The Hsp60-(p.V98I) Mutation Associated with Hereditary Spastic Paraplegia SPG13 Compromises Chaperonin Function Both in Vitro and in Vivo (original) (raw)
Abstract
We have previously reported the association of a mutation (c.292G > A/p.V98I) in the human HSPD1 gene that encodes the mitochondrial Hsp60 chaperonin with a dominantly inherited form of hereditary spastic paraplegia. Here, we show that the purified Hsp60-(p.V98I) chaperonin displays decreased ATPase activity and exhibits a strongly reduced capacity to promote folding of denatured malate dehydrogenase in vitro. To test its_in vivo_ functions, we engineered a bacterial model system that lacks the endogenous chaperonin genes and harbors two plasmids carrying differentially inducible operons with human Hsp10 and wild-type Hsp60 or Hsp10 and Hsp60-(p.V98I), respectively. Ten hours after shutdown of the wild-type chaperonin operon and induction of the Hsp60-(p.V98I)/Hsp10 mutant operon, bacterial cell growth was strongly inhibited. No globally increased protein aggregation was observed, and microarray analyses showed that a number of genes involved in metabolic pathways, some of which are essential for robust aerobic growth, were strongly up-regulated in Hsp60-(p.V98I)-expressing bacteria, suggesting that the growth arrest was caused by defective folding of some essential proteins. Co-expression of Hsp60-(p.V98I) and wild-type Hsp60 exerted a dominant negative effect only when the chaperonin genes were expressed at relatively low levels. Based on our in vivo and in vitro data, we propose that the major effect of heterozygosity for the Hsp60-(p.V98I) mutation is a moderately decreased activity of chaperonin complexes composed of mixed wild-type and Hsp60-(p.V98I) mutant subunits.
Hereditary spastic paraplegia is a heterogenous disease characterized by gait disturbance owing to spasticity and weakness of the lower limbs (1). Age of onset is typically in the third or fourth decade of life. The disease is traditionally classified into “pure” and “complicated” forms depending on the presence of additional neurological or systemic abnormalities. The major pathological feature is a progressive degeneration of the longest axons of the descending (corticospinal) and the ascending (sensory) tracts in the spinal cord (2). Presently >30 gene loci have been mapped, and disease-associated mutations have been identified in 15 genes (3). Judging from known functions of proteins encoded by mutated genes, two major pathogenetic mechanisms have been envisaged: one affecting axonal trafficking (4) and one affecting mitochondrial function and endurance (5). However, because mitochondria use the axonal transportation systems and transportation requires energy, both mechanisms are clearly not mutually exclusive.
Previously, while analyzing a French family with autosomal dominant pure hereditary spastic paraplegia SPG13, we detected heterozygosity for a mutation (c.292G > C; p.V98I) in the HSPD1 gene, which encodes the mitochondrial Hsp60 chaperonin (6). The mutation co-segregated with the disease in the large pedigree. Functional testing using a genetic complementation assay in Escherichia coli showed that the Hsp60-(p.V98I) mutant protein was functionally compromised.
Hsp60 represents the large subunit of the mitochondrial Hsp60/Hsp10 chaperonin complex, homologue of the comprehensively investigated GroEL/GroES system from E. coli (7). The functional units of these chaperonins consist of heptameric rings of the large subunit (Hsp60 or GroEL), which function as either single rings or back-to-back double rings, depending on the organism. In a cyclic reaction, Hsp60 ring complexes bind one unfolded substrate protein per ring, followed by the binding of ATP and association with the heptameric ring of the co-chaperonin Hsp10 (GroES). This leads to sequestration of the substrate protein in the inner cavity of Hsp60 where, for a certain period of time, it can fold undisturbed by other cell components. Synchronous hydrolysis of ATP in all Hsp60 subunits results in the release of the substrate protein. Null mutations in the Hsp60 gene are lethal in Saccharomyces cerevisiae, Drosophila melanogaster, and E. coli (8–11). A detailed analysis of chaperonin substrates has been performed for E. coli GroEL, where it is estimated that 10–15% of the cytosolic proteins use GroEL for folding, and ∼300 of these proteins have been identified experimentally (12,13).
To shed light on the molecular mechanisms underlying spastic paraplegia due to the Hsp60-(p.V98I) mutation, we expressed and purified the corresponding mutant protein and analyzed its properties and ability to assist folding of a model protein in vitro. In addition, we have engineered a flexible_E. coli_ experimental system that has allowed us to monitor various phenotypes in E. coli and to study potential dominant negative effects of co-expression of mutant Hsp60-(p.V98I) and wild-type Hsp60.
EXPERIMENTAL PROCEDURES
_Purification of Hsp60 and Hsp10_—Derivatives of the pET3a-derived expression plasmid encoding the Hsp60-(p.G67S) variant (14) with wild-type (glycine at position 67) and Hsp60-(p.V98I) were generated by standard methods. A fragment comprising the groES and groEL genes was amplified from E. coli and inserted into pET24d. Recombinant proteins were expressed in E. coli BL21(DE3) (15) in 1 liter of shaking flasks by inducing (2 mm IPTG)3 protein expression at _A_532 = 0.8. After expression at 37 °C for 4 h cells were harvested by centrifugation and lysed by sonication. Following pelletting of cell debris by ultracentrifugation for 30 min at 45,000 × g, the protein solution was dialyzed and fractionated by cation exchange chromatography. Hsp60- or GroEL-containing fractions were pooled and dialyzed into 0.1 m Tris-HCl, 20 mm magnesium acetate, 0.3 m NaCl, 10 mm dithiothreitol, pH 7.5. The dialyzed samples were concentrated to 10 mg/ml, reconstituted (16), centrifuged, and heptameric or tetradecameric chaperonin-purified by using an Superose 6 gel filtration (GE-Healthcare, Hillerød). Final purity, as judged by SDS-PAGE, was >95%. The co-chaperonins GroES and Hsp10 were expressed and purified according to (17) and (18), respectively.
