DsbA Plays a Critical and Multifaceted Role in the Production of Secreted Virulence Factors by the Phytopathogen Erwinia carotovora subsp. atroseptica (original) (raw)
Abstract
Erwinia carotovora subsp. atroseptica is an enterobacterial phytopathogen causing economically significant soft rot disease. Pathogenesis is mediated by multiple secreted virulence factors, many of which are secreted by the type II (Out) secretion system. DsbA catalyzes the introduction of disulfide bonds into periplasmic and secreted proteins. In this study, the extracellular proteome (secretome) of wild type E. carotovora subsp. atroseptica SCRI1043, and dsbA and_out_ mutants, was analyzed by spectral counting mass spectrometry. This revealed that dsbA inactivation had a huge impact on the secretome and identified diverse DsbA- and Out-dependent secreted proteins, representing known, predicted, and novel candidate virulence factors. Further characterization of the dsbA mutant showed that secreted enzyme activities, motility, production of the quorumsensing signal, and virulence were absent or substantially reduced. The impact of DsbA on secreted virulence factor production was mediated at multiple levels, including impacting on the Out secretion system and the virulence gene regulatory network. Transcriptome analyses revealed that the abundance of a broad, but defined, set of transcripts, including many virulence factors, was altered in the_dsbA_ mutant, identifying a new virulence regulon responsive to extracytoplasmic conditions. In conclusion, DsbA plays a crucial, multifaceted role in the pathogenesis of E. carotovora subsp.atroseptica.
The soft rot erwinias are Gram-negative, enteric bacteria and economically important plant pathogens. Erwinia carotovora subsp.atroseptica (also called Pectobacterium atrosepticum),Erwinia carotovora subsp. carotovora (also called_Pectobacterium carotovorum_), and Erwinia chrysanthemi (also called Dickeya sp.) cause soft rot disease of crop plants. E. carotovora subsp. atroseptica is exclusively a pathogen of potato, causing blackleg disease of stems and rotting of tubers (1,2). Production of secreted virulence factors is key to the pathogenesis of the soft rot erwinias. Their primary virulence characteristic is the coordinated production of large amounts of multiple secreted plant cell wall-degrading enzymes (PCWDEs),3 which leads to the breakdown of plant tissue and soft rot disease (2). The most important PCWDEs are the pectinases: primarily multiple isoforms of pectate lyase (Pel), together with isoforms of polygalacturonase (Peh), pectin methylesterase, and pectin lyase. Soft-rot erwinias also produce at least one secreted endoglucanase (cellulase, Cel) (2,3). In addition to these primary “brute force” secreted virulence factors, it is now clear that E. carotovora also produces other secreted virulence factors involved in more subtle interactions with the plant host, such as proteases, type III effectors, and the recently identified virulence factors Nip (necrosis-inducing protein) and Svx (3–6).
Consistent with this central role of protein secretion, the genome of_E. carotovora_ subsp. atroseptica SCRI1043 encodes all known major protein secretion systems (7). The type II secretion system (T2SS) of Erwinia, the Out system, is required for the secretion of the major PCWDEs, Pel, Cel, and Peh, and is thus required for virulence (3,8). The T2SS is a key conduit for secretion of virulence factors in many Gram-negative pathogens (9). Type II secretion is a two-step pathway in which proteins are exported to the periplasm via the Sec or Tat export machinery and are then secreted to the exterior of the cell by the T2SS apparatus, a complex multiprotein complex that spans the cell envelope (10). The secretion signal for T2SS substrates is poorly defined but is believed to involve recognition of folded structure motifs. In E. carotovora subsp.atroseptica, the Out T2SS is required not only for secretion of Pel and Cel but also for that of a putative proteoglycan hydrolase, ECA0852, and Svx (6). However, its full spectrum of substrates remains to be defined. Erwinia secreted proteases are secreted by a simple, one-step type I secretion system, well studied in Erwinia chrysanthemi, in which the substrate is moved directly from the cytoplasm to the cell exterior, bypassing the periplasm (11). In E. carotovora, production of PCWDEs and other secreted virulence factors is tightly regulated by N_-acyl homoserine lactone quorum sensing (AHL QS) and other environmental cues (12). QS is a mechanism of cell-cell communication in which a bacterial population coordinately regulates gene expression in response to cell density by the production and detection of chemical signals (13). In_E. carotovora subsp. atroseptica SCRI1043, QS is required for expression of PCWDEs and other secreted proteins and for virulence in potatoes (14). The AHL synthase, ExpI, produces the major AHL signal,_N_-3-oxohexanoyl-homoserine lactone (OHHL), which is freely diffusible. At high cell density, either in stationary phase in culture or at high population levels within a plant host, a threshold concentration of OHHL is achieved, and production of PCWDEs is activated. This activation is mediated by the LuxR family transcriptional regulator, VirR, which, directly or indirectly, represses expression of target genes at low OHHL levels (14).
Several studies have analyzed the extracellular proteome (secretome) of_E. carotovora_ subsp. atroseptica SCRI1043 (6,15) and E. chrysanthemi 3937 (16). These studies used two-dimensional gel-based methods to identify secreted proteins produced by the wild type under different conditions. In addition, a limited number of Out-dependent proteins (PelC, CelV, Svx, ECA0852, and ECA2220) and ExpI-dependent proteins were identified in E. carotovora subsp. atroseptica by comparison of the secretomes of Out and ExpI mutants with that of the wild type (6). However, examination of the genome sequence suggests that not all components of the E. carotovora subsp. atroseptica secretome have been identified by these studies. Flagellar motility is also a virulence factor in_Erwinia_ (1). Flagellar assembly involves movement of proteins to the exterior of the cell in a manner analogous to type III secretion, and flagellar proteins are detected at significant levels in the external milieu (17). Therefore, for the purposes of this study, flagellar proteins are also considered part of the secretome.
DsbA is a thiol-disulfide oxidoreductase that catalyzes disulfide bond formation in the periplasm. As reviewed previously (18,19), DsbA catalyzes the formation of disulfide bonds between pairs of cysteine residues in target proteins, allowing them to fold correctly. Once DsbA has donated a disulfide bond to its substrate protein, it is reoxidized by the membrane protein, DsbB, which in turn donates electrons to the electron transport chain. Incorrect disulfide bonds (which can occur in proteins with more than two Cys residues) are reshuffled by the disulfide isomerase, DsbC. DsbA is responsible for disulfide bonding in both resident periplasmic proteins and secreted proteins that access the periplasm, including some T2SS substrates. DsbA is required for the production of secreted virulence factors and/or for virulence in diverse pathogens. For example, it is required for cholera toxin assembly in_Vibrio cholerae_, secreted elastase production in Pseudomonas aeruginosa, type III secretion in Yersinia pestis, and adhesion (type IV pilus formation) in Escherichia coli (20,21). DsbA has also been implicated in virulence in Erwinia. In E. chrysanthemi, DsbA is required for the stability and secretion of CelZ and several Pels, and a_dsbA_ mutant is reduced in motility and tuber rotting (22). In Erwinia carotovora subsp. carotovora SCRI193, DsbA was shown to be required for secreted Pel and Peh activity and for virulence in potato tubers; in E. carotovora subsp. atroseptica SCRI1043, a_dsbA_ mutant was reduced in tuber rotting and motility but was not investigated further (23). However, these studies only examined the impact of DsbA on a small selection of known virulence determinants.
In this study, we undertook a global investigation into the role of DsbA in secreted protein production in E. carotovora subsp.atroseptica SCRI1043, including the use of state-of-the-art proteomic analyses to identify novel DsbA-dependent secreted proteins. Because DsbA and T2SSs can act in concert in the post-translational processing of secreted virulence factors, we also examined further the Out-dependent secretome and the relationship between DsbA and Out. We found that DsbA is required for the proper production of almost all secreted virulence factors in E. carotovora subsp. atroseptica SCRI1043 and that its effect on virulence factor production is mediated at multiple levels, from transcript to protein secretion. Moreover, we identified novel Out- and DsbA-dependent secreted proteins, representing known, predicted, and novel candidate virulence factors.
EXPERIMENTAL PROCEDURES
_Bacterial Strains, Plasmids, and Culture Conditions_—Bacterial strains and plasmids used in this study are detailed in supplemental Table 4. Erwinia strains were cultured with good aeration in Pel Minimal Medium (PMM) or Luria Broth (LB), at 25 °C (E. carotovora subsp. atroseptica) or 30 °C (E. carotovora subsp. carotovora) as described (24); overnight cultures were grown in LB. Growth was measured as optical density at 600 nm (OD600). When required, media were supplemented with antibiotics as follows: kanamycin 50 μg/ml, ampicillin 100 μg/ml, streptomycin 50 μg/ml, tetracycline 10 μg/ml, and chloramphenicol 25 μg/ml, or with synthetic OHHL (Sigma) 5 μm in dimethyl sulfoxide.
Construction of Strains and Plasmids_—_E. carotovora subsp. atroseptica chromosomal mutants defective in dsbA and_celV_ were constructed by marker (allelic) exchange using the suicide vector pKNG101 (25) as described (24). Briefly, the target gene was cloned and disrupted in pBluescript, the disrupted allele cloned into pKNG101, and the resulting plasmid introduced into E. carotovora subsp. atroseptica by conjugation. Selection on streptomycin and then high sucrose allowed isolation of mutants in which the disrupted allele had replaced the wild type copy. The primers used to amplify_dsbA_ and celV were SC80+SC81 and SC94+SC95, respectively, and plasmid details are given in supplemental Table 4. The integrity of mutants was confirmed by PCR and sequencing (data not shown). Strains SCC32 and SCC33 were made by introduction of the dsbA-uidA, CmR allele into strains TB6 and EMS6.1, respectively. For construction of the complementing plasmid, pSJC58, dsbA was amplified using primers SC117+SC118 and cloned into pQE80 on an EcoRI, BamHI fragment (DsbA is expressed from the vector promoter but is not His-tagged). Oligonucleotide primers were obtained from Sigma Genosys, and sequences are given in supplemental Table 3. Strain SCC20 was generated by transducing the_dsbA-uidA_, CmR allele from LS1A into MH1000, and strain SCC21 was generated by transducing the_carI_::Tn_phoA_′-2, TcR allele from M17 into SCC20, both using the E. carotovora subsp. carotovora generalized transducing phage φKP (26).
