Overexpression of S100B, TM4SF4, and OLFM4 Genes Is Correlated with Liver Metastasis in Taiwanese Colorectal Cancer Patients (original) (raw)
Abstract
Distant metastasis of colorectal cancer (CRC) occurs mainly in the liver and is the major cause of death. This study explored the overexpression of liver metastasis-associated mRNAs in human CRC by using a well-established, weighted enzymatic chip array platform. Analysis of 10 CRC tissue specimens compared with their normal adjacent tissues revealed that ATP2A2, ELAVL4, hTERT, KCTD2, MUC1, OLFM4, S100B, and TM4SF4 genes were upregulated (gene expression ratio of cancer tissue to paired normal tissue was >2) by microarray and bioinformatics analysis. A gene chip including eight candidate genes was constructed to investigate the circulating tumor cells in blood specimens of 103 preoperative CRC patients and further validated by reverse transcriptase–polymerase chain reaction. Liver metastasis was significantly correlated with overexpression of S100B (_p_=0.001, OR=9.217), TM4SF3 (_p_=0.011, OR=4.385), and OLFM4 (_p_=0.015, OR=3.438). These results suggest that S100B, TM4SF3, and OLFM4 overexpression may affect metastatic behavior of tumor cells in Taiwanese CRC patients.
Introduction
Colorectal cancer (CRC) is a significant public health problem worldwide. Each year, nearly 1,000,000 new cases of CRC are diagnosed, and there are 500,000 deaths from CRC (Weitz et al., 2005). In Taiwan, distant metastasis is important relative to survival for CRC patients. As many as 25%–40% of patients who undergo curative resection subsequently develop metastatic disease, suggesting that undetected micrometastasis does exist, and may play a key role in relapse (Castells et al., 1998; Ghossein et al., 1999; Ghossein and Bhattacharya, 2000). One of the major causes of relapse is the presence of disseminated tumor cells shed from the primary carcinoma into the circulation before, during, or after surgery.
Reports have described the detection of circulating tumor cells (CTCs) in the peripheral blood of CRC patients, having important prognostic and therapeutic implications (Weitz et al., 1998; Wharton et al., 1999; Yamaguchi et al., 2000; Patel et al., 2002; Wang et al., 2006a). The commonly used technique for the detection of nucleic acid material of disseminated tumor cells in the last decade has been polymerase chain reaction (PCR), reverse transcriptase–PCR (RT-PCR), or real-time quantitative PCR (Q-PCR) assays, permitting sensitive detection of CTCs in peripheral blood. Because of the heterogeneity of gene marker expression in blood, a multimarker assay would be more reliable and sensitive than a single-marker assay (Hoon et al., 1995; Racila et al., 1998; Baker et al., 2003; Sher et al., 2005). The development of biotechnology has made biochips an important tool in clinical diagnosis or drug efficacy evaluation (Burger, 2006). Our previously developed membrane array-based multimarker assay can detect CTCs in the peripheral blood of CRC patients (Chen et al., 2005; Wang et al., 2006b, 2007; Yeh et al., 2006). In fact, we have demonstrated a high correlation between real-time Q-PCR and the membrane array method in the detection of CTCs in CRC patients (Wang et al., 2006b). However, costs of the digoxigenin enzyme used for the colorimetric biochip platform were too high for routine laboratory diagnosis, and the high criteria of the operation techniques have prevented its widespread utility for clinical applications. Therefore, we have developed the next-generation biochip operation platform—weighted enzymatic chip array (EnCA) platform, which has replaced the conventional digoxigenin system with the bioton–avidin enzyme system to lower the overall cost.
The objective of this study was to explore the liver metastasis-related genes in the CTCs of peripheral blood specimens in preoperative CRC patients. Based on studying results of microarray and bioinformatics analysis in 10 paired CRC tumor specimens and paired normal adjacent tissues, there were 1089 genes indicating that CRC tumor specimens had different expression profiles from the comparative adjacent normal tissues. Initially, we selected eight upregulated candidate genes closely related to CRC carcinogenesis for further weighted EnCA construction by bioinformatics analysis. Subsequently, we evaluated the diagnostic value in detecting CTCs in the peripheral blood of 103 stage I to III preoperative CRC patients. In addition, the relationships between candidate genes and clinicopathological factors were analyzed. Results from this study demonstrated that three genes S100B, TM4SF3, and OLFM4 were significantly associated with liver metastasis in Taiwanese CRC patients. These three genes may be suitable new markers preoperatively for liver metastasis. Suitable biomarkers can predict disease prognosis and also aid in making appropriate treatment strategy decisions.
Materials and Methods
Sample collection of clinical experiments
Enrolled in this study originally were 10 American Joint Commission on Cancer/International Union Against Cancer (AJCC/UICC) stage I–III CRC patients who underwent surgical treatment in the Department of Surgery at Fooyin University Hospital for elective surgery. Patients with other malignant diseases noted in the medical history were excluded. All 10 patients underwent radical resection for a primary lesion. All paired samples including tumor and tumor-free tissues were obtained from each patient. The data of these 10 CRC patients were used for analysis of CRC-related gene expressions.