_In Vitro folding and ATPase Activity Assays_—Folding reactions were done in 0.1 m Tris-HCl (pH 7.4), 7 mm KCl, 7 mm MgCl2, 10 mm dithiothreitol (Buffer A). Porcine malate dehydrogenase (Roche Applied Science, 62.5 nm) was denatured in 10 mm HCl for 1 h and diluted 50-fold into Buffer A containing 1.3 μm chaperonin and 2.6 μm co-chaperonin. Concentrations are with respect to monomers. Following incubation at 37 °C for 5 min, folding was initiated by the addition of 0.5 mm ATP. Incubation was continued for 90–105 min. Aliquots were drawn at different time points, and the enzymatic activity was determined according to a previous study (19). ATPase activity was measured at room temperature in Buffer A containing 1.3 μm chaperonin, 2.6 μm co-chaperonin, and [γ-32P]ATP according to a previous study (20).
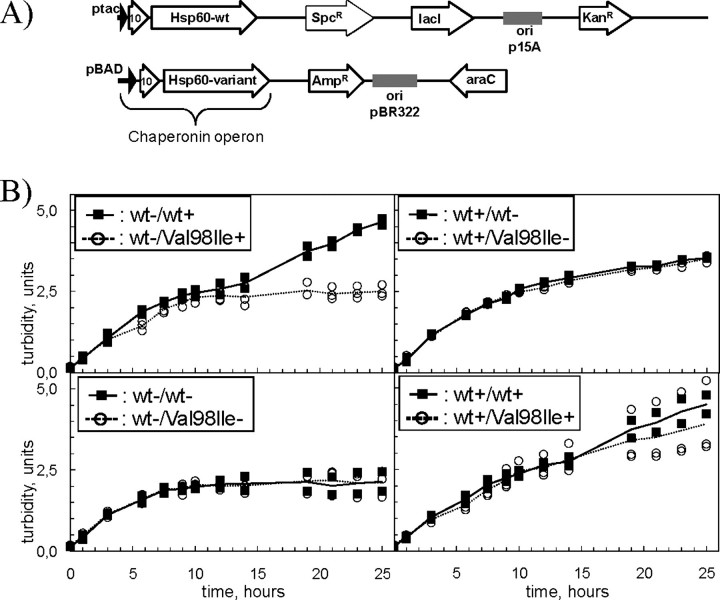
_Bacterial in Vivo System_—As starting material we used the B178 (groELS)-deleted strain that is maintained alive by the pOFX HSP60(wt)–HSP10(wt) plasmid (Hansen et al. 6). This strain was subsequently transformed with a second plasmid, a derivative of pBAD/hisA (Invitrogen), that contains an operon expressing Hsp10 and either wild-type or different mutant variants of Hsp60 under control of the arabinose-inducible BAD promoter (Fig. 2_A_).
FIGURE 2.
A, plasmids used for in vivo studies. E. coli B178 cells deleted for the groESL operon carried two independently replicating plasmids with different selection markers (KanR, kanamycin; AmpR, ampicillin; and SpcR, spectinomycin). Both plasmid types contain an operon with cDNA comprising human Hsp10 and the mature part of wild-type or mutant human Hsp60 under control of the IPTG-inducible tac promoter or the arabinose-inducible BAD promoter. The repressor genes for the tac and BAD promoters, lacI and_araC_, are also present on the respective plasmids. B, effects of mutant chaperonins on bacterial growth. B178 E. coli cells carrying a deletion of the groESL operon and transformed with different pairs of chaperonin plasmids (see Fig. 2_A_), were grown at 30 °C in medium supplemented with antibiotics and 0.1 mm IPTG. At an _A_532 ≈ 0.15 cells were harvested by centrifugation, resuspended in fresh medium, split into four aliquots, the respective inducers were added and growth was monitored for 25 h. The legend denotes induction (+) of the operons with wild-type (wt) or mutant (V98I) Hsp60 on the first/second plasmid or their repression (–).Curves link the average points of three individual transformants of cells carrying Hsp60-(p.V98I) (open circles, stippled line) and two individual transformants carrying wild-type wild-type Hsp60 (closed squares, solid line) on the second plasmid.
_Bacterial Growth Experiments_—Double-transformed cells were grown in dYT medium (21) supplemented with kanamycin (10 mg/liter), ampicillin (100 mg/liter), and IPTG (0.1 mm) at 30 °C in a shaking water bath. At_A_536 ≈ 0.1 bacteria were harvested by centrifugation at ambient temperature, resuspended in fresh medium, and split into aliquots, and the appropriate inducers were added.
_RNA and Protein Analyses_—Samples for RNA analyses were treated with RNAPROTECT (Qiagen), harvested by centrifugation, and frozen. Total RNA was purified using the RNeasy kit (Qiagen). Microarray analysis was performed using the E. coli antisense array from Affymetrix (Santa Clara, CA). cDNA synthesis, labeling, and hybridization were performed as recommended by the microarray manufacturer and normalized using GENECHIP (Affymetrix) software. Primary data sets are accessible through the Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo; accession no. GSE10974).
For qRT-PCR cDNA was synthesized from 0.5 μg of total RNA using the Advantage® RT-for-PCR kit (Clontech). Subsequent qRT-PCR analysis was performed using custom designed TaqMan® gene expression assays run on an ABI Prism® 7000 sequence detection system (Applied Biosystems). Relative transcript levels were calculated using the comparative CT method (22).
Samples for protein analysis (from 2–6 ml of culture) were harvested by centrifugation, frozen, and lysed as described previously (6). Gels were blotted to Immobilon polyvinylidene difluoride membranes (Millipore) and stained immunologically using the appropriate primary and secondary antibodies and ECLplus (GE-Healthcare) as recommended by the manufacturers. Antibodies directed against E. coli alkaline phosphatase were from ABCAM, Cambridge (ab354), and polyclonal antiserum against MetE was a generous gift of Dr. Elise Hondorp (University of Michigan).
_Measurement of Hsp60 by Immunoassay Based on xMAP Technology_—See supplemental data.