_Two-dimenstional Gel-based Secretome Analysis_—Secreted proteins were prepared from 150-ml cultures grown in PMM for 24 h at 25 °C. Secreted proteins were isolated as described (6). Briefly, following centrifugation to remove the cells, protein was precipitated from the supernatant using trichloroacetic acid to a concentration of 12.5% and collected by centrifugation. Following washing with 80% acetone, the pellet was air-dried and resuspended in 0.25 ml of CHAPS lysis buffer (8 m urea, 4% CHAPS, 5 mm magnesium acetate, 10 mm Tris-HCl, pH 8.0). Insoluble material was removed by centrifugation, and the pH of the soluble protein sample was adjusted to pH 8.5. Secreted protein samples from each strain (wild type, SCC22, and MC4) were compared using two-dimensional DiGE as described (24). 50 μg of each protein sample was labeled with a different Cy dye (Cy2, Cy3, or Cy5), and the samples were pooled and separated on a two-dimensional gel, and the fluorescent image from each channel was visualized. Three replicate gels varying the dye used for each sample gave identical results. Two-dimensional gel electrophoresis separation was performed using a linear pH 3–10 separation in the first dimension and 12% SDS-PAGE in the second. The DiGE comparison of wild type and SCC22 was performed on three independent samples with essentially identical results, and the MC4 secretome included for the three-way comparison was as reported previously (6). Proteins were identified by comparison with previous data and mass spectrometry (data not shown (6)).
_Secretome Analysis by Mass Spectrometry (Spectral Counting)_—Secreted protein samples were prepared from triplicate independent cultures of wild type, SCC22, and MC4 grown in PMM as above. Secreted protein samples were prepared as above, except that 50 μg of phosphorylase B was added to each sample, and total secreted proteins were resuspended in 0.5 ml of 2× gel sample buffer (100 mm Tris-HCl, pH 6.8, 3.2% SDS, 2.5 mm EDTA, 8% glycerol, 0.1 mg/ml bromphenol blue, 4% β-mercaptoethanol). 10 μl of each protein sample was separated on a 12% Tris-glycine SDS-PAGE mini-gel. Hence, because all cultures were harvested at the same optical density, total secreted protein from an equivalent number of cells was loaded for each strain (i.e. not normalized for amount of total protein). The gel was stained with colloidal Coomassie stain (34% methanol, 17% ammonium sulfate, 0.5% acetic acid, 0.1% Coomassie G-250), and a continuous vertical series of 20 2 × 2-mm gel pieces was cut from the middle of each lane (i.e. fractionating the protein sample by electrophoretic mobility). Ingel digestion was performed using a MassPrep station (PerkinElmer Life Sciences). Proteins within gel slices were reduced and alkylated using dithiothreitol and iodoacetamide, respectively, and then digested to peptides using trypsin. Resultant peptides were eluted from the gel pieces in 15 μl of 0.1% formic acid. 5 μl of this was injected onto a reverse phase column (15 cm, 75 μm internal diameter C18 PepMap column) attached to a 1200 liquid chromatography system (Agilent). The column eluate was sprayed into an LTQ linear ion trap mass spectrometer (Thermo, San Jose, CA). This LTQ was operated in triple play mode (for each ion of sufficient intensity, three scans were performed: a precursor scan, then a zoom scan to assign charge state, and finally an MSMS scan). Resulting data files from all gel fractions for each sample were converted to .dta format using Bioworks version 3.2 (Thermo), and the .dta files were merged to form a single .mgf file, using an inhouse script. The .mgf files were searched against a protein data base derived from the E. carotovora subsp. atroseptica SCRI1043 genome sequence (7) using MASCOT version 2.1.6 (Matrix Science, London, UK) with the following parameter settings: two miscleavages, variable methionine oxidation, carboxymethyl cysteine-fixed modification, and peptide tolerance of 1.0 Da. Results were exported from MASCOT to Excel (with significance threshold 0.05, ions score cut-off 10, standard protein scoring and require bold red), and all peptides not ranked 1 were excluded. The cutoff for genuine protein identifications, as opposed to false positives, was set by allowing 25% of identifications to score below the top Mowse score obtained when searching the output against a reversed data base. For each protein in each sample, the total number of spectra acquired (including redundant parent ions) for peptides belonging to that protein was summed to give the spectral count, a measure of relative protein abundance (27). Where the same peptide sequence was present in >1 protein, e.g. in the PelA and PelB isoforms, spectra from that peptide were excluded from the count. For each strain, the data from triplicate samples were combined; any proteins detected in only one of the three samples were excluded; and for each protein, the mean, standard error, and rank within that sample (according to abundance) were calculated.
_Spectophotometric Enzyme Activity Assays_—Supernatant samples were prepared by centrifugation to remove cells followed by careful removal of supernatant; samples were snap-frozen in liquid nitrogen if not assayed immediately. Cell-associated samples were prepared by isolating the cells from 1 ml of culture by centrifugation, resuspending the cells in 1 ml of fresh medium, sonicating for 30 s, and then snap-freezing. Protease, cellulase and pectate lyase were determined by measurement of azocasein, Ostazin Brilliant Red-cellulose, and sodium polygalacturonate breakdown, respectively, as described (24).
_Potato Virulence Assays_—Stem infection assays were performed as described (28). The stems of microplants of potato (cv. Estima) were stab-inoculated with 102 bacterial cells in 10 μl of phosphate-buffered saline, and the plants were maintained at 22 °C. Infected plants were scored over a 12-day period for symptom development, which was recorded as length of rot (mm). At least 12 replica plants were used for each strain, and Genstat for Windows version 6.1.0.200 (Lawes Agricultural Trust) was used for statistical analysis of variance, to determine the least significant difference (p < 0.05). Tuber rotting assays were performed using Maris Piper potatoes, surface-sterilized using 5% bleach, and stab-inoculated with 107 cells using a sterile pipette tip. Following inoculation, the tubers were incubated at 19 °C in a dark and moist environment for 7 days, and then the mass of rotted tissue was measured.
_Other Phenotypic Assays_—To measure OHHL levels, the pSB401-based, luminometric assay (29) was used. Cell-free supernatant samples were diluted 1/100 in LB, and 100 μl of the diluted samples were aliquoted into the wells of a black microtiter plate. The sensor strain E. coli JM109 (pSB401) was grown to an OD600 of 1, and 100 μl of this sensor culture was then added to each sample in the plate. Following incubation at 37 °C for 3 h, light production was measured using an Anthos LUCY1 luminometer. Synthetic OHHL and medium alone were used as positive and negative controls, respectively. Swimming motility was measured in motility agar (minimal medium {40 mm K2HPO4, 15 mm KH2PO4, 0.1% (NH4)2SO4, 0.4 mm MgSO4, 0.2% glucose} plus 0.1% casamino acids, 0.3% agar). Plates were inoculated with 4 μl of a 1/10 dilution (in phosphate-buffered saline) of an overnight culture, incubated at 25 °C for 18 h, and the area of the swim haloes observed. Where required, sterile copper II chloride was added to the stated final concentration. For measurement of β-glucuronidase activity, cells were permeabilized with toluene and added to 50 μl of 10 mm _p_-nitrophenyl β-d-glucuronide in a total volume of 500 μl of glucuronidase buffer, and the rate of increase of_A_405 (absorbance at 405 nm, Δ_A_405/min) at 37 °C was measured immediately. Glucuronidase buffer contained 50 mm sodium phosphate, pH 7, 1 mm EDTA, and 5 mm dithiothreitol, and glucuronidase activity was reported as Δ_A_405/min/ml/OD600.
_Anti-CelV Western Blot_—Cultures were grown for 24 h in PMM at 25 °C. Cell-associated protein samples were prepared as follows. Cells from 1 ml of culture were isolated by centrifugation, resuspended in 500 μl of 2× gel sample buffer (as above), and boiled for 10 min. Secreted proteins were precipitated from 1 ml of culture supernatant by addition of 1 ml of 1:1 chloroform/methanol, washed with 1 ml of methanol, air-dried, resuspended in 500 μl of 2× gel sample buffer, and boiled for 10 min. Protein samples (0.5 μl of cell-associated and 2 μl of secreted samples) were separated by 12% SDS-PAGE and electroblotted onto polyvinylidine fluoride (Millipore). CelV was detected by hybridization of the primary antibody, polyclonal rabbit anti-CelV (23), followed by the secondary antibody, horseradish peroxidase-conjugated goat anti-rabbit (Sigma), and then the use of an enhanced chemiluminescent detection kit (Millipore). The same results were obtained using replicate samples prepared from independent cultures.
_qRT-PCR Analysis_—RNA was prepared from four cultures of each strain grown to an OD600 of ∼3.4 in PMM at 25 °C using the Qiagen RNeasy kit and quantified using a NanoDrop spectrophotometer. cDNA synthesis and qRT-PCR analysis were performed as described (14). Data were normalized to 16 S rRNA as an internal control. All genes significantly altered were also significantly altered when dnaA (dsbA/WT in array = 1.0) was used as an alternative internal control (data not shown). Primers not described previously are shown in supplemental Table 3.
_Microarray Experiment_—The above RNA samples were also used for genome-wide transcriptional profiling using microarrays. RNA was hybridized to four E. carotovora subsp. atroseptica genomic arrays, with each array hybridizing one wild type and one dsbA mutant sample (from four independent cultures of each strain) and incorporating a dye-swap (i.e. wild type labeled with Cy3 in 2/4 and with Cy5 in 2/4). For each array, two total RNA samples (3 μg each) were indirectly labeled by reverse transcription using an 11-mer oligonucleotide mixture in the presence of amino-allyl dUTP, as described (30). Labeled cDNA samples were then coupled with Cy3 or Cy5 dye esters (GE Healthcare) and purified (30). Cy-labeled wild type and mutant cDNAs were co-hybridized to Agilent Technologies, Inc., custom ECA_11K Genome Arrays according to the manufacturer's instructions and according to Ref. 30. The probes (60-mers) include all 4521 predicted protein coding genes and 23 Lucidea Universal ScoreCard Controls (GE Healthcare), with each probe printed twice. Arrays were scanned using an ArrayWoRx Auto scanner (Applied Precision Inc.) with optimal exposure settings for each dye wavelength. Images were imported and aligned with probe position (Agilent GAL file) using GenePix Pro (version 6.1, Molecular Devices), and spot intensity data were imported into GeneSpring (version 7.3.1, Agilent Technologies Inc.). Microarray data were transformed to account for the dye-swap and normalized using default settings (LOWESS algorithm). Unreliable data (signal-background <50 in 50% replicates) were removed by filtering. Transcripts were considered to be significantly altered in abundance in the mutant if the mean normalized signal from their probes was increased or decreased by more than 2× in the mutant compared with the wild type, with a Student's t test p value <0.05 (the experiment contained four biological replicates). Microarray data has been deposited in the GEO data base with accession number GSE10647.
