Plexin B1 is repressed by oncogenic B-Raf signaling and functions as a tumor suppressor in melanoma cells (original) (raw)
. Author manuscript; available in PMC: 2011 Dec 15.
Published in final edited form as: Oncogene. 2009 Jun 1;28(30):2697–2709. doi: 10.1038/onc.2009.133
Abstract
Human melanomas show oncogenic B-Raf mutations which activate the B-Raf/MKK/ERK cascade. We screened microarrays to identify cellular targets of this pathway, and found that genes upregulated by B-Raf/MKK/ERK showed highest association with cell cycle regulators, whereas genes downregulated were most highly associated with axon guidance genes, including plexin-semaphorin family members. Plexin B1 was strongly inhibited by MAP kinase signaling in melanoma cells and melanocytes. In primary melanoma cells, plexin B1 blocked tumorigenesis as measured by growth of colonies in soft agar, spheroids in extracellular matrix, and xenograft tumors. Tumor suppression depended on residues in the C-terminal domain of plexin B1 which mediate receptor GAP activity, and also correlated with AKT inhibition. Interestingly, the inhibitory response to plexin B1 was reduced or absent in cells from a matched metastatic tumor, suggesting that changes occur in metastatic cells which bypass the tumor suppressor mechanisms. Plexin B1 also inhibited cell migration, but this was seen in metastatic cells and not in matched primary cells. Thus, plexin B1 has tumor suppressor function in early-stage cells, while suppressing migration in late-stage cells. Our findings suggest that B-Raf/MKK/ERK provides a permissive environment for melanoma genesis by modulating plexin B1.
Keywords: plexin, semaphorin, B-Raf, melanoma, tumor suppressor
Introduction
Genome sequencing studies have recently shown that oncogenic mutations of B-Raf occur in approximately 70% of malignant melanoma tumors and cell lines (1-3). Remarkably, most of these mutations specify a single amino acid substitution, V600E, which constitutively elevates B-Raf catalytic activity. Other B-Raf mutations do not affect kinase activity, but instead promote activation of c-Raf1 (4). Melanoma stages are based on clinical and histopathological features, and include dysplastic or atypical nevi, early radial growth phase (RGP) primary melanoma, advanced vertical growth phase (VGP) primary melanoma, and metastatic melanoma (5). High frequencies of B-Raf mutations have been observed in nevoid/RGP specimens as well as advanced tumors (6,7). As a result, the MKK1/2-ERK1/2 protein kinase cascade is constitutively active in most melanoma tissues and cell lines (2,3,8). B-Raf/MKK/ERK signaling promotes cell proliferation, survival, and vascular development in melanoma xenograft tumors, and several reports have demonstrated that tumor growth is suppressed upon blocking this pathway (9-11). Thus, identifying cellular targets of this pathway provides a way to elucidate molecular determinants of melanoma.
Plexins and semaphorins are receptor and ligand families which function to regulate axon guidance and neuronal growth cone collapse (12,13). Among the best characterized receptor members are plexin B1, which directly binds semaphorin 4D, and plexin A1, which complexes with class 3 semaphorins in association with neuropilins. Plexins A1 and B1 mediate cell guidance by recruiting various GTPases to distinct receptor domains. In response to ligand binding, Rho is activated by recruitment of guanine nucleotide exchange factors (GEFs) to receptors, and Rac is inhibited by direct receptor binding and sequestration (14-18). Plexins A and B also inhibit R-Ras, by binding the Rnd1 GTPase, which recruits R-Ras-GTP to the receptor, followed by GTP hydrolysis catalyzed by an intrinsic receptor GAP domain (19,20). These mechanisms have been proposed to underlie cell-cell attraction and repulsion, where R-Ras-GTP hydrolysis drives cell contraction and axonal growth cone collapse, Rac controls cell-cell attraction, and Rho causes localized increases in contractile force by stress fibers, leading to growth cone collapse and cell-cell repulsion (reviewed in 21-23).
Plexin-semaphorins have also been implicated in cancer. Plexin B1-semaphorin 4D induces invasion in many cell types, and promotes endothelial cell angiogenesis (24,25). Frequent missense mutations or overexpression of plexin B1 have been found in prostate cancers, and are associated with increased invasion (26). In several cancer cell types, plexin B1 promotes invasive growth by associating with c-Met and elevating c-Met phosphorylation (27,28). In other cells, plexin B1 blocks cell migration through c-Met by inhibiting RhoA, while activating cell migration through ErbB2 (29). Semaphorin 4D is highly expressed in malignant cells from prostate, colon, breast, and lung cancer tissues, and in head and neck squamous cell cancers mediates tumor angiogenesis via paracrine signaling to endothelial cells (30). Likewise, semaphorin 3A, 3C, and 3E are overexpressed in breast, ovarian, and lung cancer cells, and are correlated with angiogenesis and vascularization. On the other hand, overexpression of semaphorin 3F in melanoma retards angiogenesis and metastasis of mouse xenografts, and alters the tumor microenvironment to form a benign, encapsulated tumor with well-defined borders (31,32). In estrogen-receptor positive breast cancers, low plexin B1 is associated with greater malignancy and poor prognosis (33). Thus, responses to plexin-semaphorin vary widely between cancer types.
Here, we conducted a microarray screen to identify targets of constitutive B-Raf/MKK/ERK signaling in melanoma. We report that plexin B1 is repressed by MAP kinase signaling in cells isolated from human melanomas at varying stages of progression. Further experiments revealed a novel tumor suppressor role for plexin B1 in melanoma cells which is correlated with suppression of AKT, as well as a role in inhibition of cell migration. Our results demonstrate that oncogenic B-Raf promotes tumor growth and cell migration by inhibitory cross-regulation of plexin B1 signal transduction.
Results
Identifying B-Raf/MKK/ERK-regulated genes
Microarrays were used to profile responses to constitutive B-Raf/MKK/ERK signaling in melanoma. Systems-wide screening approaches often encounter a “noise” problem, where large numbers of candidate genes are altered in their mRNA or protein levels, but genes which are biologically important are difficult to identify. To aid in identifying targets of biological importance, we examined several cell lines, hypothesizing that targets regulated in common between them are likely to control responses to signaling.
Genes responsive to B-Raf/MKK/ERK were examined in eight melanoma cell lines, derived from two RGP tumors (WM35, WM1789), three VGP tumors (WM115, WM278, WM793) and three metastatic tumors (WM239A, WM1617, 1205Lu) (34). For each cell line, the B-Raf-V600E mutation was either previously reported (http://www.wistar.org/herlyn/melanoma.htm), or else verified by sequencing genomic B-Raf (data not shown). Cells were treated in the presence or absence of 10 μM U0126 for 24 h, and microarray assays were performed. Because prolonged inhibition of MKK/ERK induces apoptosis in melanoma cells (11), we monitored cell viability vs time by cell counting and caspase 3 activation. Each cell line remained >90% viable without significant caspase activation for 24 h with inhibitor (Suppl. Fig. 1). Most experiments were examined with one microarray assay. However, WM239A cells assayed in two biological replicates showed ∼7% average deviation from the mean across all genes (data not shown), indicating good reproducibility between independent experiments.
Microarray datasets were analyzed by the method of rank products, filtering significant changes by setting a false discovery rate ≤0.001. This identified 484 genes (668 probesets) decreased and 239 genes (395 probesets) increased in response to U0126. Results are shown in Suppl. Tables 1,2, and full datasets are provided in Suppl. Table 3. Genes altered significantly were then analyzed for their biological function by matching associations with KEGG pathways. This was performed using the program, “GO-Getter”, which uses a relational database to match microarray identifiers against gene ontology annotations (http://bmf2.colorado.edu/go-getter/index.psp).1 Genes are weighted by their number of associations with different functions (e.g. 1.0 for genes matching one function, 0.5 for genes matching two functions), and weighted values are summed and ranked.
Using GO-Getter, cellular pathways were ranked by the numbers of genes found responsive to MKK/ERK. Genes decreased by U0126, and thus positively regulated by B-Raf/MKK/ERK, were highly clustered among pathways related to cell cycle control (Fig. 1A). The largest subset was represented by 18 cell cycle genes (Suppl. Table 4), which included cyclin D1, cyclin kinases, and minichromosome maintenance deficient (MCM) genes. Second and third largest gene subsets contained 26 genes that function in purine and pyrimidine metabolism (Fig. 1A, Suppl. Table 4), suggesting that B-Raf promotes proliferation by inducing genes needed for DNA metabolism and S phase.
