring the resulting changes in the levels of each mRNA or protein in the cell or by evaluating other phenotypic changes, such as developmental defects, cell proliferation, organ function, lipid metabolism, or behavior. ASOs engineered to withstand degradation by extra- and intracellular nucleases can effectively inhibit miRNAs in whole animals34,35. The first ASOs used to inhibit miRNAs were composed of 22-_O_-methyl ribose-modified RNA. Such 2′-_O_-methyl oligonucleotides proved to be effective miRNA inhibitors when introduced by lipid-mediated transfection into cultured human cells or by injection in whole nematodes (Caenorhabditis elegans). Dextran-conjugated ASOs can be injected into the C. elegans germ line and block the function of a specific miRNA function in the progeny36.
“Antagomirs” were the first miRNA inhibitors demonstrated to work in mammals (Figure 3). Because the amount of pre-miRNA was unchanged by the antagomir, ASOs likely target the mature miRNA37,38. These synthetic ASOs contain 2′-_O_-methyl modified ribose sugars, terminal phosphorothioates, and at the 3′ end a cholesterol group, which helps deliver the antagomir to cells. Cholesterol conjugation causes cellular uptake of the modified nucleic acid oligonucleotide by promoting its association with high-and low-density lipoproteins that can bind cell surface membrane receptors: the Scavenger receptor BI for HDL and the LDL receptor for LDL. Intravenous injection of 80 mg of antagomir per kg mouse body weight on each of three successive days inhibited the corresponding miRNA in mouse liver, lung, kidney, heart, intestine, fat, skin, bone marrow, muscle, ovaries and adrenal glands. Nonetheless, antagomirs are unlikely to be used clinically, as they require higher doses to achieve the same efficacy as other ASO strategies.
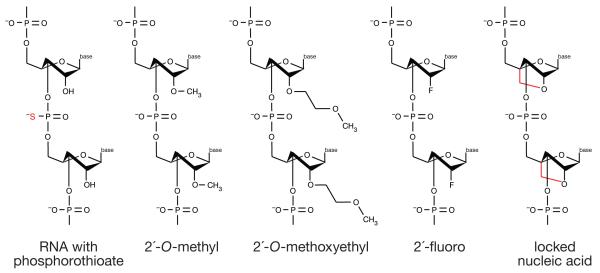
Alternative RNA chemistries (Figure 4) such as 2′-_O_-methoxyethyl (2′-MOE), 2′-fluoro, and 2′,4′ methylene (“locked nucleic acids” or LNAs) have greater affinity to bind and inhibit miRNA function in vivo than 2′-_O_-methyl RNA oligonucleotides39-41. Alternative chemistries are also more resistant to degradation. In a test of stability of modified RNAs in 10% fetal bovine serum, 2′-fluoro RNA with LNA ends was less degraded after 24 hours than a 2′-_O_-methyl RNA with LNA ends or a DNA/LNA oligonucleotide, which was degraded within 2 hours35. Phosphorothioates substitute a non-bridging oxygen atom on the phosphate group with a sulfur. Phosphorothioate bonds promote serum protein binding, thereby increasing the in vivo distribution and bioavailability of the ASO. A direct comparison of anti-miRNA oligonucleotide chemistries in vitro revealed that combining 2′-_O_-methyl and LNA with phosphorothioate ends was ~10 times more effective than the 2′-_O_-methyl or phosphorothioate modifications alone and twice as effective as the 2′-_O_-methyl with LNA modifications35.
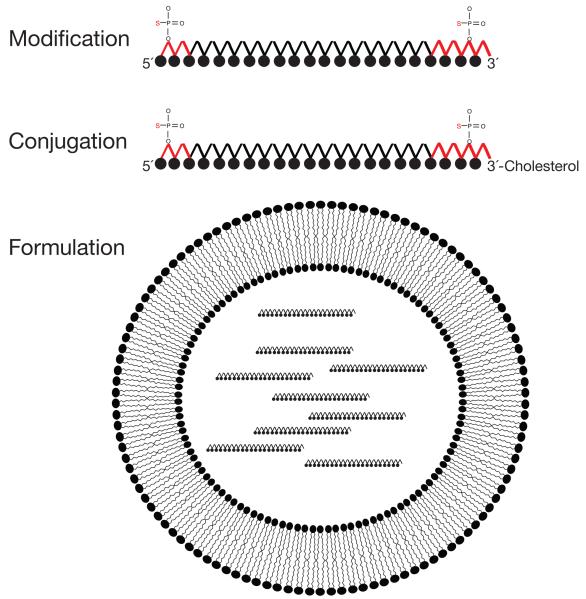
Figure 4.
