Functional association of Gdown1 with RNA polymerase II poised on human genes (original) (raw)
. Author manuscript; available in PMC: 2013 Jan 13.
Summary
Most human genes are loaded with promoter proximally paused RNA polymerase II (Pol II) molecules that are poised for release into productive elongation by P-TEFb. We present evidence that Gdown1, a protein that renders Pol II responsive to mediator, is involved in Pol II elongation control. During in vitro transcription assays Gdown1 specifically blocked elongation stimulation by TFIIF, inhibited the termination activity of TTF2, and influenced pausing factors NELF and DSIF, but did not affect the function of TFIIS or the mRNA capping enzyme. Without P-TEFb, Gdown1 led to the production of stably paused polymerases in the presence of nuclear extract. Supporting these mechanistic insights, ChIP-Seq demonstrated that Gdown1 mapped over essentially all poised polymerases across the human genome. Our results establish that Gdown1 increases the stability of poised polymerases while maintaining their responsiveness to P-TEFb and suggest that mediator overcomes a Gdown1-mediated block of initiation by allowing TFIIF function.
Introduction
Recent genome wide analyses in which Pol II was mapped by ChIP-chip and ChIP-Seq demonstrated that promoter proximal regions of most human genes and many Drosophila genes are loaded with polymerase and associated with histones containing the H3K4me3 mark associated with initiation (Gilchrist et al., 2008; Guenther et al., 2007; Hendrix et al., 2008; Muse et al., 2007; Nechaev and Adelman, 2011; Rahl et al., 2010; Zeitlinger et al., 2007). These findings are consistent with the polymerase being engaged in transcription and “poised,” before entering productive elongation and, in fact, an engaged polymerase is found on most genes examined in detail (Lee et al., 2008; Peterlin and Price, 2006; Saunders et al., 2006). An early block to elongation was demonstrated originally for the human MYC gene, the HIV provirus, and Drosophila HSP70 gene and has since been found to be an important regulatory step in the expression of many specific genes (Boettiger and Levine, 2009; Core et al., 2008; Ni et al., 2008; Peterlin and Price, 2006; Romano and Giordano, 2008; Saunders et al., 2006; Zhou and Yik, 2006).
The positive transcription elongation factor, P-TEFb, is required to release the block to productive elongation (Marshall and Price, 1995) and a number of factors have been demonstrated to play both negative and positive roles in controlling elongation (Peterlin and Price, 2006; Saunders et al., 2006). Two negative elongation factors, DSIF and NELF, contribute to the block (Lee et al., 2008; Peterlin and Price, 2006) and P-TEFb reverses their effect and allows a high rate of elongation (Cheng and Price, 2007). Although progress has been made in understanding aspects of Pol II elongation control, the mechanisms employed to generate and regulate promoter proximally polymerases are not fully understood.
Because of the prevalence of poised polymerases across the human genome and the likelihood that P-TEFb mediated reversal of this block is key to regulating gene expression, we performed biochemical studies aimed at identifying the factor(s) involved in generating promoter proximally paused polymerases. NELF and DSIF have a significant negative effect on the elongation properties of Pol II in a defined system, but a crude nuclear extract conferred even stronger negative properties to Pol II elongation complexes (Cheng and Price, 2007). In fact, crude extracts added back to isolated elongation complexes caused complete resistance to the strong positive effects of TFIIF on elongation (Cheng and Price, 2007). This result strongly suggests that additional factors work in concert with NELF and DSIF to modulate the properties of poised polymerases. In an attempt to identify the TFIIF resistance factor, a number of factors known to associate with Pol II were tested. In the present study we demonstrate that the TFIIF resistance activity is attributable to Gdown1, an Pol II binding protein that has been shown to be necessary for a mediator-dependent response to activation of transcription in vitro (Hu et al., 2006). The work presented here suggests that Gdown1 provides a link between mediator effects on initiation and regulation of transcription elongation.
Results
Gdown1 is the TFIIF resistance factor
To examine the effects of Gdown1, labeled elongation complexes were generated on an immobilized template containing the CMV promoter using a HeLa nuclear extract and early elongation complexes (EECs) containing transcripts mostly less than 25 nt in length (Figure 1A, lane 1) were extensively washed with 1.6 M salt to strip off associated factors (Cheng and Price, 2007, 2009). The resulting EECs were assembled into reactions containing defined factors or crude extracts and then chased with physiological levels of NTPs (500 μM). The effects of elongation factors present during the chase are indicated by changes in the electrophoretic pattern of resulting transcripts.
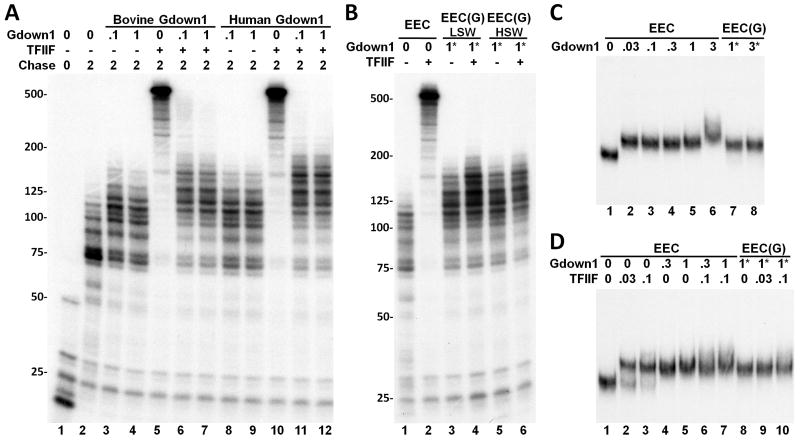
Figure 1. Gdown1 inhibits the function of TFIIF.
(A) Isolated EECs were supplemented with indicated amounts of bovine or human Gdown1 and further elongation was performed for 2 min. in the absence or presence of 0.1 pmole TFIIF. The autoradiograph shows labeled transcripts analyzed in a 6% TBE/urea gel. (B) Isolated EECs were either left untreated or incubated with 1 pmole Gdown1 for 3 min. at room temperature. After the incubation, the EEC(G)s were reisolated either with a low salt wash (LSW) or with a high salt wash (HSW) for 5 min. Then EECs or reisolated EEC(G)s were further elongated for 2 min. in the absence or presence of 0.1 pmole TFIIF. (C) Analysis of ECC•Gdown1 interaction using EC-EMSA. Isolated EECs were released from the beads through a restriction enzyme digestion and then incubated with indicated amounts of Gdown1. In lanes 7 and 8, EEC(G)s were formed, reisolated by a high salt wash, and then released by a restriction enzyme digestion. The samples were then subjected to analysis on a native gel. (D) EECs or EEC(G)s were incubated with the indicated factors and then analyzed by EC-EMSA. In lanes 6 and 7, TFIIF was added prior to the addition of Gdown1. Asterisks (*) in B, C, and D signify that the indicated amounts of Gdown1 were incubated with EECs to form EEC(G) before reisolation.
When increasing amounts of either bovine (Hu et al., 2006) or human (Figure S1) recombinant Gdown1 were added to EECs, the factor had a small positive effect on elongation evidenced by the increase in transcript length during a 2 minute chase (Figure 1A). The two amounts of Gdown1 used correspond approximately to a 5-fold and 50-fold molar excess over the Pol II in the reactions, and correspondingly the elongation stimulation effect is saturated even at the lowest level. As expected, TFIIF alone dramatically increased the length of transcripts during the chase. Strikingly, Gdown1 from either species was able to inhibit the strong stimulatory effect of TFIIF (Figure 1A). We examined the association of Gdown1 with elongation complexes by incubating the factor with EECs and then washing the immobilized complexes with low (60 mM KCl) or high (1.6 M KCl) salt. Gdown1 bound very tightly as evidenced by the slight positive effect on elongation following the washes and by the persistent resistance of these elongation complexes to TFIIF function (Figure 1B). These results strongly support the idea that Gdown1 is responsible for the TFIIF resistance activity previously described (Cheng and Price, 2007).