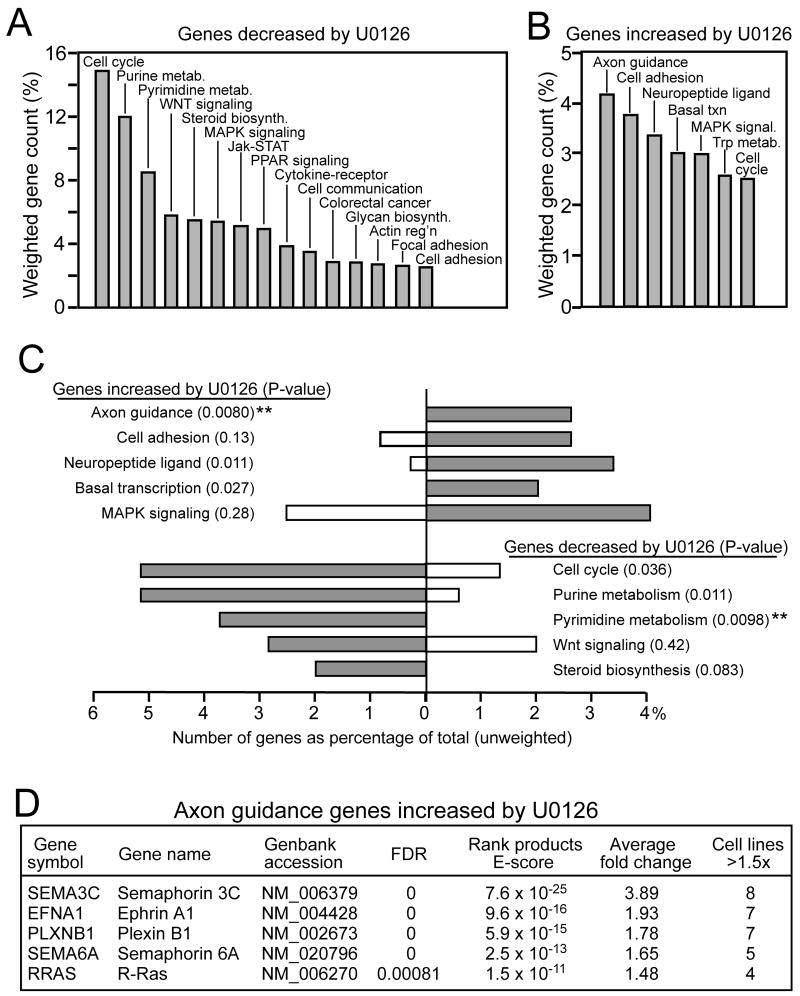
Figure 1. Axon guidance genes are repressed by B-Raf/MKK/ERK in melanoma.
(A, B) Summary of pathways containing genes significantly (A) decreased or (B) increased in response to treatment of eight melanoma cell lines with U0126. Significant responses are filtered by rank products and gene lists were sorted into KEGG pathway groups, listing those with summed weighted gene counts >2.5%. Genes decreased by U0126 are enriched in pathways required for cell cycle progression. Genes increased by U0126 are highest in axon guidance pathways. (C) Two-sample comparison of pathways that best discriminate between genes induced versus repressed by B-Raf/MKK/ERK. For each pathway, filled bars represent genes increased or decreased by U0126, while open bars represent genes respectively decreased or increased by U0126. For gene sets decreased by U0126, filled bars represent genes decreased and open bars represent genes increased by U0126. Asterisks indicate pathways that discriminate between with _P_-value<0.01. (D) Genes within the axon guidance gene group. Genes within other groups in panel C are presented in Suppl. Table 4.
Some genes were increased by U0126, and thus candidates for repression by constitutive B-Raf/MKK/ERK signaling. These were distributed among many pathway categories, with the largest cluster containing five genes previously characterized in axon guidance (Fig. 1A, Suppl. Table 4). GO-Getter features which enable two-sample comparisons were then applied to identify pathways that discriminate between genes induced vs repressed by B-Raf/MKK/ERK. The pyrimidine and purine metabolism subsets showed greatest enrichment in samples induced by B-Raf/MKK/ERK, while being weakly represented in samples repressed by B-Raf/MKK/ERK. The axon guidance genes comprised the subset showing greatest discrimination for events repressed by B-Raf/MKK/ERK (Fig. 1C).
The axon guidance genes included plexin B1, semaphorin 3C, semaphorin 6A, and ephrin A1, which are receptors or ligands controlling growth cone extension or collapse, and R-Ras, a GTPase known to mediate plexin-semaphorin signaling (Fig. 1D). Thus, clustering revealed an unprecedented cross-regulation of plexin-semaphorin pathways by B-Raf/MKK/ERK, and suggested a potential importance for guidance factor signaling in melanoma cells. We chose to explore the regulation of plexin B1.
Plexin B1 is repressed by MKK/ERK
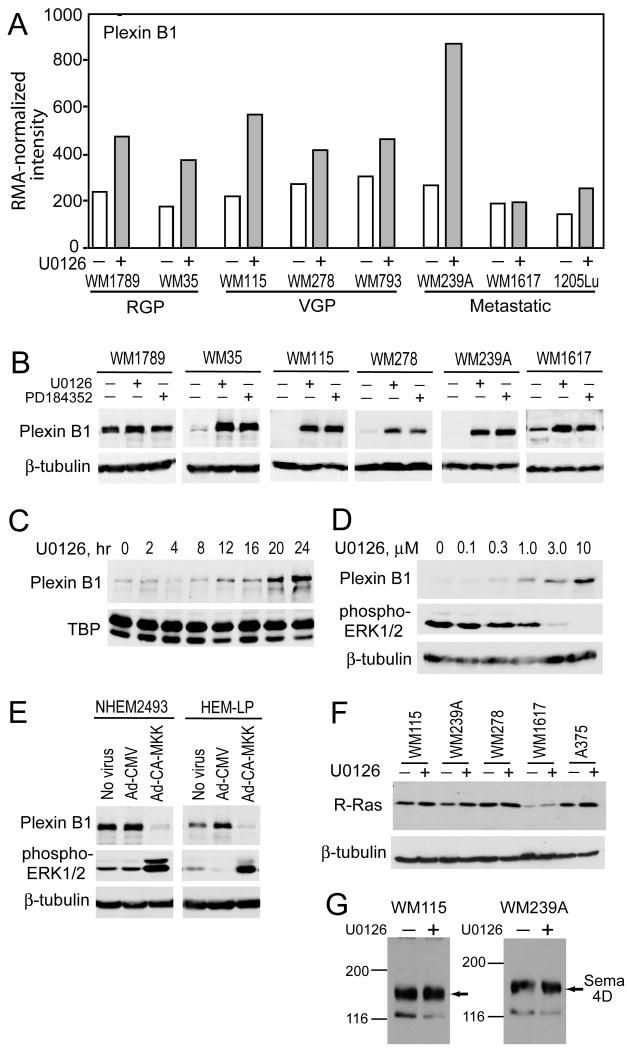
Striking changes in plexin B1 expression were observed, whereby 7 of the 8 cell lines examined showed significantly enhanced microarray signal intensity after U0126 treatment (Fig. 2A). The responses were confirmed by Western blotting, which showed elevated plexin B1 protein expression in response to U0126 as well as the MKK1/2 inhibitor, PD184352 (CI-1040) (Fig. 2B). Induction by U0126 was apparent within 12 h after drug treatment, and correlated with inhibition of phospho-ERK1/2, with EC50(U0126) ∼1 μM (Fig. 2C,D). Reciprocal regulation was observed in melanocytes, where plexin B1 expression was higher than in melanoma, and strongly suppressed by constitutively active (CA) mutant MKK1 (Fig. 2E). Thus, B-Raf/MKK/ERK appears necessary and sufficient to repress plexin B1 expression in melanoma cells. In contrast, levels of R-Ras were increased by U0126 as predicted from microarrays, but by a smaller degree than plexin B1 (Fig. 2F). The plexin B1 ligand, semaphorin 4D, was present in melanoma cells but unaffected by U0126 (Fig. 2G).
Figure 2. Plexin B1 is repressed by B-Raf/MKK/ERK in melanoma cells and melanocytes.