Strategies for delivery of anti-miRNA oligonucleotides to cells in vivo. (A) Modification. Black filled circles represent 2′-_O_-methyl, 2′-_O_-methoxyethyl, or 2′-fluoro modified nucleotides. (B) Conjugation. Antagomirs are 2′-_O_-methyl oligonucleotides conjugated to cholesterol at their 3′ ends, and contain phosphorothioate linkages between nucleotides at both ends in place of natural phosphate linkages. (C) Formulation. Lipid nanoparticles are lipid vesicles containing therapeutic RNA. The formulated lipid bilayer encapsulates the therapeutic RNA, delivering it to cells and promoting fusion with the phospholipid bilayer of cell membranes. Individual lipids within the vesicle bilayer can contain ionizable head groups that will disrupt the endosome at low pH to release the therapeutic RNA to the cytoplasm.
The 2′,4′ methylene bridge in LNAs constrains the ribose to the C3′ endo conformation present in RNA:RNA and DNA:RNA helices. (DNA:DNA helices are C2′ endo.) LNAs cannot interconvert between the C3′ endo conformation, which favors pairing with an RNA, and the C2′ endo conformation, which does not. Consequently, an LNA modification increases RNA:RNA melting temperature by 2.4□ per modification and confers high specificity for their target sequences. Moreover, locked nucleic acids are resistant to many endonucleases. ASOs containing LNAs are effective probes for accurate detection of miRNAs by Northern blotting, in situ hybridization and, most importantly, are potent miRNA inhibitors in vivo.
The unique target mRNA-binding properties of a miRNA bound to an Argonaute protein—nearly all the binding specificity comes from the seed sequence—allow antisense miRNA inhibitors to be shorter than the miRNA itself. A 16 nt LNA-modified oligonucleotide complementary to miRNA-122 injected intravenously each day for five successive days at 10 mg/kg, lowered plasma cholesterol levels for more than 20 days in African green monkeys42. This pioneering non-human primate study established that LNA-modified anti-miRNA oligonucleotides are specific, stable, and non-toxic when administered intravenously. A subsequent study showed that Hepatitis C virus (HCV) replication could be inhibited in chimpanzees by a 15 nt LNA oligonucleotide targeting miRNA-12243. Two chimpanzees were injected intravenously with 5 mg per kg of LNA inhibitor each week for 12 weeks. Two weeks after treatment ended, viral titer was 400- and 200-times lower in serum and liver, respectively. Free anti-miR-122 LNA was detected in liver for 8 weeks after treatment ceased, until week 25, at which point the drug had declined significantly and the level of miR-122 had increased. No liver toxicity was detected, and treatment was associated with improved liver histology, presumably due to prolonged suppression of viremia and normalization of the interferon pathway. No viral escape was detected by sequence analysis of the HCV RNA target sites for miR-122 at the 16th week, in contrast to treatment with an antiviral non-nucleoside polymerase inhibitor with which resistance mutations occur after 2 days of treatment. miRNA-122 inhibition by LNA-modified oligonucleotides is now being tested in humans. A successful phase 1 trial has paved the way for a phase 2 study that will assess the safety and tolerability of weekly or bi-weekly subcutaneous injections of the anti-miR-122 LNA in 55 patients with chronic HCV genotype 1 infection.
Advancements in delivery formulations reduce the effective dose of ASOs
Delivering a therapeutic RNA to its target tissue starts with the challenge of its exiting the circulatory system into a target tissue, transiting the cell membrane, and, finally, escaping from endosomal vesicles into the cytoplasm. The size of an unconjugated therapeutic RNA is 7-20 kDa. Molecules smaller than 50 kDa are filtered by the kidneys and excreted. Transfer of therapeutic RNA from the blood to the target tissue is a challenge because anything longer than 5 nm diameter, including therapeutically complexed RNA, cannot cross the capillary endothelium and will remain in circulation until filtered by the kidneys. Local delivery of therapeutic RNA by injection increases its bioavailability in target tissue and minimizes uptake in non-target tissues, but is limited to eye, skin, mucous membranes and tumors. Systemic delivery into the bloodstream is challenged by phagocytic immune cells such as macrophages and monocytes, which remove complexed RNAs from the body. Most ASOs delivered to muscle, heart and bone end up not in the cytoplasm, where they can find their target mRNAs, but in phagolysosomes. Cells of the liver, spleen and some tumors allow molecules up to 200 nm in diameter to enter and so the liver is among the most successful organs for delivering therapeutic RNAs.