An Elongation Complex-Electrophoretic Mobility Shift Assay (EC-EMSA) (Cheng and Price, 2008) was performed to determine if Gdown1 inhibits TFIIF function by physically blocking TFIIF interaction with elongation complexes. EECs were liberated from the paramagnetic beads used to isolate them by restriction enzyme digestion of the DNA linking them to the beads and were then analyzed on a native gel using the short, labeled nascent transcripts to detect the EECs. In the absence of other factors the complexes have a unique mobility (Figure 1C, lane 1). Addition of increasing amounts of Gdown1 caused a reduction of mobility consistent with specific binding of Gdown1 to the elongation complexes (Figure 1C, lane 2-6). 0.03 pmole of Gdown1 caused a complete shift and only when 100 times more Gdown1 (3 pmole) was added was there any evidence of non-specific binding (Figure 1C, lane 6). The specific shift, but not the non-specific shift, was retained when the complexes were washed with 1.6 M KCl after factor binding (Figure 1C, lanes 7 and 8). TFIIF also caused a shift in the mobility of EECs, but the combination of TFIIF and Gdown1 resulted in only one shift (Figure 1D). EECs isolated after saturation with Gdown1 and a high salt wash did not exhibit a second shift when TFIIF was added strongly suggesting that Gdown1 blocks TFIIF binding (Figure 1D, lanes 8-10). Although it is possible that TFIIF displaced Gdown1 in the EC-EMSA, this would be contradictory to the finding that Gdown1 was stably bound and inhibited TFIIF in the elongation assay (Figure 1A and 1B). The results shown in Figure 1 are consistent with Gdown1 binding stably to Pol II elongation complexes and preventing the association and function of TFIIF.
Effect of Gdown1 on DSIF and NELF, HCE and TFIIF
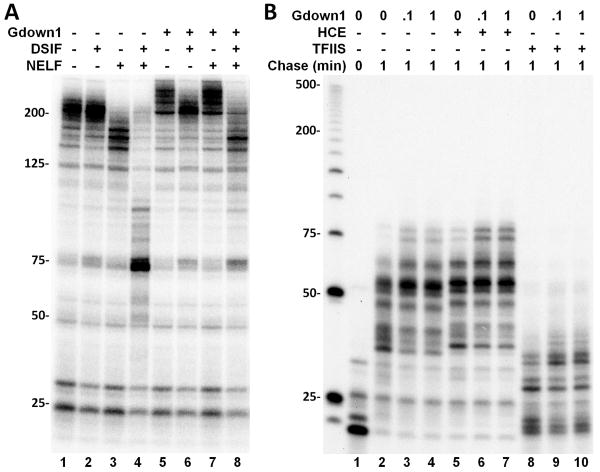
We next determined if Gdown1 would affect other factors known to functionally interact with elongation complexes. As a control EECs without Gdown1 were isolated and chased for 6 minutes either alone or in the presence of DSIF, NELF or both DSIF and NELF. DSIF had no effect, however NELF displayed a moderate negative activity and as seen before (Cheng and Price, 2007; Renner et al., 2001) the combination of DSIF and NELF was strongly negative. Different results were obtained with EECs that were converted into EEC(G)s by allowing Gdown1 to bind and then washing the excess away with 1.6 M salt as described in Figure 1B. On EEC(G)s DSIF eliminated most of the positive effect of Gdown1 (Figure 2A, lane 6) and NELF had no effect (Figure 2A, lane 7). Together NELF and DSIF had a moderately negative activity on EEC(G)s. Gdown1 modulated the activities of DSIF and NELF individually and in combination with each other. The effect of Gdown1 on other Pol II interacting factors, the human mRNA capping enzyme (HCE) and TFIIS, the transcript cleavage factor that can release Pol II blocked at arrest sites during elongation, was examined. The 5′ guanylylation of the RNA, evidenced by a shift of about 1.5 nt of transcripts longer than about 30 nt by HCE, or the negative effect exhibited by high concentrations of TFIIS (discussed more fully below) were unchanged by Gdown1 (Figure 2B). Also neither HCE nor TFIIS affected the slight positive effect of Gdown1 (Figure 2B). These data demonstrate that Gdown1 does not directly interfere with the function of all Pol II binding factors, and this supports a dramatic role of Gdown1 in completely blocking TFIIF function and influencing both DSIF and NELF alone and in combination.
Figure 2. Gdown1 affects the functions of DSIF and NELF, but not HCE or TFIIS.
(A) Isolated EECs or EEC(G)s (as described in the Figure 1 legend if Gdown1 is indicated) were supplemented with combinations of DSIF and NELF before a 6 minute chase. (B) Isolated EECs were supplemented with Gdown1 and further elongation was performed for 1 minute in the absence or presence of 1 pmole HCE or TFIIS.
Function of Gdown1 in HeLa nuclear extract
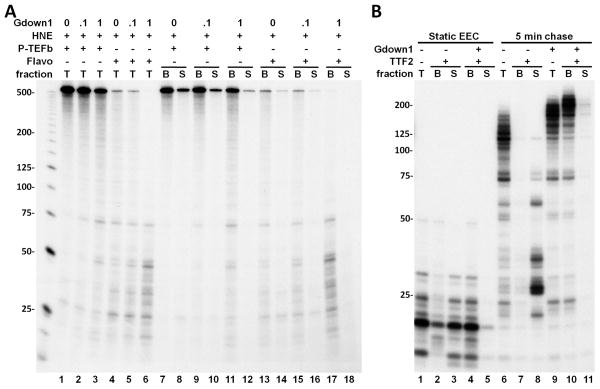
To further characterize the properties of Gdown1, HeLa nuclear extract (HNE) was added to isolated EECs and then elongation was carried out for three minutes in the presence of increasing amounts of Gdown1. When P-TEFb was functional, run-off transcripts were the predominant products generated (Figure 3A). Addition of Gdown1 had no effect at 0.1 pmole and only a slight negative effect at 1 pmole on run-off (Figure 3A, lanes 1-3). At the highest level of Gdown1, the slight loss of run-off transcripts was accompanied by an increase in short transcripts (Figure 3A, lanes 1-3). Inhibition of the P-TEFb by Flavopiridol (FP) (Chao and Price, 2001) revealed that Gdown1 blocked the appearance of the few remaining P-TEFb-independent, run-off transcripts and led to an accumulation of transcripts between 20 and 75 nt in length (Figure 3A, lanes 4-6). The appearance of the very short transcripts in the absence of added Gdown1 can be explained by the presence of limiting amounts of Gdown1 in the HNE. We hypothesize that the crude extract contains factors that work with Gdown1 to generate a strong negative effect on elongation. In addition, our results indicate that P-TEFb is able to counteract Gdown1 and all negative factors.
Figure 3. The effects of Gdown1 on Pol II elongation in the presence of HNE.
(A). Isolated EECs were supplemented with Gdown1, HNE, 1 pmole P-TEFb or 1 μM FP as indicated and further elongation was performed for 3 min. Transcription elongation was then stopped and an aliquot was taken from each reaction to show the pattern of transcripts for the total reaction (“T”). Another aliquot was separated into a supernatant fraction (“S”) and a bead bound fraction (“B”). (B) Isolated EECs were supplemented with 1 pmole Gdown1 and/or TTF2. Elongation complexes were either incubated with 0.5 mM ATP for 5 min. at room temperature (lanes 1-5) or further elongated for 5 min. upon the addition of 0.5 mM NTP (lanes 6-11). After the incubation or elongation reactions were done, the supernatant fractions in the indicated reactions were separated from the beads.
