ETV6 mutations in early immature human T cell leukemias (original) (raw)
A substantial proportion of adult T-ALL samples display gene expression and mutation characteristics of both T cell and acute myeloid leukemias; mutations in ETV6 are found exclusively within this new molecular subgroup of adult T-ALL patients.
Abstract
Early immature T cell acute lymphoblastic leukemias (T-ALLs) account for ∼5–10% of pediatric T-ALLs and are associated with poor prognosis. However, the genetic defects that drive the biology of these tumors remain largely unknown. In this study, analysis of microarray gene expression signatures in adult T-ALL demonstrated a high prevalence of early immature leukemias and revealed a close relationship between these tumors and myeloid leukemias. Many adult immature T-ALLs harbored mutations in myeloid-specific oncogenes and tumor suppressors including IDH1, IDH2, DNMT3A, FLT3, and NRAS. Moreover, we identified ETV6 mutations as a novel genetic lesion uniquely present in immature adult T-ALL. Our results demonstrate that early immature adult T-ALL represents a heterogeneous category of leukemias characterized by the presence of overlapping myeloid and T-ALL characteristics, and highlight the potential role of ETV6 mutations in these tumors.
T cell acute lymphoblastic leukemia (T-ALL) is a heterogeneous disease in which genetic lesions coordinately affect cell proliferation, differentiation, and survival of thymocytes (Ferrando, 2009; Paganin and Ferrando, 2011). T-ALL accounts for 15% of pediatric and 25% of adult ALL cases. Importantly, significant differences in outcome are present between pediatric and adult T-ALL (Pui et al., 2008). Thus, although >70% of children achieve long lasting complete remissions, only 50% of adult T-ALL patients are currently cured. In addition, pediatric and adult T-ALLs show marked differences in the frequency of specific genetic lesions. For example, chromosomal translocation and aberrant expression of the TAL1 and TLX3 oncogenes are highly prevalent in children, but rare in adults. In contrast, translocations activating TLX1 are rarely found in pediatric leukemias but represent one third of adult T-ALL cases (Ferrando et al., 2002, 2004).
RESULTS AND DISCUSSION
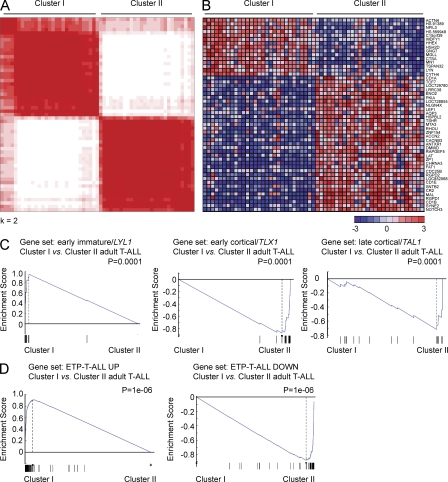
To gain more insight in the genetic and oncogenic mechanisms driving adult T-ALL, we analyzed a series of 57 T-ALL samples treated in the Eastern Cooperative Oncology Group (ECOG) E2993 protocol (Marks et al., 2009) using gene expression oligonucleotide microarrays. Unsupervised analysis and consensus clustering of microarray gene expression data revealed the presence of 2 stable gene expression clusters (Cluster I and II) encompassing approximately half of the tumor samples analyzed each (Fig. 1 A). Supervised analysis demonstrated that each of these two groups is characterized by a distinct gene expression program encompassing 365 differentially expressed unique genes (fold change >1.5; P < 0.0001; Fig. 1 B). Similar results were obtained in an independent validation set of 30 adult T-ALLs (Chiaretti et al., 2004). Thus, consensus clustering identified two robust clusters encompassing 12 and 18 samples, respectively in this series (Fig. S1). Moreover, cross analysis of clusters identified in the discovery and validation datasets using gene set enrichment analysis (GSEA) demonstrated a high level of enrichment in the expression signatures associated with clusters I and II in our discovery series in the clusters identified in this second dataset and vice versa, a high level of enrichment of a high level of enrichment in differentially expressed genes between clusters I and II in our validation series in the clusters identified in this our original dataset (Fig. S1).
Figure 1.
Gene expression profiling identifies high prevalence of early immature leukemias in adult T-ALL. (A) Consensus clustering of microarray gene expression data of 57 adult T-ALL samples. (B) Top 50 differentially expressed genes between adult T-ALL cluster I and cluster II samples. Genes in the heat map are shown in rows and each individual sample is shown in one column. The scale bar shows color-coded differential expression from the mean in standard deviation units with red indicating higher levels and blue lower levels of expression. (C) GSEA analysis of early immature/_LYL1_-positive, early cortical/_TLX1_-positive, and late cortical/_TAL1_-positive associated genes in the gene expression clusters I and II identified in adult T-ALL. (D) GSEA of genes associated with pediatric ETP-T-ALLs in adult T-ALL gene expression clusters.
Gene expression profiling of pediatric T-ALLs has defined distinct molecular subgroups associated with the activation of specific transcription factor oncogenes and unique gene expression signatures reflecting a distinct arrest during T cell development (Ferrando et al., 2002, 2004; Soulier et al., 2005; Homminga et al., 2011). Early immature/_LYL1_-positive T-ALLs show an early block at the double-negative stage of thymocyte development (Ferrando et al., 2002). In contrast, early cortical T-ALLs are characteristically CD1a, CD4, and CD8 positive and are frequently associated with translocations inducing aberrant expression of the TLX1 and TLX3 homeobox transcription factor oncogenes (Ferrando et al., 2002). Finally, late cortical thymocyte T-ALLs show expression of CD4, CD8, and CD3 and activation of the TAL1 transcription factor oncogene (Ferrando et al., 2002; Soulier et al., 2005; Homminga et al., 2011). GSEA of genes associated with these pediatric molecular groups of T-ALL (Ferrando et al., 2002) against the two clusters of adult T-ALLs identified in our series showed a highly significant enrichment of LYL1/immature T-ALL–associated genes in cluster I, whereas cluster II was associated with TLX1/early cortical and TAL1/late cortical T-ALL signatures (Fig. 1 C). Consistently, all TLX1 (n = 5) and TLX3 (n = 5), and 8/10 TAL1 high/LMO1/LMO2, expression leukemias in this series were included in cluster II.
