Factors Influencing the Microbial Composition of Metalworking Fluids and Potential Implications for Machine Operator's Lung (original) (raw)
Abstract
Hypersensitivity pneumonitis, also known as “machine operator's lung” (MOL), has been related to microorganisms growing in metalworking fluids (MWFs), especially Mycobacterium immunogenum. We aimed to (i) describe the microbiological contamination of MWFs and (ii) look for chemical, physical, and environmental parameters associated with variations in microbiological profiles. We microbiologically analyzed 180 MWF samples from nonautomotive plants (e.g., screw-machining or metal-cutting plants) in the Franche-Comté region in eastern France and 165 samples from three French automotive plants in which cases of MOL had been proven. Our results revealed two types of microbial biomes: the first was from the nonautomotive industry, showed predominantly Gram-negative rods (GNR), and was associated with a low risk of MOL, and the second came from the automotive industry that was affected by cases of MOL and showed predominantly Gram-positive rods (GPR). Traces of M. immunogenum were sporadically detected in the first type, while it was highly prevalent in the automotive sector, with up to 38% of samples testing positive. The use of chromium, nickel, or iron was associated with growth of Gram-negative rods; conversely, growth of Gram-positive rods was associated with the absence of these metals. Synthetic MWFs were more frequently sterile than emulsions. Vegetable oil-based emulsions were associated with GNR, while mineral ones were associated with GPR. Our results suggest that metal types and the nature of MWF play a part in MWF contamination, and this work shall be followed by further in vitro simulation experiments on the kinetics of microbial populations, focusing on the phenomena of inhibition and synergy.
INTRODUCTION
The term metalworking fluids (MWFs) refers to two types of products: straight oils and aqueous fluids that may be oil-in-water emulsions or water-based synthetic fluids. They are widely used in machining or grinding operations to cool and lubricate metalwork pieces and tools. Continuous recycling of these MWFs, intended to cut costs, leads to progressive contamination by metal particles and salts and potential development of microbial biomes. To prevent or to stop microbial proliferation, users may add chemical biocides to the MWFs.
Occupational exposure to MWFs through inhalation of aerosols (12) has been associated with cutaneous as well as respiratory diseases (1), i.e., asthma and hypersensitivity pneumonitis (HP) known as “machine operator's lung” (MOL) (33). Although asthma and dermatitis seem to be caused by the chemical toxicity of MWFs due to additional components (e.g., biocides, defoamers, corrosion inhibitors, or dyes) (22, 33, 38), MOL has been related to microorganisms that frequently contaminate MWFs, especially Mycobacterium immunogenum, a recently described, fast-growing mycobacterium included in the Mycobacterium chelonae-Mycobacterium abscessus complex (18, 24, 37, 48, 52, 54). Arguments in favor of this etiology are the presence of specific precipitins in MWF-related HP cases (5, 14, 32) and/or the presence of M. immunogenum in their environment (4, 18, 42, 52). The ability to induce HP in mice exposed intranasally to M. immunogenum (16, 40) is evidence of the role of this microorganism.
Since the first reports (5, 27), most described cases of MWF-related HP have been in the U.S. automotive industry (4, 18, 20, 24, 28, 32, 37, 42, 56). The first European case clusters were recently reported in automotive plants in Great Britain (8) and in France (41).
Following an investigation of the first French outbreak (41), we analyzed the microbial biomes of MWFs in two other automotive plants in which cases of HP were suspected, and we simultaneously implemented a regional survey (STEFI study on occupational health and MWF exposure) to obtain clinical and environmental data on the metalworking industry of the Franche-Comté region in eastern France.
The aims of our present study were (i) to describe the microbiological contamination in French MWFs, focusing in particular on M. immunogenum detection, and (ii) to look for chemical, physical, and environmental parameters associated with variations in microbiological patterns.
MATERIALS AND METHODS
Plants of interest. (i) STEFI survey.
Factories using reused MWFs in the Franche-Comté region were identified with the help of occupational physicians and regional employers' organizations. Nineteen plants were investigated. The fields of activity were screw-machining (n = 2 plants), cutting (n = 4), tool manufacturing (n = 1), mold manufacturing (n = 2), manufacturing of inexpensive jewelry (n = 1), ceramic product manufacturing (n = 1), electronic component manufacturing (n = 1), engine and turbine manufacturing (n = 2), gear and transmission manufacturing (n = 1), light metal foundry (n = 1), and industrial mechanics (n = 2).
(ii) Automotive plants.
Three plants (AP1, AP2, and AP3, located in northern France) were investigated because of recent proven cases of HP among their employees (13 cases, 1 case, and 1 case, respectively).
Sample collection and preparation.
Only reused MWF samples (i.e., samples that were filtered or decanted after use and then stored in sumps for future reuse) were included and analyzed in the study: (i) STEFI survey, n = 180 (122 aqueous fluids and 58 straight oils); (ii) AP1, n = 83 (aqueous fluids); (iii) AP2, n = 44 (aqueous fluids); and iv) AP3, n = 38 (aqueous fluids).
Culture and isolation conditions.
Microbiological analysis of samples from AP1 and AP2 was performed by plating 100 μl of homogenized sample onto 3 media: Mueller-Hinton agar (MHA) (Mast Group Ltd., Bootle, Merseyside, England), 3% (wt/vol) malt agar (MA) (Oxoid Ltd., Basingstoke, Hampshire, England) supplemented with 50 mg liter−1 chloramphenicol (Sigma-Aldrich Chimie, Lyon, France), and Difco actinomycete isolation agar (ACT) (BD Difco, Le Pont-De-Claix, France) supplemented with 30 μg ml−1 amphotericin B (Fungizone; Bristol-Myers, Rueil-Malmaison, France), 32 μg ml−1 fosfomycin (Fosfocina; Laboratorios ERN, S.A, Barcelona, Spain), and 32 μg ml−1 aztreonam (Azactam; Sanofi-Aventis, Paris, France). To enhance analysis performance, samples from STEFI plants and AP3 were subjected to the above-cited protocol that was improved by using Middlebrook 7H10 agar supplemented with Middlebrook oleic acid-albumin-dextrose-catalase (OADC) enrichment (BD Difco) (MBA). MHA and ACT plates were incubated for 14 days at 30°C, MBA plates for 21 days at 30°C, and MA plates for 14 days at 20°C.
