A Novel WWTR1-CAMTA1 Gene Fusion is a Consistent Abnormality in Epithelioid Hemangioendothelioma of Different Anatomic Sites (original) (raw)
. Author manuscript; available in PMC: 2012 Aug 1.
Published in final edited form as: Genes Chromosomes Cancer. 2011 May 16;50(8):644–653. doi: 10.1002/gcc.20886
Abstract
The classification of epithelioid vascular tumors remains challenging, as there is considerable morphologic overlap between tumor subtypes, across the spectrum from benign to malignant categories. A t(1;3)(p36.3;q25) translocation was reported in two cases of epithelioid hemangioendothelioma (EHE), however, no follow-up studies have been performed to identify the gene fusion or to assess its prevalence in a larger cohort of patients. We undertook a systematic molecular analysis of 17 EHE, characterized by classic morphologic and immunophenotypic features, from various anatomic locations and with different malignant potential. For comparison we analyzed 13 epithelioid hemangiomas, five epithelioid angiosarcomas and four epithelioid sarcoma-like EHE. A fluorescence in situ hybridization (FISH) positional cloning strategy, spanning the cytogenetically defined regions on chromosomes 1p36.3 and 3q25, confirmed rearrangements in two candidate genes from these loci in all EHE cases tested. None of the other benign or malignant epithelioid vascular tumors examined demonstrated these abnormalities. Subsequent RT-PCR confirmed in three EHE the WWTR1-CAMTA1 fusion product. CAMTA1 and WWTR1 have been previously shown to play important roles in oncogenesis. Our results demonstrate the presence of a WWTR1-CAMTA1 fusion in all EHE tested from bone, soft tissue and visceral location (liver, lung) in keeping with a unique and specific pathological entity. Thus, FISH or RT-PCR analysis for the presence of WWTR1-CAMTA1 fusion may serve as a useful molecular diagnostic tool in challenging diagnoses.
INTRODUCTION
Epithelioid vascular tumors encompass a wide histologic spectrum, including epithelioid hemangioma (EH), a benign tumor; epithelioid hemangioendothelioma (EHE), a low grade malignant tumor; and epithelioid angiosarcoma (E-AS), a high grade malignant tumor (Wenger and Wold, 2000; O’Connell et al., 2001; Fletcher et al., 2002). Although some of these tumors are readily diagnosed based on their characteristic histologic features, such as the formation of vascular spaces, architectural growth pattern and cytologic details, the morphologic overlap between these groups precludes a refined classification in a considerable number of cases (Folpe et al., 2001). Due to their overlapping histology and rare incidence, considerable controversy surrounds the terminology and classification that should be used (O’Connell et al., 2001; Evans et al., 2003). For example, Rosai et al suggested that the ‘histiocytoid’ (epithelioid) morphology of the endothelial cells is not pathognomonic for a specific tumor entity and could be seen in a wide spectrum of vascular lesions (Rosai et al., 1987). For this reason, he argued that not enough data were available at the time to separate EH and EHE into separate entities (Dorfman et al., 1987; Rosai et al., 1987; Weiss, 1987). On the same lines, Evans et al. argued that intra-osseous EH is not a distinct tumor entity, but rather a misdiagnosed hemangioendothelioma, and suggested that the spectrum of EHE of bone to be expanded to include a range of benign to low grade malignant lesions (Evans et al., 2003). Their argument stemmed also from the observation that the skeletal EH has significant dissimilarities from the so-called ‘angiolymphoid hyperplasia with eosinophilia (i.e. EH) of skin and soft tissue (Olsen and Helwig, 1985).
EHE of soft tissue is a rare vascular neoplasm first described by Weiss and Enzinger in 1982 (Weiss and Enzinger, 1982). Because its clinical behavior is intermediate between a benign hemangioma and a high grade angiosarcoma, the term ‘hemangioendothelioma’ was suggested. Subsequently, EHE of soft tissue was stratified into two risk groups of malignancy, classic and malignant EHE, based on mitotic activity and size, in an effort to reflect different biologic subsets (Fletcher et al., 2002; Deyrup et al., 2008).
The genetic hallmark of many vascular tumors is still under investigation. To date, only a few vascular tumors have been analyzed cytogenetically, with different chromosomal translocations being reported (Boudousquie et al., 1996; He et al., 2006; Dunlap et al., 2009). In 2001, Mendlick et al. found an identical chromosomal translocation, t(1;3)(p36.3;q25) in two cases of EHE, suggesting that this may represent a recurrent abnormality in this subgroup of tumors (Mendlick et al., 2001). In this study we investigated the genes involved in the t(1;3)(p36.3;q25) translocation and its prevalence in a large spectrum of EHEs, including lesions from various anatomic locations and different malignant potential, as well as other epithelioid vascular lesions commonly included in the differential diagnosis. We hypothesized that a better understanding of the molecular signature of vascular tumors will refine the present classification system.
MATERIAL AND METHODS
Patients and Tumor Characteristics
We retrieved 23 cases of EHE from the surgical pathology files of our institution with available tissue for molecular analysis (IRB-protocol 02-060). In each case, the diagnosis and histologic grade were confirmed by reviewing the H&E slides. All tumors included for analysis were positive for the CD31 endothelial marker. The tumors were assessed morphologically for growth pattern, vasoformative nature, epithelioid versus spindle cell composition, cellular pleomorphism, mitotic activity, and necrosis. For each case, the location of the tumor was recorded, along with the anatomic structures involved. Based on their location, the lesions were classified into four groups: bone, soft tissue, intra-thoracic, and liver.
Because EHE, shows morphologic overlap with EH and with E-AS we analyzed 13 cases of EH and 5 cases of E-AS to investigate a potential genetic link between theses entities. In addition, 4 cases of epithelioid sarcoma-like EHE, considered a rare morphologic variant of EHE, were included for analysis.
