A tumor suppressor function of Smurf2 associated with controlling chromatin landscape and genome stability through RNF20 (original) (raw)
. Author manuscript; available in PMC: 2012 Aug 1.
Published in final edited form as: Nat Med. 2012 Jan 8;18(2):227–234. doi: 10.1038/nm.2596
Abstract
In addition to allelic mutations, cancers are known to harbor alterations in their chromatin landscape. Here, we show that genomic ablation of Smurf2, a HECT-domain E3 ubiquitin ligase, results in dysregulation of DNA damage response and genomic stability, culminating to increased susceptibility to various types of cancers in aged mice. We demonstrate that Smurf2 regulates histone H2B monoubiquitination as well as histone H3 tri-methylation at K4 and K79 by targeting RNF20 to proteasomal degradation in both mouse and human cells. We further show that Smurf2 and RNF20 are co-localized at the γ-H2AX foci of double-stranded DNA breaks in the nucleus. Thus, Smurf2 has a tumor suppression function that normally maintains genomic stability by controlling the epigenetic landscape of histone modifications through RNF20.
INTRODUCTION
Ubiquitin modification controls a wide array of cellular functions by tagging proteins for proteasomal degradation or incorporation into other regulatory complexes1. Central to this system are the E3 ubiquitin ligases that function in a chain of reactions, resulting in the attachment of ubiquitin moieties to target proteins. Smad ubiquitin regulatory factor 2 (Smurf2), a HECT domain-containing E3 ligase, was initially recognized as a negative regulator of TGF-β signaling by targeting Smads and the type I receptor2–6. Subsequent studies have broadened the repertoire of Smurf2 substrates and extended its function to controlling neuronal polarity and planar cell polarity7,8,9. In human cells, Smurf2 up-regulation was linked to telomere attrition and forced expression of Smurf2 was sufficient to induce senescence in fibroblasts10. Abnormal expression of Smurf2 was also reported in subsets of esophageal squamous cell carcinomas and breast carcinomas11,12, but whether dysregulation of Smurf2 leads to tumorigenesis is not clear. In an attempt to address the physiological function of Smurf2, we serendipitously found that genomic ablation of Smurf2 leads to global changes in histone modifications and predisposition to a wide spectrum of tumors. It is well established that in addition to allelic mutations, cancer cells harbor epigenetic alterations in patterns of histone and DNA modification as well as chromatin structure13. Here, we present evidence for a tumor suppressor function of Smurf2 that controls chromatin landscape by targeting RNF20, the major E3 ligase responsible for mono-ubiquitin modification of histone H2B (ubH2B)14,15, which is actively engaged in transcription16 and also involved in DNA damage repair in nuclear foci of DNA double-stranded breaks17,18.
RESULTS
Aged Smurf2−/− mice displays a wide spectrum of tumor phenotypes
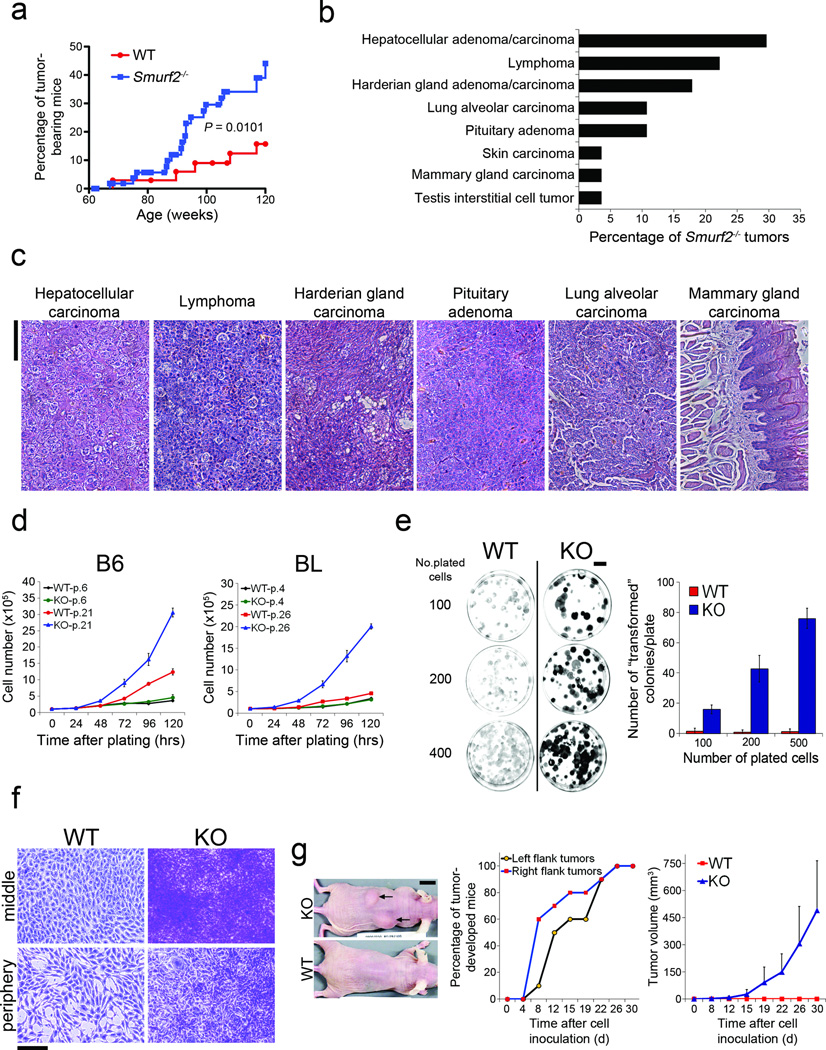
Smurf2−/− mice are relatively normal in their early lives19, however, as they aged, an unusually large number of these mice developed tumors of some sort. Eighty weeks after birth, Smurf2−/− mice showed significantly higher rate of tumor incidence than the control mice (Fig. 1a). By the end of 120 weeks, 44.1% of Smurf2−/− mice bore tumor spontaneously, compared to only 15.7% in the wild-type control group (Fig. 1a). Histopathologic examination revealed a wide spectrum of tumor types in the liver, blood, lung, pituitary, and Harderian gland (Fig. 1b,c). Occasionally, tumors were also detected in the skin, mammary gland, and testis (Fig. 1b,c).
Figure 1. Loss of Smurf2 leads to increased tumorigenicity.
(a) Spontaneous tumor incidence in wild-type (WT) (n = 35) and Smurf2−/− mice (n = 50). Statistical difference between Smurf2−/− and WT mice was assessed by Gehan-Breslow-Wilcoxon test.
(b) Distribution of tumor types in Smurf2−/− mice. Note that some animals developed multiple primary tumors.
(c) H&E staining of histological sections of representative malignancies in aged Smurf2−/− mice. Bars = 100 µm.
(d) Growth curves of wild-type (WT) and Smurf2−/− (KO) MEFs at passage 4–6 or passage 21–26 that were derived from either mixed 129/SvJ × NIH black swiss (BL) or pure C57BL/6 (B6) genetic background. Cells seeded at 105 cells per plate were counted at indicated time points. Data are presented as mean ± s.d.
(e) Colony formation assays of WT and KO MEFs (passage 57). Cells were plated at 100, 200, or 500 cells per 60 mm dish. The numbers of transformed colonies were counted after staining with crystal violet 11 d post plating, and the data are presented as mean ± s.d. Bars = 1 cm.
(f) Close-up images of crystal violet stained transformed foci. Note that WT cells were grown in monolayer only. Bars = 100 µm.
(g) Allograft tumor formation in nude mice. WT or KO MEFs at passage 57 from the B6 background were injected (n = 10). Tumor lumps and control injection sites were shown 30 d after the injection (left panel). Tumor incidence (middle panel) and average tumor volumes (right panel) were calculated. Data are presented as mean ± s.d.