In cases where localized delivery is not possible, delivery using PEGylated liposomes, lipidoids and biodegradable polymers are alternatives (Figure 5). To avoid being filtered by the kidneys and enhance intracellular delivery nucleic acids can be encapsulated in lipids forming vesicles between 50 and 500 nm. Liposomes are lipid bilayers with an aqueous core that contains the nucleic acid cargo. Lipoplexes are liposomes that contain cationic lipids that drive the interaction between the lipid bilayer and the negatively charged nucleic acid molecules. The anionic charge of the nucleic acids and their hydrophilicity is counterbalanced by the cationic lipids, resulting in a net positive charge, enabling the liposomes to bind to anionic cell surface molecules. The composition of these lipid particles can be tailored to facilitate fusion with the cytoplasmic, endosomal or nuclear membrane, as well as to promote endosomal release once inside the cell. The lipid head group, for example, can be pH sensitive, so that the liposomes interact with anionic phospholipids in the endosome, generating non-bilayer structures that disrupt the endosomal membrane, liberating the RNA.
Figure 5.
Chemical modifications that improve the stability, biodistribution, and delivery of ASOs. RNA (red; S indicates sulfur substitution of a non bridging oxygen to make a phosphorothioate linkage between nucleotides), 2′-_O_-methyl RNA contains a methyl group bound to the 2′ oxygen of the ribose; 2′-_O_-methoxyethyl RNA contains a methoxy group bound to the 2′ oxygen of the ribose; 2′-fluoro RNA contains fluorine molecule bound to the 2′ oxygen of the ribose; and locked nucleic acid (red) introduces a 2′,4′ methylene bridge in the ribose to form a bicyclic nucleotide).
In a screen of a library of ionizable cationic lipids with superior siRNA delivery capacity, a lipid nanoparticle (LNP) formulation was identified that substantially improved delivery. To test this delivery formulation, an siRNA that targets the hepatocyte mRNA transthyretin (TTR) was used because this protein has a short half-life and TTR protein levels can be easily measured in serum. The LNP delivery strategy in rodents and nonhuman primates was effective at 0.01 mg per kg and 0.1 mg per kg, respectively, administered as a single dose of TTR siRNA by 15 minute, intravenous, cephalic vein infusion at 5 ml per kg44. Forty-eight hours later, the siRNA reduced TTR mRNA levels by 30% in the livers of three Cynomolgus monkeys. Another potent delivery formula, the lipidoid, contains lipids with an amine-containing polar head group and nonpolar alkyl tails that are 12 carbons in length. When tested in nonhuman primates, this delivery formulation was effective at 0.03 mg per kg of siRNA. Forty-eight hours after infusion, the 0.03 mg per kg dose of the TTR siRNA in the lipidoid formulation reduced TTR mRNA levels by 70%45. These advanced delivery formulations are almost 100 times more effective than typical lipid-based delivery carriers, which require doses of at least 1 mg/kg of siRNA to achieve 50% gene silencing. We anticipate that such advanced lipid systems will be useful in delivering anti-miRNA ASOs to specific tissues and organs in humans.
“Decoy” transcripts can compete for miRNAs, blocking their function
ASOs act, at least in part, as competitive inhibitors of miRNAs, suggesting that miRNA-binding RNA transcripts may also be designed to sequester and thereby inhibit specific miRNAs. Such miRNA decoys could provide an inexpensive alternative to proprietary oligonucleotide chemistries and delivery formulations, enabling research laboratories to examine the consequence of inhibiting each known miRNA in a particular cultured cell46 or model animal47 or plant48. Moreover, miRNA-binding transcripts can be expressed from viral vectors, allowing the development of anti-miRNA gene therapy approaches. Ironically, the first demonstration of such transcripts, miRNA “decoys”49 or “sponges”46, preceded the discovery that plants naturally use such miRNA-binding transcripts to reduce the activity of specific miRNAs50.