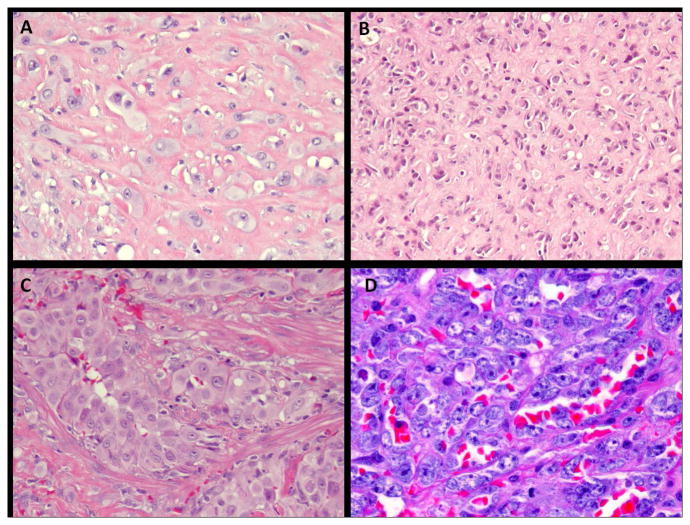
Morphologically, the most important histologic criteria applied to distinguish EHE from both EH and E-AS was the lack of well-formed vascular channel formation in EHE, with only immature, intra-cytoplasmic lumina being observed (Fig. 1A–C). Other distinctive features used for the diagnosis of EHE included the infiltrative growth pattern and the presence of a myxoid, myxochondroid or sclerotic extracellular stroma, which was seen in most, but not all cases. This feature was particularly absent in the liver EHE, possibly due to the limited biopsy samples available. In contrast, EH was defined as lobulated or well-circumscribed lesions, showing clearly vasoformative properties, forming ‘mature’ vessels with open lumina (Fig. 1C). The lesional cells had occasionally the so-called tomb-stone appearance and consistently showed abundant, glassy, eosinophilic cytoplasm. E-AS showed marked nuclear pleomorphism, high mitotic activity and areas of necrosis (Fig. 1D). Epithelioid-sarcoma-like EHE shows morphologic features more in keeping with an epithelioid sarcoma, with epithelioid cells showing dense, eosinophilic cytoplasm, but expressing endothelial markers, such as CD31 or Fli1, in addition to epithelial markers.
Figure 1.
Morphologic appearance of epithelioid vascular tumors analyzed in this study. A. Malignant EHE of the arm, with cords and single cells of epithelioid cells with moderate nuclear atypia, embedded in a hyalinized stroma (EHE#1, x200 magnification); B. Classic EHE of the liver with bland epithelioid cells with readily visible intra-cytoplasmic vacuoles (EHE#16, x 200); C. EH of the penis in a 48 year-old man, who presented as multiple cutaneous and subcutaneous nodules, and showed large epithelioid cells with abundant eosinophilic cytoplasm surrounding vascular lumina (x200); D. Radiation-induced angiosarcoma of breast, composed of predominantly epithelioid morphology and showing high grade cytologic atypia, with prominent nucleoli, as well as vascular channel formation (x200).
Fluorescence in situ hybridization (FISH) positional cloning of the t(1;3)(p36.23;q25.1)
Interphase FISH was the main analytical tool in our study, since most of the tumors were available only in archival material. FISH analysis was undertaken to explore the presence of rearrangements in the previously documented 1p36 and 3q25 chromosomal regions and to further narrow the breakpoints genomic locations, as accurately as the resolution of bacterial artificial chromosome (BAC) clones allowed. Because of the limited resolution of chromosome banding, used we extended our query regions to 1p36.33~36.11 and 3q25~27.
BAC clones were selected according to the UCSC genome browser (http://genome.ucsc.edu) and were obtained from BACPAC sources of Children’s Hospital of Oakland Research Institute (CHORI) (Oakland, CA) (http://bacpac.chori.org) (Table S1). Probe preparation and FISH procedures were carried out on paraffin-embedded 4-μm-thick tissue sections, as previously described (Antonescu et al., 2010). In brief, BAC DNA was isolated using phenol-chloroform, labeled with different fluorochromes (Enzo, PA, USA) in a nick translation reaction, and validated on normal metaphases. Probe mixtures were co-denatured, and hybridized to pretreated slides. Slides were incubated, washed and mounted with DAPI in an antifade solution. At least two hundred successive non-overlapping nuclei were examined using a fluorescence microscope. A case was confirmed as positive for rearrangement of a given gene when ≥ 20% of the nuclei examined showed a break-apart signal pattern using its respective BAC probes.
Our pilot FISH analysis showed that two probe sets within 6 MB of genomic distance of each other stayed in close association; therefore, able to distinguish a break-apart event from a non-break-apart event with relative confidence. For the first round of FISH experiments, we tested 9 probe sets on 3q25~27 and 6 probe sets on 1p36.3 (Table S1, figs. 2&3), which were evenly distributed among the spanned regions, as well as intentionally designed to flank genes previously implicated in recurrent translocations. Three-color FISH identified break-aparts in 3q25.1~25.3 and 1p36.23. In the second round of FISH analysis, new probe sets targeting the middle portion of the queried regions were designed and three-color FISH confirmed a break, and narrowed down the region in half. The same strategy was repeated until the breakpoints were narrowed down to a 200–500 kb regions.
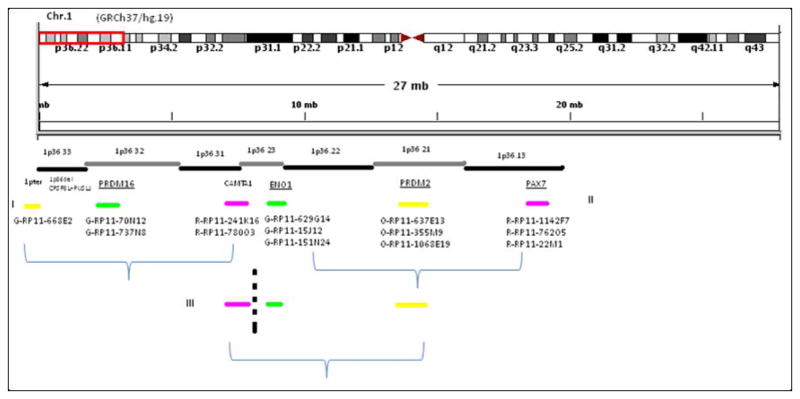
Figure 2.