_Mass Spectrometric Quantitation of Peptide Variants_—For relative quantification of the Hsp60 peptide variants the region of the lane containing Hsp60 protein was excised from a Coomassie Blue-stained SDS-polyacrylamide gel, digested with trypsin (Promega, Madison, WI), and subsequently analyzed by reverse phase (3.5 μm Kromasil C18, Eka Chemicals, Bohus) liquid chromatography coupled to an LTQ Orbitrap mass spectrometer (Thermo Fisher Scientific, Waltham, MA). The peptide, L_X_QDVANNTNEEAGDGTTTATVLAR, corresponding to the wild-type (X = V) and mutant (X = I) variant eluted at ∼20 and 21% acetonitrile, respectively. The peptides were identified by tandem MS spectra (Sequest algorithm, Thermo Fisher Scientific) and quantified by integrated peak area, from full scan peptide signal, by Genesis algorithm in the Qual browser (Thermo Fisher Scientific).
RESULTS AND DISCUSSION
_Purified Hsp60-p.V98I Is Compromised in Its Ability to Promote Refolding in Vitro_—Following recombinant expression in E. coli, wild-type Hsp60, the disease-causing Hsp60-(p.V98I) mutant protein, and the variant Hsp60-(p.G67S) were purified as described under “Experimental Procedures.” The Hsp60-(p.G67S) variant was included in the analysis as an additional wild-type control, because this variation was present in a Hsp60 cDNA clone (23) and the corresponding variant protein has been analyzed by many laboratories as wild-type Hsp60. All Hsp60 proteins were first purified as monomers and, prior to in vitro refolding assays, assembled as native heptamer complexes that were re-purified by gel filtration chromatography (15,24). Native complexes were obtained in similar amounts for all three variant proteins (data not shown), demonstrating that Hsp60-(p.V98I) subunits are efficiently assembled into chaperonin ring complexes. Wild-type Hsp10 was purified separately.
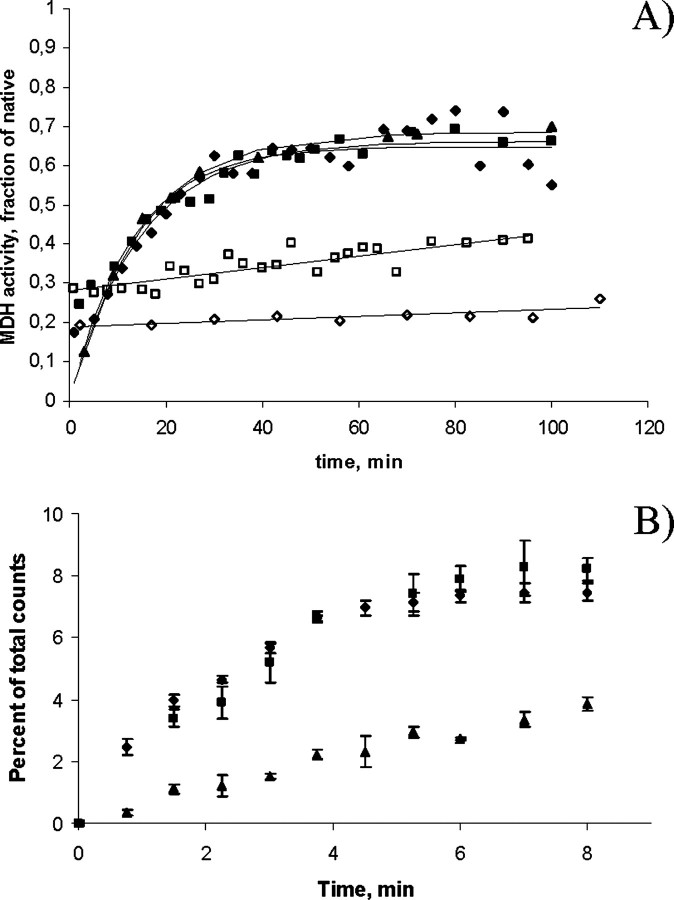
Fig. 1_A_ shows the ability of the various chaperonin complexes to refold denatured malate dehydrogenase. The refolding kinetics for wild-type Hsp60/Hsp10, Hsp60-(p.G67S)/Hsp10, and GroEL/GroES were very similar and resulted in 60–70% refolding. In contrast, Hsp60-(p.V98I)/Hsp10 displayed severely decreased refolding activity. Specifically, while the initial rate of folding was identical for GroEL/GroES, and the two wild-type Hsp60/Hsp10 controls, it was ∼100-fold lower for Hsp60-(p.V98I)/Hsp10. Furthermore, the ability to inhibit spontaneous refolding of malate dehydrogenase usually observed in the presence of surplus active chaperonins due to the binding and stabilization of the denatured protein (compare bovine serum albumin with wild-type chaperonins) was not apparent in the case of Hsp60-(p.V98I). This suggests that the conformational state of this mutant in the absence of ATP is limited in its ability to bind denatured protein. That this was not caused by differences in quaternary structure was documented by gel filtration (supplemental Fig. S1). However, the folding activity of Hsp60-(p.V98I) was not completely abolished, because the initial rate of refolding of denatured malate dehydrogenase in its presence was ∼4-fold higher than in the presence of the negative control albumin.
FIGURE 1.
A, refolding activity. Refolding of acid-denatured malate dehydrogenase by wild-type Hsp60/Hsp10 (closed diamonds), Hsp60-(p.G67S)/Hsp10 (closed boxes), GroEL/GroES (closed triangles), Hsp60-(p.V98I)/Hsp10 (open boxes), or bovine serum albumin (open diamonds) was measured as described under “Experimental Procedures.” Refolding rates were determined as the slopes of the initial linear part of the curves. B, ATPase activity. ATPase activity of wild-type Hsp60 (diamonds), Hsp60-(p.G67S) (boxes), and Hsp60-(p.V98I) (triangles) in the presence of Hsp10 was determined as described under “Experimental Procedures.” The initial rate for Hsp60-(p.V98I), as determined by the slope of the initial linear part of the curves, was 31% of that of the two wild-type controls.Error bars show the variation in five independent experiments.