Microbial identification and quantification by culture.
Plates were observed after 48 h and 5 days. When intense growth made it impossible to isolate colonies, samples were either serially diluted (the dilution factor usually was 10−3 but was 10−7 for the highest bacterial loads) or decontaminated with sodium hydroxide, malachite green, and cycloheximide, according to the method described by Wolinsky and Rynearson (55). A pilot study showed that the concentration of cultivable M. immunogenum was decreased by 2 log10 units after decontamination (data not shown). Bacteria and filamentous fungi were identified to the genus or species level using culture methods followed by visual examination, microscopy, and determination of biochemical characteristics according to reference documentation (9, 21). When identification was not possible using the usual microbiological techniques, molecular biology was used. For molecular identification, genomic DNA was extracted by boiling the bacterial or fungal colony with Chelex resin, as described previously (30). Gram-positive rods (GPR) with branching filaments and Gram-negative rods (GNR) or coccobacilli were identified by performing partial 16S rRNA gene amplification, using previously described primers fDl and rD1 for amplification (51) and p782r for sequencing (7). Rods that were morphologically consistent with the genus Mycobacterium, i.e., thin, Gram-positive or Gram-variable rods, were identified by partial hsp65 gene amplification and sequenced using the previously described primers Tb11 and Tb12 (39). Filamentous fungi were identified by partial 16S-23S internal transcribed spacer (ITS) amplification and sequencing, using primers ITS1 and ITS4 (53). The amplification reactions were processed as described previously for the hsp65 gene (39) and other targets (7). Amplicons were purified with the High Pure PCR product purification kit (Roche Diagnostics, Mannheim, Germany), and then sequenced using the BigDye Terminator v3 cycle sequencing kit and a 3130 genetic analyzer (Applied Biosystems, Courtaboeuf, France). Sequences were checked with the BioEdit software (BioEdit sequence alignment editor, version 7.0.5.3; http://www.mbio.ncsu.edu) (19) and then compared to those available in online databases using BLAST (Basic Local Alignment Search Tool; http://blast.ncbi.nlm.nih.gov) for all targets and BIBI (Bio Informatic Bacteria Identification; http://umr5558-sud-str1.univ-lyon1.fr/lebibi.lebibi.cgi) for the 16S rRNA gene. An identity of ≥99% to referenced strains belonging to a single microbial species allowed us to identify the isolate as belonging to this species. A lower percent identity or identity values of ≥99% to more than a single species led us to identify the isolate to the genus or the species complex, respectively (3, 23).
Detection and quantification of M. immunogenum by qPCR.
For all the STEFI samples and for the AP3 samples, M. immunogenum DNA concentrations were determined by quantitative real-time PCR (qPCR). DNA extraction of each 1.5-ml aliquot of MFW was performed according to a method adapted from those of Pickup et al. (29) and Veillette et al. (46). Aliquots were centrifuged (16,000 × g, 20 min), and pellets were resuspended in 500 μl phosphate-buffered saline (PBS) and then centrifuged and resuspended in 180 μl PBS. Twenty microliters of 20 mg ml−1 lysozyme (Roche Diagnostics) was added, and the mix was incubated with continuous shaking for 1 h. DNA was then extracted using the High Pure PCR template preparation kit (Roche Diagnostics) according to the manufacturer's protocol and an additional mechanical cell disruption (following lysis with 40 μl of 44 μg ml−1 proteinase K) in 2-ml tubes prefilled with 1.4-mm ceramic beads (MagNALyser Green beads; Roche Diagnostics) processed in a MagNALyser (Roche Diagnostics) at a setting of 6,000 rpm for 30 s (3 times, alternately with refrigeration steps). Each extraction series included one negative control (PBS). Extracts were stored at −20°C. The qPCR was performed on an AB 7500 Fast real-time PCR system (Applied Biosystems), using the procedure described by Rhodes et al. (31), with a few changes: the reaction mix consisted of 5 μl of extract or PBS, 10 μl of 2× Gene Expression master mix (Applied Biosystems), 0.3 μmol liter−1 primers and 0.25 μmol liter−1 6-carboxyfluorescein (FAM)-labeled probe in a total volume of 20 μl. Quantitative results were expressed by the determining quantification cycle (Cq), which marked the cycle at which fluorescence of the sample became significantly higher than the baseline signal. Two independent DNA extractions were performed for each MWF sample. Two Cq values, one for each replicate, were obtained for each sample tested. A sample was positive when both extracts showed a Cq value of <39 cycles; if only one of the two extracts had a Cq value of <39 cycles, it was “equivocal,” i.e., weakly positive. Equivocal results were interpreted as very weak concentrations or traces of DNA, with nonreproducible qPCR measurements as a consequence, according to the Poisson law. Two negative controls, one extraction control and one amplification control, were added to each run. The M. immunogenum DNA concentration in positive samples was calculated by comparison to a standard curve. Standards were serially diluted from a DNA extract of a suspension of pure M. immunogenum culture (strain DSM 45496). Cell equivalents (CE) per milliliter of MWF sample were estimated from the DNA concentration in the initial extract (assessed with a NanoDrop ND-1000 spectrophotometer; Thermo Fisher Scientific Inc., Wilmington, DE) and the approximate weight of one genome of M. immunogenum, as described by Rhodes et al. (31). Each standard was assessed in duplicate in each run. Standard curves showed a highly significant correlation coefficient (_r_2 = 0.994) and an efficiency of 85.2%.
Environmental data collection.