To investigate the underlying causes of the tumor phenotype, we isolated mouse embryonic fibroblasts (MEFs) from Smurf2−/− or wild type litter control embryos that had been bred into either mixed 129/SvJ × NIH black swiss (BL) or pure C57BL/6 (B6) genetic background, and cultured them in successive passages through immortalization following a modified 3T3 protocol20. Although indistinguishable in both morphology and proliferation rate from wild-type cells in early passages (passage 4–6), Smurf2−/− cells of either BL or B6 genetic background became notably smaller in size and grew much faster when immortalized after 21 to 26 passages (Fig. 1d, Supplementary Fig. S1), consistent with the tumor burden difference in aged mice. Concordant to the increase in proliferation, global gene expression patterns changed drastically between wild-type and Smurf2−/− cells in late passages (passage 28–33) (Supplementart Fig. S2, Table S1–3). The difference in growth and gene expression is randomly associated with disruption of p53 or p16 function as the result of immortalization21,22 (Supplementary Fig. S3a,b), indicating that the observed growth advantage is specific to the loss of Smurf2. Interestingly, although the passage-dependent increase in cell proliferation is a consequence of Smurf2 loss, re-introducing Smurf2 back into the immortalized Smurf2−/− cells did not reverse this trend (Supplementary Fig. S3c).
In the colony formation assay23, Smurf2−/− cells of passage 57 gave rise to many large foci that stained intensely with crystal violet, and appeared very dense and lacked contact inhibition (Fig. 1e,f). In the allograft tumor formation assay, Smurf2−/− cells of passage 57 began to develop tumors as early as 8 days following subcutaneous injection (Fig. 1g). The average volume of these tumors increased exponentially from day 12 to 30 after the injection to about 500 mm3 (Fig. 1g). Histopathological examination revealed that all tumor-bearing animals developed neoplasm with severe-to-moderate multifocal muscle invasion (Supplementary Fig. S3d). These findings are striking because immortalization without further oncogenic activation is not sufficient to cause transformation21,22 just as what we observed in wild-type cells of passage 57 that only gave rise to a few lightly stained foci in plates (Fig. 1e,f) and failed to generate any tumor in injected nude mice (Fig. 1g). Thus, after 57 passages Smurf2−/− cells have already gone through the oncogenic transformation.
Smurf2 regulates DNA damage response and genomic stability
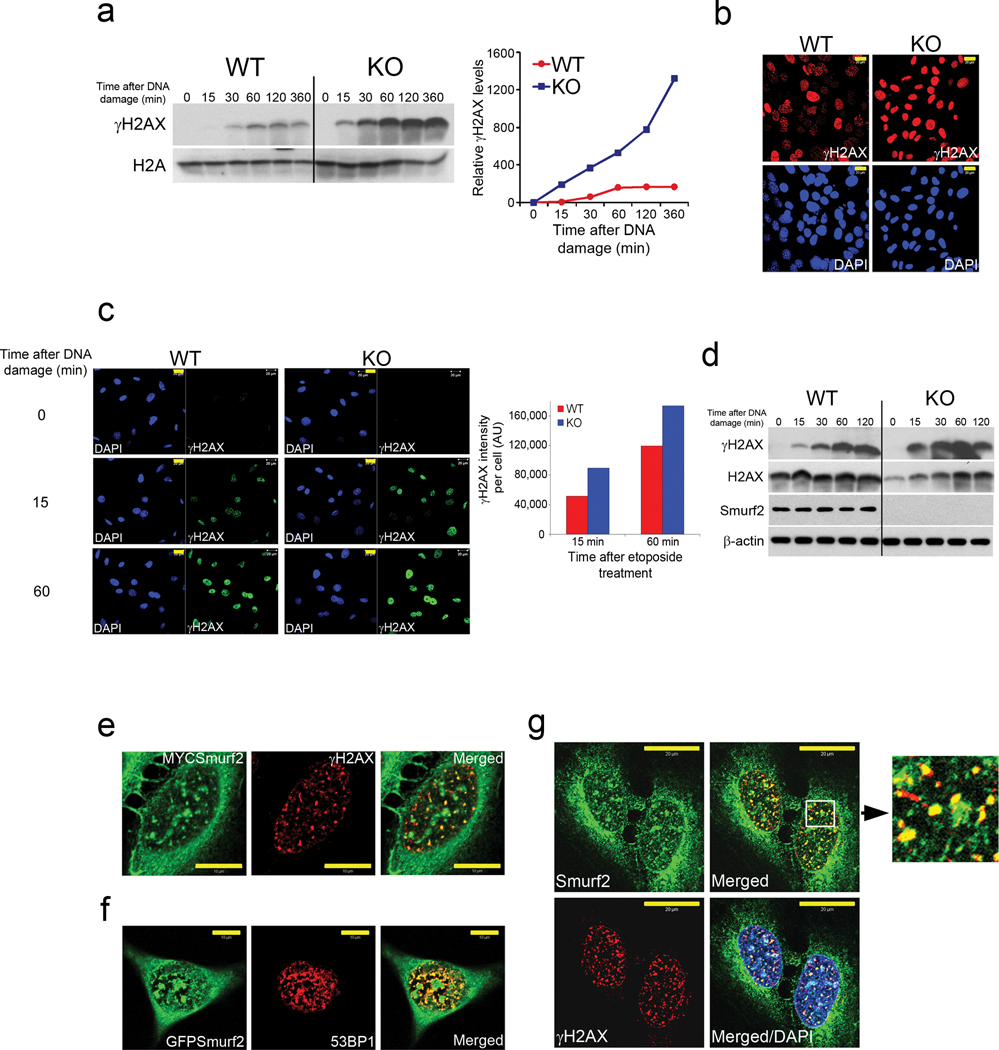
The increased tumor burden in aged Smurf2−/− mice and the accelerated Smurf2−/− cell growth in late passages described above suggest that loss of Smurf2 could set forth a series of cascading events that confer growth advantage and ultimately lead to tumorigenesis. A possible scenario would require Smurf2 to play a role in regulating DNA damage response and genomic stability; this function of Smurf2 or the lack thereof would not impinge directly on cell growth control, rather it would level the threshold to additional genetic lesions. To test this, we challenged the MEFs with etoposide, a topoisomerase II inhibitor that induces DNA double-stranded breaks24. Compared to that of wild-type cells, etoposide treatment of late passage Smurf2−/− cells led to a more rapid and robust phosphorylation of the chromatin-bound histone H2AX (γ-H2AX) (Fig. 2a), a quantitative marker for the DNA damage response at the site of double-strand breaks25. A synopsis of several key cell cycle regulators was also determined, among which the DNA damage response transducer Chk1 was up-regulated and became rapidly activated after exposure to etoposide (Supplementary Fig. S4a–b). Immunofluorescence staining also showed a more rampant appearance of γ-H2AX-positive foci in the nuclei of Smurf2−/− cells (Fig. 2b). More importantly, the enhancement of γ-H2AX response to etoposide was observed in early passage Smurf2−/− cells as well (Fig. 2c,d), implying a direct involvement of Smurf2 in DNA damage response. This previously unrecognized role of Smurf2 was further corroborated by the co-localization of transfected Smurf2 with γ-H2AX or 53BP1 nuclear foci in human U2OS cells (Fig. 2e,f, supplementary Fig. S4c) after the cells were challenged with etoposide and the co-localization of endogenous Smurf2 with γ-H2AX foci in U2OS cells (Fig. 2g). Thus, Smurf2 is directly involved in the regulation of DNA damage response.
Figure 2. Smurf2 controls DNA damage response.
(a) Western analysis of γ-H2AX in chromatin fractions of immortalized wild-type (WT) and Smurf2−/− (KO) MEFs. The cells were treated with 40 µM etoposide for durations as indicated. Quantification of the bands is shown on the right.
