Molecular basis for the inhibition of G protein-coupled inward rectifier K+ channels by protein kinase C (original) (raw)
Abstract
G protein-coupled inward rectifier K+ (GIRK) channels regulate cellular excitability and neurotransmission. The GIRK channels are activated by a number of inhibitory neurotransmitters through the G protein βγ subunit (Gβγ) after activation of G protein-coupled receptors and inhibited by several excitatory neurotransmitters through activation of phospholipase C. If the inhibition is produced by PKC, there should be PKC phosphorylation sites in GIRK channel proteins. To identify the PKC phosphorylation sites, we performed systematic mutagenesis analysis on GIRK4 and GIRK1 subunits expressed in Xenopus oocytes. Our data showed that the heteromeric GIRK1/GIRK4 channels were inhibited by a PKC activator phorbol 12-myristate 13-acetate (PMA) through reduction of single channel open-state probability. Direct application of the catalytic subunit of PKC to excised patches had a similar inhibitory effect. This inhibition was greatly eliminated by mutation of Ser-185 in GIRK1 and Ser-191 in GIRK4 that remained G protein sensitive. The PKC-dependent phosphorylation seems to mediate the channel inhibition by the excitatory neurotransmitter substance P (SP) as specific PKC inhibitors and mutation of these PKC phosphorylation sites abolished the SP-induced inhibition of GIRK1/GIRK4 channels. Thus, these results indicate that the PKC-dependent phosphorylation underscores the inhibition of GIRK channels by SP, and Ser-185 in GIRK1 and Ser-191 in GIRK4 are the PKC phosphorylation sites.
Keywords: Kir3, neurotransmitter, phosphorylation, postsynaptic
The G protein-coupled inward rectifier K+ (GIRK) channels play an important role in controlling membrane excitability and synaptic transmission (1, 2). Four members of GIRK channels have been cloned in mammals, i.e., GIRK1 through GIRK4 (Kir3.1 through Kir3.4). These channels are expressed in the heart, brain, and endocrine tissues (3, 4). Stoichiometric studies indicate that a functional GIRK channel consists of four homomeric subunits or two pairs of heteromeric subunits (3, 4). Coassembly of GIRK1 with GIRK4 forms muscarinic receptor-coupled K+ channels mainly in the heart (2, 5).
The GIRK channels are activated by certain inhibitory transmitters and hormones. On activation of Gi/o-coupled receptors by the neurotransmitters or hormones, the G protein βγ subunit (Gβγ) dissociated from the heterotrimeric Gαβγ directly interacts with cytosolic domains of GIRK channel proteins (6–9), and activates the channels through a mechanism that involves movement of pore-lining helices (10–12). Critical domains and amino acids have been identified to be responsible for the basal Gβγ-dependent and agonist-induced channel activities (6, 7, 13, 14). Furthermore, the GIRK channel activation may rely on local phosphatidylinositol-4,5-biphosphate (PIP2) in membrane microdomains (15, 16) or cytosolic Na+ (16, 17).
In addition to direct activation by Gβγ, GIRK channels are inhibited by a number of excitatory neurotransmitters or hormones, such as acetylcholine (18–20), thyroid-stimulating hormone (TSH)-releasing hormones (21), bombesin (22), substance P (SP) (23), and glutamate (24). These transmitters or hormones share common downstream signal pathways: transduction by pertussis toxin (PTX)-insensitive G protein Gq/11 (21, 25) and subsequent activation of phospholipase C (PLC) (18–22, 24, 25). It is clear that the activation of PLC results in hydrolysis of PIP2 and activation of PKC. Because PIP2 has been shown to activate GIRK, as well as other channels and transporters (15, 16, 26), PIP2 depletion has been proposed to mediate the inhibitory effects of some neurotransmitters on GIRK channels (19, 21, 27, 28) although the precise role of PIP2 in GIRK channel inhibition remains debatable (29, 30).
There is considerable evidence showing that PKC activation resulting from PLC activation underlies the inhibition of GIRK channels by a variety of neurotransmitters (18, 20, 22, 24, 31). The GIRK channels are inhibited by PKC activator phorbol 12-myristate 13-acetate (PMA) (18, 20, 22, 24, 31–34), and such inhibition is restored by protein phosphatase activation (31). Biochemical studies indicate that GIRK channel proteins are indeed phosphorylated with PKC activation (35). Because PKC activation is concomitantly accompanied with PIP2 hydrolysis, further dissection of the effect of PKC vs. PIP2 requires the demonstration of the PKC phosphorylation site(s) in the channel protein. Thus, we performed these experiments to identify the PKC phosphorylation site by systematic mutation analysis of all potential PKC phosphorylation sites. Our results indicate that Ser-185 in GIRK1 and Ser-191 in GIRK4 are likely to be the PKC sites critical for the inhibition of GIRK channels by the excitatory neurotransmitter SP.
Materials and Methods
Oocyte Preparation and Injection. Experiments were performed as described (36). In brief, frogs were anesthetized by bathing them in distilled water containing 0.3% 3-aminobenzoic acid ethyl ester. A few lobes of ovaries were removed through a small abdominal incision (≈5 mm). Then, the surgical incision was closed, and the frogs were allowed to recover from the anesthesia. Xenopus oocytes were treated with 2 mg/ml collagenase (Type IA, Sigma) in the OR2 solution (in mM) (NaCl 82/KCl 2/MgCl2 1/Hepes 5, pH 7.4) for 60 min at room temperature. After three washes (10 min each) of the oocytes with the OR2 solution, cDNAs (25–50 ng or 5–10 femtomoles in 50 nl of water) were injected into the oocytes. The oocytes were then incubated at 18°C in the ND-96 solution containing (in mM) NaCl 96, KCl 2, MgCl2 1, CaCl2 1.8, Hepes 5, and sodium pyruvate 2.5, with 100 mg/liter geneticin and 50 mg/liter tetracycline added (pH 7.4).