To determine genes related to prognosis, peripheral blood samples (5 mL) were obtained from an additional 103 stage I, II, and III CRC patients, who were admitted to the Department of Surgery at Fooyin University Hospital, Taichung Veterans General Hospital, and Kaohsiung Medical University Hospital for elective surgery. The follow-up endpoint was October 2010. CRC peripheral blood specimens were obtained within 1 week before operations for CTC analysis, and the candidate genes were further validated by the EnCA platform. The relevant data are listed in Table 1. Tumor grading was carried out and confirmed by pathologists. Each tissue sample was snap-frozen in liquid nitrogen immediately after surgery or biopsy and stored at −80°C until further use. Samples were used in experiments for real-time PCR, membrane array analysis, and immunoblotting. Postoperative surveillance consisted of comprehensive medical history, physical examination, and laboratory studies, including assessing serum carcinoembryonic antigen levels at 3-month intervals. Abdominal ultrasonography or computed tomography was performed at 6-month intervals, and chest radiography, bone scans, and total colonoscopy were performed. Patients were followed up at 3-month intervals for 2 years and at 6-month intervals thereafter. The median follow-up time was 36.4 months (range, 24–46 months). Preoperative surveillance was comprised of a comprehensive medical history, physical examination, and laboratory studies. Clinical stage and pathological features of primary tumors were defined according to the criteria of the American Joint Commission on Cancer/International Union Against Cancer (AJCC/UICC). All patients with liver metastasis development included in this study were confirmed by aspiration cytology. To avoid contamination of skin cells, sampled blood was taken via an intravenous catheter and the first few milliliters of blood were discarded. Total RNA was immediately extracted from the peripheral whole blood, which then served as a template for complementary DNA (cDNA) synthesis. The institutional review boards of the three hospitals approved the acquisition of samples as well as their subsequent use. Written informed consent was obtained from all participants.
Table 1.
Correlations Between Clinicopathological Features and Liver Metastatic Status for 103 Postoperative Colorectal Cancer Patients
| Characteristics | Total cases n (%) | Liver metastasis n (%) | Nonliver metastasis n (%) | p |
|---|---|---|---|---|
| Gender | ||||
| Male | 58 (56.3%) | 12 (20.7%) | 46 (79.3%) | 0.505 |
| Female | 45 (43.7%) | 7 (15.6%) | 38 (84.4%) | |
| Age (years) | ||||
| <60 | 62 (60.2%) | 10 (16.1%) | 52 (83.9%) | 0.456 |
| ≥60 | 41 (39.8%) | 9 (21.9%) | 32 (78.1%) | |
| Tumor sizea | ||||
| <5cm | 48 (46.6%) | 7 (15.6%) | 41 (84.4%) | 0.345 |
| ≥5cm | 55 (53.4%) | 12 (21.8%) | 43 (78.2%) | |
| Stage (UICC)b | ||||
| I | 5 (4.9%) | 0 (0.0%) | 5 (100%) | 0.535 |
| II | 28 (27.2%) | 5 (17.9%) | 23 (82.1%) | |
| III | 70 (67.9%) | 14 (20%) | 56 (80%) | |
| Depth of tumor invasion | ||||
| T1 + T2 | 9 (8.7%) | 1 (11.1%) | 8 (88.9%) | 1.000 |
| T3 + T4 | 94 (91.3%) | 18 (19.1%) | 76 (80.9%) | |
| Lymph node metastasis | ||||
| Negative | 32 (31.1%) | 5 (15.6%) | 27 (84.4%) | 0.620 |
| Positive | 71 (68.9%) | 14 (19.7%) | 57 (80.3%) | |
| Histology | ||||
| WD + MD | 86 (83.5%) | 13 (15.1%) | 73 (84.9%) | 0.050 |
| PD | 17 (16.5%) | 6 (35.3%) | 11 (64.7%) | |
| Location | ||||
| Colon | 67 (65%) | 12 (17.9%) | 55 (82.1%) | 0.848 |
| Rectum | 36 (35%) | 7 (19.4%) | 29 (80.6%) | |
| Vascular invasion | ||||
| Yes | 35 (33.9%) | 5 (14.3%) | 30 (85.7%) | 0.435 |
| No | 68 (66.1%) | 14 (20.6%) | 54 (79.4%) | |
| Perineural invasion | ||||
| Yes | 12 (11.7%) | 1 (8.3%) | 11 (91.7%) | 0.458 |
| No | 91 (88.3%) | 18 (19.8%) | 73 (80.2%) |
Total RNA extraction and first-strand cDNA synthesis
Total RNA was isolated from 10 paired CRC patient's tissues with ISOGEN™ (Nippon Gene) and the QIAmp® Mini Kit (Qiagen, Inc.), and total RNA was also extracted from the fresh whole blood of 103 postoperative CRC patients using a QIAmp RNA Blood Mini Kit (Qiagen, Inc.) according to the manufacturer's instructions. Purified RNA was quantified at OD 260 nm by a ND-1000 spectrophotometer (Nanodrop Technology) and quantitated by a Bioanalyzer 2100 (Agilent Technology). First-strand cDNA was synthesized from total RNA by using an RT-PCR kit (Promega Co.). The reverse transcription was carried out in a reaction mixture consisting of a 25 μg/mL oligo (dT) 15-mer primer, 100 mmol/L PCR Nucleotide Mix, 200 μmol/L M-MLV Reverse Transcriptase, and 25 μL of Recombinant RNasin® Ribonuclease Inhibitor (Promega). The reaction mixtures with RNA were incubated at 42°C for 2 h minimum, heated to 95°C for 5 min, and then stored at −80°C until analysis.