(b) Immunofluorescence staining of γ-H2AX in immortalized WT and KO MEFs after cells were treated with etoposide for 1 h. Bars = 20 µm.
(c) γ-H2AX immunostaining of early passage MEFs (passage 6) as in (b). Bars = 20 µm. Quantification is shown on the right.
(d) Western blot analysis of γ-H2AX and Smurf2 in early passage MEFs as in (a).
(e) Co-localization of MYC-tagged Smurf2 with γ-H2AX in U2OS cells treated with etoposide for 1 h. Bars = 10 µm.
(f) Co-localization of GFP-tagged Smurf2 with 53BP1 in U2OS cells as in (e). Bars = 10 µm.
(g) Co-localization of endogenous Smurf2 with γ-H2AX in U2OS cells as in (e). Inset shows a close-up of merged Smurf2/γH2AX staining. Bars = 20 µm.
The increase in the number of etoposide-induced γ-H2AX positive foci in Smurf2−/− cells hints a defect in the DNA damage check-point control, which would render the cells continue to grow in the presence of unrepaired DNA damage, leading to the accumulation of genetic lesions and genomic instability. Smurf2−/− cells indeed exhibited increased viability facing a spectrum of etoposide or UV-C insults compared to wild-type cells (Supplementary Fig. S4d), and underwent less apoptosis when challenged by 20 µM etoposide (Supplementary Fig. S4e). On mitotic chromosome spreads, Smurf2−/− and wild-type cells at passage 6 did not show significant difference in chromosome abnormality, which is consistent with the lack of difference in growth between these two types of cells. At passage 37, both Smurf2−/− and wild-type cells exhibited hyperploidy (data not shown), a typical trait of immortalization; however, Smurf2−/− cells at this passage contained much higher numbers of Robertsonian translocations, undefined translocations, and marker chromosomes than their wild type counterparts (Supplementary Fig. S4f). These results indicate that loss of Smurf2 caused a defect in the maintenance of genomic stability that was exacerbated in immortalized MEFs after moving through the mitotic crisis, and suggest that the compromised control of genomic stability may render Smurf2−/− mice to develop tumors under the stress of aging. The failure of exogenous Smurf2 to suppress the elevated growth of immortalized Smurf2−/− MEFs (Supplementary Fig. S3c) also corroborates with genomic instability as the root cause.
Changes in global chromatin landscape associated with Smurf2 loss
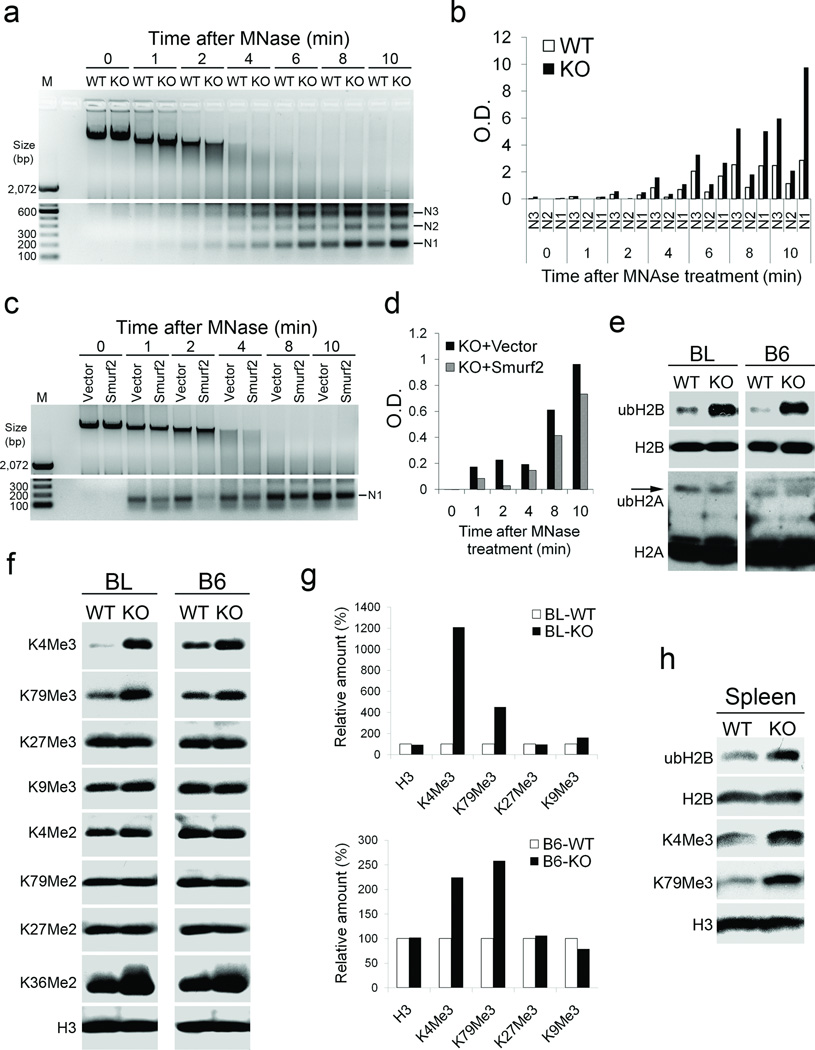
The increased differential gene activities and the sensitized DNA damage response associated with Smurf2 loss both point to possible alterations in chromatin structure and patterns of histone modifications, which collectively constitute the epigenetic landscape that regulates transcription and DNA damage responses26,27. To assess if such alterations occur as a result of Smurf2 loss, we examined the level of chromatin compaction with micrococcal nuclease (MNase) digestion, which cuts linker DNA connecting nucleosomes. The whole nuclei isolated from immortalized Smurf2−/− cells (Fig. 3a,b) or freshly prepared from Smurf2−/− dermal fibroblasts (Supplementary Fig. S5a,b) were more thoroughly digested than their wild type counter-parts within 2–10 minutes of MNase treatment, suggesting that loss of Smurf2 rendered the chromatin more loosely compacted. To ascertain if Smurf2 loss alone is responsible for loose chromatin compaction, we restored Smurf2 expression in immortalized Smurf2−/− cells and found that the MNase release of mononucleosomes was indeed impeded (Fig. 3c,d).
Figure 3. Smurf2 controls chromatin compaction and patterns of histone modification.
(a) MNase digestion of whole nuclei prepared from immortalized wild-type (WT) and Smurf2−/− (KO) MEFs. N1-N3 indicate the length of DNA wrapped around 1–3 nucleosomes.
(b) Quantification of N1-N3 in (a).
(c) MNase digestion of whole nuclei isolated from Smurf2−/− (KO) cells stably transfected with Smurf2 or a control vector.
(d) Quantification of N1 in (c).
(e) Western blot analyses of ubH2B and ubH2A in immortalized WT and KO cells from B6 and BL background.
(f) Western blot analyses of various forms of histone methylation in immortalized WT and KO cells from B6 and BL background.
(g) Quantification of histone tri-methylation level in (f).
(h) Western blot analysis of histone modifications in the spleens derived from WT and KO mice. Each sample is a mixture of 3 spleens from 3 different mice.