To determine if Gdown1 enhanced termination in the presence of HNE, bead bound elongation complexes (B) were separated from reaction supernatants (S) from an identical set of reactions (Figure 3A, lanes 7-18). As has been found before (Marshall and Price, 1992) some of the short transcripts were released from the template and appeared in the supernatant due to termination in the absence of extra added Gdown1. However, after Gdown1 was added, all transcripts remained bound to the template indicating that termination was blocked. Because TTF2 is the major termination factor present in the extract (Jiang et al., 2004), we examined the effect of Gdown1 on TTF2 using the defined system with immobilized EECs. Gdown1 dramatically blocked the release of transcripts into the supernatant by TTF2 in the absence of elongation (Figure 3B, lanes 1-5) or during elongation (Figure 3B, lanes 6-11). These results indicate that Gdown1 renders the elongation complex resistant to TTF2.
Role of TFIIS in the generation of short transcripts in vitro
In an attempt to identify negative factors that worked with Gdown1 in HNE, extract was fractionated using phosphocellulose (P-11), Mono Q, Mono S, and glycerol gradient sedimentation. Using a variety of assays four factors were identified. Representative assays and comparisons of the identified activities to known factors are shown in Figure S2 and S3. Importantly, both DSIF and NELF that were recently shown to co-map with promoter proximally paused Pol II (Gilchrist et al., 2008; Rahl et al., 2010) were rediscovered. One factor that that we call the Gdown1 Negative Accessory Factor, GNAF, was detected, but the identity of that factor or factors was not determined because its activity was lost upon further fractionation. The final negative activity identified was the transcript cleavage factor TFIIS. Although TFIIS has been thought of as a “positive” factor, it does not increase the maximum rate of elongation by Pol II (Guo and Price, 1993; Luse et al., 2011) and its ability to stimulate the intrinsic transcript cleavage activity of Pol II is an inherently negative activity. At low concentrations its activity leads to suppression of pausing and arrest, but at higher concentrations normally found in HNE this activity has an overall negative effect on elongation (Figure S3C).
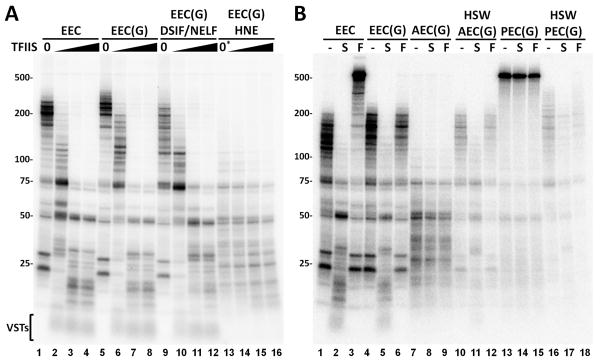
The function of TFIIS was examined by adding increasing amounts of the factor to EECs, EEC(G)s, EEC(G)s with DSIF and NELF, and to EEC(G)s in the presence of HNE (Figure 4) and allowing elongation for 6 minutes. A strong negative effect was seen on EECs that saturated at the intermediate amount of TFIIS (Figure 4A, lanes 1-4). Except for the slight positive effect on elongation due to Gdown1, similar results were found when TFIIS was added to EEC(G)s (Figure 4A, lanes 5-8). Addition of DSIF and NELF had the expected negative effect on EEC(G)s and TFIIS still acted as a negative factor (Figure 4A, lanes 9-12). Elongation of EEC(G)s in the presence of HNE without P-TEFb function gave only short transcripts as expected. TFIIS had only a modest negative effect even at the highest levels (Figure 4A, lanes 13-16). This is likely due to the fact that TFIIS is present at high levels in the extract. The TFIIS-dependent, very short transcripts that appeared in all reactions without extract (Figure 4A, VSTs) were absent when extract was present. This could be due to a TFIIS inhibitory activity present in the extract that works only on elongation complexes that have short transcripts or due to degradation of the very short transcripts that might be released from elongation complexes. Overall, these experiments and those shown earlier (see Figure 2B) suggest that TFIIS may play a role in promoter proximal pausing and demonstrate that Gdown1 does not influence TFIIS activity.
Figure 4. Negative effect of TFIIS and the influence of other factors on Gdown1.
(A) EECs or EECs with Gdown1, EEC(G)s, were incubated with factors or extract and allowed to elongate for 6 min. in the presence of increasing amounts of TFIIS (0, 0.5, 1.5 or 5 pmole). (B) The effects of TFIIS (S) and TFIIF (F) were examined on the indicated elongation complexes. Elongations complexes were EECs or EEC(G)s. Abortive elongation complexes, AEC(G)s, are EEC(G)s in the presence of a HNE with FP. Productive elongation complexes are EEC(G)s incubated with HNE, extra P-TEFb and ATP for 5 minutes and then chased in the presence of FP. Elongation complexes were washed with 1.6 M salt (HSW) before the chase. Chases were for 6 min. and contained the indicated elongation complexes alone or were supplemented with TFIIS, or TFIIF. Details are present in the Experimental Procedures and in the text. The asterisk above lane 13 in A signifies that although no TFIIS was added, the extract present does contain TFIIS.
Fate of Gdown1 during productive elongation
To determine what happens to Gdown1 during the P-TEFb-mediated transition into productive elongation a variety of elongation complexes were tested for the retention of Gdown1 based on inhibition of TFIIF, but not TFIIS. On EECs devoid of Gdown1, TFIIS and TFIIF had strong negative and positive activities respectively (Figure 4B, lanes 1-3). As demonstrated earlier, EEC(G)s responded only to TFIIS, not TFIIF (Figure 4B, lanes 4-6). Abortive elongation complexes (AECs) generated by incubating EEC(G)s in extract in which P-TEFb was inhibited, did not respond to either TFIIS or TFIIF (Figure 4B, lanes 7-9). As before, the lack of response to TFIIS may be due to the fact that TFIIS is present in the extract and is functioning already. To determine if Gdown1 remained bound after incubation with extract, elongation complexes were washed with 1.6 M salt (HSW) to remove associated factors (but not Gdown1) and the resulting complexes were allowed to elongate. TFIIS, but not TFIIF functioned on these complexes indicating that, as expected, Gdown1 remained bound (Figure 4B, lanes 10-12).
Productive elongation complexes (PECs) can be made by incubating EECs and nuclear extract with P-TEFb and ATP for 5 minutes (Cheng and Price, 2007) and here PECs generated this way with EEC(G)s were able to reach runoff during a short chase. No effect was found when reactions containing PEC(G)s were supplemented with TFIIS or TFIIF during the chase (Figure 4B, lanes 13-15). Evidently, the P-TEFb dependent transition into productive elongation makes elongation complexes resistant to the negative effect of TFIIS and causes them to exhibit properties similar to TFIIF stimulation. High salt washed PEC(G)s retained resistance to TFIIF strongly suggesting that Gdown1 remained bound (Figure 4B, lanes 16-18). The slight lengthening of transcripts see in these three lanes is due to limited elongation during the 5 minute incubation with ATP due to the presence of low levels of NTPs in the extract. These results demonstrate that elongation complexes containing Gdown1 can enter productive elongation without loss of Gdown1.