Molecular Biology. Rat GIRK1 (Kir3.1) cDNA (GenBank accession no. U01071) and rat GIRK4 (Kir3.4) cDNA (GenBank accession no. X83584) are gifts from H. Lester at California Institute of Technology (Pasadena). Rat SP receptor NK1 cDNA (GenBank accession no. J05097) was generously provided by S. Nakanishi at Kyoto University Faculty of Medicine (Kyoto). These cDNAs were subcloned into the eukaryotic expression vector pcDNA3.1 (Invitrogen) and used for Xenopus oocyte expression without cRNA synthesis. PCR was used to generate the GIRK1/GIRK4 dimer (36). Site-specific mutations were made by using a site-directed mutagenesis kit (Stratagene). Correct constructions and mutations were confirmed with DNA sequencing.
Electrophysiology. Whole-cell currents were studied on oocytes 2–4 days postinjection. Two-electrode voltage clamp was performed as we have detailed (36) by using an amplifier (Geneclamp 500, Axon Instruments, Foster City, CA) at room temperature (≈24°C). The extracellular recording solution contained 90 mM KCl, 3 mM MgCl2, and 5 mM Hepes (pH 7.4). The recording pipette was filled with 3 M KCl.
Single-channel currents were studied in cell-attached and inside-out patches by using an Axo-patch 200B amplifier (Axon Instruments). The bath solution contained 140 mM KCl, 10 mM Na2H2P2O7, 5 mM NaF, 0.1 mM Na3VO3, 0.2 mM ATP, 0.2 mM GTP, 1 mM MgCl2, 10 mM K-EGTA, and 10 mM Hepes (pH 7.4). The recording pipette was filled with the same solution (36). The pipette tip was ≈2 μm for cell-attached patch and ≈4 μm for excised patches. The open-state probability (NPo) was calculated as we have described (36) by counting all active channels from a given patch. The single channel conductance was measured by using a slope command potential from –100 to 100 mV.
Drug Treatment and Administration. PMA, 4α-phorbol-12,13-didecanoate (4α-PDD), and Gβγ were purchased from Calbiochem. The catalytic subunit of PKC, chelerythrine, calphostin-C, and SP (acetate salt) were purchased from Sigma. PMA, chelerythrine, calphostin-C, and 4α-PDD were dissolved in DMSO as stocks and mixed with a recording solution, reaching a final concentration as indicated in the text. Other chemicals were dissolved in double-distilled water or experimental solutions. Exposures to these chemicals were done after currents were stabilized.
Data Analysis. Data are presented as means ± SE. The Student t test or a single-factor ANOVA was used. Differences of chemical effects before vs. during chemical exposures were considered to be statistically significant if P ≤ 0.05.
Results
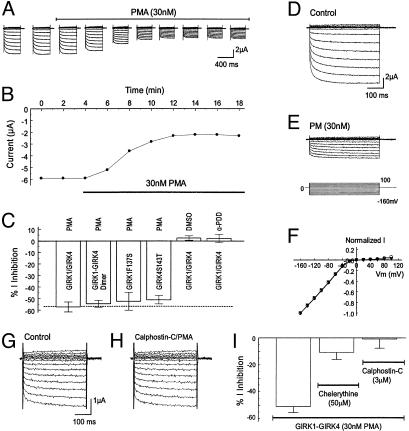
Inhibition of GIRK Channels by PKC Activation. In two-electrode voltage clamp, inward rectifying currents were recorded 2–3 days postinjection of GIRK1 with GIRK4 by using a bath solution containing 90 mM K+. These currents were sensitive to micromolar concentrations of Ba2+ (36). The GIRK currents were inhibited by 30 nM PMA, a specific and potent PKC activator (37). The inhibition started 2 min after exposure to PMA and reached a maximal effect in 8–10 min (Fig. 1 A and B). Thereafter, all PMA experiments were done with an exposure of cells to PMA for 10–20 min. At the maximal effect, 57.3 ± 4.3% (n = 6) of the inward rectifying currents were inhibited (Fig. 1_C_). The current inhibition occurred in all negative membrane potentials and did not show voltage-dependence (Fig. 1 D_–_F). Similar inhibition was seen in a tandem-dimeric GIRK1-GIRK4 channel (54.6 ± 3.0%, n = 5). The PMA effect was completely eliminated in the presence of 3 μM calphostin-C, a specific PKC blocker (Fig. 1 G and H). A similar effect was seen with another specific PKC blocker chelerythrine (50 μM), although to a less degree (Fig. 1_I_). In control experiments, 4α-PDD, an inactive PMA analog (37), and DMSO had no inhibitory effect (5.5 ± 3.0%, n = 5; 1.0 ± 2.9%, n = 5, respectively) (Fig. 1_C_), suggesting that the inhibition of GIRK1/GIRK4 channels was due to PKC activation, consistent with previous reports (18, 20, 22, 24, 31–34).
Fig. 1.
Inhibition of GIRK1/GIRK4 channels by PKC activation. (A) Whole-cell currents were studied in two-electrode voltage clamp by using a recording solution containing 90 mM K+. Inward rectifying currents were recorded from an oocyte 3 days after coinjection of the GIRK1 and GIRK4 cDNAs. Membrane potential (_V_m) was held at 0 mV. A series of command pulse potentials from –160 mV to 100 mV with a 20-mV increment was applied to the cell. Exposure to 30 nM PMA produced the inhibition of GIRK currents. (B) The current amplitude in A measured at –160 mV was plotted against time. The currents decreased rapidly after the oocyte was exposed to 30 nM PMA. At maximal inhibition, the current amplitude was inhibited by ≈50%. (C) Summary of data obtained from PMA experiments. PMA (30 nM) inhibited heteromeric, dimeric, and homomeric GIRK channels to almost the same degree whereas DMSO and 4α-PDD had no effect on the heteromeric GIRK1/GIRK4. Data are shown as means ± SE (n = 4–9). (D_–_F) Voltage independence of GIRK1/GIRK4 inhibition by PMA. (D) Currents were recorded from an oocyte in the same condition as in A. These currents were strongly inhibited by exposure to 30 nM PMA (10 min). (F) When currents in D and E were scaled to the same magnitude at –160 mV, the I/V relationship of the currents recorded in these two conditions became identical. Open circles, control; filled triangles, PMA exposure. (G and H) Blockade of the PMA effect by specific PKC blocker calphostin-C (3 μM). (I) A similar effect was seen with another specific PKC blocker chelerythrine (n = 6–9).