Posttranslational modification of histones regulates various chromatin functions including chromatin condensation, DNA damage repair, and transcription28,29. Because Smurf2 is an E3 ligase, we then examined the ubiquitin modification by Western blot analysis of acid-extracted histones. This revealed that the level of ubiquitinated histone H2B (ubH2B), but not ubH2A, was dramatically increased in Smurf2−/− cells relative to that in wild-type cells in either BL or B6 background (Fig. 3e). Concurrent to the increase in ubH2B, histone H3 tri-methylation on K4 and K79 (K4Me3 and K79Me3, respectively), which is consequentially associated with ubH2B30–33, was increased significantly (Fig. 3f,g). Di-methylation on H3K36, which is closely linked to transcriptional elongation34, was slightly increased, whereas other forms of histone modifications, including tri-methylation at K27 and K9, di-methylation at K4, K79, and K27, as well as acetylation of core histones were unchanged (Fig. 3f, Supplementary Fig. S5c). To determine the temporal origin of the changes in chromatin landscape, we isolated spleen tissues and found that the levels of ubH2B, histone H3 K4Me3, and K79Me3 were all increased in Smurf2−/− mice (Fig. 3h). Thus, the changes in histone modification patterns and DNA damage response described earlier were of immediate consequence associated with Smurf2 loss. It is possible that in the absence of Smurf2 the chromatin landscape is reshaped in a way that it alters the global gene expression patterns and weakens the control of genomic stability, which ultimately propels tumorigenesis in aged Smurf2−/− mice.
Smurf2 regulates the stability of ubH2B E3 ligase RNF20
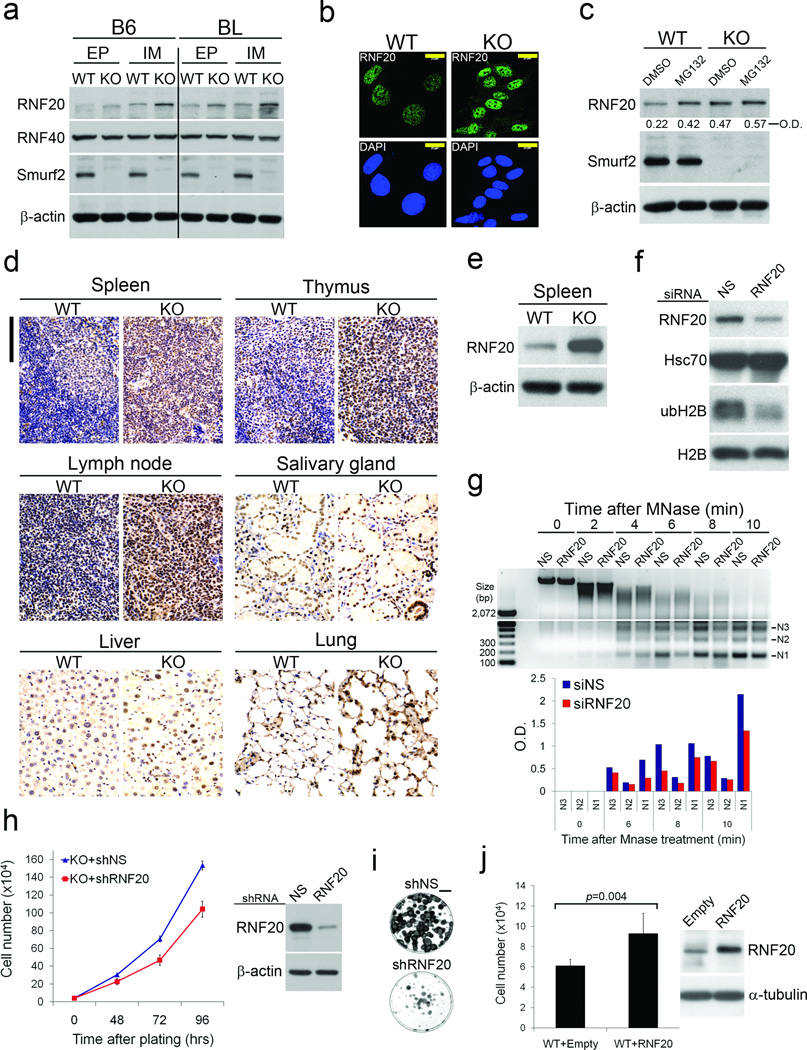
To investigate how loss of Smurf2 leads to the described changes in chromatin landscape, we turned our attention to RNF20/hBre1A, the major ubH2B-specific E3 ligase14,15. In Smurf2−/− cells, the levels of endogenous RNF20 were significantly up-regulated relative to those of wild-type cells in both early and late passages, while the levels of a closely-related RNF40/hBre1B and ubH2B deubiquitinase USP22 remained comparable (Fig. 4a, Supplementary Fig. S6a). Up-regulation of RNF20 was also observed in Smurf2−/− nuclei following immunofluorescence staining (Fig. 4b, Supplementary Fig. S6b), and was likely a post-transcriptional event since RT-PCR detected no difference in the RNF20 mRNA levels between Smurf2−/− and wild-type cells (Supplementary Fig. S6c). Treating cells with proteasome inhibitor MG132 increased the accumulation of RNF20 in wild-type cells but failed to further increase the already high level of RNF20 in Smurf2−/− cells (Fig. 4c). Moreover, blocking protein synthesis with cycloheximide showed that RNF20 had a slower turnover rate in Smurf2−/− cells than it did in wild-type cells (Supplementary Fig. S6d).
Figure 4. Smurf2 controls chromatin compaction through regulation of RNF20.
(a) Western blot analyses of RNF20 and RNF40 in early passage (EP) and immortalized (IM) wild-type (WT) and Smurf2−/− (KO) MEFs derived from the B6 or BL background.
(b) Immunofluorescence staining of endogenous RNF20 in immortalized WT and KO cells. The specificity of anti-RNF20 antibody is shown in Fig. S6b. Bars = 20 µm.
(c) Western blot analysis of RNF20 in MEF cells following MG132 treatment (20 µM; 8 h).
(d) IHC staining of RNF20 (brown) in different tissue sections prepared from WT and KO mice. The nuclei were counterstained with hematoxylin (blue). Bars = 100 µm.
(e) Western blot analysis of RNF20 in the spleen.
(f) Western blot analysis of ubH2B in Smurf2−/− MEFs following RNF20 knockdown.
(g) MNase digestion of whole nuclei isolated from Smurf2−/− MEFs following RNF20 knockdown. Bottom panel: quantification of N1-N3 nucleosome intensity.
(h) Growth curves of Smurf2−/− (KO) MEFs stably expressing shRNF20 or non-silencing shNS. The efficiency of RNF20 knockdown is shown on the right.
(i) Colony formation assays of Smurf2KO cells transfected with shRNF20 or non-silencing shNS. The cells were plated at 400 per 60 mm dish, and the colonies were visualized eleven days later after staining with crystal violet. Bars = 1 cm.
(j) Cell counts of pools WT MEFs stably expressing either RNF20 or an empty vector. The cells were seeded at 2×104 per 60mm dish and allowed to grow in culture for 72 h. Data are presented as mean ± s.d. and the levels of total RNF20 are shown on the right.
In tissue sections prepared from Smurf2−/− mice, up-regulation of RNF20 was confirmed in the spleen, thymus, lymph node, and lung by immunohistochemical (IHC) staining (Fig. 4d), and by Western blot analysis in the spleen (Fig. 4e). Moderate level of up-regulation was also seen in the salivary gland and the liver (Fig. 4d). The panoramic tissue distribution of high RNF20 expression is consistent with the wide tumor spectrum observed in Smurf2−/− mice (Fig. 1b,c). It is also correlative to the distribution of tissue origins of human tumors that express high levels of RNF20 documented in the Human Protein Atlas35. Interestingly, IHC staining showed that the tumors from Smurf2−/− mice bore the same hallmark of high RNF20 expression as did the surrounding normal tissues (Supplementary Fig. S6e, and data not show), suggesting that these tumors are likely clonal descendants of RNF20-high expressing cells.