FISH positional cloning strategy using BAC probe sets on 1p36.33~1p36.11. Three sets of experiments identified the breakpoint in 1p36.23. Underlined genes have been previously reported in other chromosomal translocations.
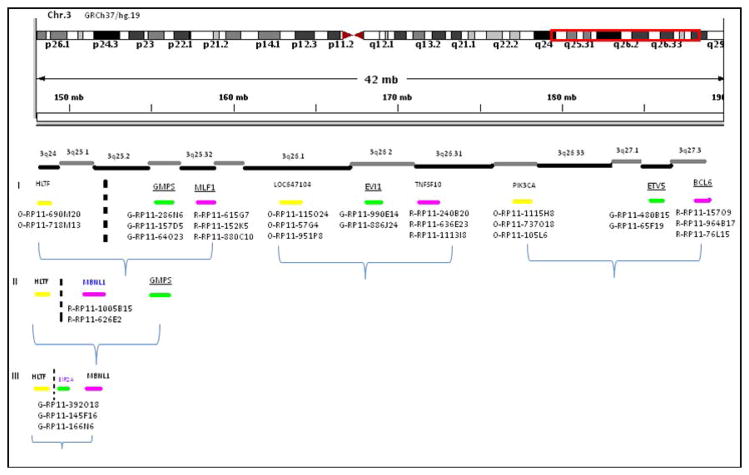
Figure 3.
Distribution of BAC probe sets tested spanning the 3q24~27 region. Three rounds of FISH experiments illustrated in this diagram were able to narrow down the break-apart region between 3q24~25.1. Underlined genes have been previously reported in other chromosomal translocations.
Reverse transcription-polymerase chain reaction (RT-PCR)
In three EHE tumors adequate RNA extracted from frozen tissue (Trizol Reagent; Invitrogen, USA) was available to investigate possible fusion transcripts. RNA quality was determined by Eukaryote Total RNA Nano Assay and cDNA was tested by RT-PCR for PGK housekeeping gene. A two-step RT-PCR was applied, with oligo(dT)20 primer under SuperScript® III system (Invitrogen, USA) used for first-strand cDNA synthesis, followed by a second-step PCR, using the HotStar Taq Master Mix (Qiagen, Valencia, CA). The RT-PCR products were analyzed by electrophoresis and the RT-PCR-amplified products were sequenced using by the Sanger method.
RESULTS
Tumor Characteristics
Of the 23 cases of EHE initially selected, six cases were subsequently excluded due to unsuccessful FISH (insufficient number of cells in core biopsies in four liver EHE and due to decalcification in two bone EHE). Thus 17 patients were included, who had equal sex distribution (8 females, 9 men) and a median age of 48 years (range 25–68) (Table 1). The tumor locations were: seven cases in the soft tissue (five in the trunk and two in the extremity), seven intra-thoracic (all multifocal, involving lung, pleura and mediastinum), two in the liver, and one in the bone (mandible). Histologically, eight tumors showed a bland morphologic appearance, with low mitotic activity, while nine tumors showed moderate to focally marked nuclear pleomorphism, readily identified mitotic activity, with or without necrosis. The latter group qualified for the ‘malignant’ EHE designation, by WHO classification. Clinical follow-up information in 15 patients showed nine patients were alive with no evidence of disease (range 4–116 months, median 20), two were alive with disease, and four died of disease.
Table 1.
Clinical, pathologic, and Molecular Findings in Epithelioid Hemangioendothelioma.
| EHE Case # | Age | Sex | Location | Malig Grade | FISH rearrangements for WWTR1 & CAMTA1 | Clinical Follow-Up (mo) |
|---|---|---|---|---|---|---|
| 1 | 68 | M | ST, arm | yes | +* | NED (16) |
| 2 | 59 | F | ST, groin | no | + | NED (116) |
| 3 | 39 | M | ST, subclavicle | yes | + | NED (14) |
| 4 | 54 | M | ST, arm | yes | +* | NED (108) |
| 5 | 52 | F | ST, abd wall, bilateral breast | yes | + | AWD (15) |
| 6 | 66 | M | ST, neck/shoulder | no | +* | NED (4) |
| 7 | 39 | F | ST, axilla | no | + | DOD (43) |
| 8 | 42 | F | IT, mediastinum, lung, SVC | yes | + | NED (23) |
| 9 | 34 | M | IT, lung, pleura | yes | + | DOD (4) |
| 10 | 32 | M | IT, pleura | yes | + | AWD (7) |
| 11 | 29 | F | IT, lung, paratracheal LNs | yes | + | LFU |
| 12 | 61 | M | IT, lung | no | + | DOD (82) |
| 13 | 65 | F | IT, lung | no | + | NED (20) |
| 14 | 55 | M | IT, lung | no | + | NED (30) |
| 15 | 41 | F | Liver | no | + | NED (7) |
| 16 | 48 | F | Liver | no | + | LFU |
| 17 | 25 | M | Bone/mandible | yes | + | DOD (24) |
In the control group, the anatomic distribution of EH locations were: eight in the bone (four in the foot, one in the hand and three in the chest wall/clavicle), two in the soft tissue (arm), two in the head and neck (parotid) and one in the penis. The five cases of E-AS tested showed a predominantly solid/epithelioid morphology and had the following clinical characteristics: two lesions from the breast (one primary and one radiation-induced), two secondary AS from soft tissue (one tumor developed in the context of chronic lymphedema and radiation for breast cancer and the other following radiation for a soft tissue sarcoma), and one in the head and neck. The four cases of epithelioid sarcoma-like EHE had the usual clinical presentation as multiple superficial nodules; tumor locations were abdominal wall, back and foot.
FISH positional cloning identified CAMTA1 on 1p36.23 and WWTR1 on 3q25.1 as candidate fusion genes
FISH analyses for 1p36.23 break-apart, 3q25.1 break-apart, and fusion between 1p36.23 and 3q25.1 were carried out for each case. Combined results confirmed the translocation t(1;3)(1p36.23;3q25.1) in all EHE cases, but in none of the EH, E-AS or epithelioid sarcoma-like EHE cases (Fig. 4, 5C). On 1p36.23, the breakpoint was mapped within one of the largest genes, CAMTA1, about 984kb. Three-color FISH showed a break-apart between green-RP11-1120I14 and Orange-RP11-338N10 in all EHE cases (Fig. 4A and inset). This suggested the breakpoint was in the region of chr.1:7626000-7768000 (hg19), between CAMTA1 exon 6 and exon 12 (ENST00000303635).