The first miRNA decoy consisted of an adenoviral vector with two sites for the muscle-specific miRNA-133 inserted in the 3′ UTR of a GFP reporter gene, under control of an RNA polymerase II promoter from cytomegalovirus (CMV). This viral vector was used to confirm that loss of miRNA-133 expression, in mouse and human disease models, leads to cardiomyocyte hypertrophy49. Unlike chemically modified anti-miRNA oligonucleotides, miRNA decoys that include GFP allow one to determine the tissues or cell types where a miRNA is produced, as GFP will be repressed where the miRNA is present. “miRZips” and “TuD RNAs” (tough decoy RNAs) are microRNA decoy targets transcribed by RNA polymerase III (H1 or U6)51,52. Their nuclear export has been optimized to achieve high cytoplasmic expression. miRZips use the RNA polymerase III H1 promoter to express a single microRNA decoy hairpin with one arm that is perfectly complementary to the microRNA. This strategy causes degradation of the microRNA. TuD RNAs use the RNA polymerase III U6 promoter to express an RNA that contains multiple microRNA binding sites between 18 bp stem regions that help prevent nuclease degradation of the RNA decoy targets. For the TuD RNAs, the binding site is perfectly complementary to the miRNA, but contains 4 nt inserted at the site of Ago2 cleavage to prevent the TuD RNA from being inactivated. Such adaptations of the miRNA sponge concept allow longer term inhibition of microRNAs than can be achieved by transient transfection of RNA polymerase II sponge reporter vectors or modified RNA oligonucleotides.
miRNA sponges can be stably integrated into chromosomes, designed to be drug-inducible or controlled by promoters whose expression is restricted to a desired cell type, tissue, or developmental stage. For example, in Drosophila, a miRNA sponge for miR-8 revealed that post-synaptic expression of miR-8 is required for proper development of the neuromuscular junction47. Further modification of the sponge concept is underway to create separate sponge-expressing lines of transgenic fruit flies for each fly miRNA. Each line can be crossed with a second fly strain producing a transcription factor that promotes sponge expression in a specific cell type or developmental time, allowing discovery of the contribution of a miRNA to development, physiology or behavior47. In the future, we anticipate that transgenic sponges will be designed to permit their expression in mice at particular developmental stages or in specific tissues, perhaps by using the well established Cre-loxP system53,54.
miRNA replacement therapy seeks to reintroduce a missing miRNA
Some diseases may be due to loss or reduced expression of a particular microRNA. Interestingly, expression of most miRNAs in cancer is lower than normal. For example, the miRNA _let_-7 represses expression of the oncogenes Ras, Myc and HMGA-2, and _let_-7 levels were found to be low and HMGA2 mRNA high in primary tumors derived from 100 patients diagnosed with ovarian cancer. _let_-7 expression was also reduced in mammosphere-derived cancer stem cells when compared with normal breast or non-selected tumor cells, indicating that let-7 may prevent proliferation of cancer-initiating stem cells25,55. p53 expression caused by DNA damage promotes transcription of the miRNA-34 miRNA family, which is deleted in some cancers. miRNA-15 and 16 are frequently deleted in B-cell lymphocytic leukemia, and their expression is reduced by 80% in prostate cancer. Other miRNA genes, including _let_-7, reside at fragile sites where chromosomes often break, leading to cancer56. Thus, many miRNAs meet the classical definition of tumor suppressor genes. Replacement of such tumor suppressor miRNAs might augment traditional cancer chemotherapy.
miRNAs whose expression is lost or reduced can be replenished by adding back the miRNA. Adding the miRNA back in a single dose may not allow sustained target regulation due to inefficient delivery or degradation, but data from multiple doses of siRNAs suggest that three-to-five doses of replacement miRNA, modified or formulated for optimal delivery, might provide sufficient miRNA for 20 to 30 days. Alternatively, cells can be infected with viral vectors encoding short hairpin RNAs (Figure 3) that are processed in the cell into mature miRNAs26,27,56. Viral delivery of miRNAs can be optimized to achieve a specific and continuous level of expression. miRNA replacement therapy must be both effective and safe. Over expression of shRNA in rats caused hepatotoxicity, organ failure and death57. Argonaute proteins and the pre-miRNA export protein, Exportin-5 limit the amount of exogenous siRNA or miRNA that a cell can tolerate57-62. shRNAs that are more pre-miRNA-like or authentic pre-miRNAs themselves will likely minimize toxicity while retaining potency for their intended targets60,63.