Oligonucleotide microarray analysis
The oligonucleotide array contains 22,500 elements designed for expression profiling (Human 1A V2; Agilent Technologies), in which over 18,000 well-characterized, full-length human genes have been defined. First-strand cDNA targets for hybridization were made by reverse transcription of the mRNA isolated from 10 paired CRC tissues and normal adjacent tissues by using SuperScript II RT (Gibco-BRL) in the presence of either Cy3- or Cy5-labeled dUTP (Amersham Pharmacia Biotech). The targets were dried down to 18 μL by a SpeedVac™ concentrator (Thermo Electron Co), and then 3.6 μL of 20X SSC, 1.8 μL of 10 mg/mL poly-A, and 0.54 μL of 10% SDS were added. Then, the mixture was heated to 100°C for 2 min and proceeded to the hybridization reaction on the Human 1A Oligo Microarray V2 array slides (Agilent Technologies) in an incubator at 60°C for 17 h. After being sequentially washed with 1X SSC, 0.2X SSC, and 0.5X SSC, hybridized microarray slides were scanned and fluorescence signals were detected using an Axon GenePix Pro 3.0™ (Axon Instruments). The acceptance criterion for a gene signal was a signal-to-noise ratio of ≥2. If either the Cy3 or Cy5 signal of a specific spot passed the criterion, the flag of its ratio was counted to be “true”. The element with the “true” flag was analyzed with GeneSpring GX7 (Silicon Genetics). The differentially expressed elements were analyzed by the two-sided statistical tolerance interval (95%).
Design of oligonucleotide probes and EnCA preparation
The design and preparation of the membrane array was based on our previous study (Chen et al., 2005). We used OMP3 (Oligonucleotide Modeling Platform, DNA software) to design oligonucleotides of eight candidate genes. The oligonucleotide sequences of probe and mRNA markers are described as follows (5′–3′): AATTCCTCTCTGGAAGATCCCAGCACCGACTACTACCAAGAGCTGCAGAG for MUC1 (mucin 1, cell surface associated); CATCGGCATCTTCGGGCAGGATGAGGACGTGACGTCAAAAGCTTTCACAG for ATP2A2 (ATPase, Ca++ transporting, cardiac muscle, slow twitch 2); GGAAAGGATACGGGACAATGAGAACAGAACTTCACAAGGCCCCGTGAAGC for KCTD2 (potassium channel tetramerization domain containing 2); CACACCGGCCAACCAGCTGTTATCTGCATGGATGAGGAACGCATTTAAAG for S100B (S100 calcium-binding protein β (neural)); CTGGTACACCTCAGGATAATCCAATGTTTTAATACTTAGGCAACACTGGC for ELAVL4 (ELAV (embryonic lethal, abnormal vision, Drosophila-like 4 (Hu antigen D)); GACTGACGATAGGTTGATGCATCCACAGATTCATTTGTTCCCGATCTGGC for TM4SF3 (transmembrane 4 superfamily member 3); AGCACAGAAGCACATCACATACACCAGCAAGGTTTCCAACTACTGCACTG for OLFM4 (olfactomedin 4); and CTCATATATTCAGTATTTTACTCCCACAGCACCTCCCCCCAATTTGACCC for hTERT (human telomerase reverse transcriptase). The newly synthesized oligonucleotides were dissolved in deionized (DI) water to a concentration of 20 mM and then placed inside a BioJet Plus 3000-nL dispensing system (BioDot, Inc.), which sequentially blotted the eight candidate genes, one house-keeping gene (β-actin), and one bacterial gene from Mycobacterium tuberculosis (50 nL per spot and 1.5 mm between spots) on Nytran® SuperCharge nylon membrane (Schleicher & Schuell) in triplicate (Fig. 1). Dimethyl sulfoxide was also dispensed onto the membrane as a blank control. After being air-dried and UV crosslinked, the membranes were heated at 80°C for 2 h and stored in a desiccator at room temperature for up to 2 weeks. The labeling of the first cDNA and operating procedures for chip hybridization were the same as previously described (Chen et al., 2006; Wu et al., 2006a, 2006b); however, the probe labeling in this study used biotin in place of digoxigenin. After hybridization, the chips were washed, blocked, and then incubated for 30 min in strepavidin–horseradish peroxidase conjugate (Amersham Pharmacia Biotech) diluted in Tris-buffered saline. The blot was processed using the 3,3′-diaminobenzidine kit (Amersham Pharmacia Biotech) according to the manufacturer's instructions. The colorimetric signal was detected with an Alpha Innotech Image system (Alpha Innotech Corp.), and the intensity of each spot on the membrane was measured using AlphaEase FC software (Alpha Innotech Corp). The expression level of each gene spot measured on oligonucleotide gene chip were quantitated and then normalized with reference gene (β-actin) density. When the normalized spot density is >2, it is defined as the overexpressed gene spot. Genes with a greater than twofold change in expression across all time points were considered to be overexpressed. The criteria for determining the overexpressed spot have been also described elsewhere (Wu et al., 2006a, 2006b).
