Effects of Tissue Handling on RNA Integrity and Microarray Measurements From Resected Breast Cancers (original) (raw)
Abstract
Background
Reliable stability and yield of RNA from breast cancer tissues are important for biobanking, clinical trials, and diagnostic testing.
Methods
Aliquots of fresh primary tumor tissue from 17 surgically resected invasive breast cancers were placed into RNAlater at room temperature after tumor removal (baseline) and up to 3 hours thereafter or were snap frozen at baseline and 40 minutes thereafter. Samples were stored at −80°C until gene expression profiling with Affymetrix Human Gene U133A microarrays. We evaluated the effects of cold ischemic time (the time from tumor specimen removal to sample preservation) and sample preservation method on RNA yield, Bioanalyzer-based RNA integrity number, microarray-based 3′/5′ expression ratios for assessing transcript integrity, and microarray-based measurement of single-gene and multigene expression signatures. The statistical significance of the effects was assessed using linear mixed effects regression models. All statistical tests were two-sided.
Results
Sample preservation in RNAlater statistically significantly improved RNA integrity compared with snap freezing as assessed by the RNA integrity number, which increased from 7.31 to 8.13 units (difference = 0.82 units, 95% confidence interval [CI] = 0.53 to 1.11 units, P < .001), and RNA yield, which increased threefold from 8.9 to 28.6 μg (difference = 19.7 μg, 95% CI = 14.1 to 25.3 μg, P < .001). Prolonged cold ischemic delay at room temperature before sample stabilization decreased the RNA integrity number by 0.12 units/h (95% CI = 0.02 to 0.23 units/h) compared with a projected average RNA integrity number of 7.39 if no delays were present (P = .008) and decreased the RNA yield by 1.5 μg/h (95% CI = 0 to 4 μg/h) from a baseline mean RNA yield of 33.5 μg if no delays were present (P = .019). Prolonged cold ischemia statistically significantly increased the 3′/5′ ratio of control gene transcripts, particularly of STAT1 (P < .001). Snap freezing statistically significantly increased the 3′/5′ ratio of three control transcripts (ACTB_,_ GAPDH, and 18S rRNA). Expression levels of single genes and multigene signatures for breast cancer were largely unaffected by sample preservation method or cold ischemia.
Conclusions
Sample preservation in RNAlater improves RNA yield and quality, whereas cold ischemia increases RNA fragmentation as measured by the 3′/5′ expression ratio of control genes. However, expression levels of single genes and multigene signatures that are of diagnostic relevance in breast cancer were mostly unaffected by sample preservation method or prolonged cold ischemic duration.
CONTEXT AND CAVEATS
Prior knowledge
Genomic profiling based on measurements of RNA expression in biopsy specimens is used to identify molecular subclasses of tumors that are associated with different prognostic outcomes or differential response to breast cancer treatments. Parameters of tumor sample collection can vary widely and may affect the quality of isolated RNA and expression measurements of single genes and multigene signatures.
Study design
A systematic evaluation of the effects of tissue preservation method (ie, RNAlater vs snap freezing) and prolonged cold ischemia on RNA quality and expression of single genes and multigene signatures across 17 primary breast tumors that represented a range of disease stages and surgical conditions.
Contribution
Samples preserved in RNAlater had improved RNA integrity and threefold greater RNA yield compared with snap-frozen samples. Exposure of tumor tissue to an additional 40 minutes at room temperature before preservation did not affect the integrity or the purity of the extracted RNA. Tissue samples that sat for up to 3 hours at room temperature before preservation in RNAlater had a small but statistically significant reduction in the RNA integrity number but no change in RNA yield or purity compared with samples preserved immediately. Nevertheless, there was no or minimal effect on the expression of single genes or multigene signatures of breast cancer.
Implications
RNAlater is better than snap freezing for collection of RNA with high yield and quality for gene expression profiling of small tissue samples from patients with breast cancer. Typical delays that could occur during tissue sampling do not adversely affect RNA yield or quality for evaluation of gene expression using microarrays, even if moderate degradation of transcripts is evident.
Limitation
The study was not designed or powered to evaluate the influence of other factors, such as anesthesia, other medications, or other stress responses related to surgery on RNA yield and/or integrity.
From the Editors
Genomic profiling has enabled the identification of molecular subclasses of tumors that are associated with different prognostic outcomes (1) or differential response to breast cancer treatments (2–4). Such multiplex molecular assays could support personalized cancer treatment (5,6). Indeed, multigene predictive or prognostic tests based on measurements of RNA expression—some of which use a microarray platform (7)—are now offered commercially. In addition, microarray-based genomic data are widely used by cancer researchers. However, information about how the biological samples were collected and preserved is usually limited in research tissue cohorts. Therefore, to inform best practices for sample collection in clinical research and practice, it is important to evaluate systematically variables of biospecimen integrity that affect the quality of isolated RNA and expression measurements of single genes and multigene signatures (8–10).
There are two main sources of variability in gene expression measurements: pre-analytic sources, which are related to the tumor sample itself and influenced by the sample collection process, and technical sources, which are related to the analytical platform. The Food and Drug Administration–sponsored MicroArray Quality Control Consortium assessment of the analytical performance of several quantitative gene expression platforms confirmed agreement in gene expression measurements generated by different commercial microarray platforms (11) and by quantitative real-time polymerase chain reaction (12). However, these studies used commercially available universal human reference RNAs and thus did not evaluate the effect of pre-analytic sources of variability. Tumor specimen collection, handling, or processing can affect the quality of RNA and distort gene expression patterns that are typically associated with a disease condition (13). Relevant factors that are associated with sample collection include warm ischemic duration (ie, the time between surgical incision and tumor specimen removal) (14–17), activation of cellular stress response induced by surgical manipulation (18,19), cold ischemic duration (ie, the time from tumor specimen removal to sample preservation) (20,21), methods of tissue processing and storage (21,22), and intratumor heterogeneity (23–25). Some types of tissue appear to be more susceptible to pre-analytic variation (13); in general, tumor specimens appear to be less prone to pre-analytic stresses compared with normal tissue from the corresponding organ or site (26).
Several groups (27,28) have proposed guidelines for human tumor specimen collection in clinical and translational research studies. Nonetheless, studies that are specifically designed to compare effects of variables in specimen handling on multigene assay predictions should inform future refinements of standard operating procedures for clinical research and testing. Such studies could prioritize the most critical and correctible variables so that their effects can be minimized by generally implementable standardized collection protocols in a clinical setting. This study was designed to evaluate the effects of post-resection cold ischemic time and tissue preservation method on RNA yield and integrity and on microarray-based measurements of gene expression in the context of breast cancer.