miRNA-directed regulation can improve traditional gene therapy approaches
Gene therapy holds great promise to replace defective protein-coding genes underlying many genetic diseases. However, ensuring expression of the therapeutic transgene in the correct tissue while minimizing its expression elsewhere remains challenging because even tissue-specific promoters can be leaky. Combining miRNA regulation with gene therapy allows targeted and potent expression of transgenes. Such “de-targeting” strategies incorporate miRNA target sites in the 3′ UTR of the therapeutic transgene, preventing its expression in cells that express the corresponding miRNA. The transgene will be expressed in the intended cell-type, where the miRNA is not expressed. For example, miRNA-122 is specific to the liver, so systemically delivered transgenes containing binding sites for miRNA-122 will be silenced in hepatocytes, but not cells elsewhere. This strategy was used to restrict the expression of a transgene in a lentiviral vector to astroglial cells64. Starting with a lentivirus engineered to preferentially infect neurons and glia, miRNA-124 target sites were inserted in the 3′ UTR to prevent transgene expression in neuronal cells, which express miRNA-124, and allow transgene expression in glial cells, which do not express miRNA-124. Injection of the vector into the hippocampus in mice produced transgene expression in astrocytes and Bergmann glial cells, but not in pyramidal neurons or Purkinje cells64. Since each site is only 21 nt long, binding sites for multiple, tissue-specific miRNAs can be incorporated in the 3′ UTR, extinguishing transgene expression in many different tissues simulataneously.
miRNA-mediated transgene detargeting has also been used to promote immune tolerance of a transgene-encoded antigen. Annoni and colleagues exploited the tissue specificity of miRNA-142, which is expressed only in hematopoietic cells, to prevent a lentiviral vector from producing transgenic protein in antigen presenting cells65. By blocking transgene expression in immune cells, they avoided the common problem of T-cells detecting and eliminating cells expressing the foreign transgenic protein. Interestingly, a control experiment to prevent expression in the liver using miRNA-122 binding sites revealed that liver expression of the transgene was required to induce antigen tolerance.
Replication-selective oncolytic viruses—genetically engineered adenoviruses that selectively infect and kill tumor cells—have been proposed as alternatives to standard chemotherapy. Avoiding expression in the liver is particularly important as adenovirus-based therapies cause liver toxicity. Since neuroendocrine tumors of the ileum can metastasize to the liver66, a key challenge is to produce the transgenic protein in the cancer cells residing in the liver, but not in untransformed hepatocytes. Whyte and colleagues proposed a clever solution to this problem. They used the chromogranin-A promoter, which is active in neuroendocrine tumors, to specifically express the E1A protein, a viral protein that activates viral and cellular genes critical for viral infection, while adding miRNA-122 binding sites to the 3′ UTR of the E1A mRNA to prevent viral replication in hepatocytes67. In a mouse model, the miRNA-regulated, oncolytic adenovirus killed tumor cells without detectable liver toxicity.
Unique miRNA expression patterns in stem cells can be exploited to select for a specific cell type from a mixed cell population, before adding cells back to the patient and as a strategy for monitoring lineage-specific differentiation of induced pluripotent and embryonic stem cells. In stem cell therapy applications, where cells are engineered to express normal genes that are mutated in the patient, it is critical to remove the pluripotent cell population from the therapeutic differentiated cells before transplanting them back into the patient to prevent unwanted proliferation and tumor development. Expression of a suicide gene or fluorescent reporter can be controlled by miRNAs whose expression is specific to a differentiated cell type in a population of pluripotent stem cells68. Differentiation-induced miRNA expression could turn off the reporter gene to allow separation of the differentiated cells from the pluripotent cells. A suicide gene can also be turned off by cell type specific miRNAs to allow differentiated cells of a specific lineage to proliferate. Additionally, by combining multiple miRNA target sites, expression of a transgene can potentially be suppressed in multiple cell types or tissues. Such a strategy requires calibrating miRNA expression and target site affinity so that the desired level of regulation of the transgene is achieved.