(A) RMA-normalized intensities for plexin B1 (probeset 215807_s_at) shows induction by U0126 (10 μM, 24 h) in 7 of 8 melanoma cell lines. (B) Western blots confirm induction of plexin B1 with 24 h treatment with U0126 (10 μM) or PD184352 (3 μM) in melanoma cell lines. (C) A time course in WM239A cells shows induction of plexin B1 by 12 h of U0126 (10 μM) treatment. (D) Dose response study in WM239A cells shows that plexin B1 induction is correlated with phospho-ERK1/2 inhibition by U0126 (24 h). (E) Reciprocal control by MKK/ERK is revealed in melanocytes, where plexin B1 is repressed upon adenoviral delivery of constitutively active MKK1. (F) Western blots show a modest induction of R-Ras by U0126, in 4 of 5 cell lines. (G) Western blots reveal the presence of semaphorin 4D in WM115 and WM239A cells, which is unaffected by treatment with U0126 (10 μM, 24 h). Equal amounts of lysates (20 μg) were loaded in all lanes.
Plexin B1 inhibits colony formation in soft agar
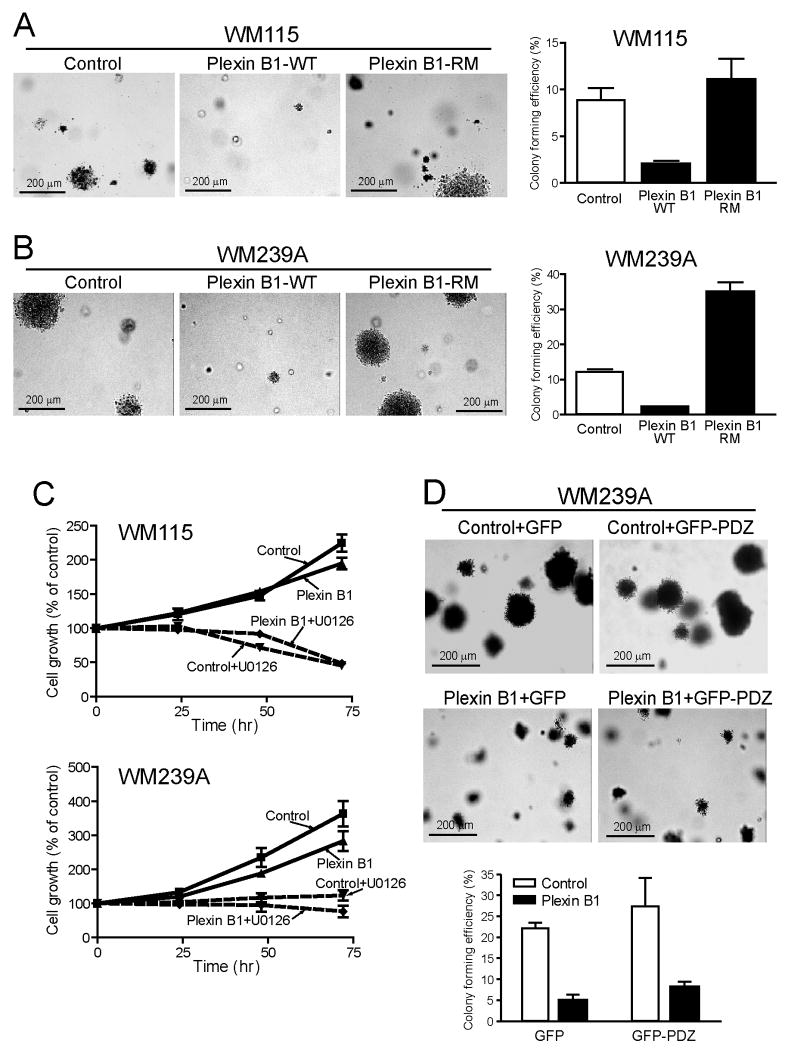
We hypothesized that melanoma responses to B-Raf/MKK/ERK might involve repression of plexin B1, and that conversely, induction of plexin B1 might block tumorigenesis. To test this, we generated stable transfectants of WM115 and WM239A cell lines, which were respectively isolated from a primary melanoma and a corresponding lymph node metastasis from the same patient (34). Both WM115 and WM239A cells readily formed colonies from single cells when suspended in soft agar (Fig. 3A,B). Stable expression of plexin B1 reduced the number of colonies formed by each cell line, favoring small colonies over large ones (Fig. 3A,B). In contrast, plexin B1 did not alter cell doubling times or ERK phosphorylation (Fig. 3C, Suppl. Fig. 2). The results demonstrate that plexin B1 inhibits the ability of melanoma cells to form 3D colonies, without affecting cell proliferation.
Figure 3. Plexin B1 inhibits colony growth in soft agar.
(A) WM115 and (B) WM239A cell lines expressing plexin B1 were resuspended in agar. Colony forming efficiency was measured after 21 days, measuring the ratio of colonies with diameter >200 μm to the total number of cells plated. Colonies formed in control lines are significantly reduced upon stable expression of plexin B1. Mutant plexin B1-RM, defective in R-Ras GAP activity, does not repress colony growth from melanoma cells. (C) Stable expression of plexin B1 has little effect on proliferation of WM115 or WM239A cells grown on 2D culture dishes. (D) Expression of GFP-PDZ in WM239A cells does not reverse the inhibitory response to plexin B1. Western blots showing expression of plexin B1-RM and GFP-PDZ are shown in Suppl. Fig. 3.
We tested the involvement of receptor domains implicated in signaling. A mutant plexin B1, defective at residues in the receptor GAP domain (R1677M/R1678M “RM”, ref. 19), was stably expressed in WM115 and WM239A cells (Suppl. Fig. 3A). Unlike plexin B1-WT, plexin B1-RM did not retard colony growth in soft agar, and instead allowed efficient colony formation (Fig. 3A,B). Interestingly, colony-forming efficiency increased in WM293A cells transfected with plexin-B1-RM (Fig. 3B). The importance of RhoA regulation was tested by stably co-expressing plexin B1 with the PDZ domain from PDZ-RhoGEF, which inhibits RhoA by competing for binding of LARG1 and PDZ-RhoGEF exchange factors to plexin B1 (24). The PDZ domain alone did not affect colony growth of WM239A cells in soft agar, and when combined with plexin B1-WT did not block the inhibitory response to plexin B1 (Fig. 3D). Together, the results indicate that plexin B1 suppresses colony formation by regulating effectors controlled by its GAP domain. The effect of plexin B1 does not seem to involve RhoA activation, although the results do not rule out the possibility that inhibition occurs by Rho inactivation.
Plexin B1 inhibits spheroid growth within an extracellular matrix environment
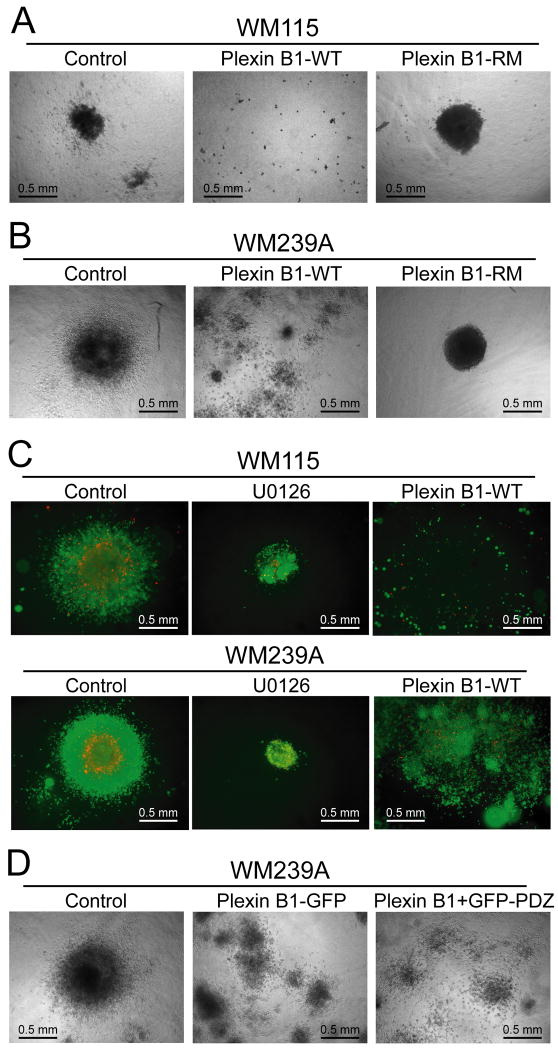
Smalley et al. described a 3D culture system where melanoma cells are implanted into collagen gels, creating spheroids that mimic the architecture and microenvironment of human skin tumors (35). Spheroids formed from WM239A and WM115 cells showed different morphologies after suspension into matrix (Fig. 4A,B). Typically, WM239A cells aggregated into a few large spheroids, each with diameter >400 μm, and significant numbers of cells with fibroblastoid morphology invaded into the extracellular matrix after 10 days (Fig. 4B, left panel). WM115 cells formed smaller spheroids with diameter 100-200 μm, and showed lower invasiveness (Fig. 4A, left panel).