RESULTS AND DISCUSSION
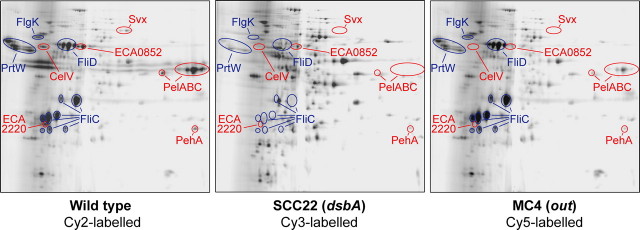
Inactivation of dsbA Has a Very Large Impact on the Secretome of E. carotovora subsp. atroseptica SCRI1043 as Visualized Using Two-dimensional DiGE_—A chromosomal insertion mutant of dsbA in E. carotovora subsp. atroseptica SCRI1043, strain SCC22, was constructed by marker exchange. As expected, this mutant showed negligible secreted Pel activity on indicator plates (data not shown) and was also impaired in motility (see below). The mutant showed no difference in growth rate compared with the wild type in several media (data not shown and see below). To assess the impact of dsbA inactivation on the extracellular protein profile (secretome) of E. carotovora subsp.atroseptica, an initial two-dimensional DiGE-based comparison of the secreted proteins of the wild type, dsbA, and out mutants was performed. MC4 is an out (T2SS) mutant of E. carotovora subsp. atroseptica SCRI1043 (6). As shown inFig. 1, all of the well known proteins in the E. carotovora subsp. atroseptica secretome were absent or greatly reduced in the dsbA mutant, e.g. PelABC, CelV, PrtW, Svx, ECA0852, and FliCD. Many of these proteins were also present at negligible levels in the out mutant e.g. PelC and Svx, but others were unaffected by Out inactivation, e.g. FliC and PrtW (6). It should be noted that many other spots only appear, or seem more abundant, in the dsbA mutant; this is because DiGE involves equal amounts of protein being labeled for each sample. In the dsbA mutant, the major secreted proteins were missing, and the total amount of secreted protein recovered was less than in the wild type, hence loading the same amount of protein in each channel artificially amplified the signal from contaminating cellular proteins. Because the secretomes of the three strains (wild type, SCC22, and MC4) were radically different, quantitative DiGE and quantitative mass spectrometry (MS) techniques, which rely on normalization over the whole sample, were judged to be inappropriate for a quantitative comparison of the three strains. In addition, secreted proteins of E. carotovora subsp.atroseptica tend to be resolved poorly by two-dimensional gels (e.g. the basic Pels streak across the gels, seeFig. 1). Hence we decided to utilize a semi-quantitative MS approach, spectral counting (27), a technique distinct from the two-dimensional gel-based methods used previously to study the secretome of Erwinia (6,15,16), to gain a more detailed description of the proteins present in the secretome of wild type E. carotovora subsp. atroseptica and in the dsbA and_out mutants.
FIGURE 1.
Inactivation of dsbA has a large impact on the secretome of_E. carotovora_ subsp. atroseptica SCRI1043 as shown by two-dimensional DiGE. Secreted proteins were prepared from wild type_E. carotovora_ subsp. atroseptica (Eca) SCRI1043 (left), SCC22 (dsbA, middle), and MC4 (out, right) and labeled with different Cy dyes as indicated. The pooled sample was then separated by two-dimensional SDS-PAGE (over pH 3–10), and the fluorescent images were visualized. Proteins absent or at greatly reduced levels in both the dsbA and the out mutants are labeled in_red_, whereas those absent or greatly reduced in the dsbA mutant but present in the out mutant are labeled in blue. Note that equal amounts of protein were labeled in each channel, although the total amount of secreted protein recovered was much lower in the dsbA mutant than the wild type.
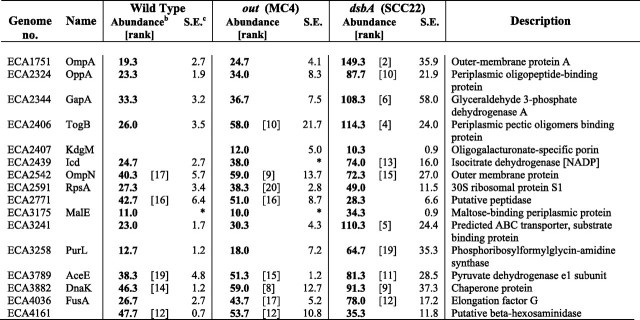
_Spectral Counting Mass Spectrometry Reveals Multiple DsbA-dependent and Out-dependent Proteins in the Secretome of E. carotovora subsp. atroseptica_—In a spectral counting approach, a complex mixture of proteins (here secretome samples) is analyzed by liquid chromatography-tandem mass spectrometry, following an initial one-dimensional gel fractionation step. For each protein, the total number of spectra acquired (total number of parent ions detected) by the mass spectrometer from peptides in that protein is counted. (Parent ions with the same peptide sequence and charge state can be counted multiple times, and the numbers of spectra from all the detectable peptides from a given protein are added together.) The total number of spectra observed for a given protein has been shown to be dependent on the relative abundance of that protein in a complex mixture (27). In this study, three independent secretome samples from each of the wild type, MC4 (out), and SCC22 (dsbA) were analyzed by spectral counting. Secreted protein samples derived from equivalent amounts of culture were compared (i.e. samples were not normalized for total protein amount). Almost 300 proteins were detected in the secretome of one or more of these strains (according to our quality criteria, see under “Experimental Procedures”) The results for all of these proteins are given in supplemental Table 1. Selected proteins, including the most abundant proteins in each strain, are detailed in Table 1.
TABLE 1.
Selected proteins present in the secretome of the wild type, out, and dsbA mutants of E. carotovora subsp. atroseptica (Eca) SCRI1043 as determined by mass spectrometry (spectral counting)a
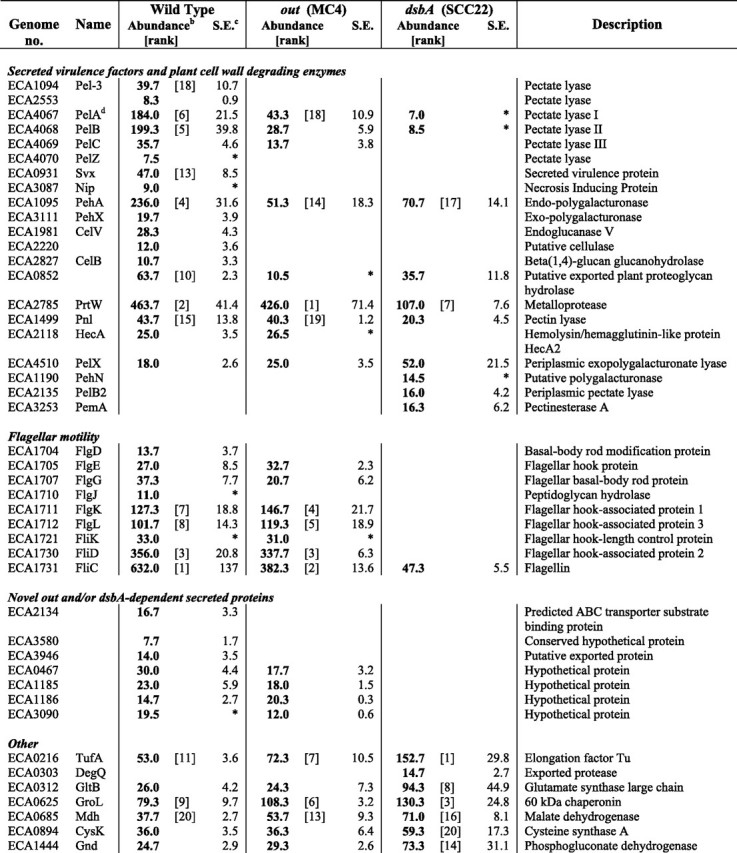
In the wild type secretome, many known or predicted secreted PCWDEs and other virulence factors were identified, including several PCWDEs detected for the first time in E. carotovora subsp. atroseptica (Table 1). The majority of the top-scoring identifications (most abundant proteins) were known or predicted secreted proteins and flagellar proteins. Several abundant periplasmic and cytoplasmic proteins were also in the top 20 identifications, assumed to be present in the “secretome” because of normal cellular lysis or leakage. Other, less abundant, known or candidate secreted proteins were also observed (Table 1). Proteins missing or greatly reduced in abundance in the out mutant revealed both new and previously described Out-dependent secreted proteins (see below). In the dsbA mutant, as suggested by the two-dimensional gel experiment (Fig. 1), all the major secreted and flagellar proteins were missing or greatly reduced; almost all (including the top-scoring) proteins identified were outer membrane, periplasmic, or cytoplasmic proteins. In general, cellular proteins were more abundant (or only seen) in the dsbA mutant compared with the other strains (Table 1). This is most likely due either to artificial amplification of these proteins during preparation (in the absence of large amounts of genuinely secreted proteins) and/or to the dsbA mutant cells being more “leaky” than the wild type. At least a partial contribution of the former effect seems likely as, in general, the out mutant (missing some but not all of the major secreted proteins) showed an intermediate level of cellular proteins between the dsbA mutant and the wild type, e.g. for RpsA and OppA.
Many secreted proteins were found to be both Out- and DsbA-dependent. Six secreted pectate lyase isoforms (PelABCZ, Pel-3, and ECA2553) were identified in the wild type secretome, and all were missing, or greatly reduced, in the_out_ mutant and missing, or negligible, in the dsbA mutant (Table 1). This confirms previous reports of secreted Pel enzymes being Out-dependent and having a requirement for DsbA-dependent disulfide bond formation for stable folding and secretion (6,22). The virulence factors, Svx and Nip, and the polygalacturonases, PehA and PehX (all with multiple Cys residues), were also shown to be Out- and DsbA-dependent. Interestingly, PehA levels were not reduced as dramatically in the dsbA mutant as the Pels and Svx (30% of wild type levels compared with < 5%), suggesting that it may not be destabilized to the same extent by loss of disulfide bonding. In addition, three known or predicted cellulases, CelV, CelB, and ECA2220, were identified in the secretome of the wild type but not that of the out and dsbA mutants, and the predicted proteoglycan hydrolase, ECA0852, was significantly decreased in the absence of Out or DsbA. This approach therefore identified multiple new substrates of the Out T2SS (Nip, PelABZ, PehAX, Pel-3, ECA2553, CelB, ECA2134, ECA3580, and ECA3496), including several that are not PCWDEs. Nip is an important virulence determinant in planta, and we hypothesized previously that its targeting might be Out-dependent (5). The cellular location of PehX has been controversial in E. chrysanthemi (16), but it was clearly secreted in an Out-dependent manner in E. carotovora subsp.atroseptica.