The following data were collected from employees or safety data sheets: MWF provider and reference, metals worked (ferrous metals, copper, aluminum, nickel, chrome, and silicon), MWF sump volume (≤500 liters, 501 to 5,000 liters, or ≥5,000 liters), type of MWF (aqueous fluid or straight oil), nature of aqueous fluid (emulsion, semisynthetic, or synthetic), and nature of the oil base (mineral or vegetable).
Statistical treatment.
Statistical associations between predicted and explanatory variables obtained from the STEFI plants and AP3 were investigated. Predicted variables were as follows: presence or absence of Gram-positive rods (GPR); presence or absence of Gram-negative rods (GNR); presence or absence of microorganisms detected by culture; and positive qPCR, positive or equivocal qPCR, equivocal or negative qPCR, or negative qPCR. Explanatory variables were environmental data, as listed above, and the following microbiological parameters: presence or absence of GPR, presence or absence of GNR, presence or absence of filamentous fungi, total concentration of microorganisms, and number of isolates identified from the sample.
Qualitative variables were processed using the chi-square test, the exact Pearson chi-square test, or Fisher's exact test. The explanatory variable “nature of aqueous fluid” was subjected to logistic regression for each predicted variable, using the category “synthetic” as a reference. Quantitative variables were compared using Student's t test. A P value of less than 0.05 was considered significant. All statistical calculations were performed using SAS software.
RESULTS
Culture results. (i) STEFI.
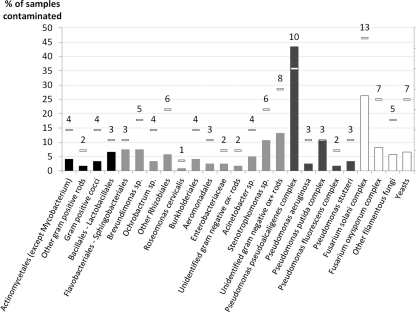
A total of 303 microbial strains were isolated, including 232 bacteria and 71 fungi. The prevalences of different microorganisms in MWF samples are provided in detail in Fig. 1. Microbial contamination was almost limited to aqueous fluids, since 84.5% of straight oils remained sterile versus 18% of aqueous fluids and only 14 strains were isolated from straight oils (average, 0.2 strain per sample) versus 289 from aqueous fluids. Microorganism concentrations ranged from 10 to 2.5 × 104 CFU ml−1 (median, 120 CFU ml−1) within straight oils, whereas they ranged from 10 to 1.6 × 108 CFU ml−1 (median, 7.0 × 105 CFU ml−1) within aqueous fluids. Bacteria were isolated from 77% of aqueous samples. GNR were isolated from 72% of aqueous samples and accounted for 71% of the 289 strains. No Mycobacterium strain was isolated (Fig. 2). The main bacterial microorganisms, isolated from 43% of aqueous samples and 10 out of 19 plants, were members of the Pseudomonas pseudoalcaligenes complex (Fig. 1). Members of the Pseudomonas pseudoalcaligenes complex were the microorganisms with the highest cumulative concentrations within the plant in 9 out of 19 plants and the most frequently isolated microorganisms (i.e., isolated from the highest number of samples within each plant) in 7 plants, regardless of the levels of contamination or microbial diversity (Table 1). Fungi were isolated from 38% of aqueous samples. The main fungal microorganisms, isolated from 26% of aqueous samples, were members of the Fusarium solani complex (Fig. 1), reflecting the predominance of the genus Fusarium (71% of fungal strains).
Fig 1.
Prevalences of microorganisms among samples from the STEFI study. Horizontal bars represent the number of plants contaminated (out of 19). Black bars, Gram-positive bacteria; gray bars, Gram-negative bacteria (dark gray, Pseudomonas spp.); white bars, fungi. To facilitate graphical representation, genera or species were grouped as follows: Acinetobacter johnsonii and Acinetobacter spp. were grouped as Acinetobacter spp.; Cellulosimicrobium spp., Cellulomonas spp., Dietzia spp., Brevibacterium spp., Gordonia spp., and Leucobacter spp. were grouped as Actinomycetales (except Mycobacterium spp.); Aeromonas spp. and Shewanella putrefaciens were grouped as Aeromonadales; Bacillus simplex, Bacillus spp., Paenibacillus glucanolyticus, and Aerococcus spp. were grouped as Bacillales and Lactobacillales; Brevundimonas diminuta and Brevundimonas spp. were grouped as Brevundimonas spp.; Alcaligenes faecalis, Achromobacter xylosidans, Comamonas aquatica, and Delftia spp. were grouped as Burkholderiales; Empedobacter brevis, Wautersiella falsenii, Sphingobacterium mizutaii, Sphingobacterium spp., and Chitinophaga spp. were grouped as Flavobacteriales and Sphingobacteriales; Pseudochrobactrum spp., Agrobacterium tumefaciens, Methylobacterium spp., and Azorhizobium spp. were grouped as other Rhizobiales; Stenotrophomonas maltophilia and Stenotrophomonas spp. were grouped as Stenotrophomonas spp.; Pseudomonas fluorescens, P. azotoformans, and P. synxantha were grouped as Pseudomonas fluorescens complex; P. pseudoalcaligenes, P. mendocina, P. oleovorans, and P. alcaliphila were grouped as Pseudomonas pseudoalcaligenes complex; P. plecoglossicida, P. monteilii, and P. putida were grouped as Pseudomonas putida complex; Absidia spp., Acremonium spp., Aspergillus fumigatus, Paecilomyces lilacinus, Lecytophora hoffmannii, and Cladosporium spp. were grouped as other filamentous fungi; and yeasts were composed of Candida parapsilosis, Candida lipolytica, Candida guilliermondii, Candida spp., and Cryptococcus laurentii.