Figure 4. Plexin B1 inhibits spheroid growth in extracellular matrix.
(A) WM115 and (B) WM239A cell lines were grown in collagen matrix for 7-10 days. Plexin B1 inhibits the formation of spheroids in both cell types, although small spheroid clusters are still apparent in WM239A cells, suggesting only partial inhibition in this cell line. In both cell types, formation of large spheroids is recovered by expression of the R-RasGAP defective mutant, plexin B1-RM. (C) WM115 and WM239A cells are cultured as spheroids in the presence of U0126 (10 μM) for 10 days, and then stained for cell viability, where green indicates live cells and red indicates dead cells. Neither U0126 treatment nor plexin B1 expression affected viability of spheroid cells. (D) WM239A cells expressing plexin B1, and either GFP (virus control) or GFP-PDZ were cultured as spheroids and implanted into collagen. Inhibiting plexin B1 activation of RhoA by expressing GFP-PDZ did not rescue spheroid growth, demonstrating that plexin B1 does not signal through RhoA.
Stable expression of plexin B1 inhibited spheroid growth and reduced the extent of cell aggregation in both WM115 and WM239A cells (Fig. 4A,B, middle panel). Inhibition was most pronounced in WM115 cells, where spheroids of both large and small diameter were strongly repressed (Fig. 4A). In contrast, plexin B1 reduced the overall diameter of large spheroids from WM239A cells, while increasing numbers of smaller cell clusters. No loss of cell viability within spheroids was observed upon expression of plexin B1 or in response to U0126 (Fig. 4C, middle and right panels).
The plexin B1-RM mutation rescued the inhibition of spheroid formation observed with plexin B1-WT (Fig. 4A,B, right panels). In both WM115 and WM239A cells, spheroids formed large cell aggregates with diameter comparable to controls, indicating that R-Ras or other effectors regulated by the receptor GAP domain are needed for spheroid growth. Curiously, cells expressing plexin B1-RM showed a marked absence of invasion into matrix compared to control transfectants (Fig. 4A,B). This suggests that although the plexin B1 GAP domain controls spheroid growth, cell invasion is inhibited through other mechanisms. Co-expression of GFP-PDZ with plexin B1-WT was unable to rescue the phenotype of plexin B1 in spheroid culture of WM239A cells (Fig. 4D, Suppl. Fig. 3B).
Together, the results show that in 3D culture, plexin B1 strongly represses tumor forming potential in primary melanoma cells and only partially represses tumor potential in cells from metastatic melanoma. In both spheroids and soft agar culture, the inhibitory mechanism can largely be attributed to signaling through the R-Ras GAP domain in plexin B1, and not through RhoA-GEF regulation.
Plexin B1 inhibits tumor formation in melanoma xenografts
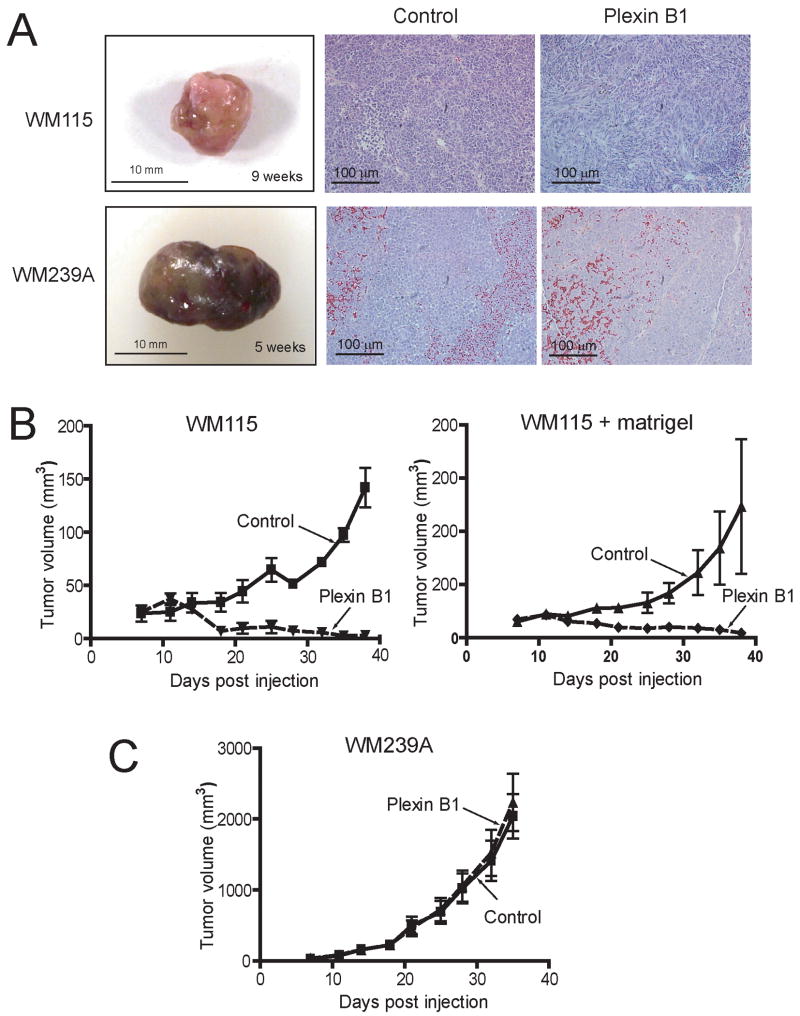
We next examined the effect of plexin B1 on growth of melanoma tumors in vivo. WM115 and WM239A cells under control vs plexin B1 expression conditions were injected intradermally into athymic nude mice. Tumors from control WM115 cells were unpigmented, and showed areas of necrosis upon gross and histological examination (Fig. 5A). In contrast, tumors from control WM239A cells, which are normally unpigmented in vitro, were more pigmented, more hemorrhagic, and more homogeneous, with fewer areas of necrosis (Fig. 5A).
Figure 5. Plexin B1 is a tumor suppressor in melanoma cells.
(A) Xenograft tumors from WM115 cells are smaller and unpigmented, whereas xenografts from WM239A cells are larger and pigmented. No differences in tumor morphology due to plexin B1 expression are apparent in histological sections (hematoxylin + eosin). (B) Xenograft tumor growth from WM115 cells injected intradermally into athymic nude mice is strongly inhibited in cells stably expressing plexin B1. Similar results were obtained in cells injected with Matrigel. (C) In contrast, plexin B1 expression does not affect xenograft growth from WM239A cells.
Stable expression of plexin B1 was maintained in xenografts tumors (Suppl. Fig. 4), and suppressed tumor growth in WM115 xenografts, but in contrast had little effect on the growth of WM239A xenografts (Fig. 5B). Tumor growth rate of WM115 xenografts was slower than WM239A cells, also observed when WM115 cells were co-injected with Matrigel to enhance growth rate. The results demonstrate that plexin B1 acts as a tumor suppressor in melanoma cells isolated from a primary tumor. In contrast, WM239A cells from a matched metastatic tumor showed no effect of plexin B1, indicating that metastatic cells somehow bypass the tumor suppressor response (Fig. 5C).
Mouse tail vein injections of control and plexin B1-expressing cells produced only one lung colony among four mice injected with WM239A cells, and no lung colonies among four mice injected with WM115 cells (data not shown). Therefore, we were unable to determine whether plexin B1 reduced metastases, although the results clearly showed that metastases were not enhanced by plexin B1.