In parallel experiments we measured the ATPase activity of all of our Hsp60 variants. We found that, although all exhibited ATPase activity, the initial rate for Hsp60-(p.V98I) was substantially reduced (31%) compared with that of the wild-type variants (Fig. 1_B_).
Taken together, the kinetic data demonstrate that the Hsp60-(p.V98I) mutant displays a reduced ATPase hydrolysis rate and a severely decreased, but not fully abolished folding activity. Valine 74 in GroEL, which corresponds to the mutated valine 98 in human Hsp60, is positioned in the equatorial domain, but distant from the ATP binding site. Therefore, the effect of the V98I mutation on ATPase activity is not due to direct interference with the ATPase active site. Rather, it may affect conformational changes in the ring that accompany ATP binding and precede ATP hydrolysis. It has been shown that GroEL subunits exhibit positive intra-ring cooperativity with respect to ATP binding and that mutations that decrease this cooperativity result in decreased folding rates (25).
_A Bacterial System for Investigating the in Vivo Effects of the Hsp60-(p. V98I) Variant_—To address the question of how the decreased chaperonin activity of the Hsp60-(p.V98I) variant protein affects a cellular system where folding of a subset of proteins depends on chaperonin activity, we engineered a well defined and easily manipulatable E. coli system in which expression of two plasmid-encoded chaperonin operons can be separately turned on and off. These cells carry a deletion of the essential endogenous groES/groEL genes and are maintained alive by a plasmid expressing an IPTG-inducible operon encoding human Hsp10 and the mature form of wild-type Hsp60 (Fig. 2_A_, upper panel). Into this strain we introduced a second plasmid, which is independently replicated and selectable, expressing Hsp10 and either wild-type or the respective mutant Hsp60 cDNA under control of an arabinose-inducible promoter (Fig. 2_A_,lower panel).
These two cell types, carrying either wild-type Hsp60 or Hsp60-(p.V98I) on the second plasmid, were grown in the presence of IPTG to specifically induce the expression of the wild-type chaperonin operon present on the first plasmid. At an optical density of ∼0.1, cultures were split into four aliquots, and the medium was replaced with fresh medium containing the appropriate inducers to specifically express the different chaperonin operons alone or in combination, or with no inducers to shut off chaperonin expression. Three independent transformants with Hsp60-(p.V98I) and two with wild-type Hsp60 on the second plasmid were investigated. When shifted to expression of the Hsp60-(p.V98I) chaperonin operon on the second plasmid, cells stopped growing after 8–10 h, whereas cells shifted to expression of the wild-type chaperonin operon on the second plasmid continued growth for at least 25 h (Fig. 2_B_, upper left panel). These results are in agreement with our previous finding that Hsp60-(p.V98I) is unable to support growth of E. coli cells lacking the endogenous chaperonin genes. Cultures depleted for chaperonin expression also stopped growing, yet at a somewhat earlier time point (Fig. 2_B_, lower left panel).
The growth curves of all investigated clones of the two cell types were very similar when only the wild-type chaperonin operon on the first plasmid was induced (Fig. 2_B_,upper right panel), showing that all wild-type constructs are fully functional and that the observed growth differences were indeed related to expression of the respective Hsp60 variant. Bacteria that were induced for both wild-type Hsp60 and Hsp60-(p.V98I) continued to grow for at least 25 h (Fig. 2_B_, lower right panel). Yet the growth properties of the three independent clones varied significantly, with two out of the three clones showing a clearly slower growth rate than the cells co-expressing two wild-type chaperonin operons.
Analysis of the protein profiles in the soluble and insoluble fractions (supplemental Fig. S2) showed distinct differences depending on the inducers added. However, there were no noticeable differences between the cells shifted to expression of the arabinose-induced Hsp60-(p.V98I) versus wild-type Hsp60 chaperonin operon. In particular, we noted no qualitative or quantitative increases in protein bands in the insoluble fraction of lysates from Hsp60-(p.V98I)-expressing cells. This indicates that no wholesale aggregation of proteins occurred in the cells expressing the Hsp60-(p.V98I) mutant protein, in contrast to the vast protein aggregation seen with a temperature-sensitive GroEL variant (26).
_Transcript Changes Related to Expression of Hsp60-(p.V98I)_—To identify regulatory responses that could give clues as to the cause(s) of the growth arrest observed in the Hsp60-(p.V98I)-expressing cells, we performed microarray analyses. We reasoned that the decreased chaperonin activity of the Hsp60-(p.V98I) protein would greatly impair or abolish folding of E. coli proteins that depend on chaperonin assistance. The resulting decreased levels of various activities may cause compensatory up-regulation or deregulation of genes. The transcriptomes of cells shifted to expression of the arabinose-inducible chaperonin operon with either the Hsp60-(p.V98I) mutant or the wild-type Hsp60 from the second plasmid were compared. RNA samples from three independent cultures expressing the mutant or two independent cultures expressing wild-type Hsp60 were pooled, respectively. We identified 61 genes (40 transcript units) that were up-regulated by a factor of ≥3 and 65 (61 transcript units) that were down-regulated by a factor of ≥3 (supplemental Tables S1 and S2).
Nine of the 61 strongly up-regulated genes have previously been characterized as essential for robust aerobic growth in rich media (27). Surprisingly, only two of the up-regulated genes (metF and _sdh_A) encode proteins that are obligatory GroEL substrates (class III), and one (gatD) represents a substrate that partially depends on GroEL (class II (13)). The metF gene and an additional 10 strongly up-regulated genes (comprising five transcription units) are controlled by the MetJ transcriptional regulator and eleven (comprising three transcriptional units) by the PhoB transcriptional activator. Strong up-regulation of the MetJ-repressor-controlled metE gene upon depletion or temperature-inactivation of GroEL has been reported in two studies from the Horwich laboratory (26,28). The MetJ protein acts both as a repressor in the presence of the downstream metabolite_S_-adenosylmethionine and as an activator in its absence. Decreased levels of MetF activity would result in reduced amounts of_S_-adenosylmethionine and cause increased transcription of MetJ/_S_-adenosylmethionine-repressed genes.