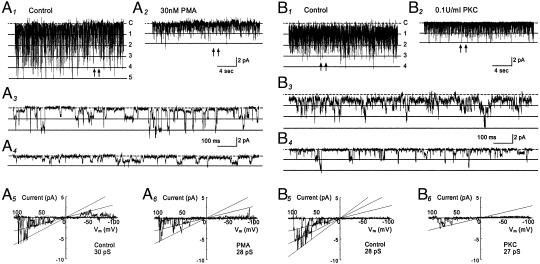
The effect of PMA on single-channel biophysical properties was studied in cell-attached patches with 150 mM K+ applied to extracellular solutions at a membrane potential of –80 mV. Inward rectifying currents with single-channel conductance of ≈30 pS were recorded from oocytes that had received a GIRK1-GIRK4 injection (Fig. 2 A1 and A3). These currents were inhibited with an exposure of the cells to 30 nM PMA (Fig. 2 A2 and A4). The current inhibition was mainly produced by a suppression of the channel open-state probability (NPo) whereas the single-channel conductance was only modestly inhibited (Fig. 2 A5 and A6). On average, 30 nM PMA produced an inhibition of the NPo by 59% (0.103 ± 0.029 vs. 0.042 ± 0.008, n = 10 patches) and a decrease in single-channel conductance by 5% (28.1 ± 0.9 pS vs. 26.7 ± 1.1 pS, n = 11).
Fig. 2.
Comparison of the effect of PMA to that of PKC. (A1–A6) GIRK1-GIRK4 currents were recorded from a cell-attached patch with 150 mM K+ applied to the extracellular solution at a membrane potential of –80 mV. Five active currents were seen at baseline with NPo 0.671 (A1). These currents were inhibited when the oocytes was exposed to 30 nM PMA for 10 min (NPo 0.076, A2). (A3 and A4) The single-channel currents are better seen with an expended time scale taken from A1 and A2 at places pointed by arrows, respectively. (A5 and A6) In contrast to the NPo, single-channel conductance was inhibited only modestly by PMA. (B1–B6) Inhibition of the GIRK1-GIRK4 currents by purified catalytic subunit of PKC. (B1) Single-channel currents were studied in an excised inside-out patch with 150 mM K+ applied to both sides of the patch membrane. Four active channels are seen at baseline with NPo 1.143. (B2) The channel activity was strongly inhibited (NPo 0.144) 10 min after an exposure of the internal surface of the patch member to PKC (0.1 units/ml). (B5 and B6) PKC had very little effect on single-channel conductance in another inside-out patch. Note that substate conductances are seen in all records.
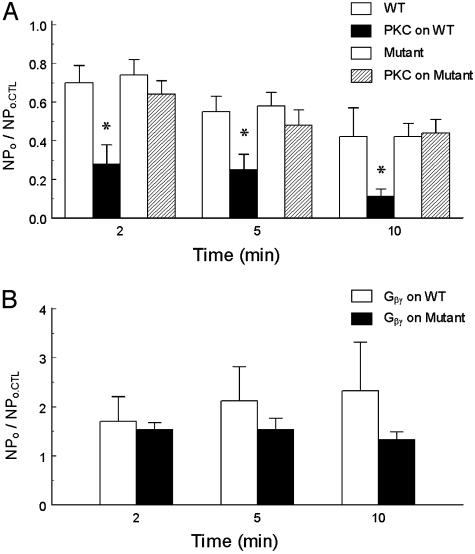
In excised inside-out patches, the GIRK1-GIRK4 currents were strongly inhibited by the catalytic subunit of PKC purified from the rat brain. Evident inhibition of the channel activity was seen 2 min after a 20-s exposure of internal patch membranes to PKC (Fig. 2 B1_–_B4). By 5 min after the exposure, the NPo was reduced by >75% (1.207 ± 0.636 vs. 0.260 ± 0.167, n = 6) when the channel activity dropped by ≈45% in the absence of PKC (0.332 ± 0.162 vs. 0.199 ± 0.114, n = 5) (see Fig. 4_A_). Similar to PMA, PKC had very little effect on the single-channel conductance (28.3 ± 0.6 pS vs. 27.5 ± 1.3 pS, n = 6; Fig. 2 B5 and B6).
Fig. 4.
Effects of catalytic subunit PKC and Gβγ on WT and mutant GIRK channels. Single-channel currents were studied in inside-out patches in the same condition as Fig. 2. (A) Exposure of the internal patch membrane to the catalytic subunit of PKC (0.1 unit/ml for 20 s) strongly inhibited the WT channels (*, P < 0.05 in comparison with the control group WT without PKC) whereas PKC did not significantly reduce activity of the GIRK1S185A/GIRK4S191A mutant in comparison with that in the absence of PKC (_P_ > 0.05, n = 4–10). NPo.CTL = baseline control NPo. (B) A 20-s exposure of the internal patch membranes to 20 nM Gβγ markedly enhanced channel activity of the GIRK1S185A/GIRK4S191A mutant in comparison with the WT GIRK1-GIRK4 in the absence of G protein in A. No statistical difference was found between WT and mutant groups although the mutant seems to run down faster (n = 5–6).