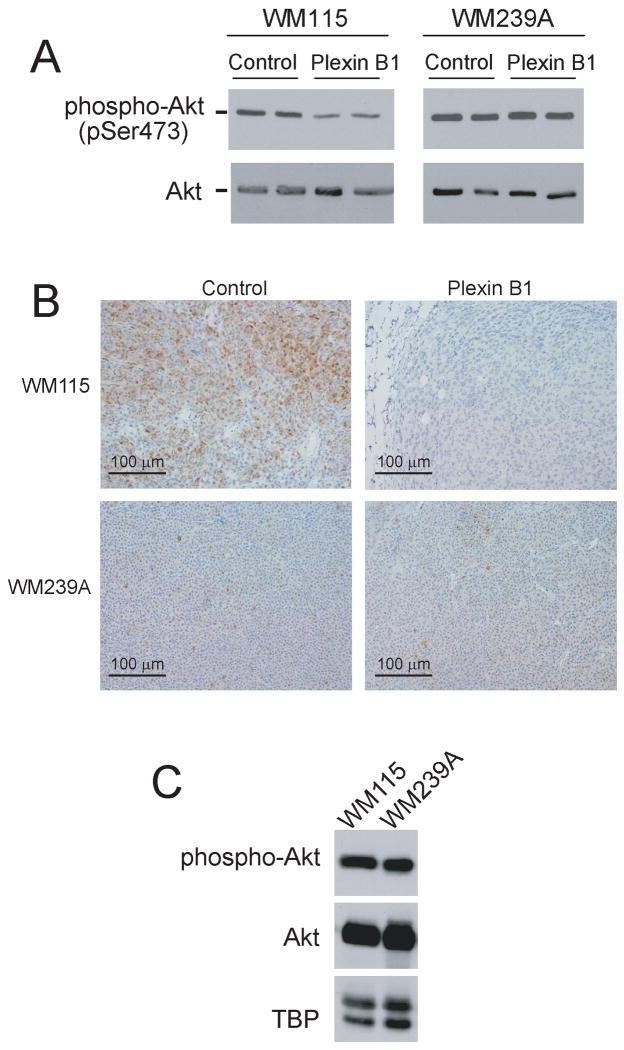
Ttumor suppressor responses correlate with inhibition of AKT
The PI-3 kinase/AKT pathway augments growth of many tumor types and is an established downstream target of R-Ras signaling (36-38). Therefore, we tested whether plexin B1 represses PI-3 kinase/AKT, and found that phospho-AKT was inhibited in WM115 cells expressing plexin B1 compared to controls (Fig. 6A). This was recapitulated in xenograft tumors, where strong phospho-AKT staining was observed in control tumors, but significantly reduced in tumors expressing plexin B1 (Fig. 6B). In contrast, WM239A cells showed no effect of plexin B1 expression on phospho-AKT, either in cultured cells or xenograft tumors (Fig. 6A,B). Although WM239A xenografts showed lower phospho-AKT reactivity overall compared to WM115 control xenografts, Western blots of xenograft tissue lysates showed comparable levels of phospho-AKT and AKT, when normalized to total protein (Fig. 6C). Thus, we believe that the differences in immunohistochemical reactivity between WM115 and WM239A xenografts were most likely due to variations in fixation, not absolute phospho-AKT signal. Overall, the repression of AKT correlated with the tumor suppressor phenotype response to plexin B1 in WM115 and WM239A cells and tumors.
Figure 6. Tumor suppressor activity of plexin B1 is correlated with repression of phospho-AKT.
(A) Western blots show that phospho-AKT is repressed in WM115 cells stably expressing plexin B1, whereas no effect is observed in WM239A cells. Two biological replicates are shown for each condition. (B) Immunohistochemistry of xenografts from WM115 cells shows that phospho-AKT is significantly repressed in tumors from cells expressing plexin B1, whereas plexin B1 has no effect on phospho-AKT in WM239A xenografts. Each image is representative of tumors from three mice. (C) Western blots of lysates from xenografts of WM115 and WM239A cells show comparable levels of phospho-AKT and total AKT, normalized to TATA binding protein (TBP). Thus, the tumor suppressive effect of plexin B1 is correlated with its inhibition of phospho-AKT.
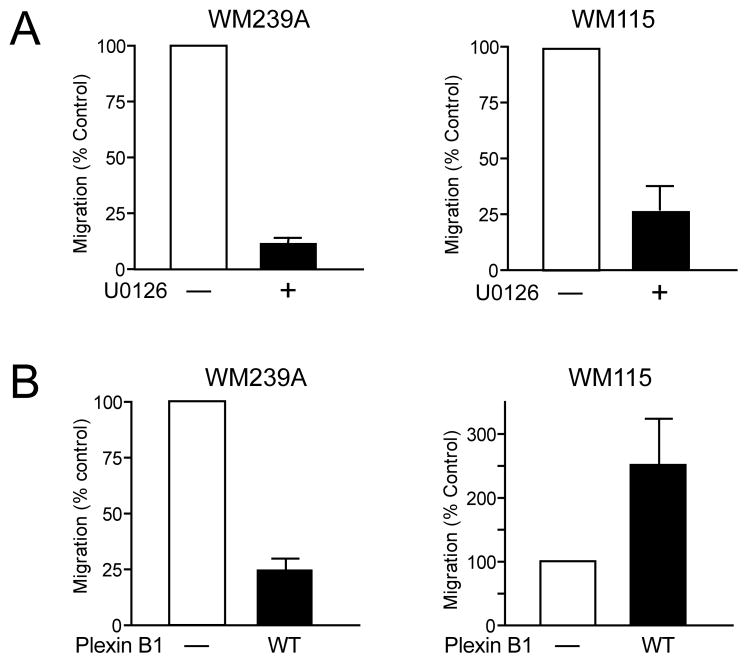
Plexin B1 inhibits migration of metastatic melanoma cells
Melanoma cells treated with MKK inhibitor showed inhibition of cell migration towards chemoattractant (FBS) (Fig. 7A), indicating that B-Raf/MKK/ERK promotes cell migration. We tested whether this response might be controlled through plexin B1. Expression of plexin B1 in WM239A cells inhibited migration compared to controls (Fig. 7B, left panel). The results indicated that plexin B1 suppresses cell migration, suggesting that plexin B1 repression may be one mechanism by which B-Raf/MKK/ERK enhances migration. Curiously, plexin B1 enhanced cell migration when expressed in WM115 cells (Fig. 7B, right panel), opposite to its effect in WM239A cells. The results suggest that B-Raf/MKK/ERK promotes migration in part by repressing plexin B1, and that this response is stronger in metastatic tumor cells. In cells isolated from a matched primary VGP tumor from the same patient, plexin B1 has the opposite effect of enhancing cell migration.
Figure 7. Plexin B1 inhibits migration in metastatic cells.
(A) U0126 inhibits transwell cell migration in WM239A and WM115 cells, indicating that migration is positively regulated by B-Raf/MKK/ERK. (B) Stable expression of plexin B1 inhibits migration of WM239A cells, mimicking the effect of U0126. In contrast, migration of WM115 cells is not inhibited, and is somewhat elevated, by plexin B1.
Discussion
In this report, we demonstrate that B-Raf signaling in melanoma suppresses axon guidance genes, including plexin-semaphorin family members. By investigating the functional importance of one of these genes, plexin B1, we demonstrate a novel tumor suppressor function for plexin B1, revealed by colony growth in soft agar, spheroid growth in collagen, and xenograft tumor growth. A tumor suppressor role for plexin B1 in melanoma was unexpected, because plexin B1 has often been described as a positive regulator of cancer progression, which enhances cell invasion through c-Met interactions (28). The direct evidence that plexin B1 functions as a tumor suppressor increases our understanding of its pleiotropic roles in cancer etiology.
An important feature of our screening approach is its survey of signaling responses for their consistency across many cell lines. This proved to be an effective strategy for eliminating spurious gene responses due to cell variability, improving the stringency of the screen. Clustering candidates by biological function revealed a previously unrecognized cross-regulatory pathway for the control of plexin-semaphorin signaling by MAP kinase pathways. Other microarray experiments in human melanoma cell lines have reported B-Raf mutation-associated gene signatures (39-43). However, prior studies have not revealed connections with axon guidance genes, indicating that additional insight can be gained using inhibitors to directly examine targets responsive to B-Raf/MKK/ERK. The axon guidance genes were in each case induced in response to MKK inhibitor, from which we infer that MAP kinase signaling inhibits plexin and semaphorin expression. In contrast, no evidence for feedback signaling from plexin B1 to ERK was observed. Although reports have shown that plexin B1 activates ERK in other cell systems (44), this appears not true in melanoma, where ERK is already constitutively activated by oncogenic B-Raf. Our dataset provides a valuable resource for discovery of signaling targets of B-Raf in melanoma.
Site-directed mutagenesis implicated the plexin B1 GAP domain in the tumor suppressor response, which has been previously shown to inhibit signaling through R-Ras. Although R-Ras is less potent in transformation than other forms of Ras, it is aberrantly expressed in gastric cancers, promotes proliferation and tumorigenicity in breast and cervical cancer cells, and facilitates invasion and migration in glioma and breast cancer cells (45-51). The signaling mechanisms are not completely understood, however R-Ras activates PI-3 kinase/AKT pathways in many cell types. We found that plexin B1 suppressed AKT phosphorylation at residues needed for kinase activation, in both cells and xenograft tumors. Thus, we speculate that the mechanism for tumor suppression may involve inhibition of PI-3 kinase/AKT, following R-Ras-GTP hydrolysis and inactivation by plexin B1.