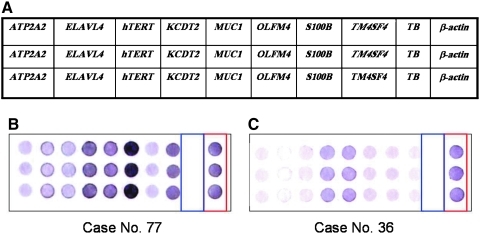
FIG. 1.
Comparison of gene expression patterns between a colorectal cancer (CRC) patient without liver metastasis (Case No. 36) and a patient with liver metastasis (Case No. 77). Oligonucleotide fragments were blotted on membrane in triplicate. A triplicate set of eight mRNA markers (i.e., ATP2A2, ELAVL4, hTERT, KCTD2, MUC1, OLFM4, S100B, and TM4SF4) for CRC was blotted on the nylon membrane. In addition, a housekeeping gene (β-actin) and a tuberculosis gene (TB) serving as positive and negative controls were also blotted on the membrane. The spots within the red rectangle of each image represent β-actin (positive control), the spots within the blue rectangle of each image represent the tuberculosis gene (negative control), and the spots within the green rectangle of each image represent the blank control. (A) Schematic representation of the membrane array with eight target genes, one housekeeping gene (β-actin), one bacterial gene (TB), and blank control. (B) Patient with liver metastasis (Case No. 77). (C) Patient without liver metastasis (Case No. 36).
Statistical analysis
All data were analyzed using the Statistical Package for the Social Sciences Version 12.0 (SPSS, Inc.). A univariate analysis of clinicopathological features and gene expression of each group (liver metastasis group versus nonliver metastasis group) were compared using the chi-square test. The correlation between gene expression and liver metastasis status was assessed using the relative risk ratio and a 95% confidence interval (CI). A _p-_value of <0.05 was considered statistically significant.
Results
Microarray analysis of colorectal tissues and identification of candidate genes by bioinformatics
We utilized 10 paired CRC tumor specimens and normal adjacent tissues for microarray analysis. All results from the experiments underwent standardized analysis and validation. Subsequently, we used GeneSpring Biological data analysis software, wherein hierarchical clustering was performed in the experimental groups to initially assess gene expression of all genes on the chip. After further analysis and validation, there were 1089 genes that displayed different expression profiles in CRC tumor specimens from the adjacent normal tissues. The 1089 genes validated in the CRC tissues were analyzed with DAVID (Database for Annotation, Visualization, and Integrated Discovery) (Dennis et al., 2003) and KEGG (Kyoto Encyclopedia of Genes and Genomes) databases (Zhong et al., 2010). Eight candidate genes of CRC including ATP2A2, ELAVL4, hTERT, KCTD2, MUC1, OLFM4, S100B, and TM4SF4 genes were upregulated in cancer tissues (gene expression ratio of cancer tissue to paired normal tissue was >2) and correlated with CRC carcinogenesis by bioinformatics analysis.
Patient characteristics
Initially, 103 preoperative CRC patients were enrolled. Of these, 58 were male (56.3%) and 45 female (43.7%). Sixty-two patients (60.2%) were aged <60 years and 41 (39.8%) were ≥60 years (range, 32–73 years; mean age, 56.43±9.35 years). The primary tumor location for 67 cases (65%) was the colon, and for 36 (35%), the primary location was the rectum. Nineteen cases (18.4%) developed liver metastasis during follow-up. In total, 5 cases (4.9%) were UICC stage I, 28 cases (27.2%) were UICC stage II, and 70 cases (67.9%) were UICC stage III. Table 1 lists the other clinicopathological characteristics of patients and tumors.
Correlation between liver metastasis and clinicopathological data
No statistical correlations existed between liver metastasis and age, sex, tumor size, depth of tumor invasion, lymph node metastasis, differentiation, tumor location, and vascular and perineural invasion (_p_-value <0.05 is significant; Table 1).
Correlation between liver metastasis and gene expression
As shown in Table 2, the correlations between gene expression and patients with or without liver metastasis were examined. Statistical analyses indicate that gene expression of S100B, TM4SF3, and OLFM4 were strongly correlated between patients with liver metastasis and nonliver metastasis (_p_=0.001, 0.011, and 0.015, respectively); however, ATP2A2, ELAVL4, hTERT, KCTD2, and MUC1 gene expressions were not correlated (all _p_>0.05). CRC patients with S100B overexpression have a risk of liver metastasis 9.217 times greater than those patients without increasing expression of S100B (_p_=0.001; 95% CI, 2.516–33.764). Overexpression of TM4SF3 indicated a 4.385 times higher risk of liver metastasis. Liver metastatic risk was 3.438 times higher with OLFM4 overexpression (all p<0.05).