To determine if relaxed chromatin compaction in Smurf2−/− cells is a direct consequence of the increased RNF20 level, we knocked down RNF20 using siRNA, which consequentially led to a reduction of ubH2B level as well (Fig. 4f). Under this condition, the numbers of mononucleosomes released by MNase digestion in siRNF20-transfected Smurf2KO cells decreased significantly (Fig. 4g). To determine if altering RNF20 level affects cell growth, we generated stable Smurf2−/− cells expressing RNF20-specific shRNA, and found that knockdown of RNF20 retarded cell growth (Fig. 4h) and curtailed the ability of shRNF20-bearing Smurf2−/− cells to undergo transformation (Fig. 4i). Conversely, stable wild-type MEFs over-expressing RNF20 grew much faster than vector control cells (Fig. 4j). These data strongly support a role of Smurf2 in the maintenance of chromatin structure through RNF20.
RNF20 is a substrate of Smurf2-mediated ubiquitination and degradation
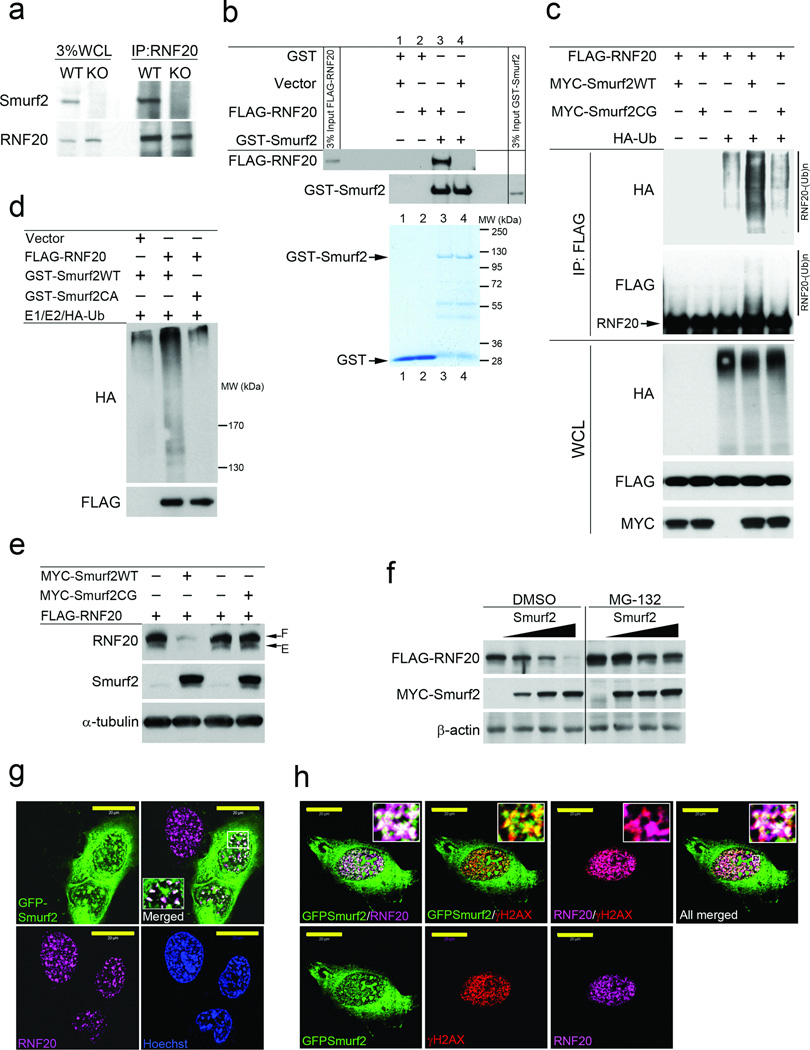
We conducted several lines of experiments to determine if RNF20 is a direct substrate of Smurf2-mediated ubiquitination and proteasome degradation. First, we demonstrated interaction between Smurf2 and RNF20 by co-IP experiments with the endogenous proteins (Fig. 5a) and the transfected proteins produced in HEK293 cells (Supplementary Fig. S7a), as well as GST pull-down experiment with purified FLAG-RNF20 and recombinant GST-Smurf2 (Fig. 5b). Second, we showed that co-expression of FLAG-RNF20, HA-ubiquitin, and MYC-Smurf2 in HEK293 cells could enhance the background level of poly-ubiquitination of RNF20, which underwent auto-ubiquitination due to its intrinsic E3 ligase activity; however, the E3 ligase defective Smurf2CG mutant4 did not support the enhancement, implying specificity (Fig. 5c). We further reconstituted the ubiquitination of RNF20 in vitro using E1, E2, HA-ubiquitin, purified FLAG-RNF20 from in vitro translation, and bacterially expressed GST-Smurf2 but not GST-Smurf2CA (Fig. 5d). Third, co-expression of FLAG-RNF20 with MYC-Smurf2 but not the mutant MYC-Smurf2CG decreased RNF20 protein level (Fig. 5e), and this Smurf2-induced degradation could be blocked by MG132 treatment (Fig. 5f). Finally, in transfected U2OS cells, GFP-Smurf2 was found to co-localize with the endogenous RNF20 and reduced its level accordingly (Fig. 5g, Supplementary Fig. S7b,c). Moreover, when the cells were challenged with etoposide, both GFP-Smurf2 and RNF20 were co-localized with γ-H2AX foci (Fig. 5h). Taken together, these results demonstrate that RNF20 is a direct substrate of Smurf2, and Smurf2 is required for maintaining histone modification pattern over the general chromatin landscape and regulating acute DNA damage response at sites of DNA double-stranded breaks by fine tuning the RNF20 level.
Figure 5. RNF20 is a direct target of Smurf2 for proteasome-dependent degradation.
(a) Co-IP analysis of endogenous Smurf2 and RNF20 interaction in MEFs. WCL, whole cell lysate.
(b) GST-pull down experiment showing direct interaction between purified GST-Smurf2 and FLAG-RNF20. FLAG-RNF20 was isolated from rabbit reticulocyte lysate after in vitro translation by FLAG immunoprecipitation and eluted with FLAG peptide. Coomassie blue staining shows purified GST and GST-Smurf2.
(c) Western blot analysis of poly-ubiquitination of RNF20 by Smurf2 in vivo. FLAG-RNF20 and HA-ubiquitin were transiently expressed in HEK293 cells with MYC-Smurf2WT or MYC-Smurf2CG. The cells were treated with MG132 (20 µM) for 7 hours before analysis. Antibodies to HA and FLAG were used to visualized ubiquitinated RNF20 following immunoprecipitation with FLAG M2 beads.
(d) Western blot analysis of poly-ubiquitination of in vitro translated RNF20 by recombinant GST-Smurf2. GST-Smurf2CA was used as a control.
(e) Overexpression of MYC-Smurf2 but not mutant Smurf2CG promotes RNF20 degradation in HEK293 cells. F: FLAG tagged, and E: endogenous RNF20.
(f) Western blot analysis shows that over-expression of MYC-Smurf2 leads to proteasome-specific degradation of FLAG-RNF20 in HEK293 cells.
(g) Confocal images showing co-localization of GFP-Smurf2 and endogenous RNF20 in the nuclei. Transient transfection was used to express GFP-Smurf2 in U2OS cells. Bars = 20 µm.
(h) Confocal images showing co-localization of GFP-Smurf2 and endogenous RNF20 at sites of DNA double-stranded breaks. U2OS cells transiently expressing GFP-Smurf2 were treated with etoposide for 1 h. Bars = 20 µm.