Prospects
The realization that the inappropriate production of individual miRNAs contributes to disease has reinvigorated antisense oligonucleotide drug development. ASOs readily inhibit miRNAs—far more reliably than they do mRNAs, and the unique properties of Argonaute proteins permits the use of remarkably short ASOs: 15 nt oligonucleotide ASO are now in clinical trials and 8 nt versions show promise in non-human primates. Many new roles for individual miRNAs in disease, aging and cancer are likely to emerge over the next five years. Once the role of a specific miRNA in disease pathogenesis is established, selecting specific anti-miRNA inhibitor chemistries and delivery strategies promises to be straightforward. Nonetheless, effective and safe delivery of anti-miRNA drugs remains difficult for many cell types such as brain and muscle. Thus, treating diseases with anti-miRNA oligonucleotides will require the development of novel modification, conjugation or formulation strategies. It is our hope that the anti-miRNA therapeutics field will soon converge on a small number of “platform” technologies that allow a rapid and safe development path from academic discovery to effective drug.
Table 1.
miRNA therapeutics in commercial development.
| Company | Diseases | Chemistries | Stage | References |
|---|---|---|---|---|
| RegulusTherapeutics | immuno-inflammatory,cardiovascular,metabolic disease,oncology, fibrosis, andhepatitis C infection | miRNA inhibitors using2′-methoxyethyl , 2′-fluoro RNA, bicyclicribose modifications | pre-clinical | Esau et al., 2006Esau et al., 2007Krutzfeldt et al., 2005Krutzfeldt et al., 2007www.regulusrx.com |
| SantarisPharma A/S | cancer andinflammatory diseases,hepatitis C infection | miRNA inhibitors usinglocked nucleic acidchemistry | miR-122 inhibitor:Phase I completed,Phase II initiated | Ørom et al., 2006Elmén et al., 2008www.santaris.com |
| miRagenTherapeutics | cardiovascular andmuscle diseases | miRNA inhibition andreplacement | pre-clinical | www.miragentherapeutics.com |
| MirnaTherapeutics | non-small cell lungcancer and prostatecancer | miRNA replacementusing siRNAs | pre-clinical | www.mirnatherapeutics.com |
References
- 1.Bartel DP. MicroRNAs: Target Recognition and Regulatory Functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chiang HR, et al. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes Dev. 2010;24:992–1009. doi: 10.1101/gad.1884710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Medina PP, Slack FJ. microRNAs and cancer: an overview. Cell Cycle. 2008;7:2485–2492. doi: 10.4161/cc.7.16.6453. [DOI] [PubMed] [Google Scholar]
- 4.Petrocca F, Lieberman J. Micromanipulating cancer: microRNA-based therapeutics? RNA Biol. 2009;6:335–340. doi: 10.4161/rna.6.3.9013. [DOI] [PubMed] [Google Scholar]
- 5.Mencia A, et al. Mutations in the seed region of human miR-96 are responsible for nonsyndromic progressive hearing loss. Nat Genet. 2009;41:609–613. doi: 10.1038/ng.355. [DOI] [PubMed] [Google Scholar]
- 6.Lewis MA, et al. An ENU-induced mutation of miR-96 associated with progressive hearing loss in mice. Nat Genet. 2009;41:614–618. doi: 10.1038/ng.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jopling CL, Schutz S, Sarnow P. Position-dependent function for a tandem microRNA miR-122-binding site located in the hepatitis C virus RNA genome. Cell Host Microbe. 2008;4:77–85. doi: 10.1016/j.chom.2008.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Umbach JL, Cullen BR. The role of RNAi and microRNAs in animal virus replication and antiviral immunity. Genes Dev. 2009;23:1151–1164. doi: 10.1101/gad.1793309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baek D, et al. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Selbach M, et al. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 12.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 14.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 15.Grimson A, et al. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rajewsky N, Socci ND. Computational identification of microRNA targets. Dev Biol. 2004;267:529–535. doi: 10.1016/j.ydbio.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 17.Cullen BR. Viral and cellular messenger RNA targets of viral microRNAs. Nature. 2009;457:421–425. doi: 10.1038/nature07757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cullen BR. Five questions about viruses and microRNAs. PLoS Pathog. 2010;6:e1000787. doi: 10.1371/journal.ppat.1000787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stern-Ginossar N, et al. Host immune system gene targeting by a viral miRNA. Science. 2007;317:376–381. doi: 10.1126/science.1140956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nachmani D, Lankry D, Wolf DG, Mandelboim O. The human cytomegalovirus microRNA miR-UL112 acts synergistically with a cellular microRNA to escape immune elimination. Nat Immunol. 2010;11:806–813. doi: 10.1038/ni.1916. [DOI] [PubMed] [Google Scholar]