Notably, transcriptionally defined early immature adult T-ALLs in cluster I showed an early arrest in T cell development and were characterized by the expression of cytoplasmic CD3 together with early hematopoietic markers such as CD34 (22/29, 76%) and CD133 (14/29, 48%) and myeloid-associated antigens such as CD13 (19/29, 65%), CD33 (16/29, 55%), and CD11b (16/29, 55%; Table S1). However, despite these common features, we could still identify three different immunophenotype categories within this group based on the expression of CD5. Thus, 10/29 cases (34%) showed complete absence of surface CD5 expression, 16/29 (55%) showed low levels of surface CD5 expression, and 3/29 (11%) were strongly CD5 positive (Table S1). Importantly, weak expression of CD5 in immature pediatric T-ALL leukemias has recently been associated with a gene expression program resembling early T cell progenitors and defines a group of childhood T-ALLs with poor prognosis (Coustan-Smith et al., 2009). To test the relationship of adult T-ALLs with early T cell precursor (ETP) pediatric T-ALLs, we performed GSEA of adult T-ALL clusters I and II using a gene set of ETP-T-ALL–associated transcripts (Coustan-Smith et al., 2009). This analysis showed a high level of enrichment of transcripts associated with this poor prognostic pediatric T-ALL group in adult immature/cluster I T-ALLs (Fig. 1 D).
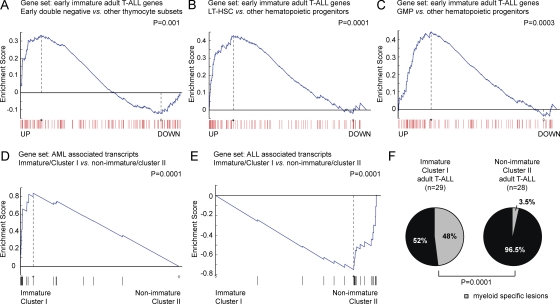
To explore the relationship between the genetic program associated with adult early immature T-ALLs and normal thymocytes, we performed GSEA analysis of gene expression signatures associated with distinct human thymocyte subsets including early double-negative (CD34+, CD1a−, CD4−, CD8−), late double-negative (CD34+, CD1a+, CD4−, CD8−), immature single-positive (CD4+, CD8−, CD3−), early double-positive (CD4+, CD8+, CD3−), late double-positive (CD4+, CD8+, CD3+), mature CD4 single-positive (CD4+, CD8−, CD3+), and mature CD8 single-positive (CD4−, CD8+, CD3+) cells. This analysis demonstrated a significant enrichment of genes characteristic for the earliest double-negative (CD34+, CD1a−, CD4−, CD8−) T cell progenitors in the early immature T-ALL group (Fig. 2 A). Notably, early double-negative thymocytes are not yet committed to T cell development and retain multilineage potential (Bhandoola and Sambandam, 2006). Therefore, we used a similar approach to analyze the relationship of early immature T-ALLs with other hematopoietic lineages and progenitor populations (Novershtern et al., 2011). Surprisingly, this analysis revealed a significant enrichment of genes up-regulated in long-term hematopoietic stem cells (Fig. 2 B) and in granulocyte-monocyte progenitors (Fig. 2 C) in this group. These results, together with the high prevalence of expression of immature (CD34, CD133) and myeloid (CD13, CD33, and CD11b) surface markers in these leukemias (Table S1) made us consider the possibility that early immature adult T-ALL tumors may be transcriptionally and genetically related to acute myeloid leukemias (AMLs).
Figure 2.
Myeloid and stem cell features of immature adult T-ALL. (A–C) GSEA of transcripts significantly up-regulated in early immature adult T-ALL in gene expression signatures obtained from human early double negative thymocytes (CD34+CD1a−CD4−CD8−; A) against all other thymocyte groups, long term hematopoietic stem cells (LT-HSC; B), and granulocyte-monocyte progenitors (GMP; C) against all other hematopoietic populations. (D and E) GSEA of AML- (D) or ALL-associated (E) transcripts in early immature (cluster I) versus other (cluster II) adult T-ALLs. (F) Differential distribution of myeloid-specific lesions in immature (cluster I) versus other (cluster II) adult T-ALLs.
To test this hypothesis, we analyzed the enrichment of lymphoid and myeloid leukemia gene expression signatures (Golub et al., 1999) in immature adult T-ALLs. This analysis revealed a highly significant enrichment of AML-associated transcripts in the early immature T-ALL cluster I group, whereas ALL associated transcripts were enriched in the T-ALL cluster II samples (Fig. 2, D and E). Based on this observation, we hypothesized that myeloid-specific genetic alterations might be uniquely present in immature adult T-ALLs. Strikingly, mutation analysis of AML oncogenes and tumor suppressor genes revealed the presence of myeloid mutations targeting IDH1, IDH2, DNMT3A, FLT3, and NRAS in 14/29 (48%) of immature adult T-ALL cases, whereas only 1/28 (3.5%) of all other adult T-ALLs showed a mutation in NRAS (P = 0.0001; Fig. 2 F and Table S2). In addition, immature adult T-ALL samples showed a lower prevalence of prototypical T-ALL genetic alterations such as activating mutations in the IL7R gene (1/29 [3.5%] immature adult T-ALLs vs. 6/28 [21.5%] of all other adult T-ALL samples; P < 0.05) and mutations in NOTCH1 and FBXW7 activating the NOTCH-signaling pathway (12/29 [41%] immature adult T-ALLs vs. 26/28 [93%] of all other adult T-ALLs; P < 0.05; Table S2). Notably, myeloid-specific gene alterations and mutations activating IL7R or NOTCH signaling were overlapping in 7/29 (24%) of all immature adult T-ALLs (Table S2). Overall, these results show that early immature adult T-ALLs are a gray zone leukemia category characterized by the presence of overlapping myeloid and T-lymphoid immunophenotypic features and genetic alterations.