The major protease, PrtW, was not Out-dependent (as expected, see Ref.6), yet was reduced to 23% of wild type levels in the dsbA mutant. Similarly, pectin lyase levels were unaffected in the out mutant but were reduced (∼50%) in the_dsbA_ mutant. Pectin lyase is not expected to be secreted in an Out-dependent manner because it does not have a classical N-terminal signal sequence. However, in E. carotovora subsp. carotovora, it has been suggested to be efficiently released by stress-induced cellular lysis and thus to be present in the supernatant at reasonable abundance (31), consistent with the observations here. The HecA2 protein (see below) was also observed in the secretome, in a DsbAbut not an Out-dependent manner. There were also a few candidate PCWDEs identified only, or more strongly, in the dsbA samples and that are thus DsbA-independent: PelX, PehN, PelB2, PemA. These are known or likely to be periplasmic enzymes.
Importantly, several proteins of unknown function, ECA2134, ECA3580, and ECA3946, were shown to be present in the secretome in an Out- and DsbA-dependent manner, similar to many known secreted virulence factors. Thus these proteins represent novel candidate secreted virulence factors. Although ECA2134 is annotated as a putative ABC transporter periplasmic iron-binding protein, its Out dependence and the lack of any genes encoding other ABC transporter components nearby suggest otherwise (7). Its similarity to iron-binding proteins raises the possibility that it may represent a novel iron-binding secreted protein involved in host interaction. Four other novel DsbA-dependent exported proteins, possibly secreted, were observed: ECA0467, ECA1185, ECA1186, and ECA3090. Interestingly, the latter three were recently reported to be induced or repressed by plant extracts in the wild type secretome of E. carotovora subsp. atroseptica SCRI1043 (15), whereas the former (designated a hypothetical protein) has been seen for the first time in this study.
Overall, the spectral counting analysis identified _dsbA_- and_out_-dependent secreted proteins, and _dsbA_-dependent,_out_-independent secreted proteins, and confirmed that dsbA inactivation affects essentially the entire complement of major secreted proteins in E. carotovora subsp. atroseptica, a much wider ranging impact than was initially anticipated. This study represents the first report, to our knowledge, of a proteomic analysis of the extracellular proteins (secretome) of a DsbA mutant. However, proteomic studies to identify periplasmic DsbA-dependent proteins in E. coli (32,33) and Salmonella (34) have been described. These studies saw a FliC defect in the dsbA mutant similar to that observed in E. carotovora subsp. atroseptica, and also reported increased OsmY in an E. coli dsbA mutant, consistent with our observation of increased osmY expression in E. carotovora subsp. atroseptica (see below).
Production of Secreted Enzyme Activities, Production of OHHL and Motility Are Reduced or Eliminated by Loss of DsbA_—To confirm the proteomic results and to investigate further the massive impact of the_dsbA mutation on the extracellular proteome, production of secreted enzyme activities and motility were examined. Measurement of secreted Pel and endoglucanase (Cel) activity throughout growth showed that both activities were expressed in a typical growth phase-dependent manner in the wild type but were negligible in the dsbA mutant, consistent with the (virtual) absence of multiple Pels and Cels from the secretome (Fig. 2_A_). Secreted protease (Prt) activity in the dsbA mutant was reduced to one-third that of the wild type, consistent with the reduced levels of the major protease, PrtW, observed in the secretome (Fig. 2_A_). Production of all three secreted enzyme activities could be restored in the dsbA mutant by expression of DsbA in trans, confirming the requirement for DsbA (Fig. 2_B_).
FIGURE 2.
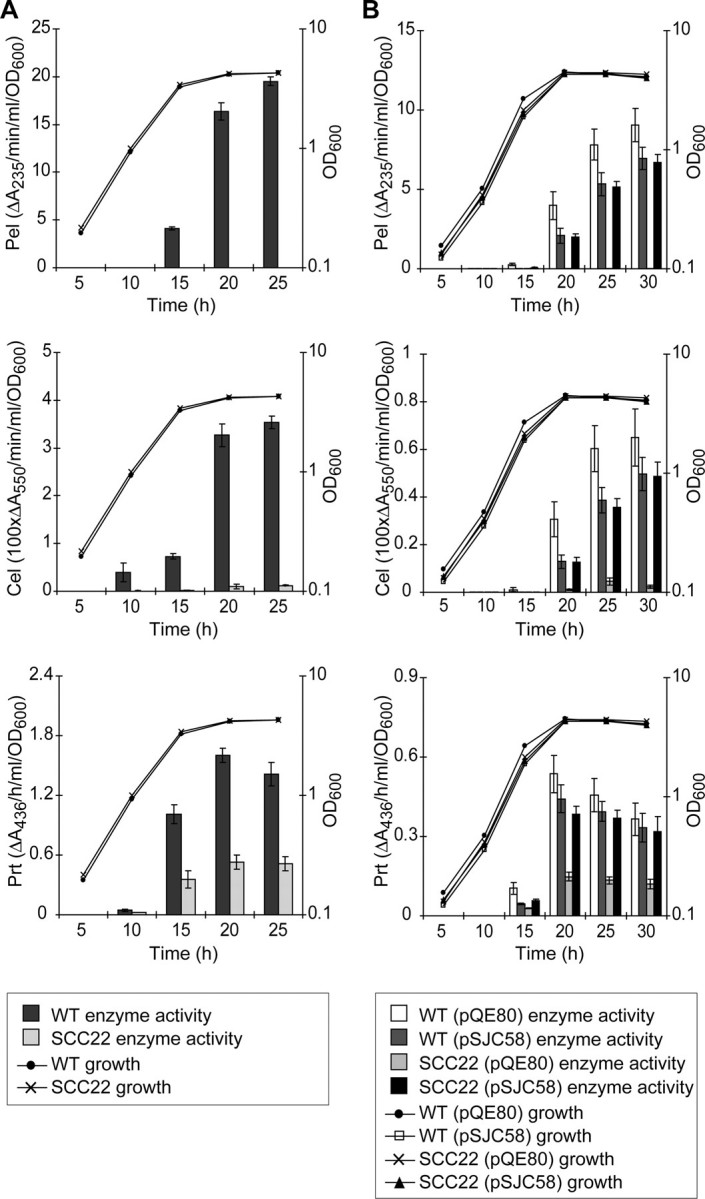
The effect of DsbA inactivation on the levels of secreted pectate lyase (Pel), endoglucanase (Cel), and protease (Prt) enzyme activity. A, levels of Pel (top), Cel (middle), and Prt (bottom) activity in the culture supernatant were determined at intervals throughout growth in PMM for wild type E. carotovora subsp. atroseptica SCRI1043 (WT, dark gray bars) and SCC22 (dsbA mutant, light gray bars).B, to demonstrate complementation of the dsbA phenotypes, levels of secreted Pel (top), Cel (middle), and Prt (bottom) were measured for WT (pQE80) (vector control, white bars), WT (pSJC58) (dsbA in trans, darker gray bars), SCC22 (pQE80) (lighter gray bars), and SCC22 (pSJC58) (black bars). Secreted enzyme activities were measured as described under “Experimental Procedures.” Growth is reported as OD600. A full key to symbols and shading is given at the base of each panel.Bars show mean ± S.E. (n ≥ 3).
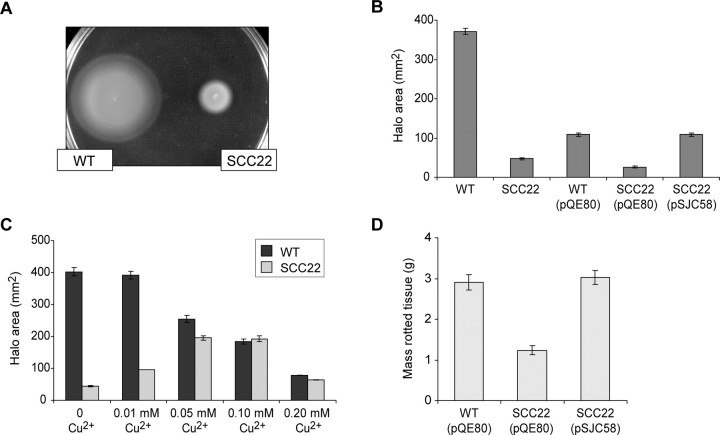
As part of the phenotypic characterization of the dsbA mutant, and because QS is central to secreted enzyme production in E. carotovora subsp. atroseptica (14), the production of the QS signal molecule, OHHL, was also measured in the dsbA mutant. Surprisingly, because the AHL synthase, ExpI, is a cytoplasmic enzyme, levels of OHHL were reduced in the dsbA mutant to approximately two-thirds that of wild type (Fig. 3_A_), and this decrease could be complemented by expression of dsbA in trans (Fig. 3_B_). Loss of swimming motility is a well known phenotype of dsbA mutants. As shown in Fig. 4, strain SCC22 failed to exhibit normal swimming motility in 0.3% agar, but motility was restored by expression of DsbA in trans. Chemical suppression of a dsbA mutation by the oxidant Cu2+ has been reported in E. coli (35). The dsbA mutation in E. carotovora subsp. atroseptica could also be chemically suppressed for the motility phenotype in a dose-dependent fashion by Cu2+ (Fig. 4_C_).
FIGURE 3.
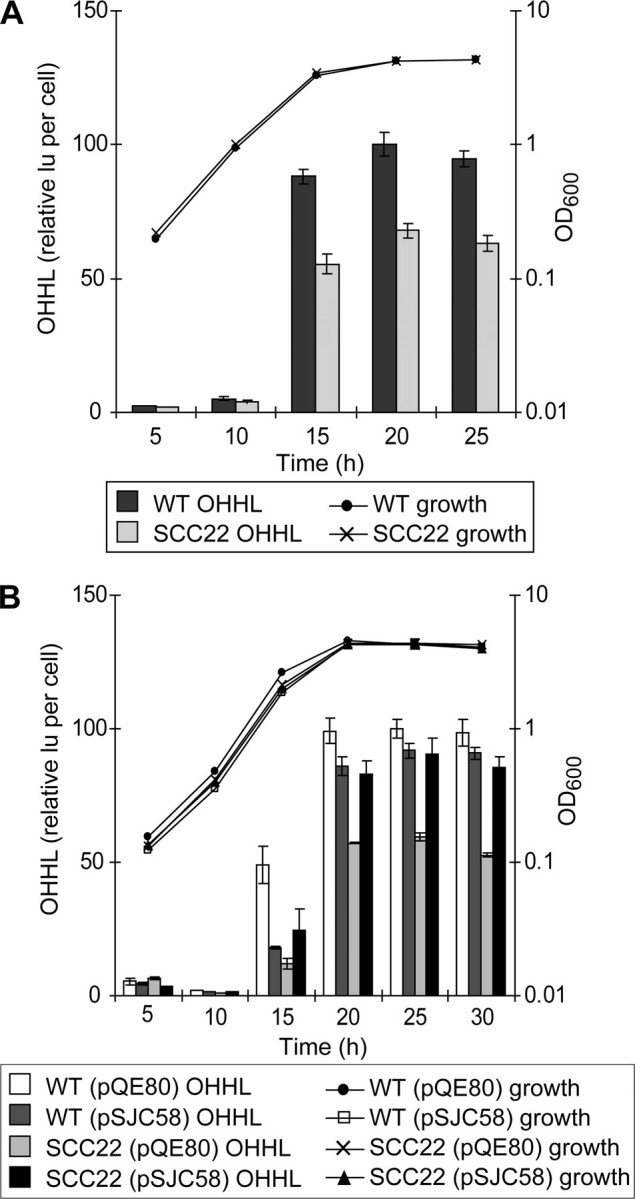
Production of OHHL is reduced in the dsbA mutant. A, levels of OHHL activity in the culture supernatant were determined at intervals throughout growth in PMM for wild type E. carotovora subsp. atroseptica SCRI1043 (WT, dark gray bars) and SCC22 (dsbA mutant, light gray bars). B, to demonstrate complementation, levels of OHHL were measured for WT (pQE80) (vector control,white bars), WT (pSJC58) (dsbA in trans, darker gray bars), SCC22 (pQE80) (lighter gray bars), and SCC22 (pSJC58) (black bars). OHHL levels were measured using the biosensor E. coli JM109 (pSB401) and are reported as light units (lu) per OD600, relative to WT at 20 h (A) or WT (pQE80) at 25 h (B) (with these values set to 100). Growth is reported as OD600. A full key to symbols and shading is given at the base of each panel. Bars show mean ± S.E. (n ≥ 3).