- 21.Murphy E, Vanicek J, Robins H, Shenk T, Levine AJ. Suppression of immediate-early viral gene expression by herpesvirus-coded microRNAs: implications for latency. Proc Natl Acad Sci U S A. 2008;105:5453–5458. doi: 10.1073/pnas.0711910105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Umbach JL, et al. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature. 2008;454:780–783. doi: 10.1038/nature07103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bellare P, Ganem D. Regulation of KSHV lytic switch protein expression by a virus-encoded microRNA: an evolutionary adaptation that fine-tunes lytic reactivation. Cell Host Microbe. 2009;6:570–575. doi: 10.1016/j.chom.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ellis-Connell AL, Iempridee T, Xu I, Mertz JE. Cellular microRNAs 200b and 429 regulate the Epstein-Barr virus switch between latency and lytic replication. J Virol. 2010;84:10329–10343. doi: 10.1128/JVI.00923-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson CD, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67:7713–7722. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- 26.Esquela-Kerscher A, et al. The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle. 2008;7:759–764. doi: 10.4161/cc.7.6.5834. [DOI] [PubMed] [Google Scholar]
- 27.Kumar MS, et al. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci U S A. 2008;105:3903–3908. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trang P, et al. Regression of murine lung tumors by the let-7 microRNA. Oncogene. 2010;29:1580–1587. doi: 10.1038/onc.2009.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kota J, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137:1005–1017. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gabriely G, et al. MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol Cell Biol. 2008;28:5369–5380. doi: 10.1128/MCB.00479-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anand S, et al. MicroRNA-132-mediated loss of p120RasGAP activates the endothelium to facilitate pathological angiogenesis. Nat Med. 2010;16:909–914. doi: 10.1038/nm.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Medina PP, Nolde M, Slack FJ. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2010;467:86–90. doi: 10.1038/nature09284. [DOI] [PubMed] [Google Scholar]
- 33.Ameres SL, et al. Target RNA-directed trimming and tailing of small silencing RNAs. Science. 2010;328:1534–1539. doi: 10.1126/science.1187058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davis ME, et al. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature. 2010;464:1067–1070. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lennox KA, Behlke MA. A direct comparison of anti-microRNA oligonucleotide potency. Pharm Res. 2010;27:1788–1799. doi: 10.1007/s11095-010-0156-0. [DOI] [PubMed] [Google Scholar]
- 36.Zheng G, Ambros V, Li WH. Inhibiting miRNA in Caenorhabditis elegans using a potent and selective antisense reagent. Silence. 2010;1:9. doi: 10.1186/1758-907X-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krutzfeldt J, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 38.Krutzfeldt J, et al. Specificity, duplex degradation and subcellular localization of antagomirs. Nucleic Acids Res. 2007;35:2885–2892. doi: 10.1093/nar/gkm024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davis S, Lollo B, Freier S, Esau C. Improved targeting of miRNA with antisense oligonucleotides. Nucleic Acids Res. 2006;34:2294–2304. doi: 10.1093/nar/gkl183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Esau C, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 41.Orom UA, Kauppinen S, Lund AH. LNA-modified oligonucleotides mediate specific inhibition of microRNA function. Gene. 2006;372:137–141. doi: 10.1016/j.gene.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 42.Elmen J, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 43.Lanford RE, et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Semple SC, et al. Rational design of cationic lipids for siRNA delivery. Nat Biotechnol. 2010;28:172–176. doi: 10.1038/nbt.1602. [DOI] [PubMed] [Google Scholar]