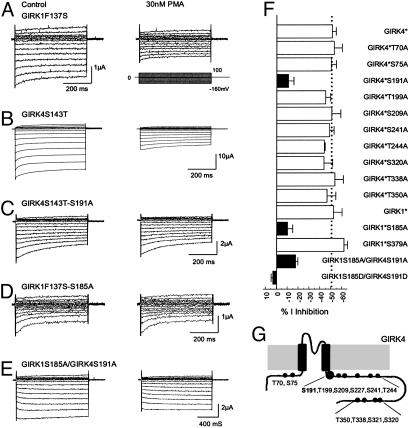
PKC Phosphorylation Sites in GIRK1 and GIRK4 Subunits. To elucidate which subunit of GIRK1 and GIRK4 was phosphorylated by PKC, experiments were conducted on homomeric GIRK1 and GIRK4 channels. It is known that GIRK1 does not form homomeric channels (38–40), and the basal currents of homomeric GIRK4 are too small to study (5). Consistent with these previous observations, we failed to record currents from the oocytes injected with GIRK1 or GIRK4. No detectable currents were seen with cRNA injection. Previous studies have shown that mutation of a single amino acid in the pore region allows homomeric expression of GIRK channels (39), which has been widely used in studying GIRK channels (10, 13, 14, 34, 36). Therefore, the F137S mutation was constructed in GIRK1 (GIRK1F137S, GIRK1*) and the S143T in GIRK4 (GIRK4S143T, GIRK4*) by using the site-directed mutagenesis technique. Consistent with the previous reports, both GIRK1* and GIRK4* expressed inward rectifying currents. These currents were inhibited by 30 nM PMA to the same degree as their heteromeric counterpart (Figs. 1_C_ and 3 A and B), suggesting that both GIRK1 and GIRK4 subunits can be phosphorylated by PKC.
Fig. 3.
PMA sensitivity of homomeric and heteromeric GIRK1 and GIRK4. (A) Currents were recorded from an oocyte in the same condition as Fig. 1_A_. The GIRK1F137S currents were markedly inhibited by 30 nM PMA. (B) Similar current inhibition was seen in another oocyte injected with the GIRK4S143T cDNA. (C) When Ser-191 was substituted with an alanine in GIRK4S143T, the GIRK4S143T-S191A currents were barely inhibited by 30 nM PMA. Similar results were found in the GIRK1F137S-S185A mutant (D) and heteromeric GIRK1S185A/GIRK4S191A channel (E). (F) Identification of PKC phosphorylation sites by alanine-scanning mutagenesis. Ser-185 in the GIRK4 and Ser-191 in the GIRK4 were shown to affect the PMA sensitivity. Mutation of any one greatly eliminated the PMA sensitivity of the homomeric channels. Joint mutations of both also significantly diminished the PMA sensitivity of heteromeric GIRK1/GIRK4 channel. Mutations of other putative PKC phosphorylation sites had no effect (P < 0.01 for all filled bars). GIRK1*, GIRK1F137S; GIRK4*, GIRK4S143T. (G) Schematic of the GRIK4 subunit and locations of all serine/threonine residues studied.
To identify the PKC phosphorylation sites, a sequence alignment of amino acids was made for GIRK channels. Twelve putative PKC phosphorylation sites were found in GIRK1, -2, and -4 that have been shown to be inhibited by PMA (34), according to the consensus sequences, i.e., R(K)X0–2T/SX0–2R(K) (41). We systematically screened all of them in the GIRK4* by mutating each of them to an alanine residue. The effects of PMA on these mutants were tested subsequently. All mutants except the GIRK4*S227A and GIRK4*S321A expressed detectable inward rectifying currents (Table 1, which is published as supporting information on the PNAS web site). The S191A mutation significantly diminished the PMA-induced channel inhibition (11.1 ± 4.8%, n = 14; P < 0.01) (Fig. 3_C_) whereas no significant change in the PMA effect was found in other mutants (Fig. 3_F_). To express detectable GIRK4*S227A and GIRK4*S321A currents, another mutation of I229L, which has been previously used to rescue nonfunctional GIRK mutants by enhancing the channel-PIP2 interaction (14, 42), was introduced into these two mutants. The GIRK4*I229L/S321A showing larger basal currents was still inhibited by PMA to the same degree as the GIRK4*I229L (59.9 ± 3.7%, n = 10 for GIRK4*I229L; and 59.6 ± 3.9%, n = 9 for GIRK4*I229L-S321A), suggesting that Ser-321 in GIRK4 is not a PKC phosphorylation site. There was no evident expression of inward rectifying currents by injection of the GIRK4*I229L/S227A mutant, either by coinjections of GIRK4*S227A with the cDNAs encoding Gβγ. We also mutated the Ser-227 to asparigine, a residue that is found at the corresponding site in Kir5.1. Still, the GIRK4*S227N failed to express detectable currents. Therefore, whether Ser-227 in GIRK4 is another PKC phosphorylation site remains to be proven.
If the Ser-191 in GIRK4 is a phosphorylation site, mutation of Ser-185 at the corresponding site in GIRK1 may also affect the PKC-induced channel inhibition. Thus, the Ser-185 in GIRK1* was mutated to alanine. The inhibition of GIRK1*S185A by PMA was significantly reduced in comparison with GIRK1* (10.3 ± 4.7%, n = 8; P < 0.01) (Fig. 3 D and F) whereas mutation of Ser-379 in GIRK1* had no effect, suggesting that Ser-185 is a PKC phosphorylation site in the GIRK1 channel.
To understand whether the Ser-191 in GIRK4 and Ser-185 in GIRK1 indeed mediate the PMA-induced inhibition of the heteromeric GIRK1/GIRK4 channel, GIRK1S185A and GIRK4S191A mutations were constructed in the WT GIRK1 and GIRK4, and coexpressed in the oocytes. The GIRK1S185A/GIRK4S191A showed baseline currents 3.7 ± 0.4 μA (n = 14) and were inhibited by 30 nM PMA by 17.8 ± 2.0%, which was significantly smaller than that of the WT GIRK1/GIRK4 (P < 0.01) (Fig. 3 E and F). More evident effects were observed in the GIRK1S185D/GIRK4S191D mutant, which had baseline currents 1.7 ± 0.1 μA (n = 14) and was not inhibited by PMA at all (Fig. 3_F_). Because the GIRK1S185A/GIRK4S191A did not completely eliminate the current inhibition by PMA, there may be another unidentified PKC site that could be the Ser-221 in GIRK1 and Ser-227 in GIRK4. Thereby, heteromeric expression of these two mutants was attempted. However, coinjections of GIRK1S221A/GIRK4S227A, GIRK1/GIRK4S227A, or GIRK1S221A/GIRK4 still led to no convincing functional expression.