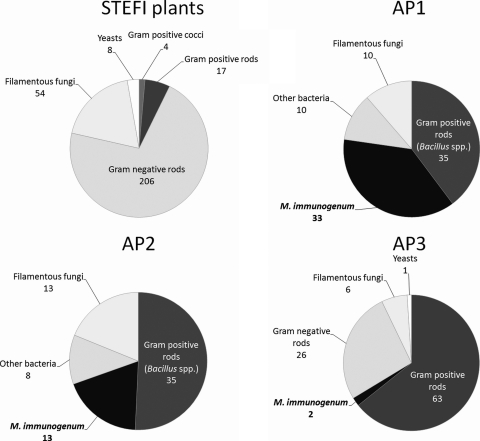
Fig 2.
Distribution of strains isolated from the 19 STEFI plants and the three automotive plants. STEFI plants, natural state with no case of HP; AP1 and AP2, natural state with cases of HP; AP3, state after use of biocides due to cases of HP.
Table 1.
Major microorganisms in each STEFI planta
| Plant | No. of samples (% of nonsterile samples) | Avg concn of microorganisms (log10 CFU ml−1) | Microorganism found at the highest concn | Diversity (avg no. of isolates per sample) | Most frequently found microorganism |
|---|---|---|---|---|---|
| M | 5 (60) | 7.5 | Pseudomonas fluorescens (complex) | 2.40 | Fusarium solani (complex)/F. oxysporum (complex) |
| U | 3 (67) | 7.3 | P. putida (complex) | 1.67 | P. putida (complex) |
| L | 2 (100) | 7.0 | Acinetobacter sp. | 3.50 | All microorganisms were isolated once |
| N | 6 (83) | 6.7 | P. pseudoalcaligenes (complex) | 3.83 | P. pseudoalcaligenes (complex) |
| G | 9 (100) | 6.6 | P. pseudoalcaligenes (complex) | 1.22 | P. pseudoalcaligenes (complex) |
| E | 41 (90) | 6.4 | P. pseudoalcaligenes (complex) | 1.88 | P. pseudoalcaligenes (complex) |
| I | 4 (200) | 6.3 | P. pseudoalcaligenes (complex) | 3.63 | Fusarium solani (complex) |
| S | 3 (100) | 6.3 | Stenotrophomonas maltophilia | 3 | S. maltophilia |
| Q | 4 (100) | 6.2 | S. maltophilia | 2.25 | Rhizopus sp. |
| D | 2 (100) | 6.0 | P. pseudoalcaligenes (complex) | 5.00 | P. pseudoalcaligenes (complex)/F. solani (complex) |
| O | 2 (100) | 6.0 | Acinetobacter sp. | 4.00 | F. solani (complex) |
| C | 5 (100) | 5.8 | P. pseudoalcaligenes (complex) | 2.60 | P. pseudoalcaligenes (complex) |
| H | 11 (45) | 5.5 | P. pseudoalcaligenes (complex) | 1.09 | P. pseudoalcaligenes (complex)/S. maltophilia |
| T | 2 (100) | 5.2 | Brevundimonas diminuta | 3 | Brevundimonas diminuta /F. solani (complex) |
| A | 13 (77) | 4.9 | P. pseudoalcaligenes (complex) | 1.20 | P. pseudoalcaligenes (complex) |
| B | 4 (100) | 4.4 | P. pseudoalcaligenes (complex) | 2.50 | Fusarium solani (complex) |
| F | 3 (0) | 0 | |||
| K | 2 (0) | 0 |
(ii) AP1.
Eighty-eight strains were isolated (average, 1.2 per sample), including 78 bacterial strains and 10 fungal strains. Bacteria were isolated from 40% of samples. Thirty-three M. immunogenum strains were isolated from 33 samples. The main bacterial microorganisms, isolated from 42% of aqueous samples, were Bacillus spp., which accounted for 40% of the 88 strains (Fig. 2). Fungi were isolated from 11% of samples. Gram-negative bacteria (excluding Pseudomonas spp.) were present in fewer than 12% of samples.
(iii) AP2.
Sixty-nine strains were isolated, including 56 bacterial strains and 13 fungal strains. Bacteria were isolated from 98% of samples. M. immunogenum was isolated from 13 MWFs (30%). The main bacterial microorganisms, isolated from 80% of aqueous samples, consisted of Bacillus spp., which accounted for 51% of the 69 strains (Fig. 2). Fungi were isolated from 27% of samples. Gram-negative bacteria were present in fewer than 7% of samples.
(iv) AP3.
Of the 38 samples, 4 (10.5%) remained sterile. A total of 98 strains were isolated, including 91 bacterial strains and 7 fungal strains (Fig. 2). Microorganism concentrations ranged from 10 to 5.6 105 CFU ml−1 (median, 200 CFU ml−1). Bacteria were isolated from 82% of samples. GPR were the main bacterial microorganisms. They were isolated from 71% of aqueous samples and accounted for 68% of isolates. M. immunogenum was isolated from 2 samples. GNR were isolated from 47% of samples. Members of the P. pseudoalcaligenes complex were isolated from 42% of samples, accounting for 65% of isolates, and were the microorganisms with the highest cumulated concentrations (9.5 × 105 CFU ml−1 for GNR versus 2.0 × 105 CFU ml−1 for GPR). Few fungi (7 isolates from 7 samples) were found, and these included various genera.
qPCR results.
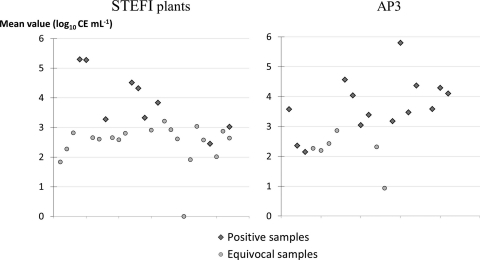
The qPCR results are shown in Fig. 3. When measured concentrations were above 103 CE ml−1, both duplicates were detected with a Cq of <39 cycles (“positive” samples). “Equivocal” PCR results (where only one of the duplicates was positive) were observed for lower concentrations.
Fig 3.
Quantitative real-time PCR analysis of the 28 positive or equivocal samples from STEFI plants and the 21 positive or equivocal samples from AP3. CE ml−1, cell equivalents per milliliter (1 CE is the amount of DNA contained in one bacterium).