Smurf2 regulates RNF20 stability in various human tumor cells and tissues
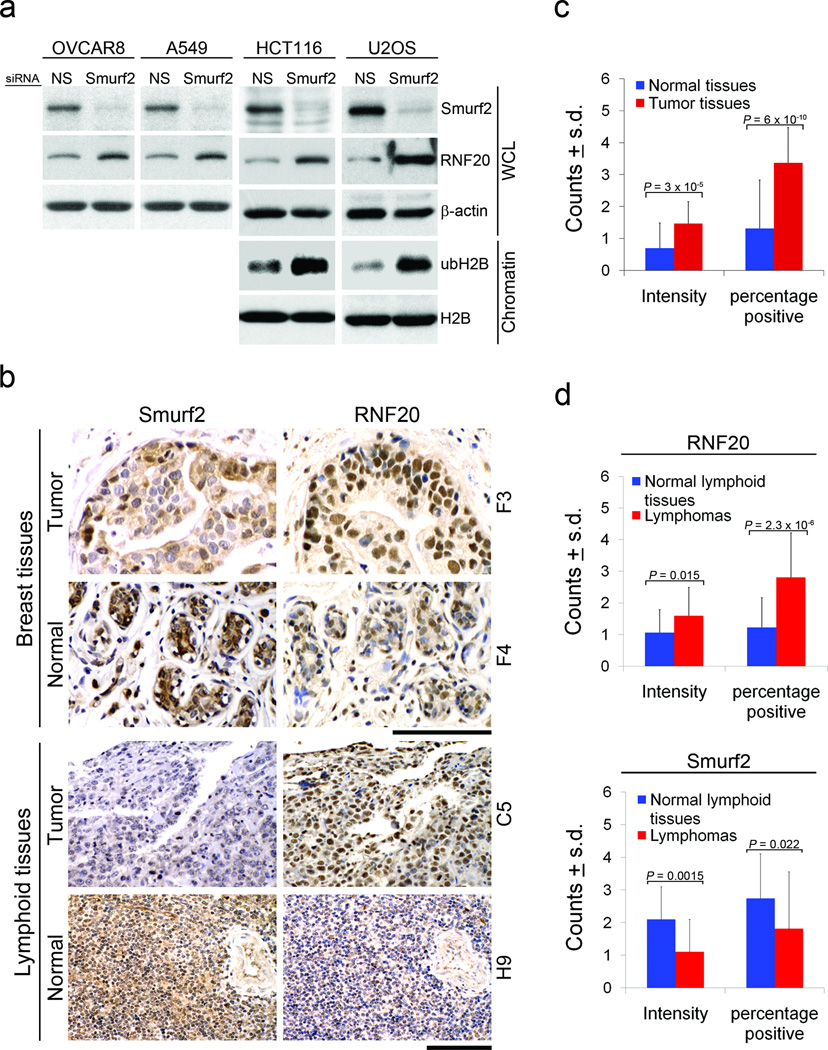
The RNF20-mediated alteration of chromatin landscape offers an attractive model to account for the increased tumor burden in aged Smurf2−/− mice. To address if this is also a feature of human tumors, we analyzed a number of established (immortalized) human tumor cell lines. Without exception, knockdown of Smurf2 with siRNA in OVCAR8, A549, HCT116 and U2OS cells dramatically increased RNF20 expression (Fig. 6a). We also extracted chromatin from HCT116 and U2OS cells after Smurf2 knockdown and found that the level of ubH2B was also increased (Fig. 6a). On the other hand, knockdown of RNF20 in U2OS or HCT116 cells reduced the extent of MNase digestion of isolated whole nuclei, resulting in increased chromatin compaction (Supplementary Fig. S8). Thus, decreasing RNF20 expression causes chromatin to be more tightly compacted in human cancer cells as well. These results are in agreement with our observation that increasing the RNF20 levels in Smurf2-deficient cells relaxed chromatin structure (Fig. 3a,b, Supplementary Fig. S5a,b).
Figure 6. Smurf2-RNF20 relationship is preserved in different human cells and cancer tissues.
(a) Western blot analysis of Smurf2, RNF20, and ubH2B following siRNA-mediated knock-down of Smurf2 in HCT116 (colon carcinoma), U2OS (osteosarcoma), OVCAR8 (ovarian carcinoma), and A549 (lung adenocarcinoma) human cancer cells. The level of ubH2B was assessed on acid-extracted histones.
(b) IHC staining of Smurf2 and RNF20 in breast cancer or lymphoma and their matching normal tissues. F3 and F4 (for breast) and C5 and H9 (for lymphoid) are coordinates of samples in the tissue array. Bars = 100 µm.
(c) RNF20 expression in normal and tumor breast tissues array (n = 40). The staining intensity and the percentage of positively stained cells were quantified as described (Supplementary Table S4). _p_-value was calculated using unpaired two-tail Student’s t-test.
(d) Expression of RNF20 and Smurf2 in lymphomas (n = 55) and normal lymphoid tissues (n= 18) of the lymphoma tissues array. The staining intensity and the percentage of positively stained cells were quantified as described (Supplementary Table S5). _p_-value was calculated using unpaired two-tail Student’s t-test.
Previously, we showed a switch of Smurf2 distribution from high in the nucleus in normal tissues to high in the cytoplasm in breast cancer cells in an array of 40 human breast cancer and matching normal tissues, despite a mixed range of Smurf2 expression12 (also Fig. 6b). It is possible that the nuclear function of Smurf2 is partially impaired in these cancer cells. In this same array and using a scoring matrix that takes into account both the intensity in a positive cell and the percentage of positive cells in a given field, we found a strong correlation between high RNF20 expression and tumor formation, with 32 tumors exhibited high whereas only 5 exhibited low RNF20 expression compared to that in their corresponding normal breast tissues (Fig. 6b,c, Supplementary Table S4). Reciprocal to the decreased nuclear staining of Smurf2 in tumor tissues, staining of RNF20 in the nucleus was increased in tumors (Fig. 6b). These results suggest a correlative relationship from impairment of Smurf2 nuclear function to high RNF20 to tumor formation. Similar inverse relationship between Smurf2 and RNF20 was also observed in lymphomas; we found by IHC staining that Smurf2 expression was significantly decreased while RNF20 increased in an array of 55 human lymphoma tissues (Fig. 6b,d, Supplementary Table S5). Thus, the chromatin landscape in human tumors is clearly sensitive to fluctuations in the RNF20 level, and loss of Smurf2 is likely to be one of the contributing events that cause genome instability in certain human cancers, too.
DISCUSSION
In the literature, there are only a few sporadic reports describing the roles of Smurf2 in tumorigenesis ranging from senescence to metastasis10–12, 36. In light of the current genetic cancer model study, this could be simply because altering Smurf2 expression has no immediate impact on cell growth, just like classical “caretaker” tumor suppressor gene MSH2 and MLH1 of the DNA repair pathway37. Through mechanistic analyses, we identified RNF20 as a direct target of Smurf2 in its regulation of chromatin landscape. Ubiquitination of histone H2B has been known to play an important role in controlling chromatin landscape and transcription29. This modification interferes with chromatin compaction due to specific interactions between the ubiquitin moiety and the nucleosomal surface38. Recent reports have clearly implicated ubH2B in regulating DNA double strand break repair by homologous recombination and non-homologous end-joining, and revealed the co-localization of RNF20 and γH2AX foci as we describe here17,18. These data argue that dysregulation of ubH2B could open the access of the chromatin to recombinatorial exchanges, causing excessive chromosomal breaks and translocations. Thus, loss of Smurf2 could promote tumorigenesis by altering the pattern of histone modification and the extent of chromatin compaction, which consequently introduces instability into the genome and changes gene expression profile (Supplementary Fig. S9); ultimately, this could lead to cancer.
METHODS
Animals and histopathology
For spontaneous tumor development, Smurf2−/− mice in mixed 129/SvJ × NIH Black Swiss and pure C57BL/6 background19 were monitored twice weekly, moribund mice or mice with obvious tumors were euthanized and subjected to necropsy. After 28 month of age, all mice were euthanized and necropsied.
To assess tumorigenecity of immortalized MEFs in vivo, 4–6-week-old female athymic nu/nu mice were subcutaneously injected with immortalized wild type or Smurf2−/− MEFs at two hind flanks (2×106 cells/flank).
Tumor histology was read by a board-certified veterinary pathologist in the National Cancer Institute core facility. All mice were maintained and handled according to protocols approved by the Animal Care and Use Committee of the US National Cancer Institute.