Interestingly, the tumor inhibitory response to plexin B1 is reduced or absent in cells isolated from a metastatic tumor compared to cells from a matched primary tumor. Thus, the tumor suppressor role of plexin B1 occurs in cells from early-stage melanomas, and is somehow bypassed in cells from late stage tumors. The absence of tumor suppression by plexin B1 in WM239A xenografts correlated with its lack of effect on phospho-AKT, raising the possibility that alternative pathways in late-stage melanoma may activate AKT independently of plexin B1. For example, c-Met is elevated in many metastatic cancers, and could conceivably supercede responses to plexin B1. This is consistent with the behavior of melanoma cell lines, where c-Met protein is enhanced in metastatic cells compared to cells from matching primary tumors (Suppl. Fig. 5).
On the other hand, plexin B1 suppressed cell migration in cell lines derived from a metastatic tumor, but not a matched primary tumor. Conceivably, alterations in metastatic cells might enable plexin B1 to switch from a tumor suppressor role in primary melanomas, towards a pro-migratory role as tumors progress to metastasis. Similar effects on signaling have been reported in other systems. For example, TGFβ signaling has a tumor suppressor role in early breast cancer cells but switches to a pro-metastatic role as the cells adopt an aggressive metastatic phenotype (52-53). Thus, responses to signaling pathways may change, resulting in selective advantage for a subset of cells depending on the microenvironment or stage of cancer progression. Our findings on plexin B1 provide a preliminary indication that stage-dependent events might likewise bypass tumor suppressor mechanisms in advanced melanomas.
Materials and Methods
Cell lines
NHEM693 and NHEM2493 (Cambrex) were grown in Medium154+Human Melanocyte Growth Supplement (Cascade Biologics). Melanoma cell lines were purchased from ATCC or were a gift from Meenhard Herlyn, Wistar Institute (34). Melanoma cell lines were grown in RPMI (Invitrogen), 10% fetal bovine serum (FBS, Gemini) and penicillin-streptomycin (100 U/mL). Media for WM1789 cells included insulin (5 μg/mL) and bFGF (7 ng/mL, R&D).
Expression constructs
Adenovirus for expressing constitutively active (CA) human MKK1 (“R4F”Δ(32-51)/S281E/S222D) was a gift of Andrey Sorokin, Medical College of Wisconsin. VSV-tagged human plexin B1 (WT and RM: R1677M/R1678M) in pCDNA3 were gifts of Kun-liang Guan, U. California, San Diego. Lentivirus constructs for expression of GFP-PDZ-RhoGEF and GFP were gifts of J. Silvio Gutkind, NIH. A retroviral vector for GFP expression (pREX-GFP) was a gift of Xuedong Liu, U. Colorado, Boulder. WM115 and WM239A cells were transfected with pCDNA3-VSV-plexin B1, then selected in RPMI-10% FBS containing 0.4 μg/mL G418 (Invitrogen). Plexin B1-expressing cells labeled with anti-VSV antibody (Roche) and anti-mouse FITC were enriched twice by FACS, each time collecting the top half of FITC-positive cells. Plexin B1 knockdown was performed using SmartPool RNAi with siScramble or lamin-A/C RNAi controls (Dharmacon), transfecting cells in suspension with DMRIE-C (Invitrogen). Knockdown was maximal after 1-2 days and sustained for up to 7 days.
Microarrays
Melanoma cells were treated one day after plating with RPMI-0.01% FBS containing 0.1% DMSO, 10 μM U0126 (Promega) or 3 μM PD184352 (CI-1040, a generous gift of Pfizer, Inc.). After 24 h, cells were washed with PBS and total RNA was isolated (RNeasy, Qiagen) and stored in aliquots at -80°C. cDNA synthesis, transcription of biotin-labeled cRNA, and fragmentation were carried out following standard protocols from the Affymetrix Technical Manual (http://www.affymetrix.com). Samples were hybridized onto U133A GeneChips and processed at the CU-Boulder Microarray Core Facility. Data were background corrected using MicroArray Suite 5.0 and normalized by robust multi-array average quantile normalization (RMAExpress v.0.1). Data were analyzed using Rank Products software from the University of Glasgow (54), calculating the log ratio, log2(A/B) = log2A-log2B for each cell line, where A and B correspond to control and U0126-treated conditions. RANK output files were filtered using false discovery rate [(false positives)÷(false positives+true positives)]≤0.001, yielding less than one false positive feature per file.
Western blots
Cell extracts in 2% SDS, 20 mM Tris pH 7.5, 10 mM dithiothreitol were boiled for 30 min. Protein was quantified by DC assay (Bio-Rad), and lysates normalized by total protein. Western blots were transferred overnight at 4°C, membranes were blocked with 5% nonfat milk and washed in Tris-buffered saline+0.1% Tween-20. Primary antibodies recognized plexin B1 (Santa Cruz, sc-25642, 1: 500), semaphorin 4D (BD Transduction Laboratories, 610670, 1:500), phospho-Akt (Cell Signaling, 9271, 1:1000), Akt (Cell Signaling, 9272, 1:2000), phospho-ERK1/2 (Sigma, M8159, 1:12,000), ERK2 (Santa Cruz, sc-154, 1:5000), and c-Met (Santa Cruz, sc-10, 1:1000). Blots were probed with secondary antibody coupled to horseradish peroxidase (Jackson ImmunoResearch, 1:10,000) and visualized by enhanced chemiluminescence (Amersham).
Cell migration
25,000 cells were plated into Transwell chambers (Falcon 353097) in serum-free RPMI, using RPMI-10% FBS as the bottom chamber chemoattractant. Cells on the top membrane were trypsinized and counted on a hemacytometer, after scrubbing cells from the bottom membrane. Cells on the bottom membrane were quantified by fixing cells in methanol and staining with hematoxylin after scrubbing the top membrane, manually counting the entire membrane. Cell migration=(#cells on bottom)÷(#top + #bottom).
Proliferation
Growth of cells was quantified by incubating cells in 96-wells with 4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT; Molecular Probes) for 4 h prior to harvesting. MTT-formazan was solubilized in 100 μL DMSO and quantified at 540 nm using a microplate reader. Background corrections were performed by subtracting absorbance measurements from wells without cells.
Xenografts
Stably transfected WM115 cells (“plexin B1”=VSV-plexin B1+GFP or “control”=GFP alone) were trypsinized and resuspended to 4×107 cells/mL in RPMI-10%FCS or the same medium mixed 1:1 with Matrigel. Stably transfected WM239A cells were resuspended to 2×107 cells/mL in RPMI-10%FCS. Each cell line (100 μL) was injected intradermally to the right posterior flank of six female athymic nu/nu mice (NIH; 8-9 weeks old), and bidimensional tumor measurements were taken for 4-12 weeks. Tumor volume equalled (π×short diameter2×long diameter)÷6. Prior to harvest, mice were injected intraperitoneally with bromodeoxyuridine solution (30 μg/g), and sacrificed 2 h later by cervical dislocation. Tumors were harvested and fixed overnight in 10% neutral buffered formalin (Sigma). Fixed tissues were embedded and sectioned at the UCHSC Histopathology Laboratory. Lungs were dissected and fixed overnight in 4% formaldehyde+PBS+30% sucrose. Fixed tissues were frozen in freezing medium (TFM-C, Triangle Biomedical) and sectioned on a cryostat. Eight μm sections were taken every 100 μm of tissue, dried overnight and mounted with Prolong Gold Antifade Reagent containing DAPI (Molecular Probes). Housing and care of animals was in accordance with UCHSC-IACUC guidelines.
Immunohistochemistry
Slides were deparaffinized, rehydrated and treated for 30 min with 0.3% H2O2 to block endogenous peroxidase activity. Antigen retrieval was performed in a decloaking chamber (Biocare) with antigen retrieval solution (DakoCytomation). Slides were blocked for 1 h in 2% BSA, incubated with primary antibody overnight at 4°C, incubated with anti-rabbit EnVision-system labeled polymer-HRP (DakoCytomation) for 30 min, r.t., and developed with DAB+solution (DakoCytomation). Slides were counterstained with hematoxylin (Anatech).
3D culture
Spheroids were formed and grown in collagen as described (34). Cells were plated in triplicate on 1.5% Noble agar for 3-5 days. Spheroids were collected and centrifuged (2000 rpm) to remove media, resuspended in 2.5 mg/mL bovine dermal collagen (PureCol, INAMED) in RPMI-10% FBS, L-glutamine, pyruvate and penicillin-streptomycin, and overlaid on a pre-solidified layer of the same collagen solution. Collagen/cell suspensions were allowed to solidify for 1 h, then overlaid with RPMI-10% FBS. Spheroids were grown for 7-10 days to monitor invasion before fixing with neutral buffered formalin.