Patients and Methods
Study Design and Tumor Specimen Collection
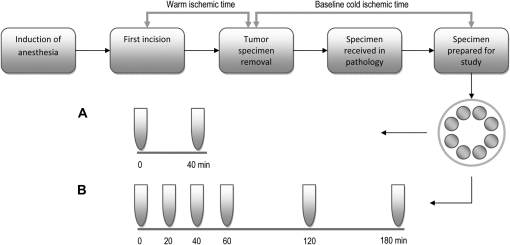
Tumor tissue samples from 11 previously untreated breast cancer patients were collected at the time of intraoperative pathology assessment of surgical resection specimens at the University of Texas M.D. Anderson Cancer Center (MDACC). All patients provided written informed consent. The study was approved by the Institutional Review Board of MDACC. The times at which the first surgical incision was made and the specimen was removed from the patient were recorded in the operating room. Surgical resection specimens were immediately transported at room temperature to the adjacent Pathology Department for immediate pathological evaluation, including measurement, inking, slicing, and macroscopic evaluation of the tumor and margins, with radiological specimen evaluation, if required. Thereafter, a pathologist obtained a piece of tumor tissue for this study if the tumor was grossly obvious, measured more than 2.0 cm in diameter, and was considered by the pathologist on duty to not be required for diagnostic or other consented research use. To minimize intratumoral heterogeneity, the piece of tumor tissue was minced into fragments of 1–2 mm in a Petri dish using a sterile blade, and those fragments were stirred and divided into eight grossly equal portions. A portion of the tissue fragments was placed into 1.5 mL of RNAlater RNA stabilization reagent at room temperature (Ambion Inc, Austin, TX) immediately after mincing (defined as 0 minutes or baseline), and the remaining portions were placed in 1.5 mL of room temperature RNAlater after being held at room temperature for 20, 40, 60, 120, or 180 minutes, or snap frozen in dry ice in a prechilled sample vial immediately (0 minutes or baseline) and after 40 minutes at room temperature (Figure 1). The time that each portion of tissue fragments was placed into contact with RNAlater solution or in the prechilled vial was recorded in the Pathology Department. The lid of the Petri dish was closed during the additional time that the sample was held at room temperature to prevent drying or contamination of tissue. All stabilized samples were stored at −80°C until RNA extraction. For each sample, we recorded the time that it was placed in the −80°C freezer and the time that it was removed from the freezer for RNA extraction. For the first 11 samples, the selection of which tissue aliquot to collect from the Petri dish at each given time was informal (ie, in no particular sequence) because the aliquots were grossly similar. We subsequently extended the study with an additional six breast cancers that were processed as described above, except that to address any potential bias due to size of the tissue portions, the grossly similar tissue portions were assigned to the different conditions of preservation and cold ischemia as follows: The aliquots were placed on prenumbered sections in the Petri dish and each aliquot number was preassigned to a treatment according to a randomized scheme. RNA was extracted from all 17 samples; microarray profiles were generated from the first 11 samples only.
Figure 1.
Study design. Tissue specimen preparation involved mincing tumor tissue in a Petri dish, mixing the pieces, and dividing the mixture into eight grossly identical portions. Randomly selected portions were then snap frozen in dry ice in a prechilled sample vial immediately (baseline, 0 minutes) or after 40 minutes at room temperature (A), or placed into RNAlater immediately (baseline, 0 minutes) or after 20, 40, 60, 120, or 180 minutes at room temperature (B).
RNA Isolation and Quality Assessment
Total RNA was extracted from breast tumor tissue with the use of an RNeasy kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. The concentration of total RNA (ng/μL) was determined at an absorbance of 260 nm (A260) using a Nanodrop ND-1000 spectrophotometer (Thermo Scientific, Wilmington, DE) and was used to calculate the total RNA yield (μg); RNA purity was assessed by measuring absorbance at 260 nm and at 280 nm (A280) and was based on the A260/A280 ratio. Samples with total RNA yield of at least 1 μg and an A260/A280 ratio of at least 1.8 were considered to be of sufficient quality for further analysis. The RNA was analyzed using the Agilent 2100 Bioanalyzer RNA 6000 Nano LabChip (Agilent Technologies, Palo Alto, CA) to produce an electrophoresis trace from which the RNA integrity number was calculated using the 2100 Expert Software (Agilent Technologies). RNA integrity number values range from 10 for intact RNA to 1 for totally degraded; values greater than 6 are generally considered to be of acceptable integrity for gene expression measurement (29).
Analysis of Gene Expression by Microarray Hybridization
We generated gene expression profiles for the first set of 11 tumor samples (MD40–MD71; Table 1). Complementary DNA (cDNA) synthesis and complementary RNA (cRNA) generation were performed as described previously (4). Briefly, first-strand cDNA synthesis was performed by using 1 μg of total RNA and Superscript II (Invitrogen, Carlsbad, CA). Second-strand cDNA synthesis was performed in the presence of DNA polymerase I, DNA ligase, and RNase H (Invitrogen). The double-stranded cDNA was then transcribed into cRNA in the presence of biotin-labeled ribonucleotides with the use of a BioArray HighYield RNA Transcript Labeling kit (Enzo Life Sciences, Plymouth Meeting, PA). The amplified, biotin-labeled cRNA was purified on RNeasy columns (Invitrogen), quantified spectrophometrically, and incubated in the presence of fragmentation buffer (×1 tris acetate, magnesium acetate, potassium acetate) (31) at 94°C for 35 minutes. The resulting fragmented cRNA (10 μg) was hybridized to Affymetrix human genome U133A gene chips (Affymetrix Inc, Santa Clara, CA) at 42°C for 16 hours according to the manufacturer's instructions. Slides containing the hybridized gene chips were scanned using a GeneChip scanner 3000 (Affymetrix Inc), and the scanned images were analyzed using GeneChip Operating Software (Affymetrix Inc) to produce the raw gene expression data files. Datasets from this study are available at the Gene Expression Omnibus repository (http://www.ncbi.nlm.nih.gov/geo/) under accession ID GSE25011.
Table 1.