Table 2.
The Relationship of Candidate Genes and Liver Metastasis in 103 Postoperative Colorectal Cancer Patients
| Overexpression | Total cases | Liver metastasis | (%) | OR | 95% CI | p_-Value_ |
|---|---|---|---|---|---|---|
| ATP2A2 | ||||||
| Positive | 16 | 5 | (31.2) | 2.370 | 0.713–7.884 | 0.151 |
| Negative | 87 | 14 | (16.1) | |||
| ELAVL4 | ||||||
| Positive | 55 | 13 | (23.6) | 2.167 | 0.752–6.239 | 0.146 |
| Negative | 48 | 6 | (12.5) | |||
| hTERT | ||||||
| Positive | 52 | 9 | (17.3) | 1.165 | 0.430–3.158 | 0.763 |
| Negative | 51 | 10 | (19.6) | |||
| KCTD2 | ||||||
| Positive | 14 | 3 | (21.4) | 1.244 | 0.311–4.978 | 0.719 |
| Negative | 89 | 16 | (17.9) | |||
| MUC1 | ||||||
| Positive | 12 | 4 | (33.3) | 2.533 | 0.676–9.501 | 0.227 |
| Negative | 91 | 15 | (16.5) | |||
| OLFM4 | ||||||
| Positive | 35 | 11 | (31.4) | 3.438 | 1.232–9.594 | 0.015a |
| Negative | 68 | 8 | (11.8) | |||
| S100B | ||||||
| Positive | 12 | 7 | (58.3) | 9.217 | 2.516–33.764 | 0.001a |
| Negative | 91 | 12 | (13.2) | |||
| TM4SF3 | ||||||
| Positive | 14 | 6 | (42.9) | 4.385 | 1.306–14.718 | 0.011a |
| Negative | 89 | 13 | (14.6) |
Correlation between gene expression and clinicopathological data
This study assessed correlations between the expression of three liver metastasis-associated genes (including S100B, TM4SF3, and OLFM4) and clinicopathological features of 103 postoperative CRC patients (Table 3). No statistically significant correlations existed between genotype distributions and sex, tumor location, depth of tumor invasion, lymph node metastasis, cancer stage, or histology (all _p_>0.05).
Table 3.
Correlation Between OLFM4, S100B, and TM4SF3 Gene Expression and Clinicopathological Properties of 103 Postoperative Colorectal Cancer Patients
| | | OLFM4 overexpressiona | S100B overexpressiona | TM4SF3 overexpressiona | | | | | | ----------------------- | ------------------------------- | ------------------------------- | -------------------------------- | ---------- | ---------- | ---------- | ---------- | | Variables | Total cases | +35 | −68 | +12 | −91 | +14 | −89 | | Gender | | | | | | | | | Male | 58 (56.3%) | 22 (37.9%) | 36 (62.1%) | 8 (13.8%) | 50 (86.2%) | 9 (15.5%) | 49 (84.5%) | | Female | 45 (43.7%) | 13 (28.9%) | 32 (71.1%) | 4 (8.9%) | 41 (91.1%) | 5 (11.1%) | 40 (88.9%) | | | _p-_Value | 0.337 | 0.544 | 0.518 | | | | | | Age (years) | | | | | | | | | <60 | 62 (60.2%) | 19 (30.6%) | 43 (69.4%) | 4 (6.5%) | 58 (93.5%) | 6 (9.7%) | 56 (90.3%) | | ≥60 | 41 (39.8%) | 16 (39.0%) | 25 (61.0%) | 8 (19.5%) | 33 (80.5%) | 8 (19.5%) | 33 (80.5%) | | | _p-_Value | 0.379 | 0.060 | 0.154 | | | | | | Tumor Size | | | | | | | | | <5cm | 48 (46.6%) | 16 (33.3%) | 32 (66.7%) | 7 (14.6%) | 41 (85.4%) | 9 (18.8%) | 39 (81.3%) | | ≥5cm | 55 (53.4%) | 19 (34.5%) | 36 (65.5%) | 5 (9.1%) | 50 (90.9%) | 5 (9.1%) | 50 (90.9%) | | | _p_-Value | 0.897 | 0.386 | 0.154 | | | | | | Stage (UICC) | | | | | | | | | I | 5 (4.9%) | 2 (40.0%) | 3 (60.0%) | 0 (0%) | 5 (100.0%) | 1 (20.0%) | 4 (80.0%) | | II | 28 (27.2%) | 10 (35.7%) | 18 (64.3%) | 4 (14.3%) | 24 (85.7%) | 5 (17.9%) | 23 (82.1%) | | III | 70 (67.9%) | 23 (32.9%) | 47 (67.1%) | 8 (11.4%) | 62 (88.6%) | 8 (11.4%) | 62 (88.6%) | | | _p_-Value | 0.924 | 0.653 | 0.642 | | | | | | Depth of tumor invasion | | | | | | | | | T1+T2 | 9 (8.7%) | 3 (33.3%) | 6 (66.7%) | 0 (0%) | 9 (100%) | 1 (11.1%) | 8 (88.9%) | | T3+T4 | 94 (91.3%) | 32 (34.0%) | 62 (66.0%) | 12 (12.8%) | 82 (87.2%) | 13 (13.8%) | 81 (86.2%) | | | _p_-Value | 1.000 | 0.594 | 1.000 | | | | | | Lymph node metastasis | | | | | | | | | Negative | 32 (31.1%) | 10 (31.2%) | 22 (68.8%) | 3 (9.4%) | 29 (90.6%) | 4 (12.5%) | 28 (87.5%) | | Positive | 71 (68.9%) | 25 (35.2%) | 46 (64.8%) | 9 (12.7%) | 