(i) STEFI.
Of the 58 straight oils, no sample was positive (there were 55 negative samples and 3 equivocal samples [data not shown]), whereas of the 122 aqueous fluids, 9 samples (7%), originating from 3 plants, were positive and 18 (15%) were equivocal.
(ii) AP3.
Of the 38 aqueous MWFs, 13 aqueous samples (34%) were positive and 8 (21%) were equivocal.
Correlation between chemical or physical characteristics of MWFs and microbial biome.
Significantly different distributions of samples are shown in Table 2. The presence of GPR or GNR or the absence of microorganisms detected by culture correlated with the nature of the aqueous fluid, the nature of the oily component of emulsions, some types of metals, and the presence of filamentous fungi. In addition, logistic regression showed that (i) the probability of GNR contamination was higher with emulsions than with synthetic fluids (odds ratio = 3.3; 95% confidence interval, 1.2 to 8.9) and (ii) the probability of an absence of microorganisms was higher with synthetic fluids than with emulsions (odds ratio = 4.2; 95% confidence interval, 1.3 to 13.0). Other predicted variables gave no statistical associations with any of the explanatory variables.
Table 2.
Correlation between chemical or physical characteristics of MWFs and microbial biome
| Explanatory variable | Predicted variables | ||||
|---|---|---|---|---|---|
| Samples with GNR | Samples with GPR | Sterile samples | |||
| %a | P value | % | P value | % | P value |
| Nature of aqueous fluid | |||||
| Emulsion | 71 | =0.04 | 11 | =0.03 | |
| Semisynthetic fluid | 59 | 22 | |||
| Synthetic fluid | 43 | 33 | |||
| Nature of oil base (emulsions) | |||||
| Mineral origin | 61.5 | =0.03 | 36.5 | =0.05 | |
| Vegetable origin | 88.5 | 15.4 | |||
| Metals worked | |||||
| Chrome | 87 | ≤0.001 | 8.7 | ≤0.001 | |
| No chrome | 56 | 34.5 | |||
| Nickel | 88 | ≤0.001 | 9.3 | =0.002 | |
| No nickel | 57 | 33.6 | |||
| Ferrous metal | 71 | =0.01 | |||
| No ferrous metal | 49 | ||||
| Filamentous fungi | |||||
| Presence | 84 | =0.01 | |||
| Absence | 57 |
DISCUSSION
The main microbiological conclusions of this work are the major prevalence of GNR and the near absence of M. immunogenum in nonautomotive plant (STEFI) samples, in contrast with results from automotive plants, where M. immunogenum was found in about one-third of samples and was associated mainly with other GPR.
Culture methods seemed to be essential to analyze fluid microorganisms because they allowed us to appreciate the diversity of this microbial biome, using nonselective media (MHA and MA) as well as media targeting fast-growing mycobacteria and other bacteria responsible for other HPs (mesophilic actinomycetes involved in farmer's lung disease). However, culture methods can lead to a gross underestimation of the size and diversity of microorganism populations in MWFs (26, 45), because they are not suitable for detection of dead, noncultivable, or slow-growing microorganisms, which are inhibited by fast-growing ones. qPCR allowed rapid, specific, and sensitive quantification of M. immunogenum. Sensitivity was evaluated using a water-based suspension of pure mycobacteria; our culture method was able to detect 6% of M. immunogenum quantified by qPCR at best (data not shown).
Analysis of STEFI MWFs first showed very little microbial contamination in straight oils, which is clearly due to the lack of water in their composition. This finding, which is consistent with the absence of described HP cases related to straight oils, calls us to focus on aqueous fluids only and is useful for identifying plants with a low hazard of MOL, i.e., plants using only straight MWFs.
In STEFI plants, no sample was found to be positive for M. immunogenum using culture methods, and few samples (7% of aqueous fluids) were found to be positive with M. immunogenum qPCR, whereas high prevalences of M. immunogenum were found in automotive plants by culture methods (AP1 and AP2) or qPCR (AP3). The low proportion of positive samples determined by using culture methods in AP3 is probably related to the intensive use of biocides before MWF sampling.
In STEFI plants, Gram-negative rods were the main bacterial type and consisted mostly of the genus Pseudomonas, especially _Pseudomonas pseudoalcaligenes_-affiliated bacteria. This qualitative finding is consistent with several previous published MWF analyses, in which Pseudomonas species (including P. pseudoalcaligenes) were found, mainly where MOL had not occurred (10, 15, 24, 26, 37). These bacteria seem to be remarkably well adapted to aqueous MWFs, which can be explained by (i) the ubiquity of Pseudomonas spp. and their ability to grow in a large variety of media, especially water, (ii) a tolerance to the generally elevated pH of MWFs (34), and (iii) their ability to degrade hydrocarbons. Indeed, although a wide range of microorganisms have been found to utilize hydrocarbons as a sole source of carbon, most of the strains isolated in such media were Pseudomonas (43). Furthermore, Pseudomonas oleovorans has been shown to be able to grow even on toxic linear _n_-alkanes (25, 43). In contrast, few studies have reported microbial biomes where the main microorganisms were non-Pseudomonas bacteria, notably Ochrobactrum spp. (15), Klebsiella pneumoniae (6), and Pantoea agglomerans and Citrobacter freundii (44). The rather low biodiversity (2.6 isolates per sample) is consistent with previous data revealing highly conserved microbial communities in MWFs from different machines and applications (44). Quantitatively, the observed concentration range is similar to published values (10, 15), despite one study finding higher maximum concentrations (15).
Besides these bacteria, we found that fungi, especially Fusarium spp., were common in MWFs from the STEFI study, which is consistent with previous MWF studies (15, 24). These fungi were identified as constituents of biofilms in MWF sumps (13).