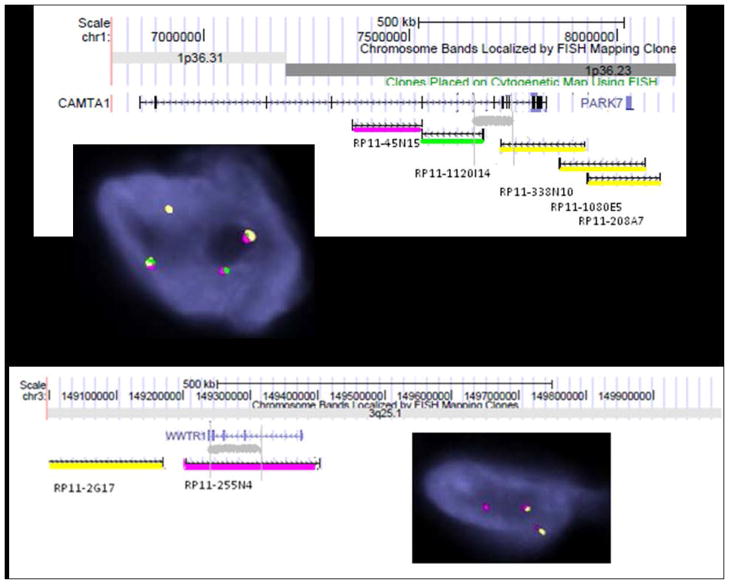
Figure 4.
Identification of candidate genes on 1p36.23 and 3q25 by FISH.
A. Grey area showed 1p36.23 breakpoint location within CAMTA1 gene. Three-color FISH showed a break-apart between green-RP11-1120I14 and Orange-RP11-338N10 (inset). B. Two-color FISH (orange-RP11-2G17 and Red-580-RP11-255N4) identified a split red signal associated with the orange signal (inset). This pattern narrowed the breakpoint at chr.3: 149270000-149360000 (hg.19), which localized in WWTR1 exon 4 to exon 8 (ENST00000465804) (Scale was downloaded from UCSC genome browser for accuracy consideration).
Figure 5.
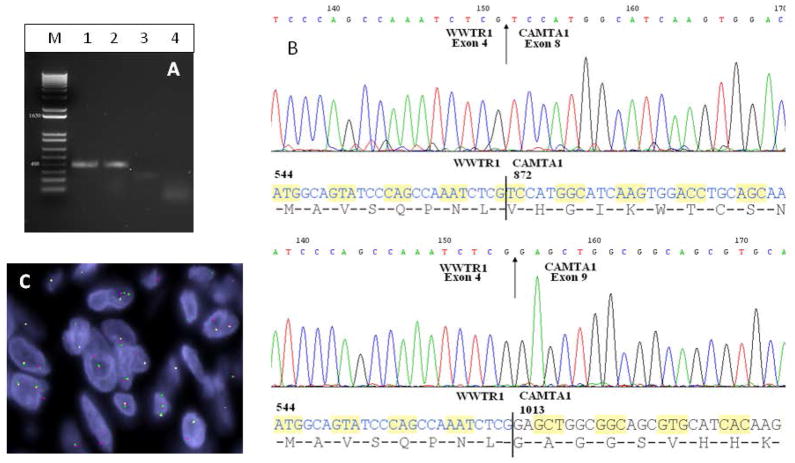
RT-PCR detection of WWTR1-CAMTA1 fusion transcript variants and FISH demonstration of fused CAMTA1 and WWTR1 signals. A. Gel electrophoresis showing amplified products in lanes 1–3, of two distinct sizes (M, size marker, lane 1, EHE#4, lane 2, EHE#6, lane 3, EHE#1, lane 4, negative control no-RNA). B. Sequencing of the 3 amplicons identified two molecular variants, with exon 4 of WWTR1 being fused in-frame to either exon 8 (variant 1, upper panel) or exon 9 (variant 2, lower panel) of CAMTA1. C. FISH demonstration of fused signals, using probes centromeric to CAMTA1 and telomeric to WWTR1.
On 3q25, a two-color FISH showed a split red signal (RP11-255N4) associated with the orange signal (RP11-2G17), which located the breakpoint in the region of chr.3: 149270000-149360000 (hg19), between WWTR1 exon 4 and exon 8 (ENST00000465804) (Fig. 4B and inset).
RT-PCR confirmation of 5′WWTR1-CAMTA1 3′fusion transcripts in EHE
Three tumors with available frozen tissue were analyzed by RT-PCR with different combinations of WWTR1 forward primers exons 3–7 and CAMTA1 reverse primers exons 7–9 (listed in Table S2). Two different WWTR1-CAMTA1 variant transcripts were amplified from three EHE cases. The 5′WWTR1 showed a consistent breakpoint within intron 4 in all three cases, while two variants were seen in the 3′CAMTA1 breakpoint. Exon 4 of WWTR1 was fused to CAMTA1 exon 8 (variant 1, EHE#4&6; see Table 1) and to CAMTA1 exon 9 (variant 2, EHE#1)(Fig. 5).
DISCUSSION
Histologically, epithelioid vascular tumors show remarkable similarities and diagnostic criteria to differentiate them still involve significant subjectivity. Particularly problematic is the wide recognition of epithelioid hemangioma within bone, which historically has been lumped under the ‘hemangioendothelioma’ group of tumors, and only recently has been accepted as a distinct entity from EHE (O’Connell et al., 2001). To complicate issues further, the neoplastic etiology of the cutaneous counterpart, a.k.a. ‘angiolymphoid hyperplasia with eosinophilia’, has been debated, with some authors speculating that it may represent a reactive process, due to its prominent inflammatory infiltrate and angiocentric distribution around a larger vessel with evidence of mural damage (Fetsch and Weiss, 1991). Difficulties exist at the malignant end of the spectrum as well, the distinction between the so-called malignant EHE and a high grade E-AS can be quite challenging in a small sample or when the angiosarcoma lacks vaso-formative properties. Clinical behavior and, consequently, treatment and prognosis, vary significantly among vascular tumors; thus it is paramount to effectively distinguish them from each other. The significant morphologic overlap between these entities across the benign to malignant spectrum of epithelioid vascular neoplasms underscore the lack of objective genetic hallmarks needed for an accurate classification.