FIGURE 4.
The impact of DsbA inactivation on motility and virulence in potato tubers. A, motility plate (0.3% agar) showing the swim haloes formed by wild type E. carotovora subsp. atroseptica SCRI1043 (WT) and SCC22 (dsbA) after 18 h of incubation.B, motility of WT (pQE80) (vector control), SCC22 (pQE80), and SCC22 (pSJC58) (dsbA in trans). C, motility of wild type (dark gray bars) and SCC22 (light gray bars) in varying concentrations of Cu2+. B and C, motility is reported as halo area after 18 h of incubation; bars show mean ± S.E. (n ≥ 3). D, potato tuber virulence assay measuring the amount of rotted tissue generated by WT (pQE80), SCC22 (pQE80), and SCC22 (pSJC58) after 7 days of incubation (inoculum of 107 cells); bars show mean ± S.E. (n = 20).
The Virulence of the dsbA Mutant Is Severely Reduced in Planta_—The virulence in planta of strain SCC22 was compared with that of the wild type using a potato stem infection assay, designed to mimic Blackleg disease (28). The_dsbA mutant was found to be greatly reduced in lesion formation compared with the wild type. The overall wild type score was 27 mm, whereas that of SCC22 was 7.7 mm (the difference between the strains is statistically significant; least significant difference 5.7, p < 0.05). This is consistent with the impaired secretion of PCWDEs and other virulence proteins and lack of motility observed for this mutant in vitro. Interestingly, the dsbA mutant was not totally avirulent. The residual lesions could be due to slight environmental suppression of the_dsbA_ phenotype in an oxidative host environment,dsbA_-independent virulence factors such as the product of the up-regulated cfa gene cluster (see below), and/or small amounts of pectin lyase and other “leaking” cellular enzymes. The in planta virulence defect of the dsbA mutant was similarly observed in potato tubers and could be complemented by the expression of_dsbA in trans (Fig. 4_D_).
The Out Secretion System Is Impaired in the DsbA Mutant of E. carotovora subsp. atroseptica_—Next we investigated the cause(s) of the wide ranging impact of dsbA mutation on the levels of multiple extracellular proteins. The simplest reason for DsbA dependence is that the protein is a direct target of DsbA; it requires disulfide bond formation in the periplasm for stability and function, as has been shown for Pels in E. chrysanthemi (22). Good candidates for direct DsbA dependence identified in this study include the following: five secreted Pels, PehA, PehX, Svx, Nip, and ECA2220, all of which have multiple Cys residues and are Out-dependent (therefore will exist as periplasmic intermediates). Consistent with this, in addition to the loss of secreted Pel proteins and secreted Pel activity, negligible Pel activity was observed in the cell-associated fraction of the dsbA mutant (data not shown). However, many proteins reduced or absent in the secretome of the_dsbA mutant do not possess any cysteine residues and so cannot require disulfide bonding for stability and/or activity. These include CelV, CelB, ECA2134, and ECA0852, all of which were also Out-dependent. In fact, it was apparent from the secretomic data that all secreted proteins that were Out-dependent were also DsbA-dependent (Table 1). This strongly suggested that the Out secretion machinery was itself DsbA-dependent. To confirm this hypothesis, the major endoglucanase, CelV, was used as an example of a DsbA-dependent secreted protein lacking Cys residues.
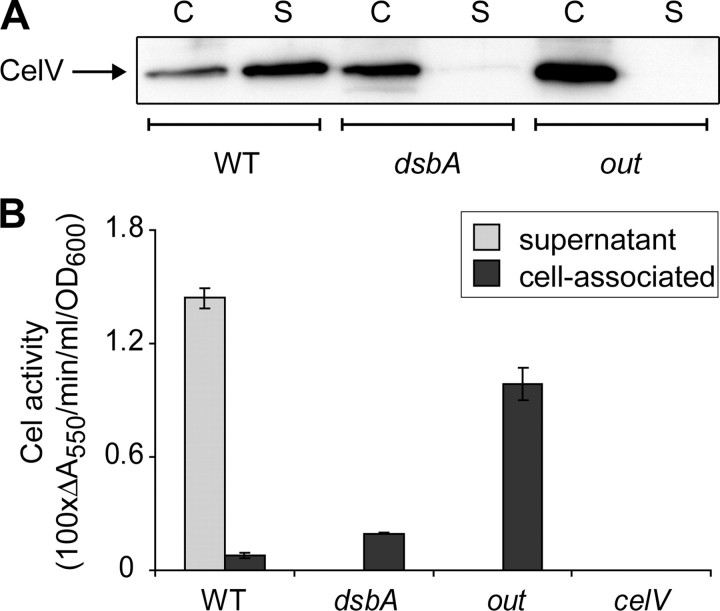
Western blotting with an anti-CelV antibody (36) was used to detect CelV in the supernatant and cell-associated (including cytoplasm and periplasm) fractions of wild type, dsbA, and out cultures grown to stationary phase in PMM. CelV was detectable in the supernatant of the wild type but not the dsbA or out mutants, confirming the proteomic findings. In contrast, CelV protein was detectable in the cell-associated fractions of all three strains (Fig. 5_A_). CelV was present in the cell-associated fraction at higher levels in the out mutant than the wild type, as expected from a mutant unable to secrete the protein. Similarly, in the dsbA mutant, CelV was present in the cell-associated fraction at higher levels than in the wild type, but it was not present in the supernatant, demonstrating a secretion defect in this mutant. Spectrophotometric Cel activity assays were performed on supernatant and cell-associated samples of wild type, dsbA, out, and_celV_ cultures. A defined celV mutant, SCC29 (see “Experimental Procedures” for construction), showed undetectable Cel activity, confirming that CelV was responsible for all detectable Cel activity in E. carotovora subsp. atroseptica under these conditions. Similar to the Western blot, Cel activity was detected in the supernatant only in the wild type strain but was detectable in the cell-associated fraction of the wild type, out, and dsbA strains; also more Cel activity was detectable in the cellular fractions of the out and dsbA mutants than in the wild type (Fig. 5_B_). Thus the cell-associated CelV in the dsbA and out mutants showed enzymatic activity, again consistent with a secretion defect. The reason why less cellular Cel/CelV was observed in the dsbA mutant than in the_out_ mutant is not clear, but it could be due to slightly reduced transcription (see below) and/or to greater activity of proteases in the “stressed” periplasm of the dsbA mutant.
FIGURE 5.
CelV is not secreted to the medium in the dsbA mutant. A, anti-CelV Western blot of proteins in the supernatant (S) and the cell-associated (C) fractions of wild type E. carotovora subsp. atroseptica SCRI1043 (WT), SCC22 (dsbA), and MC4 (out). Protein samples were prepared from equal volumes of culture (and resuspended in equal volumes of sample buffer) for each fraction and each strain (see “Experimental Procedures”). Protein samples were separated by 12% SDS-PAGE and immunoblotted with anti-CelV antibody; four times the amount of protein sample was loaded for the supernatant than cell-associated samples. B, endoglucanase (Cel) activity was determined for supernatant and cell-associated culture samples prepared from wild type E. carotovora subsp. atroseptica SCRI1043, SCC22, MC4, and SCC29 (celV), after growth for 24 h in PMM.Bars show mean ± S.E. (n = 3).
The Pul T2SS of Klebsiella oxytoca is also DsbA-dependent (37), and similarly, the Out system of E. chrysanthemi is unable to secrete the Cys-free substrate, RhiE, in a dsbA mutant (38). In contrast, we concluded previously that the Out system in E. carotovora subsp.carotovora SCRI193 was not DsbA-dependent, because some CelV, also lacking Cys residues, could be secreted in the dsbA mutant (23). However, in this earlier study, less than half the total CelV produced was secreted, in contrast with 80% secretion in the wild type, suggesting that Out function was impaired, although not eliminated. The reason why Out function was not eliminated in the earlier study is likely to be because it was performed in media containing yeast extract, whereas in this study a minimal medium was used. It has been reported that oxidizing compounds (e.g. cystine) in rich media can at least partially suppress some phenotypes of dsbA mutants by nonspecific oxidation (32). Indeed we observed that the motility defect of the E. carotovora subsp. atroseptica dsbA mutant was much less severe when the swim agar contained tryptone (data not shown). However, we are unable to rule out the possibility of a difference in behavior between the subspecies. Examination of the predicted Out protein sequences in E. carotovora subsp. atroseptica SCRI1043 revealed that only two Out proteins, the minor pseudopilin, OutK, and the secretin “pilot” protein, OutS, have Cys residues predicted to be suitably located in the periplasm. Thus these two proteins are candidates for DsbA-dependent disulfide bonding. Consistent with this prediction, both PulK and PulS, the corresponding T2SS components in K. oxytoca, are disulfide-bonded, and at least the disulfide bond in PulK is required for T2SS function (39).