Cultures from automotive plant (AP) samples provided a description of a completely different microbial biome. The majority of the isolates were GPR, and the prevalence of samples containing GPR was higher than that of samples containing GNR. The few published descriptions of microbial biomes similar to those of our APs came from plants with cases of HP (24). Notably, a 6-month follow-up also showed the persistence of a Gram-positive microbial biome (rods, cocci, and mycobacteria, including M. immunogenum) in an MWF system associated with HP cases, in spite of dumping, cleaning, and recharging (47).
Our results indicate a low level of exposure to M. immunogenum in nonautomotive plants in the Franche-Comté region, which can be considered at low risk of MOL. This conclusion is strengthened by the clinical part of the STEFI study: evaluation of the respiratory function of employees working in the investigated plants showed no established or suspected HP diagnosis among 379 subjects exposed to MWFs (I. Thaon, unpublished data). In contrast, the risk of HP was clinically proven for the APs and was supported by a significant presence of M. immunogenum. As STEFI samples contained mainly GNR (frequently the Pseudomonas genus and the P. pseudoalcaligenes species), with no GPR and few mycobacteria, these MWFs seem to fit with the concept of an indigenous pattern of MWF microbial biome, which was previously proposed by several authors (6, 10, 24). The corollary of this concept is the existence of an alternative microbial biome that may be responsible for MOL. This alternative microbial biome was described as the association of GPR, mycobacteria, and fungi in a study of a hundred cases of MOL (24). The microbial biomes from our three APs (with a predominance of GPR and with cultivable M. immunogenum or DNA frequently detected) clearly resemble this description (24), although our results lead us to include filamentous fungi, especially the Fusarium species, in the indigenous microbial biome. However, this could also be explained by fungal colonization of surfaces above sumps and pipes, suggesting an external source rather than an active contamination of the MWFs.
The contrast between the results from the STEFI study and from the APs could be due to methodological biases and should therefore be carefully interpreted. AP3 sampling took place after intensive use of biocides following the discovery of a single HP case; this could explain the lower numbers of genera and species observed and the lower microorganism concentrations compared with STEFI results. However, the use of biocides is not necessarily a confounding factor, since it is assumed to be one of the factors leading to alteration of the microbial biome through the selection of resistant bacteria. Indeed, adding biocide can result in an increase in bacterial load, with maintenance of pathogenic, highly resistant bacteria (Enterobacteriaceae, Moraxella spp., and P. aeruginosa) (10), while mycobacteria, including M. immunogenum, were shown to exhibit reduced susceptibility to some biocides (35, 49), possibly due to biofilms (11).
Interestingly, there seems to be no described case of MOL outside the automotive industry. Therefore, this industrial field might be considered the ecological niche of M. immunogenum. Alternatively, the total microbial load could be a key point, as the reduction in competition due to the use of biocides in such industries would favor the proliferation of M. immunogenum (11, 24, 36).
We demonstrated associations between GNR detected by culture and the use of chromium, nickel, or iron, while silicon was associated with the absence of GNR. In contrast, growth of GPR was associated with the absence of chromium, nickel, and iron. Engineered materials may play a part, since it has been shown that some metals, such as zinc or copper, can modulate activity of bacteria and viruses (2) and of fungi (2, 50). More generally, copper tends to be considered an alternative antimicrobial surface (17). The category of aqueous MWFs seems to play a role, as well as the nature of the oily component of MWF. Synthetic MWFs are indeed more frequently sterile, while emulsions are frequently contaminated by GNR. In these emulsions, vegetable oil-based MWFs were more frequently contaminated with GNR, while the mineral ones were associated with the presence of GPR.
In this work, we highlight the existence of two types of MWF microbial biomes: the first type, which was associated with a lack of MOL cases, was found in the nonautomotive industry in Franche-Comté; the second one was observed in the automotive industry, where cases of MOL had occurred. M. immunogenum, the likely causative agent of MOL, was sporadically found only as DNA traces in STEFI samples, while it was highly prevalent in the automotive sector. The relationships identified between constituents of microbial biomes and some characteristics of the industrial environment lead us to propose several working hypotheses: (i) the presence of a biome based on GNR and filamentous fungi in high concentrations decreases the growth of M. immunogenum; (ii) synthetic MWFs inhibit microbial growth of GNR compared to emulsions; (iii) chromium, nickel, and iron promote the growth of GNR; and (iv) mineral oil promotes the growth of GPR, while vegetable oil promotes that of GNR. This is a rationale for further in vitro simulation experiments concerned with the kinetics of microbial populations, focusing on inhibition or synergic phenomena. These experiments could involve controlled variations of microbial concentrations, metal types, and MWF characteristics.
ACKNOWLEDGMENTS
This work was financially supported by a research grant from the University Hospital of Besançon (API-CHU 2008), the national health insurance fund for employees (Caisse Nationale d'Assurance Maladie des Travailleurs Salariés), and the University of Franche-Comté.
We thank Charlène Gaudin, Isabelle Vieille, Florence Skana, and Karine Humbert for their excellent technical assistance and Fabrice Poncet at the Plateforme de Séquençage, IFR 133, University of Franche-Comté, Besançon, France, for his help with sequencing. We thank Frances Sheppard (Clinical Investigation Center [CIC Inserm], University Hospital Besancon, France) for her editorial assistance.