To investigate the prevalence of the t(1;3)(p36.3;q25) previously identified in two cases of EHE arising from liver and soft tissue (Mendlick et al., 2001), we undertook a molecular analysis of a large number of EHE from different anatomic locations and with different degrees of malignancy. FISH targeting to narrow-down the breakpoints from the previously mapped chromosomal regions identified rearrangements in CAMTA1 on 1p36.23 and WWTR1 on 3q25.1 in all 17 EHE tumors tested, but in none of the EH, E-AS or epithelioid sarcoma-like EHE tumors. RT-PCR demonstrated a fusion transcript, in which exon 4 of WWTR1 was fused in-frame with either exons 8 or 9 of CAMTA1.
To our knowledge this is the first time that either CAMTA1 or WWTR1 has been implicated in a recurrent chromosomal translocation. However, both genes have been previously shown to play important roles in oncogenesis (Barbashina et al., 2005; Henrich et al., 2006; Lei et al., 2008; Chan et al., 2009; Zhang et al., 2009).
CAMTA1 belongs to a recently described family of calmodulin-binding transcription activators family of proteins (Bouche et al., 2002). CAMTA1 primary structure contains a nuclear localization signal, two DNA-binding domains (CG-1 and TIG), calmodulin-binding motifs and ankyrin repeats. CAMTA1 is a transcription activator potentially involved in cell cycle regulation (Nakatani et al., 2004). It may interact with Ca2+/calmodulin and be engaged in Ca2+ signaling (Bouche et al., 2002). In mammalian cells, Ca2+ and the Ca2+-receptor calmodulin are involved in the regulation of gene transcription. For example, Ca2+/calmodulin is required to activate calcineurin, which dephosphorylates many proteins including the transcription factor NFAT (nuclear factor of activated cells) and induces its translocation to the nucleus (Lipskaia and Lompre, 2004; Munaron, 2006). In addition, Ca2+/calmodulin is required for CaMK to phosphorylate the cAMP-responsive element binding protein (CREB) (Lipskaia and Lompre, 2004; Munaron, 2006). As expression of CAMTA1 parallels that of TP53 in neuroblastoma cell lines, the authors speculated that CAMTA1 could be involved in cell cycle regulation, similar to TP53 (Nakatani et al., 2004). Using the single CAMTA protein in Drosophila, Gong et al demonstrated that CAMTA may function as dimers, and that the CG-1 domain may mediate this dimerization of CAMTA transcription factors. Therefore, in organisms with multiple CAMTAs, the possibility of homo- and heterodimerization exists with further functional implications (Finkler et al., 2007; Gong et al., 2007).
Loss of genetic material on the short arm of chromosome 1 occurs in many human cancers (Ragnarsson et al., 1999). However, the most extensive 1p deletion mapping in search of tumor suppressors has been done in neuroblastomas, about 30% of which have 1p losses (Maris et al., 2000; Attiyeh et al., 2005; White et al., 2005). Low CAMTA1 expression seems to be significantly associated with poor outcome in neuroblastoma (Attiyeh et al., 2005; Henrich et al., 2006). In addition, CAMTA1 gene was located within the commonly deleted region of neuroblastoma (Katoh, 2003). The potential role of CAMTA1 in tumor development is also supported by Barbashina et al., who showed that a deleted region on 1p36 involved CAMTA1 in a subset of gliomas (Barbashina et al., 2005). Identifying CAMTA1 downstream target genes and interacting proteins are among the major tasks ahead. Such studies should provide important information to elucidate the role of CAMTA1 in oncogenesis and, consequently, improve diagnostic tools and therapies.
WWTR1, also known as TAZ, is a transcriptional co-activator with PDZ-binding motif, initially identified by its ability to interact with 14-3-3 proteins. Sharing amino acid sequence homology with YAP (Yes-associated protein), WWTR1 contains a conserved WW domain capable to interact with the PDZ domain (Kanai et al., 2000). In mammals, WWTR1 and YAP are downstream effectors of the Hippo Pathway, which controls organ size and contact inhibition by regulating cell proliferation and apoptosis (Chan et al., 2011a). Within the Hippo pathway, WWTR1 is negatively regulated by LATS tumor suppressor kinase (Justice et al., 1995; Xu et al., 1995; Tapon et al., 2002); phosphorylation by LATS inactivates WWTR1 by inducing 14-3-3 binding and translocation from the nucleus to the cytoplasm. The Hippo pathway is conserved from fly to human and its deregulation in mammals often leads to tumorigenesis (Chan et al., 2011a).
WWTR1 itself has no DNA-binding domain, and thus it must bind to DNA-binding transcription factors to stimulate expression of target genes (Lei et al., 2008). In mammals, major interacting partners of WWTR1 gene are the transcription factors TEAD1-4 (TEADs). When WWTR1 is not inhibited by the Hippo pathway and remains in the nucleus, it interacts with TEADs and activates expression of genes such as CTGF, IGFBP3, ITGB2, Birc5/Survivin, Gli2, and Axl (Chan et al., 2011a). In addition, interaction of WWTR1 with TEADs was recently reported to promote nuclear retention of WWTR1 (Chan et al., 2009). WWTR1 has been also shown to interact with other proteins, such as EphrinB1 (Xing et al., 2010), Cbfa1/Runx2 (Hong et al., 2005), Wbp2 (Chan et al., 2011c), and PAX3 (Murakami et al., 2006). The identification of a myriad of WWTR1-interacting transcription factors participating in various cellular and development processes raises an important question as to which protein is most relevant to the oncogenic function of WWTR1 and what is the underlying molecular mechanism (Chan et al., 2009). Chan et al. (2009) presented evidence that the nuclear accumulation mediated by TEADs promotes oncogenic transformation. In addition, angiomotin, a novel regulator of endothelial cell migration (Troyanovsky et al., 2001), was shown to interact with WWTR1, leading to its cytoplasmic retention and inhibiting its transcriptional output and oncogenic property, independent of the Hippo pathway (Chan et al., 2011b).