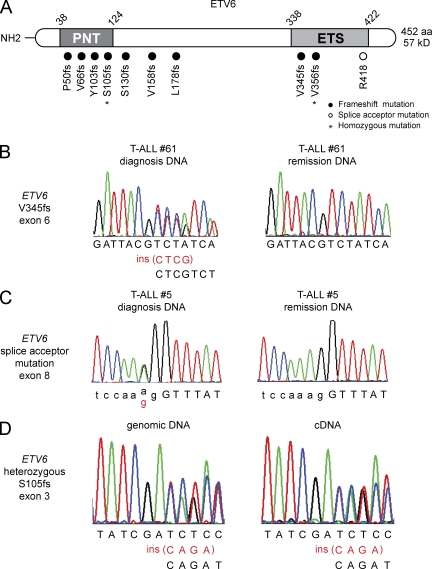
Notably, a similar cooccurrence of both T lymphoid and myeloid characteristics has been previously documented in rare leukemias harboring the translocation t(8;12)(q13;p13), which results in the fusion of the ETV6 tumor suppressor and the NCOA2 gene (Strehl et al., 2008; Homminga et al., 2011). Given the prominent role of ETV6 in hematopoietic stem cell homeostasis (Hock et al., 2004) and to test a possible broader role of ETV6 in the pathogenesis of T-ALL, we screened all coding exons of ETV6 for the presence of somatic mutations in an extended series of adult leukemia cases. This analysis revealed the presence of ETV6 mutations in 10 out of 82 (12%) adult T-ALL samples analyzed (Fig. 3, A–C; Table 1). Strikingly, ETV6 mutations were exclusively found in early immature adult T-ALLs with expression of the early hematopoietic marker CD34, cytoplasmic CD3, and the myeloid-associated antigen CD33 in the absence of CD4 and CD8 (Table S3). The overall incidence of ETV6 mutations in immature adult T-ALL in this series was 10 out of 41 (24%). In seven cases, ETV6 mutations corresponded to frameshift insertions or deletions in the N-terminal part of the protein, whereas the three remaining alleles corresponded to C-terminal truncating alleles (2 frameshift insertions and deletions and an exon 8 splice acceptor site mutation; Fig. 3 A). Analysis of bone marrow remission genomic DNA confirmed the somatic origin of ETV6 mutations (three frameshift and one splice site) in each of the four patient samples with available material (Fig. 3, B and C; Table 1). ETV6 mutations found in T-ALL were heterozygous in 8 out of 10 cases (80%) with two patient samples showing copy neutral loss of heterozygosity resulting in homozygous N-terminal and C-terminal truncating alleles (Table 1). RT-PCR analysis confirmed expression of the remaining wild-type ETV6 allele in heterozygous ETV6 mutant adult T-ALL samples (Fig. 3 D). In addition, and in contrast with the lower incidence of NOTCH1 mutations present in the immature group, 8/10 (80%) of ETV6 mutant T-ALL cases showed mutations in NOTCH1 (Table 1), suggesting a specific interaction between the oncogenic programs activated by NOTCH1 and mutational loss of ETV6 in the pathogenesis of adult immature T cell lymphoblastic leukemia.
Figure 3.
ETV6 mutations in early immature adult T-ALL. (A) Schematic representation of the structure of the ETV6 protein. The N-terminal pointed (PNT) homodimerization domain and the C-terminal DNA-binding domain (ETS-domain) are indicated. ETV6 mutations identified in primary adult T-ALL samples are shown. Filled circles represent frameshift mutations, whereas the splice acceptor mutation in the exon 8 splice acceptor sequence is depicted as an open circle. (B and C) Representative DNA sequencing chromatograms of paired diagnosis and remission genomic T-ALL DNA samples showing a somatic frameshift mutation in exon 6 (B) and a splice acceptor mutation in exon 8 of ETV6 (C). (D) Sequence analysis of paired ETV6 genomic DNA and cDNA shows biallelic expression of wild-type and mutant ETV6 transcripts.
Table 1.
ETV6 mutations in early immature adult T-ALL
| ETV6 mutations | T-ALL specific lesions Myeloid specific lesions | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Sample ID | Exon | Predicted amino acid change | Homozygous/heterozygous | Somatic/germline | NOTCH1 | FBXW7 | NRAS | IDH2 | DNMT3A |
| 3 | 3 | S105fs | het | NA | MUT | WT | MUT | WT | WT |
| 5 | 8 | R417fs | het | somatic | MUT | WT | WT | WT | WT |
| 18 | 3 | Y103fs | homo (LOH) | somatic | MUT | WT | WT | WT | WT |
| 24 | 4 | S130fs | het | somatic | MUT | WT | WT | WT | WT |
| 27 | 2 | P50fs | het | NA | MUT | WT | MUT | WT | WT |
| 58 | 5 | L178fs | het | NA | WT | WT | WT | MUT | MUT |
| 59 | 5 | V158fs | het | NA | MUT | WT | WT | WT | MUT |
| 60 | 3 | V66fs | het | NA | WT | WT | WT | WT | WT |
| 61 | 6 | V345fs | het | somatic | MUT | WT | WT | WT | WT |
| 62 | 6 | N356fs | homo (LOH) | NA | MUT | WT | WT | WT | WT |
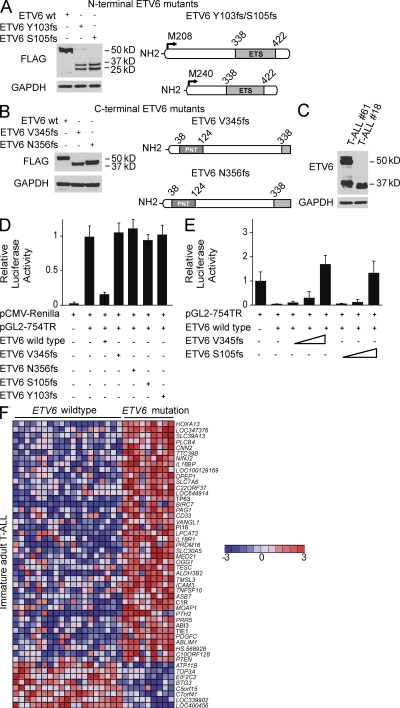
The ETV6 tumor suppressor gene is frequently translocated in lymphoid and myeloid hematopoietic tumors and encodes a transcriptional repressor with an N-terminal pointed (PNT) homodimerization domain and a C-terminal ETS DNA-binding domain (Bohlander, 2005). Transfection of plasmids driving the expression of N-terminal truncating ETV6 mutations (Y103fs, S105fs) found in adult T-ALL resulted in the activation of the M208 and M240 internal translation initiation sites (Barjesteh van Waalwijk van Doorn-Khosrovani et al., 2005) and the expression of N-terminal truncated protein products (23 and 28 kD) devoid of the PNT domain and retaining the DNA-binding domain (Fig. 4 A). C-terminal ETV6 truncating mutations (V345fs, N356fs) resulted in the expression of C-terminal truncated polypeptides (42 and 44 kD, respectively; Fig. 4 B). Consistently, Western blot analysis of a primary patient sample harboring a homozygous N-terminal truncating mutation detected the presence of shorter mutant ETV6 isoforms (23 and 28 kD; Fig. 4 C), whereas analysis of a sample with a heterozygous C-terminal mutation revealed the expression of both wild-type (50 and 57 kD) and truncated (42 kD) ETV6 proteins (Fig. 4 C).