Surprisingly, genes that are strongly induced by heat shock (29), high level expression of a misfolded protein (30), or oxidative stress (31) were not among the ≥3-fold up-regulated genes in the cells shifted to Hsp60-(p.V98I) expression. An ∼2-fold up-regulation of the chaperone gene clpB and the protease gene lon and a moderate (<2-fold) up-regulation of the dnaK/dnaJ chaperone system and the hslU protease subunit genes were observed (data not shown). Nevertheless, this represents a much lower induction level than that observed by other authors upon heat shock or expression of a misfolding protein (29,30).
The ≥3-fold down-regulated genes encode a number of enzymes, transport proteins and regulators (supplemental Table S2). Interestingly, the regulators for the strongly up-regulated genes are mostly distinct from those of the strongly down-regulated ones. A number of both up- and down-regulated genes are controlled by the global regulator CRP (32), with CRP functioning as a repressor for the down-regulated genes and as an inducer for the respective up-regulated genes. Furthermore, transcript levels of several genes involved in response to acid stress (33,34) were strongly decreased in cells expressing Hsp60-(p.V98I). PhoP, which is a transcriptional regulator of this response, has been classified as a partially GroEL-dependent protein. Taken together, our transcript analyses suggest that the observed growth arrest is most likely due to impaired folding of a small number of proteins that are essential for robust growth, and it appears to occur before misfolded proteins accumulate.
_Effects of Hsp60-(p.V98I) Co-expression_—We determined the relative concentrations of total soluble Hsp60 protein in samples taken 10 h after inducer shift (Table 1). Hsp60 levels in the depleted cells dropped to 2–3 μg of Hsp60 per milligram of total protein, demarcating the lower threshold level for growth of E. coli. Cells only induced for the IPTG-inducible wild-type chaperonin operon displayed 3- to 4-fold higher Hsp60 levels, whereas cells only induced for the arabinose-inducible chaperonin operon displayed 10- to 30-fold higher levels. The values for the relative Hsp60 concentrations must be taken with caution as total protein concentration was determined with bovine serum albumin as standard likely leading to an overestimation of the values given by up to a factor of 2. Comparing the obtained values we noticed that in cells induced for both chaperonin operons the relative Hsp60 concentration was substantially higher than the sum of the levels in cells induced for each operon alone. The reason for this unexpected result is unknown.
TABLE 1.
Measurement of relative Hsp60 expression levels
Hsp60 levels in soluble extracts from the bacterial cells were measured by a quantitative immunoassay. Average concentrations and the range of concentrations in the individual samples (2 wt/wt and 3 wt/Val98Ile lysates, respectively) are given. The “genotype” with respect to the Hsp60 variant present on the first/second plasmid is indicated, and induction (+) or repression (–) of the chaperonin operon on the first/second plasmid is denoted.
| Induction | wt/wt, average (range) | wt/V981, average (range) |
|---|---|---|
| μ_g/mg_ | ||
| -/+ | 32.7 (21.7-43.6) | 96.0 (40.6-124) |
| +/- | 8.97 (5.35-12.6) | 9.40 (6.25-13.2) |
| -/- | 2.60 (2.45-2.75) | 2.16 (1.0-3.15) |
| +/+ | 213 (141-286) | 296 (165-385) |
To determine the fractions of wild-type Hsp60 and Hsp60-(p.V98I) protein in cells co-expressing both variants we performed mass spectrometry and quantitation of tryptic peptides in which the V98I mutation is situated (Table 2). Assuming that the two Hsp60 species associate as well with each other as with themselves, then the calculated relative amounts of heptameric chaperonin complexes consisting of wild-type subunits only were 12.27 μg/mg total soluble protein (Table 2). This is in the same range as in cells induced for wild-type Hsp60, and there may thus be a sufficient amount of chaperonin complexes consisting of wild-type subunits only to support growth.
TABLE 2.
Fraction of wild-type Hsp60 in cells carrying Hsp60-(p.V98I) on the second plasmid
The fraction of wild-type subunits was measured by mass spectrometry as described under “Experimental Procedures.” The relative amounts of complexes consisting of wild-type Hsp60 subunits only was calculated by multiplying the total Hsp60 protein concentration with the fraction of wild-type subunits taken to the power of seven.
| Induction | Fraction of wild-type Hsp60, average (range) | Calculated levels of pure wt complexes, average (range) |
|---|---|---|
| μ_g/mg soluble protein_ | ||
| -/+ | 0.138 (0.0936-0.160) | 6.35 × 10-4 (1.03 × 10-5-9.89 × 10-4) |
| +/+ | 0.620 (0.568-0.657) | 12.27 (3.22-18.43) |
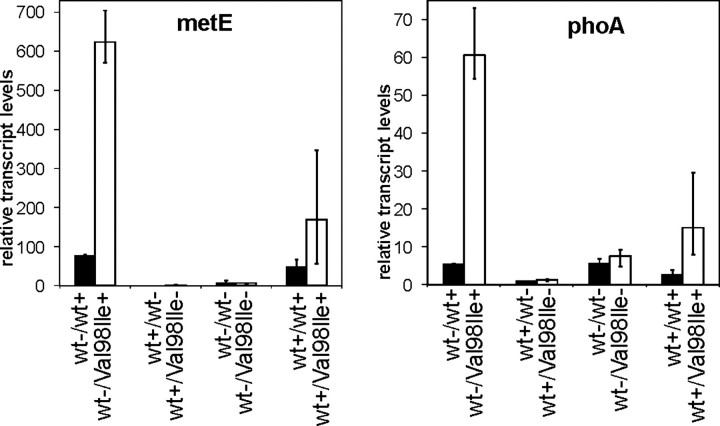
We chose the MetJ-controlled metE gene and the PhoB-regulated_phoA_ gene as representatives of genes strongly up-regulated in response to expression of malfunctioning Hsp60-(p.V98I) and determined their transcript levels in the individual cell cultures by qRT-PCR. Both_metE_ and pho_A transcripts were strongly up-regulated in all three cultures shifted to expression of the Hsp60-(p.V98I) operon (Fig. 3). Cultures depleted for chaperonins or cultures expressing only the wild-type chaperonin operon on the first plasmid displayed low levels of metE and phoA transcripts, and in these cases there was no significant difference between the cells harboring either wild-type or mutant Hsp60 on the second plasmid. Interestingly, cells in which wild-type Hsp60 and Hsp60-(p.V98I) were co-expressed had moderately increased levels of both metE and_phoA transcripts compared with cells expressing two wild-type chaperonin operons. This suggests that co-expression of Hsp60-(p.V98I) mutant subunits has some impact on the cells, in accordance with the moderate growth phenotype differences described above.