Distribution of Pol II and Gdown1 across the human genome
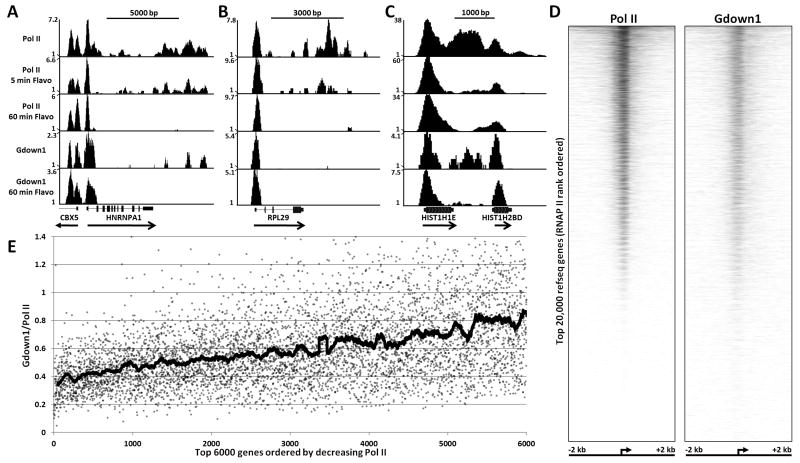
The biochemical analyses described above provided mechanistic insights into the function of Gdown1 as a factor regulating the properties of poised polymerases and because of this we reasoned that Gdown1 should be associated with poised polymerases in vivo. ChIP-Seq was used to map Gdown1 occupancy relative to Pol II in control HeLa cells or cells treated with FP. The following examples illustrate key aspects of what was found. In control cells, for the highly transcribed HNRNPA1 gene there was a peak of Pol II density centered about 75 bp downstream of the annotated transcription start site (TSS), low levels of Pol II over the rest of the region encoding the mRNA, and high levels throughout an approximately 3 kbp region downstream of the Poly(A) addition site (Figure 5A). After 5 minutes of FP treatment the only significant change in Pol II density was a loss of density in the region several kbp downstream of the large peak at the 5′ end of the gene. After 60 minutes of FP treatment, only Pol II over the 5′ end of the gene remained. Two peaks of such polymerases were found over the 5′ end of the CBX5 gene (encoding HP1) that is transcribed in the opposite direction from HNRNPA1 (Figure 5A). One of the peaks corresponds to a site 50 bp downstream of the annotated promoter and the other is about 400 bp further downstream. EST data indicates that the second peak is likely generated by initiation from an unannotated promoter. Results similar to those found for HNRNPA1 were found for the RPL29 gene (Figure 5B). Interestingly, for histone genes, exemplified by HIST1H1E, in addition to a very strong peak of poised polymerases, a high density of Pol II was found downstream of the mature 3′ end even though different 3′ end processing machinery is used on histone mRNAs. Those polymerases terminated and were not replaced during a 5 minute FP treatment indicating that P-TEFb is needed for histone mRNA production and that termination occurs within 5 minutes. This quick turnover is probably due to the fact that the 3′ end polymerases are less than 1000 bp downstream of the TSS and because histone mRNA 3′ processing is rapid (Adamson and Price, 2003). Overall, the results from the control Pol II ChIP-Seq dataset compared to two datasets from cells treated with FP for 5 or 60 minutes support the idea that P-TEFb is needed for most mRNA production (Chao and Price, 2001; Rahl et al., 2010).
Figure 5. ChIP-Seq analyses of Pol II and Gdown1.
Pol II and Gdown1 occupancy in HeLa cells was determined by ChIP-Seq analysis as described in Experimental Procedures. Vertical axis indicates occupancy normalized for the total number of reads obtained (counts per 1 million reads). (A), (B), and (C) show density of Pol II and Gdown1 across indicated human genes. Pol II occupancy was determined in cells treated with FP for 0, 5, or 60 minutes. Gdown1 occupancy was determined in cells treated with FP for 0 or 60 minutes. (D) Pol II and Gdown1 binding from -2 kb to +2 kb around the TSS of top 20,000 RefSeq genes without TSSs within 1000 bp of any other TSS, rank ordered from most Pol II bound to least Pol II bound. The high resolution heatmaps for Pol II and Gdown1 were generated as described in Experimental Procedures. (E) Ratio of Gdown1 to Pol II for the region from -500 to +500 around the TSS of 5000 genes rank ordered by the amount of Pol II. Dots are individual ratios for each gene and the thick line is a running average (100 points each).
Because the in vitro evidence presented above strongly suggested that Gdown1 plays a role in the generation and stability of poised polymerases, we used an affinity purified Gdown1 antibody (Figure S1) to map the genomic regions associated with Gdown1 using ChIP-Seq. As predicted, in both control and FP treated HeLa cells, Gdown1 mapped over the promoter proximally paused polymerases in all three examples shown (Figure 5A-5C). Gdown1 was found in the same region as 3′ Pol II on HNRNPA1 and the HIST1H1E histone gene (Figure 5A and 5C), however, it was notably absent from that region on RPL29 (Figure 5B). Variable amounts of Gdown1 were also found in regions occupied by promoter proximally paused polymerase. An example of this is seen in the HNRNPA1 gene that has two positions of poised polymerases with different Pol II occupancy (Figure 5A, 60 minute FP treatment). The second smaller peak has about 80% less Pol II, but an equal amount of Gdown1. Pol II binding was plotted from -2kb to +2kb centered around the TSS for top 20,000 human RefSeq genes, rank ordered by the amount of Pol II detected in the region (Figure 5D). Gdown1 occupancy was plotted based on the Pol II rank order, demonstrating a strong spatial correlation between Gdown1 and Pol II occupancy at the 5′ end of most genes (Figure 5D). In this analysis the intensity of Pol II gradually declined (as dictated by the rank ordering). The amount of Gdown1 also gradually declined for the bottom half of genes, but was relatively constant over the top half of genes. The gene set in which all genes with TSSs within 1000 bp of another were eliminated contains 20,286 genes. Of these, 9366 have at least 5 sequences in the region from -500 to +500 for the control Pol II and 92% of these also have at least 5 sequences from the control Gdown1 dataset. To quantitatively assess the relative distribution of Pol II and Gdown1, the ChIP-Seq data was further analyzed by looking at the ratio of Gdown1 to Pol II on 1 kb regions centered on TSSs of the 6000 genes with the highest levels of Pol II. Although a scatter plot of the data had a wide spread (Figure 5E, dots) a running average of the points showed a clear trend of increasing Gdown1 to Pol II ratio as the amount of polymerase decreased (Figure 5E, line). Both analyses suggest that Gdown1 is preferentially associated with promoter proximally paused polymerases on genes with lower Pol II occupancy.
Metagene analyses
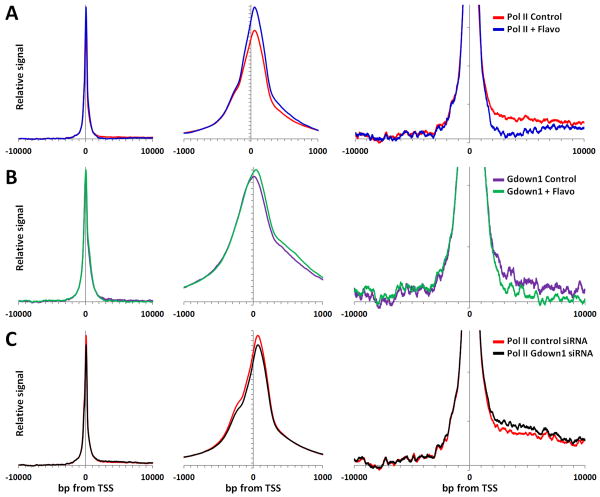
To generate a more global view, the densities of Pol II and Gdown1 were compiled for the regions from -10,000 to +10,000 relative to the TSS of 20,286 RefSeq genes that do not have TSSs within 1000 bp of another TSS. The signals from the lowest 10% of the 20,000 data points were averaged and subtracted from each point to remove the background and then datasets were normalized so that the area under each distribution was equal (Figure S4 and Figure 6). This normalization appropriately emphasizes the positional information inherent in ChIP-Seq data and does not attempt to quantitatively compare datasets except for relative differences in positional information. Using this background subtracted and normalized data the distribution of Pol II was found to peak about 45 bp downstream of the TSS with a shoulder centered about 250 bp upstream of the TSS and lower levels in the 2000 - 10,000 bp region downstream of the TSS (Figure 6A). The shoulder likely represents divergent transcription as has been recently described (Seila et al., 2009). Comparison with Pol II ChIP-Seq data from FP treated cells demonstrated that the position of the peak of Pol II was shifted only a few bp downstream, and that as expected, transcription of far downstream regions was almost eliminated (Figure 6A). Interestingly, there was an increase in Pol II in the region from +300 to +1000 after FP treatment. These polymerases may have “crept” into this region during the hour without P-TEFb function. Identical analysis of Gdown1 datasets gave a slightly broader peak of Gdown1 around the TSS that included the upstream shoulder at -250 and the +300 to +1000 region just mentioned. Significantly, a downstream shift in the main peak to more closely align with the peak of Pol II occurred in the data from FP treated cells (Figure 6B, center panel). The pattern for Gdown1 in the region downstream of the TSS was very similar to that seen with Pol II including the reduction of signal in the region after FP treatment (Figure 6B, right). This supports the in vitro finding that Gdown1 is associated with Pol II and that a significant fraction of the productive elongation complexes have Gdown1.