Reduced PrtW and OHHL Are Because of a Transcriptional Effect of dsbA Mutation_—The impairment of Out secretion in the dsbA mutant is able to explain the absence of certain Cys-free proteins from the secretome of the dsbA mutant, e.g. CelV. However, other Cys-free, Out-independent proteins were also affected in the dsbA mutant. For example, levels of the major secreted protease, PrtW, and secreted Prt activity were significantly reduced in the dsbA mutant (Figs.1 and2;Table 1). PrtW contains no cysteines, is Out-independent, and by analogy with E. chrysanthemi, is predicted to be secreted by the adjacent type I secretion system (whose periplasmic exposed components, PrtE and PrtF, also lack Cys residues) and hence would not access the periplasm. To determine whether the PrtW defect was at the level of transcript abundance, qRT-PCR was used to compare the level of_prtW mRNA between the wild type and the dsbA mutant. The abundance of prtW mRNA was indeed reduced 4-fold in the dsbA mutant compared with the wild type (Table 2), consistent with the observed 3–4-fold reduction in secreted PrtW/Prt activity (Fig. 2_A_; Table 1). The cytoplasmic AHL synthase, ExpI, would also not depend on DsbA for activity, yet levels of OHHL were reduced in the dsbA mutant to 66% of the wild type (Fig. 3). qRT-PCR analysis of the expI transcript showed that it was reduced to 64% of wild type levels in the dsbA mutant (Table 2), providing an explanation for the observed decrease in OHHL levels. Given these results, and because altered transcription of three secreted enzyme genes in a_dsbA_ mutant had been suggested previously (23), we examined other related genes by qRT-PCR, including those encoding dsbA_-dependent secreted proteins. As shown in Table 2, the transcript levels of pelC, pehA, svx, and_celB, were decreased 2–5 -fold in the dsbA mutant compared with the wild type. The transcripts encoding the transcriptional regulator, Hor, and the dsbA_-dependent conserved hypothetical protein, ECA2134, were also significantly decreased in the dsbA mutant compared with the wild type. In addition, nip and_celV were decreased to 50 and 75% of wild type levels, respectively, although the results did not meet the p < 0.05 significance threshold. Control genes, including rpoS, recA_, and dnaA, were not significantly affected in the dsbA mutant (data not shown), although rpoA was found to be >2× increased in the_dsbA mutant (Table 2).
TABLE 2.
Transcripts altered in abundance in the dsbA mutant of E. carotovora subsp. atroseptica SCRI1043 compared with the wild type by quantitative RT-PCR analysis
| Gene | Genome identifier | Relative abundance (dsbA/WT)a | p valueb |
|---|---|---|---|
| prtW | ECA2785 | 0.25 | 0.0003 |
| celB | ECA2827 | 0.22 | 0.0006 |
| pelC | ECA4069 | 0.47 | 0.0163 |
| pehA | ECA1095 | 0.39 | 0.0410 |
| svx | ECA0931 | 0.35 | 0.0004 |
| expI | ECA0105 | 0.64 | 0.0354 |
| hor | ECA1931 | 0.36 | 0.0150 |
| ECA2134 | ECA2134 | 0.37 | 0.0006 |
| fliC | ECA1731 | 0.039 | 0.0038 |
| rpoA | ECA4006 | 2.3 | 0.0024 |
| cfl | ECA0609 | 19.9 | 0.0009 |
| nip | ECA3087 | 0.50 | 0.0790 |
| celV | ECA1981 | 0.75 | 0.0684 |
| dnaA | ECA4441 | 1.1 | 0.1480 |
Most of the above genes found to have reduced transcript levels in the_dsbA_ mutant are known to be regulated by QS in E. carotovora subsp. atroseptica (6,14). It was therefore possible that the reduced transcript levels of these genes (and hence also the reduced secreted PrtW) was due simply to the decreased OHHL levels observed in the_dsbA_ mutant (Fig. 3). However, addition of 5 μm OHHL to the culture medium had no effect on either the reduced levels of secreted Prt or the reduced transcript levels in the dsbA mutant (data not shown). We noted that not only did the set of genes influenced by dsbA and expI in E. carotovora appear to overlap (this study and Refs.14,23) but also that the QS dependence of dsbA expression had never been tested in the model_E. carotovora_ subsp. carotovora strain SCRI193. To examine further a possible relationship between dsbA and expI in_Erwinia_, the QS dependence of dsbA expression was determined in both E. carotovora subsp. carotovora SCRI193 and E. carotovora subsp. atroseptica SCRI1043 using dsbA-uidA transcriptional fusions. In both E. carotovora subsp.carotovora and E. carotovora subsp. atroseptica, expression of dsbA-uidA throughout growth was modestly reduced in the QS mutant and could be restored by the addition of OHHL (Fig. 6). Interestingly, expression of an expI-lacZ fusion was unaffected in the dsbA mutant of either strain (data not shown), suggesting that the dsbA impact on expI transcript levels (Table 2) might be post-transcriptional. Although the transcriptional impact of dsbA inactivation was independent of QS, modulation of OHHL levels in the presence of functional DsbA might be expected to “fine-tune” the regulatory system in vivo. Similarly, the modulation of dsbA expression by QS might provide a means of “topping-up” DsbA protein levels to cope with the simultaneous increase in substrate expression.
FIGURE 6.
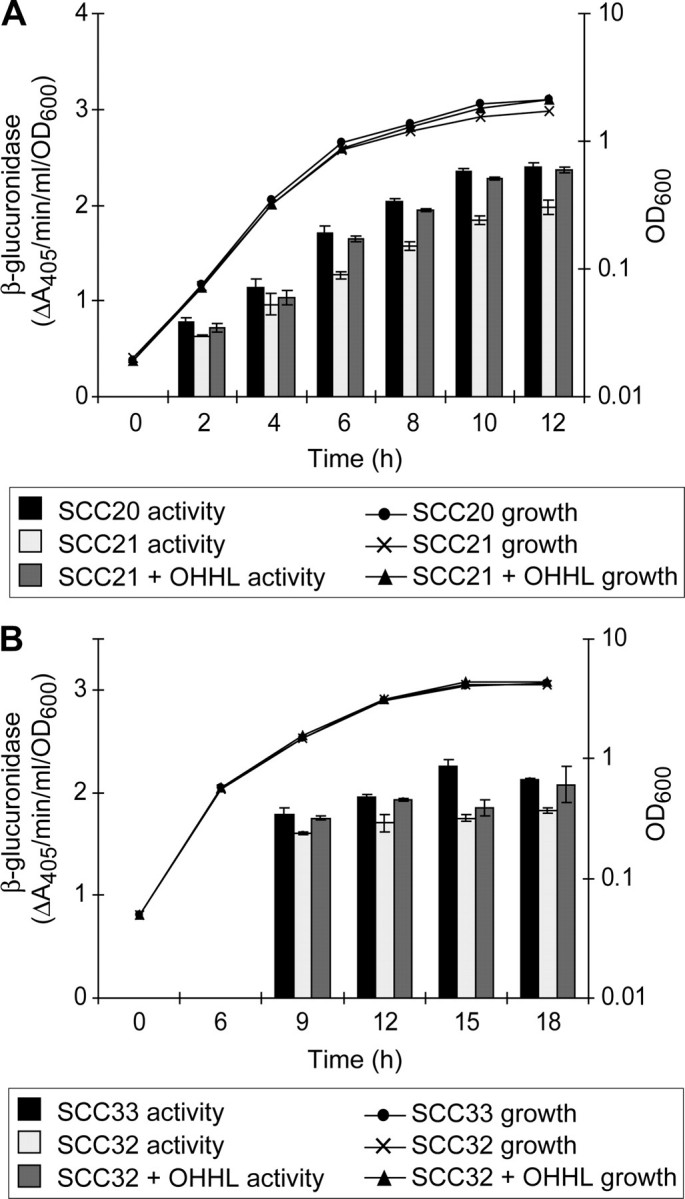
Expression of a dsbA-uidA fusion is modulated by quorum sensing. A, expression of dsbA-uidA in E. carotovora subsp. carotovora SCRI193 was measured during growth in LB in a wild type background (SCC20, dsbA-uidA, black bars), a_carI_ (expI) background (SCC21, dsbA-uidA, carI, light gray bars), and in SCC21 with exogenous 5 μm OHHL (SCC21 + OHHL, dark gray bars). B, expression of dsbA-uidA in E. carotovora subsp. atroseptica SCRI1043 was measured in PMM in a wild type background (SCC33, dsbA-uidA, black bars), an_expI_ background (SCC32, dsbA-uidA, expI, light gray bars), and in SCC32 with 5 μm OHHL added (SCC21 + OHHL, dark gray bars). Expression of dsbA-uidA was measured as β-glucuronidase activity per cell and growth reported as OD600. A full key to symbols and shading is given at the base of the figure. Bars show mean ± S.E. (n = 3).
_Microarray Analysis Confirms a Pleiotropic Transcriptional Impact of DsbA Inactivation_—The qRT-PCR experiments, targeting specific genes of interest, showed that absence of DsbA produced a transcriptional effect on multiple genes. This suggested that DsbA might influence the transcription of an even larger set of genes. To test this hypothesis, a genome-wide transcriptional profiling experiment to compare the dsbA mutant with the wild type was performed using an E. carotovora subsp.atroseptica SCRI1043 microarray. This confirmed that DsbA does indeed exert a wide ranging but defined transcriptional influence. In total, 184 genes showed significantly altered transcript levels (>2× change,p < 0.05) in the dsbA mutant compared with the wild type, with 91 decreased and 93 increased in the mutant (supplemental Table 2). Details of selected genes altered in the dsbA mutant are given inTable 3.
TABLE 3.