Figure 4.
Functional analysis of truncating ETV6 mutant alleles. (A and B) Western blot analysis of FLAG-tagged N-terminal (Y103fs, S105fs) (A) and C-terminal (V345fs, N356fs) (B) ETV6 mutants expressed in HEK293T cells. The FLAG epitope was introduced in the C terminus of the N-terminal Y103fs and S105fs mutants and in the N terminus of the C-terminal ETV6 V345fs and N356fs mutants. Blots were probed with anti-FLAG or anti-GAPDH as loading control. (C) Lysates from primary adult T-ALL samples harboring a heterozygous C-terminal (V345fs, #61) and a homozygous N-terminal (Y103fs,#18) ETV6 mutation were analyzed by Western blot. Wild-type ETV6 proteins are detected as 50 and 57 kD anti-ETV6 immunoreactive bands and anti-GAPDH as loading control. (D) Effects of ETV6 mutant alleles in the activity an ETV6-responsive reporter plasmid (pGL2-754TR). Luciferase activity is shown relative to empty vector and normalized using an internal control plasmid expressing Renilla luciferase. Experiments were performed in triplicate and repeated at least twice. (E) Luciferase assay analyzing the effects of increasing amounts (indicated by wedges) of C-terminal (V345fs) and N-terminal (S105fs) ETV6 mutants on the activity of wild-type ETV6. Experiments were performed in triplicate and repeated at least twice. (F) Supervised analysis of ETV6 mutant versus ETV6 wild-type gene expression signatures in early immature adult T-ALLs (P < 0.001). Top 50 differentially expressed genes are shown. Genes in the heat map are shown in rows, each individual sample is shown in one column. The scale bar shows color-coded differential expression from the mean in standard deviation units, with red indicating higher levels and blue lower levels of expression.
To elucidate the functional consequences of ETV6 alterations, we performed luciferase reporter assays using an ETV6-responsive reporter construct (pGL2-754TR) derived from the stromelysin-1 gene (Fenrick et al., 2000). In line with the role of ETV6 as transcriptional repressor, expression of wild-type ETV6 strongly decreased luciferase activity (Fig. 4 D). In contrast, C-terminal (V345fs; N356fs) and N-terminal (Y103fs; S105fs) ETV6 mutants were functionally inactive with no transcriptional repression activity (Fig. 4 D). Moreover, expression of C-terminal (V345fs) and N-terminal (S105fs) ETV6 mutants abolished the transrepression effects of wild-type ETV6 (Fig. 4 E), suggesting that these mutant ETV6 alleles harbor dominant-negative activity. Consistently, supervised analysis of microarray gene expression signatures of ETV6 wild-type (n = 19) and _ETV6_-mutated (n = 9) immature adult T cell leukemias showed that _ETV6_-mutated cases have a characteristic gene expression signature dominated by up-regulated transcripts (Fig. 4 F and Fig. S2), which included the myeloid marker CD33 and genes involved in leukemia development and stem cell homeostasis, such as HOXA13 (Su et al., 2006; Bach et al., 2010), TP63 (Bernassola et al., 2005; Crum and McKeon, 2010), PTEN (Yilmaz et al., 2006; Zhang et al., 2006), and PRDM16 (Du et al., 2005; Chuikov et al., 2010), which is a gene translocated to the ETV6 locus in AML (Duhoux et al., 2011; Fig. 4 F). Notably, all ETV6 mutant T-ALL samples in our series were characteristically CD33 positive (5/5 [100%] immature ETV6 mutant T-ALLs vs. 11/24 [46%] immature ETV6 wild-type T-ALLs; P < 0.05; Table S3).
After these results, and to test the transcriptional effects of ETV6 inactivation in immature T-ALL, we analyzed the gene expression changes induced upon small interfering RNA (siRNA)-mediated knockdown of ETV6 in LOUCY cells, a T-ALL cell line with a transcriptional program highly related to that of immature T-ALLs (Fig. 5 A, Fig. S3). Notably, GSEA analysis of this ETV6 knockdown signature showed a significant enrichment in genes that are characteristically up-regulated in ETV6 mutant immature T-ALLs, including HOXA13, PRDM16, PTEN, and CD33 (P = 0.016; Fig. 5 B).
Figure 5.