Patient clinical and pathological characteristics and details of sample collection*
| Patient ID | Age at diagnosis, y | Histology at diagnosis | ER status | PR status | HER2 status | Pathological lymph node status | Tumor size, cm | Histological grade | AJCC stage† | Surgical procedure | Total duration of surgery, min | Warm ischemic time‡, min | Baseline cold ischemic time§, min |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MD40 | 78 | IDC | N | N | N | P | 2.8 | 3 | IIB | SM, ALND | 145 | 21 | 35 |
| MD49 | 52 | IDC, ILC | P | P | N | P | 2.8 | 2 | IIB | SM, SNB, ALND | 105 | 27 | 23 |
| MD50 | 47 | IDC | P | P | N | P | 2.3 | 2 | IIB | SM, SLNB | 195 | 40 | 35 |
| MD52 | 49 | IDC | P | P | N | N | 2.3 | 2 | IIA | TM, SLNB | 187 | 67 | 37 |
| MD53 | 57 | IDC | N | N | N | N | 3 | 3 | IIA | SM, SLNB | 92 | 36 | 35 |
| MD57 | 64 | ILC | P | P | N | N | 4.6 | 2 | IIA | TM, ALND | 122 | 71 | 27 |
| MD64 | 82 | IDC | N | N | P | N | 2.8 | 3 | IIA | SM, SLNB | 129 | 39 | 44 |
| MD66 | 57 | IDC | P | P | N | N | 2.6 | 2 | IIA | TM, ALND | 355 | 75 | 45 |
| MD67 | 44 | IDC, ILC | P | P | N | P | 3 | 2 | IIB | SM, SNB, ALND | 148 | 12 | 45 |
| MD69 | 60 | IDC, ILC | P | P | N | N | 2.9 | 2 | IIA | TM, SLNB | 121 | 59 | 34 |
| MD71 | 48 | IDC | P | P | N | P | 2.9 | 3 | IIB | SM, ALND | 174 | 20 | 46 |
| MD115 | 61 | IDC | P | N | N | P | 3 | 3 | IIB | SM, SNB | 296 | 110 | 48 |
| MD117 | 66 | IDC | P | N | N | N | 2.1 | 3 | IIB | TM, SNB | 89 | 41 | 50 |
| MD118 | 70 | IDC, FLC | P | P | N | N | 2.4 | 2 | IIA | TM, SNB | 184 | 118 | 65 |
| MD119 | 46 | IDC | P | P | N | N | 2.1 | 2 | IIA | SM, SNB | 165 | 59 | 55 |
| MD120 | 63 | IDC, FLC | P | P | N | P | 2.6 | 2 | IIB | TM, SNB, ALND | 180 | 55 | 68 |
| MD122 | 57 | ISCC | N | N | N | P | 5.5 | 2 | IIIA | TM, SNB | 111 | 53 | 56 |
Microarray Processing and Statistical Analysis
The raw intensity data (CEL) files from human genome U133A Gene Chip microarrays were processed using the Affymetrix Microarray Suite 5.0 algorithms as implemented in the software package affy on the R/Bioconductor platform (www.bioconductor.org) (32) to generate probe-level intensities that were normalized to an array-wide median array intensity of 600 units. Expression values were log2–transformed and subsequently scaled by the expression levels of 1322 breast cancer reference genes to reference values that had been established as the median expression of these genes in an independent reference cohort of invasive breast cancers (N = 444) (33,34). The quality of gene chip hybridization and microarray profiling was assessed based on a set of four metrics that compare the expression level of the reference genes in each sample with the historical reference values before and after scaling. The quality metrics include the median deviation, the interquartile range of deviations, the Kolmogorov–Smirnov statistic for equality of the distributions, and the P value of the Kolmogorov–Smirnov statistic. Dimensionality was reduced through a principal component analysis model of the eight metrics, which were further summarized in two multivariable statistics, the Hotteling _T_2 and the sum of squares of the residuals or Q statistic (35). Control limits for Q and _T_2 for sample acceptance were established from historical human genome U133A microarray profiles that had passed the Affymetrix data quality guidelines (36).
We evaluated the effect of time until sample preservation on five standard Affymetrix RNA integrity metrics, defined as the 3′/5′ expression ratios and calculated from the ratio of the raw (pre-normalization) expression levels of probe sets from the 3′-most end over from the 5′-most end of each of the following gene transcripts: ACTB, GAPDH, STAT1, 18S ribosomal RNA (rRNA), and 28S rRNA (36); on a set of single gene expression measures (ESR1: probe set 205225_at; ERBB2: probe set 216836_s_at; MKI67: probe set 212021_s_at); and on four multigene expression indices [the sensitivity to endocrine therapy (SET) index (33), the genomic grade index (GGI) (37,38), the intrinsic subtypes predictor PAM50 (39), and a microarray approximation of the recurrence score computed as described previously (2,40)]. All multigene indices were evaluated as numeric (continuous) measurements, except for PAM50, for which the effect of the experimental factors was evaluated qualitatively based on the agreement in subtype assignment within each tumor sample.
Linear mixed-effects (LME) regression models with a random within-group intercept were used to estimate the effects of sample stabilization method and time delay until sample stabilization (0 vs 40 minutes), adjusting for a batch effect due to the two sets of samples (first set of 11 samples vs second set of six samples). The statistical significance of the fixed-effect coefficients and the random-effects interaction was evaluated by using the likelihood ratio test to compare the full model with a reduced model that did not include the term of interest (41). The effect of sample stabilization delay (cold ischemic time) was assessed using similar LME models with a fixed slope representing the cold ischemic time effect on each index and a random intercept to account for biological variation among tumors. The tumor-level effects of warm ischemic time (time between first surgical incision and tumor specimen removal) and of baseline cold ischemic time (time from surgical removal of tumor specimen to immediate sample stabilization) were modeled as group-level fixed effects in the random intercept fixed-slope LME model (42). We used a simple scalar within-group error covariance structure for this experimental design because different tissue aliquots were used at the different time points and, therefore, the potential for serial correlation in the response within each tumor sample was minimized. Statistical significance of the tumor-level predictors was assessed by using the likelihood ratio test to compare the full model with a reduced model without the tumor-level predictor (41). Details of the statistical model are provided in Supplementary Statistical Methods (available online). Statistical computations were performed in R (version 2.10.1) (43) using the software package lme4 for LME regression analysis (44). Statistical significance of the difference in ischemic times between different groups was based on the Mann–Whitney test. All reported confidence intervals (CIs) are symmetric and based on the normal distribution. All statistical tests were two-sided.
Results
Patient Clinical Characteristics and Sample Collection Details
All of the 17 breast cancer samples had sufficient RNA yield and purity for inclusion in this study. The median age of the patients at diagnosis was 57 years (range = 44–82 years), and 13 of the breast tumors were estrogen receptor positive, three were triple negative, and one was HER2 positive (Table 1). Eight patients underwent total mastectomy, and nine had segmental mastectomy. The median warm ischemic time—defined as the time between first surgical incision and tumor specimen removal—was 53 minutes (range = 12–118 minutes) and was statistically significantly longer for patients who underwent total mastectomy compared with patients who underwent segmental mastectomy (67 vs 40 minutes, difference = 27 minutes, 95% CI = 0 to 54 minutes, P = .016 [Mann–Whitney test]). The median baseline cold ischemic time—defined as the time from tumor specimen removal until the sample was stabilized by placing it in RNAlater or by snap freezing—was 45 minutes (range = 23–68 minutes) and included the time required to transfer the tumor specimens from the operating room to the room where specimen sectioning for pathological evaluation was performed (median = 5.5 minutes, range = 4–8 minutes). Samples in RNAlater solution were held at room temperature for median duration of 62 minutes before they were placed into the −80°C freezer for storage until RNA processing.
Sample Preservation in RNAlater vs Snap Freezing
To assess whether collection of a sample into RNAlater solution helps preserve RNA integrity, we compared the effects of immediate freezing of tumor samples in prechilled sterile vials plunged into dry ice vs stabilization of the samples in RNAlater at room temperature before freezing. We compared the quality of RNA from the 17 tumor samples stabilized either by immediate freezing or by placement in RNAlater and after holding the samples for 40 minutes at room temperature before freezing or placement in RNAlater. The yield and quality of RNA isolated from each sample by each stabilization method according to stabilization delay are given in Supplementary Table 1 (available online). RNA from samples that were snap frozen at baseline or after 40 minutes at room temperature was more fragmented compared with RNA from the same samples stabilized in RNAlater, as evidenced by the Bioanalyzer electrophoresis traces (Supplementary Figure 1, available online). The first three snap-frozen samples that we processed (MD53, MD57, and MD64) showed extensive RNA fragmentation (Supplementary Figure 1, available online), possibly due to a short thawing period that occurred before the first solution of the RNA extraction kit was added. The protocol was subsequently modified to add the buffer to the frozen tissue immediately upon removal from the freezer, and this change improved the quality of RNA from the snap-frozen samples, as suggested by improved traces from the subsequent samples.