Figure 6. Global ChIP-Seq analysis of Pol II and Gdown1.
Average gene data for a customized set of 24,160 RefSeq genes, from which all genes with TSSs within 1000 bp of each other have been excluded, were analyzed as described in the text and the Experimental Procedures. The three views depict relative signals in the indicated regions around the TSSs after averaging, background subtraction, and normalization of the entire -10,000 to +10,000 region (see Figure S4). The vertical scale in right hand panel was expanded 10-fold compared to other two panels. (A) Indicated regions for Pol II from control (red) and FP treated (blue) cells. (B) Indicated regions for Gdown1 from control (purple) and FP treated (green) cells. (C) Indicated regions for Pol II from control siRNA (red) and Gdown1 siRNA treated (black) cells.
The effect of Gdown1 knockdown on the distribution of Pol II was examined next. Gdown1 is difficult to knock down due to toxic effects of reducing the protein. One siRNA was able to reduce the level of Gdown1 to about 20% after 2 days (Figure S1) and after 3 days there were fewer cells and those still contained about 20% of their normal level of Gdown1. Pol II ChIP-Seq from control and Gdown1 siRNA treated cells (48 hr.) was performed and analyzed as above. Essentially no change was found in the position of the promoter proximally paused polymerases (Figure 6C, center panel), but partial knock down of Gdown1 caused an increase in polymerases downstream of the poised polymerases (Figure 6C, right panel). The significance of the change in downstream polymerases is indicated by the lack of a change in the upstream signals and from the fact that the Gdown1 siRNA curve did not cross the control siRNA curve in the 1000 to 8000 region (each of which is comprised of 7000 data points). These results support a role of Gdown1 is helping to stabilize Pol II in promoter proximal regions.
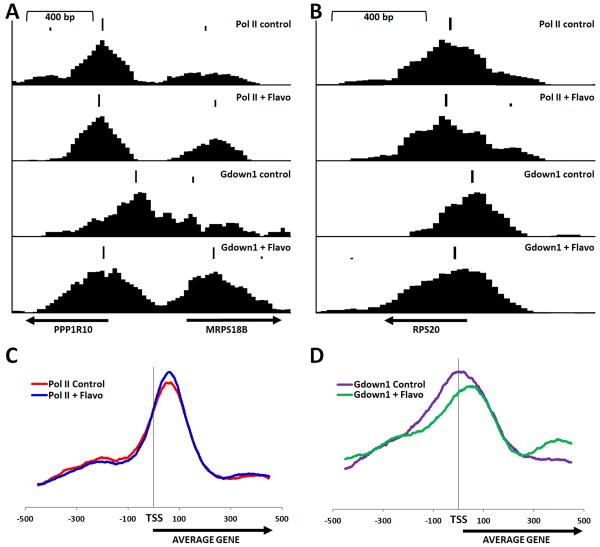
To follow up on the observation that the position of the peak of Gdown1 around the TSS shifted downstream after FP treatment of cells, a program was created to identify the precise position of peaks. The output of the program can be used to generate a track on the Genome Browser that displays both the size and position of peaks. The FP-dependent downstream shift can be seen in the three examples in Figure 7A and 7B in which the program output is displayed over each track. While the peaks of Pol II remained relatively unchanged after FP treatment, the peaks of Gdown1 shifted downstream to more closely coincide with Pol II. A genome wide average of the position (but not size) of peaks within 500 bp of the TSSs for Pol II and Gdown1 before and after treatment with FP was generated. Because information about peak height was not used, each gene is weighed equally in this average instead of genes with the most Pol II or Gdown1 being over-emphasized. The resulting distributions again demonstrated that the peak of Gdown1 but not Pol II shifted downstream after FP treatment (Figure 7C and 7D). To determine if the shift of Gdown1 was related to the relative expression of genes, two sets of genes were compared. One contained the 200 most highly expressed genes based on RNA-Seq analysis of HeLa RNA and the other was a group of 1000 genes with lower levels of Pol II found in promoter regions. As expected, analysis of the first group showed a significant effect of FP and the second had essentially no effect (Figure S5). Importantly, the shift of Gdown1 was more than double for the active genes (+45 bp) compared to the inactive genes (+20 bp)(Figure S5). Regardless of the method of analysis it is clear, overall, that the position of Gdown1 shifts downstream to more closely coincide with the poised Pol II after treatment with FP and this shift was most dramatic on genes that experience more productive elongation.
Figure 7. Effect of FP on position of Gdown1.
Peaks of Pol II and Gdown1 were identified using the program ChIP-Seq Peak and displayed above the individual tracks for Pol II and Gdown1 from control cells or FP treated cells as indicated for the promoter regions of (A) PPP1R10 and MRPS18B, and (B) RPS20. An average position for identified peaks for 25,530 RefSeq genes was compiled as described in Experimental Procedures for (C) Pol II and (D) Gdown1 from control or FP treated cells.
Discussion
Our in vitro results using a defined transcription system provided strong evidence for the involvement of Gdown1 in the Pol II elongation control process. The striking set of properties demonstrated here for Gdown1 is not duplicated by any other known elongation factor. Interestingly, Gdown1 allowed a normal response to P-TEFb in the presence of HeLa nuclear extract (HNE), but when P-TEFb was inhibited, the Gdown1 modified elongation complexes gave rise to only very short transcripts. The pattern of transcripts seen in the crude system without P-TEFb could be partially, but not completely, mimicked by the presence of only DSIF, NELF and TFIIS. An extensive search for Gdown1 Negative Accessory Factors (GNAFs) yielded evidence for an additional GNAF that functions with NELF, DSIF and TFIIS, however, we have been unable to purify it to homogeneity so far. Somewhat surprisingly, Gdown1 containing productive elongation complexes maintained their resistance to the action of TFIIF and TTF2 in the presence of HNE. One of the most obvious characteristics of productive elongation is the high elongation rate achieved, therefore, resistance to TFIIF was not expected because the factor is known to stimulate the elongation rate significantly more than any other (Price et al., 1989). It is possible that this result was dependent on the exact conditions of the experiments performed. Further evidence for the role of TFIIF in productive elongation is needed. Most significantly, we showed here that when P-TEFb was inhibited the combination of Gdown1 with negative factors in HNE caused Pol II elongation complexes to become trapped on the template, unable to terminate.
Mapping of the location of Gdown1 across the human genome by ChIP-Seq provided additional strong evidence for a role of the factor in promoter proximal pausing. Gdown1 mapped over essentially all poised polymerases and while there was a general correlation of the amount of Gdown1 with the amount of Pol II, detailed analyses indicated that the ratio of Gdown1 to Pol II was not uniform for polymerases in different genes or for different polymerases in different regions of individual genes. The highest levels of Gdown1 relative to Pol II were found over poised polymerases on genes with relatively lower levels of Pol II occupancy and these genes experienced less productive elongation. Dramatically differential loading (or removal) of Gdown1 was observed for many promoter proximally paused polymerases close to each other on the same genes. A role for Gdown1 in stabilizing poised polymerases was supported by the finding that the highest concentrations of Gdown1 were found over those polymerases and from the finding that partial knockdown of Gdown1 led to an increase in downstream polymerases.