WWTR1 is highly expressed in a wide spectrum of human cancer cell lines and various primary tumors, suggesting that this protein has oncogenic potential (Chan et al., 2008; Balasenthil et al., 2011). Ectopic WWTR1 expression also induced cell proliferation, overcame contact inhibition, and led to tumorigenesis in nude mice (Lei et al., 2008).
Following chromosomal rearrangement, the resulting fusion transcript is produced from the 14-3-3 binding protein and WW domains of WWTR1 and the transcription factor immunoglobulin (TIG)-like DNA-binding domain, ankyrin (ANK) repeats and IQ domains of CAMTA1 (Fig. S1). The WW domain of WWTR1 is capable of interacting with PPXY motifs (Pro-Pro-X-Tyr) and a coiled-coil C-terminal domain that recruits core components of the transcriptional machinery. A number of transcription factors contain PPXY motifs within their activation domains. Thus, interaction of WWTR1 with these transcription factors may mediate their transcriptional effects (Hong et al., 2005). Runx2 and peroxisome proliferator-activated receptor γ (PPARγ) are two key transcription factors that drive mesenchymal stem cells (MSC) differentiation to differentiate into either osteoblasts or adipocytes, respectively. WWTR1 may influence cell fate during the MSC process by inducing Runx2 and repressing PPARγ (Hong et al., 2005). Furthermore, the 14-3-3 binding protein domain can inhibit its transcriptional co-activation function by localizing WWTR1 in the cytoplasmic compartment (Kanai et al., 2000).
The conserved regions of CAMTA1 in the transcript fusion include TIG, ANK and IQ motifs (Fig. S1). The TIG domain has been implicated in transcription factor activity and protein dimerisation (Finkler et al., 2007). The C-terminal part of the gene contains IQ motifs that can bind calmodulin (Bouche et al., 2002). In fly, CAMTA transcription activity is stimulated in vivo by direct interaction with calmodulin through the IQ motif region (Gong et al., 2007).
In summary, our results confirm that t(1;3)(p36.23;q25.1) is a consistent genetic abnormality in EHEs of different anatomic locations and grades of malignancy. The translocation fuses CAMTA1 on 1p36.23 to WWTR1 on 3q25.1. This recurrent translocation has not been detected in any of the morphologic mimics of EHE, such as EH, E-AS or epithelioid sarcoma-like EHE, and thus can serve as a useful molecular diagnostic tool in challenging cases. Particularly important is the lack of CAMTA1 and WWTR1 rearrangements in EH, an under-recognized benign tumor, that is often mis-diagnosed as EHE, resulting in overtreatment.
Supplementary Material
Table S1 and S2
Figure S1. Schematic diagram of WWTR1, CAMTA1 and the projected fused WWTR1-CAMTA1 oncoprotein motifs. Preserved in the fusion include the 14-3-3 binding protein and WW domains of WWTR1 and the TIG (transcription factor immunoglobulin -like DNA-binding domain), ANK (ankyrin) and IQ (calmodulin-binding) motifs of CAMTA1.
Acknowledgments
PO1 CA047179-15A2 (SS,CRA), P50 CA 140146-01 (SS,CRA), Cycle for Survival (CRA), Associazione per la Ricerca e la Cura dei Tumori dell’Apparato Locomotore (CE).
We are grateful to Jesse Galle for help in collecting clinical data, and Milagros Soto and Lionel Santibañez for their editorial assistance.
References
- Antonescu CR, Zhang L, Chang NE, Pawel BR, Travis W, Katabi N, Edelman M, Rosenberg AE, Nielsen GP, Dal Cin P, Fletcher CD. EWSR1-POU5F1 fusion in soft tissue myoepithelial tumors. A molecular analysis of sixty-six cases, including soft tissue, bone, and visceral lesions, showing common involvement of the EWSR1 gene. Genes Chromosomes Cancer. 2010;49:1114–1124. doi: 10.1002/gcc.20819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attiyeh EF, London WB, Mosse YP, Wang Q, Winter C, Khazi D, McGrady PW, Seeger RC, Look AT, Shimada H, Brodeur GM, Cohn SL, Matthay KK, Maris JM. Chromosome 1p and 11q deletions and outcome in neuroblastoma. N Engl J Med. 2005;353:2243–2253. doi: 10.1056/NEJMoa052399. [DOI] [PubMed] [Google Scholar]
- Balasenthil S, Chen N, Lott ST, Chen J, Carter J, Grizzle WE, Frazier ML, Sen S, Killary AM. A migration signature and plasma biomarker panel for pancreatic adenocarcinoma. Cancer Prev Res (Phila) 2011;4:137–149. doi: 10.1158/1940-6207.CAPR-10-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbashina V, Salazar P, Holland EC, Rosenblum MK, Ladanyi M. Allelic losses at 1p36 and 19q13 in gliomas: correlation with histologic classification, definition of a 150-kb minimal deleted region on 1p36, and evaluation of CAMTA1 as a candidate tumor suppressor gene. Clin Cancer Res. 2005;11:1119–1128. [PubMed] [Google Scholar]
- Bouche N, Scharlat A, Snedden W, Bouchez D, Fromm H. A novel family of calmodulin-binding transcription activators in multicellular organisms. J Biol Chem. 2002;277:21851–21861. doi: 10.1074/jbc.M200268200. [DOI] [PubMed] [Google Scholar]
- Boudousquie AC, Lawce HJ, Sherman R, Olson S, Magenis RE, Corless CL. Complex translocation [7;22] identified in an epithelioid hemangioendothelioma. Cancer Genet Cytogenet. 1996;92:116–121. doi: 10.1016/s0165-4608(96)00175-6. [DOI] [PubMed] [Google Scholar]