The effects of stabilization method and cold ischemic delay on the yield and integrity of isolated RNA were evaluated in a 2 × 2 factorial set up (ie, two preservation protocols and two time points [0 and 40 minutes]) using a mixed-effects model to account for intertumor variation. The first three samples that were processed (mentioned above) were excluded from this analysis. Compared with snap freezing, sample preservation in RNAlater statistically significantly improved RNA integrity as assessed by the RNA integrity number, which increased from 7.31 to 8.13 units (difference = 0.82 units, 95% CI = 0.53 to 1.11 units, P < .001), and RNA yield, which increased threefold from 8.9 to 28.6 μg (difference = 19.7 μg, 95% CI = 14.1 to 25.3 μg, P < .001), even after we adjusted for the fact that the second set of samples had a statistically significantly higher RNA yield compared with the first set of samples (RNA yield for first vs second set: 8.9 vs 23.1 μg, difference = 14.2 μg, 95% CI = 0.7 to 27.7 μg, P = .04) (Table 2). The additional 40-minute delay at room temperature before sample stabilization did not statistically significantly affect RNA integrity or yield. In addition, the estimated model intercept for the A260/A280 ratio of 2.06 (95% CI = 2.04 to 2.08), which represents the average ratio for snap-frozen samples from the first set of samples at zero delay (Table 2), suggested high RNA purity, regardless of stabilization method or processing delay. The estimated intraclass correlation coefficient for RNA integrity number was 0.824, which suggests that 82% of the observed variation not explained by preservation method or cold ischemia was due to variation between tumors, and leaving only 18% of variation attributable to other causes. However, the intraclass correlation coefficients for RNA yield and A260/A280 ratio of samples from the same tumor were 0.533 and 0.406, respectively, suggesting a larger role for other causes of variability of RNA yield and A260/A280 ratio.
Table 2.
Mixed-effects analysis of the 2 × 2 study for the effect of sample preservation method and time delay on RNA quality metrics*
| Model and effect | RIN† (N = 13) | RNA yield, μg (N = 17) | RNA A260/A280 (N = 17) | |||
|---|---|---|---|---|---|---|
| Estimate (95% CI) | P‡ | Estimate (95% CI) | P‡ | Estimate (95% CI) | P‡ | |
| Fixed effects | ||||||
| Intercept | 7.31 (6.41 to 8.21) | NA | 8.89 (−0.16 to 17.9) | NA | 2.06 (2.04 to 2.08) | NA |
| Stabilization (RNAlater vs snap freezing) | 0.82 (0.53 to 1.11) | <.001 | 19.7 (14.1 to 25.3) | <.001 | −0.010 (−0.02 to 0.004) | .16 |
| Time delay (40 vs 0 min) | −0.14 (−0.42 to 0.16) | .35 | −1.98 (−7.5 to 3.6) | .48 | −0.009 (−0.02 to 0.004) | .17 |
| Block§ (2 vs 1) | 0.16 (−1.13 to 1.46) | .79 | 14.2 (0.7 to 27.7) | .04 | −0.001 (−0.03 to 0.03) | .94 |
| Random effects | ||||||
| Between-tumor SD | 1.16 (NA) | NA | 12.3 (NA) | NA | 0.024 (NA) | NA |
| Within-tumor SD | 0.53 (NA) | NA | 11.5 (NA) | NA | 0.028 (NA) | NA |
| ICC║ | 0.824 (NA) | NA | 0.533 (NA) | NA | 0.406 (NA) | NA |
Effect of Prolonged Cold Ischemia on RNA Integrity
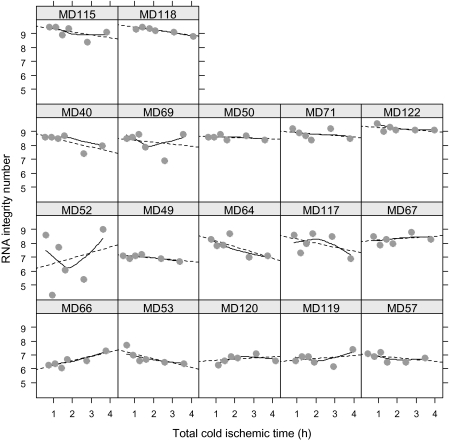
We next evaluated the effects of increasing the total cold ischemic time on the integrity of RNA isolated from breast cancer tissue samples stabilized in RNAlater (Figure 1, B). A tissue aliquot was placed into RNAlater RNA stabilization reagent at room temperature immediately after mincing (defined as 0 minutes or baseline), and additional aliquots were placed in RNAlater after being held at room temperature for 20, 40, 60, 120, or 180 minutes. Although there was considerable variation in RNA electrophoresis traces among the baseline samples (ie, those preserved in RNAlater with no delay), we observed only minor changes in the traces with longer cold ischemic time within each series of samples (Supplementary Figure 2, available online). Overall, prolonged cold ischemia had a modest effect on RNA integrity as assessed by the RNA integrity number (Figure 2). However, four samples had noticeable fragmentation at baseline, particularly in the low molecular weight (ie, 5S) region of the trace (samples MD49, MD53, MD57, and MD66; Supplementary Figure 2, available online), suggesting that partial degradation may be from intratumoral necrosis or may have occurred during tumor excision, specimen collection, and processing. Indeed, three of these samples (MD53, MD57, and MD66) had the longest recorded warm ischemic times (Table 1), but their RNA integrity numbers were within acceptable limits (>6.3; Supplementary Table 1, available online). RNA fragmentation increased with prolonged cold ischemia in two samples (accumulation in the region of the trace between the 18S and 28S peaks for sample MD52 and/or in the 5S region for sample MD66; Supplementary Figure 2, available online). Both of these patients underwent total mastectomies, and, consequently, their samples had longer warm ischemic times compared with the median warm ischemic time (67 and 75 minutes vs 53 minutes; Table 1). One tumor (MD52) showed extensive variability in the RNA integrity number across all times points (Figures 2 and 3) and substantially more RNA degradation (Supplementary Figure 2, available online) compared with other samples. This sample was from an invasive ductal carcinoma with mucinous features. Therefore, it is possible that the high mucopolysaccharide content within the tumor itself could have interfered with tissue penetration by RNA preservation solution or the Bioanalyzer assay results. This tumor was excluded from statistical analyses for RNA integrity number.
Figure 2.
Effect of total cold ischemic time on the RNA integrity number (RIN) of RNA isolated from 17 breast cancer samples. Total cold ischemic time was defined as the combined baseline cold ischemic time and additional delay at room temperature. Circles represent the RIN of RNA extracted from a tissue aliquot preserved at the indicated time (single measurement estimated from the microcapillary electrophoresis traces shown in Supplementary Figure 2, available online), dotted lines represent the linear regression trend, and solid lines represent a locally weighted polynomial regression (LOESS smoother). Case subjects have been ordered from bottom to top in terms of increasing median RIN values.
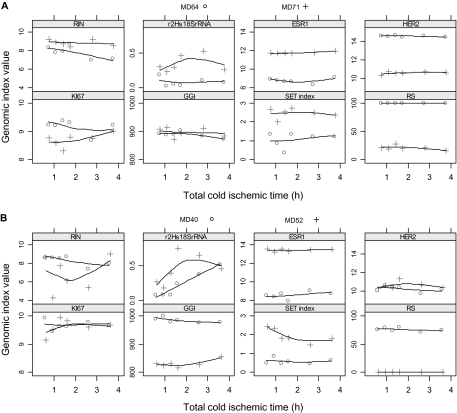
Figure 3.