62 (87.3%) | 10 (14.1%) | 61 (85.9%) | | | _p_-Value | 0.694 | 0.750 | 1.000 | | | | | | Histology | | | | | | | | | WD+MD | 86 (83.5%) | 29 (33.7%) | 57 (66.3%) | 9 (10.5%) | 77 (89.5%) | 11 (12.8%) | 75 (87.2%) | | PD | 17 (16.5%) | 6 (35.3%) | 11 (64.7%) | 3 (17.6%) | 14 (82.4%) | 3 (17.6%) | 14 (82.4%) | | | _p_-Value | 0.900 | 0.413 | 0.698 | | | | | | Location | | | | | | | | | Colon | 67 (65%) | 22 (32.8%) | 45 (67.2%) | 7 (10.4%) | 60 (89.6%) | 8 (11.9%) | 59 (88.1%) | | Rectum | 36 (35%) | 13 (36.1%) | 23 (63.9%) | 5 (13.9%) | 31 (86.1%) | 6 (16.7%) | 30 (83.3%) | | | _p_-Value | 0.738 | 0.604 | 0.505 | | | | | | Vascular invasion | | | | | | | | | Yes | 35 (33.9%) | 10 (28.6%) | 25 (71.4%) | 4 (11.4%) | 31 (88.6%) | 4 (11.4%) | 31 (88.6%) | | No | 68 (66.1%) | 25 (36.8%) | 43 (63.2%) | 8 (11.8%) | 60 (88.2%) | 10 (14.7%) | 58 (85.3%) | | | _p_-Value | 0.406 | 1.000 | 0.768 | | | | | | Perineural invasion | | | | | | | | | Yes | 12 (11.7%) | 3 (25.0%) | 9 (75.0%) | 1 (8.3%) | 11 (91.7%) | 1 (8.3%) | 11 (91.7%) | | No | 91 (88.3%) | 32 (35.2%) | 59 (64.8%) | 11 (12.1%) | 80 (87.9%) | 13 (14.3%) | 78 (85.7%) | | | _p_-Value | 0.747 | 1.000 | 1.000 | | | | |
Discussion
The recent identification of gene overexpressed in CRC, combined with advances in molecular biology techniques, permits opportunities to establish more sensitive, specific, and cost-effective ways of identifying local recurrence or metastasis. Currently, one of the most compelling possibilities is the development of a highly sensitive molecular diagnostic procedure that permits detection of tumor cells in different tissues and biological fluids, especially peripheral blood. Efforts have been focused on identifying markers that can detect disseminated cancer cells in the circulation, which will aid early diagnosis of malignant cell metastasis. However, the heterogeneity of the expression of tumor genes and the variable performance of these assays have posed major problems for the detection of CTCs.
A panel of molecular markers is necessary to increase the sensitivity of CTCs detection (Racila et al., 1998). Since 2005, our research group has developed membrane arrays to detect CTC oncogenes with a sensitivity of 83.7% and a specificity of 90.9%, and we will continue to improve the detection sensitivity and specificity of the membrane array method (Chen et al., 2005). A membrane array has been able to simultaneously detect a panel of informative molecular markers for the presence of CTCs in CRC patients undergoing curative resection, with time-saving and cost advantages (Wang et al., 2007).
In the present investigation, we found that CRC liver metastasis was significantly correlated with S100B, TM4SF3, and OLFM4 gene overexpression in CTCs of CRC patient blood specimens, which were analyzed by a constructed EnCA, including eight candidate genes. The technical difference between the EnCA system and the conventional membrane array includes replacing digoxigenin by biotin. Moreover, this EnCA assay is a high-throughput assay that saves time and is cost-effective compared with PCR and sequencing techniques.
At the preclinical and clinical levels, the presence of S100B protein expression is an independent predictive marker of poor prognosis of CRC (Hwang et al., 2011). Moreover, postoperative CRC patients who demonstrated S100B overexpression had the highest risk ratio of liver metastasis in this study. TM4SF3 has been reported to be overexpressed in esophageal carcinomas and was involved in cancer metastasis (Zhou et al., 2008). OLFM4 was correlated with CRC carcinogenesis (Koshida et al., 2007). In the present study, the S100B, TM4SM3, and OLFM4 gene overexpressions are independent CRC liver metastasis-related biomarkers.