Transcriptional program regulated by ETV6 in T-ALL. (A) RT-PCR (top) and Western blot (bottom) analysis of ETV6 expression in LOUCY cells expressing control siRNA or one of two ETV6-specific siRNAs. (B) GSEA of the early immature ETV6 mutant T-ALL–associated transcripts in control siRNA versus ETV6 knockdown LOUCY cells. Heatmap of genes up-regulated in early immature ETV6 mutant T-ALL most differentially expressed in control siRNA versus ETV6 knockdown LOUCY cells. The myeloid marker CD33 and genes involved in leukemia development and stem cell homeostasis are shown in red. Genes in the heat map are shown in rows and each individual sample is shown in one column. The scale bar shows color-coded differential expression from the mean in standard deviation units, with red indicating higher levels and blue lower levels of expression.
Overall, our results demonstrate a high prevalence of early immature T-ALLs in adult T-ALL, which show increased heterogeneity in their immunophenotypes compared with pediatric ETP-TALLs, mixed lymphoid and myeloid characteristics, and frequent truncating mutations in the ETV6 tumor suppressor gene.
MATERIALS AND METHODS
Patient samples and cell lines.
Leukemic DNA and cryopreserved lymphoblast samples were provided by the Eastern Cooperative Oncology Group (ECOG). All samples were collected in clinical trials E2993 (Marks et al., 2009) and C10403. Informed consent to use leftover material for research purposes was obtained from all the patients at trial entry according to the Declaration of Helsinki. T cell phenotype was confirmed by flow cytometry. In total, 82 adult T-ALL samples were used in this study, including 57 samples used for the gene expression profiling study and mutation analysis and 25 additional cases used for mutation screening.
The LOUCY cell line was cultured in RPMI-1640 media supplemented with 10% fetal bovine serum, 100 U/ml penicillin G, and 100 µg/ml streptomycin at 37°C in a humidified atmosphere under 5% CO2. HEK293T cells were cultured under similar conditions in DME media.
Sorting of human thymocyte populations from human thymus.
Thymus samples were obtained as surgical tissue discards from 3 pediatric patients (age from 7 d to 6 mo) undergoing cardiac surgery at the New York Presbyterian Hospital. Single-cell suspensions were prepared by dissecting the thymus into small pieces and passing the tissue through a cell strainer (diameter 70 μm), and mononuclear cells were isolated by Ficoll density centrifugation (GE Healthcare). CD34+ human thymocyte populations were obtained by positive selection using magnetically activated cell sorting using the CD34 MicroBead kit (Miltenyi Biotec) and by negative selection using the CD3 MicroBead kit (Miltenyi Biotec). Magnetically isolated CD34+ cells were then labeled with fluorophore-conjugated monoclonal antibodies against CD34 (CD34-APC) and CD1a (CD1-PE; BD). Early double negative thymocytes (CD34+CD1a−CD3−CD4−CD8−) and late double negative thymocytes (CD34+CD1a+CD3−CD4−CD8−) were isolated by FACS. Immature single-positive thymocytes (CD4+CD8−CD3−), were isolated by FACS after immunomagnetic depletion of total thymocytes with the CD3 MicroBead kit (Miltenyi Biotec) and immunostaining with CD4-PE (BD) and CD34-FITC antibodies. Early double-positive cells (CD4+CD8+CD3−) were isolated by FACS from CD3 immunodepleted cells after staining with CD4-PE and CD8-FITC (BD) antibodies. Late double-positive (CD4+CD8+CD3+) and mature single-positive CD4 (CD4+CD3+) and CD8 (CD8+CD3+) thymocytes were FACS after triple immunolabeling with CD4-PE, CD8-FITC, and CD3-APC (BD) antibodies. Cell sorting was performed with a FACSDiva cell sorter (BD), and the purity of all sorted populations was >95%.
Microarray gene expression profiling of normal T cell subsets and primary adult T-ALL samples.
RNA isolation from sorted subsets of human thymocytes was performed in triplicate using the RNeasy Micro kit (QIAGEN) according to manufacturer’s protocol. Next, 10 ng of RNA from each T cell subset was amplified using the Ovation PicoSL WTA System (NuGen), labeled by the Encore BiotinIL Module (NuGen), and hybridized to the HumanHT-12 v4 Expression BeadChip (Illumina) according to manufacturer’s protocol. Similarly, RNA isolation from lymphoblast obtained from adult T-ALL patient samples was achieved using the RNeasy plus mini kit (QIAGEN) according to manufacturer’s protocol. Next, 500 ng of RNA was amplified and biotinylated using the TotalPrep RNA Amplification kit (Invitrogen) and hybridized to the HumanHT-12 v4 Expression BeadChip (Illumina).
Gene expression profiling data from 57 adult T-ALL patients and normal T cell subsets were normalized using quantile normalization without background subtraction. Unsupervised consensus clustering was performed with K = 2, as previously described (Monti et al., 2003), using the GenePattern application (Gould et al., 2006). Genes differentially expressed between cluster I and cluster II adult T-ALL cases were identified using the Comparative Marker Selection application in GenePattern (Gould et al., 2006) using a nonparametric P value calculation with 1,000 permutations.
A validation series of 30 previously reported adult T-ALL microarray data (Chiaretti et al., 2004) was normalized with GC-RMA using the open-source Bioconductor project (www.bioconductor.org) within the statistical programming language R (http://cran.r-project.org/). In this series, we selected the top 250 most variable probes from the unsupervised consensus clustering analysis of the 57 ECOG T-ALL expression samples in our discovery series matched by gene names for consensus clustering as described in the previous paragraph. The relationship of clusters identified in the discovery series and validation series was established by cross GSEA analysis (Subramanian et al., 2005) of the top up- and down-regulated genes (Student’s t test, P < 0.0001) in clusters I and cluster II of one series against the list of genes ranked by the t-score (cluster I vs. cluster II) in the other.
Enrichment of early immature/_LYL1_-positive, early cortical/TLX1, and late cortical/_TAL1_-associated genes in the gene expression clusters identified in our adult T-ALL series was analyzed by GSEA using the t test metric and 1,000 permutations of the genes. In addition, enrichment of myeloid- and lymphoid-associated genes was studied by a similar GSEA analysis using myeloid and lymphoid (Golub et al., 1999) gene sets. Similarly, we analyzed the relationship of the early immature adult T-ALL signature with that of different sorted human hematopoietic populations (Novershtern et al., 2011; Gene Expression Omnibus [GEO] accession no. GSE24759], the gene expression profiles generated for the specific populations of sorted human thymocytes, and gene sets associated with ETP T-ALL (Coustan-Smith et al., 2009), as described above. GEO accession nos. for adult T-ALL and normal thymocyte microarray data analyzed here are GSE33469 and GSE33470, respectively.