Effect of increasing total cold ischemic time on the RNA integrity number (RIN) and microarray-based gene expression indices. Total cold ischemic time was defined as the baseline cold ischemic time plus delay at room temperature. The _y_-axis of the plots (labeled Genomic index value) shows the scaled and log2-transformed expression levels for single genes (ESR1, HER2, KI67), the 3′/5′ expression ratio for 18S ribosomal RNA (r2Hs18S rRNA), the values of the multigene genomic indices (GGI, SET index, RS), and the value of the RIN. Shown are the data from four breast cancer case subjects that represent typical breast cancer phenotypes defined by the combined estrogen receptor and HER2 status from routine pathology testing. Symbols represent measurements derived from a single microarray profile that was generated from RNA extracted from the tissue aliquot preserved at the indicated time. A) A grade 3 estrogen receptor–positive and HER2-negative breast cancer (MD71, crosses) and a grade 3 estrogen receptor–negative and HER2-positive breast cancer (MD64, circles). B) A grade 3 estrogen receptor–negative and HER2-negative cancer (MD40, circles) and a grade 2 estrogen receptor–positive and HER2-negative mucinous breast cancer (MD52, crosses). Solid lines indicate a smooth local regression line. ESR1 = log2-transformed expression of estrogen receptor 1 gene; GGI = genomic grade index; HER2 = log2-transformed expression of v-erb-b2 erythroblastic leukemia viral oncogene homolog 2 gene; KI67 = log2-transformed expression of proliferation antigen identified by monoclonal antibody Ki-67 gene; r2Hs18S rRNA = 3′/5′ ratio for 18S ribosomal RNA; RS = microarray-based recurrence score; SET index = sensitivity to endocrine therapy index.
Statistical regression analysis based on random intercept fixed-slope LME models confirmed that additional cold ischemic delay at room temperature before sample stabilization decreased the RNA integrity number by 0.12 units/h (95% CI = 0.02 to 0.23 units/h) compared with a projected average RNA integrity number of 7.39 if no delays were present (P = .008) and decreased the RNA yield by 1.5 μg/h (95% CI = 0 to 4 μg/h) from a baseline mean RNA yield of 33.5 μg if no delays were present (P = .019) (Table 3). It is interesting that neither of the tumor-level covariates (ie, warm ischemia or minimal cold ischemia) had any effect on the RNA quality metrics or yield. When surgery type and tumor size were added as group-level variables, total mastectomy was associated with statistically significantly lower RNA yield (P = .001), and larger tumor size was associated with a statistically significantly higher RNA yield (P < .001; data not shown). These covariates did not have an effect on RNA integrity number (data not shown). Finally, the block factor (first 11 samples vs subsequent six samples) did not have a statistically significant effect or interaction with the extended cold ischemic delay, implying that the effect of cold ischemic delay was similar in both batches of samples.
Table 3.
Mixed-effects analysis of the effect of cold and warm ischemic times on RNA quality metrics*
| Model and effect | RIN (N = 16) | RNA yield, μg (N = 17) | RNA A260/A280 (N = 17) | |||
|---|---|---|---|---|---|---|
| Estimate (95% CI) | P† | Estimate (95% CI) | P† | Estimate (95% CI) | P† | |
| Fixed effects | ||||||
| Intercept | 7.39 (4.56 to 10.2) | NA | 33.5 (−14.4 to 81.4) | NA | 2.03 (1.95 to 2.11) | NA |
| Warm ischemia‡ (1/h) | 0.13 (−1.14 to 1.41) | .81 | −8.0 (−28.9 to 12.8) | .39 | −0.006 (−0.04 to 0.03) | .68 |
| Baseline cold ischemia§ (1/h) | 0.63 (−3.54 to 4.81) | .73 | −3.1 (−73.8 to 67.5) | .92 | 0.013 (−0.101 to 0.126) | .80 |
| Cold ischemic delay║ (1/h) | −0.12 (−0.23 to −0.02) | .01 | −1.5 (−4.0 to 0.95) | .02 | 0.002 (−0.01 to 0.01) | .66 |
| Random effects | ||||||
| Between-tumor SD | 1.05 (NA) | NA | 17.4 (NA) | NA | 0.025 (NA) | NA |
| Within-tumor SD | 0.43 (NA) | NA | 10.7 (NA) | NA | 0.033 (NA) | NA |
| Intraclass correlation coefficient | 0.856 (NA) | NA | 0.727 (NA) | NA | 0.379 (NA) | NA |
Effect of Tissue Preservation Method on Stability of Single Gene Biomarkers and Multigene Indices
We extended the study to evaluate the effect of tissue preservation on gene expression measurements derived from microarrays. Samples of tumors from the first 11 patients (MD40–MD71; Table 1) were subjected to gene expression profiling by use of Affymetrix U133A microarrays. Of the 86 microarrays profiled (ie, six different time points in RNAlater and two time points snap frozen per sample, except for MD40, for which snap-frozen samples were not collected), only four failed microarray-based quality control tests (all originated from snap-frozen samples), whereas seven had RNA integrity number less than 6, suggesting a rather weak association between RNA integrity number and microarray-based quality control. Single gene measurements and multigene indices were generally not affected by extended cold ischemic delays at room temperature for samples preserved in RNAlater, as shown in Figure 3 for four tumor samples from representative breast cancer phenotypes, including a carcinoma with mucinous features (from patient MD52) that had variable RNA integrity numbers. For example, the tumor sample from patient MD40 displayed a decrease in the RNA integrity number and an increase in the 3′/5′ transcript ratio (an indicator of mRNA degradation) for 18S rRNA with increasing cold ischemic time. Both of these findings represent increasing RNA fragmentation, but gene expression measurements for ESR1, ERBB2, and MKI67 and the GGI and SET index were unaffected (Figures 2 and 3).
LME regression analysis of the cold ischemic time series from the first set of 11 samples suggested that prolonged cold ischemic time, in addition to reducing RNA integrity number, was also associated with a large and statistically significant increase in the 3′/5′ ratio for STAT1 RNA (P < .001), moderate but statistically significant increases in the 3′/5′ ratios for beta actin RNA (P = .02) and 18S rRNA (P = .006), and a statistically significant reduction in the 3′/5′ ratio for 28S rRNA (P = .006) (Table 4). Based on the size of the estimated effects, the 3′/5′ ratios for STAT1 RNA and 28S rRNA appear to be potentially sensitive markers of cold ischemic effect; however, both ratios had intraclass correlation coefficient less than 0.5, suggesting that a larger within-tumor variation is associated with these metrics than with the RNA integrity number. The warm ischemic time or the minimum cold ischemic time associated with each tumor specimen did not have a statistically significant effect on the 3′/5′ ratio of any of the control genes evaluated (data not shown). It is interesting that samples that were preserved in RNAlater had statistically significantly lower 3′/5′ transcript ratios compared with samples preserved by snap freezing, suggesting that they had less mRNA fragmentation (ACTB, P < .001; GAPDH, P < .001; STAT1, P = .014; 18S rRNA, P < .001; Supplementary Table 2, available online). On the other hand, preservation in RNAlater resulted in statistically significantly lower expression of ESR1 (P = .003) and ERBB2 (P = .016) compared with snap freezing of samples, but the magnitude of the reduction in the expression level of these genes was rather small and thus unlikely to affect assignment of estrogen receptor and HER2 status based on microarray measurements. In addition, multigene indices were not affected by the preservation method of the tumor samples (Supplementary Table 2, available online).