Even S100B, TM4SF3, and OLFM4 expression levels positively correlate with liver metastasis in this study. There were few liver metastasis patients without S100B, TM4SF3, and OLFM4 genes overexpression. These findings revealed that other carcinogenesis-related genes might be responsible for those cases and further effort to explore more genes related to cancer metastasis will be needed in the future.
Conclusion
In conclusion, this study outlined the creation of the EnCA method, which was performed with simple devices and materials using a straightforward procedure. The EnCAs were designed to simultaneously detect multiple gene overexpression of CRC liver metastasis-associated genes. Identifying these risk genes can be useful in predicting the genetic risk of CRC liver metastasis. These results also highlight the need for prompt further studies on a larger number of CRC cases to validate and standardize testing procedures before their use in clinical practice.
Acknowledgments
This work was supported by grants from the National Science Council of the Republic of China (NSC 96-2320-B-037-010-MY3, NSC 99-2320-B-037-014-MY3) and by an Excellence for Cancer Research Center Grant (DOH100-TD-C-111-002) through funding by the Department of Health, Executive Yuan, Taiwan (ROC).
Disclosure Statement
No competing financial interests exist.
References
- Baker M.K. Mikhitarian K. Osta W. Callahan K. Hoda R. Brescia F. Kneuper-Hall R. Mitas M. Cole D.J. Gillanders W.E. Molecular detection of breast cancer cells in the peripheral blood of advanced-stage breast cancer patients using multimarker real-time reverse transcription-polymerase chain reaction and a novel porous barrier density gradient centrifugation technology. Clin Cancer Res. 2003;9:4865–4871. [PubMed] [Google Scholar]
- Burger J. [Biochips—tools of 21st century medicine] Versicherungsmedizin. 2006;58:9–13. [PubMed] [Google Scholar]
- Castells A. Bessa X. Daniels M. Ascaso C. Lacy A.M. Garcia-Valdecasas J.C. Gargallo L. Novell F. Astudillo E. Filella X. Pique J.M. Value of postoperative surveillance after radical surgery for colorectal cancer: results of a cohort study. Dis Colon Rectum. 1998;41:714–723. doi: 10.1007/BF02236257. discussion 723–724. [DOI] [PubMed] [Google Scholar]
- Chen C.C. Chang T.W. Chen F.M. Hou M.F. Hung S.Y. Chong I.W. Lee S.C. Zhou T.H. Lin S.R. Combination of multiple mRNA markers (PTTG1, Survivin, UbcH10 and TK1) in the diagnosis of Taiwanese patients with breast cancer by membrane array. Oncology. 2006;70:438–446. doi: 10.1159/000098557. [DOI] [PubMed] [Google Scholar]
- Chen Y.F. Wang J.Y. Wu C.H. Chen F.M. Cheng T.L. Lin S.R. Detection of circulating cancer cells with K-ras oncogene using membrane array. Cancer Lett. 2005;229:115–122. doi: 10.1016/j.canlet.2004.12.026. [DOI] [PubMed] [Google Scholar]
- Dennis G., Jr. Sherman B.T. Hosack D.A. Yang J. Gao W. Lane H.C. Lempicki R.A. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:3. [PubMed] [Google Scholar]
- Ghossein R.A. Bhattacharya S. Molecular detection and characterisation of circulating tumour cells and micrometastases in solid tumours. Eur J Cancer. 2000;36:1681–1694. doi: 10.1016/s0959-8049(00)00152-0. [DOI] [PubMed] [Google Scholar]
- Ghossein R.A. Bhattacharya S. Rosai J. Molecular detection of micrometastases and circulating tumor cells in solid tumors. Clin Cancer Res. 1999;5:1950–1960. [PubMed] [Google Scholar]
- Hoon D.S. Wang Y. Dale P.S. Conrad A.J. Schmid P. Garrison D. Kuo C. Foshag L.J. Nizze A.J. Morton D.L. Detection of occult melanoma cells in blood with a multiple-marker polymerase chain reaction assay. J Clin Oncol. 1995;13:2109–2116. doi: 10.1200/JCO.1995.13.8.2109. [DOI] [PubMed] [Google Scholar]
- Hwang C.C. Chai H.T. Chen H.W. Tsai H.L. Lu C.Y. Yu F.J. Huang M.Y. Wang J.Y. S100B protein expressions as an independent predictor of early relapse in UICC Stages II and III colon cancer patients after curative resection. Ann Surg Oncol. 2011;18:139–145. doi: 10.1245/s10434-010-1209-7. [DOI] [PubMed] [Google Scholar]
- International Union Against Cancer. TNM Classification of Malignant Tumors. 6th. Wiley-Liss; New York: 2002. [Google Scholar]