Microarray data from human T-ALL cell lines (Palomero et al., 2007; GEO accession no. GSE5682) were analyzed for their relationship with cluster I (early immature) and cluster II T-ALL signatures by GSEA using the Student’s t test metric and 1,000 permutations of the genes.
Sequence analysis.
All exon sequences from ETV6 were amplified from genomic DNA by PCR and analyzed by direct dideoxynucleotide sequencing. Mutational hotspot regions for FLT3, DNMT3A, IDH1, IDH2, NOTCH1, IL7R, and FBXW7 were sequenced. Primer sequences used for FLT3 (Van Vlierberghe et al., 2005), DNMT3A (Ley et al., 2010), IDH1 (Andersson et al., 2011), IDH2 (Andersson et al., 2011), NOTCH1 (Weng et al., 2004), IL7R (Zenatti et al., 2011), and FBXW7 (Thompson et al., 2007) have been previously described. Primer sequences used for ETV6 sequencing are shown in Table S4.
Luciferase reporter assays.
We generated N-terminal or C-terminal FLAG-tagged wild-type and mutant ETV6 constructs in the pCDNA3.1 (-) plasmid by direct PCR cloning from cDNA obtained from primary ETV6 wild-type and mutant adult T-ALL samples. A FLAG epitope was introduced into the N terminus of the C-terminal ETV6 V345fs and N356fs mutants and in the C terminus of the N-terminal Y103fs and S105fs mutants. We tested the ETV6 transcriptional repression activity in a luciferase reporter assay using the pGL2-T574 reporter construct (Fenrick et al., 2000). In these assays, we transfected HEK293T cells with 50 ng of wild-type pcDNA3-ETV6 or different types of N-terminal and C-terminal ETV6 mutants, together with a plasmid driving the expression of the Renilla luciferase gene (pCMV-Renilla) used as transfection control. Alternatively, we transfected HEK293T cells with a constant amount of wild-type pcDNA3-ETV6 in combination with increased concentrations of N-terminal and C-terminal ETV6 mutant constructs at 1:1, 1:2, and 1:3 ratios. We measured luciferase activity 48 h after transfection with the Dual-Luciferase Reporter assay kit (Promega).
Western blot.
Western blot analysis was performed using a mouse monoclonal antibody against ETV6 (1:5,000; Abnova), a rabbit antibody against FLAG epitope (1:1,000; Cell Signaling Technology), and a mouse monoclonal antibody GAPDH (1:1,000; Santa Cruz Biotechnology, inc.) using standard procedures.
ETV6 siRNA knockdown.
For siRNA-mediated knockdown of ETV6 in the LOUCY T-ALL cell line, we performed electroporation of 50 nM _ETV6_-specific siRNAs (Applied Biosystems) or 50 nM of scramble siRNAs as control, using an exponential decay pulse (300 V, 1000 µF; Genepulser MxCell, Bio-Rad). After 72 h, RNA was isolated using the RNeasy plus mini kit (QIAGEN) according to manufacturer’s protocol. Knockdown of ETV6 was evaluated by RT-PCR and Western blot analysis, after which the siRNA showing the highest ETV6 knockdown (siRNA ETV6#1) was selected for further analysis. Finally, gene expression profiling of biological replicates for LOUCY siRNA scramble control and LOUCY siRNA ETV6#1 was performed using the HumanHT-12 v4 Expression BeadChip as described previously for the primary leukemia samples.
The GEO accession code for the microarray data of LOUCY cells upon siRNA ETV6 knockdown analyzed here is GSE33820.
Statistical analysis.
The Fisher’s exact test was used to compare the frequency of specific genomic lesions between of immature and other adult T-ALL patients.
Online supplemental material.
Fig. S1 shows a consensus clustering analysis of a validation series of 30 adult T-ALLs and cross GSEA analysis between clusters identified in the discovery and validation cohorts. Fig. S2 shows a heatmap of the supervised analysis from ETV6 mutant versus ETV6 wild-type gene expression signatures in early immature adult T-ALLs (P < 0.001; FC > 1.25). Fig. S3 shows GSEA analysis of transcripts significantly up-regulated in early immature adult T-ALL in gene expression signatures obtained from a panel of T-ALL cell lines. Table S1 shows an overview of the immunophenotypic features of the adult T-ALL samples included in this study (n = 57). Table S2 shows an overview of the myeloid and T-lymphoid specific genetic alterations in adult T-AL. Table S3 shows the immunophenotypic features of the ETV6 mutant adult T-ALL samples (n = 10). Table S4 shows the primer sequences used for ETV6 mutations analysis. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20112239/DC1.
Acknowledgments
This study was supported by the Fund for Scientific Research Flanders (P. Van Vlierberghe); the ECOG tumor bank; the National Institutes of Health (R01CA120196 to A. F. and U24 CA114737 to E.P.); the Stand Up To Cancer Innovative Research Award (A.F.), the Chemotherapy Foundation (A.F.); the Rally Across America Foundation (A.F); and the Swim Across America Foundation (A.F.). P. Van Vlierberghe is an ASH Scholar and A.F. is a Leukemia & Lymphoma Society Scholar.
The authors declare that they have no competing financial interests.
P. Van Vlierberghe performed experiments and wrote the manuscript. A.A. performed bioinformatic analyses including analysis of gene expression profiling data and GSEA. A.P. performed experiments. J.E.H performed luciferase assays. I.R. isolated RNA and performed gene expression profiling of primary T-ALL samples. M.H. performed sequencing analysis of myeloid specific lesions. M.R. performed statistical analysis. E.P., J.R., P.H.W, S.M.L. and J.M.R provided samples and correlative clinical and immunophenotypic data from ECOG. A.F. designed the studies, directed research and wrote the manuscript.