The sensitivity of the GIRK1S185A/GIRK4S191A to the catalytic subunit of PKC was studied in inside-out patches. No detectable inhibition was found in comparison with this mutant studied in the absence of PKC (P > 0.05) (Fig. 4_A_). Like its WT counterpart, the GIRK1S185A/GIRK4S191A remained to be stimulated by the Gβγ subunit in excised patches although the channel rundown seemed to be faster than the WT channels (Fig. 4_B_).
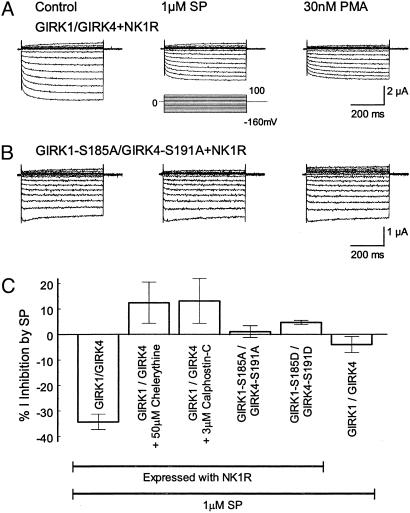
PKC Phosphorylation in GIRK Inhibition by SP. SP has been shown to excite neurons through suppression of GIRK channel activity (23, 43, 44), which is believed to be mediated by activation of the Gq-PLC-PKC signaling pathway (25, 31, 45). If this is the case, mutation of the PKC phosphorylation sites should abolish the GIRK inhibition by SP. To test this idea, WT and mutant GIRK channels were coexpressed with an SP receptor NK1R. Consistent with previous reports (18, 19, 46), we observed a transient increase followed by a steady inhibition in the GIRK currents on exposure to 1 μM SP. The former was not seen in a Cl– free bath solution suggesting that it is likely caused by activation of oocyte-endogenous Cl– channels as shown previously (46–48). The latter inhibition was long lasting and more remarkable. At maximal effect, the GIRK currents were inhibited by 34.3 ± 3.0% (n = 8) (Fig. 5 A and C). SP had no significant effect on the GIRK currents without NK1R (4.0 ± 3.2%, n = 5) (Fig. 5_C_).
Fig. 5.
Mediation of substance P-induced inhibition of GIRK channels by PKC. (A) The GIRK1/GIRK4 channel coexpressed with NK1 receptor was inhibited by 1 μM SP. After the inhibition, 30 nM PMA had no further inhibitory effect. (B) Mutation of Ser-185 in the GIRK4 and Ser-191 in the GIRK4 to alanine eliminated the SP- and PMA-induced channel inhibition. (C) SP inhibited the heteromeric GIRK1/GIRK4 channel coexpressed with NK1 receptor. The inhibition was not seen in the mutants with alanine or aspartate at these putative phosphorylation sites, in the presence of specific PKC blockers, or in the GIRK1/GIRK4 expressed without the NK1 (n = 4–12).
After the inhibition, PMA (30 nM) produced only small and insignificant additional inhibition (6.1 ± 3.8%, n = 3, P > 0.05), suggesting that the preexposure of GIRK channels to SP prevents the inhibition of GIRK channels by further activation of PKC. The SP-induced inhibition was abolished with specific PKC inhibitors or PKC-site mutations (Fig. 5 B and C). Neither the GIRK1S185A/GIRK4S191A nor the GIRK1S185D/GIRK4S191D was inhibited by SP (1.1 ± 2.3%, n = 12; and 4.7 ± 0.8%, n = 6, respectively). We also examined the SP sensitivity of the GIRK4*I229L expressed with the NK1R. The currents retained to be inhibited by 1 μM SP (–20.3 ± 2.1%, n = 4; Fig. 6, which is published as supporting information on the PNAS web site). Thus, these data indicate that the PKC phosphorylation sites identified in the present study are involved in the modulation of GIRK channels by the neurotransmitter SP.
Discussion
We have presented evidence for the molecular mechanism underlying the inhibition of heteromeric GIRK1/GIRK4 channels by PKC. The PKC-dependent inhibition of GIRK channels relies on Ser-185 in GIRK1 and Ser-191 in GIRK4. Their phosphorylation accounts for the inhibition of GIRK channels by excitatory transmitter SP.
Inhibition of GIRK Channels by PKC. Modulation of ion channels by protein phosphorylation and dephosphorylation is an important regulatory mechanism (49). Previous studies have shown that GIRK channels are targets of protein kinase A (35, 50, 51), PKC (18, 20, 22, 24, 31), tyrosine kinase (34, 52), Ca2+-calmodulin-dependent kinase II (35), and phosphatases (30, 35, 51), although the molecular mechanisms underlying the GIRK channel phosphorylation are mostly unknown (except tyrosine kinase phosphorylation; see ref. 34). GIRK channels have been shown to be inhibited by PKC activation in both native cells (30–32) and heterologous expression systems (18, 20, 22, 24, 33, 34). Other studies suggest that GIRK channel inhibition by certain excitatory neurotransmitters may be mediated by PIP2 depletion (19, 21, 27, 28). In addition, PMA may affect endocytosis changing GIRK channel activity through GIRK protein turnover (53, 54). Therefore, the demonstration of PKC sites in the channel proteins becomes critical for the understanding of cell-signaling pathways responsible for the GIRK channel inhibition. In the present study, we have shown that heteromeric GIRK1/GIRK4 as well as homomeric GIRK1* and GIRK4* are inhibited by PMA at nanomolar concentrations. The effect of PMA is specific, as (i) such an effect is abolished in the presence of a specific PKC blocker, (ii) the catalytic subunit of PKC has the same effect, (iii) 4α-PDD, a PMA analog that is incapable of activating PKC (37), has no effect, and (iv) most importantly, the inhibitory effect by PKC and PMA is eliminated by replacing potential PKC phosphorylation sites in the GIRK channel proteins. Because PMA inhibits the GIRK4*I229L [a mutant defective in PIP2 sensitivity (42)] and because PMA at μM concentrations decreases PIP2 (55), PIP2 depletion may have rather small contribution to the GIRK channel inhibition by nM PMA. Although there may be other mechanisms, these results indicate that PKC phosphorylation plays a major role in the GIRK channel inhibition.