- Koshida S. Kobayashi D. Moriai R. Tsuji N. Watanabe N. Specific overexpression of OLFM4(GW112/HGC-1) mRNA in colon, breast and lung cancer tissues detected using quantitative analysis. Cancer Sci. 2007;98:315–320. doi: 10.1111/j.1349-7006.2006.00383.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel H. Le Marer N. Wharton R.Q. Khan Z.A. Araia R. Glover C. Henry M.M. Allen-Mersh T.G. Clearance of circulating tumor cells after excision of primary colorectal cancer. Ann Surg. 2002;235:226–231. doi: 10.1097/00000658-200202000-00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racila E. Euhus D. Weiss A.J. Rao C. McConnell J. Terstappen L.W. Uhr J.W. Detection and characterization of carcinoma cells in the blood. Proc Natl Acad Sci USA. 1998;95:4589–4594. doi: 10.1073/pnas.95.8.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sher Y.P. Shih J.Y. Yang P.C. Roffler S.R. Chu Y.W. Wu C.W. Yu C.L. Peck K. Prognosis of non-small cell lung cancer patients by detecting circulating cancer cells in the peripheral blood with multiple marker genes. Clin Cancer Res. 2005;11:173–179. [PubMed] [Google Scholar]
- Wang J.Y. Lin S.R. Wu D.C. Lu C.Y. Yu F.J. Hsieh J.S. Cheng T.L. Koay L.B. Uen Y.H. Multiple molecular markers as predictors of colorectal cancer in patients with normal perioperative serum carcinoembryonic antigen levels. Clin Cancer Res. 2007;13:2406–2413. doi: 10.1158/1078-0432.CCR-06-2054. [DOI] [PubMed] [Google Scholar]
- Wang J.Y. Wu C.H. Lu C.Y. Hsieh J.S. Wu D.C. Huang S.Y. Lin S.R. Molecular detection of circulating tumor cells in the peripheral blood of patients with colorectal cancer using RT-PCR: significance of the prediction of postoperative metastasis. World J Surg. 2006a;30:1007–1013. doi: 10.1007/s00268-005-0485-z. [DOI] [PubMed] [Google Scholar]
- Wang J.Y. Yeh C.S. Chen Y.F. Wu C.H. Hsieh J.S. Huang T.J. Huang S.Y. Lin S.R. Development and evaluation of a colorimetric membrane-array method for the detection of circulating tumor cells in the peripheral blood of Taiwanese patients with colorectal cancer. Int J Mol Med. 2006b;17:737–747. [PubMed] [Google Scholar]
- Weitz J. Kienle P. Lacroix J. Willeke F. Benner A. Lehnert T. Herfarth C. von Knebel Doeberitz M. Dissemination of tumor cells in patients undergoing surgery for colorectal cancer. Clin Cancer Res. 1998;4:343–348. [PubMed] [Google Scholar]
- Weitz J. Koch M. Debus J. Hohler T. Galle P.R. Buchler M.W. Colorectal cancer. Lancet. 2005;365:153–165. doi: 10.1016/S0140-6736(05)17706-X. [DOI] [PubMed] [Google Scholar]
- Wharton R.Q. Jonas S.K. Glover C. Khan Z.A. Klokouzas A. Quinn H. Henry M. Allen-Mersh T.G. Increased detection of circulating tumor cells in the blood of colorectal carcinoma patients using two reverse transcription-PCR assays and multiple blood samples. Clin Cancer Res. 1999;5:4158–4163. [PubMed] [Google Scholar]
- Wu C.H. Lin S.R. Hsieh J.S. Chen F.M. Lu C.Y. Yu F.J. Cheng T.L. Huang T.J. Huang S.Y. Wang J.Y. Molecular detection of disseminated tumor cells in the peripheral blood of patients with gastric cancer: evaluation of their prognostic significance. Dis Markers. 2006a;22:103–109. doi: 10.1155/2006/281315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.H. Lin S.R. Yu F.J. Wu D.C. Pan Y.S. Hsieh J.S. Huang S.Y. Wang J.Y. Development of a high-throughput membrane-array method for molecular diagnosis of circulating tumor cells in patients with gastric cancers. Int J Cancer. 2006b;119:373–379. doi: 10.1002/ijc.21856. [DOI] [PubMed] [Google Scholar]
- Yamaguchi K. Takagi Y. Aoki S. Futamura M. Saji S. Significant detection of circulating cancer cells in the blood by reverse transcriptase-polymerase chain reaction during colorectal cancer resection. Ann Surg. 2000;232:58–65. doi: 10.1097/00000658-200007000-00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh C.S. Wang J.Y. Wu C.H. Chong I.W. Chung F.Y. Wang Y.H. Yu Y.P. Lin S.R. Molecular detection of circulating cancer cells in the peripheral blood of patients with colorectal cancer by using membrane array with a multiple mRNA marker panel. Int J Oncol. 2006;28:411–420. [PubMed] [Google Scholar]
- Zhong H. Yang X. Kaplan L.M. Molony C. Schadt E.E. Integrating pathway analysis and genetics of gene expression for genome-wide association studies. Am J Hum Genet. 2010;86:581–591. doi: 10.1016/j.ajhg.2010.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z. Ran Y.L. Hu H. Pan J. Li Z.F. Chen L.Z. Sun L.C. Peng L. Zhao X.L. Yu L. Sun L.X. Yang Z.H. TM4SF3 promotes esophageal carcinoma metastasis via upregulating ADAM12m expression. Clin Exp Metastasis. 2008;25:537–548. doi: 10.1007/s10585-008-9168-0. [DOI] [PubMed] [Google Scholar]