Our in vitro results suggest that promoter proximally paused polymerases with Gdown1 associated may be relatively stably bound instead of constantly turning over. The lifetime of these polymerases cannot be measured in vivo using ChIP methods which just determine where polymerases are on average. However, the Lis lab has found using FRET that polymerases are stopped in vivo on the Drosophila HSP70 gene after induction, but in the absence of P-TEFb function are stable for up to the three minutes that are addressable by their assay (Ni et al., 2008). Our results suggest that the dwell time of some poised polymerases may be significantly longer. For highly expressed genes the dwell time is limited by the number of mRNAs produced per hour. For genes that are only rarely transcribed the turnover of poised polymerases may only be limited by the occasional function of P-TEFb and in the extreme case by the cell cycle. For dividing cells it is likely that replication fork movement is not compatible with stably poised polymerases and during mitosis all polymerases terminate. Because TTF2 is responsible for termination during mitosis (Jiang et al., 2004) and it is inhibited by Gdown1, there must be a mechanism to reverse the effects of Gdown1 during mitosis.
The only other previous biochemical study of Gdown1 found that the protein was strongly associated with less than half of the Pol II purified from calf thymus and that transcription reactions reconstituted with the polymerase containing Gdown1 were dependent on mediator to observe strong effects of activators (Hu et al., 2006). The authors concluded that Gdown1 played a negative role in transcription that was overcome by mediator. The run-off assay used in that study required both initiation and elongation for a significant distance. However, because DSIF, NELF, TFIIS and GNAF, as well as P-TEFb were not present in their defined system, the inhibitory role of Gdown1 in that study was likely a negative effect on initiation. In support of this, we have observed that a titration of Gdown1 into HNE leads to a direct inhibition of initiation (data not shown). Our results demonstrate that Gdown1 blocks the binding of TFIIF and suggests that their results could be explained by mediator dependent removal or modification of Gdown1 that would allow TFIIF to bind and Pol II to initiate. A role for Gdown1 in regulating TFIIF binding to Pol II in the absence of nucleic acids is supported by EM structures of mammalian Pol II with either Gdown1 or the large subunit of TFIIF bound. In that study part of the density of Gdown1 overlaps the binding site of TFIIF providing a structural explanation for the exclusion of TFIIF by Gdown1 (Weihau Chang, personal communication).
A model can be created that is consistent with all of the information gathered to date on the function of Gdown1 (Figure S6). Because Gdown1 was found on 30% of the polymerases isolated from calf thymus and 50% of polymerases from pig liver (Hu et al., 2006), it is possible that transcription cycles can be started with either form. In the absence of Gdown1, Pol II forms a preinitiation complex (PIC) with TFIIF and rapidly initiates (Figure S6A). The early elongation complexes come under the control of DSIF, NELF and TFIIS which keep the poised polymerase close to the promoter waiting for a short time for P-TEFb to trigger the transition into productive elongation. Poised polymerases in the absence of Gdown1 are transient due to their ability to be terminated by TTF2 (Jiang et al., 2004; Marshall and Price, 1992). If a PIC forms with Pol II containing Gdown1, initiation is blocked (Figure S6B). Mediator is then needed to “remodel” Gdown1 so that TFIIF can bind and initiation can occur. The poised polymerases are now controlled by DSIF, NELF, TFIIS, Gdown1 and GNAF. These polymerases are still responsive to P-TEFb, but differ from those lacking Gdown1 in that their elongation is more restricted and they are resistant to termination and, thereby, stably poised. Importantly, the model provides an explanation of why a downstream shift of Gdown1 in FP treated cells is seen. Since it is likely that poised polymerases preclude formation of PICs due to promoter occlusion (Kornberg, 2007), in normal, untreated cells, loss of a poised polymerase to productive elongation would allow a new PIC to form. The steady state density of Gdown1 over the promoter region that is observed in control cells could be due to new PICs, some of which could contain Gdown1. In FP treated cells the P-TEFb-dependent transition into productive elongation is blocked and, therefore, PIC formation would be blocked and less Gdown1 would be found over the promoter. The only Gdown1 detected under this condition would be in the region of the poised polymerase and in the region centered 250 bp upstream of the TSS potentially from divergent transcription and in the region 300-1000 bp downstream of the TSS, which is what was found. This model does not take into account the possibility that Gdown1 might be associated with factors other than Pol II (like mediator). If this is the case then the shift could be due to transfer of Gdown1 from other factors to Pol II, but it is not obvious how this could be affected by FP treatment.
Previous studies have elucidated many aspects of the function of TFIIS, and although most have emphasized the suppression of pause and arrest sites, the negative role of TFIIS demonstrated in this study is not contradictory. The ability of TFIIS to affect elongation is manifest through its ability to stimulate the intrinsic transcript cleavage activity of Pol II (Guo and Price, 1993; Izban and Luse, 1993; Reines, 1992). The structure of TFIIS bound to Pol II is known (Kettenberger et al., 2004) and recent structural work on the backtracked state demonstrates details of the TFIIS•Pol II•nascent RNA interactions found at pause and arrest sites (Cheung and Cramer, 2011). At low concentrations the inherently negative activity of TFIIS has the effect of giving paused and arrested polymerases a chance to encounter the block a number of times and this increases the number of polymerases molecules that pass through the site, thereby providing an overall positive effect on the average elongation rate. Even at the optimal concentration, TFIIS does not increase the rate of elongation of the fastest moving polymerases (Guo and Price, 1993; Luse et al., 2011). As we have demonstrated here, at slightly higher concentrations TFIIS negatively impacts elongation. Both outcomes are derived from ability of TFIIS to stimulate transcript cleavage, and the only difference is the balance between the NTP driven forward motion of the polymerase and transcript cleavage. Evidently, at higher concentrations, transcript cleavage dominates. Concerning Pol II elongation control, on the positive side, the stimulation of nascent transcript cleavage has been shown to aid restarting of paused polymerases in Drosophila (Adelman et al., 2005). Based on in vitro experiments, we hypothesize that TFIIS may also have a role in helping keep Pol II poised in promoter proximal positions. Supporting a negative role of TFIIS in vivo, it was recently demonstrated using global RNA-Seq that short nascent transcripts were slightly extended in cells when TFIIS was knocked down (Nechaev et al., 2010).
We have presented strong evidence that incorporation of Gdown1 into elongation complexes generates significantly more stably poised Pol II molecules with properties in vitro that mimic poised polymerases in vivo. This could be one of the main reasons that Pol II is found promoter proximally paused on the majority of mammalian genes regardless of the level of expression of those genes. At genes where expression is low due to lack of recruitment of P-TEFb, a high fraction of polymerases contain Gdown1. This could be due to the fact that those polymerases do not terminate and over time become predominant. The model for incorporation of Gdown1 into poised polymerases requires that mediator be present at all genes with poised polymerases. Evidence for mediator over the promoters (slightly upstream of the poised polymerase) can be easily found in the mediator ChIP-Seq datasets obtained from mouse embryonic stem cells (Kagey et al., 2010). Both highly expressed genes and genes that are not highly expressed contain mediator slightly upstream of the poised polymerase (Figure S7). Stably poised polymerases would help keep these promoters open. A role for Gdown1 in promoter proximal pausing might explain why knockdown of NELF did not relieve all blocks in Drosophila (Gilchrist et al., 2008) or mouse (Rahl et al., 2010) if Gdown1 and GNAF functioned as least partly independently of NELF. Supporting a conserved role for Gdown1 in metazoans, potential Gdown1 homologues are present though out vertebrates and in Drosophila. However, the genetic linkage of Gdown1 with a glutamate receptor like protein in humans (Roginski et al., 2004) is not conserved. One study has shown that DSIF is needed to observe effects of mediator on activation of transcription in vitro (Malik et al., 2007), but details of the functional interactions between mediator, Gdown1, GNAF, NELF, DSIF, TFIIS, and P-TEFb action remain to be elucidated.