Selected genes with transcripts altered in abundance in the dsbA mutant of E. carotovora subsp. atroseptica SCRI1043 compared with the wild type by microarray analysis
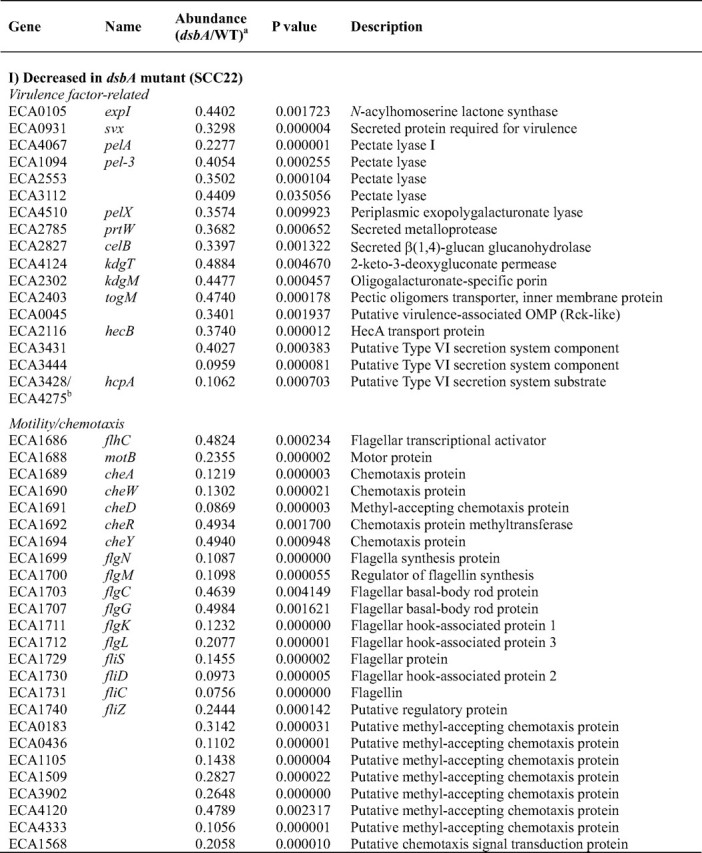
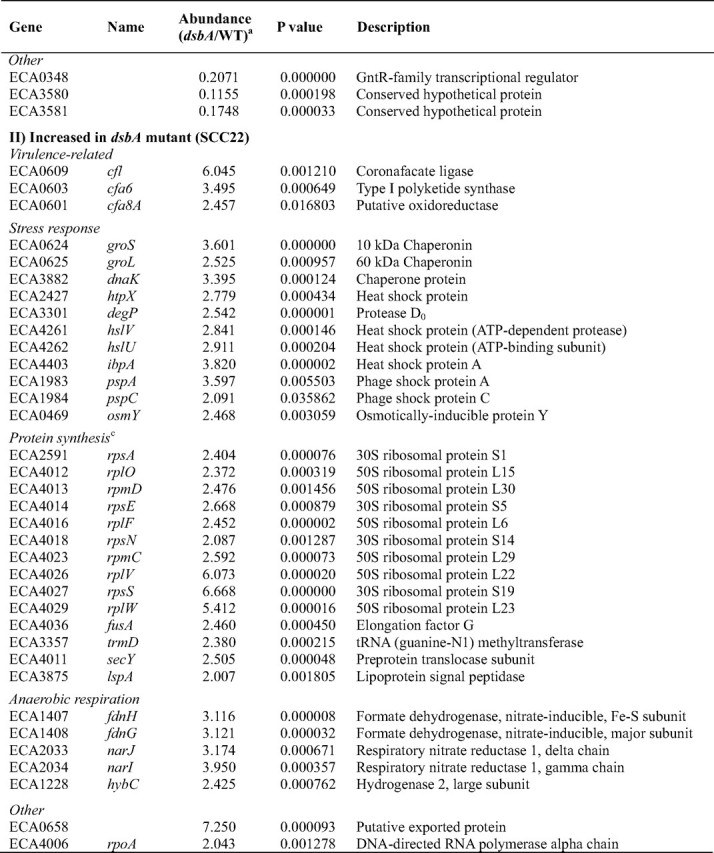
The expression of very many known or predicted virulence-related genes was reduced in the dsbA mutant (Table 3). Consistent with the qRT-PCR results, transcript levels of_expI, svx, pelA, prtW_, and celB were significantly decreased in the mutant (and rpoA was significantly increased). The_pehA_ and nip transcripts were also decreased in the_dsbA_ mutant (p < 0.05) but missed the stringent 2× cutoff (relative abundance values of 0.77 and 0.57 respectively; data not shown). Many other secreted virulence factor-related genes were also identified as being decreased in expression in the dsbA mutant,e.g. four other predicted Pels, genes involved in pectin breakdown product uptake (kdgT, kdgM, and togM), a predicted virulence-related outer membrane protein, and several putative type VI secretion-related genes, including the putative secretion substrate HcpA (40). Pectin lyase was decreased by ∼50% in the secretome of the dsbA mutant (Table 2), and the corresponding transcript was decreased in the dsbA mutant (p < 0.05) but just missed the 2× cutoff (relative abundance 0.56; data not shown).
Motility is an important virulence determinant in bacterial pathogens. The microarray experiment revealed that expression of 25 genes involved in motility and chemotaxis, or predicted to encode methyl-accepting chemotaxis proteins, was significantly decreased in the dsbA mutant (Table 3). For example, the major flagellin (fliC), the flagellar hook-associated protein-2 (fliD), and the anti-σ factor (flgM) were reduced at least 10× in the dsbA mutant. To verify this result, qRT-PCR analysis of fliC was performed and gave a significant 25× decrease in fliC transcript levels in the dsbA mutant (Table 2). These transcriptomic data are consistent with the virtual absence in the dsbA mutant of all the flagellar proteins observed in the secretome of the wild type (Table 1) and with the lack of motility of this mutant (Fig. 4). Flagellar gene expression is sequential and hierarchical, in concert with the ordered assembly of the organelle (41–43). In a dsbA mutant, the P-ring, and thus the flagellar basal body, cannot be assembled properly because of a lack of disulfide bonding in FlgI (44). Because late gene expression is dependent on successful completion of hook-basal body assembly, it is unsurprising that late/FliA-dependent genes (e.g. fliC, motB, cheA, and fliD) were reduced in expression. However, all types of flagellar transcripts, early (flhC) and middle (e.g. flgC, flgM, flgK, and fliD), as well as late, were reduced in the_dsbA_ mutant. Thus the entire flagellar regulon is down-regulated in response to DsbA inactivation. This response is likely to be mediated via the master transcriptional regulators, FlhDC, that are required for the expression of all the other flagellar genes and are regulated by multiple other regulatory systems (43). (As well as flhC, the flhD transcript was also decreased in the_dsbA_ mutant by 1.7× (p < 0.05); data not shown.)
The coordinated down-regulation of many virulence-related genes in the_dsbA_ mutant suggests the existence of an important pleiotropic virulence regulator(s), which is responsive to periplasmic redox status and/or the presence of misfolded proteins and can influence the expression of multiple genes in response to this cue. Presumably such a system allows the expression of virulence determinants to be coordinately “dampened down” if their post-translational processing cannot proceed efficiently. The predicted regulatory protein(s) responsible for sensing the absence of DsbA activity in the periplasm and relaying this signal back to gene expression in the cytoplasm is not yet known. The regulatory system may involve previously identified regulators of virulence factor production (e.g. PecSM and AepA) or of virulence and motility (e.g. HexA) in Erwinia (3,7,45), and/or components of known periplasmic stress response pathways, e.g. Cpx and σE (46,47). Alternatively, it may involve novel regulatory proteins, such as the predicted GntR family transcriptional regulator, ECA0348, whose expression was decreased 5× in the dsbA mutant. A 3× reduction was also seen in the_slyA_ family virulence regulator, hor. To detect the presence/absence of DsbA activity, we predict that the primary sensor in such a signal cascade will be a protein that is structurally disulfide-bonded, redox-regulated, or sensitive to misfolded proteins.
The microarray also identified a set of transcripts increased in abundance in the dsbA mutant (Table 3). Many of these represented known stress-response genes, consistent with a cellular response to protein misfolding in the periplasm, including cytoplasmic heat shock proteins (e.g. groLS and_dnaK_) and periplasmic stress-response proteins degP, pspA, and osmY (47). In addition, 25 genes involved in protein synthesis were up-regulated. This is also likely to be a response to protein misfolding and/or mistargeting. Unexpectedly, three genes in the coronafacic acid synthesis cluster (cfl, cfa6, and cfa8A) were also significantly up-regulated in the_dsbA_ mutant. The increase in cfl transcript levels was verified by qRT-PCR analysis, where an increase of almost 20× was seen (Table 2). The cfa gene cluster is predicted to direct the biosynthesis of coronafacic acid, the polyketide precursor of the phytotoxin coronatine, and cfa has been implicated as an important virulence determinant (7). The increased expression of cfl/cfa may be simply due to the misfolding and inactivation of a periplasmic negative regulator of cfa in the absence of DsbA. Alternatively, this finding may provide the first evidence that cfa expression is stress-induced. Other transcripts increased in abundance in the mutant include those of genes involved in anaerobic respiration and a putative exported protein (ECA0658), increased 7× in the dsbA mutant (Table 3).
DsbA has been shown to exert an effect on specific virulence gene expression at the transcript level in other organisms. For example, in P. aeruginosa, transcription of the type III secretion regulator,exsA, and the downstream effector, exoT, were greatly decreased in a dsbA mutant (48). However, in this study we have demonstrated a genome-wide transcriptional response to dsbA inactivation, including diverse virulence genes. This impact of DsbA on transcript levels could be via altered transcription rates and/or via altered mRNA stability.
Because the regulon of genes requiring dsbA for full expression is clearly populated by key virulence factors, novel genes identified as being part of this regulon are therefore good candidates for new virulence factors, for example, ECA3580, ECA3581, ECA2134, and the predicted transcriptional regulator ECA0348. This is clearly even more true for those that represent both DsbA-dependent transcripts and also Out/DsbA-dependent secreted proteins, for example ECA2134 (see above) and ECA3580. HecA, secreted by a type V two-partner mechanism, has a role in virulence in E. chrysanthemi (49). In E. carotovora subsp. atroseptica, hecA2 is one of two adjacent open reading frames that show homology to E. chrysanthemi HecA but were judged likely to be pseudogenes (7). Surprisingly, this study revealed that HecA2 protein is expressed and that expression of hecB (encoding its putative transporter) is DsbA-dependent, in common with other virulence factors. The role of HecAB in E. carotovora subsp.atroseptica clearly merits further investigation. The transcript of another candidate virulence factor, the putative type VI secretion system component, ECA3444, was also found to be decreased >10× in the_dsbA_ mutant. During manuscript preparation, ECA3444 was confirmed to indeed be required for full virulence in planta in E. carotovora subsp. atroseptica SCRI1043 (50).
_Conclusions_—DsbA is required for the proper expression of very many, if not all, secreted virulence factors in the phytopathogen, E. carotovora subsp. atroseptica SCRI1043. The Out T2SS is responsible for the secretion of a wider range of virulence determinants than just PCWDEs. DsbA affects the production of multiple virulence factors at one or more levels: transcript abundance, protein stability, and/or protein secretion. The transcriptional impact of dsbA mutation implies that a feedback regulatory system controls expression of a broad spectrum of virulence genes in response to extracytoplasmic conditions. Novel candidate virulence factors have been identified through their presence in the Out- and DsbA-dependent secretome and/or membership of the DsbA-dependent regulon.
Supplementary Material
Supplemental Data
Acknowledgments
We thank Rita Monson, Tom Burr, and Jenny Morris for assistance and advice with qRT-PCR and microarray analysis; Jim Metcalfe for qRT-PCR facilities; Ian Foulds for technical support, and Sonia Humphris for assistance with plant tests. The contribution of Svenja Hester in performing mass spectrometry analysis is also gratefully acknowledged.