Cell Culture and transfections
Primary MEFs and mouse dermal fibroblasts were derived as described19,39. To establish immortalized MEFs, MEFs derived from three different Smurf2−/− embryos and two different wild type embryos from 129SvJ × NIH Black Swiss background were mixed at passage 4 according their genotype to derive BL-Smurf2−/− and BL-WT cells, respectively. These cells then passaged on a 3T3 protocol20 to generate immortalized cells. Simultaneously, MEFs derived from a Smurf2−/− and a sibling wild-type embryo in C57BL/6 background were also passaged according to the above 3T3 protocol to generate immortalized B6-Smurf2−/− and B6-WT cells, respectively. Human U2OS, Hela and HEK293 cells were obtained from the American Type Culture Collection. HCT116 and A549 cells were from the DCTD Tumor Repository of the US National Cancer Institute. Fugene 6 (Roche), Lipofectamine 2000 and oligofectamine (Invitrogen) reagents were used according to manufacturer’s suggestion to transfect plasmid DNA and siRNA. Smurf2-specific siRNA and control NS siRNA were described previously12. The target siRNA sequences used for RNF20 were: 5’-CCTTGCTGTAACATGCGTAAA-3’ (murine)39; and 5’-CCGTATCATCCTTAAACGTT-3’ (human).
For reconstitution of Smurf2 in Smurf2−/− cells, immortalized MEFs were infected with retroviral particles containing pBabe-FLAG-Smurf2-puro vector12. For generation of WT MEFs overexpressing RNF20, cells were infected with retroviruses containing pBabe-FLAG-RNF20-puro vector.
Immunofluorescence, immunohistochemistry, immunoprecipitation
Immunofluorescence was conducted as described40,41. Immunohistochemistry of tissue sections with anti-RNF20 (ab32629) and anti-Smurf2 (H-50) antibodies was performed using enzymatic ABC-DAB staining (Vector Lab). Nuclei were counterstained with hematoxylin. All comparative images were obtained under identical microscope and camera settings. Breast cancer tissue array BR801 with matching normal tissues, and lymphoma tissue array 801 were from US BioMax, Inc. Immunohistochemistry of the array was analyzed and quantified by a board certified pathologist in the US National Cancer Institute.
For immunoprecipitation experiments, cells were lysed either in NP-40 (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 0.5%NP-40) or in FT lysis buffer (600 mM KCl, 20 mM Tris-HCL, pH 7.8; 20% Glycerol, EDTA-free protease and phosphatase inhibitors) after repetitive freezing-thawing cycles and subsequently treated with 250U Benzonase (Novagen) as described42. The lysates were then clarified by centrifugation (10,000g, 20 min; 4°C) and diluted four times with FT dilution buffer (45 mM Tris-HCl, pH 7.6, 2.25 mM EDTA, 0.1% NP-40). All immunoprecipitaions were conducted overnight at 4°C. Soluble chromatin fractions were prepared as described40. Nucleosome core histones were extracted from the cells using a modified acid-based extraction protocol43.
Micrococal nuclease digestion of chromatin
The assay was performed as described44 with a few modifications. In brief, cell nuclei were isolated using hypotonic buffer (10 mM Hepes, pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.34 M Sucrose, 10% Glycerol, 1mM DTT) supplemented with EDTA-free protease and phosphatase inhibitors and 0.1% Triton-X. After washing in the hypotonic buffer without Triton-X, an aliquot of the nuclei was re-suspended in 1N NaOH and normalized to ~ 1.5 mg ml−1 DNA concentration in the reaction buffer (15 mM Tris-HCl, pH 7.4, 60 mM KCl, 0.25 M sucrose, 1 mM CaCl2, 0.5 mM DTT). The digestion was carried out with micrococcal nuclease (MNase; Roche) at 5U MNase per 250 µl of reaction buffer at 25°C for indicated period of time. The reaction was terminated by adding equal volume of 2×TNESK buffer (20 mM Tris-HCl, pH 7.4, 0.2M NaCl, 2 mM EDTA, 2%SDS) with freshly added proteinase K (0.2 mg ml−1). The samples were then incubated overnight at 37°C, and genomic DNA was purified as described45.
GST fusion protein, pull-down assays and in vitro ubiquitination assays
GST fusion proteins were prepared from E.coli using glutathione-Sepharose beads (Amersham Pharmacia Biotech). In vitro translated (TNT kit, Promega) FLAG-RNF20 in rabbit reticulocyte lysate was immunoprecipitated using FLAG-M2 beads and eluted from the beads using FLAG peptide (Sigma). The purified FLAG-RNF20 was incubated with GST or GST-Smurf2 fusion proteins for 1 hr at 4°C in the binding buffer (50 mM Tris-HCl, pH 7.5, 120 mM NaCl, 2 mM EDTA, 0.1% NP40). RNF20 proteins that bound to GST-fusion proteins were retrieved on glutathione-Sepharose beads, and subjected to immunoblot analyses. The above purified FLAG-RNF20 and GST-Smurf2 fusion proteins were also used in vitro ubiquitination assay as described46.
Supplementary Material
1
ACKNOWLEDGEMENTS
We thank M. Anver for pathology service, V. Barr for assistance with microscope; X. Wu for microarray experiments, N. Morris for animal experiments, N. Teja for assistance with cell culture. We also thank K. Sixt for comments on the manuscript. This research is supported by the Intramural Research Program of the US National Cancer Institute/NIH, Center for Cancer Research. M.Y. was partially supported by JSPS grant #21689053.
Footnotes
Author Contributions
M.Y. and Y. T. maintained mouse colonies and generated primary MEFs and MDFs. M.Y., S.Y.C and Y.E.Z. observed and analyzed spontaneous tumor formation in mice. S.S.B. performed karyotyping analysis. Y.E.Z. analyzed microarray data. M.B. performed all other experiments described in the manuscript. M.B. and Y.E.Z. conceived the study, analyzed the data, and wrote the paper.
Conflict of Interest
The authors declare that they have no conflict of interest.