Experimental Procedures
(more detailed procedures can be found in supplemental information)
Materials
Preparation of HeLa nuclear extract (Adamson et al., 2003) and purification of P-TEFb (Cheng and Price, 2007), DSIF (Renner et al., 2001), NELF (Renner et al., 2001), HCE (Moteki and Price, 2002), TFIIS (Palangat et al., 2005), and TFIIF (Peng et al., 1998) were as described earlier. Gdown1 antibodies were generated in sheep and affinity purified.
Expression and purification of Gdown1 proteins
Human and bovine Gdown1 expression plasmids were constructed using pET151 vector (Invitrogen) and the bovine protein was purified as described previously (Hu et al., 2006). The recombinant His-tagged human Gdown1 protein was expressed in Escherichia coli and purified on Ni-NTA resin (Qiagen) as previously described (Byers et al., 2005) and Mono Q as detailed in the supplementary information. SDS PAGE and silver staining for one of the final eluted fractions is shown in Figure S1.
In vitro transcription assays
The generation and isolation of early elongation complexes using immobilized templates and in vitro transcription assays were as described earlier (Cheng and Price, 2007, 2009). Basically, elongation complexes containing RNA mostly less than 25 nt in length were generated by initiation with a 30 second pulse at 500 μM A,U, and GTP and 1 μM 32P-CTP. The elongation complexes were washed with 1.6 M salt and then chased with added factors and 500 μM NTPs. Except where indicated bovine Gdown1 was used in add back assays. Labeled transcripts were analyzed in denaturing RNA gels using autoradiography or phosphorimaging.
Termination assays of stalled or elongating complexes were accomplished by incubation with the indicated amount of TTF2 for 5 minutes in the presence of only 500 μM ATP or all NTPs. In both cases, the reactions were stopped by addition of EDTA and then the reactions were magnetically separated into beads and supernatant fractions. Elongation complex electrophoretic mobility shift assays were carried out using the protocol described previously (Cheng and Price, 2008).
Chromatin immunoprecipitation (ChIP) and sequencing
ChIP assays were performed using the protocol described by Lee et al. (Lee et al., 2006). HeLa cells were grown to 90% confluence and treated for one hour with FP (final concentration 1 μM with 0.1% DMSO) or 0.1% DMSO alone. For each immunoprecipitation, 5 × 107-1 × 108 cells were used. Cells were crosslinked with 1% paraformaldehyde for 15 minutes before being stopped with 125 mM glycine. Cells were washed twice with cold PBS and harvested by scraping. Cells for each immunoprecipitation were pelleted in PBS and incubated for 10 minutes each in 10 ml of Lysis Buffer 1 (50 mM HEPES-KOH, pH 7.5, 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40, and 0.25% Triton X-100), followed by 10 ml of Lysis Buffer 2 (10 mM Tris-HCl, pH 8.0, 100 mM NaCl and 1 mM EDTA), and finally sonicated in Lysis buffer 3 (10 mM Tris-HCl, pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.1% Sodium Deoxycholate, and 0.5% Sarkosyl) on ice using a Fisher Model 550 Sonic Dismembrator (Fisher Scientific) at a setting of 4 for eighteen 20-second pulses with 1 minute rests between pulses. After sonication, 1/10 volume of 10% Triton X-100 was added and then the samples were spun at 20,000 g for 10 minutes at 4°C.
For each immunoprecipitation 100 μl of Protein G Dynabeads (Invitrogen) were washed with Block Solution (0.5% BSA in 1× PBS) three times and resuspended in 250 μl Block Solution. The beads were incubated with 10 μg of Pol II (Santa Cruz, sc-899) or Gdown1 antibody at 4°C overnight. After 3 washes in Block Solution the beads were incubated with the sonicated cell lysate at 4°C overnight. The beads were then washed four times with RIPA buffer (50 mM HEPES-KOH, pH7.5, 100 mM NaCl, 1 mM EDTA, 500 mM LiCl, 1% [v/v] NP-40, 0.7% Sodium Deoxycholate), once with a buffer containing TE and 50 mM NaCl. Immunocomplexes were eluted for 30 minutes at 65°C with Elution buffer (1% SDS, 50 mM Tris-HCl, pH 8.0, 10 mM EDTA) and the eluted material incubated in Elution buffer overnight at 65oC. DNA fragments were size selected from an agarose gel, blunt-ended, ligated to the Solexa adaptors, and sequenced using the Illumina 1G Genome Analyzer as described previously (Barski et al., 2007; Rahl et al., 2010).
Data analysis
Raw sequences generated from Illumina/Solexa sequencer were aligned using ELAND software to NCBI Build 36.1 (UCSC hg18) of the human genome. To illustrate the entire DNA fragment, the 3′ end of each read was extended 200 bp. The reference genome was partitioned into 25 bp bins and the total reads (including partial reads) in each bin were summed and used to generate the visualization file in wiggle (WIG) format.
For Figures 5E, 6 and 7, a custom annotated RefSeq gene list was generated by merging the all TSSs for each gene that were within 500 bases of each other. Then genes with TSSs within 1000 bp of another TSS (6% of the total number) were removed from the list. After extension of each sequence by 200 bases the number of reads within 10,000 bases of the TSS of each gene in the custom gene list was tabulated. Similar analysis was applied to the location of the center of peaks generated from the peak finding algorithm for the region within 500 bp relative to the TSS. Heat maps were generated using the program R (www.r-project.org). Genes were rank ordered based on the sequence density for Pol II from -2K to +2K from the TSS. A peak finding algorithm (ChIP-Seq Peak) was designed to determine precise position and height of each significant peak (Li and Price, manuscript in preparation).
Supplementary Material
01
Acknowledgments
We wish to thank Peter Nagy, Lori Wallrath, Miles Pufall, and members of the Price, Young and Gnatt labs and for suggestions on the manuscript. We also thank R.G. Roeder for the initial suggestion that Gdown1 and TFIIF might compete for binding to Pol II. This work was supported by NIH grants GM35500 and AI074392 to D.H.P., NHGRI grant HG002668-05 to R.A.Y., and GM64474 to A.G.
Footnotes
Accession Numbers: The ChIP-Seq datasets are deposited in GEO under accession number GSE32442.