FIGURE 3.
Quantitative RT-PCR analysis of metE and phoA transcript levels. RNA purified from samples taken 10 h after inducer shift from the growth experiment shown inFig. 2_A_ were subjected to qRT-PCR as described under “Experimental Procedures.”metE (left) and phoA (right) transcript levels are given in relation to the endogenous reference gene yeeX and normalized to the average expression levels of cells carrying wild-type Hsp60 on the second plasmid and induced with IPTG only. Columns show the average values obtained from the three cultures harboring Hsp60-(p.V98I) (open columns, V98I) and two cultures harboring wild-type Hsp60 (closed columns, wt) cDNA, respectively, on the second plasmid. The legend denotes induction (+) of the operons with wild-type (wt) or mutant (V98I) Hsp60 on the first/second plasmid or their repression (–). Error bars denote the range of values measured in the biological replicates.
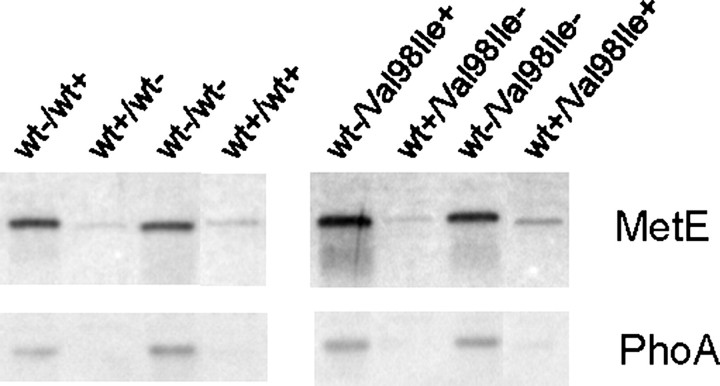
Analysis of MetE and PhoA protein levels by Western blotting showed that both proteins were indeed present at higher levels in the soluble extracts from cells shifted to expression of Hsp60-(p.V98I) (Fig. 4). However, scanning of the Western blots (data not shown) showed that the protein levels were increased to a lesser extent than the corresponding transcript levels.
FIGURE 4.
Western blot analysis of MetE and PhoA proteins. Soluble extracts were prepared from samples taken 10 h after inducer shift in the growth experiment shown in Fig. 2_B_. Aliquots corresponding to 0.5 μgof total soluble protein were subjected to SDS-PAGE followed by Western analysis with anti-MetE or anti-PhoA antibodies. Results from one representative of each cell type are shown. Induction (+) or repression (–) of the operons with wild-type (wt) or mutant (V98I) Hsp60 on the first/second plasmid are indicated below the lower graph.
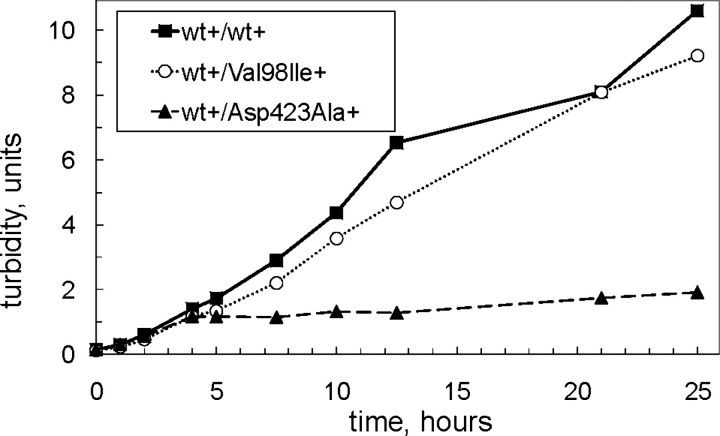
_Assessment of a Potential Dominant Negative Effect_—To further explore the impact of the Hsp60-(p.V98I) mutant, we monitored bacterial growth of cells co-expressing wild-type Hsp60 and Hsp60-(p.V98I) and cells co-expressing wild-type and an ATPase-deficient Hsp60 mutant (Hsp60-(p.D423A). The ATPase-deficient mutant was constructed in analogy to the D398A mutant of E. coli GroEL that alters a residue in the ATPase active site and has been shown to form chaperonin complexes that possess only ∼2% ATPase activity (35). Co-expression of the Hsp60-(p.D423A) active site mutant with wild-type Hsp60 resulted in a severe growth defect with characteristics comparable to those of chaperonin depletion (Fig. 5). In contrast to the active site mutant, co-expression of Hsp60-(p.V98I) with wild-type Hsp60 only moderately affected bacterial growth (see also growth curves from the experiment shown in Fig. 2_A_).
FIGURE 5.
Assessment of dominant negative effect of mutant co-expression. E. coli B178 cells carrying a deletion of the groESL genes and co-transformed with plasmid pairs carrying chaperonin operons with Hsp10 and wild-type Hsp60 on the first and Hsp10 and Hsp60-(p.V98I), Hsp10-Hsp60-(p.D423A) or wild-type Hsp60, respectively, on the second plasmid were grown up at 30 °C in medium supplemented with 0.1 mm IPTG. At _A_532 ≈ 0.15 cells were harvested by centrifugation and resuspended in fresh medium supplemented with both arabinose and IPTG. The legend denotes induction (+) or repression (–) of the indicated Hsp60 operons on the first/second plasmid.