PKC Phosphorylation Sites in GIRK1 and GIRK4. By systematic mutation analysis, two PKC phosphorylation sites have been identified in the present study. Substitutions of these sites with a nonphosphorylatable residue greatly eliminate the channel inhibition by PKC and PMA whereas similar mutations of other potential PKC phosphorylation sites have no effect. The PMA sensitivity is completely eliminated in the GIRK1S185D/GIRK4S191D mutant with a much smaller baseline current, suggesting that the negative charges at these sites may mimic the effect of phosphorylation. Rogalski et al. (34) have shown that the homomeric GIRK2S146T channel is inhibited by PMA. Because this serine residue is conserved in all GIRK channels, it (Ser-196) might be involved in GIRK2 modulation by PKC as well. We have noticed that the GIRK1S185A/GIRK4S191A mutant is still modestly inhibited by PMA. The remaining effect is likely produced by Ser-227 in GIRK4 and Ser-221 in GIRK1 because the corresponding site in Kir1 channels has been identified to be a PKC phosphorylation site (56). We have spent enormous efforts on this residue but failed to express functional channels. Therefore, whether it is the missing phosphorylation site remains to be known. Nonetheless, our results are consistent with previous deletional and chimerical studies implying that phosphorylation sites are located in the proximal C terminus (18, 35). Because this region is known to be critical for Kir channel gating (10, 57), our observations suggest that the PKC-dependent phosphorylation of GIRK channels plays a role in GIRK channel gating.
PKC-Dependent Phosphorylation in Transmitter-Induced Inhibition of GIRK Channels. In addition to the activation by neurotransmitters through PTX-sensitive G protein Gi/o, GIRK channels are inhibited by numerous neurotransmitters (18, 20, 22, 24, 25). The GIRK channel inhibition by these neurotransmitters involves activation of PTX-insensitive Gq/11 and PLC pathways (25). The downstream signaling pathways have not been well demonstrated due to the complex consequences of PLC activation. Some reports suggest that PIP2 depletion is important (19, 21, 28, 29) whereas others show that PKC activation is critical (18, 20, 22, 24, 31–33). SP is one of the neurotransmitters that activate Gq/11 signaling pathways (25, 58) and modulate cellular excitability through reducing inward rectifier K+ currents in neurons (23, 44). Our results show that the mutation of PKC phosphorylation sites as well as PKC inhibitors abolish the inhibition of GIRK1/GIRK4 channels by SP. Also, SP inhibits the GIRK4*I229L mutant. These results thus provide direct evidence for PKC-dependent phosphorylation of GIRK channel in neurotransmitter actions (18, 20, 22–24). This observation, however, does not rule out the regulation of local PIP2 in the control of ion channel functions (26) because it is also possible that the GIRK channels are excessively regulated by different intracellular mechanisms.
Functional Implications. It is known that the inhibitory transmitters activate PTX-sensitive Gi/o-coupled signaling pathways whereas the excitatory transmitters initiate Gq-PLC pathways, creating opposing pathways for modulation of GIRK channel activity. It has been demonstrated that the opposite modulation of GIRK channels by the two kinds of transmitters plays an important role in regulating cellular excitability and neurotransmission. The activation of GIRK channels by Gβγ-dependent transmitters can be interfered with or blocked by excitatory transmitters that activate PKC (23, 32). The GIRK channel phosphorylation may disrupt the interaction of Gβγ with GIRK channels and antagonize the receptor-mediated GIRK channel activation observed in neurons in a manner similar to the voltage-gated Ca2+ channels. In contrast to GIRK channels, the N-type Ca2+ channels in presynaptic membranes are activated by PKC phosphorylation (59, 60) and inhibited by direct binding of the Gβγ to the channel protein (61–63). The activation effect by PKC is dominant when both signaling pathways are simultaneously initiated (62, 63). As a result, the PKC-dependent phosphorylation hinders the Gβγ-mediated inhibition of Ca2+ channel (62, 64). Thus, the PKC-dependent phosphorylation may act as a switch controlling the channel activity modulated by Gβγ, an effect that is currently believed to act directly and to be subject to only moderate regulation in cell signaling. Such apparently opposite effects may allow neurons to integrate presynaptic signals according to their temporal and spatial placement.
Supplementary Material
Supporting Information
Acknowledgments
We thank Dr. Henry A. Lester at California Institute of Technology for the Kir3.1 and Kir3.4 cDNA, and Dr. Shigetada Nakanishi at Kyoto University Faculty of Medicine for the NK1 receptor cDNA. This work was supported by National Institutes of Health Grants HL58410 and HL67890 and by American Diabetes Association Grant 1-01-RA-12.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: GIRK, G protein-coupled inward rectifier K+; PIP2, phosphatidylinositol-4,5-biphosphate; SP, substance P; PTX, pertussis toxin; PLC, phospholipase C; PMA, phorbol 12-myristate 13-acetate; 4α-PDD, 4α-phorbol-12,13-didecanoate; Gβγ, G protein βγ subunit.