REFERENCES
- 1.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 2.Izzi L, Attisano L. Regulation of the TGFbeta signalling pathway by ubiquitin-mediated degradation. Oncogene. 2004;23:2071–2078. doi: 10.1038/sj.onc.1207412. [DOI] [PubMed] [Google Scholar]
- 3.Lonn P, Moren A, Raja E, Dahl M, Moustakas A. Regulating the stability of TGFbeta receptors and Smads. Cell Res. 2009;19:21–35. doi: 10.1038/cr.2008.308. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Y, Chang C, Gehling DJ, Hemmati-Brivanlou A, Derynck R. Regulation of Smad degradation and activity by Smurf2, an E3 ubiquitin ligase. Proc Natl Acad Sci U S A. 2001;98:974–979. doi: 10.1073/pnas.98.3.974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin X, Liang M, Feng XH. Smurf2 is a ubiquitin E3 ligase mediating proteasome-dependent degradation of Smad2 in transforming growth factor-beta signaling. J Biol Chem. 2000;275:36818–36822. doi: 10.1074/jbc.C000580200. [DOI] [PubMed] [Google Scholar]
- 6.Kavsak P, et al. Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGF beta receptor for degradation. Mol Cell. 2000;6:1365–1375. doi: 10.1016/s1097-2765(00)00134-9. [DOI] [PubMed] [Google Scholar]
- 7.Li H, Seth A. An RNF11: Smurf2 complex mediates ubiquitination of the AMSH protein. Oncogene. 2004;23:1801–1808. doi: 10.1038/sj.onc.1207319. [DOI] [PubMed] [Google Scholar]
- 8.Schwamborn JC, Muller M, Becker AH, Puschel AW. Ubiquitination of the GTPase Rap1B by the ubiquitin ligase Smurf2 is required for the establishment of neuronal polarity. Embo J. 2007;26:1410–1422. doi: 10.1038/sj.emboj.7601580. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Narimatsu M, et al. Regulation of planar cell polarity by Smurf ubiquitin ligases. Cell. 2009;137:295–307. doi: 10.1016/j.cell.2009.02.025. [DOI] [PubMed] [Google Scholar]
- 10.Zhang H, Cohen SN. Smurf2 up-regulation activates telomere-dependent senescence. Genes Dev. 2004;18:3028–3040. doi: 10.1101/gad.1253004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fukuchi M, et al. High-level expression of the Smad ubiquitin ligase Smurf2 correlates with poor prognosis in patients with esophageal squamous cell carcinoma. Cancer Res. 2002;62:7162–7165. [PubMed] [Google Scholar]
- 12.Jin C, et al. Smad ubiquitination regulatory factor 2 promotes metastasis of breast cancer cells by enhancing migration and invasiveness. Cancer Res. 2009;69:735–740. doi: 10.1158/0008-5472.CAN-08-1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnstone SE, Baylin SB. Stress and the epigenetic landscape: a link to the pathobiology of human diseases? Nat Rev Genet. 2010;11:806–812. doi: 10.1038/nrg2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim J, Hake SB, Roeder RG. The human homolog of yeast BRE1 functions as a transcriptional coactivator through direct activator interactions. Mol Cell. 2005;20:759–770. doi: 10.1016/j.molcel.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 15.Zhu B, et al. Monoubiquitination of human histone H2B: the factors involved and their roles in HOX gene regulation. Mol Cell. 2005;20:601–611. doi: 10.1016/j.molcel.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 16.Minsky N, et al. Monoubiquitinated H2B is associated with the transcribed region of highly expressed genes in human cells. Nat Cell Biol. 2008;10:483–488. doi: 10.1038/ncb1712. [DOI] [PubMed] [Google Scholar]
- 17.Moyal L, et al. Requirement of ATM-Dependent Monoubiquitylation of Histone H2B for Timely Repair of DNA Double-Strand Breaks. Mol Cell. 2011;41:529–542. doi: 10.1016/j.molcel.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakamura K, et al. Regulation of Homologous Recombination by RNF20-Dependent H2B Ubiquitination. Mol Cell. 2011;41:515–528. doi: 10.1016/j.molcel.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 19.Tang LY, et al. Ablation of Smurf2 reveals an inhibition in TGF-beta signalling through multiple mono-ubiquitination of Smad3. Embo J. 2011 doi: 10.1038/emboj.2011.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Todaro GJ, Green H. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J Cell Biol. 1963;17:299–313. doi: 10.1083/jcb.17.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harvey DM, Levine AJ. p53 alteration is a common event in the spontaneous immortalization of primary BALB/c murine embryo fibroblasts. Genes Dev. 1991;5:2375–2385. doi: 10.1101/gad.5.12b.2375. [DOI] [PubMed] [Google Scholar]
- 22.Sherr CJ. Tumor surveillance via the ARF-p53 pathway. Genes Dev. 1998;12:2984–2991. doi: 10.1101/gad.12.19.2984. [DOI] [PubMed] [Google Scholar]
- 23.Franken NA, Rodermond HM, Stap J, Haveman J, van Bree C. Clonogenic assay of cells in vitro. Nat Protoc. 2006;1:2315–2319. doi: 10.1038/nprot.2006.339. [DOI] [PubMed] [Google Scholar]
- 24.Montecucco A, Biamonti G. Cellular response to etoposide treatment. Cancer Lett. 2007;252:9–18. doi: 10.1016/j.canlet.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 25.Bonner WM, et al. GammaH2AX and cancer. Nat Rev Cancer. 2008;8:957–967. doi: 10.1038/nrc2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Falk M, Lukasova E, Kozubek S. Chromatin structure influences the sensitivity of DNA to gamma-radiation. Biochim Biophys Acta. 2008;1783:2398–2414. doi: 10.1016/j.bbamcr.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 27.Murga M, et al. Global chromatin compaction limits the strength of the DNA damage response. J Cell Biol. 2007;178:1101–1108. doi: 10.1083/jcb.200704140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu Rev Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- 29.Weake VM, Workman JL. Histone ubiquitination: triggering gene activity. Mol Cell. 2008;29:653–663. doi: 10.1016/j.molcel.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 30.Sun ZW, Allis CD. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002;418:104–108. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- 31.Laribee RN, et al. BUR kinase selectively regulates H3 K4 trimethylation and H2B ubiquitylation through recruitment of the PAF elongation complex. Curr Biol. 2005;15:1487–1493. doi: 10.1016/j.cub.2005.07.028. [DOI] [PubMed] [Google Scholar]
- 32.Lee JS, et al. Histone crosstalk between H2B monoubiquitination and H3 methylation mediated by COMPASS. Cell. 2007;131:1084–1096. doi: 10.1016/j.cell.2007.09.046. [DOI] [PubMed] [Google Scholar]
- 33.Kim J, et al. RAD6-Mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell. 2009;137:459–471. doi: 10.1016/j.cell.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kizer KO, et al. A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3 K36 methylation with transcript elongation. Mol Cell Biol. 2005;25:3305–3316. doi: 10.1128/MCB.25.8.3305-3316.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berglund L, et al. A genecentric Human Protein Atlas for expression profiles based on antibodies. Mol Cell Proteomics. 2008;7:2019–2027. doi: 10.1074/mcp.R800013-MCP200. [DOI] [PubMed] [Google Scholar]
- 36.Fukunaga E, et al. Smurf2 induces ubiquitin-dependent degradation of Smurf1 to prevent migration of breast cancer cells. J Biol Chem. 2008;283:35660–35667. doi: 10.1074/jbc.M710496200. [DOI] [PubMed] [Google Scholar]
- 37.Haber D, Harlow E. Tumour-suppressor genes: evolving definitions in the genomic age. Nat Genet. 1997;16:320–322. doi: 10.1038/ng0897-320. [DOI] [PubMed] [Google Scholar]
- 38.Fierz B, et al. Histone H2B ubiquitylation disrupts local and higher-order chromatin compaction. Nat Chem Biol. 2011;7:113–119. doi: 10.1038/nchembio.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hogan B, Beddington R, Constantini F, Lacy E. Manipulating the Mouse Embryo: A Laboratory Manual. 2 edn. Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 40.Blank M, Lerenthal Y, Mittelman L, Shiloh Y. Condensin I recruitment and uneven chromatin condensation precede mitotic cell death in response to DNA damage. J Cell Biol. 2006;174:195–206. doi: 10.1083/jcb.200604022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Galanty Y, et al. Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature. 2009;462:935–939. doi: 10.1038/nature08657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rudolph C, Adam G, Simm A. Determination of copy number of c-Myc protein per cell by quantitative Western blotting. Anal Biochem. 1999;269:66–71. doi: 10.1006/abio.1999.3095. [DOI] [PubMed] [Google Scholar]
- 43.Shechter D, Dormann HL, Allis CD, Hake SB. Extraction, purification and analysis of histones. Nat Protoc. 2007;2:1445–1457. doi: 10.1038/nprot.2007.202. [DOI] [PubMed] [Google Scholar]
- 44.Ziv Y, et al. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat Cell Biol. 2006;8:870–876. doi: 10.1038/ncb1446. [DOI] [PubMed] [Google Scholar]
- 45.Zaret K. Micrococcal nuclease analysis of chromatin structure. Chapter 21, Unit 21. Curr Protoc Mol Biol. 2005;21 doi: 10.1002/0471142727.mb2101s69. [DOI] [PubMed] [Google Scholar]
- 46.Yamashita M, et al. Ubiquitin ligase Smurf1 controls osteoblast activity and bone homeostasis by targeting MEKK2 for degradation. Cell. 2005;121:101–113. doi: 10.1016/j.cell.2005.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1