Growth curves of cells co-expressing wild-type Hsp60 and a doubly mutated Hsp60-(p.V98I-D423A) variant protein showed defective growth in the same fashion as in cells co-expressing the wild-type Hsp60 and the single ATPase mutant (supplemental Fig. S3). This is consistent with the notion that Hsp60 subunits with the V98I mutation are freely incorporated into heterocomplexes with wild-type Hsp60 subunits.
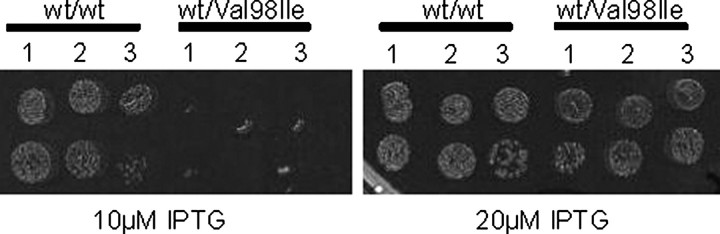
_In Vivo Demonstration of a Dosage Effect_—If high level co-expression of both wild-type Hsp60 and Hsp60-(p.V98I) operons from the two plasmids results in relatively moderate effects, it should be possible to identify in vivo conditions where the combined expression levels of two wild-type chaperonin operons can still support growth, whereas the corresponding combined levels of a wild-type and Hsp60-(p.V98I) mutant operon cannot. To find such in vivo conditions, we systematically varied both the growth temperature and the levels of wild-type Hsp60 and Hsp60-(p.V98I) by carefully titrating the two inducers. We found that, at a concentration of 0.04% arabinose and 20 μm IPTG, both cell types formed colonies at 40 °C (Fig. 6). However, upon a further reduction of the IPTG concentration to 10 μm, i.e. a further down-regulation of the IPTG-induced wild-type Hsp60 operon in both cell types, only the cells possessing an arabinose-induced wild-type chaperonin operon on the second plasmid formed colonies. This “dosage effect” is consistent with the notion that Hsp60 heterocomplexes possess decreased chaperonin activity such that at relatively low levels of wild-type chaperonin, a “poisonous” effect of the V98I mutant can be readily detected.
FIGURE 6.
Failure of cells co-expressing Hsp60-(pV98I) to form colonies under conditions of limiting Hsp60 levels. The groESL operon was deleted by phage transduction/recombination in E. coli LMG190 cells transformed with the plasmid carrying the IPTG-inducible chaperonin operon with wild-type Hsp60 as described previously (6). One of the selected clones was subsequently transformed with a second plasmid carrying arabinose-inducible chaperonin operons with either wild-type Hsp60 or Hsp60-(p.V98I). Three colonies of each cell type were grown up, and a dilution series was spotted on agar plates containing either 20 μm or 10 μm IPTG as indicated and 0.04% arabinose to partially induce the IPTG/arabinose inducible chaperonin operons. Plates were incubated overnight at 40 °C and photographed.
_Conclusions_—Based on our findings the V98I mutation affects the ATPase activity and results in a dramatically decreased folding activity of complexes consisting of the mutant subunits only. However, in contrast to an artificially constructed ATPase mutant, the V98I mutation exerts no salient dominant negative effect when co-expressed with wild-type Hsp60. Because no effect of the mutations on complex assembly was observed, we assume that heptameric complexes with all possible combinations and arrangements of subunits are formed. Incorporation of only one or two ATPase mutant subunits has a dramatic effect, whereas complexes composed of Hsp60-(p.V98I) and wild-type Hsp60 subunits appear to possess reduced but residual activity.
Regarding the situation in mitochondria of SPG13 patients that contain both wild-type and Hsp60-(p.V98I) mutant protein, our results suggest that subtly reduced chaperonin activity is the major effect. Such subtly reduced chaperonin activity will cause reduced folding and premature degradation of a subset of proteins, resulting among other things in decreased activities of certain enzymes. This subtle defect may be partially compensated in most tissues by up-regulation of the chaperonin genes, tuning of the transcription of other genes involved in mitochondrial protein quality control, or by increased expression of genes encoding the affected proteins. However, in mitochondria that are situated in the distal part of the body's longest axons, these compensatory mechanisms may be greatly limited due to the long distance (up to 1 meter) between axonal mitochondria and the cell body with the nucleus. The validity of our hypothesis will have to be eventually tested by studying animal models in which the appropriate tissues are accessible.
Supplementary Material
Supplemental Data
Acknowledgments
We thank Dr. Elise Hondorp, University of Michigan, for the kind gift of MetE antiserum and helpful discussions.
*
This work was supported by grants from the Ludvig og Sara Elsass Fond, the Lundbeck Foundation, the EU 6th Framework Program, the NOVO Nordisk Foundation, and the Swiss National Fund. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “_advertisement_” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available athttp://www.jbc.org) contains supplemental Figs. S1–S3, Tables S1 and S2, and additional text and references.
Footnotes
3
The abbreviations used are: IPTG, isopropyl 1-thio-β-d-galactopyranoside; qRT, quantitative reverse transcription.