Footnotes
Published ahead of print 4 November 2011
REFERENCES
- 1.Awosika-Olumo AI, Trangle KL, Fallon LF. 2003. Microorganism-induced skin disease in workers exposed to metalworking fluids. Occup. Med. (Lond.) 53:35–40 [DOI] [PubMed] [Google Scholar]
- 2.Babich H, Stotzky G. 1978. Toxicity of zinc to fungi, bacteria, and coliphages: influence of chloride ions. Appl. Environ. Microbiol. 36:906–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balajee SA, et al. 2009. Sequence-based identification of Aspergillus, Fusarium, and Mucorales species in the clinical mycology laboratory: where are we and where should we go from here? J. Clin. Microbiol. 47:877–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beckett W, Kallay M, Sood A, Zuo Z, Milton D. 2005. Hypersensitivity pneumonitis associated with environmental mycobacteria. Environ. Health Perspect. 113:767–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernstein DI, Lummus ZL, Santilli G, Siskosky J, Bernstein IL. 1995. Machine operator's lung. A hypersensitivity pneumonitis disorder associated with exposure to metalworking fluid aerosols. Chest 108:636–641 [DOI] [PubMed] [Google Scholar]
- 6.Chazal PM. 1995. Pollution of modern metalworking fluids containing biocides by pathogenic bacteria in France. Reexamination of chemical treatments accuracy. Eur. J. Epidemiol. 11:1–7 [DOI] [PubMed] [Google Scholar]
- 7.Chun J, Goodfellow M. 1995. A phylogenetic analysis of the genus Nocardia with 16S rRNA gene sequences. Int. J. Syst. Bacteriol. 45:240–245 [DOI] [PubMed] [Google Scholar]
- 8.Dawkins P, et al. 2006. An outbreak of extrinsic alveolitis at a car engine plant. Occup. Med. (Lond.) 56:559–565 [DOI] [PubMed] [Google Scholar]
- 9.de Hoog GS. 2000. Atlas of clinical fungi, 2nd ed Centraalbureau voor Schimmelcultures, Utrecht, Netherlands [Google Scholar]
- 10.Dilger S, Fluri A, Sonntag H-G. 2005. Bacterial contamination of preserved and non-preserved metal working fluids. Int. J. Hyg. Environ. Health 208:467–476 [DOI] [PubMed] [Google Scholar]
- 11.Falkinham JO. 2009. Effects of biocides and other metal removal fluid constituents on Mycobacterium immunogenum. Appl. Environ. Microbiol. 75:2057–2061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Falkinham JO. 2003. Mycobacterial aerosols and respiratory disease. Emerg. Infect. Dis. 9:763–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fishwick D, et al. 2005. Respiratory symptoms, immunology and organism identification in contaminated metalworking fluid workers. What you see is not what you get. Occup. Med. (Lond.) 55:238–241 [DOI] [PubMed] [Google Scholar]
- 14.Fox J, et al. 1999. Metal working fluid-associated hypersensitivity pneumonitis: an outbreak investigation and case-control study. Am. J. Ind. Med. 35:58–67 [DOI] [PubMed] [Google Scholar]
- 15.Gilbert Y, Veillette M, Duchaine C. 2010. Metalworking fluids biodiversity characterization. J. Appl. Microbiol. 108:437–449 [DOI] [PubMed] [Google Scholar]
- 16.Gordon T, Nadziejko C, Galdanes K, Lewis D, Donnelly K. 2006. Mycobacterium immunogenum causes hypersensitivity pneumonitis-like pathology in mice. Inhal Toxicol. 18:449–456 [DOI] [PubMed] [Google Scholar]
- 17.Grass G, Rensing C, Solioz M. 2011. Metallic copper as an antimicrobial surface. Appl. Environ. Microbiol. 77:1541–1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gupta A, Rosenman KD. 2006. Hypersensitivity pneumonitis due to metal working fluids: sporadic or under reported? Am. J. Ind Med. 49:423–433 [DOI] [PubMed] [Google Scholar]
- 19.Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95–98 [Google Scholar]
- 20.Hodgson MJ, et al. 2001. Hypersensitivity pneumonitis in a metal-working environment. Am. J. Ind. Med. 39:616–628 [DOI] [PubMed] [Google Scholar]
- 21.Holt JG. (ed). 1994. Bergey's manual of determinative bacteriology, 9th ed Williams & Wilkins, Baltimore, MD [Google Scholar]
- 22.Howell JK, Lucke WE, Steigerwald JC. 1996. Metalworking fluids: composition and use, p 13–20 In Proceedings of the Industrial Metalworking Environment—Assessment and Control, 13 to 16 November 1995, Dearborn, MI [Google Scholar]
- 23.Janda JM, Abbott SL. 2007. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. J. Clin. Microbiol. 45:2761–2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kreiss K, Cox-Ganser J. 1997. Metalworking fluid-associated hypersensitivity pneumonitis: a workshop summary. Am. J. Ind. Med. 32:423–432 [DOI] [PubMed] [Google Scholar]
- 25.Lee M, Chandler AC. 1941. A study of the nature, growth and control of bacteria in cutting compounds. J. Bacteriol. 41:373–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lonon MK, Abanto M, Findlay RH. 1999. A pilot study for monitoring changes in the microbiological component of metalworking fluids as a function of time and use in the system. Am. Ind. Hyg. Assoc. J. 60:480–485 [DOI] [PubMed] [Google Scholar]
- 27.Muilenberg ML, Berge HA, Sweet T. 1993. Hypersensitivity pneumonitis and exposure to acid-fast bacilli in coolant aerosols. J. Allergy Clin. Immunol. 91:311 [Google Scholar]
- 28.O'Brien DM. 2003. Aerosol mapping of a facility with multiple cases of hypersensitivity pneumonitis: demonstration of mist reduction and a possible dose/response relationship. Appl. Occup. Environ. Hyg. 18:947–952 [DOI] [PubMed] [Google Scholar]
- 29.Pickup RW, et al. 2005. Mycobacterium avium subsp. paratuberculosis in the catchment area and water of the River Taff in South Wales, United Kingdom, and its potential relationship to clustering of Crohn's disease cases in the city of Cardiff. Appl. Environ. Microbiol. 71:2130–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Provost F, Laurent F, Uzcategui LR, Boiron P. 1997. Molecular study of persistence of Nocardia asteroides and Nocardia otitidiscaviarum strains in patients with long-term nocardiosis. J. Clin. Microbiol. 35:1157–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rhodes G, et al. 2008. Detection of Mycobacterium immunogenum by real-time quantitative Taqman PCR. J. Microbiol. Methods 73:266–268 [DOI] [PubMed] [Google Scholar]