Footnotes
Abbreviations used:
AML
acute myeloid leukemia
ETP
early T cell precursor
GSEA
gene set enrichment analysis
siRNA
small interfering RNA
T-ALL
T cell acute lymphoblastic leukemia
References
- Andersson A.K., Miller D.W., Lynch J.A., Lemoff A.S., Cai Z., Pounds S.B., Radtke I., Yan B., Schuetz J.D., Rubnitz J.E., et al. 2011. IDH1 and IDH2 mutations in pediatric acute leukemia. Leukemia. 25:1570–1577 10.1038/leu.2011.133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach C., Buhl S., Mueller D., García-Cuéllar M.P., Maethner E., Slany R.K. 2010. Leukemogenic transformation by HOXA cluster genes. Blood. 115:2910–2918 10.1182/blood-2009-04-216606 [DOI] [PubMed] [Google Scholar]
- Barjesteh van Waalwijk van Doorn-Khosrovani S., Spensberger D., de Knegt Y., Tang M., Löwenberg B., Delwel R. 2005. Somatic heterozygous mutations in ETV6 (TEL) and frequent absence of ETV6 protein in acute myeloid leukemia. Oncogene. 24:4129–4137 [DOI] [PubMed] [Google Scholar]
- Bernassola F., Oberst A., Melino G., Pandolfi P.P. 2005. The promyelocytic leukaemia protein tumour suppressor functions as a transcriptional regulator of p63. Oncogene. 24:6982–6986 10.1038/sj.onc.1208843 [DOI] [PubMed] [Google Scholar]
- Bhandoola A., Sambandam A. 2006. From stem cell to T cell: one route or many? Nat. Rev. Immunol. 6:117–126 10.1038/nri1778 [DOI] [PubMed] [Google Scholar]
- Bohlander S.K. 2005. ETV6: a versatile player in leukemogenesis. Semin. Cancer Biol. 15:162–174 10.1016/j.semcancer.2005.01.008 [DOI] [PubMed] [Google Scholar]
- Chiaretti S., Li X., Gentleman R., Vitale A., Vignetti M., Mandelli F., Ritz J., Foa R. 2004. Gene expression profile of adult T-cell acute lymphocytic leukemia identifies distinct subsets of patients with different response to therapy and survival. Blood. 103:2771–2778 10.1182/blood-2003-09-3243 [DOI] [PubMed] [Google Scholar]
- Chuikov S., Levi B.P., Smith M.L., Morrison S.J. 2010. Prdm16 promotes stem cell maintenance in multiple tissues, partly by regulating oxidative stress. Nat. Cell Biol. 12:999–1006 10.1038/ncb2101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coustan-Smith E., Mullighan C.G., Onciu M., Behm F.G., Raimondi S.C., Pei D., Cheng C., Su X., Rubnitz J.E., Basso G., et al. 2009. Early T-cell precursor leukaemia: a subtype of very high-risk acute lymphoblastic leukaemia. Lancet Oncol. 10:147–156 10.1016/S1470-2045(08)70314-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crum C.P., McKeon F.D. 2010. p63 in epithelial survival, germ cell surveillance, and neoplasia. Annu. Rev. Pathol. 5:349–371 10.1146/annurev-pathol-121808-102117 [DOI] [PubMed] [Google Scholar]
- Du Y., Jenkins N.A., Copeland N.G. 2005. Insertional mutagenesis identifies genes that promote the immortalization of primary bone marrow progenitor cells. Blood. 106:3932–3939 10.1182/blood-2005-03-1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duhoux F.P., Ameye G., Montano-Almendras C.P., Bahloula K., Mozziconacci M.J., Laibe S., Wlodarska I., Michaux L., Talmant P., Richebourg S., et al. ; on behalf of the Groupe Francophone de Cytogénétique Hématologique (GFCH) and of the Belgian Cytogenetic Group for Haematology and Oncology (BCG-HO) 2011. PRDM16 (1p36) translocations define a distinct entity of myeloid malignancies with poor prognosis but may also occur in lymphoid malignancies. Br. J. Haematol. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- Fenrick R., Wang L., Nip J., Amann J.M., Rooney R.J., Walker-Daniels J., Crawford H.C., Hulboy D.L., Kinch M.S., Matrisian L.M., Hiebert S.W. 2000. TEL, a putative tumor suppressor, modulates cell growth and cell morphology of ras-transformed cells while repressing the transcription of stromelysin-1. Mol. Cell. Biol. 20:5828–5839 10.1128/MCB.20.16.5828-5839.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrando A.A. 2009. The role of NOTCH1 signaling in T-ALL. Hematology (Am Soc Hematol Educ Program). 1:353–361 10.1182/asheducation-2009.1.353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrando A.A., Neuberg D.S., Staunton J., Loh M.L., Huard C., Raimondi S.C., Behm F.G., Pui C.H., Downing J.R., Gilliland D.G., et al. 2002. Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer Cell. 1:75–87 10.1016/S1535-6108(02)00018-1 [DOI] [PubMed] [Google Scholar]
- Ferrando A.A., Neuberg D.S., Dodge R.K., Paietta E., Larson R.A., Wiernik P.H., Rowe J.M., Caligiuri M.A., Bloomfield C.D., Look A.T. 2004. Prognostic importance of TLX1 (HOX11) oncogene expression in adults with T-cell acute lymphoblastic leukaemia. Lancet. 363:535–536 10.1016/S0140-6736(04)15542-6 [DOI] [PubMed] [Google Scholar]
- Golub T.R., Slonim D.K., Tamayo P., Huard C., Gaasenbeek M., Mesirov J.P., Coller H., Loh M.L., Downing J.R., Caligiuri M.A., et al. 1999. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 286:531–537 10.1126/science.286.5439.531 [DOI] [PubMed] [Google Scholar]
- Gould J., Getz G., Monti S., Reich M., Mesirov J.P. 2006. Comparative gene marker selection suite. Bioinformatics. 22:1924–1925 10.1093/bioinformatics/btl196 [DOI] [PubMed] [Google Scholar]