- 45.Love KT, et al. Lipid-like materials for low-dose, in vivo gene silencing. Proc Natl Acad Sci U S A. 2010;107:1864–1869. doi: 10.1073/pnas.0910603106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Loya CM, Lu CS, Van Vactor D, Fulga TA. Transgenic microRNA inhibition with spatiotemporal specificity in intact organisms. Nat Methods. 2009;6:897–903. doi: 10.1038/nmeth.1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Todesco M, Rubio-Somoza I, Paz-Ares J, Weigel D. A collection of target mimics for comprehensive analysis of microRNA function in Arabidopsis thaliana. PLoS Genet. 2010;6:e1001031. doi: 10.1371/journal.pgen.1001031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Care A, et al. MicroRNA-133 controls cardiac hypertrophy. Nat Med. 2007;13:613–618. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- 50.Franco-Zorrilla JM, et al. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet. 2007;39:1033–1037. doi: 10.1038/ng2079. [DOI] [PubMed] [Google Scholar]
- 51.Haraguchi T, Ozaki Y, Iba H. Vectors expressing efficient RNA decoys achieve the long-term suppression of specific microRNA activity in mammalian cells. Nucleic Acids Res. 2009;37:e43. doi: 10.1093/nar/gkp040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sauer B, Henderson N. Cre-stimulated recombination at loxP-containing DNA sequences placed into the mammalian genome. Nucleic Acids Res. 1989;17:147–161. doi: 10.1093/nar/17.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stern P, et al. A system for Cre-regulated RNA interference in vivo. Proc Natl Acad Sci U S A. 2008;105:13895–13900. doi: 10.1073/pnas.0806907105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu F, et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109–1123. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 56.Bonci D, et al. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med. 2008;14:1271–1277. doi: 10.1038/nm.1880. [DOI] [PubMed] [Google Scholar]
- 57.Grimm D, et al. Argonaute proteins are key determinants of RNAi efficacy, toxicity, and persistence in the adult mouse liver. J Clin Invest. 2010;120:3106–3119. doi: 10.1172/JCI43565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Diederichs S, et al. Coexpression of Argonaute-2 enhances RNA interference toward perfect match binding sites. Proc Natl Acad Sci U S A. 2008;105:9284–9289. doi: 10.1073/pnas.0800803105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McBride JL, et al. Artificial miRNAs mitigate shRNA-mediated toxicity in the brain: implications for the therapeutic development of RNAi. Proc Natl Acad Sci U S A. 2008;105:5868–5873. doi: 10.1073/pnas.0801775105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Boudreau RL, Monteys AM, Davidson BL. Minimizing variables among hairpin-based RNAi vectors reveals the potency of shRNAs. RNA. 2008;14:1834–1844. doi: 10.1261/rna.1062908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Boudreau RL, Martins I, Davidson BL. Artificial microRNAs as siRNA shuttles: improved safety as compared to shRNAs in vitro and in vivo. Mol Ther. 2009;17:169–175. doi: 10.1038/mt.2008.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Beer S, et al. Low-level shRNA cytotoxicity can contribute to MYC-induced hepatocellular carcinoma in adult mice. Mol Ther. 2010;18:161–170. doi: 10.1038/mt.2009.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bauer M, et al. Prevention of interferon-stimulated gene expression using microRNA-designed hairpins. Gene Ther. 2009;16:142–147. doi: 10.1038/gt.2008.123. [DOI] [PubMed] [Google Scholar]
- 64.Colin A, et al. Engineered lentiviral vector targeting astrocytes in vivo. Glia. 2009;57:667–679. doi: 10.1002/glia.20795. [DOI] [PubMed] [Google Scholar]
- 65.Annoni A, et al. In vivo delivery of a microRNA-regulated transgene induces antigen-specific regulatory T cells and promotes immunologic tolerance. Blood. 2009;114:5152–5161. doi: 10.1182/blood-2009-04-214569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Leja J, et al. A novel chromogranin-A promoter-driven oncolytic adenovirus for midgut carcinoid therapy. Clin Cancer Res. 2007;13:2455–2462. doi: 10.1158/1078-0432.CCR-06-2532. [DOI] [PubMed] [Google Scholar]
- 67.Leja J, et al. Double-detargeted oncolytic adenovirus shows replication arrest in liver cells and retains neuroendocrine cell killing ability. PLoS One. 2010;5:e8916. doi: 10.1371/journal.pone.0008916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brown BD, et al. Endogenous microRNA can be broadly exploited to regulate transgene expression according to tissue, lineage and differentiation state. Nat Biotechnol. 2007;25:1457–1467. doi: 10.1038/nbt1372. [DOI] [PubMed] [Google Scholar]