Table 4.
Summary of mixed-effects analysis of the effect of cold ischemic delay on microarray-based RNA transcript quality metrics and gene expression measurements*
| Variable | No. of tumors | Estimate (1/h) (95% CI) | P† | ICC |
|---|---|---|---|---|
| RNA integrity metric | ||||
| RIN‡ | 16 | −0.12 (−0.23 to 0.02) | .008 | 0.856 |
| 3′/5′ ratio | ||||
| ACTB | 11 | 0.08 (0.01 to 0.14) | .02 | 0.646 |
| GAPDH | 11 | 0.03 (−0.03 to 0.08) | .34 | 0.552 |
| STAT1 | 11 | 0.94 (0.45 to 1.44) | <.001 | 0.560 |
| 18S rRNA | 11 | 0.04 (0.01 to 0.07) | .006 | 0.499 |
| 28S rRNA | 11 | −1.78 (−3.02 to −0.54) | .006 | 0.340 |
| Gene or gene signature | ||||
| ESR1 | 11 | −0.04 (−0.12 to 0.03) | .25 | 0.972 |
| ERBB2 | 11 | −0.06 (−0.13 to 0.01) | .10 | 0.949 |
| MKI67 | 11 | 0.02 (−0.05 to 0.08) | .63 | 0.642 |
| SET index | 11 | −0.08 (−0.16 to 0.01) | .08 | 0.937 |
| GGI | 11 | −5.39 (−9.23 to −1.55) | .007 | 0.956 |
| Recurrence score§ | 8 | 0.15 (−1.47 to 1.76) | .86 | 0.865 |
LME regression analysis of the first set of 11 samples revealed that increasing cold ischemic time had no statistically significant effect on the expression levels of single genes (ESR1, ERBB2, and MKI67) or multigene indices, except for GGI which was statistically significantly reduced (P = .007); however, the magnitude of the effect (−5.4 units/h of cold ischemia) corresponds to less than 1% of the range of the GGI and thus is unlikely to affect tumor grade assignment. The assignment of breast cancer subtypes (PAM50) was not affected by cold ischemic delay in nine of the 11 tumors studied (Supplementary Table 3, available online) and was similar in both snap frozen samples in all 10 tumors studied. Tumors MD49 and MD71, for which subtype assignment varied with cold ischemic delay, were probably not sufficiently representative of any of the breast cancer subtypes, and therefore the observed variation was more likely the result of equivocal subtype assignment rather than loss of RNA integrity (Supplementary Table 3, available online). The large intraclass correlation coefficients for genes and genomic indices (Table 4) suggest that by far the largest source of the residual (unexplained) variation in these indices across all samples is the between-tumor variation, which is consistent with the fact that the selected genes and multigene indices are strongly associated with the distinct breast cancer phenotypes of the tumors used in this study (32,35–38) (Table 1). Warm ischemic time had no statistically significant effect on any of the genomic measures examined (data not shown).
Discussion
In this study, we systematically evaluated the effects of tissue preservation method and prolonged cold ischemia on RNA quality and microarray indices across 17 primary breast tumors that represented a range of disease stages and surgical conditions. Contrary to previous studies that reported no major differences in RNA integrity between samples preserved in RNAlater and samples preserved by snap freezing (21,22), we found that samples preserved in RNAlater had improved RNA integrity and considerably (ie, threefold) greater RNA yield compared with snap-frozen samples. We also found that exposure of tumor tissue to an additional 40 minutes at room temperature—a realistic delay that could occur during routine examination and tissue sampling—did not affect the integrity or the purity of the extracted RNA. Analysis of the time course data suggested that allowing tissue samples to sit in a closed Petri dish at room temperature for up to 3 hours before preservation in RNAlater caused a small but statistically significant reduction in the RNA integrity number but had no effect on RNA yield or purity.
Several factors could have contributed to the almost tripled RNA yield and higher RNA integrity number for the samples preserved in RNAlater compared with those preserved by snap freezing. The most likely explanation is that nucleic acids in small pieces of tissue are better preserved in RNAlater than by snap freezing. Indeed, small pieces of tissue have high ratio of surface area to volume, which might preferentially favor tissue penetration by a preservative solution but accelerate tissue desiccation or degradation when tissue is snap frozen. It is also possible that the kit-based RNA extraction procedure that we used is better suited to samples that are preserved in RNAlater. Nonetheless, our results are likely to be relevant for clinical research studies that use small tumor samples from core or endoscopic biopsy specimens and rely on standard protocols for sample handling and purification of RNA (45).
In addition to cold ischemic duration and sample preservation method, we also evaluated the effect of intraoperative duration on tumor sample integrity. We have used the term “warm ischemic time” to describe the interval between first surgical excision and surgical removal of the tumor specimen from the patient. However, we recognize that in breast surgery there is a variable delay between first incision and surgical compromise of the vascular supply to the tumor, that tumor ischemia is not defined by a single moment of vascular clamping, and that the operative duration may include axillary surgery. Indeed, the surgical interruption of blood supply is likely to be progressively incremental during the surgical procedure, be variable between operations, and depends on the anatomy of vascular supply to each tumor. We observed that samples from total mastectomies had longer warm ischemic durations and statistically significantly lower RNA yields compared with samples from segmental mastectomies, but they displayed no difference in RNA integrity number. Overall, the duration of warm ischemia did not statistically significantly affect RNA integrity or yield in this study. However, a limitation of this small study is that we cannot exclude the possibility that other factors such as anesthesia, other medications, or other stress responses related to surgery might have an influence on RNA yield and/or integrity.
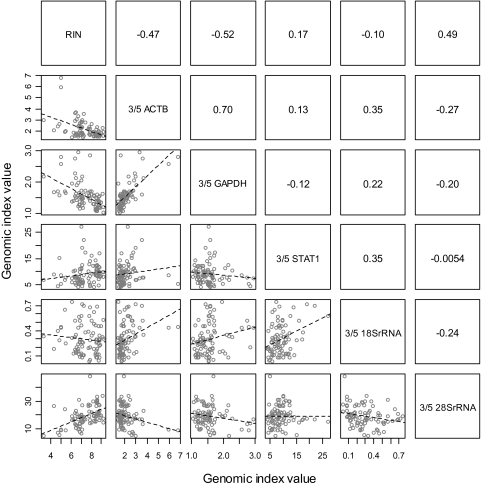
A key question for quality assurance of clinical tumor samples is whether the RNA integrity number or the microarray-based RNA integrity measures provide valuable information about the suitability of a sample for microarray analysis**.** We observed that the RNA integrity number was inversely associated with the extent of transcript degradation (assessed by an increase in 3′/5′ expression ratio) of two of the housekeeping genes studied**,** GAPDH and ACTB (Figure 4). This finding is consistent with a previous report (11). Furthermore, prolonged cold ischemia reduced the RNA integrity number and increased transcript degradation (increased 3′/5′ ratio) of three of the housekeeping genes studied but did not affect the mRNA transcripts for ESR1, ERBB2, and MKI67 (Table 4). This finding suggests a weak association between the RNA integrity number and the integrity of the mRNA transcripts evaluated here.
Figure 4.