- Chan SW, Lim CJ, Chen L, Chong YF, Huang C, Song H, Hong W. The Hippo pathway in biological control and cancer development. J Cell Physiol. 2011a;226:928–939. doi: 10.1002/jcp.22435. [DOI] [PubMed] [Google Scholar]
- Chan SW, Lim CJ, Chong YF, Pobbati AV, Huang C, Hong W. Hippo Pathway-independent Restriction of TAZ and YAP by Angiomotin. J Biol Chem. 2011b;286:7018–7026. doi: 10.1074/jbc.C110.212621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SW, Lim CJ, Guo K, Ng CP, Lee I, Hunziker W, Zeng Q, Hong W. A role for TAZ in migration, invasion, and tumorigenesis of breast cancer cells. Cancer Res. 2008;68:2592–2598. doi: 10.1158/0008-5472.CAN-07-2696. [DOI] [PubMed] [Google Scholar]
- Chan SW, Lim CJ, Huang C, Chong YF, Gunaratne HJ, Hogue KA, Blackstock WP, Harvey KF, Hong W. WW domain-mediated interaction with Wbp2 is important for the oncogenic property of TAZ. Oncogene. 2011c;30:600–610. doi: 10.1038/onc.2010.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SW, Lim CJ, Loo LS, Chong YF, Huang C, Hong W. TEADs mediate nuclear retention of TAZ to promote oncogenic transformation. J Biol Chem. 2009;284:14347–14358. doi: 10.1074/jbc.M901568200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deyrup AT, Tighiouart M, Montag AG, Weiss SW. Epithelioid hemangioendothelioma of soft tissue: a proposal for risk stratification based on 49 cases. Am J Surg Pathol. 2008;32:924–927. doi: 10.1097/pas.0b013e31815bf8e6. [DOI] [PubMed] [Google Scholar]
- Dorfman HD, Tsuneyoshi M, Bauer TW. Reply to letter. Am J Surg Pathol. 1987;11:653–654. doi: 10.1097/00000478-198611000-00002. [DOI] [PubMed] [Google Scholar]
- Dunlap JB, Magenis RE, Davis C, Himoe E, Mansoor A. Cytogenetic analysis of a primary bone angiosarcoma. Cancer Genet Cytogenet. 2009;194:1–3. doi: 10.1016/j.cancergencyto.2009.04.008. [DOI] [PubMed] [Google Scholar]
- Evans HL, Raymond AK, Ayala AG. Vascular tumors of bone: A study of 17 cases other than ordinary hemangioma, with an evaluation of the relationship of hemangioendothelioma of bone to epithelioid hemangioma, epithelioid hemangioendothelioma, and high-grade angiosarcoma. Hum Pathol. 2003;34:680–689. doi: 10.1016/s0046-8177(03)00249-1. [DOI] [PubMed] [Google Scholar]
- Fetsch JF, Weiss SW. Observations concerning the pathogenesis of epithelioid hemangioma (angiolymphoid hyperplasia) Mod Pathol. 1991;4:449–455. [PubMed] [Google Scholar]
- Finkler A, Ashery-Padan R, Fromm H. CAMTAs: calmodulin-binding transcription activators from plants to human. FEBS Lett. 2007;581:3893–3898. doi: 10.1016/j.febslet.2007.07.051. [DOI] [PubMed] [Google Scholar]
- Fletcher CD, Unni K, Mertens F. WHO. Tumours of Soft Tissue and Bone. Lyon: IARC Press; 2002. Pathology & Genetics. [Google Scholar]
- Folpe AL, Chand EM, Goldblum JR, Weiss SW. Expression of Fli-1, a nuclear transcription factor, distinguishes vascular neoplasms from potential mimics. Am J Surg Pathol. 2001;25:1061–1066. doi: 10.1097/00000478-200108000-00011. [DOI] [PubMed] [Google Scholar]
- Gong P, Han J, Reddig K, Li HS. A potential dimerization region of dCAMTA is critical for termination of fly visual response. J Biol Chem. 2007;282:21253–21258. doi: 10.1074/jbc.M701223200. [DOI] [PubMed] [Google Scholar]
- He M, Das K, Blacksin M, Benevenia J, Hameed M. A translocation involving the placental growth factor gene is identified in an epithelioid hemangioendothelioma. Cancer Genet Cytogenet. 2006;168:150–154. doi: 10.1016/j.cancergencyto.2006.02.010. [DOI] [PubMed] [Google Scholar]
- Henrich KO, Fischer M, Mertens D, Benner A, Wiedemeyer R, Brors B, Oberthuer A, Berthold F, Wei JS, Khan J, Schwab M, Westermann F. Reduced expression of CAMTA1 correlates with adverse outcome in neuroblastoma patients. Clin Cancer Res. 2006;12:131–138. doi: 10.1158/1078-0432.CCR-05-1431. [DOI] [PubMed] [Google Scholar]
- Hong JH, Hwang ES, McManus MT, Amsterdam A, Tian Y, Kalmukova R, Mueller E, Benjamin T, Spiegelman BM, Sharp PA, Hopkins N, Yaffe MB. TAZ, a transcriptional modulator of mesenchymal stem cell differentiation. Science. 2005;309:1074–1078. doi: 10.1126/science.1110955. [DOI] [PubMed] [Google Scholar]
- Justice RW, Zilian O, Woods DF, Noll M, Bryant PJ. The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev. 1995;9:534–546. doi: 10.1101/gad.9.5.534. [DOI] [PubMed] [Google Scholar]
- Kanai F, Marignani PA, Sarbassova D, Yagi R, Hall RA, Donowitz M, Hisaminato A, Fujiwara T, Ito Y, Cantley LC, Yaffe MB. TAZ: a novel transcriptional co-activator regulated by interactions with 14-3-3 and PDZ domain proteins. EMBO J. 2000;19:6778–6791. doi: 10.1093/emboj/19.24.6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh M. Identification and characterization of FLJ10737 and CAMTA1 genes on the commonly deleted region of neuroblastoma at human chromosome 1p36.31-p36.23. Int J Oncol. 2003;23:1219–1224. [PubMed] [Google Scholar]