Viability
Spheroid-collagen cultures were rinsed with PBS and incubated for 2 h with 100 μL 10 μM calcein AM + 20 μM ethidium homodimer-1 (Live/Dead Viability/Cytotoxicity Kit, Molecular Probes). Cell images were collected with an Olympus IX81 microscope, DP70-CCD camera, and pseudo-colored using Adobe Photoshop.
Soft agar
0.6% Noble agar in RPMI-10%FBS was added to 6 well plates (2 mL/well) and allowed to solidify. Cells (25,000) were trypsinized and plated in triplicate, resuspended in 1.5 mL of 0.35% Noble agar in RPMI-10%FBS. After the agar solidified, 1 mL RPMI-10%FBS was added, changing media every 3-4 days. After 10-21 days, colonies were fixed, stained with crystal violet, and counted.
Caspase 3
Cells were lysed in 40 mM PIPES pH 7.2, 200 mM NaCl, 2 mM EDTA, 20% sucrose, 0.2 % CHAPS, 20 mM DTT. Lysates (50 μg) were aliquoted in triplicate into 96 well plates. Reactions were initiated by addition of colorimetric caspase substrate Ac-DEVD-pNA (QuantiPak, Biomol, AK-004) at 37 °C, reading 405 nm absorbance at 30 and 50 min.
Supplementary Material
1
2
3
4
5
Acknowledgments
We thank Meenhard Herlyn for providing melanoma cell lines used in this study, and Silvio Gutkind, Kun-liang Guan, Xuedong Liu, and Andrey Sorokin for expression plasmids. We also thank Norma Aumen for tumor sectioning, Helen Marshall for microarray analysis, and Micah Hamady and Rob Knight for providing access to GO-Getter. This work was supported by NIH awards R01-CA118972 (N.G.A.) and P50-CA058187 (D.C.), and fellowship awards F32-CA105796 (G.M.A.), ACS-PF-04-152 (C.H.C.), and T32-GM008759 (K.L.C.).
Footnotes
1
Hamady, M. and Knight, R. “GO-Getter: Exploiting Gene Ontology and other functional classifications for high-throughput genome, transcriptome, and proteome analyses” In preparation.
References
- 1.Dhomen N, Marais R. New insight into BRAF mutations in cancer. Curr Opin Genet Dev. 2007;17:31–39. doi: 10.1016/j.gde.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 2.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 3.Brose MS, Volpe P, Feldman M, et al. BRAF and RAS mutations in human lung cancer and melanoma. Cancer Res. 2002;62:6997–7000. [PubMed] [Google Scholar]
- 4.Wan PT, Garnett MJ, Roe SM, et al. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116:855–867. doi: 10.1016/s0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- 5.Bogenrieder T, Herlyn M. Cell-surface proteolysis, growth factor activation and intercellular communication in the progression of melanoma. Crit Rev Oncol Hematol. 2002;44:1–15. doi: 10.1016/S1040-8428(01)00196-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Omholt K, Platz A, Kanter L, Ringborg U, Hansson J. NRAS and BRAF mutations arise early during melanoma pathogenesis and are preserved throughout tumor progression. Clin Cancer Res. 2003;9:6483–6488. [PubMed] [Google Scholar]
- 7.Pollock PM, Harper UL, Hansen KS, et al. High frequency of BRAF mutations in nevi. Nat Genet. 2003;33:19–20. doi: 10.1038/ng1054. [DOI] [PubMed] [Google Scholar]
- 8.Zhuang L, Lee CS, Scolyer RA, et al. Activation of the extracellular signal regulated kinase (ERK) pathway in human melanoma. J Clin Pathol. 2005;58:1163–1169. doi: 10.1136/jcp.2005.025957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hingorani SR, Jacobetz MA, Robertson GP, Herlyn M, Tuveson DA. Suppression of BRAF(V599E) in human melanoma abrogates transformation. Cancer Res. 2003;63:5198–5202. [PubMed] [Google Scholar]
- 10.Sharma A, Trivedi NR, Zimmerman MA, Tuveson DA, Smith CD, Robertson GP. Mutant V599EB-Raf regulates growth and vascular development of malignant melanoma tumors. Cancer Res. 2005;65:2412–2421. doi: 10.1158/0008-5472.CAN-04-2423. [DOI] [PubMed] [Google Scholar]
- 11.Koo HM, VanBrocklin M, McWilliams MJ, Leppla SH, Duesbery NS, Woude GF. Apoptosis and melanogenesis in human melanoma cells induced by anthrax lethal factor inactivation of mitogen-activated protein kinase kinase. Proc Natl Acad Sci U S A. 2002;99:3052–3057. doi: 10.1073/pnas.052707699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Negishi M, Oinuma I, Katoh H. Plexins: axon guidance and signal transduction. Cell Mol Life Sci. 2005;62:1363–1371. doi: 10.1007/s00018-005-5018-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujisawa H. Discovery of semaphorin receptors, neuropilin and plexin, and their functions in neural development. J Neurobiol. 2004;59:24–33. doi: 10.1002/neu.10337. [DOI] [PubMed] [Google Scholar]
- 14.Vikis HG, Li W, Guan KL. The plexin-B1/Rac interaction inhibits PAK activation and enhances Sema4D ligand binding. Genes Dev. 2002;16:836–845. doi: 10.1101/gad.966402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perrot V, Vazquez-Prado J, Gutkind JS. Plexin B regulates Rho through the guanine nucleotide exchange factors leukemia-associated Rho GEF (LARG) and PDZ-RhoGEF. J Biol Chem. 2002;277:43115–43120. doi: 10.1074/jbc.M206005200. [DOI] [PubMed] [Google Scholar]
- 16.Swiercz JM, Kuner R, Behrens J, Offermanns S. Plexin-B1 directly interacts with PDZ-RhoGEF/LARG to regulate RhoA and growth cone morphology. Neuron. 2002;35:51–63. doi: 10.1016/s0896-6273(02)00750-x. [DOI] [PubMed] [Google Scholar]
- 17.Driessens MH, Hu H, Nobes CD, et al. Plexin-B semaphorin receptors interact directly with active Rac and regulate the actin cytoskeleton by activating Rho. Curr Biol. 2001;11:339–344. doi: 10.1016/s0960-9822(01)00092-6. [DOI] [PubMed] [Google Scholar]
- 18.Aurandt J, Vikis HG, Gutkind JS, Ahn N, Guan KL. The semaphorin receptor plexin-B1 signals through a direct interaction with the Rho-specific nucleotide exchange factor, LARG. Proc Natl Acad Sci U S A. 2002;99:12085–12090. doi: 10.1073/pnas.142433199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oinuma I, Ishikawa Y, Katoh H, Negishi M. The Semaphorin 4D receptor Plexin-B1 is a GTPase activating protein for R-Ras. Science. 2004;305:862–865. doi: 10.1126/science.1097545. [DOI] [PubMed] [Google Scholar]
- 20.Oinuma I, Katoh H, Negishi M. Molecular dissection of the semaphorin 4D receptor plexin-B1-stimulated R-Ras GTPase-activating protein activity and neurite remodeling in hippocampal neurons. J Neurosci. 2004;24:11473–11480. doi: 10.1523/JNEUROSCI.3257-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Charest PG, Firtel RA. Big roles for small GTPases in the control of directed cell movement. Biochem J. 2007;401:377–390. doi: 10.1042/BJ20061432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dickson BJ. Rho GTPases in growth cone guidance. Curr Opin Neurobiol. 2001;11:103–110. doi: 10.1016/s0959-4388(00)00180-x. [DOI] [PubMed] [Google Scholar]
- 23.Fukata M, Nakagawa M, Kaibuchi K. Roles of Rho-family GTPases in cell polarisation and directional migration. Curr Opin Cell Biol. 2003;15:590–597. doi: 10.1016/s0955-0674(03)00097-8. [DOI] [PubMed] [Google Scholar]
- 24.Basile JR, Barac A, Zhu T, Guan KL, Gutkind JS. Class IV semaphorins promote angiogenesis by stimulating Rho-initiated pathways through plexin-B. Cancer Res. 2004;64:5212–5224. doi: 10.1158/0008-5472.CAN-04-0126. [DOI] [PubMed] [Google Scholar]