References
- 1.Luscher, C., Jan, L. Y., Stoffel, M., Malenka, R. C. & Nicoll, R. A. (1997) Neuron 19**,** 687–695. [DOI] [PubMed] [Google Scholar]
- 2.Wickman, K., Nemec, J., Gendler, S. J. & Clapham, D. E. (1998) Neuron 20**,** 103–114. [DOI] [PubMed] [Google Scholar]
- 3.Dascal, N. (1997) Cell Signalling 9**,** 551–573. [DOI] [PubMed] [Google Scholar]
- 4.Yamada, M., Inanobe, A. & Kurachi, Y. (1998) Pharmacol. Rev. 50**,** 723–760. [PubMed] [Google Scholar]
- 5.Krapivinsky, G., Gordon, E. A., Wickman, K., Velimirovic, B., Krapivinsky, L. & Clapham, D. E. (1995) Nature 374**,** 135–141. [DOI] [PubMed] [Google Scholar]
- 6.Huang, C. L., Slesinger, P. A., Casey, P. J., Jan, Y. N. & Jan, L. Y. (1995) Neuron 15**,** 1133–1143. [DOI] [PubMed] [Google Scholar]
- 7.Kunkel, M. T. & Peralta, E. G. (1995) Cell 83**,** 443–449. [DOI] [PubMed] [Google Scholar]
- 8.Reuveny, E., Slesinger, P. A., Inglese, J., Morales, J. M., Iniguez-Lluhi, J. A., Lefkowitz, R. J., Bourne, H. R., Jan, Y. N. & Jan, L. Y. (1994) Nature 370**,** 143–146. [DOI] [PubMed] [Google Scholar]
- 9.Slesinger, P. A., Reuveny, E., Jan, Y. N. & Jan, L. Y. (1995) Neuron 15**,** 1145–1156. [DOI] [PubMed] [Google Scholar]
- 10.Jin, T., Peng, L., Mirshahi, T., Rohacs, T., Chan, K. W., Sanchez, R. & Logothetis, D. E. (2002) Mol. Cell 10**,** 469–481. [DOI] [PubMed] [Google Scholar]
- 11.Sadja, R., Smadja, K., Alagem, N. & Reuveny, E. (2001) Neuron 29**,** 669–680. [DOI] [PubMed] [Google Scholar]
- 12.Yi, B. A., Lin, Y. F., Jan, Y. N. & Jan, L. Y. (2001) Neuron 29**,** 657–667. [DOI] [PubMed] [Google Scholar]
- 13.He, C., Zhang, H., Mirshahi, T. & Logothetis, D. E. (1999) J. Biol. Chem. 274**,** 12517–12524. [DOI] [PubMed] [Google Scholar]
- 14.He, C., Yan, X., Zhang, H., Mirshahi, T., Jin, T., Huang, A. & Logothetis, D. E. (2002) J. Biol. Chem. 277**,** 6088–6096. [DOI] [PubMed] [Google Scholar]
- 15.Huang, C. L., Feng, S. & Hilgemann, D. W. (1998) Nature 391**,** 803–806. [DOI] [PubMed] [Google Scholar]
- 16.Sui, J. L., Petit-Jacques, J. & Logothetis, D. E. (1998) Proc. Natl. Acad. Sci. USA 95**,** 1307–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ho, I. H. & Murrell-Lagnado, R. D. (1999) J. Biol. Chem. 274**,** 8639–8648. [DOI] [PubMed] [Google Scholar]
- 18.Hill, J. J. & Peralta, E. G. (2001) J. Biol. Chem. 276**,** 5505–5510. [DOI] [PubMed] [Google Scholar]
- 19.Kobrinsky, E., Mirshahi, T., Zhang, H., Jin, T. & Logothetis, D. E. (2000) Nat. Cell Biol. 2**,** 507–514. [DOI] [PubMed] [Google Scholar]
- 20.Leaney, J. L., Dekker, L. V. & Tinker, A. (2001) J. Physiol. (London) 534**,** 367–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lei, Q., Talley, E. M. & Bayliss, D. A. (2001) J. Biol. Chem. 276**,** 16720–16730. [DOI] [PubMed] [Google Scholar]
- 22.Stevens, E. B., Shah, B. S., Pinnock, R. D. & Lee, K. (1999) Mol. Pharmacol. 55**,** 1020–1027. [PubMed] [Google Scholar]
- 23.Velimirovic, B. M., Koyano, K., Nakajima, S. & Nakajima, Y. (1995) Proc. Natl. Acad. Sci. USA 92**,** 1590–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharon, D., Vorobiov, D. & Dascal, N. (1997) J. Gen. Physiol 109**,** 477–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takano, K., Yasufuku-Takano, J., Kozasa, T., Singer, W. D., Nakajima, S. & Nakajima, Y. (1996) J. Neurophysiol. 76**,** 2131–2136. [DOI] [PubMed] [Google Scholar]
- 26.Hilgemann, D. W., Feng, S. & Nasuhoglu, C. (2001) Sci. STKE 2001**,** RE19. [DOI] [PubMed] [Google Scholar]
- 27.Cho, H., Nam, G. B., Lee, S. H., Earm, Y. E. & Ho, W. K. (2001) J. Biol. Chem. 276**,** 159–164. [DOI] [PubMed] [Google Scholar]
- 28.Meyer, T., Wellner-Kienitz, M. C., Biewald, A., Bender, K., Eickel, A. & Pott, L. (2001) J. Biol. Chem. 276**,** 5650–5658. [DOI] [PubMed] [Google Scholar]
- 29.Cho, H., Hwang, J. Y., Kim, D., Shin, H. S., Kim, Y., Earm, Y. E. & Ho, W. K. (2002) J. Biol. Chem. 277**,** 27742–27747. [DOI] [PubMed] [Google Scholar]
- 30.Kim, D. & Bang, H. (1999) J. Physiol. (London) 517**,** 59–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takano, K., Stanfield, P. R., Nakajima, S. & Nakajima, Y. (1995) Neuron 14**,** 999–1008. [DOI] [PubMed] [Google Scholar]
- 32.Andrade, R., Malenka, R. C. & Nicoll, R. A. (1986) Science 234**,** 1261–1265. [DOI] [PubMed] [Google Scholar]
- 33.Chen, Y. & Yu, L. (1994) J. Biol. Chem. 269**,** 7839–7842. [PubMed] [Google Scholar]