Scatter plot matrix for the correlation between RNA integrity number (RIN) and microarray-based mRNA transcript integrity metrics (ratio of the expression level of probe sets from the 3′ over the 5′ end of the transcript) for actin B, GAPDH, and STAT1 RNAs and 18S and 28S ribosomal RNAs obtained from 86 microarray profiles. The lower part of the matrix shows a series of pairwise scatter plots (the pair of variables plotted in each plot is identified through the labels on the diagonal; eg, the first plot on the top left has RIN on the _x_-axis and the 3′/5′ ratio for ACTB RNA on the _y_-axis). The upper part of the matrix shows the corresponding pairwise Pearson correlation coefficients (eg, the correlation coefficient for the 3′/5′ratio for ACTB RNA vs RIN is −0.47).
The RNA integrity number is derived from the relative expression and integrity of 28S rRNA and 18S rRNA transcripts. However, as discussed by Fleige et al. (46), structural differences between ribosomal and messenger RNA species, the more rapid degradation of 28S rRNA than 18S rRNA, and the greater similarity of 18S rRNA to average-length mRNAs all lead to uncertainty about the relationship between RNA integrity number and mRNA integrity, except when there is marked sample degradation. Indeed, moderate degradation of RNA (as determined from RNA integrity number) does not preclude the accurate measurement of normalized gene expression levels from a sample (26).
We found that 28S rRNA and 18S rRNA transcript integrity had different patterns of association with sample integrity. Prolonged cold ischemic duration was associated paradoxically with statistically significantly increased integrity (lower 3′/5′ expression ratio) of 28S rRNA transcripts and statistically significantly decreased integrity (higher 3′/5′ expression ratio) of 18S rRNA transcripts (Table 4). Although it is possible that 28S rRNA degradation occurs preferentially at the 5′ end of the transcript, thus reducing expression of the 5′ probe set and resulting in higher 3′/5′ ratio, we do not know of any biological precedent for this explanation. However, the association between the integrity of samples and the integrity of 18S rRNA transcripts is consistent with our observations for the other housekeeping gene transcripts (ACTB, STAT1, and GAPDH).
We were surprised that perturbations to sample integrity had little effect on the normalized gene expression measurements of ESR1, ERBB2, and MKI67, three key genes used for molecular assessment of breast cancer. However, we also note that expression of ESR1 and ERBB2 was statistically significantly lower in RNAlater-preserved samples compared with snap-frozen samples, but these differences (<3% of range) would be too small to alter positive vs negative receptor status in most tumors (Supplementary Table 2, available online).
De Cecco et al. (20) suggested that multigene signatures might provide more robust results from partially compromised samples than would single genes, but we did not observe such a difference. We found that the GGI showed a small yet statistically significant decline with prolonged cold ischemia. We also observed variability of intrinsic subtype assignment in two of 11 tumors using the multigene PAM50 signature. This variability is probably not due to RNA integrity but is rather more likely to be due to the mathematical method that is used to assign a gene profile to the closest subtype, wherein certain ambiguous profiles might not be assigned consistently.
In conclusion, this study demonstrated that RNAlater was better than snap freezing for collection of RNA with high yield and quality for gene expression profiling of small tissue samples from patients with breast cancer. Overall, prolonged cold ischemic time of samples in a closed Petri dish at room temperature had no substantial effect on mRNA yield and integrity or the expression of relevant genes for breast cancer assessment, if samples were collected and preserved in RNAlater solution. We believe that these findings are highly relevant for biobanking and for clinical trials that plan to analyze the transcriptome of breast cancer samples, especially those obtained from small biopsy specimens. However, a broader investigation to identify genes whose expression might be more susceptible to ischemic stresses, and possibly expression indices developed from such genes, may provide useful tools to assess more subtle effects of ischemic stress on sample integrity and microarray profiles.
Funding
This work was supported in whole or in part with Federal Funds from the National Cancer Institute, National Institutes of Health, under contract no. HHSN261200800001E to WFS and CH.
Supplementary Material
Supplementary Data
Footnotes
The authors would like to thank Dr Daniel Silver (Dana Farber Cancer Institute) for helpful advice during the course of this study. The sponsors reviewed and approved the design of the study and monitored its conduct by quarterly teleconferences led by Dr Helen Moore, Director, Biospecimen Research Network, National Cancer Institute. The sponsors did not have a role in the collection of samples, analysis, interpretation of the data, writing of the article, or decision to submit the article. C. Hatzis holds stock in Nuvera Biosciences which has developed gene-based assays to predict treatment response. W. F. Symmans is a cofounder of and equity holder in Nuvera Biosciences and has received a royalty from patents on the gene-based assays. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government.
References
- 1.Sorlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98(19):10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Loi S, Haibe-Kains B, Desmedt C, et al. Definition of clinically distinct molecular subtypes in estrogen receptor-positive breast carcinomas through genomic grade. J Clin Oncol. 2007;25(10):1239–1246. doi: 10.1200/JCO.2006.07.1522. [DOI] [PubMed] [Google Scholar]
- 3.Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351(27):2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 4.Rouzier R, Perou CM, Symmans WF, et al. Breast cancer molecular subtypes respond differently to preoperative chemotherapy. Clin Cancer Res. 2005;11(16):5678–5685. doi: 10.1158/1078-0432.CCR-04-2421. [DOI] [PubMed] [Google Scholar]
- 5.Dobbe E, Gurney K, Kiekow S, et al. Gene-expression assays: new tools to individualize treatment of early-stage breast cancer. Am J Health Syst Pharm. 2008;65(1):23–28. doi: 10.2146/ajhp060352. [DOI] [PubMed] [Google Scholar]
- 6.Sotiriou C, Pusztai L. Gene-expression signatures in breast cancer. N Engl J Med. 2009;360(8):790–800. doi: 10.1056/NEJMra0801289. [DOI] [PubMed] [Google Scholar]
- 7.Ross JS, Hatzis C, Symmans WF, et al. Commercialized multigene predictors of clinical outcome for breast cancer. Oncologist. 2008;13(5):477–493. doi: 10.1634/theoncologist.2007-0248. [DOI] [PubMed] [Google Scholar]
- 8.Abdullah-Sayani A, Bueno-de-Mesquita JM, van de Vijver MJ. Technology Insight: tuning into the genetic orchestra using microarrays—limitations of DNA microarrays in clinical practice. Nat Clin Pract Oncol. 2006;3(9):501–516. doi: 10.1038/ncponc0587. [DOI] [PubMed] [Google Scholar]
- 9.Ioannidis JP. Is molecular profiling ready for use in clinical decision making? Oncologist. 2007;12(3):301–311. doi: 10.1634/theoncologist.12-3-301. [DOI] [PubMed] [Google Scholar]
- 10.Sparano JA, Solin LJ. Defining the clinical utility of gene expression assays in breast cancer: the intersection of science and art in clinical decision making. J Clin Oncol. 2010;28(10):1625–1627. doi: 10.1200/JCO.2009.25.2882. [DOI] [PubMed] [Google Scholar]