References
- 1.Fink, J. K. (2006) Curr. Neurol. Neurosci. Rep. 6 65–76 [DOI] [PubMed] [Google Scholar]
- 2.Deluca, G. C., Ebers, G. C., and Esiri, M. M. (2004) Neuropathol. Appl. Neurobiol. 30 576–584 [DOI] [PubMed] [Google Scholar]
- 3.Depienne, C., Stevanin, G., Brice, A., and Durr, A. (2007) Curr. Opin. Neurol. 20 674–680 [DOI] [PubMed] [Google Scholar]
- 4.Crosby, A. H., and Proukakis, C. (2002) Am. J. Hum. Genet. 71 1009–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bross, P., Rugarli, E. I., Casari, G., and Langer, T. (2004) in Mitochondrial Function and Genetics (Koehler, C., and Bauer, M., eds) pp. 97–121, Springer, Heidelberg
- 6.Hansen, J. J., Dürr, A., Cournu-Rebeix, I., Georgopoulos, C., Ang, D., Nielsen, M. N., Davoine, C. S., Brice, A., Fontaine, B., Gregersen, N., and Bross, P. (2002) Am. J. Hum. Genet. 70 1328–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horwich, A. L., Farr, G. W., and Fenton, W. A. (2006) Chem. Rev. 106 1917–1930 [DOI] [PubMed] [Google Scholar]
- 8.Cheng, M. Y., Hartl, F. U., Martin, J., Pollock, R. A., Kalousek, F., Neupert, W., Hallberg, E. M., Hallberg, R. L., and Horwich, A. L. (1989) Nature 337 620–625 [DOI] [PubMed] [Google Scholar]
- 9.Hallberg, E. M., Shu, Y. M., and Hallberg, R. L. (1993) Mol. Cell. Biol. 13 3050–3057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perezgasga, L., Segovia, L., and Zurita, M. (1999) FEBS Lett. 456 269–273 [DOI] [PubMed] [Google Scholar]
- 11.Fayet, O., Ziegelhoffer, T., and Georgopoulos, C. (1989) J. Bacteriol. 171 1379–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ewalt, K. L., Hendrick, J. P., Houry, W. A., and Hartl, F. U. (1997) Cell 90 491–500 [DOI] [PubMed] [Google Scholar]
- 13.Kerner, M. J., Naylor, D. J., Ishihama, Y., Maier, T., Chang, H. C., Stines, A. P., Georgopoulos, C., Frishman, D., Hayer-Hartl, M., Mann, M., and Hartl, F. U. (2005) Cell 122 209–220 [DOI] [PubMed] [Google Scholar]
- 14.Studier, F. W., Rosenberg, A. H., Dunn, J. J., and Dubendorff, J. W. (1990) Methods Enzymol. 185 60–89 [DOI] [PubMed] [Google Scholar]
- 15.Viitanen, P. V., Lorimer, G., Bergmeier, W., Weiss, C., Kessel, M., and Goloubinoff, P. (1998) Methods Enzymol. 290 203–217 [DOI] [PubMed] [Google Scholar]
- 16.Kamireddi, M., Eisenstein, E., and Reddy, P. (1997) Protein Expr. Purif. 11 47–52 [DOI] [PubMed] [Google Scholar]
- 17.Dickson, R., Larsen, B., Viitanen, P. V., Tormey, M. B., Geske, J., Strange, R., and Bemis, L. T. (1994) J. Biol. Chem. 269 26858–26864 [PubMed] [Google Scholar]
- 18.Miller, A. D., Maghlaoui, K., Albanese, G., Kleinjan, D. A., and Smith, C. (1993) Biochem. J. 291 139–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carlier, M. F., and Pantaloni, D. (1981) Biochemistry 20 1918–1924 [DOI] [PubMed] [Google Scholar]
- 20.Miller, J. H. (1972) Experiments in Molecular Genetics, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
- 21.Corydon, T. J., Bross, P., Holst, H. U., Neve, S., Kristiansen, K., Gregersen, N., and Bolund, L. (1998) Biochem. J. 331 309–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Livak, K. J., and Schmittgen, T. D. (2001) Methods 25 402–408 [DOI] [PubMed] [Google Scholar]
- 23.Venner, T. J., Singh, B., and Gupta, R. S. (1990) DNA Cell Biol. 9 545–552 [DOI] [PubMed] [Google Scholar]
- 24.Nielsen, K. L., and Cowan, N. J. (1998) Mol. Cell 2 93–99 [DOI] [PubMed] [Google Scholar]
- 25.Danziger, O., Rivenzon-Segal, D., Wolf, S. G., and Horovitz, A. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 13797–13802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chapman, E., Farr, G. W., Usaite, R., Furtak, K., Fenton, W. A., Chaudhuri, T. K., Hondorp, E. R., Matthews, R. G., Wolf, S. G., Yates, J. R., Pypaert, M., and Horwich, A. L. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 15800–15805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gerdes, S. Y., Scholle, M. D., Campbell, J. W., Balazsi, G., Ravasz, E., Daugherty, M. D., Somera, A. L., Kyrpides, N. C., Anderson, I., Gelfand, M. S., Bhattacharya, A., Kapatral, V., D'Souza, M., Baev, M. V., Grechkin, Y., Mseeh, F., Fonstein, M. Y., Overbeek, R., Barabasi, A. L., Oltvai, Z. N., and Osterman, A. L. (2003) J. Bacteriol. 185 5673–5684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horwich, A. L., Low, K. B., Fenton, W. A., Hirshfield, I. N., and Furtak, K. (1993) Cell 74 909–917 [DOI] [PubMed] [Google Scholar]
- 29.Richmond, C. S., Glasner, J. D., Mau, R., Jin, H., and Blattner, F. R. (1999) Nucleic Acids Res. 27 3821–3835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lesley, S. A., Graziano, J., Cho, C. Y., Knuth, M. W., and Klock, H. E. (2002) Protein Eng. 15 153–160 [DOI] [PubMed] [Google Scholar]
- 31.Zheng, M., Wang, X., Templeton, L. J., Smulski, D. R., LaRossa, R. A., and Storz, G. (2001) J. Bacteriol. 183 4562–4570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martinez-Antonio, A., and Collado-Vides, J. (2003) Curr. Opin. Microbiol. 6 482–489 [DOI] [PubMed] [Google Scholar]
- 33.Bearson, S., Bearson, B., and Foster, J. W. (1997) FEMS Microbiol. Lett. 147 173–180 [DOI] [PubMed] [Google Scholar]
- 34.Zwir, I., Shin, D., Kato, A., Nishino, K., Latifi, T., Solomon, F., Hare, J. M., Huang, H., and Groisman, E. A. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 2862–2867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rye, H. S., Burston, S. G., Fenton, W. A., Beechem, J. M., Xu, Z. H., Sigler, P. B., and Horwich, A. L. (1997) Nature 388 792–798 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data