- 32.Robertson W, et al. 2007. Clinical investigation of an outbreak of alveolitis and asthma in a car engine manufacturing plant. Thorax 62:981–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosenman KD. 2009. Asthma, hypersensitivity pneumonitis and other respiratory diseases caused by metalworking fluids. Curr. Opin. Allergy Clin. Immunol. 9:97–102 [DOI] [PubMed] [Google Scholar]
- 34.Saha R, Donofrio RS, Bagley ST. 2010. Development of a real-time TaqMan assay to detect mendocina sublineage Pseudomonas species in contaminated metalworking fluids. J. Ind. Microbiol. Biotechnol. 37:843–848 [DOI] [PubMed] [Google Scholar]
- 35.Selvaraju SB, Khan IUH, Yadav JS. 2005. Biocidal activity of formaldehyde and nonformaldehyde biocides toward Mycobacterium immunogenum and Pseudomonas fluorescens in pure and mixed suspensions in synthetic metalworking fluid and saline. Appl. Environ. Microbiol. 71:542–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Selvaraju SB, Khan IUH, Yadav JS. 2008. Differential biocide susceptibility of the multiple genotypes of Mycobacterium immunogenum. J. Ind. Microbiol. Biotechnol. 35:197–203 [DOI] [PubMed] [Google Scholar]
- 37.Shelton BG, Flanders WD, Morris GK. 1999. Mycobacterium sp. as a possible cause of hypersensitivity pneumonitis in machine workers. Emerg. Infect. Dis. 5:270–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suuronen K, Aalto-Korte K, Piipari R, Tuomi T, Jolanki R. 2007. Occupational dermatitis and allergic respiratory diseases in Finnish metalworking machinists. Occup. Med. (Lond.) 57:277–283 [DOI] [PubMed] [Google Scholar]
- 39.Telenti A, et al. 1993. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J. Clin. Microbiol. 31:175–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thorne PS, Adamcakova-Dodd A, Kelly KM, O'Neill ME, Duchaine C. 2006. Metalworking fluid with mycobacteria and endotoxin induces hypersensitivity pneumonitis in mice. Am. J. Respir. Crit. Care Med. 173:759–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tillie-Leblond I, et al. 2011. Hypersensitivity pneumonitis and metalworking fluids contaminated by mycobacteria. Eur. Respir. J. 37:640–647 [DOI] [PubMed] [Google Scholar]
- 42.Trout D, et al. 2003. Evaluation of hypersensitivity pneumonitis among workers exposed to metal removal fluids. Appl. Occup. Environ. Hyg. 18:953–960 [DOI] [PubMed] [Google Scholar]
- 43.van Beilen JB, Wubbolts MG, Witholt B. 1994. Genetics of alkane oxidation by Pseudomonas oleovorans. Biodegradation 5:161–174 [DOI] [PubMed] [Google Scholar]
- 44.van der Gast CJ, Knowles CJ, Wright MA, Thompson IP. 2001. Identification and characterisation of bacterial populations of an in-use metal-working fluid by phenotypic and genotypic methodology. Int. Biodeterior. Biodegrad. 47:113–123 [Google Scholar]
- 45.van der Gast CJ, Whiteley AS, Lilley AK, Knowles CJ, Thompson IP. 2003. Bacterial community structure and function in a metal-working fluid. Environ. Microbiol. 5:453–461 [DOI] [PubMed] [Google Scholar]
- 46.Veillette M, Pagé G, Thorne PS, Duchaine C. 2008. Real-time PCR quantification of Mycobacterium immunogenum in used metalworking fluids. J. Occup. Environ. Hyg. 5:755–760 [DOI] [PubMed] [Google Scholar]
- 47.Veillette M, Thorne PS, Gordon T, Duchaine C. 2004. Six month tracking of microbial growth in a metalworking fluid after system cleaning and recharging. Ann. Occup. Hyg. 48:541–546 [DOI] [PubMed] [Google Scholar]
- 48.Wallace RJ, Zhang Y, Wilson RW, Mann L, Rossmoore H. 2002. Presence of a single genotype of the newly described species Mycobacterium immunogenum in industrial metalworking fluids associated with hypersensitivity pneumonitis. Appl. Environ. Microbiol. 68:5580–5584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Watt WD. 2003. Observations on the relationship between triazines and mycobacteria in metal removal fluids. Appl. Occup. Environ. Hyg. 18:961–965 [DOI] [PubMed] [Google Scholar]
- 50.Weaver L, Michels HT, Keevil CW. 2010. Potential for preventing spread of fungi in air-conditioning systems constructed using copper instead of aluminium. Lett. Appl. Microbiol. 50:18–23 [DOI] [PubMed] [Google Scholar]
- 51.Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weiss L, et al. 2002. Respiratory illness in workers exposed to metalworking fluid contaminated with nontuberculous mycobacteria—Ohio, 2001. JAMA 287:3073–3074 [PubMed] [Google Scholar]
- 53.White TJ, Bruns T, Lee S, Taylor JW. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p 315–322_In_Innis MA, Gelfand DH, Sninsky JJ, White TJ. (ed), PCR protocols: a guide to methods and applications. Academic Press, San Diego, CA [Google Scholar]
- 54.Wilson RW, et al. 2001. Mycobacterium immunogenum sp. nov., a novel species related to Mycobacterium abscessus and associated with clinical disease, pseudo-outbreaks and contaminated metalworking fluids: an international cooperative study on mycobacterial taxonomy. Int. J. Syst. Evol. Microbiol. 51:1751–1764 [DOI] [PubMed] [Google Scholar]
- 55.Wolinsky E, Rynearson TK. 1968. Mycobacteria in soil and their relation to disease-associated strains. Am. Rev. Respir. Dis. 97:1032–1037 [DOI] [PubMed] [Google Scholar]
- 56.Zacharisen MC, et al. 1998. The spectrum of respiratory disease associated with exposure to metal working fluids. J. Occup. Environ. Med. 40:640–647 [DOI] [PubMed] [Google Scholar]