- Lei QY, Zhang H, Zhao B, Zha ZY, Bai F, Pei XH, Zhao S, Xiong Y, Guan KL. TAZ promotes cell proliferation and epithelial-mesenchymal transition and is inhibited by the hippo pathway. Mol Cell Biol. 2008;28:2426–2436. doi: 10.1128/MCB.01874-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipskaia L, Lompre AM. Alteration in temporal kinetics of Ca2+ signaling and control of growth and proliferation. Biol Cell. 2004;96:55–68. doi: 10.1016/j.biolcel.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Maris JM, Weiss MJ, Guo C, Gerbing RB, Stram DO, White PS, Hogarty MD, Sulman EP, Thompson PM, Lukens JN, Matthay KK, Seeger RC, Brodeur GM. Loss of heterozygosity at 1p36 independently predicts for disease progression but not decreased overall survival probability in neuroblastoma patients: a Children’s Cancer Group study. J Clin Oncol. 2000;18:1888–1899. doi: 10.1200/JCO.2000.18.9.1888. [DOI] [PubMed] [Google Scholar]
- Mendlick MR, Nelson M, Pickering D, Johansson SL, Seemayer TA, Neff JR, Vergara G, Rosenthal H, Bridge JA. Translocation t(1;3)(p36.3;q25) is a nonrandom aberration in epithelioid hemangioendothelioma. Am J Surg Pathol. 2001;25:684–687. doi: 10.1097/00000478-200105000-00019. [DOI] [PubMed] [Google Scholar]
- Munaron L. Intracellular calcium, endothelial cells and angiogenesis. Recent Pat Anticancer Drug Discov. 2006;1:105–119. doi: 10.2174/157489206775246502. [DOI] [PubMed] [Google Scholar]
- Murakami M, Tominaga J, Makita R, Uchijima Y, Kurihara Y, Nakagawa O, Asano T, Kurihara H. Transcriptional activity of Pax3 is co-activated by TAZ. Biochem Biophys Res Commun. 2006;339:533–539. doi: 10.1016/j.bbrc.2005.10.214. [DOI] [PubMed] [Google Scholar]
- Nakatani K, Nishioka J, Itakura T, Nakanishi Y, Horinouchi J, Abe Y, Wada H, Nobori T. Cell cycle-dependent transcriptional regulation of calmodulin-binding transcription activator 1 in neuroblastoma cells. Int J Oncol. 2004;24:1407–1412. [PubMed] [Google Scholar]
- O’Connell JX, Nielsen GP, Rosenberg AE. Epithelioid vascular tumors of bone: a review and proposal of a classification scheme. Adv Anat Pathol. 2001;8:74–82. doi: 10.1097/00125480-200103000-00003. [DOI] [PubMed] [Google Scholar]
- Olsen TG, Helwig EB. Angiolymphoid hyperplasia with eosinophilia. A clinicopathologic study of 116 patients. J Am Acad Dermatol. 1985;12:781–796. doi: 10.1016/s0190-9622(85)70098-9. [DOI] [PubMed] [Google Scholar]
- Ragnarsson G, Eiriksdottir G, Johannsdottir JT, Jonasson JG, Egilsson V, Ingvarsson S. Loss of heterozygosity at chromosome 1p in different solid human tumours: association with survival. Br J Cancer. 1999;79:1468–1474. doi: 10.1038/sj.bjc.6690234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosai J, Gold J, Landy R. Vascular disorder. Am J Surg Pathol. 1987;11:651–654. doi: 10.1097/00000478-198708000-00011. [DOI] [PubMed] [Google Scholar]
- Tapon N, Harvey KF, Bell DW, Wahrer DC, Schiripo TA, Haber DA, Hariharan IK. salvador Promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell. 2002;110:467–478. doi: 10.1016/s0092-8674(02)00824-3. [DOI] [PubMed] [Google Scholar]
- Troyanovsky B, Levchenko T, Mansson G, Matvijenko O, Holmgren L. Angiomotin: an angiostatin binding protein that regulates endothelial cell migration and tube formation. J Cell Biol. 2001;152:1247–1254. doi: 10.1083/jcb.152.6.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss SW. Reply to letter. Am J Surg Pathol. 1987;11:654. [Google Scholar]
- Weiss SW, Enzinger FM. Epithelioid hemangioendothelioma: a vascular tumor often mistaken for a carcinoma. Cancer. 1982;50:970–981. doi: 10.1002/1097-0142(19820901)50:5<970::aid-cncr2820500527>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- Wenger DE, Wold LE. Malignant vascular lesions of bone: radiologic and pathologic features. Skeletal Radiol. 2000;29:619–631. doi: 10.1007/s002560000261. [DOI] [PubMed] [Google Scholar]
- White PS, Thompson PM, Gotoh T, Okawa ER, Igarashi J, Kok M, Winter C, Gregory SG, Hogarty MD, Maris JM, Brodeur GM. Definition and characterization of a region of 1p36.3 consistently deleted in neuroblastoma. Oncogene. 2005;24:2684–2694. doi: 10.1038/sj.onc.1208306. [DOI] [PubMed] [Google Scholar]
- Xing W, Kim J, Wergedal J, Chen ST, Mohan S. Ephrin B1 regulates bone marrow stromal cell differentiation and bone formation by influencing TAZ transactivation via complex formation with NHERF1. Mol Cell Biol. 2010;30:711–721. doi: 10.1128/MCB.00610-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Wang W, Zhang S, Stewart RA, Yu W. Identifying tumor suppressors in genetic mosaics: the Drosophila lats gene encodes a putative protein kinase. Development. 1995;121:1053–1063. doi: 10.1242/dev.121.4.1053. [DOI] [PubMed] [Google Scholar]
- Zhang H, Liu CY, Zha ZY, Zhao B, Yao J, Zhao S, Xiong Y, Lei QY, Guan KL. TEAD transcription factors mediate the function of TAZ in cell growth and epithelial-mesenchymal transition. J Biol Chem. 2009;284:13355–13362. doi: 10.1074/jbc.M900843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 and S2
Figure S1. Schematic diagram of WWTR1, CAMTA1 and the projected fused WWTR1-CAMTA1 oncoprotein motifs. Preserved in the fusion include the 14-3-3 binding protein and WW domains of WWTR1 and the TIG (transcription factor immunoglobulin -like DNA-binding domain), ANK (ankyrin) and IQ (calmodulin-binding) motifs of CAMTA1.