Supplementary Information: This document includes seven supplementary figures and complete experimental procedures.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adamson TE, Price DH. Cotranscriptional processing of Drosophila histone mRNAs. Mol Cell Biol. 2003;23:4046–4055. doi: 10.1128/MCB.23.12.4046-4055.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamson TE, Shore SM, Price DH. Analysis of RNA polymerase II elongation in vitro. Methods Enzymol. 2003;371:264–275. doi: 10.1016/S0076-6879(03)71019-2. [DOI] [PubMed] [Google Scholar]
- Adelman K, Marr MT, Werner J, Saunders A, Ni Z, Andrulis ED, Lis JT. Efficient release from promoter-proximal stall sites requires transcript cleavage factor TFIIS. Mol Cell. 2005;17:103–112. doi: 10.1016/j.molcel.2004.11.028. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Boettiger AN, Levine M. Synchronous and stochastic patterns of gene activation in the Drosophila embryo. Science. 2009;325:471–473. doi: 10.1126/science.1173976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers SA, Price JP, Cooper JJ, Li Q, Price DH. HEXIM2, a HEXIM1-related protein, regulates positive transcription elongation factor b through association with 7SK. J Biol Chem. 2005;280:16360–16367. doi: 10.1074/jbc.M500424200. [DOI] [PubMed] [Google Scholar]
- Chao SH, Price DH. Flavopiridol inactivates P-TEFb and blocks most RNA polymerase II transcription in vivo. J Biol Chem. 2001;276:31793–31799. doi: 10.1074/jbc.M102306200. [DOI] [PubMed] [Google Scholar]
- Cheng B, Price DH. Properties of RNA polymerase II elongation complexes before and after the P-TEFb-mediated transition into productive elongation. J Biol Chem. 2007;282:21901–21912. doi: 10.1074/jbc.M702936200. [DOI] [PubMed] [Google Scholar]
- Cheng B, Price DH. Analysis of factor interactions with RNA polymerase II elongation complexes using a new electrophoretic mobility shift assay. Nucleic Acids Res. 2008;36:e135. doi: 10.1093/nar/gkn630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng B, Price DH. Isolation and functional analysis of RNA polymerase II elongation complexes. Methods. 2009;48:346–352. doi: 10.1016/j.ymeth.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung AC, Cramer P. Structural basis of RNA polymerase II backtracking, arrest and reactivation. Nature. 2011;471:249–253. doi: 10.1038/nature09785. [DOI] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist DA, Nechaev S, Lee C, Ghosh SK, Collins JB, Li L, Gilmour DS, Adelman K. NELF-mediated stalling of Pol II can enhance gene expression by blocking promoter-proximal nucleosome assembly. Genes Dev. 2008;22:1921–1933. doi: 10.1101/gad.1643208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Price DH. Mechanism of DmS-II-mediated pause suppression by Drosophila RNA polymerase II. J Biol Chem. 1993;268:18762–18770. [PubMed] [Google Scholar]
- Hendrix DA, Hong JW, Zeitlinger J, Rokhsar DS, Levine MS. Promoter elements associated with RNA Pol II stalling in the Drosophila embryo. Proc Natl Acad Sci U S A. 2008;105:7762–7767. doi: 10.1073/pnas.0802406105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Malik S, Negroiu CC, Hubbard K, Velalar CN, Hampton B, Grosu D, Catalano J, Roeder RG, Gnatt A. A Mediator-responsive form of metazoan RNA polymerase II. Proc Natl Acad Sci U S A. 2006;103:9506–9511. doi: 10.1073/pnas.0603702103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izban MG, Luse DS. SII-facilitated transcript cleavage in RNA polymerase II complexes stalled early after initiation occurs in primarily dinucleotide increments. J Biol Chem. 1993;268:12864–12873. [PubMed] [Google Scholar]
- Jiang Y, Liu M, Spencer CA, Price DH. Involvement of transcription termination factor 2 in mitotic repression of transcription elongation. Mol Cell. 2004;14:375–385. doi: 10.1016/s1097-2765(04)00234-5. [DOI] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettenberger H, Armache KJ, Cramer P. Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol Cell. 2004;16:955–965. doi: 10.1016/j.molcel.2004.11.040. [DOI] [PubMed] [Google Scholar]
- Kornberg RD. The molecular basis of eukaryotic transcription. Proc Natl Acad Sci U S A. 2007;104:12955–12961. doi: 10.1073/pnas.0704138104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Li X, Hechmer A, Eisen M, Biggin MD, Venters BJ, Jiang C, Li J, Pugh BF, Gilmour DS. NELF and GAGA factor are linked to promoter-proximal pausing at many genes in Drosophila. Mol Cell Biol. 2008;28:3290–3300. doi: 10.1128/MCB.02224-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Johnstone SE, Young RA. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nature protocols. 2006;1:729–748. doi: 10.1038/nprot.2006.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luse DS, Spangler LC, Ujvari A. Efficient and rapid nucleosome traversal by RNA polymerase II depends on a combination of transcript elongation factors. The Journal of biological chemistry. 2011;286:6040–6048. doi: 10.1074/jbc.M110.174722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S, Barrero MJ, Jones T. Identification of a regulator of transcription elongation as an accessory factor for the human Mediator coactivator. Proc Natl Acad Sci U S A. 2007;104:6182–6187. doi: 10.1073/pnas.0608717104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall NF, Price DH. Control of formation of two distinct classes of RNA polymerase II elongation complexes. Mol Cell Biol. 1992;12:2078–2090. doi: 10.1128/mcb.12.5.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall NF, Price DH. Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J Biol Chem. 1995;270:12335–12338. doi: 10.1074/jbc.270.21.12335. [DOI] [PubMed] [Google Scholar]
- Moteki S, Price D. Functional coupling of capping and transcription of mRNA. Mol Cell. 2002;10:599–609. doi: 10.1016/s1097-2765(02)00660-3. [DOI] [PubMed] [Google Scholar]
- Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechaev S, Adelman K. Pol II waiting in the starting gates: Regulating the transition from transcription initiation into productive elongation. Biochim Biophys Acta. 2011;1809:34–45. doi: 10.1016/j.bbagrm.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechaev S, Fargo DC, dos Santos G, Liu L, Gao Y, Adelman K. Global analysis of short RNAs reveals widespread promoter-proximal stalling and arrest of Pol II in Drosophila. Science. 2010;327:335–338. doi: 10.1126/science.1181421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Z, Saunders A, Fuda NJ, Yao J, Suarez JR, Webb WW, Lis JT. P-TEFb is critical for the maturation of RNA polymerase II into productive elongation in vivo. Mol Cell Biol. 2008;28:1161–1170. doi: 10.1128/MCB.01859-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palangat M, Renner DB, Price DH, Landick R. A negative elongation factor for human RNA polymerase II inhibits the anti-arrest transcript-cleavage factor TFIIS. Proc Natl Acad Sci U S A. 2005;102:15036–15041. doi: 10.1073/pnas.0409405102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Liu M, Marion J, Zhu Y, Price DH. RNA polymerase II elongation control. Cold Spring Harb Symp Quant Biol. 1998;63:365–370. doi: 10.1101/sqb.1998.63.365. [DOI] [PubMed] [Google Scholar]
- Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Price DH, Sluder AE, Greenleaf AL. Dynamic interaction between a Drosophila transcription factor and RNA polymerase II. Mol Cell Biol. 1989;9:1465–1475. doi: 10.1128/mcb.9.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reines D. Elongation factor-dependent transcript shortening by template-engaged RNA polymerase II. J Biol Chem. 1992;267:3795–3800. [PMC free article] [PubMed] [Google Scholar]
- Renner DB, Yamaguchi Y, Wada T, Handa H, Price DH. A highly purified RNA polymerase II elongation control system. J Biol Chem. 2001;276:42601–42609. doi: 10.1074/jbc.M104967200. [DOI] [PubMed] [Google Scholar]
- Roginski RS, Mohan Raj BK, Birditt B, Rowen L. The human GRINL1A gene defines a complex transcription unit, an unusual form of gene organization in eukaryotes. Genomics. 2004;84:265–276. doi: 10.1016/j.ygeno.2004.04.004. [DOI] [PubMed] [Google Scholar]
- Romano G, Giordano A. Role of the cyclin-dependent kinase 9-related pathway in mammalian gene expression and human diseases. Cell Cycle. 2008;7:3664–3668. doi: 10.4161/cc.7.23.7122. [DOI] [PubMed] [Google Scholar]
- Saunders A, Core LJ, Lis JT. Breaking barriers to transcription elongation. Nat Rev Mol Cell Biol. 2006;7:557–567. doi: 10.1038/nrm1981. [DOI] [PubMed] [Google Scholar]
- Seila AC, Core LJ, Lis JT, Sharp PA. Divergent transcription: a new feature of active promoters. Cell Cycle. 2009;8:2557–2564. doi: 10.4161/cc.8.16.9305. [DOI] [PubMed] [Google Scholar]
- Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, Young RA. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet. 2007;39:1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Yik JH. The Yin and Yang of P-TEFb regulation: implications for human immunodeficiency virus gene expression and global control of cell growth and differentiation. Microbiol Mol Biol Rev. 2006;70:646–659. doi: 10.1128/MMBR.00011-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01