- 34.Rogalski, S. L., Appleyard, S. M., Pattillo, A., Terman, G. W. & Chavkin, C. (2000) J. Biol. Chem. 275**,** 25082–25088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Medina, I., Krapivinsky, G., Arnold, S., Kovoor, P., Krapivinsky, L. & Clapham, D. E. (2000) J. Biol. Chem. 275**,** 29709–29716. [DOI] [PubMed] [Google Scholar]
- 36.Mao, J., Li, L., McManus, M., Wu, J., Cui, N. & Jiang, C. (2002) J. Biol. Chem. 277**,** 46166–46171. [DOI] [PubMed] [Google Scholar]
- 37.Monroe, J. G., Niedel, J. E. & Cambier, J. C. (1984) J. Immunol. 132**,** 1472–1478. [PubMed] [Google Scholar]
- 38.Hedin, K. E., Lim, N. F. & Clapham, D. E. (1996) Neuron 16**,** 423–429. [DOI] [PubMed] [Google Scholar]
- 39.Vivaudou, M., Chan, K. W., Sui, J. L., Jan, L. Y., Reuveny, E. & Logothetis, D. E. (1997) J. Biol. Chem. 272**,** 31553–31560. [DOI] [PubMed] [Google Scholar]
- 40.Wischmeyer, E., Doring, F., Wischmeyer, E., Spauschus, A., Thomzig, A., Veh, R. & Karschin, A. (1997) Mol. Cell. Neurosci. 9**,** 194–206. [DOI] [PubMed] [Google Scholar]
- 41.Kennelly, P. J. & Krebs, E. G. (1991) J. Biol. Chem. 266**,** 15555–15558. [PubMed] [Google Scholar]
- 42.Zhang, H., He, C., Yan, X., Mirshahi, T. & Logothetis, D. E. (1999) Nat. Cell Biol. 1**,** 183–188. [DOI] [PubMed] [Google Scholar]
- 43.Gray, P. A., Rekling, J. C., Bocchiaro, C. M. & Feldman, J. L. (1999) Science 286**,** 1566–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stanfield, P. R., Nakajima, Y. & Yamaguchi, K. (1985) Nature 315**,** 498–501. [DOI] [PubMed] [Google Scholar]
- 45.Yamada, K. & Akasu, T. (1996) J. Physiol. (London) 496**,** 439–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Choi, S., Lee, J. H., Kim, Y. I., Kang, M. J., Rhim, H., Lee, S. M. & Nah, S. Y. (2003) Eur. J. Pharmacol. 468**,** 83–92. [DOI] [PubMed] [Google Scholar]
- 47.Gereau, R. W. & Heinemann, S. F. (1998) Neuron 20**,** 143–151. [DOI] [PubMed] [Google Scholar]
- 48.Parker, I., Gundersen, C. B. & Miledi, R. (1985) Proc. R. Soc. London Ser. B Biol. Sci. 223**,** 279–292. [DOI] [PubMed] [Google Scholar]
- 49.Levitan, I. B. (1994) Annu. Rev. Physiol. 56**,** 193–212. [DOI] [PubMed] [Google Scholar]
- 50.Mullner, C., Vorobiov, D., Bera, A. K., Uezono, Y., Yakubovich, D., Frohnwieser-Steinecker, B., Dascal, N. & Schreibmayer, W. (2000) J. Gen. Physiol. 115**,** 547–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mullner, C., Yakubovich, D., Dessauer, C. W., Platzer, D. & Schreibmayer, W. (2003) Biophys. J. 84**,** 1399–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang, X. Y., Morielli, A. D. & Peralta, E. G. (1993) Cell 75**,** 1145–1156. [DOI] [PubMed] [Google Scholar]
- 53.van Balkom, B. W., Savelkoul, P. J., Markovich, D., Hofman, E., Nielsen, S., van der Sluijs, P. & Deen, P. M. (2002) J. Biol. Chem. 277**,** 41473–41479. [DOI] [PubMed] [Google Scholar]
- 54.Beron, J., Forster, I., Beguin, P., Geering, K. & Verrey, F. (1997) Mol. Biol. Cell 8**,** 387–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nasuhoglu, C. Feng, S., Mao, Y., Shammat, I., Yamamato, M., Earnest, S., Lemmon, M. & Hilgemann, D. W. (2002) Am. J. Physiol. 283**,** C223–C234. [DOI] [PubMed] [Google Scholar]
- 56.Lin, D., Sterling, H., Lerea, K. M., Giebisch, G. & Wang, W. H. (2002) J. Biol. Chem. 277**,** 44278–44284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Perozo, E. (2002) Structure (Cambridge, MA) 10**,** 1027–1029. [DOI] [PubMed] [Google Scholar]
- 58.Nakajima, Y., Nakajima, S. & Inoue, M. (1988) Proc. Natl. Acad. Sci. USA 85**,** 3643–3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Swartz, K. J., Merritt, A., Bean, B. P. & Lovinger, D. M. (1993) Nature 361**,** 165–168. [DOI] [PubMed] [Google Scholar]
- 60.Yang, J. & Tsien, R. W. (1993) Neuron 10**,** 127–136. [DOI] [PubMed] [Google Scholar]
- 61.Mintz, I. M. & Bean, B. P. (1993) Neuron 10**,** 889–898. [DOI] [PubMed] [Google Scholar]
- 62.Zamponi, G. W., Bourinet, E., Nelson, D., Nargeot, J. & Snutch, T. P. (1997) Nature 385**,** 442–446. [DOI] [PubMed] [Google Scholar]
- 63.De Waard, M., Liu, H., Walker, D., Scott, V. E., Gurnett, C. A. & Campbell, K. P. (1997) Nature 385**,** 446–450. [DOI] [PubMed] [Google Scholar]
- 64.Hamid, J., Nelson, D., Spaetgens, R., Dubel, S. J., Snutch, T. P. & Zamponi, G. W. (1999) J. Biol. Chem. 274**,** 6195–6202. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information