- 25.Conrotto P, Valdembri D, Corso S, et al. Sema4D induces angiogenesis through Met recruitment by Plexin B1. Blood. 2005;105:4321–4329. doi: 10.1182/blood-2004-07-2885. [DOI] [PubMed] [Google Scholar]
- 26.Wong OG, Nitkunan T, Oinuma I, Zhou C, Blanc V, Brown RS, Bott SR, Naruculam J, Box G, Munson P, Constantinou J, Feneley MR, Klocker H, Eccles SA, Negishi M, Freeman A, Masters JR, Williamson M. Plexin-B1 mutations in prostate cencer. Proc Natl Acad Sci USA. 2007;104:19040–19045. doi: 10.1073/pnas.0702544104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giordano S, Corso S, Conrotto P, et al. The semaphorin 4D receptor controls invasive growth by coupling with Met. Nat Cell Biol. 2002;4:720–724. doi: 10.1038/ncb843. [DOI] [PubMed] [Google Scholar]
- 28.Conrotto P, Corso S, Gamberini S, Comoglio PM, Giordano S. Interplay between scatter factor receptors and B plexins controls invasive growth. Oncogene. 2004;23:5131–5137. doi: 10.1038/sj.onc.1207650. [DOI] [PubMed] [Google Scholar]
- 29.Swiercz JM, Worzfeld T, Offermanns S. ErbB-2 and Met reciprocally regulate cellular signaling via plexin-B1. J Biol Chem. 2008;283:1893–1901. doi: 10.1074/jbc.M706822200. [DOI] [PubMed] [Google Scholar]
- 30.Basile JR, Castilho RM, Williams VP, Gutkind JS. Semaphorin 4D provides a link between axon guidance processes and tumor-induced angiogenesis. Proc Natl Acad Sci USA. 2006;103:9017–9022. doi: 10.1073/pnas.0508825103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bielenberg DR, Hida Y, Shimizu A, et al. Semaphorin 3F, a chemorepulsant for endothelial cells, induces a poorly vascularized, encapsulated, nonmetastatic tumor phenotype. J Clin Invest. 2004;114:1260–1271. doi: 10.1172/JCI21378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kessler O, Shraga-Heled N, Lange T, et al. Semaphorin-3F is an inhibitor of tumor angiogenesis. Cancer Res. 2004;64:1008–1015. doi: 10.1158/0008-5472.can-03-3090. [DOI] [PubMed] [Google Scholar]
- 33.Rody A, Holtrich U, Gaetje R, et al. Poor outcome in estrogen receptor-positive breast cancers predicted by loss of plexin B1. Clin Cancer Res. 2007;13:1115–1122. doi: 10.1158/1078-0432.CCR-06-2433. [DOI] [PubMed] [Google Scholar]
- 34.Hsu MY, Elder DE, Herlyn M. Melanoma: The Wistar melanoma (WM) cell lines. Human Cell Culture. 1999;1:259–274. [Google Scholar]
- 35.Smalley KS, Haass NK, Brafford PA, Lioni M, Flaherty KT, Herlyn M. Multiple signaling pathways must be targeted to overcome drug resistance in cell lines derived from melanoma metastases. Mol Cancer Ther. 2006;5:1136–1144. doi: 10.1158/1535-7163.MCT-06-0084. [DOI] [PubMed] [Google Scholar]
- 36.Marte BM, Rodriguez-Viciana P, Wennstrom S, Warne PH, Downward J. R-Ras can activate the phosphoinositide 3-kinase but not the MAP kinase arm of the Ras effector pathways. Curr Biol. 1997;7:63–70. doi: 10.1016/s0960-9822(06)00028-5. [DOI] [PubMed] [Google Scholar]
- 37.Osada M, Tolkacheva T, Li W, et al. Differential roles of Akt, Rac, and Ral in R-Ras-mediated cellular transformation, adhesion, and survival. Mol Cell Biol. 1999;19:6333–6344. doi: 10.1128/mcb.19.9.6333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ito Y, Oinuma I, Katoh H, Kaibuchi K, Negishi M. Sema4D/plexin-B1 activates GSK-3beta through R-Ras GAP activity, inducing growth cone collapse. EMBO Rep. 2006;7:704–709. doi: 10.1038/sj.embor.7400737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johansson P, Pavey S, Hayward N. Confirmation of a BRAF mutation-associated gene expression signature in melanoma. Pigment Cell Res. 2007;20:216–221. doi: 10.1111/j.1600-0749.2007.00375.x. [DOI] [PubMed] [Google Scholar]
- 40.Pavey S, Johansson P, Packer L, et al. Microarray expression profiling in melanoma reveals a BRAF mutation signature. Oncogene. 2004;23:4060–4067. doi: 10.1038/sj.onc.1207563. [DOI] [PubMed] [Google Scholar]
- 41.Hoek KS, Schlegel NC, Brafford P, et al. Metastatic potential of melanomas defined by specific gene expression profiles with no BRAF signature. Pigment Cell Res. 2006;19:290–302. doi: 10.1111/j.1600-0749.2006.00322.x. [DOI] [PubMed] [Google Scholar]
- 42.Bloethner S, Chen B, Hemminki K, et al. Effect of common B-RAF and N-RAS mutations on global gene expression in melanoma cell lines. Carcinogenesis. 2005;26:1224–32. doi: 10.1093/carcin/bgi066. [DOI] [PubMed] [Google Scholar]
- 43.Shields JM, Thomas NE, Cregger M, et al. Lack of extracellular signal-regulated kinase mitogen-activated protein kinase signaling shows a new type of melanoma. Cancer Res. 2007;67:1502–1512. doi: 10.1158/0008-5472.CAN-06-3311. [DOI] [PubMed] [Google Scholar]
- 44.Basile JR, Gavard J, Gutkind JS. Plexin-B1 utilizes RhoA and Rho kinase to promote the integrin-dependent activation of Akt and ERK and endothelial cell motility. J Biol Chem. 2007;282:34888–34895. doi: 10.1074/jbc.M705467200. [DOI] [PubMed] [Google Scholar]
- 45.Cox AD, Brtva TR, Lowe DG, Der CJ. R-Ras induces malignant, but not morphologic, transformation of NIH3T3 cells. Oncogene. 1994;9:3281–3288. [PubMed] [Google Scholar]
- 46.Nishigaki M, Aoyagi K, Danjoh I, et al. Discovery of aberrant expression of R-RAS by cancer-linked DNA hypomethylation in gastric cancer using microarrays. Cancer Res. 2005;65:2115–2124. doi: 10.1158/0008-5472.CAN-04-3340. [DOI] [PubMed] [Google Scholar]
- 47.Yu Y, Feig LA. Involvement of R-Ras and Ral GTPases in estrogen-independent proliferation of breast cancer cells. Oncogene. 2002;21:7557–7568. doi: 10.1038/sj.onc.1205961. [DOI] [PubMed] [Google Scholar]
- 48.Mora N, Rosales R, Rosales C. R-Ras promotes metastasis of cervical cancer epithelial cells. Cancer Immunol Immunother. 2007;56:535–544. doi: 10.1007/s00262-006-0205-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rincon-Arano H, Rosales R, Mora N, Rodriguez-Castaneda A, Rosales C. R-Ras promotes tumor growth of cervical epithelial cells. Cancer. 2003;97:575–585. doi: 10.1002/cncr.11093. [DOI] [PubMed] [Google Scholar]
- 50.Nakada M, Niska JA, Tran NL, McDonough WS, Berens ME. EphB2/R-Ras signaling regulates glioma cell adhesion, growth, and invasion. Am J Pathol. 2005;167:565–576. doi: 10.1016/S0002-9440(10)62998-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Keely PJ, Rusyn EV, Cox AD, Parise LV. R-Ras signals through specific integrin alpha cytoplasmic domains to promote migration and invasion of breast epithelial cells. J Cell Biol. 1999;145:1077–1088. doi: 10.1083/jcb.145.5.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tang B, Vu M, Booker T, et al. TGF-beta switches from tumor suppressor to prometastatic factor in a model of breast cancer progression. J Clin Invest. 2003;112:1116–1124. doi: 10.1172/JCI18899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tian F, DaCosta Byfield S, Parks WT, et al. Reduction in Smad2/3 signaling enhances tumorigenesis but suppresses metastasis of breast cancer cell lines. Cancer Res. 2003;63:8284–8292. [PubMed] [Google Scholar]
- 54.Breitling R, Armengaud P, Amtmann A, Herzyk P. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 2004;573:83–92. doi: 10.1016/j.febslet.2004.07.055. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2
3
4
5