*
This work was supported by the Biotechnology and Biological Sciences Research Council. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “_advertisement_” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Microarray data have been deposited in the GEO data base with accession number GSE10647.
The on-line version of this article (available athttp://www.jbc.org) contains supplemental Tables 1–4 and additional references.
Footnotes
3
The abbreviations used are: PCWDE, plant cell wall degrading enzyme; Pel, pectate lyase; qRT, quantitative RT; PMM, Pel Minimal Medium; DiGE, difference in gel electrophoresis; OHHL, _N_-3-oxohexanoyl-homoserine lactone; CHAPS, 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid; Cel, cellulase; T2SS, type II secretion system; WT, wild type; Prt, protease; AHL,_N_-acyl homoserine lactone; QS, quorum sensing.
References
- 1.Perombelon, M. C. M. (2002) Plant Pathol. (Oxf.) 51 1–12 [Google Scholar]
- 2.Toth, I. K., Bell, K. S., Holeva, M. C., and Birch, P. R. J. (2003) Mol Plant Pathol. 4 17–30 [DOI] [PubMed] [Google Scholar]
- 3.Thomson, N. R., Thomas, J. D., and Salmond, G. P. (1999) Methods Microbiol. 29 347–426 [Google Scholar]
- 4.Toth, I. K., and Birch, P. R. (2005) Curr. Opin. Plant Biol. 8 424–429 [DOI] [PubMed] [Google Scholar]
- 5.Pemberton, C. L., Whitehead, N. A., Sebaihia, M., Bell, K. S., Hyman, L. J., Harris, S. J., Matlin, A. J., Robson, N. D., Birch, P. R., Carr, J. P., Toth, I. K., and Salmond, G. P. (2005) Mol. Plant-Microbe Interact. 18 343–353 [DOI] [PubMed] [Google Scholar]
- 6.Corbett, M., Virtue, S., Bell, K., Birch, P., Burr, T., Hyman, L., Lilley, K., Poock, S., Toth, I., and Salmond, G. (2005) Mol. Plant-Microbe Interact. 18 334–342 [DOI] [PubMed] [Google Scholar]
- 7.Bell, K. S., Sebaihia, M., Pritchard, L., Holden, M. T., Hyman, L. J., Holeva, M. C., Thomson, N. R., Bentley, S. D., Churcher, L. J., Mungall, K., Atkin, R., Bason, N., Brooks, K., Chillingworth, T., Clark, K., Doggett, J., Fraser, A., Hance, Z., Hauser, H., Jagels, K., Moule, S., Norbertczak, H., Ormond, D., Price, C., Quail, M. A., Sanders, M., Walker, D., Whitehead, S., Salmond, G. P., Birch, P. R., Parkhill, J., and Toth, I. K. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 11105–11110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reeves, P. J., Whitcombe, D., Wharam, S., Gibson, M., Allison, G., Bunce, N., Barallon, R., Douglas, P., Mulholland, V., Stevens, S., Walker, D., and Salmond, G. P. C. (1993) Mol. Microbiol. 8 443–456 [DOI] [PubMed] [Google Scholar]
- 9.Sandkvist, M. (2001) Infect. Immun. 69 3523–3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Filloux, A. (2004) Biochim. Biophys. Acta 1694 163–179 [DOI] [PubMed] [Google Scholar]
- 11.Delepelaire, P. (2004) Biochim. Biophys. Acta 1694 149–161 [DOI] [PubMed] [Google Scholar]
- 12.Whitehead, N. A., Byers, J. T., Commander, P., Corbett, M. J., Coulthurst, S. J., Everson, L., Harris, A. K., Pemberton, C. L., Simpson, N. J., Slater, H., Smith, D. S., Welch, M., Williamson, N., and Salmond, G. P. (2002) Antonie Leeuwenhoek 81 223–231 [DOI] [PubMed] [Google Scholar]
- 13.Whitehead, N. A., Barnard, A. M., Slater, H., Simpson, N. J., and Salmond, G. P. (2001) FEMS Microbiol. Rev. 25 365–404 [DOI] [PubMed] [Google Scholar]
- 14.Burr, T., Barnard, A. M., Corbett, M. J., Pemberton, C. L., Simpson, N. J., and Salmond, G. P. (2006) Mol. Microbiol. 59 113–125 [DOI] [PubMed] [Google Scholar]
- 15.Mattinen, L., Nissinen, R., Riipi, T., Kalkkinen, N., and Pirhonen, M. (2007) Proteomics 7 3527–3537 [DOI] [PubMed] [Google Scholar]
- 16.Kazemi-Pour, N., Condemine, G., and Hugouvieux-Cotte-Pattat, N. (2004) Proteomics 4 3177–3186 [DOI] [PubMed] [Google Scholar]
- 17.Macnab, R. M. (2004) Biochim. Biophys. Acta 1694 207–217 [DOI] [PubMed] [Google Scholar]
- 18.Kadokura, H., Katzen, F., and Beckwith, J. (2003) Annu. Rev. Biochem. 72 111–135 [DOI] [PubMed] [Google Scholar]
- 19.Nakamoto, H., and Bardwell, J. C. (2004) Biochim. Biophys. Acta 1694 111–119 [DOI] [PubMed] [Google Scholar]
- 20.Yu, J., and Kroll, J. S. (1999) Microbes Infect. 1 1221–1228 [DOI] [PubMed] [Google Scholar]
- 21.Lasica, A. M., and Jagusztyn-Krynicka, E. K. (2007) FEMS Microbiol. Rev. 31 626–636 [DOI] [PubMed] [Google Scholar]
- 22.Shevchik, V. E., Bortoli-German, I., Robert-Baudouy, J., Robinet, S., Barras, F., and Condemine, G. (1995) Mol. Microbiol. 16 745–753 [DOI] [PubMed] [Google Scholar]
- 23.Vincent-Sealy, L. V., Thomas, J. D., Commander, P., and Salmond, G. P. (1999) Microbiology 145 1945–1958 [DOI] [PubMed] [Google Scholar]
- 24.Coulthurst, S. J., Lilley, K. S., and Salmond, G. P. (2006) Mol. Plant Pathol. 7 31–35 [DOI] [PubMed] [Google Scholar]
- 25.Kaniga, K., Delor, I., and Cornelis, G. R. (1991) Gene (Amst.) 109 137–141 [DOI] [PubMed] [Google Scholar]
- 26.Toth, I., Perombelon, M., and Salmond, G. (1993) J. Gen. Microbiol. 139 2705–2709 [Google Scholar]
- 27.Liu, H., Sadygov, R. G., and Yates, J. R., III (2004) Anal. Chem. 76 4193–4201 [DOI] [PubMed] [Google Scholar]
- 28.Holeva, M. C., Bell, K. S., Hyman, L. J., Avrova, A. O., Whisson, S. C., Birch, P. R., and Toth, I. K. (2004) Mol. Plant-Microbe Interact. 17 943–950 [DOI] [PubMed] [Google Scholar]
- 29.Winson, M. K., Swift, S., Fish, L., Throup, J. P., Jorgensen, F., Chhabra, S. R., Bycroft, B. W., Williams, P., and Stewart, G. S. (1998) FEMS Microbiol. Lett. 163 185–192 [DOI] [PubMed] [Google Scholar]
- 30.Venkatesh, B., Babujee, L., Liu, H., Hedley, P., Fujikawa, T., Birch, P., Toth, I., and Tsuyumu, S. (2006) J. Bacteriol. 188 3088–3098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chatterjee, A., McEvoy, J. L., Chambost, J. P., Blasco, F., and Chatterjee, A. K. (1991) J. Bacteriol. 173 1765–1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hiniker, A., and Bardwell, J. C. (2004) J. Biol. Chem. 279 12967–12973 [DOI] [PubMed] [Google Scholar]
- 33.Vertommen, D., Depuydt, M., Pan, J., Leverrier, P., Knoops, L., Szikora, J. P., Messens, J., Bardwell, J. C., and Collet, J. F. (2008) Mol. Microbiol. 67 336–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Agudo, D., Mendoza, M. T., Castanares, C., Nombela, C., and Rotger, R. (2004) Proteomics 4 355–363 [DOI] [PubMed] [Google Scholar]
- 35.Hiniker, A., Collet, J. F., and Bardwell, J. C. (2005) J. Biol. Chem. 280 33785–33791 [DOI] [PubMed] [Google Scholar]
- 36.Walker, D. S., Reeves, P. J., and Salmond, G. P. (1994) Mol. Plant-Microbe Interact. 7 425–431 [Google Scholar]
- 37.Sauvonnet, N., and Pugsley, A. P. (1998) Mol. Microbiol. 27 661–667 [DOI] [PubMed] [Google Scholar]
- 38.Laatu, M., and Condemine, G. (2003) J. Bacteriol. 185 1642–1649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pugsley, A. P., Bayan, N., and Sauvonnet, N. (2001) J. Bacteriol. 183 1312–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mougous, J. D., Cuff, M. E., Raunser, S., Shen, A., Zhou, M., Gifford, C. A., Goodman, A. L., Joachimiak, G., Ordonez, C. L., Lory, S., Walz, T., Joachimiak, A., and Mekalanos, J. J. (2006) Science 312 1526–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Berg, H. C. (2003) Annu. Rev. Biochem. 72 19–54 [DOI] [PubMed] [Google Scholar]
- 42.Aldridge, P., and Hughes, K. T. (2002) Curr. Opin. Microbiol. 5 160–165 [DOI] [PubMed] [Google Scholar]
- 43.McCarter, L. L. (2006) Curr. Opin. Microbiol. 9 180–186 [DOI] [PubMed] [Google Scholar]
- 44.Dailey, F. E., and Berg, H. C. (1993) Proc. Natl. Acad. Sci. U. S. A. 90 1043–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harris, S. J., Shih, Y. L., Bentley, S. D., and Salmond, G. P. (1998) Mol. Microbiol. 28 705–717 [DOI] [PubMed] [Google Scholar]
- 46.Raivio, T. L. (2005) Mol. Microbiol. 56 1119–1128 [DOI] [PubMed] [Google Scholar]
- 47.Duguay, A. R., and Silhavy, T. J. (2004) Biochim. Biophys. Acta 1694 121–134 [DOI] [PubMed] [Google Scholar]
- 48.Ha, U. H., Wang, Y., and Jin, S. (2003) Infect. Immun. 71 1590–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rojas, C. M., Ham, J. H., Deng, W. L., Doyle, J. J., and Collmer, A. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 13142–13147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu, H., Coulthurst, S. J., Pritchard, L., Hedley, P., Ravensdale, M., Burr, T., Takle, G., Brurberg, M. B., Birch, P. R., Salmond, G. P., and Toth, I. K. (2008) PLoS Pathogens 10.1371/journal.ppat.1000093 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data