- 11.Shi L, Reid LH, Jones WD, et al. The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat Biotechnol. 2006;24(9):1151–1161. doi: 10.1038/nbt1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Canales RD, Luo Y, Willey JC, et al. Evaluation of DNA microarray results with quantitative gene expression platforms. Nat Biotechnol. 2006;24(9):1115–1122. doi: 10.1038/nbt1236. [DOI] [PubMed] [Google Scholar]
- 13.Jewell SD, Srinivasan M, McCart LM, et al. Analysis of the molecular quality of human tissues: an experience from the Cooperative Human Tissue Network. Am J Clin Pathol. 2002;118(5):733–741. doi: 10.1309/VPQL-RT21-X7YH-XDXK. [DOI] [PubMed] [Google Scholar]
- 14.Dash A, Maine IP, Varambally S, et al. Changes in differential gene expression because of warm ischemia time of radical prostatectomy specimens. Am J Pathol. 2002;161(5):1743–1748. doi: 10.1016/S0002-9440(10)64451-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Erickson HS, Josephson JW, Vira M, et al. Influence of hypoxia induced by minimally invasive prostatectomy on gene expression: implications for biomarker analysis. Am J Transl Res. 2010;2(3):210–222. [PMC free article] [PubMed] [Google Scholar]
- 16.Huang J, Qi R, Quackenbush J, et al. Effects of ischemia on gene expression. J Surg Res. 2001;99(2):222–227. doi: 10.1006/jsre.2001.6195. [DOI] [PubMed] [Google Scholar]
- 17.Spruessel A, Steimann G, Jung M, et al. Tissue ischemia time affects gene and protein expression patterns within minutes following surgical tumor excision. Biotechniques. 2004;36(6):1030–1037. doi: 10.2144/04366RR04. [DOI] [PubMed] [Google Scholar]
- 18.Febbo PG, Kantoff PW. Noise and bias in microarray analysis of tumor specimens. J Clin Oncol. 2006;24(23):3719–3721. doi: 10.1200/JCO.2006.06.7942. [DOI] [PubMed] [Google Scholar]
- 19.Lin DW, Coleman IM, Hawley S, et al. Influence of surgical manipulation on prostate gene expression: implications for molecular correlates of treatment effects and disease prognosis. J Clin Oncol. 2006;24(23):3763–3770. doi: 10.1200/JCO.2005.05.1458. [DOI] [PubMed] [Google Scholar]
- 20.De Cecco L, Musella V, Veneroni S, et al. Impact of biospecimens handling on biomarker research in breast cancer. BMC Cancer. 2009;9:409. doi: 10.1186/1471-2407-9-409. doi:10.1186/1471-2407-9-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mutter GL, Zahrieh D, Liu C, et al. Comparison of frozen and RNALater solid tissue storage methods for use in RNA expression microarrays. BMC Genomics. 2004;5(1):88. doi: 10.1186/1471-2164-5-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Micke P, Ohshima M, Tahmasebpoor S, et al. Biobanking of fresh frozen tissue: RNA is stable in nonfixed surgical specimens. Lab Invest. 2006;86(2):202–211. doi: 10.1038/labinvest.3700372. [DOI] [PubMed] [Google Scholar]
- 23.Bakay M, Chen YW, Borup R, et al. Sources of variability and effect of experimental approach on expression profiling data interpretation. BMC Bioinformatics. 2002;3:4. doi: 10.1186/1471-2105-3-4. doi:10.1186/1471-2105-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barry WT, Kernagis DN, Dressman HK, et al. Intratumor heterogeneity and precision of microarray-based predictors of breast cancer biology and clinical outcome. J Clin Oncol. 2010;28(13):2198–2206. doi: 10.1200/JCO.2009.26.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blackhall FH, Pintilie M, Wigle DA, et al. Stability and heterogeneity of expression profiles in lung cancer specimens harvested following surgical resection. Neoplasia. 2004;6(6):761–767. doi: 10.1593/neo.04301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schoor O, Weinschenk T, Hennenlotter J, et al. Moderate degradation does not preclude microarray analysis of small amounts of RNA. Biotechniques. 2003;35(6):1192–1196. doi: 10.2144/03356rr01. 1198-1201. [DOI] [PubMed] [Google Scholar]
- 27.TABPW Group. Expression profiling—best practices for data generation and interpretation in clinical trials. Nat Rev Genet. 2004;5(3):229–237. doi: 10.1038/nrg1297. [DOI] [PubMed] [Google Scholar]
- 28.Leyland-Jones BR, Ambrosone CB, Bartlett J, et al. Recommendations for collection and handling of specimens from group breast cancer clinical trials. J Clin Oncol. 2008;26(34):5638–5644. doi: 10.1200/JCO.2007.15.1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schroeder A, Mueller O, Stocker S, et al. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol. 2006;7:3. doi: 10.1186/1471-2199-7-3. doi:10.1186/1471-2199-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greene FL, Page DL, Fleming ID, et al. Manual for Staging Cancer. 6th ed. New York: Springer-Verlag; 2002. [Google Scholar]
- 31.Affymetrix. GeneChip Expression Analysis: Technical Manual. Santa Clara, CA: Affymetrix: 2004. [Google Scholar]
- 32.Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Symmans WF, Hatzis C, Sotiriou C, et al. Genomic index of sensitivity to endocrine therapy for breast cancer. J Clin Oncol. 2010;28(27):4111–4119. doi: 10.1200/JCO.2010.28.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hatzis C, Pusztai L, Valero V, et al. A genomic predictor of response and survival following taxane-anthracycline chemotherapy for invasive breast cancer. JAMA. 2011;305(18):1873–1881. doi: 10.1001/jama.2011.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jackson JE, Mudholkar GS. Control procedures for residuals associated with principal components analysis. Technometrics. 1979;21(3):341–349. [Google Scholar]
- 36.Affymetrix. Gene Chip Expression Analysis: Data Analysis Fundamentals. Santa Clara, CA: Affymetrix: 2004. [Google Scholar]
- 37.Liedtke C, Hatzis C, Symmans WF, et al. Genomic grade index is associated with response to chemotherapy in patients with breast cancer. J Clin Oncol. 2009;27(19):3185–3191. doi: 10.1200/JCO.2008.18.5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sotiriou C, Wirapati P, Loi S, et al. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst. 2006;98(4):262–272. doi: 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]
- 39.Parker JS, Mullins M, Cheang MC, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27(8):1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fan C, Oh DS, Wessels L, et al. Concordance among gene-expression-based predictors for breast cancer. N Engl J Med. 2006;355(6):560–569. doi: 10.1056/NEJMoa052933. [DOI] [PubMed] [Google Scholar]
- 41.Pinheiro JC, Bates DM. Mixed-Effects Models in S and S-Plus. New York, NY: Springer-Verlag; 2000. [Google Scholar]
- 42.Gelman A, Hill J. Data Analysis Using Regression and Multilevel/Hierarchical Models. New York, NY: Cambridge University Press; 2007. [Google Scholar]
- 43.R Development Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2009. [Google Scholar]
- 44.Bates DM, Maechler M. lme4: Linear Mixed-Effects Models Using S4 Classes. R package version 0.999375-32. 2009. http://CRAN.R-project.org/package=lme4. [Google Scholar]
- 45.Loi S, Symmans WF, Bartlett JM, et al. Proposals for uniform collection of biospecimens from neoadjuvant breast cancer clinical trials: timing and specimen types. Lancet Oncol. 2011 doi: 10.1016/S1470-2045(11)70117-6. [published online ahead of print June 20, 2011] doi:10.1016/S1470-2045(11)70117-6. [DOI] [PubMed] [Google Scholar]
- 46.Fleige S, Pfaffl MW. RNA integrity and the effect on the real-time qRT-PCR performance. Mol Aspects Med. 2006;27(2–3):126–139. doi: 10.1016/j.mam.2005.12.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data