- Hock H., Meade E., Medeiros S., Schindler J.W., Valk P.J., Fujiwara Y., Orkin S.H. 2004. Tel/Etv6 is an essential and selective regulator of adult hematopoietic stem cell survival. Genes Dev. 18:2336–2341 10.1101/gad.1239604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homminga I., Pieters R., Langerak A.W., de Rooi J.J., Stubbs A., Verstegen M., Vuerhard M., Buijs-Gladdines J., Kooi C., Klous P., et al. 2011. Integrated transcript and genome analyses reveal NKX2-1 and MEF2C as potential oncogenes in T cell acute lymphoblastic leukemia. Cancer Cell. 19:484–497 10.1016/j.ccr.2011.02.008 [DOI] [PubMed] [Google Scholar]
- Ley T.J., Ding L., Walter M.J., McLellan M.D., Lamprecht T., Larson D.E., Kandoth C., Payton J.E., Baty J., Welch J., et al. 2010. DNMT3A mutations in acute myeloid leukemia. N. Engl. J. Med. 363:2424–2433 10.1056/NEJMoa1005143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks D.I., Paietta E.M., Moorman A.V., Richards S.M., Buck G., DeWald G., Ferrando A., Fielding A.K., Goldstone A.H., Ketterling R.P., et al. 2009. T-cell acute lymphoblastic leukemia in adults: clinical features, immunophenotype, cytogenetics, and outcome from the large randomized prospective trial (UKALL XII/ECOG 2993). Blood. 114:5136–5145 10.1182/blood-2009-08-231217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monti S., Tamayo P., Mesirov J.P., Golub T. 2003. Consensus Clustering: A Resampling-Based Method for Class Discovery and Visualization of Gene Expression Microarray Data. Journal Machine Learning. 52:91–118 10.1023/A:1023949509487 [DOI] [Google Scholar]
- Novershtern N., Subramanian A., Lawton L.N., Mak R.H., Haining W.N., McConkey M.E., Habib N., Yosef N., Chang C.Y., Shay T., et al. 2011. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell. 144:296–309 10.1016/j.cell.2011.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paganin M., Ferrando A. 2011. Molecular pathogenesis and targeted therapies for NOTCH1-induced T-cell acute lymphoblastic leukemia. Blood Rev. 25:83–90 10.1016/j.blre.2010.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palomero T., Sulis M.L., Cortina M., Real P.J., Barnes K., Ciofani M., Caparros E., Buteau J., Brown K., Perkins S.L., et al. 2007. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nat. Med. 13:1203–1210 10.1038/nm1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pui C.H., Robison L.L., Look A.T. 2008. Acute lymphoblastic leukaemia. Lancet. 371:1030–1043 10.1016/S0140-6736(08)60457-2 [DOI] [PubMed] [Google Scholar]
- Soulier J., Clappier E., Cayuela J.M., Regnault A., García-Peydró M., Dombret H., Baruchel A., Toribio M.L., Sigaux F. 2005. HOXA genes are included in genetic and biologic networks defining human acute T-cell leukemia (T-ALL). Blood. 106:274–286 10.1182/blood-2004-10-3900 [DOI] [PubMed] [Google Scholar]
- Strehl S., Nebral K., König M., Harbott J., Strobl H., Ratei R., Struski S., Bielorai B., Lessard M., Zimmermann M., et al. 2008. ETV6-NCOA2: a novel fusion gene in acute leukemia associated with coexpression of T-lymphoid and myeloid markers and frequent NOTCH1 mutations. Clin. Cancer Res. 14:977–983 10.1158/1078-0432.CCR-07-4022 [DOI] [PubMed] [Google Scholar]
- Su X., Drabkin H., Clappier E., Morgado E., Busson M., Romana S., Soulier J., Berger R., Bernard O.A., Lavau C. 2006. Transforming potential of the T-cell acute lymphoblastic leukemia-associated homeobox genes HOXA13, TLX1, and TLX3. Genes Chromosomes Cancer. 45:846–855 10.1002/gcc.20348 [DOI] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 102:15545–15550 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson B.J., Buonamici S., Sulis M.L., Palomero T., Vilimas T., Basso G., Ferrando A., Aifantis I. 2007. The SCFFBW7 ubiquitin ligase complex as a tumor suppressor in T cell leukemia. J. Exp. Med. 204:1825–1835 10.1084/jem.20070872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Vlierberghe P., Meijerink J.P., Stam R.W., van der Smissen W., van Wering E.R., Beverloo H.B., Pieters R. 2005. Activating FLT3 mutations in CD4+/CD8- pediatric T-cell acute lymphoblastic leukemias. Blood. 106:4414–4415 10.1182/blood-2005-06-2267 [DOI] [PubMed] [Google Scholar]
- Weng A.P., Ferrando A.A., Lee W., Morris J.P., IV, Silverman L.B., Sanchez-Irizarry C., Blacklow S.C., Look A.T., Aster J.C. 2004. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 306:269–271 10.1126/science.1102160 [DOI] [PubMed] [Google Scholar]
- Yilmaz O.H., Valdez R., Theisen B.K., Guo W., Ferguson D.O., Wu H., Morrison S.J. 2006. Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature. 441:475–482 10.1038/nature04703 [DOI] [PubMed] [Google Scholar]
- Zenatti P.P., Ribeiro D., Li W., Zuurbier L., Silva M.C., Paganin M., Tritapoe J., Hixon J.A., Silveira A.B., Cardoso B.A., et al. 2011. Oncogenic IL7R gain-of-function mutations in childhood T-cell acute lymphoblastic leukemia. Nat. Genet. 43:932–939 10.1038/ng.924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Grindley J.C., Yin T., Jayasinghe S., He X.C., Ross J.T., Haug J.S., Rupp D., Porter-Westpfahl K.S., Wiedemann L.M., et al. 2006. PTEN maintains haematopoietic stem cells and acts in lineage choice and leukaemia prevention. Nature. 441:518–522 10.1038/nature04747 [DOI] [PubMed] [Google Scholar]